Abstract
The turnover of most mRNAs in Saccharomyces cerevisiae begins with deadenylation followed by decapping and 5′→3′ exonucleolytic digestion. An important question involves the mechanisms that allow particular mRNAs to exhibit different rates of both deadenylation and decapping. Since the cap structure plays a critical role in the assembly of translation initiation factors, we hypothesized that the status of the cytoplasmic cap binding complex would affect the rate of decapping. To test this hypothesis, we examined mRNA decay rates in yeast strains that were defective in several translation initiation factors that are part of the cap binding complex. These experiments yielded three significant observations. First, any mutation known to inhibit translation initiation also increased the rate of decapping. Second, decapping still occurred only after deadenylation, suggesting that the ability of the poly(A) tail to inhibit decapping does not require efficient translation of the transcript. Third, mutants with defects in translation initiation factors also showed an increase in the rate of deadenylation, suggesting that the rate of deadenylation may be controlled primarily by the translation status of the transcript. These results argue that the nature of the translation initiation complex is a critical factor in determining the mRNA half-life. This view also implies that some cis-acting sequences that modulate mRNA decay rate do so by affecting the translation status of the transcript.
The differential turnover of mRNAs is an important aspect of the modulation of gene expression in the eukaryotic cell (for reviews, see references 9, 15, 29, and 45). It is now clear that, at least in Saccharomyces cerevisiae, mRNAs with different decay rates are degraded primarily by a single general pathway of mRNA turnover. In this pathway, mRNAs are first deadenylated, which allows the transcript to become a substrate for a decapping reaction (18, 28, 40–42), and once decapped, they are susceptible to 5′-to-3′ exonucleolytic degradation by the Xrn1p exoribonuclease (28, 41, 42). Several observations suggest that a similar pathway of degradation is likely to exist in mammalian cells. For example, deadenylation can be the first step in mammalian mRNA turnover (see, e.g., references 48 and 53). Moreover, deadenylated decapped intermediates of the decay process can be detected (17), and mammalian homologs of the yeast Xrn1p exoribonuclease (7) and the yeast-decapping enzyme (encoded by the DCP1 gene [10]) have been identified (51).
The basis for differential decay rates of individual yeast mRNAs is that the transcripts differ in their rates of deadenylation and decapping. For example, at 24°C the unstable MFA2 transcript (t1/2 = 4 min) deadenylates more rapidly (∼13 A’s/min) than does the stable PGK1 transcript (t1/2 = 45 min; ∼4 A’s/min) (18). In addition, the MFA2 transcript is decapped rapidly after deadenylation whereas the PGK1 transcript is decapped slowly (41, 42). Given these differences, in order to understand differential mRNA degradation it will be critical to determine the sequences and properties of mRNAs that modulate the rates of deadenylation and decapping.
In addition to being the site of decapping, the cap structure is crucial for the ability of a mRNA to initiate translation efficiently (5, 44). This dual role of the cap structure has led to the hypothesis that the rate of decapping is specified by the nature of the cap binding complex or by proteins that interact with this complex of proteins. Given this view, a likely set of proteins to affect decapping are those that make up the cytoplasmic cap binding complex, also referred to as the eIF-4F complex (22, 49). In addition, a second set of proteins known as eIF-3 may play a role in decapping, since this complex recruits the 40S ribosomal subunit to the 5′ end of the mRNA by binding to both the eIF-4F complex and the mRNA itself (25, 43). An important issue is how these proteins influence the ability of a mRNA to be decapped.
Prior examination on the effects of mutations in translation initiation factors on mRNA stability have been mixed. For example, mutations in the Prt1 protein, which is a component of the eIF-3 complex, have been reported to accelerate the decay of the SSA1 and Ip mRNAs under specific conditions (6, 16). Conversely, specific mutations in the translation initiation factor eIF-5A have been reported to block 5′-to-3′ exonucleolytic digestion (55). Finally, it has been reported that mutations in the eIF-4E protein that lead to a partial decrease in translation in vivo have no effect on the degradation rate of the PGK1 mRNA in yeast (37), which is known to be degraded primarily by deadenylation-dependent decapping (42). This observation has raised the possibility that the eIF-4E protein and possibly other initiation factors do not play a major role in mRNA turnover (37). However, it is possible that the eIF-4E alleles examined, which were partial-loss-of-function alleles, were simply not strong enough to show an effect.
To examine more completely the role of the translation initiation factors in controlling mRNA turnover, we have examined the stability of the MFA2 and PGK1 mRNAs in strains carrying alleles of several of the components of eIF-4F and eIF-3 that were known to cause a defect in translation initiation. These two mRNAs were ideal for comparison in this analysis for several reasons. First, they represent the range of mRNA stability in yeast, since the MFA2 mRNA is unstable (t1/2 = 4 min) and the PGK1 transcript is stable (t1/2 = 45 min) (27). Most importantly, the decay of these mRNAs has been well characterized, and both are degraded by the deadenylation-dependent decapping pathway of mRNA turnover (41, 42). Moreover, the different rates of mRNA degradation of these two transcripts can be attributed to specific differences in deadenylation and decapping (18, 41, 42). We observed that mutations in either the eIF-4F complex or the eIF-3 complex can influence the rate of mRNA turnover by increasing both the rate of deadenylation and the rate of decapping. These observations have implications for understanding the control of mRNA turnover and suggest that the status of the translation initiation complex on the mRNA dictates the rates of both deadenylation and decapping.
MATERIALS AND METHODS
Yeast strains.
The genotypes of all the strains used are listed in Table 1, and the strains were grown in standard media. All strains except yRP1323 and yRP1324 have GAL1 upstream activating sequence-regulated PGK1pG and MFA2pG genes as well as the LEU2 gene, collectively termed LEU2pm, integrated at the CUP1 locus (26). The CAF20 and eIF-4G2 disruption plasmids were obtained from Michael Altmann and used to disrupt the CAF20 and eIF-4G2 loci with a URA3 gene in the yRP841 background (23, 34). Both integrations were verified by Southern blot analysis (data not shown). The eIF-4G1Δ strain and the cdc33-42 strains were obtained from Nahum Sonenberg and repeatedly crossed into the same genetic background by using strain yRP841 (2, 23). The prt1-63 strain was obtained from Alan Hinnebusch and repeatedly crossed to yRP841 (20, 21). The mutant eIF-4A (SS8-3A) strain was obtained from Patrick Linder (47) and transformed with pRP469 and pRP485 for mRNA analysis. The yeast strains were transformed by standard techniques, and plasmids were maintained when necessary by growth in selective media.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| yRP841 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 26 |
| yRP1314 | MATa leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [CDC33/TRP1] | This study |
| yRP1315 | MATa leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [CDC33/URA3] | This study |
| yRP1316 | MATα leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1317 | MATα his4 leu2 lys2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1318 | MATα leu2-3,112 lys2-201 trp1 ura3-52 caf20::URA3 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1319 | MATa leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG tif4632::URA3 | This study |
| yRP1320 | MATa his3Δ1 leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG tif4631::LEU2 | This study |
| yRP1321 | MATa leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | This study |
| yRP1322 | MATa leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | This study |
| yRP1323 | MATa ade2 his3 leu2 ura3 tif1::HIS3 tif2::ADE2 [tif1/LEU2 MFA2pG/URA3] | 47 |
| yRP1324 | MATa ade2 his3 leu2 ura3 tif1::HIS3 tif2::ADE2 [tif1/LEU2 PGK1pG/URA3] | 47 |
| yRP1326 | MATα leu2 prt1-63 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yRP1327 | MATα leu2 prt1-63 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG [cdc33-42/TRP1] | This study |
| yRP1328 | MATa his4 leu2 lys2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 | This study |
| yRP1329 | MATa his4 leu2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 [cdc33-42/TRP1] | This study |
| yRP1330 | MATa leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG xrn1::URA3 | This study |
| yRP1331 | MATa his4 leu2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG xrn1::URA3 | This study |
| yRP1332 | MATα his4 leu2 lys2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG xrn1::URA3 [cdc33-42/TRP1] | This study |
| yRP1333 | MATa his4 leu2 lys2 trp1 ura3 cdc33::LEU2 cup1::LEU2/PGK1pG/MFA2pG xrn1::URA3 [cdc33-42/TRP1] | This study |
| yRP1373 | MATa his4 leu2 lys2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG upf1::URA3 | This study |
| yRP1374 | MATa his4 leu2 lys2 prt1-63 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG upf1::URA3 | This study |
mRNA analysis.
Steady-state transcriptional shutoff experiments were performed as described previously with slight modifications (13). Briefly, the cells were grown to mid-log phase in galactose medium, shifted to 38°C for 1 h, harvested, and shifted to medium containing dextrose to inhibit transcription, with an aliquot removed at each time point (specified on figures).
Transcriptional pulse-chase experiments used to track a synchronous pool of mRNAs were done as previously described with slight modifications (18, 41). Briefly, cells were grown to mid-log phase in medium containing raffinose, shifted to 38°C for 1 h, harvested, and resuspended in medium containing galactose to induce the transcripts; finally dextrose was added to shut off transcription.
mRNA was isolated as described previously (13). Agarose Northern assays were done with 10 μg of RNA, and RNase H and polyacrylamide Northern (RNA) assays were done with 40 μg of RNA as previously described (40). Northern assay results were quantitated on a Molecular Dynamics PhosphorImager and standardized to 7S RNA, a polymerase III transcript (13).
In vivo translation.
In vivo [35S]methionine incorporation was performed as previously described (2) with some modifications. Briefly, yeast strains were grown to mid-log phase (optical density at 600 nm ≈ 0.4) at 24°C in medium containing dextrose, at which point half of each culture was shifted to 38°C for 1 h. The cells were then harvested, resuspended in prewarmed methionine-minus medium containing dextrose, and supplemented with 25 μCi of [35S]methionine. At various time points, aliquots were removed and the cells were harvested and resuspended in 1 ml of ice-cold 10% trichloroacetic acid. The samples were heated at 95°C for 5 min and cooled on ice. The precipitated protein was collected on glass fiber filters, washed with 10 ml of 5% trichloroacetic acid, and washed again with 10 ml of 70% ethanol and finally with 1 ml of acetone. The filters were air dried for 30 min and then counted in a liquid scintillation counter.
RESULTS
Defects in the cytoplasmic cap binding complex (eIF-4F) lead to higher mRNA decay rates.
To test the hypothesis that translation initiation factors binding to the 5′ end of a mRNA play a role in controlling decapping, we first examined the effects of mutations in the components of the cap binding complex (eIF-4F) on mRNA turnover. This complex in yeast is generally described as consisting of eIF-4E (cdc33), the cap binding protein (1), and an eIF-4G subunit, which is encoded by two genes in yeast, TIF4631, which encodes eIF-4G1, and TIF4632, which encodes eIF-4G2 (23). In addition, we examined the effects of mutations in eIF-4A, a DEAD box helicase required for translation (47) and known to physically associate with eIF-4F in mammals (19, 24), and p20, encoded by the CAF20 gene, that is thought to bind the cap binding protein in a negative regulatory role (3). If deletion of a given gene was lethal, temperature-sensitive mutants were used in this analysis. All temperature-sensitive alleles used were alleles that had previously been shown to have effects on translation rate in vivo (2, 20, 21, 47) (see below). If a strain with deletion of a particular gene was viable, strains with disruptions of the gene were used.
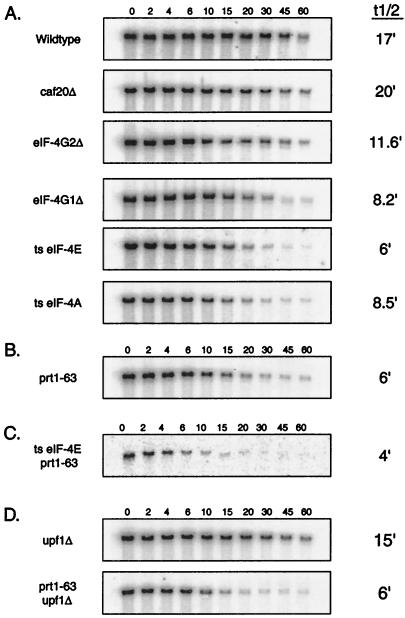
The translation initiation mutations could be placed into two categories based on their effects on mRNA half-life. For example, disruption of the CAF20 gene had little or no consequence on the decay rate of either the PGK1 mRNA or the MFA2 mRNA (Fig. 1A and Table 2). Since this protein does not significantly affect growth rate or translation rate (34), it was not surprising that a caf20Δ mutation failed to alter mRNA decay rates.
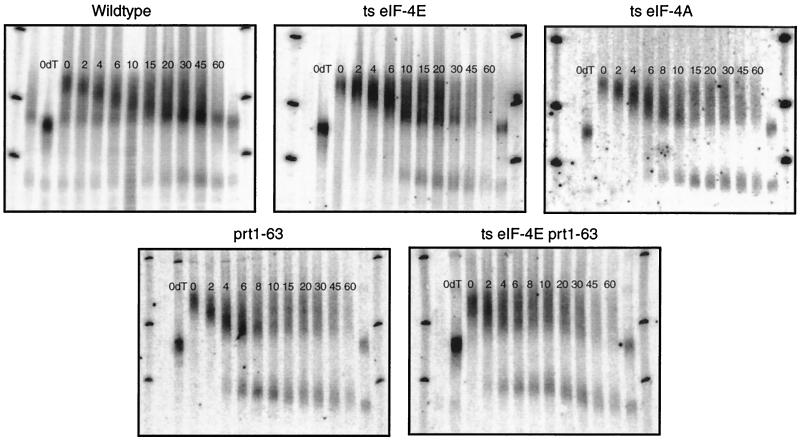
FIG. 1.
Disruption of translation initiation factor function leads to a decrease in the half-lives of mRNAs. (A) mRNA half-life measurements from strains containing mutations in translation initiation factors from the eIF-4F complex. (B) mRNA half-life measurements from strains containing a mutation in PRT1, a translation initiation factor from the eIF-3 complex. (C) mRNA half-life measurements from a strain containing mutations in both eIF-4E and PRT1. (D) mRNA half-life measurements from strains containing mutations in both PRT1 and UPF1. Steady-state half-life measurements of the PGK1pG mRNA were determined from agarose Northern gels. The numbers above the lanes indicate minutes after transcriptional repression. A representative gel for each strain is shown, and the average value (minutes) of the half-life for the full-length transcript from at least three experiments in each strain is given on the right. The experimental variation for the half-life measurements of each strain was less than ±15%. Northern gels were probed with oligonucleotide oRP141 (5′-AATTGATCTATCGAGGAATTCC-3′), which is complementary to the poly(G) and flanking 3′ sequence in the PGK1pG mRNA. ts, temperature sensitive.
TABLE 2.
Effects of translation initiation factors on mRNA half-life and deadenylation of PGK1 and MFA2
| Strain | PGK1 RNA half-lifea (min) | PGK1 deadenylation rateb (A’s/min) | MFA2 RNA half-lifea (min) |
|---|---|---|---|
| Wild type | 17.0 | 3.1 | 4.0 |
| caf20Δ | 20.0 | NDc | 4.4 |
| eIF-4G2Δ | 11.6 | ND | 2.6 |
| eIF-4G1Δ | 8.0 | 3.7 | 2.0 |
| ts eIF-4E cdc33-42 | 6.0 | 4.1 | 3.0 |
| ts eIF-4A | 8.5 | 4.5 | 3.5 |
| gcd10-505 | 4.9 | ND | 2.3 |
| prt1-63 | 6.0 | 4.0 | 3.0 |
| ts eIF-4E prt1-63 | 4.0 | 5.1 | 2.0 |
Comparative half-life measurements for PGK1 and MFA2 mRNA based on a minimum of three transcription shutoff experiments. The experimental variation for the half-life measurements of each strain was less than ±15%.
Deadenylation rates of PGK1 are given for all the translation initiation mutants based on measurements taken from transcriptional shutoff mRNAs run on acrylamide Northern gels. The deadenylation rates are based on a minimum of two transcription shutoff experiments. The experimental variation for the half-life measurements of each strain was less than ±15%.
ND, not determined.
In contrast, following a shift to the restrictive temperature for 1 h, temperature-sensitive mutations in eIF-4E and eIF-4A and a deletion of eIF-4G1 all led to higher rates of mRNA degradation. For example, in wild-type cells the PGK1 mRNA decayed with an average half-life of 17 min at 38°C (Fig. 1A). It should be noted that this is a shorter half-life than has been previously reported at lower temperatures and presumably reflects the faster decay of yeast mRNAs that is seen at high temperatures (27). Notably, the half-life of the PGK1 mRNA from a strain containing the temperature-sensitive mutation in eIF-4E/cdc33-42, was decreased to about 6 min, almost a threefold reduction compared to that of the wild-type strain at 38°C (Fig. 1A). Similarly, the PGK1 mRNA decayed faster in the temperature-sensitive eIF-4A (t1/2 = 8.5 min) and eIF-4G1Δ (t1/2 = 8.2 min) mutants (Fig. 1A and Table 2), although the magnitude of the change was not as great. Control experiments done at the permissive temperature showed that PGK1 and MFA2 mRNA from the conditional eIF-4E/cdc33-42 allele decayed with wild-type kinetics. In contrast, the eIF-4G1Δ showed an increased decay rate at high and low temperatures.
Faster mRNA turnover was also observed for the unstable MFA2 mRNA in translation initiation mutant strains. In a wild-type strain at 38°C, the MFA2 half-life was 4 min (Table 2), whereas in strains containing the temperature-sensitive eIF-4E/cdc33-42, temperature-sensitive eIF-4A, or eIF-4G1Δ mutations, the MFA2 half-life was decreased to 3, 3.5, and 2 min respectively. In addition, the MFA2 mRNA showed accelerated decay in an eIF-4G2Δ strain, even though the decay of the PGK1 mRNA in this strain was only slightly affected (Table 2). These smaller changes in the MFA2 mRNA decay rate were extremely reproducible, but because of their smaller magnitude, they should be interpreted with caution. Moreover, it might be difficult to accelerate the decay rate of the MFA2 transcript greatly since this mRNA is already highly unstable.
The conclusion that the mRNA decay rate was higher in the various translation initiation mutants was also supported by the observation that the total levels of individual mRNAs in strains containing translation initiation mutations were decreased. For example, after 1 h at the restrictive temperature, the level of the full-length PGK1 mRNA in the eIF-4E/cdc33-42 strain was approximately 28% of the level in a wild-type strain under similar conditions (data not shown). In combination, these results indicated that lesions in the cap binding complex can accelerate mRNA degradation for yeast transcripts and suggest a role for these proteins in modulating mRNA turnover rates (see Discussion).
Defects in the eIF-3 complex lead to higher mRNA decay rates.
In addition to the cap binding complex, a second complex of translation factors, termed eIF-3, plays a crucial role in interacting with the eIF-4F complex and the 40S–eIF-2 complex to initiate assembly at the 5′ end of the mRNA. The eIF-3 complex is made up of about eight proteins that physically interact with both the mRNA and the eIF-4F complex (33, 38). These interactions place eIF-3 in the immediate vicinity of the 5′ cap structure. For this reason, components of the eIF-3 complex were also tested for a role in mRNA turnover. The Prt1 protein was tested for its effect on mRNA turnover by using a temperature-sensitive mutation of the protein.
An interesting result was that a mutation in the Prt1 protein also gave faster degradation of the PGK1 and MFA2 transcripts (Table 2 and Fig. 1B). For example, the PGK1 half-life was decreased from a wild-type value of 17 min to 6 min in prt1-63 mutant strains. Mutations in the Gcd10 protein, which has been hypothesized to be a component of the eIF-3 complex in addition to having other functions (21), gave similar results (data not shown). These results showed that defects in the eIF-3 complex can lead to an acceleration of mRNA turnover.
The above experiments indicated that the PGK1 and MFA2 mRNAs were degraded more rapidly in strains that carried lesions in several different translation initiation factors. The decrease in mRNA stability corresponded to lesions known to decrease translation initiation. To magnify the effect on mRNA decay rate, we reasoned that a double mutant defective in both eIF-4F and eIF-3 function might show an even stronger phenotype with respect to mRNA decay rates. To test this hypothesis, we constructed a strain carrying both the temperature-sensitive eIF-4E/cdc33-42 and the prt1-63 mutations. This strain showed an increased sensitivity to higher temperatures and failed to grow above 24°C, whereas either single mutant strain grew well at 30°C. The half-lives of both PGK1 and MFA2 mRNAs in the double mutant were shorter than those in either single mutant, suggesting that blocking multiple steps of translation initiation leads to a more severe mRNA turnover defect (Fig. 1C). These results demonstrate that lesions that alter the normal assembly and function of the translation initiation complex on the 5′ end of the transcript result in faster mRNA degradation.
The effect of mutations on the mRNA decay rate generally corresponds to their effect on the translation rate.
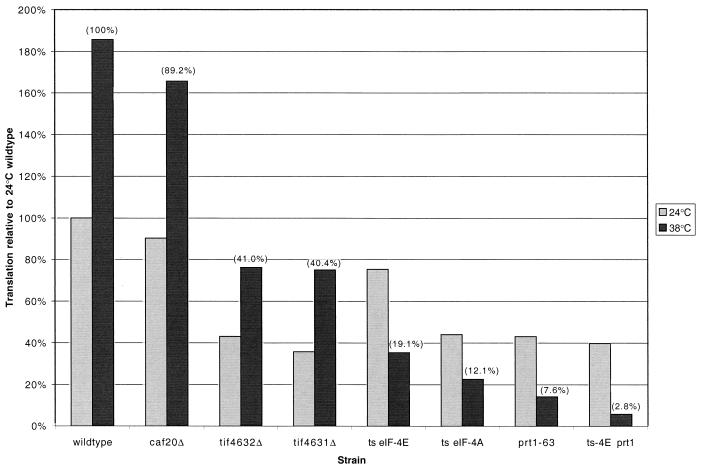
Our analysis of the effect of various translation initiation mutations on mRNA decay compared several mutations whose effects on translation rate have been individually determined by a number of different laboratories using different types of assays. To relate our observations on the effects on mRNA decay, it was necessary to compare the effects of all these mutations on translation rate by using the same experimental protocol. To do this, we measured the rate of incorporation of [35S]methionine into trichloroacetic acid-precipitable counts in the various strains both at 24°C and after a shift to 38°C for 1 h (see Materials and Methods). As can be seen in Fig. 2, with the exception of the caf20Δ mutants, all the mutants had a decreased translation rate to some extent. The rates for the tif4631Δ and tif4632Δ strains were decreased to approximately 40% of wild-type rates at both 24 and 38°C. All of the temperature-sensitive strains showed a temperature-dependent inhibition of translation rate, with the residual translation at 38°C being 19, 12, 7.5, and 3% for the eIF-4E/cdc33-42, temperature-sensitive eIF-4A, prt1-63, and eIF-4E/cdc33-42 prt1-63 mutants, respectively. These results indicate a strong correlation between translation rate and mRNA stability (see Discussion).
FIG. 2.
Relative rates of translation in various translation initiation mutants. The rate of [35S]methionine incorporation was measured either at 24°C or after a 1-h temperature shift to 38°C. The rates of translation are given relative to the wild-type strain at 24°C (where rates were originally measured as the rate of incorporation per optical density unit after 30 min). The rate of translation for each strain is the average of at least two independent experiments. The numbers in parentheses are the rates of incorporation of the strains at 38°C relative to the wild-type strain at 38°C. ts, temperature sensitive.
The faster mRNA turnover seen in translation initiation mutants occurs by the normal 5′→3′ pathway of decay.
An important question was whether the faster decay seen in the various translation initiation mutants was due to an acceleration of the normal pathway of deadenylation-dependent decapping or occurred because the transcript was being degraded rapidly by alternative degradation mechanisms, which are known to exist in yeast (4). To interpret the effects of the lesions in the translation initiation factors, we determined if the accelerated decay required the Dcp1p decapping enzyme and the Xrn1p 5′-to-3′ exoribonuclease, which are known to function in the normal degradation of these mRNAs (10, 41, 42). To this end, strains were made which combined the temperature-sensitive eIF-4E lesions with a dcp1Δ or a xrn1Δ mutation.
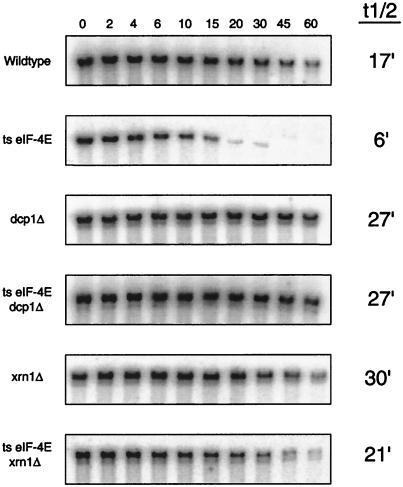
We observed that the deletion of either the DCP1 gene or the XRN1 gene prevented the faster degradation seen in the temperature-sensitive eIF-4E/cdc33-42 strain. For example, deletion of the DCP1 gene in combination with the cdc33-42 lesion led to a stabilization of the mRNA, since the decapping step was prevented, giving the PGK1 mRNA an increased half-life of 27 min (Fig. 3). The eIF-4E/cdc33-42 dcp1Δ double mutant had a half-life identical to that of the dcp1Δ mutant, i.e., four to five times longer than that seen in the temperature-sensitive eIF-4E/cdc33-42 mutant alone. Analysis of the mRNA from the temperature-sensitive eIF-4E/cdc33-42 xrn1Δ double-mutant strains showed very similar results to the temperature-sensitive eIF-4E/cdc33-42 dcp1Δ double mutant (Fig. 3), although the mRNA from the temperature-sensitive eIF-4E/cdc33-42 xrn1Δ double mutant was not quite as stable as the mRNA from the xrn1Δ mutant, presumably due to residual alternative 5′-to-3′ exonuclease activity subsequent to the decapping step (42). We interpreted these results to suggest that the faster decay seen in the various translation initiation mutants was due to an increase in the rates of specific steps in the 5′-to-3′ mRNA decay pathway.
FIG. 3.
mRNAs from strains containing a mutation in eIF-4E undergo turnover through the general 5′→3′ pathway. mRNA half-lives were measured in strains containing mutations in both eIF-4E and DCP1 and in strains containing mutations in both eIF-4E and XRN1. Steady-state half-life measurements of the PGK1pG mRNA were determined from agarose Northern gels. The numbers above the lanes indicate minutes after transcriptional repression. The average value (minutes) of the half-life for the full-length transcript from at least three experiments in each strain is given on the right. Northern gels were probed with oligonucleotide oRP141. ts, temperature sensitive.
Defects in translation initiation increase the rate of both deadenylation and decapping.
To determine the basis for the faster mRNA decay seen in the various translation initiation mutants, it was important to determine what step in mRNA decay was being accelerated. In principle, the higher observed decay rate could be due either to an acceleration of deadenylation and/or decapping or to a bypass of the requirement for deadenylation before decapping (41). In the following experiments, we examined the rates of deadenylation and decapping in the various mutant strains.
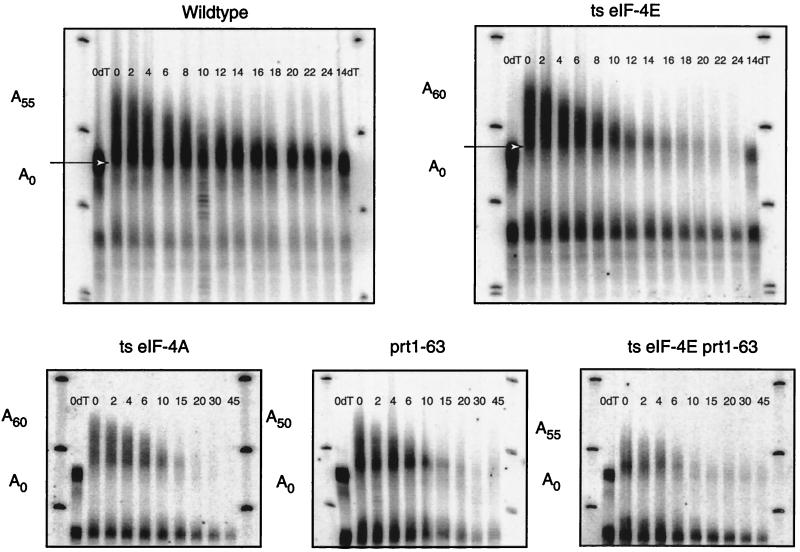
One method for measuring the rates of deadenylation and decapping is to monitor the changes in poly(A) tail length and mRNA levels following repression of transcription from a culture at steady state (40). Two significant observations were made from this examination. First, measurement of the rate of deadenylation, as assessed by the shortening of the longest poly(A) tails in the population, indicated that the temperature-sensitive eIF-4E/cdc33-42, temperature-sensitive eIF-4A, eIF-4G1Δ (data not shown), and prt1-63 strains all showed higher rates of deadenylation (Fig. 4; summarized in Table 2). For example, measurements of the wild-type strain showed that deadenylation of the mRNA present as full-length mRNA proceeded at a rate of about 3.1 adenylate residues per min (Fig. 4 and Table 2); whereas the deadenylation of the full-length mRNA from the temperature-sensitive eIF-4E/cdc33-42 mutant strain proceeded at a rate of 4.1 adenylate residues per min. The largest change in deadenylation rate was seen in the eIF-4E/cdc33-42 prt1-63 double mutant, wherein the poly(A) tail of the PGK1 mRNA now shortened at 5 to 6 adenylate residues per min. Similar results were obtained by measuring deadenylation rates in transcriptional pulse-chase experiments, where the deadenylation rate is measured by monitoring the poly(A) tail length from a synchronous population of mRNAs (see below) (data not shown). Based on these observations, we concluded that lesions in the various translation initiation factors promoted faster deadenylation. This has implications for understanding the control of the deadenylation rate (see Discussion).
FIG. 4.
The rate of deadenylation is increased when translation initiation is impaired. Polyacrylamide Northern gels of transcriptional repression experiments examining the deadenylation of PGK1 and accumulation of oligoadenylated mRNAs in various translation initiation mutants are shown. Minutes following transcription repression are given directly above each sample. The top fragment was produced by cleavage of the full-length mRNA with RNase H and oligonucleotide oRP70 (5′-CGGATAAGAAAGCAACACCTGG-3′). This cleavage shortens the mRNA enough to visualize small differences at the 3′ end. The bottom fragment is the decay intermediate stabilized by the poly(G) insertion in the 3′ UTR. The arrow indicates the size of the oligoadenylated mRNA. The numbers after the A’s in the cartoons represent the range of poly(A) tail sizes found on each mRNA species as determined by comparison with the oligo(dT) lanes, in which the poly(A) tails have been completely removed by cleavage with RNase H, oligonucleotide oRP70, and oligo(dT). The blots were probed with oRP141. ts, temperature sensitive.
The second important observation from examining the decay of the mRNA following transcriptional repression was that the deadenylated form of the PGK1 mRNA, which is relatively stable in wild-type strains due to the low rate of decapping for this transcript, decays rapidly in all the mutant strains except the eIF-4G2Δ and the caf20Δ strains (Fig. 4 and data not shown). A clear example of this phenomenon is seen by comparing the wild-type and temperature-sensitive eIF-4E strains (compare panels in Fig. 4). Since decapping is a prerequisite for 5′-to-3′ exonucleolytic digestion, these observations indicate that defects in the translation initiation complex increase the rate of decapping (see Discussion).
Decapping remains dependent on prior deadenylation in translation initiation mutants.
In the 5′-to-3′ mRNA decay pathway, mRNA turnover proceeds sequentially by deadenylation followed by decapping and exonucleolytic decay. To determine if the rapid decay seen in the translation initiation mutants still required deadenylation, we examined the decay in these strains by using a transcriptional pulse-chase method (18). These experiments used the regulatable GAL1 upstream activating sequence to create a synchronous pulse of mRNA that can be monitored throughout the decay pathway, thereby allowing an examination of the relationship between deadenylation and decay.
Transcriptional pulse-chase experiments with the various mutants provided two observations that argued that decapping still required prior deadenylation. First, the mRNA levels did not begin to drop until after the poly(A) tail had been shortened to an oligo(A) length. This is characteristic of mRNAs that require deadenylation before decay (18) and is not seen with mRNAs that are degraded independently of poly(A) tail shortening, which occurs in response to premature translation termination (41). The second observation, that the rapid decay seen in the translation initiation mutants still required deadenylation, came from the analysis of an mRNA decay intermediate stabilized by the insertion of a poly(G) tract in the 3′ untranslated region (UTR), which serves to block the Xrn1p exoribonuclease (18, 52). Consistent with earlier work on transcripts that require deadenylation before decay (18), this mRNA fragment was produced in the various mutant strains, including the temperature-sensitive eIF-4E/cdc33-42 prt1-63 double-mutant strain, only after a substantial fraction of the mRNA had been deadenylated to an oligo(A) length (Fig. 5). Moreover, these mRNA decay intermediates had oligo(A) tails, even at times when the mRNA population consisted of transcripts with both long and short poly(A) tails (Fig. 5), arguing that only oligoadenylated transcripts are substrates for the next step in decay. These results are in strong contrast to cases where decapping is independent of deadenylation, wherein this mRNA fragment is produced before poly(A) shortening has reached a oligo(A) tail and so the mRNA fragment itself has long poly(A) tails (41).
FIG. 5.
The rates of both deadenylation and decapping are increased when translation initiation is impaired. Transcriptional pulse-chase analysis of the PGK1pG mRNA in various translation initiation mutant strains is used to examine deadenylation and fragment production. Time points used after a 6-min transcriptional induction and subsequent repression are shown above each lane. The top fragment was produced by cleavage of the full-length mRNA with RNase H and oligonucleotide oRP70. This cleavage shortens the mRNA enough to visualize small differences at the 3′ end. The bottom fragment is the decay intermediate stabilized by the poly(G) insertion in the 3′ UTR. The blots were probed with oRP141. ts, temperature sensitive.
These observations indicate that the ability of the poly(A) tail to act as an inhibitor of decapping is not prevented even if there is a strong inhibition to translation. This is inconsistent with the simple model that the mechanism by which poly(A) tails inhibit decapping is by bringing the poly(A) binding protein to the mRNA and thereby, through its interaction with eIF-4G, stabilizing the cap binding complex.
The increased rate of mRNA turnover in translation initiation mutants is not UPF1 dependent.
The above results showed that defects in the Prt1 protein led to faster decay of the PGK1 and MFA2 mRNAs, by promoting increases in the rates of both deadenylation and decapping. It has also been recently shown that prt1 mutations can affect the levels of the SSA1 and SSA2 mRNAs in yeast, suggesting that in prt1 mutants these mRNAs might also be degraded more rapidly (6). Interestingly, the faster turnover seen in the prt1 mutant strains was dependent on the Upf1 protein, which is known to be required for the degradation of mRNAs containing early nonsense codons (36). Given this requirement for the Upf1p in the faster decay seen in the prt1 mutants, we determined if the faster decay we observed in the prt1 mutants for the PGK1 and MFA2 transcripts was also UPF1 dependent.
Strains that contained mutations in both the eIF-4E/CDC33 gene and the UPF1 gene or in the PRT1 gene and the UPF1 gene were created. Transcriptional shutoffs were performed as previously described to determine the mRNA half-lives of PGK1 and MFA2 in the mutant strains. In both the temperature-sensitive double-mutant eIF-4E/cdc33-42 upf1Δ and the prt1-63 upf1Δ strains, the half-lives resembled those of the single-translation-initiation mutant, indicating that the Upf1 protein was not required for the faster decay of these mRNAs in response to mutations in the translation initiation factors (Fig. 1D).
These results indicated that the faster decay observed for the PGK1 and MFA2 mRNAs in response to the mutations in the PRT1 or eIF-4E/CDC33 gene is somehow different from the proposed faster decay seen with the SSA1 and SSA2 mRNAs in the PRT1 strain. Unfortunately, since the actual mechanism by which the SSA1 and SSA2 transcripts are degraded is not known, it is difficult to evaluate these differences. It is possible that the SSA1 or SSA2 mRNAs are subjected to a different mechanism of turnover and therefore give different results. Alternatively, since SSA1 and SSA2 encode heat shock proteins, these mRNAs might have methods for enhancing their translation during heat shock and as such might have a different dependence on the Upf1p.
DISCUSSION
Mutations in translation initiation factors increase the rate of decapping.
Several observations indicated that defects in the translation initiation factors eIF-4E, eIF-4G, eIF-4A, and Prt1 (a component of eIF-3) led to an increase in the rate of decapping. First, in strains carrying mutations in these translation initiation factors, the decay rate of the PGK1 and MFA2 transcripts was higher. Second, oligoadenylated PGK1 mRNA species produced following deadenylation were degraded rapidly in the translation initiation mutants (Fig. 4 and 5). In contrast, such oligoadenylated PGK1 transcripts persisted in the wild-type strain due to the normally low rate of decapping for this mRNA. These results support the conclusion that the ability of the decapping enzyme to cleave a mRNA substrate is affected by the ability of the translation initiation complex to form or to persist on the transcript. This interpretation is also supported by the prior observation that altering the translation initiation process in cis by the insertion of secondary structures in the 5′ UTR increases the rate of decapping of the PGK1 mRNA (42).
Comparison of translation rates in the various mutants allowed us to relate the effects on mRNA turnover to changes in the translation rate. In general, the stronger the inhibition of observed translation rate, the higher the observed mRNA decay rate. For example, the tif4631Δ and tif4632Δ mutations had the smallest effects on PGK1 mRNA decay, and these strains showed the highest level of residual translation. It is interesting and notable that the tif4632Δ mutation caused a decrease in the translation rate without significantly affecting the decay rate of the PGK1 mRNA, although the MFA2 mRNA did show accelerated decay in this strain (Table 2 and Fig. 2). This suggests that there will be some mRNA-specific effects of mutations in particular translation initiation factors. In the other extreme, the strongest defect in the translation rate was seen in the eIF-4E/cdc33-42 prt1-63 double mutant, which showed the highest mRNA decay rates (Table 2 and Fig. 2).
The increase in decapping rate we observed in a defective allele of the eIF-4E protein is seemingly different from reports that small N- and C-terminal deletions of the eIF-4E protein had no effect on the half-lives of the mRNAs in strains containing those mutant proteins (37). To compare our results with this prior work, we examined the turnover of mRNAs from two of these eIF-4E deletion mutants (cdc33Δ7/209 and cdc33Δ202) and also found no change in the half-lives of PGK1 and MFA2 mRNA (data not shown). This suggests that these particular mutations in eIF-4E are competent for proper mRNA turnover, possibly due to their partial ability to assemble a cap binding complex. The mutation used in the current work, temperature-sensitive eIF-4E/cdc33-42, leads to a more severe defect in cap binding than do the nonlethal eIF-4E truncation mutations and also leads to a more significant decrease in translation of mRNAs at the restrictive temperature (2). This more severe defect in eIF-4E function manifests itself as an increase in the rate of mRNA turnover.
A straightforward mechanistic interpretation of these observations is that decapping requires the destabilization or disassembly of the cap binding complex, to allow the decapping enzyme to gain access to the cap structure. In the simplest view, the cap binding complex and the decapping enzyme could be viewed as being in direct competition for the 5′ cap structure. However, it is not unreasonable to expect that there will be specific transitions or states of the cap binding complex that promote interactions of the decapping enzyme with this complex and ultimately lead to decapping of the transcript. It should be noted that this view of decapping predicts that there should be specific alleles of translation initiation factors that lead to an inhibition of the decapping rate. However, one possibility that cannot be formally ruled out is that translation initiation indirectly protects the mRNA from decapping. For example, the mere presence of elongating ribosomes bound to the mRNA could be hypothesized to prevent decapping.
Mutations in translation initiation factors increase the rate of deadenylation.
We also observed that the same lesions in translation initiation factors that gave higher rates of decapping also led to an increase in the rate of deadenylation. The critical experiment was that direct measurement of deadenylation rate in wild-type and mutant strains showed that in strains containing mutations in translation initiation factors (eIF-4E, eIF-4A, Prt1 [Fig. 4 and 5], and eIF-4G1 [data not shown]), deadenylation was faster than in a wild-type strain. The most extreme example of this effect was observed in the temperature-sensitive eIF-4E/cdc33-42 prt1-63 double mutant, whose deadenylation rate was increased threefold over that of the wild-type strain (Table 2). These observations led us to propose that the status of the translation initiation complex plays a major role in modulating the deadenylation rate for yeast mRNAs.
The conclusion that the status of the translation initiation complex plays a significant role in determining the deadenylation rate is supported by several prior observations in the literature. First, insertion of a stem-loop into the PGK1 5′ UTR to inhibit ribosome assembly leads to a higher rate of deadenylation (42). Second, alterations of the PGK1 5′ UTR and AUG context which decrease the translation initiation rate also increase the rate of deadenylation (31). Third, the AU-rich elements known to promote deadenylation in mammalian cells have also been observed to inhibit translation initiation rate (30). Similarly, the human ferritin L-chain mRNA also shows changes in poly(A) tail length based on the translational status of the mRNA (39). Finally, deadenylation of the c-myc mRNA is inhibited by the addition of the translation elongation inhibitor cycloheximide (32).
Why do changes in translation initiation affect the deadenylation rate? The simplest hypothesis is that the rate of translation initiation affects the nature of the physical interaction between the 5′ and 3′ ends of the mRNA and that changes in the nature of this 5′-3′ interaction in turn affect the rate of deadenylation by altering the availability of the poly(A) tail to the deadenylating nuclease(s). There are two ways in which this could be occurring. In one model, the poly(A) binding protein (PAB) is an inhibitor of deadenylation and must dissociate from the poly(A) tail for deadenylation to occur. This view is suggested by the observation that in both mammalian cell extracts and Xenopus oocytes PAB appears to be an inhibitor of deadenylation (11, 54). Interestingly, PAB interacts with the cap binding complex in several systems (50), and such an interaction can increase the ability of PAB to bind the poly(A) tail (35). This suggests that decreases in the stability of the cap binding complex could lead to destabilization of PAB bound to the poly(A) tail and thereby activate deadenylation. It should be noted that this model of deadenylation occurring in the absence of Pab1p is at odds with the observation that yeast strains deficient for PAB have lower rates of deadenylation (14, 46). This observation in yeast leads to an alternative model in which PAB bound to the poly(A) tail is required for proper deadenylation but that poly(A) shortening is slowed by the process of translation. Transient dissociation between the PAB protein and translation initiation factors, possibly between rounds of translation initiation, allows for periods of deadenylation to occur. This model explains both the pab1Δ phenotypes and the increase in deadenylation seen in yeast translation initiation mutants. Further experiments are required to resolve these issues.
What determines the mRNA half-life?
Our results imply that the relative translational efficiency of a transcript will be a major determinant of the mRNA half-life. In strains carrying strong defects in various translation initiation factors, the mRNA half-lives were significantly decreased, and both mRNAs examined showed relatively similar decay rates. For example, in a wild-type strain there is about a fivefold difference in mRNA half-life between PGK1 and MFA2, but in the translation initiation mutants there is only about a twofold difference in half-life between the two mRNAs, implying that stability differences may arise due to differences in the translational efficiency of the individual mRNAs. This observation suggests that the PGK1 mRNA is stable in the wild-type strain because it is efficiently translated and that the MFA2 mRNA is highly unstable because it is poorly translated. Consistent with this hypothesis, replacing the context of the PGK1 AUG with the MFA2 AUG context, which decreases the translation initiation rate of the PGK1 mRNA, also leads to faster mRNA degradation (31). Moreover, inhibiting PGK1 translation with a stem-loop leads to faster decay, due to the loss of efficient translation (42). In contrast, insertion of a stem-loop into the MFA2 5′ UTR has little effect on the decay rate, presumably because this mRNA is already so poorly translated (8).
The view that the translation efficiency of a mRNA is a major determinant of the decay rate has important implications for understanding the function of sequences that ultimately dictate differential rates of mRNA degradation. This hypothesis leads to the prediction that at least some sequences that promote the mRNA decay rate will function by decreasing the rate of translation initiation, with corresponding downstream changes on the decapping and deadenylation rates. Interestingly, several cis-acting elements that can destabilize yeast transcripts have been described as affecting the rates of both deadenylation and decapping (15, 31, 40, 42). This effect of such instability sequences on both steps in decay can now be easily understood if they primarily affect the rate of translation initiation.
ACKNOWLEDGMENTS
We thank Michael Altmann, Alan Hinnebusch, Patrick Linder, and Nahum Sonenberg for providing many of the yeast strains used in this work. In addition, we thank the members of the Parker laboratory for their support and contributions in the preparation of the manuscript.
This work was supported by the Howard Hughes Medical Institute and by a grant to R.P. from the National Institute of Health (GM45443).
REFERENCES
- 1.Altmann M, Handschin C, Trachsel H. mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:998–1003. doi: 10.1128/mcb.7.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmann M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae. Initiation factor 4E-dependent cell-free system. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J S J, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SK12 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee A K. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Bacteriol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes C A. Upf1 and Upf2 proteins mediate normal yeast mRNA degradation when translation initiation is limited. Nucleic Acids Res. 1998;26:2433–2441. doi: 10.1093/nar/26.10.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashkirov V I, Scherthan H, Solinger J A, Buerstedde J, Heyer W. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beelman C A, Parker R. Differential effects of translational inhibition in cis and trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 9.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 10.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for the normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein P, Peltz S W, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caponigro G, Muhlrad D, Parker R. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae. A stimulatory role for rare codons. Mol Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 15.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cereghino G P, Atencio D P, Saghbini M, Beiner J, Scheffler I E. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol Biol Cell. 1995;6:1125–1143. doi: 10.1091/mbc.6.9.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 19.Edery I, Humbelin M, Darveau A, Lee K A, Milburn S, Hershey J W, Trachsel H, Sonenberg N. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem. 1983;258:11398–11403. [PubMed] [Google Scholar]
- 20.Evans D R H, Rasmussen C, Hanic-Joyce P J, Johnston G C, Singer R A, Barnes C A. Mutational analysis of the PRT1 protein subunit of yeast translation initiation factor 3. Mol Cell Biol. 1995;15:4525–4535. doi: 10.1128/mcb.15.8.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Barrio M T, Naranda T, Vazquez de Aldana C R, Cuesta R, Hinnebusch A G, Hershey J W B, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 22.Goyer C, Altmann M, Trachsel H, Sonenberg N. Identification and characterization of cap-binding proteins from yeast. J Biol Chem. 1989;264:7603–7610. [PubMed] [Google Scholar]
- 23.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Jr, Trachsel H, Sonenberg N. TIF4631 and TIF4632. Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grifo J A, Tahara S M, Morgan M A, Shatkin A J, Merrick W C. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- 25.Hannig E M. Protein synthesis in eukaryotic organisms: new insights into the function of translation initiation factor eIF-3. Bioessays. 1995;17:915–919. doi: 10.1002/bies.950171103. [DOI] [PubMed] [Google Scholar]
- 26.Hatfield L, Beelman C A, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 30.Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- 31.LaGrandeur T, Parker R. The context of the translation start codon, in combination with other mRNA features, contributes to mRNA decay rates in Saccharomyces cerevisiae. RNA. 1999;5:420–433. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laird-Offringa I A, De Witt C L, Elfferich P, van der Eb A J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990;10:6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eIF4G with picornaviral proteases. Implication for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 34.Lanker S, Müller P P, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 35.Le H, Tanguay R L, Balasta M L, Wei C, Browning K S, Metz A M, Goss D J, Gallie D R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Bio Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 36.Leeds P, Wood J M, Lee B-S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linz B, Koloteva N, Vasilescu S, McCarthy J E G. Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:9131–9140. doi: 10.1074/jbc.272.14.9131. [DOI] [PubMed] [Google Scholar]
- 38.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors, 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muckenthaler M, Gunkel N, Stripecke R, Hentze M W. Regulated poly(A) tail shortening in somatic cells mediated by cap-proximal translational repressor proteins and ribosome association. RNA. 1997;3:983–995. [PMC free article] [PubMed] [Google Scholar]
- 40.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 41.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ to 3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 42.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naranda T, MacMillan S E, Hershey J W B. Purified yeast translation factor eIF3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- 44.Rhoads R E. Cap recognition and entry of mRNA into the protein synthesis initiation cycle. Trends Biochem Sci. 1988;13:52–56. doi: 10.1016/0968-0004(88)90028-x. [DOI] [PubMed] [Google Scholar]
- 45.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 47.Schmid S R, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae. Analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–234. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 49.Sonenberg N, Morgan M A, Merrick W C, Shatkin A J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci USA. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarun S, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 51.Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;15:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vreken P, Raue H A. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol Cell Biol. 1992;12:2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson T, Triesman R. Removal of poly(A) and consequent degradation of the c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 54.Wormington M, Searfoss A M, Hurney C A. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- 55.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]