HMG FUNCTIONAL MOTIFS
The orderly progression of most DNA-related activities such as transcription, replication, recombination, and repair involves changes in the structure of the DNA and in the organization of the chromatin fiber. Some of these structural changes are facilitated by a family of ubiquitous and abundant nonhistone nuclear proteins known as the high-mobility-group (HMG) proteins. In the narrowest traditional sense, the HMG protein family consists of six proteins and is subdivided into three subfamilies: the HMG-1/-2 subfamily, the HMG-I/Y subfamily and the HMG-14/-17 subfamily. These three HMG subfamilies are similar in several physical characteristics (detailed reviews on these proteins are found in references 10, 12, 14, 28, and 54); however, each of the subfamilies has a unique protein signature and a characteristic functional sequence motif. These functional sequence motifs are the main site of interaction between the HMG proteins and the DNA or chromatin targets. The HMG-1 domain (often referred to as the HMG-1 box) is the functional motif of the largest HMG subfamily, the HMG-1/-2 proteins; the AT hook is the functional motif of the HMG-I/Y group, and the nucleosomal binding domain is the functional motif of the HMG-14/-17 subfamily. Significantly, all of these functional motifs bind to specific structures in DNA or in chromatin, with little if any specificity for the target DNA sequence. All the HMG proteins are considered to function as architectural elements that modify the structure of DNA and chromatin to generate a conformation that facilitates and enhances various DNA-dependent activities.
The functional motifs characteristic of the HMG-1 (8, 10, 45, 61, 63) and HMG-I/Y (3, 51) subfamilies have been identified in numerous nuclear proteins that interact with DNA and chromatin. However, it is important to clearly distinguish the archetypal, or canonical, HMG proteins from the proteins containing these HMG motifs embedded in their primary sequence. The former are ubiquitous in all the cells of higher eukaryotes, are relatively abundant, and bind to DNA in a sequence-independent fashion, while the latter are cell-type specific, are not abundant, bind to DNA in a sequence-specific fashion, and frequently contain additional, distinct non-HMG functional motifs.
In considering the biological importance of the HMG motifs, it is important to take into account their relative abundance in the nucleus. The cellular levels of HMG fluctuate; however, on the average, the amount of HMG-1/-2 in a cell is about 10-fold lower than that of a histone, the amount of HMG-14/-17 is 10-fold-lower than that of HMG-1/-2, and the amount of HMG-I/Y is 10-fold lower than that of HMG-14/-17 (54). The amount of HMG-14/-17 in the average cell, about 105 molecules, is sufficient to bind to 1% of the nucleosomes, i.e., to approximately 100,000 nucleosomes. Thus, even small fluctuations in the cellular levels of these abundant proteins may have significant biological consequences, since the expression of certain genes can be affected by structural changes in a single nucleosome (118, 119).
STRUCTURE AND BINDING TARGETS OF HMG MOTIFS
Each HMG functional motif has distinct, identifiable features and induces characteristic changes in the structure of its binding target.
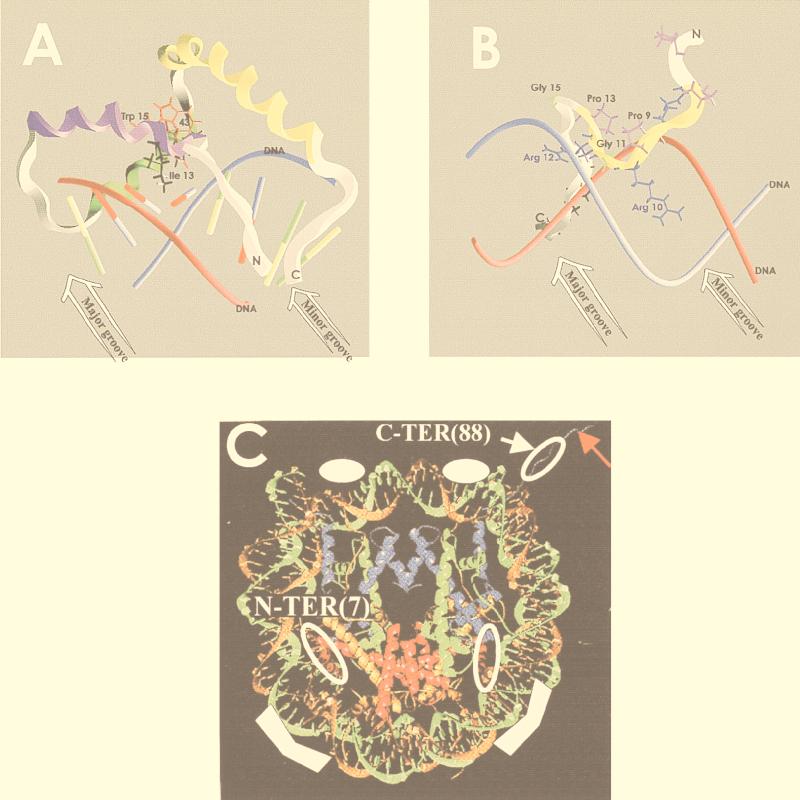
The HMG-1 domain binds to and bends the minor groove of the DNA.
The HMG-1 domain consists of approximately 80 amino acids and has a characteristic, twisted, L-shaped fold formed by three α-helical segments (10, 87, 112). Homology model building experiments suggest that the overall structure of the domain is conserved to a greater extent than that indicated by amino acid sequence homology comparison (7). The HMG-1 domain binds the DNA exclusively through the minor groove (69, 113). A wedge of hydrophobic amino acids protruding from the concave surface of the protein partially intercalates between the DNA bases, expanding the minor groove, thereby significantly unwinding and bending the DNA (Fig. 1A). The various HMG-1 domains, including those found in the archetypal HMG-1/-2 proteins, produce specific changes in the structure of the target DNA. The amino acid sequence in the helical regions of the L-shaped HMG-1 domain provides specificity in DNA binding, while the type of intercalating amino acids and the angle of the L-shaped fold affect the degree to which the DNA is unwound and bent. The interactions between the HMG-1 domain and its target are highly specific and affected by single point mutations (29, 40, 69, 70, 98, 113, 117; see references 10 and 14 for additional references). A particularly striking example is the SRY protein, in which single point mutations in the HMG-1 domain disrupt the development of the male phenotype (83, 84, 113). Additional factors that determine the binding specificity of this motif are the amino acid sequences adjacent to the HMG-1 domain (68) and the number of the domains in a protein (44).
FIG. 1.
Interaction of HMG functional motifs with their binding site. (A) HMG-1 domain. Shown is a representation of the interaction of the HMG-1 domain in LEF-1 with a 16-bp oligonucleotide containing its cognate sequence. The red and blue tubes represent the DNA, the colored sticks represent the base pairs, and the ribbon represents the polypeptide backbone. Helices I, II, and III of the HMG-1 domain are shown in violet, green, and yellow, respectively. The view illustrates the distortion caused by Ile13, the hydrophobic pocket generated by the region containing Trp15 and Trp43, and the distortion of the base pair stacking in the minor groove. (B) AT hook. Shown is a surface representation of the interaction of the second DNA binding domain of HMG-I with a region of the PRDII element of the IFN-β enhancer. The red and blue tubes represent the DNA backbone, the ribbon represents the polypeptide backbone, and the AT hook domain is shown in yellow. Note the central Arg 10-Gly 11-Arg 12 sequence positioned in the minor DNA groove. (C) Nucleosomal binding domain. Shown is a model of the HMG-14 binding sites in nucleosome cores. The DNA and histones in the core particles are shown as ribbon traces (71). The solid white symbols in the two major grooves flanking the dyad axis and approximately 25 bp from the end of the DNA indicate the regions where HMG-14/-17 proteins protect the DNA from hydroxyl radical cleavage. The open white symbols represent the approximate location of the cross-links between the N terminus and C terminus of the HMG. The red arrow points to the amino-terminal region of histone H3. Histones H2B and H3 are represented by red and blue ribbons, respectively. Panels A and B were generated by David Landsman using GRASP software (76); panel C is reproduced from reference 105. The coordinates of the structures in panels A to C are available in the protein data bank at Brookhaven National Laboratory (PDB) under 2EZE, 1LEF, and 1AOI, respectively.
The ability of the HMG-1 domain to induce site-specific DNA deformations is an important aspect of its biological function (see below). An additional fundamental property is the ability of this domain to recognize and bind to altered DNA conformations, such as stem-loops, four-way junctions, and specifically kinked or underwound DNA (see references 10 and 14 for references). This type of binding has been considered to be biologically significant; however, recent results suggest that in the nucleus, under physiological conditions, these interactions do not occur to a significant degree (81, 82).
The AT hook tethers proteins to and unbends the minor groove of the DNA.
The AT hook motif is a positively charged stretch of 9 amino acids containing the invariant tripeptide GlyArgPro (GRP), usually flanked by arginine residues (89). Pattern analysis of the amino acid residues flanking the conserved GRP tripeptide indicates that the AT hook motifs can be further subdivided into three classes; these bind to their DNA targets with varying affinities but with very little specificity for the DNA sequence (3, 51). The AT hook binds to the DNA through the minor groove with the optimal DNA binding site centered at the sequence AA(T/A)T (3, 72). The AT hook has a narrow DNA recognition surface which is devoid of hydrophobic amino acids and does not significantly distort the B-form DNA structure (Fig. 1B). In fact, detailed studies on the interaction of HMG-I protein with the beta interferon (IFN-β) enhancer sequence suggest that the AT hook could reverse and prevent intrinsic distortions in DNA conformation (31, 51, 123). These structural changes facilitate the binding of additional transcription factors to the enhancer region (31, 51, 123). In the archetypal HMG-I/Y and the closely related HMGI-C (43), the regions outside the AT hook motifs also contribute to DNA binding (35). The specificity of these regions is further increased by posttranslational modifications, some of which are cell cycle regulated (88, 92). The archetypal AT hook proteins contain three copies of this motif; however, numerous nuclear proteins contain this motif in either single or multiple copies (3). These AT hook proteins frequently contain other known functional domains, such as histone folds, zinc fingers, and homeodomains. These regions and the distance between adjacent AT hooks affect the DNA sequence specificity of the proteins (35, 51, 123). The AT hook motifs themselves may serve to tether these proteins to the minor grooves of their binding sites and induce conformational changes that promote their cellular function.
The nucleosomal binding domain anchors HMG-14 and HMG-17 proteins to nucleosomes.
The third functional HMG motif, the nucleosomal binding domain, has been identified in the canonical HMG-14/-17 proteins, in a set of proteins found in trout tissues (24, 54), and in the tissue-specific thyroid hormone receptor interactor Trip7 (64). HMG-14 and HMG-17 are archetypal HMG proteins in that they are relatively abundant and found in the nuclei of all higher eukaryotes. They are the only nuclear proteins known to specifically recognize the generic structure of the 146-bp nucleosome core, i.e., the building block of the chromatin fiber (14). The two proteins bind to nucleosomes cooperatively and form complexes containing either two molecules of HMG-14 or two molecules of HMG-17 but not complexes containing one molecule of each protein (86). As shown in Fig. 1C, the major sites of interaction between these proteins and the nucleosomal core DNA are located 25 bp from the end of the DNA and in the two major grooves flanking the nucleosomal dyad axis (2). The nucleosomal binding domain motif is a positively charged stretch of approximately 30 amino acids with a bipartite structure: the amino-terminal region is extremely conserved and enriched in arginine residues, while the carboxy-terminal region is highly enriched in lysine and proline residues (14). This domain serves as the primary site of interaction between the HMG-14/-17 proteins and the nucleosome core. The nucleosomal footprint of the isolated nucleosomal binding domain is very similar to that of the entire HMG molecule; however, neither the structure of the free domain nor that of the nucleosome-bound domain is known. The binding of HMG-14/-17 may induce specific changes in the conformation of the nucleosome core particle (86); it is not known whether these changes can also be induced by the isolated nucleosomal binding domain. Although the target of HMG-14/-17 is the nucleosome, the main function of these proteins is to change the architecture of the higher-order chromatin structure (see below). These changes are mediated through the C-terminal region of the proteins (26, 106). The nucleosomal binding domain anchors these HMG proteins to the nucleosome cores to facilitate HMG-14/-17-dependent changes in chromatin structure.
BIOLOGICAL FUNCTION AND MECHANISM OF ACTION
The unifying functional theme of the HMG motifs is their ability to modify the structure of their target site and induce structural changes that facilitate the progression of a wide range of DNA-dependent activities. The structural alterations induced by the HMG motifs embedded in sequence-specific proteins affect only the function of their target genes. In contrast, the archetypal HMGs have multiple targets and coregulate many types of DNA-binding activities through both specific and nonspecific sequence manipulation of DNA structure. What is the mechanism whereby these evolutionarily conserved proteins, which bind to their target in a sequence-independent manner, regulate a wide array of activities?
Multiple modes of sequence recognition and DNA bending by the HMG-1 domain.
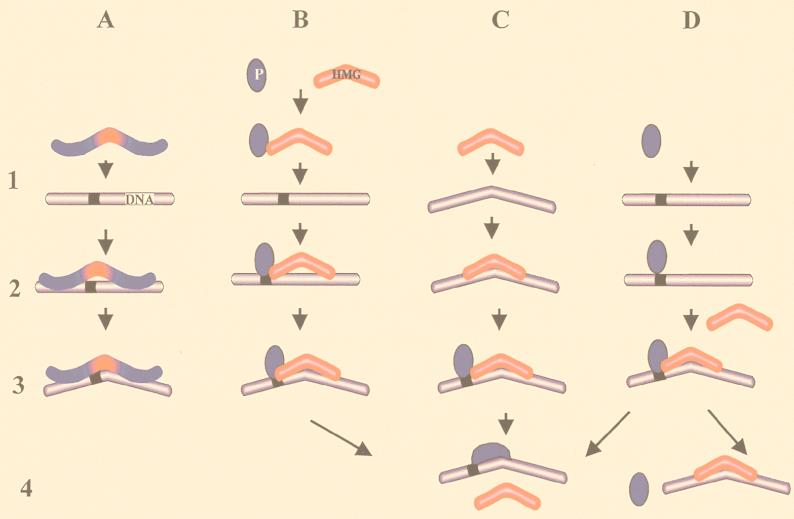
A common theme unifying the mechanism of action of the archetypal, non-sequence-specific HMG-1/-2 proteins and the sequence specific HMG-1 domain proteins is that all use a protein module necessary for sequence-specific DNA recognition and a module involved in DNA bending. The HMG-1 domain proteins recognize their targets in a sequence-specific fashion. The broad-range effects of the archetypal HMG-1/-2 proteins could be explained if it is assumed that these proteins have multiple modes of action and can function as independent units that are mobilized by sequence-specific proteins to induce a conformational change at a specific locus. As indicated in Fig. 2, the binding of an HMG-1 domain to its target can be arbitrarily divided into three major steps. In step 1 a protein targets a DNA region, in step 2 the protein binds to that region, and in step 3 the formation of a complex containing bent DNA, a DNA recognition module, and a DNA-bending protein module occurs. The conformational changes in the structure of the DNA induced by these proteins (represented by step 3 in all the schemes) juxtapose distantly bound proteins and facilitate the assembly of multiprotein complexes.
FIG. 2.
Multiple modes of sequence recognition and DNA bending by the HMG-1 domain. The HMG-1 domains are shown in red, the DNA recognition elements are shown in blue, and the black ribbon on the DNA represents a specific oligonucleotide sequence. The numbers on the left indicate four arbitrarily chosen steps involved in the interaction of an HMG-1 domain with its target. Step 1 represents targeting of a molecule to its binding site, step 2 represents binding, step 3 represents the induction of a conformational change in the target DNA, and step 4 represents possible changes in the protein contacts in the DNA-protein complex. The letters represent four different binding modes. (A) Binding by a sequence-specific HMG-1 domain protein. The HMG-1 domain targets the protein to a specific sequence and induces a conformational change in that locus. (B to D) Binding by the archetypal HMG-1/-2 proteins. (B) A preformed binary complex of a sequence-specific protein or protein complex (P) and an HMG molecule bind to a specific sequence. (C) The HMG-1/-2 protein targets distorted DNA structures. In step 3 a specific protein may target the DNA-HMG complex. (D) A sequence-specific protein binds to its cognate site and triggers the subsequent binding of an HMG-1/-2 protein. The arrows point out possible dissociation pathways of the resulting DNA-protein complexes. For further details, see the text.
A close parallel between the binding of a sequence-specific HMG-1 domain protein and the archetypal HMG-1/-2 protein becomes obvious when comparing Fig. 2A and B. In Fig. 2A, the DNA recognition module is covalently linked to or is part of the HMG-1 domain. This scheme depicts the binding of the sequence-specific HMG-1 domain proteins such as LEF-1 (lymphoid enhancer factor 1), (69), SRY (113), and more than 100 sequence-specific DNA-binding proteins containing the HMG-1 domain motifs. In Fig. 2B, the archetypal HMG-1/-2 proteins combine with a sequence-specific protein (i.e., DNA recognition module) prior to the interaction with DNA. The binary complex formed by the association of a DNA recognition module with a DNA-bending module can induce a conformational change at a specific locus (Fig. 2B, step 3), much like the sequence-specific HMG-1 domain proteins. These types of functional interactions have been shown for the POU domain of Oct 1 and Oct 2 (130), for HOXD9 (125), for the human TATA-binding protein (36, 97), and for the adeno-associated virus replication protein (19). In a variation of this scheme, a binary complex of HMG-2 and the splice variant SP100B is recruited by a homologue of the heterochromatin protein HP1 to induce transcriptional silencing (66). In fact, the schemes in Fig. 2A and B are even more similar than the figure implies, since even the sequence-specific HMG-1 domain proteins are sometimes targeted to their binding sites by forming binary or ternary complexes with other proteins (55).
Alternatively, either the HMG-1 domain (Fig. 2C) or a sequence-specific protein (Fig. 2D) could first bind to the target DNA independently and facilitate the subsequent binding of the complementary module. The first two steps of the scheme in Fig. 2C represent the binding of the canonical HMG-1 proteins to distorted DNA conformations, such as four-way junctions or cis-platinum-modified DNA (see references 10 and 14 for references). It is not clear whether these interactions occur under physiological conditions (81); however, if they do, they may have significant biological effects. Indirectly, these interactions could affect the binding of sequence-specific HMG-1 domain proteins, such as SRY, LEF-1, or UBF (59). These proteins are classed as sequence specific; however, they also bind with relatively high affinity, in a sequence-independent manner, to any bent or distorted DNA (33, 41, 45). The abundant canonical HMG-1/-2 protein competes for these sites, thereby increasing the effective cellular concentration of the sequence-specific HMG-1 domain proteins. In addition, the structural alteration induced by the binding of HMG-1/-2 proteins to distorted DNA could be necessary for the subsequent binding of additional factors, as in the case of the NS1 endonuclease involved in the rolling-circle-type DNA replication of parvoviruses (20) or in the HMG-1-mediated stimulation of the binding of the estrogen receptor to its response element, which occurs only when HMG-1 is preincubated with the DNA probe (109).
Conversely, as illustrated in Fig. 2D, the binding of a sequence-specific element to its target may facilitate the subsequent binding of the archetypal HMG-1 protein, as was shown for the effect of HMG-2 on basal transcription (96) or the effect of HMG-1/-2 on the binding of steroid receptors to target DNA (11). Presumably, the HMG-1/-2 proteins are recruited into the complex both by receptor-mediated bending of the target DNA and by protein-protein interactions. A similar situation could account for the effect of HMG-1/-2 on the RAG-1- and RAG-2-dependent V(D)J cleavage (91, 108). HMG-1 and HMG-2 stimulate V(D)J cleavage on templates in which the binding sites for RAG-1 and RAG-2 are separated by 23 bp but have no effect on templates in which the two binding sites are separated by only 12 bp. Presumably, by mediating bending of the longer template, HMG-1 promotes contact between the two RAG proteins and the assembly of a complex active in DNA cleavage and recombination. It is highly significant that HMG-1 affects V(D)J recombination not only on naked DNA but also at the level of individual, specific nucleosomes (60).
Often, the details of the mechanism whereby HMG-1/-2 proteins stimulate the activities of DNA-binding proteins are not fully understood, as in the case of the HMG-1-mediated activation of p53 (53) which could occur by any or even a combination of the mechanisms illustrated in Fig. 2B to D. A complicating factor in these studies is the difficulty of directly demonstrating the formation of a ternary complex containing HMG-1/-2, a regulatory protein, and a target DNA sequence (53, 78, 108, 109, 125, 130). Most likely, many of the interactions are weak and the HMG-1/-2 proteins can be easily lost from the complex (illustrated by step 4 in Fig. 2).
The multiple pathways whereby HMG-1/-2 proteins can cause distortions in DNA structure may provide an explanation for the wide range of molecular interactions affected by these proteins. In addition, in some instances the generic HMG-1/-2 proteins can replace sequence-specific DNA-binding proteins (93), and even the sequence-specific HMG-1 domains are interchangeable (39). At the same time, each of the HMG-1 domains found in the HMG-1/-2 proteins binds to DNA in a characteristic way (124), and many of the interactions involving the HMG-1 domain are highly specific. For example HMG-1 binds to the homeobox of HOXD9 but not to that of HOXD8 (125), which interacts with TATA-binding protein in a tissue-specific fashion (97), and stimulates the DNA binding of steroid receptors without affecting other nuclear receptors such as VDR or RAR (retinoic acid receptor) (11).
In addition, HMG-1/-2 proteins stimulate overall transcription from chromatin templates (see reference 14 for references). The stimulation could be due to some change in the overall structure of chromatin, perhaps mediated by the negatively charged C terminus (1), or as illustrated above, by affecting the activity of a specific transcription factor.
The AT hook motifs facilitate the function of proteins that bind to DNA in either a sequence-specific or -nonspecific manner.
The AT hook motif serves as the predominant functional motif of the archetypal HMG-I/Y (and HMGI-C) proteins and is also found embedded in numerous nuclear proteins (3). Many of these nonarchetypal AT hook proteins contain additional domains related to proteins that are involved in chromatin functions, such as the globular region of histone H1, histone acetylase, and subunits of the chromatin remodeling complexes (3). In these proteins, the AT hook may serve as a contact which affects the specificity and affinity of the DNA-binding protein.
Although some sequence specificity is present in the AT hook itself, the main function of this motif is to anchor the proteins to the minor groove of the DNA, near sequences targeted by other regions of the AT hook proteins. Thus, similarly to the HMG-1 domain, a common denominator of the mechanism of action of the sequence-specific proteins and the archetypal AT hook proteins is the combined action of a module that recognizes a specific DNA sequence and a module that changes the conformation of the DNA (Fig. 2).
The archetypal HMG-I/Y proteins are involved in several types of nuclear functions. Thus, HMG-I/Y proteins seem to be major structural elements of metaphase chromosomes, involved in the formation of the classic chromosomal banding pattern (90). Indeed, expression of a synthetic protein containing multiple AT hooks blocks mitotic chromosome assembly and, if properly targeted, suppresses position effect variegation of specific genes (42, 46). Likewise, by displacing histone H1 from scaffold-associated regions, the HMG-I/Y may antagonize the H1-mediated general repression of transcription (126). By interfering with the binding of homoedomain proteins they may affect embryonic development (4). The HMG-I/Y proteins also function as general host factors necessary for the formation of human immunodeficiency virus type 1 and Moloney murine leukemia virus preintegration complexes (32, 67).
In addition, the HMG-I/Y proteins modulate the expression of specific genes such as the inducible form of nitric oxide synthase (80), tumor necrosis factor beta, IFN-β, interleukin 2 receptor α, E-selectin, interleukin 4, human GP91-PHOX, ɛ-immunoglobulin G (see reference 14 for references), and T-cell receptor alpha (6). In several cases the effects of HMG-I/Y and of HMGI-C are synergistic with those of NF-κB (73, 99) and other regulatory factors. The conformational changes induced by HMG-I/Y at specific DNA sequences may promote the assembly of either repressors or activators near regulatory regions. The mechanisms whereby HMG-I/Y are targeted to specific regions may be similar to those delineated for HMG-1/2 and involve both specific protein-protein and protein-DNA interactions. For example, HMG-I/Y enhances the activity of the SRF (serum response factor) by interacting directly with SRF rather than by changing the conformation of the DNA (17). The best-studied example of the effect of HMG-I/Y on gene expression is provided by the virus-inducible enhancer of IFN-β. Binding of HMG-I/Y and at specific positive DNA regulatory elements leads to DNA unbending; recruitment of the transcription factors NF-κB, ATF2/c-Jun, and IRF-1; and the formation of a stable nucleoprotein structure termed the enhanceosome (100). Formation of the enhanceosome is dependent both on DNA unbending and on specific contacts between HMG-I/Y and the transcription factors. The pivotal role of HMG-I/Y proteins in the assembly of the enhanceosome and in IFN-β gene expression had been recently demonstrated by the finding that specific acetylation of HMG-I by CBP, but not by PCAF, disassembles the enhanceosome and turns IFN-β gene expression off (74).
HMG-I/Y/C proteins are preferentially expressed in early development. Disregulation of their expression is associated with alteration in cellular growth and differentiation (14, 37, 38, 127). Lack of HMGI-C protein expression leads to the mouse pygmy phenotype (128). HMG-I/Y/C expression is significantly upregulated in many types of cancer (14, 16, 43, 127), leading to the suggestion that elevated levels of these proteins can be used as a diagnostic feature for transformed cells. Especially striking is the correlation between rearrangement in the chromosomal region containing the HMGI-C gene, and to a lesser degree the HMG-I/Y gene, and very common benign mesenchymal tumors, such as lipomas, uterine liomas, and endometrial polyps (5, 56–58, 102, 127). In some cases these translocations fuse the AT hook to novel transcription regulatory domains, leading to DNA mistargeting (5). Although it is not clear whether the rearrangements in the HMG-I/Y/C gene are primary or secondary events in the etiology of these benign cancers, it is generally accepted that the upregulated expression of these genes is an important factor in this process (102, 114). The widespread involvement of the AT hook motif in many types of cancers makes this motif and its binding site a potential target for anticancer drugs (43, 51).
HMG-14/-17 proteins decompact chromatin.
The nucleosomal binding domain motif may facilitate the binding of HMG-14/-17 variants, such as Trip7 (64), to chromatin subunits. In the HMG-14/-17 proteins, this motif serves as the main nucleosomal binding site. HMG-14/-17 proteins enhance transcription and replication but only from chromatin and not from DNA templates (22, 26, 27, 79, 103, 104, 106, 110). Thus, the proteins act as modifiers of the chromatin structure rather than polymerase-specific factors. The enhancement of the DNA-dependent activities is associated with a decompaction of the nucleosome array in the chromatin fiber (26, 104, 106, 110). Both transcriptional activation and chromatin decompaction are mediated by the negatively charged C-terminal domain of HMG-14/-17 proteins (26, 106). The isolated nucleosomal binding domain or mutants lacking the C-terminal domain inhibit transcription and fail to decompact chromatin. Specific displacement of HMG-14/-17 from chromatin by incubation with the nucleosomal binding domain inhibits transcription both in vitro (106) and in vivo (50). The carboxy-terminal domain forms specific contacts with the amino-terminal tail of histone H3, near the lysine residues which serve as targets for histone acetyltransferases (105). In fact, the presence of HMG-14/-17 inhibits the PCAF-mediated acetylation of H3 (47). Trypsinized nucleosome cores, lacking the amino-terminal tails of the core histones, interact poorly with HMG-14/-17 (21). The nucleosomal footprint of HMG-14/-17 partially overlaps with that of histone H1 (2), and HMG-14 relieves the H1-mediated transcriptional inhibition and compaction of simian virus 40 minichromosomes (26). Thus, HMG-14/-17 proteins decompact chromatin by targeting the two main elements known to be involved in chromatin compaction: histone H1 and the amino termini of the core histones. These interactions may weaken internucleosomal contacts mediated by the amino termini of the histone cores or perhaps change the entry or exit angle of the nucleosomal DNA which is affected by the presence of histone H1 (9). These two modes of action are not mutually exclusive, in fact they could act synergistically.
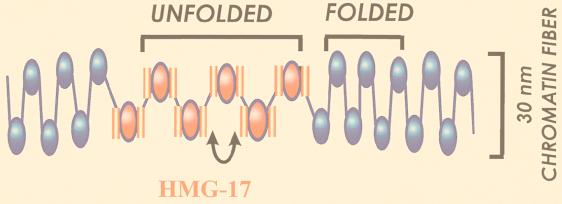
In chromatin, nucleosomes containing HMG-17 are clustered into domains which on the average consist of six contiguous nucleosomes (Fig. 3) (85). Thus, the conformational changes induced by HMG-14/-17 may be spread over a long stretch of oligonucleosomes and are not confined to the vicinity of a single nucleosome. The mechanism whereby HMG-14/-17 proteins are incorporated into chromatin are not fully understood. In some cases it is possible that these proteins are assembled into nascent chromatin during replication (15). However, recent results suggest that these nucleosomal binding proteins are not always associated with chromatin (49) and that their intranuclear location is dependent on transcriptional activity. In actively transcribing cells the proteins are distributed throughout the nucleus in numerous foci, while in transcriptionaly quiescent cells the proteins relocate and accumulate in or near clusters of interchromatin granulae (50). In this scenario, the HMG-14/-17 proteins are actively recruited, by a mechanism yet to be determined, to sites of transcription. Thus, HMG-14/-17 may be incorporated into preassembled chromatin. An attractive but yet-untested possibility is that nucleosome remodeling complexes (120) play a role in this process. The nucleosomal binding domain anchors the proteins to nucleosomes, and the carboxy-terminal domain interacts with the amino termini of the core histones and perhaps with histone H1. These interactions reduce chromatin compaction and provide increased access of various factors to the nucleosome, i.e., to the primary level of DNA packing in chromatin.
FIG. 3.
Model of the organization of HMG-17 in chromatin. Nucleosomes containing two molecules of HMG-17 (pink) are clustered into a domain whose average size is six contiguous nucleosomes. In this domain the entry-exit angles of the linker DNA are altered (double-headed arrow) and the higher-order structure of the chromatin is unfolded. HMG-17 may unfold the higher-order structure by interacting with the amino termini of the core histone, with histone H1, or with both. The structure of the 30-nm chromatin fiber is not known. The model presented was inspired by cryoelectron microscope studies, which suggested a zigzag organization of nucleosomes in the 30-nm fiber (9). However, it is equally possible that the 30-nm chromatin fiber has a solenoidal structure (34). The effect of HMG-14/-17 on the higher-order chromatin structure is the same for both the zigzag and solenoid organizations of nucleosomes.
The interaction of HMG-14/-17 proteins with nucleosome core particles stabilizes the structure of the nucleosome subunit (see reference 14 for references), a situation which is inconsistent with enhancement of transcription or replication. It is possible that in the larger context of chromatin these proteins do not stabilize the structure of the chromatin subunit. Alternatively, posttranslational modifications may weaken the interaction of the proteins with nucleosome cores and dissociate the proteins from chromatin (47). Conceivably, the dynamic interaction of HMG-14/-17 with chromatin proceeds in several steps. In the first step the proteins are recruited to specific sites, in the second step they bind to nucleosomes and unfold the higher-order chromatin structure, and in the third step they are modified and dissociate from chromatin.
ARE HMG CHROMOSOMAL PROTEINS A SPECIFIC CLASS OF NUCLEAR PROTEINS?
The HMG motifs are found in numerous nuclear proteins; however, on a quantitative basis, most of these domains are present in the archetypal HMG proteins. These chromosomal proteins have been grouped into a specific class of nonhistone proteins based on their ubiquitous distribution in the nuclei of all higher eukaryotes, on their association with isolated chromatin, on common physical properties (54), and on the observation that all these proteins act as architectural elements that affect DNA-related activities, supposedly in the context of chromatin (14). These proteins are also similar in that their carboxy-terminal regions contain significantly more negative charges than the amino-terminal regions. Even the transcripts coding for the proteins are similar in that all contain long, AT-rich 3′ untranslated regions, perhaps indicative of similarity in posttranscriptional regulation of protein levels. However, in view of the structural and functional specificity of the motifs characteristic of the three HMG subfamilies, it is important to reexamine whether these proteins should indeed be considered a specific family of nonhistones.
Unique proteins and specific functions.
As elaborated in the previous sections, each of the HMG families has a unique functional motif, induces specific changes in their target binding sites, and performs unique and distinguishable cellular functions. HMG-14/-17 proteins target the nucleosome and act by modifying the higher-order chromatin structure. HMG-1/-2 and HMG-I/Y target the minor groove of the DNA and induce specific local distortions in DNA conformation. With the exception of HMG-I/Y, each of the HMG proteins is encoded by a unique gene and all the HMG proteins are found in most higher eukaryotic cells. The ubiquitous distribution of all the HMG proteins argues that each of the archetypal HMG proteins is involved in a distinct and important cellular function. It is not clear to what degree there is functional redundancy even among the members of an HMG subfamily, which structurally are very similar. Thus, HMG-4 (107) and HMGI-C (43), which are extremely similar to HMG-1/2 and HMG-I/Y, respectively, are expressed specifically in early development or in certain types of neoplastic tissues. Depletion of HMG-2 by antisense technology affects the rate of cell proliferation (121); clearly the closely related HMG-1 cannot substitute for the missing HMG-2. Likewise, deletion of the HMG-1 gene causes lethal hypoglycemia; the HMG-2 gene does not compensate for the missing HMG-1 gene (15a). Knocking out the HMGI-C gene results in a pygmy mouse phenotype (128); the closely related HMG-I/Y cannot compensate for the missing protein. Preliminary studies with chimeric mice suggest that the converse may also be true; i.e., HMGI-C may not be able to compensate for a reduction in the amount of HMG-I/Y protein (35a). Thus, the archetypal proteins have specific functions and participate in a diverse range of nuclear activities.
Common targets for HMG proteins.
In spite of the above-mentioned differences between the various HMG proteins, there are certain similarities in their target binding sites, and all may affect the binding of the linker histone H1 to chromatin. All the HMG proteins and histone H1 preferentially target AT-rich sequences. The HMG-1 domain and the AT hook are two of only four known examples of protein motifs that bind to and deform the DNA by interacting with the minor groove (51). Likewise, several basic motifs in the termini of histone H1 also seem to interact strongly with the AT-rich region in the minor groove of the DNA (18). HMG-1/-2 and HMG-I/Y compete among themselves and with histone H1 for binding to certain distorted DNA structures, such as four-way junctions or cis-platinum-modified DNA (48, 122), and HMG-I/Y competes with H1 for binding to the AT-rich nuclear matrix elements (126). The contacts of HMG-14 and HMG-17 in the major grooves flanking the nucleosomal dyad axis partially overlap those of histone H1 (2, 111), and HMG-14 enhances polymerase II-dependent transcriptional elongation by counteracting the repressive activities of histone H1 (26). In the linker region between adjacent cores, the H1 contacts partially overlap those of HMG-1/-2 proteins. An inverse quantitative relation between HMG-1 and histone H1 during Xenopus laevis (25, 77) and Drosophila melanogaster (75) development, which correlates with a switch in the rate of transcription, has also been reported. Although this correlation has not been observed in mouse embryonic development (95), results from several laboratories suggest an interplay between HMG-1/-2 and histone H1 (77, 116, 129). Thus, a common chromatin target for all HMG proteins could involve some structural aspect linked to histone H1.
Are HMG proteins intrinsically associated with chromatin?
Because HMG proteins were found associated with purified chromatin preparations they were considered to be a special class of nonhistone proteins, whose interaction with chromatin is more stable and permanent than that of regulatory factors that bind only transiently to chromatin (14, 54). Indeed, HMG-14/-17 proteins bind to isolated nucleosome cores, and HMG-I/Y may be an important structural element of metaphase chromosomes. However, by definition, HMG proteins can be extracted from chromatin and nuclei with 0.35 M NaCl; this ionic strength is very close to that used to prepare nuclear extracts enriched in transcription, replication, or other regulatory factors. In fact HMG-14/-17, the only nuclear proteins that specifically interact with the nucleosome core particle, migrate and redistribute among nucleosomes at significantly lower ionic strength (62). Chromatin is often isolated at a low ionic strength, conditions which may lead to nonspecific adsorption of nuclear proteins to the nucleohistone fiber. It is likely that originally the highly charged HMG proteins were found in chromatin simply because they are relatively abundant and therefore easier to detect. Indeed, more sensitive staining techniques reveal that most chromatin preparations contain numerous nonhistone proteins. In many respects the intranuclear organization of the HMG proteins is similar to that of many other nonhistone nuclear proteins. Both the HMG-14/-17 class (49) and the HMG-1/-2 class (30, 52) transiently dissociate from chromatin during mitosis and are not found in mitotic chromosomes. Like most nuclear proteins, HMG-14/-17 proteins reenter the nucleus by facilitated transport, only after the formation of the nuclear membrane (49). A nuclear localization signal has also been experimentally defined in HMG-2 (94), and nuclear entry of HMG-1 requires a dephosphorylation step (115). In some cells a significant portion of HMG-1/-2 remains in the cytoplasm throughout the cell cycle (13). Furthermore, recent results suggesting that the intranuclear organization of HMG-14/-17 is dependent on transcriptional activity (50) raise the possibility that the proteins are actively recruited to transcription sites and are not permanently associated with chromatin. Likewise, even though the HMG-I/Y proteins seem to be a structural element of the metaphase chromosome, these proteins are actively recruited to the enhancer region of specific genes (100). Thus, the intranuclear organization of at least part of the HMG proteins is dynamic rather than static, and not all the molecular species are always associated with chromatin.
Flexible modules in multiprotein complexes.
Recent studies on the mechanisms of action of the archetypal HMG proteins raise the possibility that all of them function as modular units in the context of multiprotein complexes. As illustrated in Fig. 2, there are several mechanisms whereby either HMG-1/-2 or HMG-I/Y could be recruited by other components to affect the local DNA or chromatin conformation at specific target sites. Likewise, the finding that the nuclear organization of HMG-14/-17 is dynamic raises the possibility that these HMG proteins are recruited by components of the transcription or replication complex to unfold chromatin at specific sites. In this respect, the HMG proteins resemble other regulatory factors, which also function in the context of dynamic multiprotein complexes. As already pointed out by others (23, 65, 101), the composition of these complexes could be governed by the specificity of the DNA binding site and by the physiologic status of a cell. The final specificity and affinity of these regulatory complexes are the result of multiple protein-protein and protein-DNA interactions, each of which occurs with relatively low specificity and affinity. Often, the specificity of the interaction between the various components of the multiprotein complexes is determined by only a few amino acid side chains (23). Thus, in spite of a high degree of sequence conservation, the HMG proteins could specifically interact with both tissue-specific regulatory factors and components of the basal transcription or replication machinery. In fact, the high degree of sequence conservation may be indicative of a requirement to form multiple complementary interactions in many types of multiprotein complexes. In these complexes, the HMG proteins are the functional module necessary for inducing specific changes in the conformation of the targeted binding site.
SUMMARY AND PERSPECTIVE
The prevalence of the three HMG functional motifs among DNA-binding proteins and the abundance of the HMG proteins in the nucleus suggest that these motifs play a major role in facilitating the orderly progression of many DNA-related activities. Aberrant expression of HMG proteins or of the functional HMG motifs may alter the cellular phenotype and is associated with the etiology of certain cancers. On a quantitative basis most of these motifs are found in the archetypal HMG proteins. This set of nuclear proteins has been classed as a specific family based on common physical characteristics. In fact, each HMG subfamily is a distinct set of proteins with identifiable structural characteristics and a specific type of targets. Each subfamily induces characteristic changes in the structure of its binding site. Recent results suggest that a common mechanistic feature of all the HMG proteins is their ability to function as flexible modules in the context of many types of multiprotein complexes. If this is indeed true, quantitative changes in these proteins could be expected to have pleiotropic effects and to affect many cellular activities. Thus, an important future direction is to elucidate the mechanisms whereby these proteins are recruited into the various complexes. Conceivably, the archetypal HMG proteins could even influence the activity of sequence-specific DNA-binding proteins containing functional motifs related to the HMG proteins. As a group, the HMG proteins seem to affect many types of DNA-related activities, in both a sequence-dependent and -independent fashion. An important future goal is to determine which of the effects observed in the various test systems are truly biologically significant. In the nucleus of the cell, most of the DNA-related activities occur in the context of chromatin. The data available suggest a functional interplay between the linker histone H1 and the HMG proteins. It is necessary to understand how these proteins, and their related functional motifs, bind to their targets and modify the conformation of their binding sites in the context of chromatin.
ACKNOWLEDGMENTS
I thank Marco Bianchi and Raymond Reeves for discussions on topics covered in this review and David Landsman, Dolph Hatfield, Yuri Postnikov, Michael Bergel, Julio Herrera, and Hitoshi Shirakawa for critical comments on the manuscript.
REFERENCES
- 1.Aizawa S, Nishino H, Saito K, Kimura K, Shirakawa H, Yoshida Y. Stimulation of transcription in cultured cells by high mobility group protein 1: essential role of the acidic carboxyl-terminal region. Biochemistry. 1994;33:14690–14695. doi: 10.1021/bi00253a006. [DOI] [PubMed] [Google Scholar]
- 2.Alfonso P J, Crippa M P, Hayes J J, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlotta P, Rustighi A, Mantovani F, Manfioletti G, Giancotti V, Tell G, Damante G. High mobility group I proteins interfere with the homeodomains binding to DNA. J Biol Chem. 1997;272:29904–29910. doi: 10.1074/jbc.272.47.29904. [DOI] [PubMed] [Google Scholar]
- 5.Ashar H R, Cherath L, Przybysz K M, Chada K. Genomic characterization of human HMGIC, a member of the accessory transcription factor family found at translocation breakpoints in lipomas. Genomics. 1996;31:207–214. doi: 10.1006/geno.1996.0033. [DOI] [PubMed] [Google Scholar]
- 6.Bagga R, Emerson B M. An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- 7.Baxevanis A D, Bryant S H, Landsman D. Homology model building of the HMG-1 box structural domain. Nucleic Acids Res. 1995;23:1019–1029. doi: 10.1093/nar/23.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxevanis A D, Landsman D. The HMG-1 box protein family: classification and functional relationship. Nucleic Acids Res. 1995;23:1604–1613. doi: 10.1093/nar/23.9.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bednar J, Horowitz R A, Grigoryev S A, Carruthers L M, Hansen J C, Koster A J, Woodcock C L. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi M. The HMG-box domain. In: Lilley D M, editor. DNA-protein: structural interactions: frontiers in molecular biology. Oxford, United Kingdom: IRL Press; 1995. pp. 177–200. [Google Scholar]
- 11.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M E, Taraseviciene L, Nordeen S K, Allegretto E A, Edwards D P. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin M, Lehn D A, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 13.Bustin M, Neihart N. Antibodies against chromosomal HMG protein stain the cytoplasm of mammalian cells. Cell. 1979;16:181–189. doi: 10.1016/0092-8674(79)90199-5. [DOI] [PubMed] [Google Scholar]
- 14.Bustin M, Reeves R. High mobility group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 15.Bustin M, Trieschmann L, Postnikov Y V. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin Cell Biol. 1995;6:247–255. doi: 10.1006/scel.1995.0033. [DOI] [PubMed] [Google Scholar]
- 15a.Calogero, S., F. Grassi, A. Aguzzi, T. Voigtlander, P. Ferrier, and M. Bianchi. The lack of chromosomal protein HMG1 does not disrupt cell growth, but causes lethal hypoglycaemia in newborn mice. Nat. Genet., in press. [DOI] [PubMed]
- 16.Chiappetta G, Tallini G, De Biasio M C, Manfioletti G, Martinez-Tello F J, Pentimalli F, de Nigris F, Mastro A, Botti G, Fedele M, Berger N, Santoro M, Giancotti V, Fusco A. Detection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinoma. Cancer Res. 1998;58:4193–4198. [PubMed] [Google Scholar]
- 17.Chin M T, Pellacani A, Wang H, Lin S S, Jain M K, Perrella M A, Lee M E. Enhancement of serum-response factor-dependent transcription and DNA binding by the architectural transcription factor HMG-I(Y) J Biol Chem. 1998;273:9755–9760. doi: 10.1074/jbc.273.16.9755. [DOI] [PubMed] [Google Scholar]
- 18.Churchill M E, Travers A A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991;16:92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- 19.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotmore S F, Tattersall P. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J Virol. 1998;72:8477–8884. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crippa M P, Alfonso P J, Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J Mol Biol. 1992;228:442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- 22.Crippa M P, Trieschmann L, Alfonso P J, Wolffe A P, Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993;12:3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davie J R, Delcuve G P. Characterization and chromatin distribution of the H1 histones and high-mobility-group non-histone chromosomal proteins of trout liver and hepatocellular carcinoma. Biochem J. 1991;280:491–497. doi: 10.1042/bj2800491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrov S, Wolffe A P. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histone H1 and H1(0) from chromatin and the acquisition of transcriptional competence. EMBO J. 1996;15:5897–5906. [PMC free article] [PubMed] [Google Scholar]
- 26.Ding H F, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region of chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding H F, Rimsky S, Batson S C, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 28.Einck L, Bustin M. The intracellular distribution and function of the high mobility group chromosomal proteins. Exp Cell Res. 1985;156:295–310. doi: 10.1016/0014-4827(85)90539-7. [DOI] [PubMed] [Google Scholar]
- 29.Falciola L, Murchie A I, Lilley D M, Bianchi M. Mutational analysis of the DNA binding domain A of chromosomal protein HMG1. Nucleic Acids Res. 1994;22:285–292. doi: 10.1093/nar/22.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falciola L, Spada F, Calogero S, Langst G, Viot R, Grummt I, Bianchi M. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J Cell Biol. 1997;137:19–26. doi: 10.1083/jcb.137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 32.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 33.Ferrari S, Harley V R, Pontiggia A, Goodfellow P N, Lovell B R, Bianchi M E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finch J T, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank O, Schwanbeck R, Wisniewski J R. Protein footprinting reveals specific binding modes of a high mobility group protein I to DNAs of different conformation. J Biol Chem. 1998;273:20015–20020. doi: 10.1074/jbc.273.32.20015. [DOI] [PubMed] [Google Scholar]
- 35a.Fusco, A. Personal communication.
- 36.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 37.Giancotti V, Berlingieri M T, DiFiore P P, Fusco A, Vecchio G, Crane-Robinson C. Changes in nuclear proteins on transformation of rat epithelial thyroid cells by a murine sarcoma retrovirus. Cancer Res. 1985;45:6051–6057. [PubMed] [Google Scholar]
- 38.Giancotti V, Pani B, D’Andrea P, Berlingieri M T, Di F P, Fusco A, Vecchio G, Philp R, Crane R C, Nicolas R H, et al. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras and v-mos oncogenes and by cotransfection with c-myc and polyoma middle T genes. EMBO J. 1987;6:1981–1987. doi: 10.1002/j.1460-2075.1987.tb02461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 40.Giese K, Pagel J, Grosschedl R. Distinct DNA-binding properties of the high mobility group domain of murine and human SRY sex-determining factors. Proc Natl Acad Sci USA. 1994;91:3368–3372. doi: 10.1073/pnas.91.8.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci USA. 1997;94:12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girard F, Bello B, Laemmli U K, Gehring W J. In vivo analysis of scaffold-associated regions in drosophila: a synthetic high-affinity SAR binding protein suppresses position effect variegation. EMBO J. 1998;17:2079–2085. doi: 10.1093/emboj/17.7.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodwin G. The high mobility group protein, HMGI-C. Int J Biochem Cell Biol. 1998;30:761–766. doi: 10.1016/s1357-2725(98)00016-8. [DOI] [PubMed] [Google Scholar]
- 44.Grasser K D, Teo S H, Lee K B, Broadhurst R W, Rees C, Hardman C H, Thomas J O. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1) Eur J Biochem. 1998;253:787–795. doi: 10.1046/j.1432-1327.1998.2530787.x. [DOI] [PubMed] [Google Scholar]
- 45.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 46.Hart C M, Laemmli U K. Facilitation of chromatin dynamics by SARs. Curr Opin Genet Dev. 1998;8:519–525. doi: 10.1016/s0959-437x(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 47.Herrera J E, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill D A, Reeves R. Competition between HMG-I(Y), HMG-1 and histone H1 on four way junction DNA. Nucleic Acids Res. 1997;25:3523–3531. doi: 10.1093/nar/25.17.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hock R, Scheer U, Bustin M. Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J Cell Biol. 1998;143:1427–1436. doi: 10.1083/jcb.143.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. EMBO J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huth J R, Bewley C A, Nissen M S, Evans J N S, Reeves R, Gronenborn A M, Clore G M. The solution structure of an HMGI(Y)-DNA complex defined a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 52.Isackson P, Bidney J, Reeck G, Niehart N, Bustin M. High mobility group chromosomal protein isolated from nuclei and cytosol of cultured hepatoma cells are similar. Biochemistry. 1980;19:4466–4471. doi: 10.1021/bi00560a013. [DOI] [PubMed] [Google Scholar]
- 53.Jayaraman L, Moorthy N C, Murthy K G, Manley J L, Bustin M, Prives C. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 1998;12:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johns E W. The HMG chromosomal proteins. London, United Kingdom: Academic Press; 1982. [Google Scholar]
- 55.Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox 1, 2, and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 56.Kazmierczak B, Bullerdiek J, Pham K H, Bartnitzke S, Wiesner H. Intron 3 of HMGIC is the most frequent target of chromosomal aberrations in human tumors and has been conserved basically for at least 30 million years. Cancer Genet Cytogenet. 1998;103:175–177. doi: 10.1016/s0165-4608(97)00348-8. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 57.Kazmierczak B, Dal Cin P, Wanschura S, Borrmann L, Fusco A, Van den Berghe H, Bullerdiek J. HMGIY is the target of 6p21.3 rearrangements in various benign mesenchymal tumors. Genes Chromosomes Cancer. 1998;23:279–285. [PubMed] [Google Scholar]
- 58.Kazmierczak B, Rosigkeit J, Wanschura S, Meyer-Bolte K, Van de Ven W J M, Kayser K, Krieghoff B, Kastendiek H, Bartnitzke S, Bullerdiek J. HMGI-C rearrangements as the molecular basis for the majority of pulmonary chondroid hamartomas: a survey of 30 tumors. Oncogene. 1996;12:515–521. [PubMed] [Google Scholar]
- 59.Kuhn A, Voit R, Stefanovsky V, Evers R, Bianchi M, Grummt I. Functional differences between the two splice variants of the nucleolar transcription factor UBF: the second HMG box determines specificity of DNA binding and transcriptional activity. EMBO J. 1994;13:416–424. doi: 10.1002/j.1460-2075.1994.tb06276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwon J, Imbalzano A N, Matthews A, Oettinger M A. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 61.Landsman D, Bustin M. A signature for the HMG-1 box proteins. Bioessays. 1993;15:539–546. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- 62.Landsman D, Mendelson E, Druckmann S, Bustin M. Exchange of proteins during immunofractionation of chromatin. Exp Cell Res. 1986;163:95–102. doi: 10.1016/0014-4827(86)90561-6. [DOI] [PubMed] [Google Scholar]
- 63.Laudet V, Stehelin D, Clevers H. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 1993;21:2493–2501. doi: 10.1093/nar/21.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 65.Lefstin J A, Yamamoto K R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 66.Lehming N, Le Saux A, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Farnet C M, Anderson W F, Bushman F D. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lnenicek-Allen M, Read C M, Crane-Robinson C. The DNA bend angle and binding affinity of an HMG box increased by the presence of short terminal arms. Nucleic Acids Res. 1996;24:1047–1051. doi: 10.1093/nar/24.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 70.Lovell-Badge R. The HMG family of proteins. Nature. 1995;376:725. doi: 10.1038/376725a0. [DOI] [PubMed] [Google Scholar]
- 71.Luger K, Madner A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome at 2.8A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 72.Maher J F, Nathans D. Multivalent DNA-binding properties of the HMG-I protein. Proc Natl Acad Sci USA. 1996;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mantovani F, Covaceuszach S, Rustighi A, Sgarra R, Heath C, Goodwin G H, Manfioletti G. NF-λB mediated transcriptional activation is enhanced by the architectural factor HMGI-C. Nucleic Acids Res. 1998;26:1433–1439. doi: 10.1093/nar/26.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 75.Ner S S, Travers A A. HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 1994;13:1817–1822. doi: 10.1002/j.1460-2075.1994.tb06450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 77.Nightingale K, Dimitrov S, Reeves R, Wolffe A P. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 78.Oñate S A, Prendergast P, Wager J P, Nissen M, Reeves R, Pettijohn D E, Edwards D P. The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol Cell Biol. 1994;14:3376–3391. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paranjape S M, Krumm A, Kadonaga J T. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 80.Pellacani A, Chin M, Wiesel P, Ibanez M, Foster L, Patel A, Yet S, Hsieh C, Paulauskis J, Reeves R, Lee M, Perrella M. Induction of high mobility group-I(Y) protein by endotoxin and interleukin-1β in vascular smooth muscle cells: role in activation of inducible nitric oxide synthetase. J Biol Chem. 1999;274:1525–1532. doi: 10.1074/jbc.274.3.1525. [DOI] [PubMed] [Google Scholar]
- 81.Pohler J, Norman D G, Bramham J, Bianchi M E, Lilley D M. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pohler J R, Lilley D M. The interaction of HMG-box proteins with the four-way DNA junction. Biochem Soc Trans. 1997;25:S647. doi: 10.1042/bst025s647. [DOI] [PubMed] [Google Scholar]
- 83.Pontiggia A, Rimini R, Harley V R, Goodfellow P N, Lovell-Badge R, Bianchi M E. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pontiggia A, Whitfield S, Goodfellow P N, Lovell-Badge R, Bianchi M E. Evolutionary conservation in the DNA-binding and -bending properties of HMG-boxes from SRY proteins of primates. Gene. 1995;154:277–280. doi: 10.1016/0378-1119(94)00853-k. [DOI] [PubMed] [Google Scholar]
- 85.Postnikov Y V, Herrera J E, Hock R, Scheer U, Bustin M. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J Mol Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]
- 86.Postnikov Y V, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 87.Read C M, Cary P D, Crane-Robinson C, Driscoll P C, Norman D G. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993;21:3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reeves R, Nissen M. Cell cycle regulation and functions of HMG-I(Y) Prog Cell Cycle Res. 1995;1:339–349. doi: 10.1007/978-1-4615-1809-9_28. [DOI] [PubMed] [Google Scholar]
- 89.Reeves R, Nissen M S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 90.Saitoh Y, Laemmli U. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 91.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwanbeck R, Wisniewski J R. Cdc2 and mitogen-activated protein kinases modulate DNA binding properties of the putative transcriptional regulator Chironomus high mobility group protein I. J Biol Chem. 1997;272:27476–27483. doi: 10.1074/jbc.272.43.27476. [DOI] [PubMed] [Google Scholar]
- 93.Segall A C, Goodman S D, Nash H A. Architectural elements in nucleoprotein complexes: interchangeability of specific and non-specific DNA binding proteins. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shirakawa H, Tanigawa T, Sugyama S, Kobayashi M, Terashima T, Yoshida K, Arai T, Yoshida M. Nuclear accumulation of HMG2 is mediated by a basic region interspaced with a long DNA-binding sequence, and retention within the nucleus requires the acidic carboxyl terminus. Biochemistry. 1997;36:5992–5999. doi: 10.1021/bi962487n. [DOI] [PubMed] [Google Scholar]
- 95.Spada F, Brunet A, Mercier Y, Bianchi M E, Thompson E M. High mobility group 1 (HMG1) protein in mouse preimplantation embryos. Mech Dev. 1998;76:57–66. doi: 10.1016/s0925-4773(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 96.Stelzer G, Goppelt A, Lottspeich F, Meisterernst M. Repression of basal transcription by HMG2 is counteracted by TFIIH-associated factors in an ATP-dependent process. Mol Cell Biol. 1994;14:4712–4721. doi: 10.1128/mcb.14.7.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sutrias-Grau M, Bianchi M E, Bernues J. HMG-1 interacts species-specifically with the core domain of human TBP and interferes with TFIIB within the pre-initiation complex. J Biol Chem. 1999;274:1628–1634. doi: 10.1074/jbc.274.3.1628. [DOI] [PubMed] [Google Scholar]
- 98.Teo S H, Grasser K D, Thomas J O. Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex-determining factor SRY. Eur J Biochem. 1995;230:943–950. doi: 10.1111/j.1432-1033.1995.tb20640.x. [DOI] [PubMed] [Google Scholar]
- 99.Thanos D, Maniatis T. NF-λB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 100.Thanos D, Maniatis T. Virus induction of IFN(beta) gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 101.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 102.Tkachenko A, Ashar H R, Meloni A M, Sandberg A A, Chada K K. Misexpression of disrupted HMGI architectural factors activates alternative pathways of tumorigenesis. Cancer Res. 1997;57:2276–2280. [PubMed] [Google Scholar]
- 103.Tremethick D J, Hyman L. High mobility group proteins 14 and 17 can prevent the close packing of nucleosomes by increasing the strength of protein contacts in the linker DNA. J Biol Chem. 1996;271:12009–12016. doi: 10.1074/jbc.271.20.12009. [DOI] [PubMed] [Google Scholar]
- 104.Trieschmann L, Alfonso P J, Crippa M P, Wolffe A P, Bustin M. Incorporation of chromosomal proteins HMG-14/-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc Natl Acad Sci USA. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Trieschmann L, Postnikov Y, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14/-17: definition of a transcriptional activation domain distinct from the nucleosomal binding domain. Mol Cell Biol. 1995;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vaccari T, Beltrame M, Ferrari S, Bianchi M E. Hmg4, a new member of the Hmg1/2 gene family. Genomics. 1998;49:247–252. doi: 10.1006/geno.1998.5214. [DOI] [PubMed] [Google Scholar]
- 108.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Verrier C S, Roodi N, Yee C J, Bailey L R, Jensen R A, Bustin M, Parl F F. High-mobility group (HMG) protein HMG-1 and TATA-binding protein-associated factor TAF(II)30 affect estrogen receptor-mediated transcriptional activation. Mol Endocrinol. 1997;11:1009–1019. doi: 10.1210/mend.11.8.9962. [DOI] [PubMed] [Google Scholar]
- 110.Vestner B, Bustin M, Gruss C. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J Biol Chem. 1998;273:9409–9414. doi: 10.1074/jbc.273.16.9409. [DOI] [PubMed] [Google Scholar]
- 111.Vignali M, Workman J L. Location and function of linker histones. Nat Struct Biol. 1998;5:1025–1028. doi: 10.1038/4133. [DOI] [PubMed] [Google Scholar]
- 112.Weir H M, Kraulis P J, Hill C S, Raine A R, Laue E D, Thomas J O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Werner M H, Huth J R, Gronenborn A M, Clore G M. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- 114.Williams A J, Powell W L, Collins T, Morton C C. HMGI(Y) expression in human uterine leiomyomata. Involvement of another high-mobility group architectural factor in a benign neoplasm. Am J Pathol. 1997;150:911–918. [PMC free article] [PubMed] [Google Scholar]
- 115.Wisniewski J, Schultze E, Sapetto B. DNA binding and nuclear translocation of insect HMG-1 proteins are inhibited by phosphorylation. Eur J Biochem. 1994;225:687–693. doi: 10.1111/j.1432-1033.1994.00687.x. [DOI] [PubMed] [Google Scholar]
- 116.Wisniewski J R, Grossbach U. Structural and functional properties of linker histones and high mobility group proteins in polytene chromosomes. Int J Dev Biol. 1996;40:177–187. [PubMed] [Google Scholar]
- 117.Wisniewski J R, Hessler K, Claus P, Zechel K. Structural and functional consequences of mutations within the hydrophobic cores of the HMG1-box domain of the Chironomus high-mobility-group protein 1a. Eur J Biochem. 1997;243:151–159. doi: 10.1111/j.1432-1033.1997.0151a.x. [DOI] [PubMed] [Google Scholar]
- 118.Wolffe A P, Kurumizaka H. The nucleosome: a powerful regulator of transcription. Prog Nucleic Acid Res Mol Biol. 1998;61:379–422. doi: 10.1016/s0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 119.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 120.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 121.Yamazaki F, Nagatsuka Y, Shirakawa H, Yoshida M. Repression of cell cycle progression by antisense HMG2 RNA. Biochem Biophys Res Commun. 1995;210:1045–1051. doi: 10.1006/bbrc.1995.1762. [DOI] [PubMed] [Google Scholar]
- 122.Yaneva J, Leuba S H, van Holde K, Zlatanova J. The major chromatin protein histone H1 binds preferentially to cis-platinum-damaged DNA. Proc Natl Acad Sci USA. 1997;94:13448–13451. doi: 10.1073/pnas.94.25.13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yie J, Liang S, Merika M, Thanos D. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol Cell Biol. 1997;17:3649–3662. doi: 10.1128/mcb.17.7.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoshioka K, Saito K, Tanabe T, Yamamoto A, Ando Y, Nakamura Y, Shirakawa H, Yoshida M. Differences in DNA recognition and conformational change activity between boxes A and B in HMG2 protein. Biochemistry. 1999;38:589–595. doi: 10.1021/bi981834l. [DOI] [PubMed] [Google Scholar]
- 125.Zappavigna V, Falciola L, Citterich M H, Mavillo F, Bianchi M E. HMG-1 interacts with HOX proteins and enhances their DNA binding and transcription activation. EMBO J. 1996;15:4981–4988. [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao K, Kas E, Gonzalez E, Laemmli U. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou X, Chada K. HMGI family proteins: architectural transcription factors in mammalian development and cancer. Keio J Med. 1998;47:73–77. doi: 10.2302/kjm.47.73. [DOI] [PubMed] [Google Scholar]
- 128.Zhou X J, Benson K F, Ashar H R, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 129.Zlatanova J, van Holde K. Linker histones versus HMG1/2: a struggle for dominance? Bioessays. 1998;20:584–588. doi: 10.1002/(SICI)1521-1878(199807)20:7<584::AID-BIES10>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 130.Zwilling S, Koenig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]