Abstract
Background
Granulomatous mastitis (GM) is a rare chronic inflammatory disease of the breast. The current therapeutic effects of the antibiotic therapy and surgical or immunomodulatory (steroid) treatment are normally poor due to the unclear etiology.
Method
This study aimed to identify the differentially expressed mRNAs in GM tissues using RNA sequencing and further explored the functions of differentially expressed mRNAs resulting in GM. Moreover, we revealed the relationship between GM and breast cancer by shared highly expressed genes in GM tissues and breast cancer tissues.
Results
A total of 12,115 mRNAs were analyzed in the whole expression profile, and 207 mRNAs (136 upregulated and 71 downregulated mRNAs) were differently expressed between the GM tissues and normal tissues. The enrichment analysis showed that the differentially expressed mRNAs were enriched in the biological processes and played a significant role in the immune system. Besides, the genes expressed significantly highly in breast cancer tissues are found to be enriched with GM genes, which may explain the similar clinical features between breast cancer and GM. We also found that the HSD11B1 gene which was differentially expressed in GM was used as drug target of prednisone, which is a common treatment for GM.
Conclusion
This study is the first to use sequencing technology to elucidate the genetic mechanisms of GM. The finding of this study may have potential value in GM diagnosis and also provides potential drug targets for GM treatment.
Keywords: Granulomatous mastitis, RNA sequencing, Expression profiles, Etiology
Introduction
Granulomatous mastitis (GM), a rare chronic inflammatory breast disease of unclear etiology, was first described as a specific entity by Kessler and Wolloch in 1972 [1]. GM caused massive deformities of the breast, which may contribute to serious anxiety and depressive disorder of the patients [2]. Accurate diagnosis of GM remains a challenge to discriminate between GM and carcinoma, as it often clinically and radiographically mimics breast malignancy [3]. The great difficulties for a definitive diagnosis of GM hinder an effective treatment. Thus, identification of biomarkers for GM is essential.
Furthermore, there is no effective treatment for GM. The current treatment for GM includes conservative management [4], in which antibiotics, anti-inflammatory drugs, and topical or systemic corticosteroids were recommended. A primary curative therapy treatment is surgical excision [5]. However, the therapeutic effect of the empirical antibiotic therapy alone is normally poor, and the surgical or immunomodulatory (steroid) treatment is implemented with varying degrees of success [1, 2]. To develop an effective treatment for GM, improving the understanding of the etiology of GM is necessary.
The existing mechanistic theories of GM include both infectious and autoimmunity-mediated inflammatory mechanisms. Regarding the infectious etiology, an association between corynebacterium infection and GM was first proposed by a clinical pathological review of 34 patients in New Zealand in 2003 [6]. Subsequently, more and more researchers indicated that corynebacterium infection has a close relationship with the pathogenesis of GM [7, 8, 9]. However, the pathogenic mechanism due to corynebacterium is still not clear.
The autoimmune mechanism is supported by the response of GM to steroid treatment [10, 11]. The autoimmune reaction process is considered to be triggered by the extravasated protein secretions from mammary ducts secondary to local trauma, oral contraceptives, hyperprolactinemia and breast-feeding. However, the autoimmune etiology hypothesis still remains controversial [12]. In 2000, Erhan et al. [13] reported a predominance of T cells in 14 of their 18 cases using the primary antibody and concluded that a reactive T-cell-mediated inflammation with the formation of centrilobular granulomas is the pathophysiological mechanism of GM, further supporting the autoimmune etiology hypothesis. However, Asoglu et al. [14] found the serological tests for antinuclear and rheumatoid factor autoimmune antibodies to be negative, which are evidence of an autoimmune phenomenon.
Since many possible causes of GM were revealed, it is reasonable to surmise that potential alterations and expression changes in genes may initiate GM. Bercot et al. [15] reported the first case of GM due to Corynebacterium kroppenstedtii linked to strongly impaired Nod2 function which revealed the potential changes in the NOD2 gene in GM. Destek et al. [16] analyzed the gene polymorphisms associated with GM and found that mutations in MTHFR-C 677 T, PAI-1, and ACE genes reported in breast cancer were also found in GM in 1 case. However, no reports have been published so far regarding the RNA expression profiles associated with GM.
We thus present 3 cases with demonstrated histological features of GM, and this study is the first to identify differentially expressed RNAs in the biopsy samples of granulomatous tissue with that of normal tissue. The enrichment analysis of breast cancer genes in GM was also examined to identify the difference and relationship between GM and breast cancer. This study will facilitate a precise treatment for this disease entity.
Case Report
In the study, 3 patients diagnosed with chronic GM by histopathology were enrolled in the period of 2016–2017. This study was approved by the Institutional Ethics Board of Beijing Tiantan Hospital, and written informed consent was obtained from all of the recruited patients. The participants in this study had never received an antimicrobial treatment. The detailed clinical characteristics of the patients are given in Table 1. The average age of our patients was 31 years. All of them had a unilateral breast lump. Ultrasonography was performed in all patients, and the predominant findings were the presence of a hypoechoic pattern as given in Table 1; the ultrasonography of the 3 patients is given in the online supplementary material (for all online suppl. material, see www.karger.com/doi/10.1159/000507474). The tuberculosis antibody and antituberculosis antibody were negative in all patients. The patients were given levofloxacin 200 mg/day/injection for 1 week before they underwent surgery. In all patients, no recurrence has occurred so far.
Table 1.
The clinical characteristics, ultrasonography, and laboratory examination results of patients with GM
| Characteristics | Patient 1 | Patient 2 | Patient 3 |
|---|---|---|---|
| Drug-taking history | 5 days levofloxacin 500 mg/day | 5 days levofloxacin 500 mg/day | − |
| Location | Right | Left | Left |
| Clinical features | |||
| Breast lump size, cm | 5×7 | 2×2 | 5.4×7.5 |
| Swelling | No | Yes | Yes |
| Breast abscess | No | Yes | No |
| Symptoms | |||
| Fever | No | Yes | No |
| Axillary lymphadenopathy | Right | Left | Left |
| Erythema | No | No | Yes |
| Arthritis | No | No | No |
| Breast ultrasonography | |||
| Location | Peripheral | Peripheral | Subareolar |
| Hypoechoic internal echo extent, cm | 4.8×6.5 | 2.4×1.57 | 4.6×7.0 |
| Axillary lymph node location | Right | Left | Left |
| Axillary lymph node size, cm | 2.3×0.7 | 2.2×0.97 | 1.3×0.8 |
| Laboratory examination results | |||
| White blood cells (normal 4–10), n ×109/L | 8.33 | 7.83 | 8.29 |
| Prolactin (normal 1.9–25), ng/L | 5.85 | 30.4 | 10.8 |
| Secretion culture | Negative | Pseudomonas aeruginosa | Negative |
| Tuberculosis antibody | Negative | Negative | Negative |
| Antituberculosis antibody | Negative | Negative | Negative |
“–” means the patient did not have the drug-taking history, “yes” that the patient had the corresponding symptom, and “no” that he had not.
Methods
Three samples from both the GM and normal tissue were collected for RNA sequencing. Extracted RNA was assayed for integrity quality control using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA samples were used for deep RNA sequencing when RNA integrity numbers were equal to or greater than 7.0. Sequencing libraries were prepared by the NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA), following the manufacturer's recommendations. The qualities of the libraries were assessed on the Agilent Bioanalyzer 2100 system. Libraries were quantified by qPCR using the KAPA Library Quantification Kit for Illumina sequencing platforms (KAPA Biosystems). Quality control of library pools to establish equimolarity of individual libraries was performed by Illumina MiSeq (SR 1 × 50 bp). Libraries were sequenced using an Illumina HiSeq2500 (PE 2 × 300 bp).
The FASTQ files were analyzed using the STAR package [17] with the HG19 human reference genome. RSEM software [18] was used to count mapped sequencing reads on each gene transcript, and the differently expressed genes were identified by edgeR. edgeR is designed to identify changes between two or more groups by the trimmed mean of M-values normalization method and is based on the hypothesis that most genes are not expressed differently [19]. The p value was calculated for each gene and adjusted as q value by the Benjamini and Hochberg approach for controlling the false discovery rate. Genes with q values <0.05 were considered as differently expressed genes.
Gene Function Analysis
The protein-protein interaction networks of differentially expressed genes were assessed using the STRING database (http://string-db.org/) [20]. Gene ontology (GO) enrichment analyses of differentially expressed genes were completed with g: profiler web server (https://biit.cs.ut.ee/gprofiler/gost) [21]. GO terms such as biological processes (BP), cellular components (CC), and molecular functions (MF) with a corrected p value of <0.05 were considered significantly enriched in the differentially expressed genes.
Enrichment Analysis of Breast Cancer Genes
Breast cancer genes were collected from the Oncomine database (http://www.oncomine.org). Initially, we used the following filters: (a) analysis type: cancer versus normal analysis; (b) cancer type: breast cancer; (c) data set type: TCGA data sets, and (d) sample type: clinical specimen. In total, 593 samples and 20,423 measured genes were involved. Then overexpressed genes associated with 8 breast cancer types were displayed separately. These breast cancer types are intraductal cribriform, invasive breast carcinoma, invasive ductal and lobular, invasive ductal breast, invasive lobular breast, male breast carcinoma, mixed lobular and ductal, and mucinous breast carcinoma. For each breast cancer type, the top 1% overexpressed genes associated with breast cancer were selected.
Results
Gene Expression Profiles of Differentially Expressed Genes
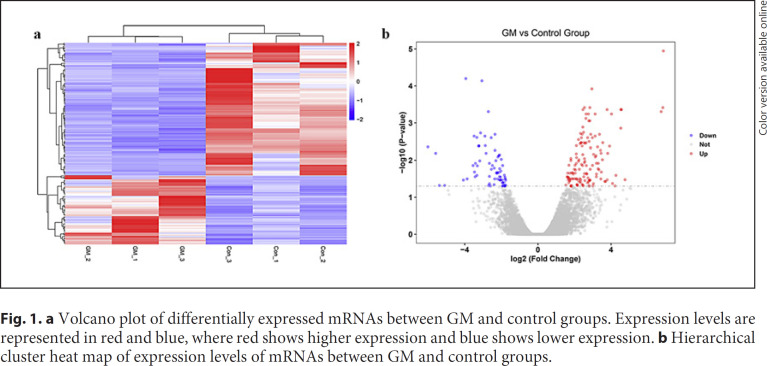
The whole expression profiles were detected on a total of 12,115 mRNAs. Among them, 207 mRNAs were differently expressed between the GM tissues and con-trol tissues, with 136 upregulated and 71 downregulated mRNAs. Volcano plots were used to determine the statistical significance of differently expressed mRNAs between the GM and control group in Figure 1. DNAJC27-AS1 (6.86067-fold change) was the highest upregulated mRNA, and TRAV12–1(−6.0142698-fold change) was the lowest downregulated mRNA. Besides, the expression levels of differently expressed mRNAs were used to generate a heat map in unsupervised hierarchical clustering analysis in Figure 1. It can be seen that the expression levels clearly self-segregated into GM and control groups.
Fig. 1.
a Volcano plot of differentially expressed mRNAs between GM and control groups. Expression levels are represented in red and blue, where red shows higher expression and blue shows lower expression. b Hierarchical cluster heat map of expression levels of mRNAs between GM and control groups.
Interestingly, among the genes expressed significantly differently between GM tissues and the controls, HSD11B1 with low expression was the drug target of prednisone. The prednisone treatment is demonstrated to be effective [22, 23]. Sakurai et al. [22] stated that all 7 of the cases who received prednisone (an initial dose of 60 mg/day) achieved a cure without surgery. Besides, medical treatment with prednisone was also reported to reduce the lesion size and extent and the rate of recurrence [24]. Thus, other genes identified by this study may implicate potential drug targets for GM.
Functional Analysis of Differentially Expressed Genes
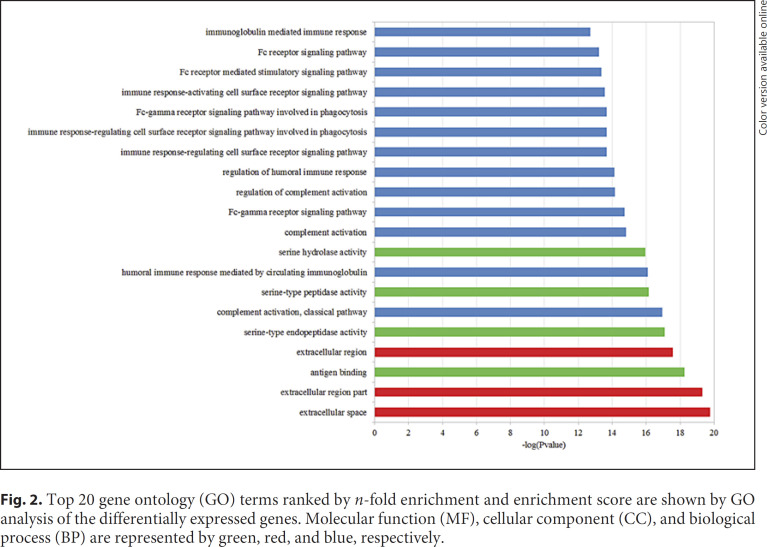
By using the p value cutoff of 0.05, GO annotation found that 67 BP terms, 15 CC terms, and 9 MF terms were significant. Figure 2 displays the top 20 generally changed GO terms ranked by p value. Among the top 20 GO terms, 6 BP terms were related to the immune system. That means the differentially expressed mRNAs play a significant role in the immune system.
Fig. 2.
Top 20 gene ontology (GO) terms ranked by n-fold enrichment and enrichment score are shown by GO analysis of the differentially expressed genes. Molecular function (MF), cellular component (CC), and biological process (BP) are represented by green, red, and blue, respectively.
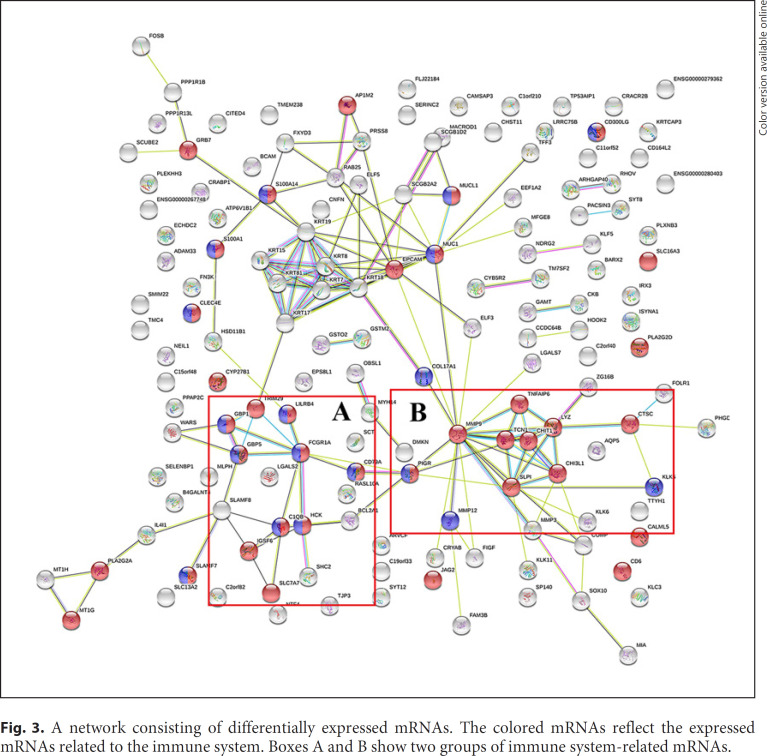
Another purpose of gene analysis is to identify the gene-gene interaction network related with GM. Here, we used the genes expressed significantly differently in GM tissues compared to those in the normal tissues to construct the network. These genes (207 genes) were input into the STRING database. A network containing 151 nodes was constructed. From this network, we identified the hub genes by considering the number of connections to other genes. Figure 3 shows the network of differentially expressed mRNAs. The network comprised 151 network nodes, in which 40 mRNAs related to the immune system were colored. The immune system-related mRNAs can be classified into 2 groups presented in box A and box B in Figure 3. The FCGR1A was the hub gene in network A, which was linked to 9 other genes such as TRIM29, GBP1, GBP5, LILRB4, HCK, C1QB, SLC7A7, IGSF6, and CD79A. The hub gene FCGR1A was associated with protein binding (GO: 0005515) and IgG binding (GO: 0019864). Similarly, the TCN1 was the hub gene in network B, and there were 9 other genes (MMP9, TNFAIP6, CHIT1, LYZ, SLPI, CHI3L1, CTSC, PIGR, and MMP12) in network B. The TCN1 was correlated with cobalamin binding (GO: 0031419). Thus, these two networks may represent the pathways associated with immune mechanisms of GM. These findings may explain the effective steroid treatment of GM, which enrich the understanding of the immune mechanisms of GM. Besides, the hub genes FCGR1A and TCN1 were the drug targets of methylaminolevulinate and hydroxycobalamin, respectively [25, 26]. Methylaminolevulinate is an FDA-approved anticancer drug, and hydroxycobalamin is mainly used therapeutically to treat vitamin B12 deficiency.
Fig. 3.
A network consisting of differentially expressed mRNAs. The colored mRNAs reflect the expressed mRNAs related to the immune system. Boxes A and B show two groups of immune system-related mRNAs.
Genes Related to Breast Cancer
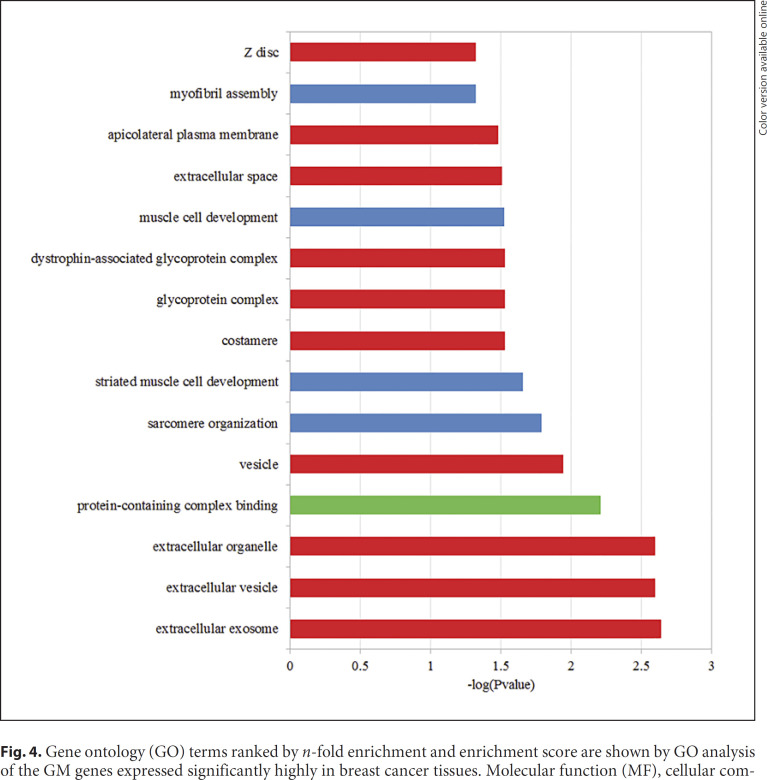
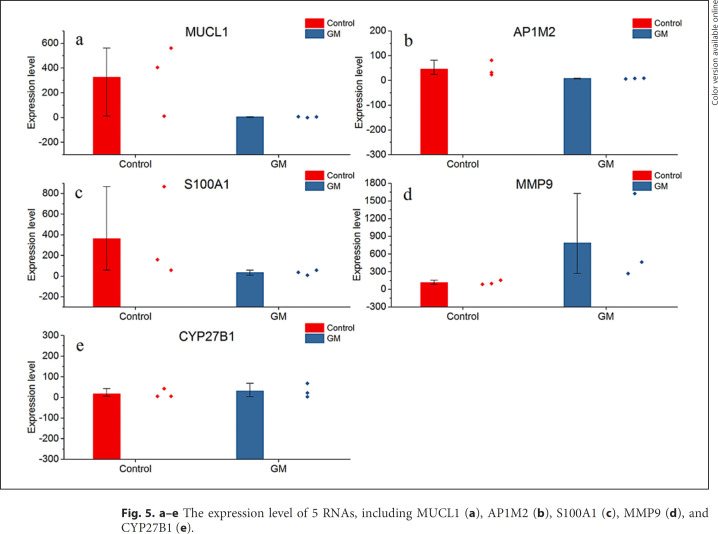
Breast cancer-related genes were collected from the Oncomine database (http://www.oncomine.org). Totally, 207 genes were selected as the overexpressed genes in the cancer tissues. Out of them, 19 breast cancer-related genes were retrieved. The genes expressed significantly highly in breast cancer tissues were significantly enriched in GM (p = 0.03746, one-sided binomial test). Figure 4 shows the function enrichment analysis of 19 GM genes significantly high in breast cancer tissues, and the results show that they are enriched in 16 GO terms including 10 CC terms, 4 BP terms and 1 MF term. The top 3 GO terms were extracellular exosome (GO: 0070062), extracellular vesicle (GO: 1903561), and extracellular organelle (GO: 0043230). There are 14 (73.6%) GM genes expressed significantly highly both in breast cancer and GM tissues. The expression level of the top 5 genes ranked by p value among 19 breast cancer genes are displayed in Figure 5. The expression levels of all chosen genes in Figure 5 were significantly different compared to the control group.
Fig. 4.
Gene ontology (GO) terms ranked by n-fold enrichment and enrichment score are shown by GO analysis of the GM genes expressed significantly highly in breast cancer tissues. Molecular function (MF), cellular component (CC), and biological process (BP) are represented by green, red, and blue, respectively.
Fig. 5.
a–e The expression level of 5 RNAs, including MUCL1 (a), AP1M2 (b), S100A1 (c), MMP9 (d), and CYP27B1 (e).
Discussion and Conclusion
From the results, GM is a disease caused by dysfunctions of the immune system. That is consistent with the favored hypothesis that GM is an autoimmune disease supported by effective steroid treatment of GM [10, 12]. This study also explained the reason for symptom similarity between breast cancer and GM. In this study, we identified that genes expressed significantly highly in breast cancer tissues are enriched with GM genes. Among them, 73.6% of the genes were highly expressed in GM tissues. Destek et al. [16] analyzed genetic polymorphisms in breast cancer and GM and found that mutations in MTHFR-C 677 T, PAI-1, and ACE genes were found in both breast cancer and GM. There are many researches demonstrating similar clinical and radiological findings between GM and breast cancer [27], but there are quite a few studies about the similarity in gene expression between GM and breast cancer. Thus, this result may fill gaps in our understanding of the genetic similarity between breast cancer and GM.
In this study, we identified two possible networks associated with the immune mechanisms of GM. There are 7 (43%) drug targets in these two networks. The HSD11B1 gene was used as drug target of prednisone, which is a common treatment for GM [27, 28]. The finding of this study provides potential drug targets for GM treatment. Further study is necessary to discover possible drugs by using the networks given in this study.
In summary, findings of this study are valuable for treatment design and diagnosis of GM. Meanwhile, this study illustrated the possible genetic relationship between GM and breast cancer to support the symptom similarity between them.
Statement of Ethics
The study was approved by the Institutional Ethics Board of Beijing Tiantan Hospital. All patients provided written informed consent.
Disclosure Statement
The authors declare that there is no conflict of interest.
Funding Sources
There has been no funding involvement neither in the study design or collection, analysis, and interpretation of data nor in the writing or submission of the paper for publication.
Author Contributions
Mr. Zhu designed the study and wrote the manuscript. Ms. Wang collected and analyzed the data. Prof. Wang reviewed and edited the paper.
Supplementary Material
Supplementary data
Acknowledgments
The authors would like to thank their patients for providing the valuable samples.
References
- 1.Aghajanzadeh M, Hassanzadeh R, Alizadeh Sefat S, Alavi A, Hemmati H, Esmaeili Delshad MS, et al. Granulomatous mastitis: Presentations, diagnosis, treatment and outcome in 206 patients from the north of Iran. Breast. 2015 Aug;24((4)):456–60. doi: 10.1016/j.breast.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Barreto DS, Sedgwick EL, Nagi CS, Benveniste AP. Granulomatous mastitis: etiology, imaging, pathology, treatment, and clinical findings. Breast Cancer Res Treat. 2018 Oct;171((3)):527–34. doi: 10.1007/s10549-018-4870-3. [DOI] [PubMed] [Google Scholar]
- 3.Lai EC, Chan WC, Ma TK, Tang AP, Poon CS, Leong HT. The role of conservative treatment in idiopathic granulomatous mastitis. Breast J. 2005 Nov-Dec;11((6)):454–6. doi: 10.1111/j.1075-122X.2005.00127.x. [DOI] [PubMed] [Google Scholar]
- 4.Kiyak G, Dumlu EG, Kilinc I, Tokaç M, Akbaba S, Gurer A, Ozkardes AB, Kilic M. Management of idiopathic granulomatous mastitis: dilemmas in diagnosis and treatment. BMC Surg. 2014;14((1)):66–66. doi: 10.1186/1471-2482-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yabanoğlu H, Çolakoğlu T, Belli S, Aytac HO, Bolat FA, Pourbagher A, et al. A Comparative Study of Conservative versus Surgical Treatment Protocols for 77 Patients with Idiopathic Granulomatous Mastitis. Breast J. 2015 Jul-Aug;21((4)):363–9. doi: 10.1111/tbj.12415. [DOI] [PubMed] [Google Scholar]
- 6.Taylor GB, Paviour SD, Musaad S, Jones WO, Holland DJ. A clinicopathological review of 34 cases of inflammatory breast disease showing an association between corynebacteria infection and granulomatous mastitis. Pathology. 2003 Apr;35((2)):109–19. [PubMed] [Google Scholar]
- 7.Troxell ML, Gordon NT, Doggett JS, Ballard M, Vetto JT, Pommier RF, et al. Cystic Neutrophilic Granulomatous Mastitis: Association With Gram-Positive Bacilli and Corynebacterium. Am J Clin Pathol. 2016 May;145((5)):635–45. doi: 10.1093/ajcp/aqw046. [DOI] [PubMed] [Google Scholar]
- 8.Poojary I, Kurian A, v A J, Devapriya J D, M A T. Corynebacterium species causing breast abscesses among patients attending a tertiary care hospital in Chennai, South India. Infect Dis (Lond) 2017 Jul;49((7)):528–31. doi: 10.1080/23744235.2017.1296184. [DOI] [PubMed] [Google Scholar]
- 9.Renshaw AA, Derhagopian RP, Gould EW. Cystic neutrophilic granulomatous mastitis: an underappreciated pattern strongly associated with gram-positive bacilli. Am J Clin Pathol. 2011 Sep;136((3)):424–7. doi: 10.1309/AJCP1W9JBRYOQSNZ. [DOI] [PubMed] [Google Scholar]
- 10.Maffini F, Baldini F, Bassi F, Luini A, Viale G. Systemic therapy as a first choice treatment for idiopathic granulomatous mastitis. J Cutan Pathol. 2009 Jun;36((6)):689–91. doi: 10.1111/j.1600-0560.2008.01102.x. [DOI] [PubMed] [Google Scholar]
- 11.Bes C, Soy M, Vardi S, Sengul N, Yilmaz F. Erythema nodosum associated with granulomatous mastitis: report of two cases. Rheumatol Int. 2010 Sep;30((11)):1523–5. doi: 10.1007/s00296-009-1109-y. [DOI] [PubMed] [Google Scholar]
- 12.Altintoprak F, Karakece E, Kivilcim T, Dikicier E, Cakmak G, Celebi F, et al. Idiopathic granulomatous mastitis: an autoimmune disease? ScientificWorldJournal. 2013 Sep;2013:148727. doi: 10.1155/2013/148727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erhan Y, Veral A, Kara E, Özdemir N, Kapkac M, Özdedeli E, et al. A clinicopthologic study of a rare clinical entity mimicking breast carcinoma: idiopathic granulomatous mastitis. Breast. 2000 Feb;9((1)):52–6. doi: 10.1054/brst.1999.0072. [DOI] [PubMed] [Google Scholar]
- 14.Asoglu O, Ozmen V, Karanlik H, Tunaci M, Cabioglu N, Igci A, et al. Feasibility of surgical management in patients with granulomatous mastitis. Breast J. 2005 Mar-Apr;11((2)):108–14. doi: 10.1111/j.1075-122X.2005.21576.x. [DOI] [PubMed] [Google Scholar]
- 15.Bercot B, Kannengiesser C, Oudin C, Grandchamp B, Sanson-le Pors MJ, Mouly S, et al. First description of NOD2 variant associated with defective neutrophil responses in a woman with granulomatous mastitis related to corynebacteria. J Clin Microbiol. 2009 Sep;47((9)):3034–7. doi: 10.1128/JCM.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Destek S, Gul VO, Ahioglu S. A variety of gene polymorphisms associated with idiopathic granulomatous mastitis. J Surg Case Rep. 2016 Sep;2016((9)):156. doi: 10.1093/jscr/rjw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkel DH, Gerstein GL, Moore GP. Neuronal spike trains and stochastic point processes. I. The single spike train. Biophys J. 1967 Jul;7((4)):391–418. doi: 10.1016/S0006-3495(67)86596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12((1)):323–323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26((1)):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013 Jan;41((Database issue)):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reimand J, Arak T, Vilo J. g: Profiler—a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011 Jul;39((Web Server issue suppl_2)):W307-15. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakurai K, Fujisaki S, Enomoto K, Amano S, Sugitani M. Evaluation of follow-up strategies for corticosteroid therapy of idiopathic granulomatous mastitis. Surg Today. 2011 Mar;41((3)):333–7. doi: 10.1007/s00595-009-4292-2. [DOI] [PubMed] [Google Scholar]
- 23.Hugon-Rodin J, Plu-Bureau G, Hugol D, Gompel A. Management of granulomatous mastitis: a series of 14 patients. Gynecol Endocrinol. 2012 Nov;28((11)):921–4. doi: 10.3109/09513590.2012.683075. [DOI] [PubMed] [Google Scholar]
- 24.Karanlik H, Ozgur I, Simsek S, Fathalizadeh A, Tukenmez M, Sahin D, et al. Can Steroids plus Surgery Become a First-Line Treatment of Idiopathic Granulomatous Mastitis? Breast Care (Basel) 2014 Oct;9((5)):338–42. doi: 10.1159/000366437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakharkar MK, Li P, Zhong Z, Sakharkar KR. Quantitative analysis on the characteristics of targets with FDA approved drugs. Int J Biol Sci. 2007 Dec;4((1)):15–22. doi: 10.7150/ijbs.4.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins D, Rosenblatt DS, Fowler B. Berlin, Heidelberg: Springer; 2016. Disorders of cobalamin and folate transport and metabolism//Inborn Metabolic Diseases; pp. pp. 385–99. [Google Scholar]
- 27.Marrazzo A, Taormina P, Marrazzo E, Palumbo VD, Damiano G, Maione C, et al. The sentinel node biopsy is not contraindicated in multifocal breast carcinoma. Eur J Oncol. 2011;16:105–10. [Google Scholar]
- 28.Postolova A, Troxell ML, Wapnir IL, Genovese MC. Methotrexate in the Treatment of Idiopathic Granulomatous Mastitis. J Rheumatol. 2019 doi: 10.3899/jrheum.181205. [DOI] [PubMed] [Google Scholar]
- 29.Takiguchi H, Chen V, Obeidat ME, Hollander Z, FitzGerald JM, McManus BM, Ng RT, Sin DD. Effect of short-term oral prednisone therapy on blood gene expression: a randomised controlled clinical trial. Resp Res. 2019;20((1)) doi: 10.1186/s12931-019-1147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data