Abstract
Caudal Type Homeobox 2 (CDX2) is a key regulator of trophectoderm formation and maintenance in preimplantation embryos. We previously demonstrated that supplementation of exogenous follistatin, during in vitro culture of bovine IVF embryos, upregulates CDX2 expression, possibly, via alteration of the methylation status of CDX2 gene. Here, we further investigated the effects of exogenous follistatin supplementation on developmental competence and CDX2 methylation in bovine somatic cell nuclear transfer (SCNT) embryos. SCNT embryos were cultured with or without follistatin for 72h, then transferred into follistatin free media until d7 when blastocysts were collected and subjected to CDX2 gene expression and DNA methylation analysis for CDX2 regulatory regions by bisulfite sequencing. Follistatin supplementation significantly increased both blastocyst development as well as blastocyst CDX2 mRNA expression on d7. Three different CpG rich fragments within the CDX2 regulatory elements; proximal promoter (fragment P1, -1644 to -1180; P2, -305 to +126) and intron 1 (fragment I, + 3030 to + 3710) were identified and selected for bisulfite sequencing analysis. This analysis showed that follistatin treatment induced differential methylation (DM) at specific CpG sites within the analyzed fragments. Follistatin treatment elicited hypomethylation at six CpG sites at positions -1374, -279, -163, -23, +122 and +3558 and hypermethylation at two CpG sites at positions -243 and +20 in promoter region and first intron of CDX2 gene. Motif analysis using MatInspector revealed that differentially methylated CpG sites are putative binding sites for key transcription factors (TFs) known to regulate Cdx2 expression in mouse embryos and embryonic stem cells including OCT1, AP2F, KLF and P53, or TFs that have indirect link to CDX2 regulation including HAND and NRSF. Collectively, results of the present study together with our previous findings in IVF embryos support the hypothesis that alteration of CDX2 methylation is one of the epigenetic mechanisms by which follistatin may regulates CDX2 expression in preimplantation bovine embryos.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12958-021-00829-7.
Keywords: Bovine preimplantation embryos, SCNT, CDX2, DNA methylation, Follistatin
Introduction
Somatic cell nuclear transfer (SCNT) is a powerful tool used to propagate valuable livestock species and to generate genetically modified animals that can be used in biomedical research and therapeutic cloning [1, 2]. Despite the successful cloning of numerous species, the application of the technology is still limited due to several complications that arise from using somatic cells as nuclei donors. These difficulties include poor embryonic development, placental abnormalities, higher abortion rates, larger birth weights and perinatal mortalities [3–5]. These abnormalities are likely caused by incomplete reprogramming of the somatic cell nucleus that impact normal transcriptional programming during preimplantation development [6–8].
A previous study demonstrated that exogenous follistatin supplementation during in vitro embryo culture increased blastocyst development and trophectoderm (TE) cell number in bovine SCNT embryos [9]. Follistatin is a binding protein for specific members of the transforming growth factor β (TGF-β) super family. Follistatin protein and mRNA are upregulated in good quality oocytes [10, 11]. Numerous studies in our laboratory showed that exogenous follistatin supplementation during in vitro embryo culture improves the developmental competence of bovine preimplantation embryos produced under different conditions. These include conventional IVF [12–14], IVF using poor quality oocytes [11], and IVF using sex sorted semen [15]. In general, follistatin treatment increases blastocyst rates, CDX2 expression and trophectoderm (TE) cell number on day 7 of development. Most recently, we demonstrated that follistatin supplementation during the initial 72h of in vitro embryo culture (IVC1) upregulated CDX2 expression in IVF blastocysts that was tightly associated with DNA methylation at specific CpG sites within the regulatory elements of CDX2 gene including the proximal promoter and intron 1. These CpG sites contain putative binding sites for several key transcription factors implicated in regulating Cdx2 expression in mouse embryos and trophoblast stem cells [16]. Importantly, increased CDX2 expression in bovine blastocysts is associated with improved pregnancy rates after embryo transfer [17]. Therefore, the objectives of the present study were to analyze the beneficial effects of follistatin on developmental competence and CDX2 expression in bovine SCNT embryos and to investigate whether these effects are linked to changes in DNA methylation as we previously reported in IVF embryos [16]. To address this, we cultured SCNT embryos with or without follistatin (10ng/ml) during the first 72h of culture, then at blastocyst stage, we analyzed DNA methylation at CDX2 regulatory regions. Results show that follistatin treatment induces changes in DNA methylation at specific CpG sites within CDX2 regulatory regions in SCNT blastocysts, these changes are associated with upregulation of CDX2 expression.
Materials and methods
Chemicals and supplies
All reagents were purchased from Sigma Aldrich (Saint Louis, MO) unless mentioned otherwise.
Oocyte collection, In vitro maturation, Enucleation, and somatic cell nuclear transfer
Bovine cumulus oocytes complexes (COCs) were retrieved from ovaries collected at local slaughterhouse in the state of Michigan. Oocyte selection and in vitro maturation (IVM) were carried out as we previously described [11, 12]. Somatic cell nuclear transfer (SCNT) was performed as reported elsewhere [18]. Briefly, in vitro matured oocytes were denuded from cumulus cells and MII oocytes with extruded first polar bodies were selected for enucleation. Bovine fibroblasts were used as donor cells and injected into the perivitelline space of each enucleated oocyte. The SCNT couplets were immediately fused in sorbitol fusion medium by a single electric pulse (2 kV/cm for 15 μs) and cultured for 0.5-1h. After fusion, the couplets were activated by 5 mM ionomycin in HH medium for 5 min then transferred to equilibrated KSOM medium containing 2 mM of 6-DMAP for 4h. Afterwards, SCNT embryos were washed and transferred into KSOM culture medium.
In vitro embryo culture and follistatin supplementation
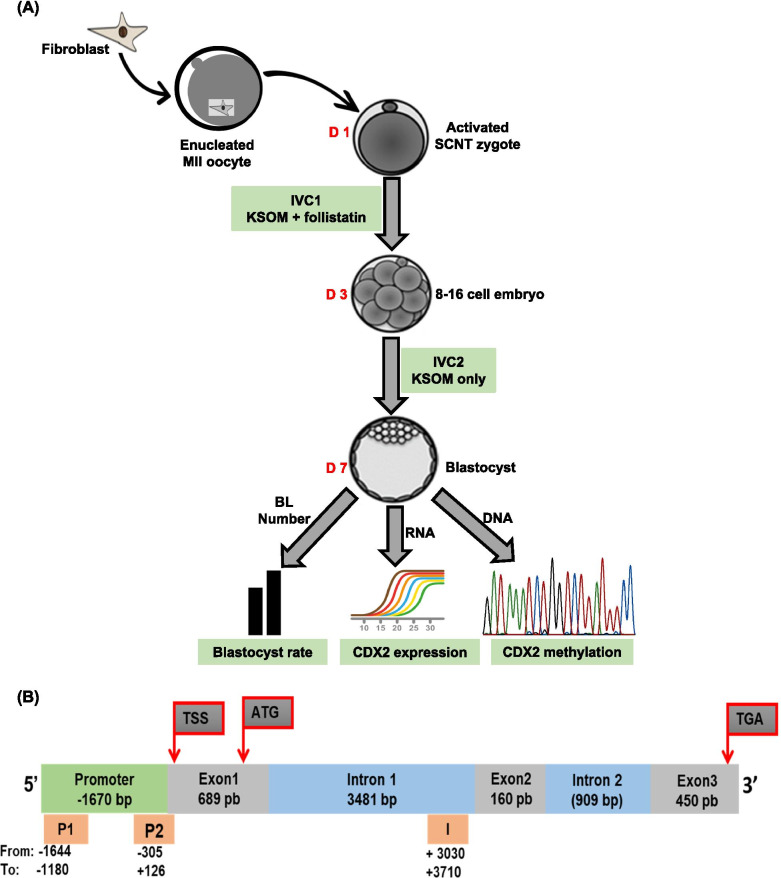
The activated SCNT embryos were cultured in KSOM media with or without the maximal stimulatory dose of follistatin, 10 ng/ml (R&D Systems, Minneapolis, MN) for 72h [14]. Then, 8-16 cell embryos were cultured in follistatin free media until d7. At the end of the culture, blastocysts were collected and properly stored until used for simultaneous isolation of genomic DNA, for bisulfite sequencing and total RNA, for CDX2 gene expression analysis. Figure 1A provides a schematic illustration of the study design.
Fig. 1.
Study design and bovine CDX2 gene structure. A To investigate the effects of follistatin on early development of somatic cell nuclear transfer (SCNT) embryos and CDX2 methylation status in blastocyst, activated SCNT zygotes were cultured with or without follistatin (10 ng/ml) during IVC1, then 8-16 cell embryos were cultured in follistatin free media until day 7, when percentage of embryos developing to blastocyst stage were recorded. Then blastocysts were used for extraction of total RNA for CDX2 gene expression analysis and genomic DNA for bisulfite sequencing analysis. B Schematic illustration of bovine CDX2 gene showing the regulatory elements of the gene (proximal promoter and intron 1) and position of the fragments selected for analysis (P1, P2 and I)
CDX2 gene expression analysis
Total RNA isolation, cDNA synthesis and qRT-PCR were performed as previously described [16]. Briefly, SCNT blastocysts from control and follistatin treated groups (n = 15 blastocysts/group, n = 6 replicates) were used for isolation of genomic DNA and total RNA using Allprep DNA/RNA micro kit (Qiagen, Valencia, CA). The extracted total RNA was used for cDNA synthesis using iScript cDNA synthesis kit (BioRad, Hercules, CA) according to the manufacturer’s protocol. Quantification of the relative transcript abundance of CDX2 gene was done by the 2−ΔΔCt method as described elsewhere [19]. CDX2 transcript abundance was normalized relative to the abundance of bovine ribosomal protein S18 (RPS18) as an endogenous control and expressed as fold change relative to the control group. Primer sequences and GenBank accession numbers were previously reported [16].
Bisulfite sequencing analysis
Analysis of CpG content by MethPrimer software [20] revealed that the bovine CDX2 gene (GenBank ID: NC_037339, Region 32081960-32087314) contains multiple CpG rich islands upstream and downstream of the transcription start site (TSS; +1) (genomic Position 32083604). Given the CDX2 regulatory elements include the promoter region and intron 1 [21, 22], three different CpG rich fragments within the promoter region (fragment P1; -1644 to -1180; P2, -305 to +126) and intron 1 (fragment I + 3030 to + 3710) were selected for bisulfite-sequencing analysis. Figure 1B provides a schematic illustration of CDX2 gene structure and position of selected fragments. Bisulfite treatment, DNA amplification, cloning and Sanger sequencing were performed following our previously reported protocols [16]. Briefly, bisulfite conversion of genomic DNA was performed using EpiTect Bisulfite Kit (Qiagen) followed by PCR amplification using GoTaq DNA Polymerase (Promega, Madison, WI). Each PCR product was extracted and purified, then cloned directly into a pCR4-TOPO vector using a TOPO-TA cloning kit for Sequencing (Invitrogen, Grand Island, NY). Cloned products were transformed into One Shot TOP10 chemically competent cells (Invitrogen). After overnight growth on selective agar plates, colony PCR was performed using M13 primers to select positive clones. Eight positive clones per replicate were selected and submitted for Sanger sequencing at Michigan State University genomic core facility. For detailed information of the bisulfite primers and different reactions conditions please see our previous publication [16].
Analysis and presentation of bisulfite sequencing data
Raw bisulfite sequencing data were aligned to the CDX2 reference sequence using BioEdit Sequence Alignment Editor (Ibis biosciences, Carlsbad, CA). Aligned sequences with sequence identity≥ 90 % and conversion rate ≥ 95 were analyzed by Bisulfite Sequencing DNA Methylation Analysis (BISMA) Software [23]. Individual CpG sites with ≥ 20 % methylation in either control or follistatin treated groups were analyzed by Fisher’s exact test to determine the differentially methylated CpG sites (DMS) [24, 25]. Methylation grids were generated by Methylation plotter web tool [26].
Transcription factor motif analysis
Motif analysis was performed by MatInspector (Genomatix) to identify putative transcription factors that bind to the differentially methylated CpG sites within the regulatory regions of CDX2 gene. A vigilant literature search was carried out to uncover any role for these transcription factors in regulating CDX2 expression.
Statistical analysis
Student T test was used to analyze embryo development and CDX2 gene expression data. Arcsine transformation was applied to the percentage data before analysis. Data are presented as untransformed mean ± standard error of mean (SEM), P < 0.05. For DNA methylation data, after initial analysis by BISMA, individual CpG sites with ≥ 20 methylation in either control or follistatin treated groups were analyzed by Fisher’s exact test to determine the differentially methylated CpG sites.
Results
Follistatin supplementation improves early development and upregulates CDX2 expression in bovine SCNT embryos
Follistatin supplementation (10ng/ml) during the initial 72h of in vitro embryo culture (IVC) triggered a significant increase in the proportion of SCNT embryos that reached the blastocyst stage at d7 (31.34 ± 0.97% Vs 20.04%± 0.85%) for follistatin and control groups, respectively (P<0.05; Fig. 2A). Moreover, the mRNA expression level of CDX2 was significantly increased in d7 blastocysts derived from follistatin treated group, compared to untreated controls (P<0.05; Fig. 2B).
Fig. 2.
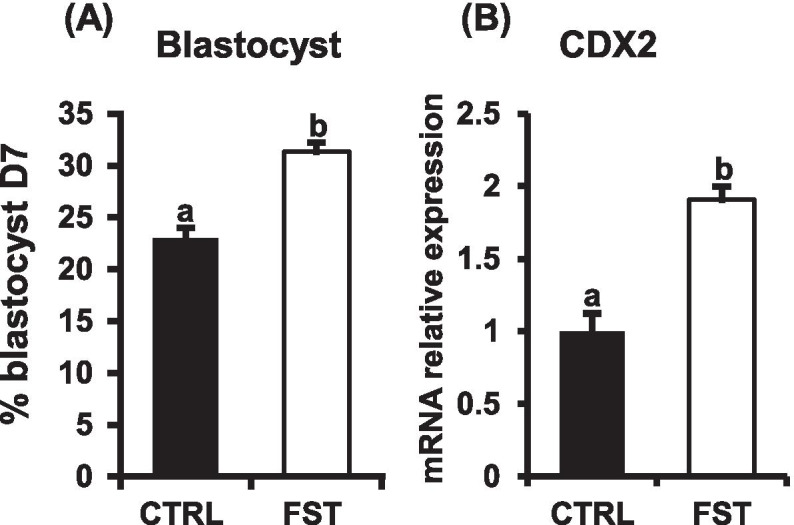
Follistatin supplementation improves blastocyst development and upregulates CDX2 expression in bovine SCNT embryos. Activated SCNT zygotes were cultured with or without follistatin (10 ng/ml) for 72h. Then, embryos were cultured in follistatin free media until d 7. A Percentage of embryos that developed to the blastocyst stage. B QRT-PCR analysis of CDX2 mRNA abundance in blastocysts in response to follistatin treatment. CDX2 mRNA expression was normalized relative to the abundance of RPS18 as a housekeeping gene. Data are expressed as mean ± SEM. Values with different superscripts indicate significant differences (P < 0.05)
Follistatin treatment induces changes in DNA methylation within CDX2 gene promoter in bovine SCNT blastocysts
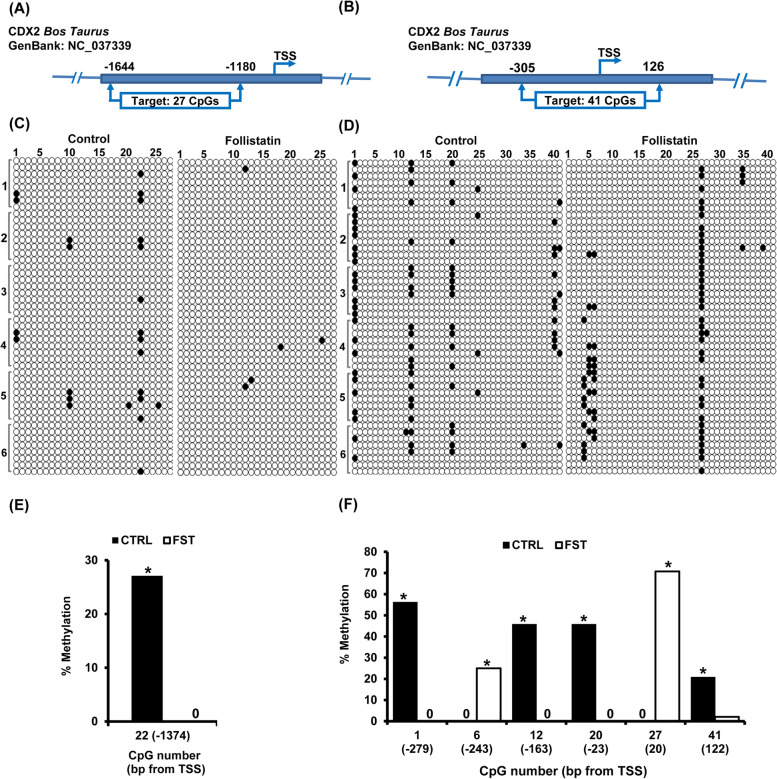
Bisulfite sequencing analysis was performed to investigate whether follistatin treatment (10 ng/ml), during the first 72h of IVC modified the methylation status of regulatory regions in the CDX2 promoter in SCNT blastocysts. Therefore, we analyzed the CpG content of CDX2 using MethPrimer software [20] (Fig. 1B). Two CpG rich fragments in the promoter region; named P1 (-1644 to -1180) and P2 (-126 to +305) were identified and selected for analysis.
For fragment P1, the target sequence was 464 bp in length (-1644 to -1180) and included 27 CpG sites (Fig. 3A). The overall methylation status of this fragment was 1.9 ± 0.45 % and 0.4 ± 0.16% for control and follistatin treated groups, respectively (Fig. 3C). Analysis of individual CpG sites, revealed that eight CpG sites were methylated in 2.1 - 27.1 % of the sequenced clones (Fig. 3C). Further analysis by Fisher’s exact test, to determine the differentially methylated CpG sites amongst the sites with ≥ 20% methylation in either control or follistatin treated groups, demonstrated a single CpG site at position -1374 that met the analysis threshold, this specific CpG site was hypomethylated in response to follistatin treatment (P<0.05; Fig 3E).
Fig. 3.
Follistatin treatment induces changes in DNA methylation within the CDX2 gene promoter in bovine SCNT blastocysts. A, B CDX2 gene map showing the genomic position of fragments P 1 ( -1644 to -1180) and P2(-305 to +126). C, D Individual CpG methylation profile in fragments P1 and P2, genomic DNA, from untreated control and follistatin treated groups (n= 6 replicates, n= 15 blastocysts/group), was subjected to bisulfite treatment followed by PCR amplification, cloning, transformation and finally colony PCR to select positive clones (n= 8 clones/replicate) for Sanger sequencing. In the methylation grid, rows represent the sequenced clones (Clones are arranged according to their replicates, replicate number are shown in the left) and columns represent CpG sites. Unmethylated CpG sites are represented by open circles while methylated CpG sites are represented by closed circle. E, F Individual CpG sites with ≥ 20% methylation, at least in one group. Values show the methylation percentage of each CpG site and those values marked with an asterisk are significantly different (*P < 0.05)
For fragment P2, the target sequence was 431 bp in length (-126 to +305) and contained 41 CpG sites (Fig. 3B). The overall methylation rate was 4.4 ± 0.31 and 3.6 ±0.31% for control and follistatin treated embryos, respectively (Fig. 3D). Analysis of individual CpG methylation sites showed that only twelve CpG sites were methylated in 2.1-70.8% of the sequenced clones (Fig. 4D), with six CpG sites meeting the criteria for analysis (≥ 20 % methylation in either control or follistatin treated groups). Statistical analysis by Fisher’s exact test revealed that four CpG sites at positions -279, -163 and – 23, 122 were hypomethylated and two other sites at positions -243 and +20 were hypermethylated in response to follistatin supplementation (P<0.05; Fig. 3F).
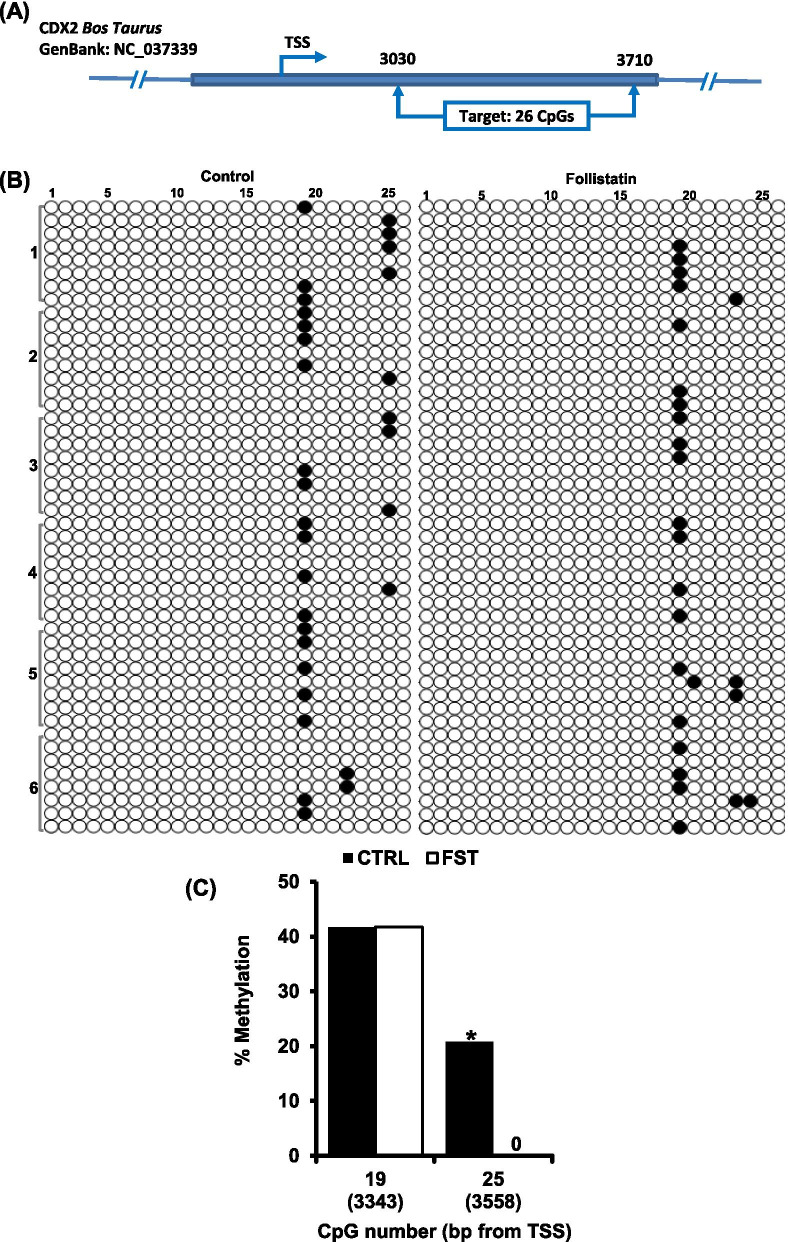
Fig. 4.
Follistatin treatment triggers changes in DNA methylation within intron 1 of CDX2 gene in SCNT blastocysts. A CDX2 gene map showing the genomic position of fragment I (+3030 to +3710). B Individual CpG methylation profile in fragment I, genomic DNA, from untreated control and follistatin treated groups (n= 6 replicates, n= 15 blastocysts/group), was subjected to bisulfite treatment followed by PCR amplification, cloning, transformation and finally colony PCR to select positive clones (n= 8 clones/replicate) for Sanger sequencing. In the methylation grid, rows represent the sequenced clones (Clones are arranged according to their replicates, replicate number are shown in the left) and columns represent CpG sites. Unmethylated CpG sites are represented by open circles while methylated CpG sites are represented by closed circle. C Individual CpG sites with ≥ 20% methylation, at least in one group. Values show the methylation percentage of each CpG site, and those values marked with an asterisk are significantly different (*P < 0.05)
Follistatin treatment triggers changes in DNA methylation within intron 1 of CDX2 gene in SCNT blastocysts
CDX2 expression is regulated via multiple regulatory regions including the proximal promoter region and intron 1 [21, 22], therefore, a CpG rich fragment from intron 1 was selected for bisulfite sequencing analysis. The target sequence of the selected fragment (I) was 680 bp in length (+3030 to + 3710) and contained 26 CpG sites (Fig. 4A). The overall methylation rate was 2.5 ± 0.26 and 2.1 ± 0.32% for control and follistatin treated groups, respectively (Fig. 4B). Analysis of methylation of individual CpG sites showed that five CpG sites were methylated in 2.1 - 41.7% of the sequenced clones, with two CpG sites at positions +3343 and +3558 meeting the threshold for analysis, ≥ 20 % methylation in either control or follistatin treated groups (Fig. 4B). Further analysis by Fisher’s exact test revealed that CpG site at position +3558 was hypomethylated in response to follistatin treatment (P<0.05; Fig. 4C), whereas the CpG site at position +3343 was not differentially methylated; no statistical difference was observed between the two groups.
Differentially methylated CpG sites are putative binding motifs for key transcription factors implicated in regulating CDX2 expression
Transcription factor motif analysis combined with a literature search revealed several binding motifs in CDX2 regulatory regions. These included key transcription factors implicated in CDX2 transcriptional regulation. For example, in fragment P1, the differentially methylated CpG site (n=1) is putative binding site for Octamer-binding transcription factor 1 (OCT1) and Heart and neural crest derivatives expressed protein (HAND). In fragment P2, the hypomethylated CpG sites (n=4) contained putative binding sites for OCT1, Activating Enhancer-Binding Protein 2 family (AP2F), and p53 protein whereas the hypermethylated CpG sites (n=2) contained binding sites for Neuron-restrictive silencer factor (NRSF), a negative regulator of gene transcription. In fragment I of intron 1, there was a single hypomethylated CpG site in follistatin treated SCNT embryos which included a putative binding site for Krüppel-like transcription factor (KLF). Table 1 provides a full list of the transcription factors that are expected to bind to DM CpG sites in the CDX2 promoter region and intron 1. Here, we focused only on TFs that are known to regulate CDX2 expression in mouse embryos (OCT1, AP2F, KLF), and TFs that may have an indirect link to CDX2 regulation (HAND, NRSF and P53). Collectively, these results indicate that follistatin treatment induces changes in DNA methylation at specific CpG sites within proximal promoter and intron 1 of CDX2 in bovine SCNT blastocysts. These DM CpG sites contain binding motifs for key transcription factors implicated in CDX2 regulation.
Table 1.
Transcription factor motif analysis of methylated CpG sites effected by follistatin treatment
| Fragment | Location (bp from TSS) |
CpG methylation in response to FST |
Putative transcription factorsa |
|---|---|---|---|
| P1 | -1374 | ↓ |
Oct1 RXRF HICF HAND HESF MYOD DEAF |
| P2 | -279 | ↓ |
OCT1 STEM |
| -243 | ↑ |
ZF5F E2FF |
|
| -163 | ↓ |
AP2F ZFXY |
|
| -23 | ↓ | P53F | |
| 20 | ↑ |
HNFP NRSF |
|
| 122 | ↓ |
WHNF ZTRE |
|
| I | 3558 | ↓ |
KLFS ZF02 SAL BED EGR GLIF NDPK |
Abbreviations: bp base pair, TSS transcription start site, FST follistatin
aPutative transcription factor binding sites that overlap differentially methylated CpG sites
Discussion
A growing body of evidence supports an embryotrophic role of follistatin in IVF embryos [11–15, 27, 28]. Exogenous follistatin supplementation (10ng/ml) during IVC1, improved blastocyst rates of bovine SCNT embryos [9]. Here, we report that follistatin supplementation increases blastocyst rates as well as CDX2 gene expression in bovine SCNT embryos. Previously, we demonstrated that follistatin treatment of bovine IVF embryos augments CDX2 expression, at least in part, by modulation of the methylation status of CDX2 gene. Follistatin induced hypomethylation of several CpG sites within the proximal promoter and intron 1 of CDX2 gene in IVF blastocysts. These sites contain putative binding motifs for transcription factors implicated in regulation of CDX2 expression in mouse embryos and trophoblast cells [16]. Therefore, we speculated that the effects of follistatin on CDX2 expression may be conserved in SCNT embryos. To address this, we analyzed the methylation status of CDX2 regulatory elements, proximal promoter, and intron 1 following the same procedures we utilized in IVF embryos [16]. Bisulfite sequencing analysis of selected fragments revealed that CDX2 is generally hypomethylated which is in line with a previous study by Zhang and colleagues [29] who reported that CXD2 is hypomethylated in bovine SCNT blastocysts.
Similar to our previous study in IVF embryos [16], follistatin treatment altered DNA methylation at specific CpG sites across the proximal promoter and intron 1 in SCNT embryos, however, some of the DM CpG sites are conserved in IVF and SCNT embryos, whereas others are exclusive for either IVF or SCNT embryos (Supplemental Table 1). In all cases, motif analysis revealed that DM sites contained putative motifs for transcription factors that are known to regulate CDX2 expression including OCT transcription factors, AP2 family, and the KLF family. In Frag P1, follistatin treatment triggered hypomethylation of one CpG site -1374 in SCNT embryos compared to hypomethylation of 2 sites −1384 and−1283 and hypermethylation of one CpG site −1501 in IVF embryos. In Frag P2, Five DM CpG sites (-279, -243, -163, -23, 20) are conserved in SCNT and IVF embryos in addition to one DM CpG site in each group, -250 in IVF and 122 in SCNT embryos. In intron region (I) follistatin reduced methylation at one CpG site in SCNT embryos (3558) compared to five hypomethylated sites (3060, 3105, 3219, 3270, 3545) in IVF embryos.
In the present study, Motif analysis revealed two putative binding sites for OCT1 were found in fragment P1 (-1374) and P2 (-279), one binding site for AP2 family is in fragment P2 (-163) and one binding site for KLFS in fragment I (+3358). Follistatin also increased methylation at a specific CpG (20) site in fragment P2 that contained a putative binding site for the transcriptional co-repressor, neuron-restrictive silencing factor (NRSF). Notably, these results are consistent with our previous findings in IVF embryos [16] which supports the hypothesis that follistatin regulates CDX2 expression via modulation of DNA methylation. Interestingly, follistatin treatment reduced methylation at putative binding sites for Heart and neural crest derivatives expressed protein (HAND) in fragment P1 and P53 protein in fragment P2. Although, to the best of our knowledge, there are no direct links between these transcription factors and CDX2 regulation in embryos, it has been shown that HAND1 is essential for trophoblast differentiation in mice [30], while activation of P53 induced Cdx2 expression in mouse embryonic stem cells [31].
Conclusions
Results of the present study demonstrate that follistatin treatment during the first 72h of IVC enhances the developmental competence of bovine SCNT embryos and upregulates CDX2 expression in blastocysts. The positive effects of follistatin on CDX2 transcription are likely linked to alterations in DNA methylation within key regulatory regions of the CDX2 gene. Collectively, these data provide additional insights into how follistatin treatment augments CDX2 expression in bovine preimplantation embryos. Further epigenetic studies are required to elucidate the role of follistatin in bovine SCNT embryos.
Supplementary Information
Additional file 1: Supplemental Table 1. Transcription factor motif analysis of methylated CpG sites effected by follistatin treatment in IVF and SCNT embryo
Acknowledgements
Not applicable
Abbreviations
- ATG
Translation initiation codon (methionine)
- CDX2
Caudal type homeobox transcription factor 2
- COCs
Cumulus oocytes complexes (COCs)
- DM
Differential Methylation
- FST
Follistatin
- H
Hour
- IVC
In Vitro Culture
- IVF
In Vitro Fertilization
- IVM
In Vitro Maturation
- SCNT
Somatic Cell Nuclear Transfer
- TE
Trophectoderm
- TGA
Translation termination codon
- TSS
Transcription Start Site
Authors’ contributions
MA contributed to study design and conceptualization; acquisition, analysis, and interpretation of data; prepared the original draft and substantially revised the manuscript. CY contributed to embryo production. SKR and JKF contributed to study design, methodology, data analysis and interpretation. JGK and GWS contributed to study design, data analysis and interpretation and fund acquisition. JGK reviewed and critically edited the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Institute of Child Health and Human Development of the National Institutes of Health (grant numbers R01HD072972 and T32HD087166) and Michigan State University AgBioResearch. The funding agencies had no role either in study design, data collection, analysis, or interpretation or in the writing of the report or in the decision to submit the paper for publication.
Availability of data and materials
All data generated or analyzed during this study are included in this published article
Declarations
Ethics approval and consent to participate
SCNT embryos were generated using oocytes harvested from ovaries collected at a local slaughterhouse within the state of Michigan. Ethical approval was not required by the Michigan State University institutional animal care and use committee (IACUC).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mohamed Ashry and Chunyan Yang contributed equally to this work.
Contributor Information
Jason G. Knott, Email: knottj@msu.edu
George W. Smith, Email: smithge7@msu.edu
References
- 1.Niemann H, Lucas-Hahn A. Somatic cell nuclear transfer cloning: practical applications and current legislation. Reprod Domest Anim. 2012;47(Suppl 5):2–10. doi: 10.1111/j.1439-0531.2012.02121.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72:1175–1184. doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 4.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 5.Schnieke AE, Kind AJ, Ritchie WA, Mycock K, Scott AR, Ritchie M, Wilmut I, Colman A, Campbell KH. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278:2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- 6.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong L, Lako M, Dean W, Stojkovic M. Epigenetic modification is central to genome reprogramming in somatic cell nuclear transfer. Stem Cells. 2006;24:805–814. doi: 10.1634/stemcells.2005-0350. [DOI] [PubMed] [Google Scholar]
- 8.Song X, Liu Z, He H, Wang J, Li H, Li J, Li F, Jiang Z, Huan Y. Dnmt1s in donor cells is a barrier to SCNT-mediated DNA methylation reprogramming in pigs. Oncotarget. 2017;8:34980–34991. doi: 10.18632/oncotarget.16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K-B, Woo J-S, Lee B-M, Park K-S, Han K-W, Kim MK. Potential functional roles of follistatin on bovine somatic cell nuclear transfer embryos. Korean J Agric Sci. 2013;40:353–358. doi: 10.7744/cnujas.2013.40.4.353. [DOI] [Google Scholar]
- 10.Patel OV, Bettegowda A, Ireland JJ, Coussens PM, Lonergan P, Smith GW. Functional genomics studies of oocyte competence: evidence that reduced transcript abundance for follistatin is associated with poor developmental competence of bovine oocytes. Reproduction. 2007;133:95–106. doi: 10.1530/rep.1.01123. [DOI] [PubMed] [Google Scholar]
- 11.Ashry M, Lee K, Mondal M, Datta TK, Folger JK, Rajput SK, Zhang K, Hemeida NA, Smith GW. Expression of TGFbeta superfamily components and other markers of oocyte quality in oocytes selected by brilliant cresyl blue staining: relevance to early embryonic development. Mol Reprod Dev. 2015;82:251–264. doi: 10.1002/mrd.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashry M, Rajput SK, Folger JK, Knott JG, Hemeida NA, Kandil OM, Ragab RS, Smith GW. Functional role of AKT signaling in bovine early embryonic development: potential link to embryotrophic actions of follistatin. Reprod Biol Endocrinol. 2018;16:1. doi: 10.1186/s12958-017-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajput SK, Yang C, Ashry M, Folger JK, Knott JG, Smith GW. Role of bone morphogenetic protein signaling in bovine early embryonic development and stage specific embryotropic actions of follistatin†. Biol Reprod. 2020;102:795–805. doi: 10.1093/biolre/ioz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KB, Bettegowda A, Wee G, Ireland JJ, Smith GW. Molecular determinants of oocyte competence: potential functional role for maternal (oocyte-derived) follistatin in promoting bovine early embryogenesis. Endocrinology. 2009;150:2463–2471. doi: 10.1210/en.2008-1574. [DOI] [PubMed] [Google Scholar]
- 15.Ashry M, Lee K, Folger JK, Rajput SK, Smith GW. Follistatin supplementation during in vitro embryo culture improves developmental competence of bovine embryos produced using sex-sorted semen. Reprod Biol. 2018;18:267–273. doi: 10.1016/j.repbio.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashry M, Rajput SK, Folger JK, Yang C, Knott JG, Smith GW. Follistatin treatment modifies DNA methylation of the CDX2 gene in bovine preimplantation embryos. Mol Reprod Dev. 2020;87(9):998–1008. doi: 10.1002/mrd.23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Sayed A, Hoelker M, Rings F, Salilew D, Jennen D, Tholen E, Sirard MA, Schellander K, Tesfaye D. Large-scale transcriptional analysis of bovine embryo biopsies in relation to pregnancy success after transfer to recipients. Physiol Genomics. 2006;28:84–96. doi: 10.1152/physiolgenomics.00111.2006. [DOI] [PubMed] [Google Scholar]
- 18.Ross PJ, Cibelli JB. Bovine somatic cell nuclear transfer. Methods Mol Biol. 2010;636:155–177. doi: 10.1007/978-1-60761-691-7_10. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 21.Benahmed F, Gross I, Gaunt SJ, Beck F, Jehan F, Domon-Dell C, Martin E, Kedinger M, Freund JN, Duluc I. Multiple regulatory regions control the complex expression pattern of the mouse Cdx2 homeobox gene. Gastroenterology. 2008;135:1238–1247. doi: 10.1053/j.gastro.2008.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Wang WC, Shashikant CS. Evidence for positive and negative regulation of the mouse Cdx2 gene. J Exp Zool B Mol Dev Evol. 2007;308:308–321. doi: 10.1002/jez.b.21154. [DOI] [PubMed] [Google Scholar]
- 23.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sproul D, Nestor C, Culley J, Dickson JH, Dixon JM, Harrison DJ, Meehan RR, Sims AH, Ramsahoye BH. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci U S A. 2011;108:4364–4369. doi: 10.1073/pnas.1013224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warden CD, Lee H, Tompkins JD, Li X, Wang C, Riggs AD, Yu H, Jove R, Yuan YC. COHCAP: an integrative genomic pipeline for single-nucleotide resolution DNA methylation analysis. Nucleic Acids Res. 2013;41:e117. doi: 10.1093/nar/gkt242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallona I, Diez-Villanueva A, Peinado MA. Methylation plotter: a web tool for dynamic visualization of DNA methylation data. Source Code Biol Med. 2014;9:11. doi: 10.1186/1751-0473-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VandeVoort CA, Mtango NR, Lee YS, Smith GW, Latham KE. Differential effects of follistatin on nonhuman primate oocyte maturation and pre-implantation embryo development in vitro. Biol Reprod. 2009;81:1139–1146. doi: 10.1095/biolreprod.109.077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhenhua G, Rajput SK, Folger JK, Di L, Knott JG, Smith GW. Pre- and Peri-/Post-Compaction Follistatin Treatment Increases In Vitro Production of Cattle Embryos. PLoS One. 2017;12:e0170808. doi: 10.1371/journal.pone.0170808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Chen X, Wang F, An X, Tang B, Zhang X, Sun L, Li Z. Aberrant DNA methylation reprogramming in bovine SCNT preimplantation embryos. Sci Rep. 2016;6:30345. doi: 10.1038/srep30345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott IC, Anson-Cartwright L, Riley P, Reda D, Cross JC. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol Cell Biol. 2000;20:530–541. doi: 10.1128/MCB.20.2.530-541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maimets T, Neganova I, Armstrong L, Lako M. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27:5277–5287. doi: 10.1038/onc.2008.166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Table 1. Transcription factor motif analysis of methylated CpG sites effected by follistatin treatment in IVF and SCNT embryo
Data Availability Statement
All data generated or analyzed during this study are included in this published article