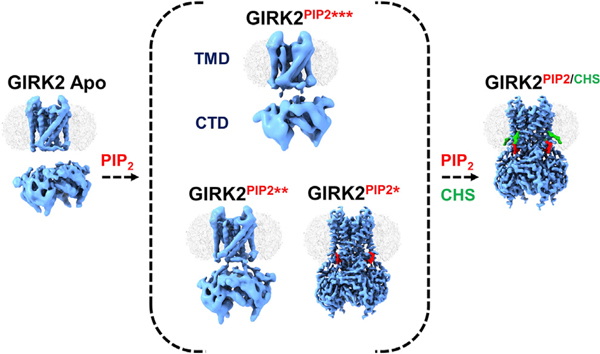
SUMMARY
G-protein-gated inwardly rectifying potassium (GIRK) channels are important for determining neuronal excitability. In addition to G proteins, GIRK channels are potentiated by membrane cholesterol, which is elevated in the brains of people with neurodegenerative diseases such as Alzheimer’s dementia and Parkinson’s disease. The structural mechanism of cholesterol modulation of GIRK channels is not well understood. In this study, we present cryo-electron microscopy (cryoEM) structures of GIRK2 in the presence and absence of the cholesterol analog cholesteryl hemisuccinate (CHS) and phosphatidylinositol 4,5-bisphosphate (PIP2). The structures reveal that CHS binds near PIP2 in lipid-facing hydrophobic pockets of the transmembrane domain. Our structural analysis suggests that CHS stabilizes PIP2 interaction with the channel and promotes engagement of the cytoplasmic domain onto the transmembrane region. Mutagenesis of one of the CHS binding pockets eliminates cholesterol-dependent potentiation of GIRK2. Elucidating the structural mechanisms underlying cholesterol modulation of GIRK2 channels could facilitate the development of therapeutics for treating neurological diseases.
Graphical Abstract
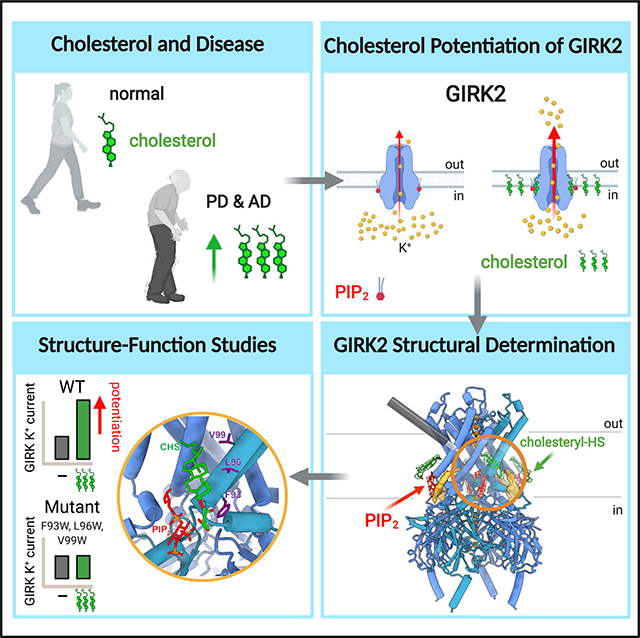
In brief
Ion channels are important in determining neuronal excitability. Elevated cholesterol levels found in some neurodegenerative diseases can affect the function of ion channels. Mathiharan et al. take a structural and functional approach to identifying physical sites for cholesterol, and they provide details on how cholesterol potentiates ion channel activity.
INTRODUCTION
G-protein-gated inwardly rectifying potassium (GIRK) channels provide a major source of inhibition in the brain and have been implicated in a variety of neurological disorders (Lüscher and Slesinger, 2010). Loss of GIRK channels leads to hyperexcitability and susceptibility to seizures, changes in alcohol consumption, and increases in sensitivity to psychostimulants (Signorini et al., 1997; Hill et al., 2003; Clarke et al., 2011; Munoz and Slesinger, 2014; Rifkin et al., 2018). Activation of G-protein-coupled receptors (GPCRs) that signal through Gαi/o G protein, such as GABAB, D2 dopamine, and m2 muscarinic receptors, leads to liberation of G protein Gβγ subunits that open GIRK channels through direct protein-protein interactions (Logothetis et al., 1987; Reuveny et al., 1994; Wickman et al., 1994).
Interestingly, some GIRK channels are found in lipid rafts enriched in cholesterol (Delling et al., 2002), and GIRK channels have been shown to be potentiated by increases in membrane cholesterol (Bukiya and Rosenhouse-Dantsker, 2017; Glaaser and Slesinger, 2017; Rosenhouse-Dantsker, 2019). Consistent with these findings, purified GIRK2 channels reconstituted into lipid membranes with phosphatidylinositol 4,5-bisphosphate (PIP2) are potentiated by cholesterol, demonstrating that cholesterol, similar to alcohol, can directly activate GIRK channels (Glaaser and Slesinger, 2017). Cholesterol levels in the brain are quite high, achieved mainly through local de novo synthesis, as transport of lipoproteins across the blood-brain barrier is not very efficient (Zhang and Liu, 2015; Jin et al., 2019). Elevated cholesterol in the brain has been recently implicated in people with neurodegenerative diseases, such as Alzheimer’s dementia (AD) and Parkinson’s disease (PD). Postmortem AD brains and patients with AD showed dysregulation of cholesterol metabolism, with presenilin 1 (PS1) mutations associated with elevations in cholesterol (Cho et al., 2019). Elevated levels of cholesterol and homocysteine in blood have been linked with the pathology of Parkinson’s disease (Paul et al., 2018). Hypercholesterolemia induces dopamine neuronal loss, where GIRK2 homotetramers are expressed, leading to reduced DA in the striatum (Paul et al., 2017b). The molecular mechanism underlying cholesterol regulation of these potassium channels is poorly understood. Mutagenesis studies and simulations have indirectly implicated multiple regions of inwardly rectifying potassium channels in cholesterol modulation, including the cytoplasmic domain (CTD), the PIP2 binding site at the inner leaflet of the membrane, and the transmembrane domain (TMD) (Epshtein et al., 2009; Rosenhouse-Dantsker et al., 2010, 2013; Bukiya et al., 2017). The precise physical location of the cholesterol sites in GIRK channels remains to be determined.
To better understand the structural basis for cholesterol modulation of GIRK channels, we employed cryo-electron microscopy (cryoEM) to visualize GIRK2 under different conditions. We present structures of the GIRK2 channel in the presence and absence of the cholesterol derivative cholesteryl hemisuccinate (CHS) and PIP2, revealing their effects on mechanistic aspects of GIRK2 gating. In addition, we provide evidence with site-directed mutagenesis and functional expression of GIRK2 channels that link the structural site for CHS with cholesterol potentiation of GIRK2 channels.
RESULTS
CHS potentiates GIRK2 channel activity
Previously, we showed that GIRK2 channels are potentiated by cholesterol in the absence of G proteins (Glaaser and Slesinger, 2017). To better understand the mechanism of cholesterol modulation, we sought to determine the site of interaction of a cholesterol analog, CHS, in GIRK2 channels. To verify the functional effects of CHS, we expressed Mus musculus GIRK2 in Pichia pastoris and extracted from membranes with n-dodecyl β-D-maltoside (DDM) detergent in the presence or absence of CHS (Figure S1A). Purified GIRK2 tetrameric channels were reconstituted into liposomes containing 1% brain PIP2 alone, which is required for channel function (Huang et al., 1998; Glaaser and Slesinger, 2017), and either 2.5% CHS or 5% cholesterol. We measured K+ permeation through GIRK2 channels using a flux assay (Figure 1A), as described previously (Whorton and MacKinnon, 2013; Glaaser and Slesinger, 2017). Upon addition of carbonyl cyanide m-chlorophenyl hydrazone (CCCP), the fluorescence emitted from 9-amino-6-chloro-2-methoxyacridine (ACMA) pre-loaded into proteoliposomes containing GIRK2 channels and 1% brain PIP2 (GIRK2PIP2) quenches to ~0.5 F/F0 (fluorescence intensity ratio) under basal conditions (Figures 1A and 1B) and quenches to ~0.2 F/F0 with proteoliposomes also containing CHS (GIRK2PIP2/CHS), indicating CHS-dependent potentiation of GIRK2 channel activity (Figures 1A and 1B). To ensure that the effect of CHS was via specific interactions with GIRK2, and not due to indirect effects via changes in membrane permeability, we reconstituted liposomes containing 1% PIP2 with and without 2.5% CHS (empty liposomes) and measured flux as with liposomes containing GIRK2. Representative traces of empty liposomes with and without 2.5% CHS (Figure 1C) show virtually no quenching, with the normalized relative K+ flux of CHS containing empty liposomes (~7.4% ± 1.3%; n = 4) (Figure 1F). The GIRK2 channel-specific inhibitor 2-hydroxyethyl methanethiosulfonate (MTS-HE) (Glaaser and Slesinger, 2017) reduced the extent of quenching, indicating inhibition of the K+ conductance. We used the inhibition with MTS-HE to quantify changes in quenching under different conditions and converted this to a percentage of relative K+ flux. Under basal conditions with proteoliposomes containing GIRK2PIP2, the relative K+ flux was 44.0% ± 2.3% (n = 18) (Figures 1D–1F). Both CHS and cholesterol significantly increased the relative K+ flux to 65.2% ± 0.4% (n = 10) and 69.4% ± 0.3% (n = 17), respectively, compared to basal conditions (Figures 1D–1F). Taken together, these results demonstrate that CHS, similar to cholesterol, potentiates basal PIP2-dependent GIRK2 channel activity (Glaaser and Slesinger, 2017).
Figure 1. Cholesterol and CHS potentiate GIRK2 channels reconstituted in liposomes with PIP2.
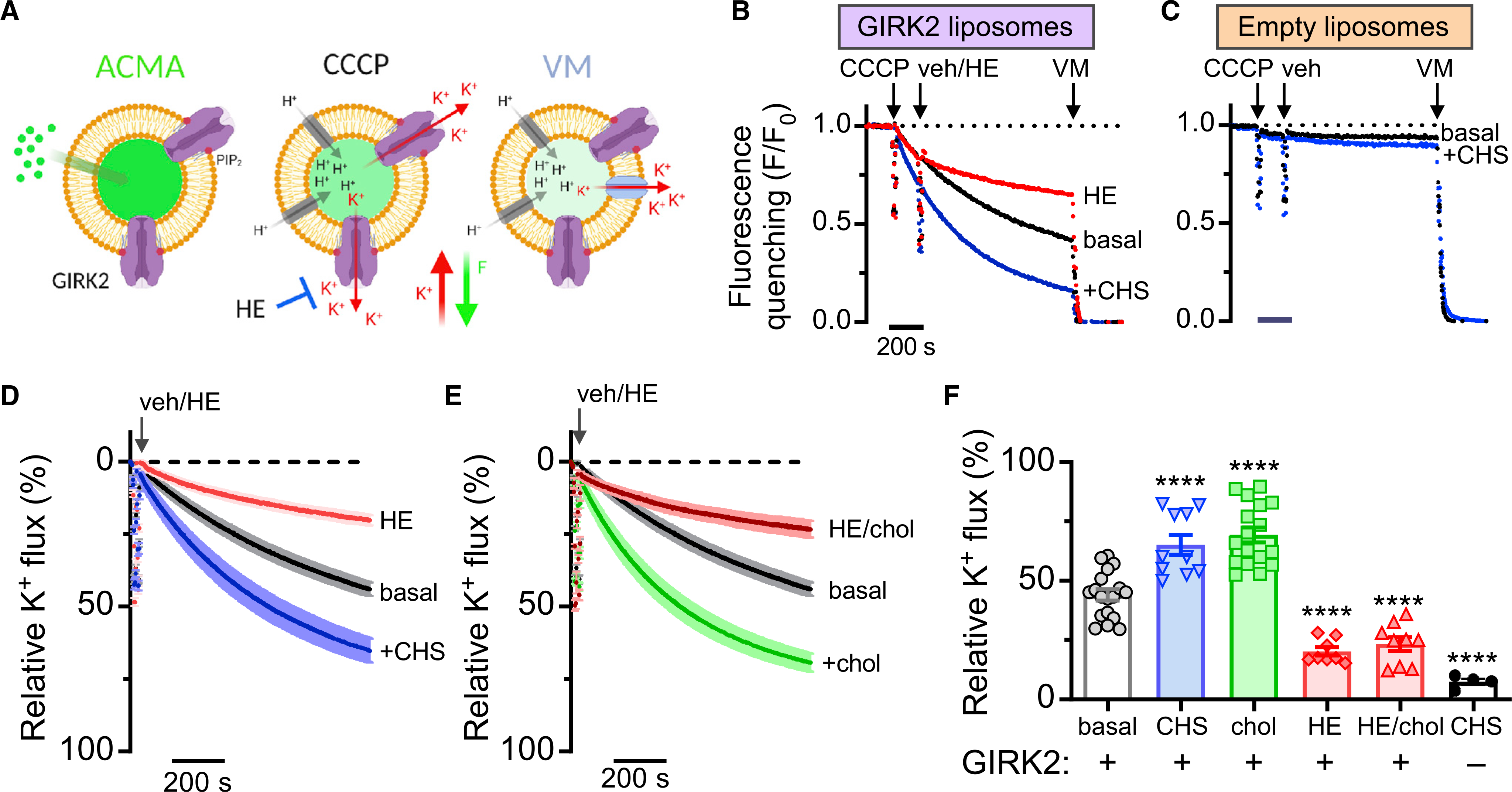
(A) Schematic shows the K+ flux assay. (Left) Proteoliposomes pre-loaded with ACMA at baseline. (Middle) Fluorescence quenches with H+ influx upon addition of CCCP when GIRK channels are open. (Right) Dissipation of the K+ gradient upon addition of valinomycin (VM) indicates vesicle capacity.
(B) Example of normalized fluorescent measurements of GIRK2-containing liposomes with 1% brain PIP2. Fluorescence quenching is measured upon addition of CCCP, then addition of the GIRK2 channel inhibitor MTS-HE (HE, 100 μM) or vehicle (veh), and lastly terminated by the addition of VM. Note increase in quenching in the presence of CHS (blue trace), as compared to basal (black trace) and HE inhibited (red trace).
(C) Example of normalized fluorescence measurements of empty liposomes with 1% PIP2. Fluorescence quenching is measured as in (B). Quenching is similarly small both in the presence of 2.5% CHS (blue traces) and with no CHS (black traces).
(D and E) Plot shows mean % relative K+ flux (±SEM) for basal (black trace, n = 18) and +CHS (blue trace, n = 10) (D). Acute application of HE shows inhibition of basal (red trace, n = 8). (E) Plot shows mean relative K+ flux (%) (±SEM) for basal (black, same as C). Acute application of HE shows inhibition of basal + 5% cholesterol (red trace, n = 9).
(F) Bar graph shows the mean (±SEM; n indicated on graph) relative K+ flux for the indicated conditions. Both CHS and cholesterol significantly increased the relative K+ flux (****p < 0.0001 versus basal using one-way ANOVA with Dunnett’s multiple comparisons post hoc test; F(4, 57) = 48.02). HE (red bar) significantly inhibited GIRK2 channels. Empty liposomes with CHS contain little K+ flux (black circles/white bar) There was no statistical difference between HE and chol/HE conditions.
See also Figure S1.
Structure of GIRK2 in the presence of CHS and PIP2
To gain insights into CHS binding in GIRK2, we proceeded with optimizing the sample for cryoEM studies. The sample quality was assessed by size-exclusion chromatography (Figure S1A) and further evaluated by negative stain electron microscopy (Figure S1B) to identify conditions with predominantly well-formed GIRK2 tetramers (Peisley and Skiniotis, 2015). To ensure the presence of CHS and PIP2 (GIRK2PIP2/CHS), the sample was purified in the presence of CHS and incubated with 2 mM diC8-PIP2 prior to cryoEM grid preparation.
We subsequently employed cryoEM and determined the structure of GIRK2PIP2/CHS at a global indicated resolution of 3.5 Å (Figure S2). The structure revealed an overall architecture similar to those solved by X-ray crystallography (Whorton and MacKinnon, 2011, 2013), with two distinct domains, the TMD and CTD, and the pore along the four-fold axis with a well-ordered inner-helix bundle crossing and G-loop gates (Figures 2A and 2B). PIP2 is bound at its canonical site near inter-domain linkers, as also observed in other inwardly rectifying potassium channels (Hansen et al., 2011; Whorton and MacKinnon, 2011, 2013) (Figure 2B). Interestingly, we observed a planar density in “site A” on the opposite side of the PIP2-channel interface that is consistent with CHS. Initial modeling of CHS in the EM density was performed with the GemSpot pipeline (Robertson et al., 2020), followed by manual refinement in Coot (Emsley and Cowtan, 2004). This approach yielded largely identical poses for CHS in good agreement to the EM map and chemically plausible interactions with GIRK2 transmembrane helices that had subtle differences in the orientation of the isooctyl tail (Figures 2B and 2C; Figure S3A). The head group of CHS is in position to form a salt bridge with R92, while its sterane rings and isooctyl tail are stacked against the TMD, surrounded by hydrophobic residues F93, L95, L96, and V99 of the M1 helix from one subunit, and I175, V178, and L179 of the M2 helix from the adjacent subunit (Figures 2B and 2C; Figure S3B). Another CHS molecule could be modeled in “site B” near the N terminus surrounded by hydrophobic residues that include V72, L79, I82, L86, L89, I97, V101, and F186 (Figure 2C; Figure S3C). We cannot rule out other putative CHS sites, as additional densities are observed near the TMD-CTD interface, but they are poorly resolved (Figures S3D–S3F). We note, however, that the positions of CHS in site A and site B in the cryoEM GIRK2 structures are different from those regions identified with computational molecular docking (cf. Bukiya et al., 2017).
Figure 2. Structure of GIRK2 with PIP2 and cholesteryl hemisuccinate (CHS).
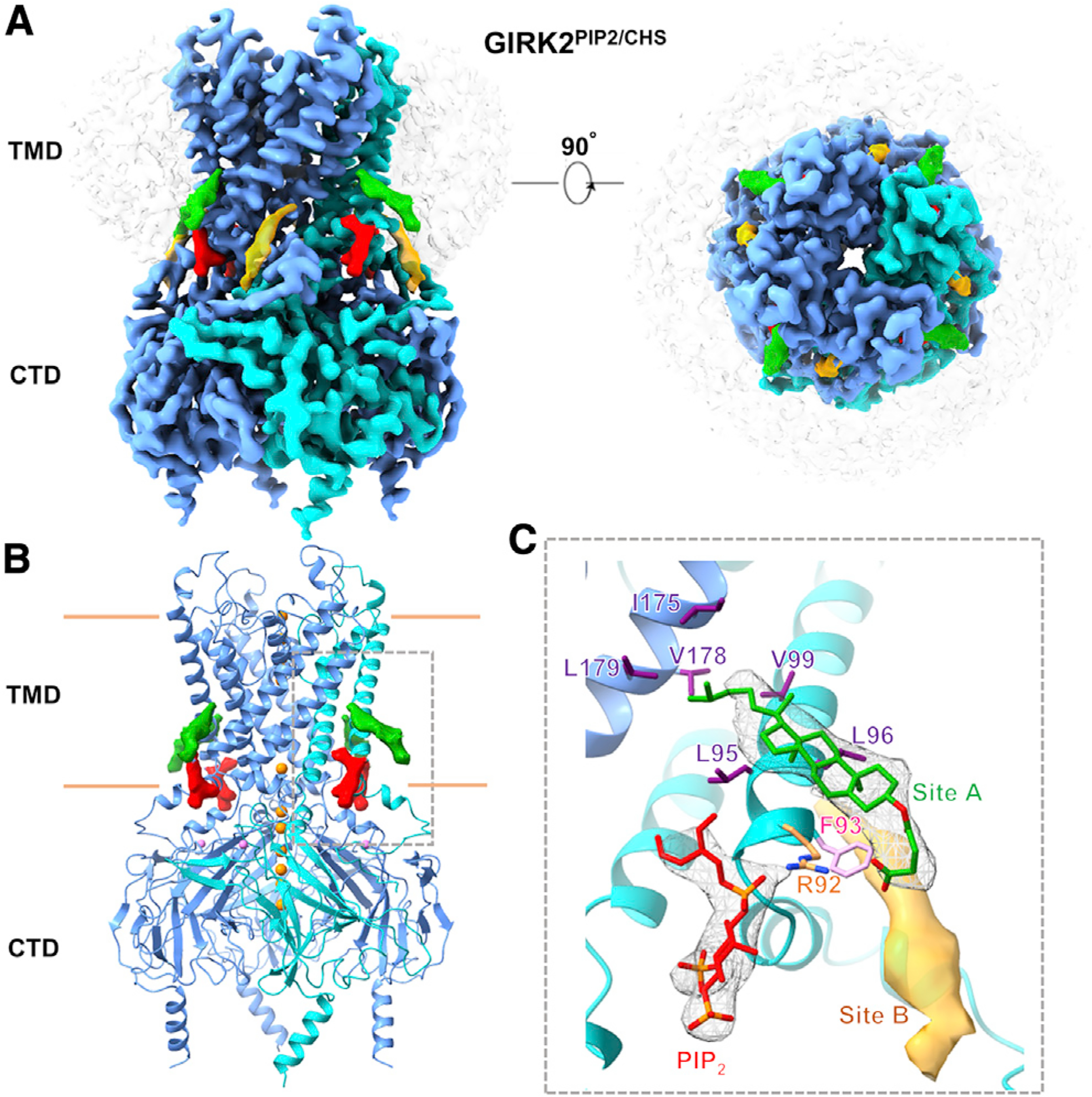
(A) GIRK2PIP2/CHS cryoEM map; one CHS (green, at “site A”) molecule per subunit is modeled into planar densities on the opposite side of the PIP2 (red)-channel interface; another planar density at “site B” near the slide helix is shown in orange. GIRK2 subunits are in blue with one highlighted in cyan; DDM micelle is shown in transparency. The top right panel shows top-bottom view from extracellular side.
(B) Model of GIRK2PIP2/CHS with the transmembrane (TMD) and cytoplasmic (CTD) domains. K+ ions (orange, sphere) are shown along the pore, and one Na+ ion (pink, sphere) is modeled per subunit as seen in X-ray crystal structures. The densities of CHS in site A and the anionic head group of PIP2 are shown at the TMD-CTD interface. The CHS pocket is outlined by a dashed box.
(C) CHS (green, density shown as mesh, at site A) modeled into a density near PIP2 (red, density shown as mesh) is surrounded by positively charged R92 (orange) and hydrophobic residues including F93 (pink), L95, L96, and V99 (purple) from one protomer (cyan), and I175, V178, and L179 (purple) from nearby subunit (blue). Another planar density (orange, at site B) adjacent to this is represented as surface. Densities of the anionic head group of PIP2, CHS, and site B are contoured at a threshold level of 0.0161, 0.0073, and 0.0073, respectively in ChimeraX, while the GIRK2 subunits are at a threshold of 0.0161 in ChimeraX.
See also Figures S1–S3 and S8 and Table S1.
Structures of GIRK2PIP2 in the absence of CHS
To assess whether CHS alters the interaction of PIP2, we next determined the structure of GIRK2PIP2 in the absence of CHS during the purification. Interestingly, cryoEM analysis resolved several different conformers of GIRK2PIP2 (Figure S4). The predominant conformer, referred to as GIRK2PIP2*, accounted for ~39% of well-defined particles and its structure was obtained at a global resolution of 3.2 Å (Figures S4C–S4F). This structure is comparable to GIRK2PIP2/CHS (Figure 3), with four PIP2 molecules bound at equivalent sites, but importantly lacked densities for CHS in site A and site B (Figures S3D–S3F). A second conformer, GIRK2PIP2**, accounting for ~27% of projections, was determined at 4.8 Å global resolution (Figures S4C and S4G). Comparison of this conformation with GIRK2PIP2* revealed that the CTD is rotated from 8° to 12° around the four-fold axis and is partially disengaged from the TMD by 0–1 Å (as evaluated by the distance between Cα atoms of T80 and L229), with a stretched tether helix (residues 197–203, Figures S5A–S5C).
Figure 3. Comparison of PIP2 binding pocket and pore.
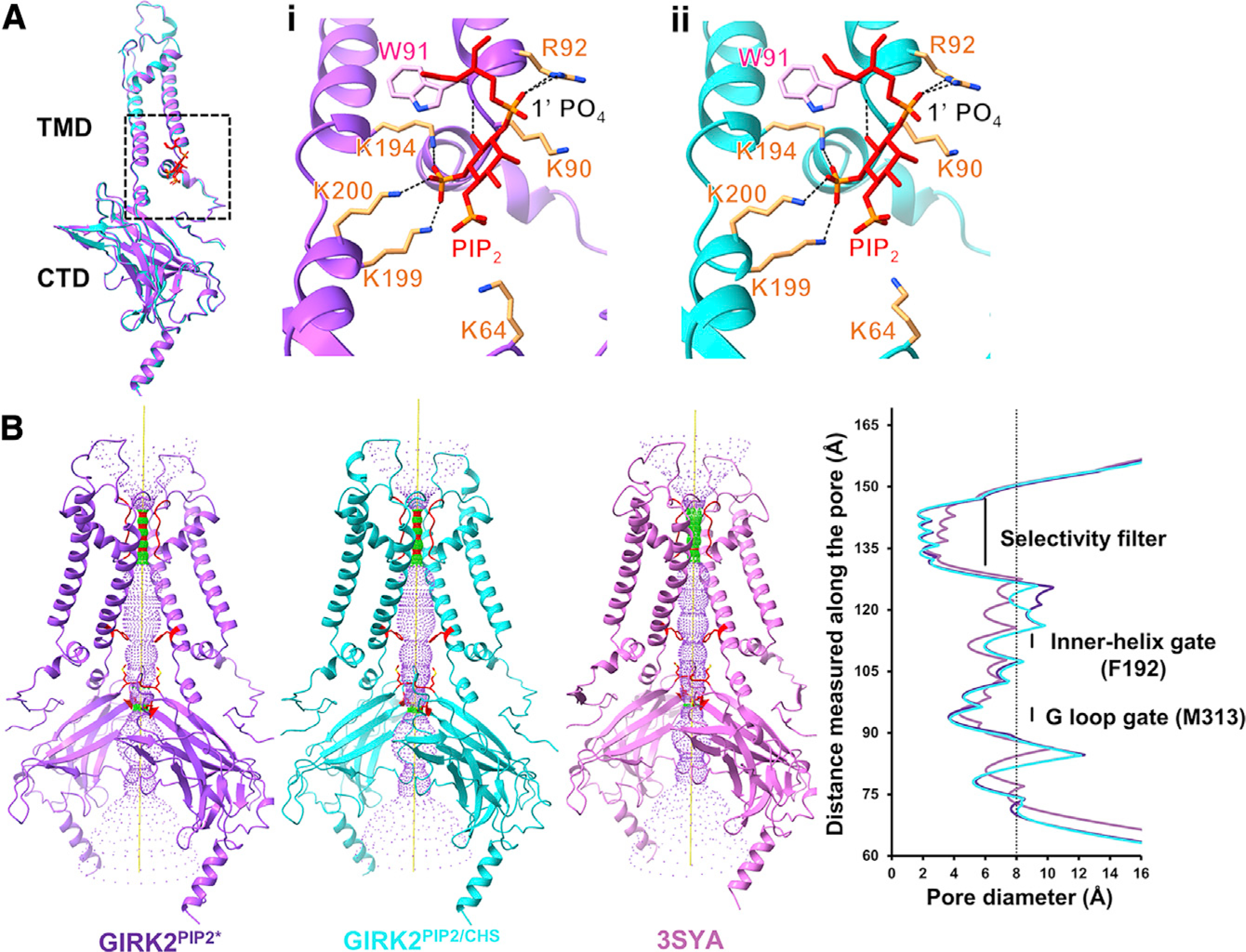
(A) Comparison of a subunit from GIRK2PIP2* (purple) and GIRK2PIP2/CHS (cyan) structures; the RMSD between them is around 0.4 Å. Insets (i) and (ii) represent PIP2 (red) coordination in GIRK2PIP2* (purple) and GIRK2PIP2/CHS (cyan), respectively; PIP2 anionic head group is surrounded by positively charged residues (orange), and the 1′ PO4 of PIP2 is coordinated by the side chain of R92.
(B) Pore comparison using HOLE software for cryoEM structure GIRK2PIP2* (purple), GIRK2PIP2/CHS (cyan), and X-ray crystal structure of GIRK2 with only PIP2 (PDB: 3SYA, pink); only two subunits are shown for clarity.
See also Figure S7.
The disengagement of the CTD is more pronounced in two additional conformers, GIRK2PIP2*** and GIRK2PIP2****, as the distance between Cα atoms of T80 and L229 is increased by 1–8 Å, stretching further the tether helix (Figures S5A and S5B). These conformers appear very similar and were obtained at low overall resolution (~7.7 Å), primarily due to the relative flexibility between TMDs and CTDs. Nevertheless, rigid body docking of TMDs and CTDs was sufficient in revealing the overall configurations. Interestingly, although the CTD detachment is different with respect to each subunit (i.e., distance), its C4 symmetry appears nearly preserved in all of the conformers. We next examined whether the interaction of PIP2 in its binding pocket was altered by the presence of CHS (Figure 3A). In the high-resolution GIRK2PIP2* map, the 1′ PO4 of the PIP2 anionic head group is coordinated by the side chain of positively charged R92, while the 5′ PO4 is coordinated with K194, K199, K200, and the O6 atom with the backbone amide group of W91 (Figure 3Ai). Positively charged residues K64 and K90 are also in proximal distance to the anionic head group of PIP2. This configuration of PIP2 looks similar to that in GIRK2PIP2/CHS (Figure 3Aii), but it has lower occupancy. Taken together, these observations suggest that the inter-domain interface of GIRK2PIP2 is relatively less stable than that of GIRK2PIP2/CHS (Figures S5A and S5B).
In both the GIRK2PIP2* and GIRK2PIP2/CHS maps, we observe densities that are compatible with Na+ and K+ ions. A Na+ ion is coordinated by D228, as shown previously in X-ray crystal structures (Whorton and MacKinnon, 2011). Similarly, densities for K+ ions are found near the selectivity filter, proximal to Y266, M319, G318, and E236 as previously determined (Whorton and MacKinnon, 2011, 2013), but also observed at positions near G158, F192, and M313. We also note the presence of additional, less well-resolved densities along the pore near N184, V188, E236, and M319. The pore diameter at the inner helix (F192) and G-loop (M313) gates is ~6.5 Å and 4 Å, respectively, which is too small for hydrated K+ (~8 Å) to permeate (Figure 3B). However, previous studies utilizing cross-linked KirBac3.1 channels (Black et al., 2020) and MD simulations of GIRK channels (Bernsteiner et al., 2019) suggest this extent of opening at the inner-helix gate may be sufficient to pass partially hydrated or transiently non-hydrated K+.
GIRK2 apo structure (no PIP2)
Lastly, we examined the structure of GIRK2 apo, which lacks both PIP2 and CHS. CryoEM maps of GIRK2 apo determined at an overall 4.8 Å resolution revealed that the CTD is detached from the TMD (Figures 4 and 5). This configuration of the CTD is quite different from the GIRK2 apo structure determined by X-ray crystallography, which showed the CTD engaged with the TMD (Whorton and MacKinnon, 2011) (Figure S5D). Inter-domain linkers consisting of tether helix and N-terminal residues 67–78 appear mostly disordered, suggesting that this interface is unstable in the absence of PIP2 and CHS (Figure 5; Figure S6; and Videos S1). The CTD has moved toward the TMD by about 2–8 Å to be engaged in the GIRK2PIP2 structure (Figure 4). Interestingly, the detached CTD appears tilted below the membrane, reflecting a “wobble” in this structure and an overall asymmetry in the absence of PIP2 and CHS. Our classification showed additional 3D classes with different orientations of detached CTD, and we used 3D variability analysis in cryoSPARC (Punjani and Fleet, 2021) to further visualize the CTD wobbling (Video S1).
Figure 4. CryoEM structures of GIRK2 with and without PIP2.
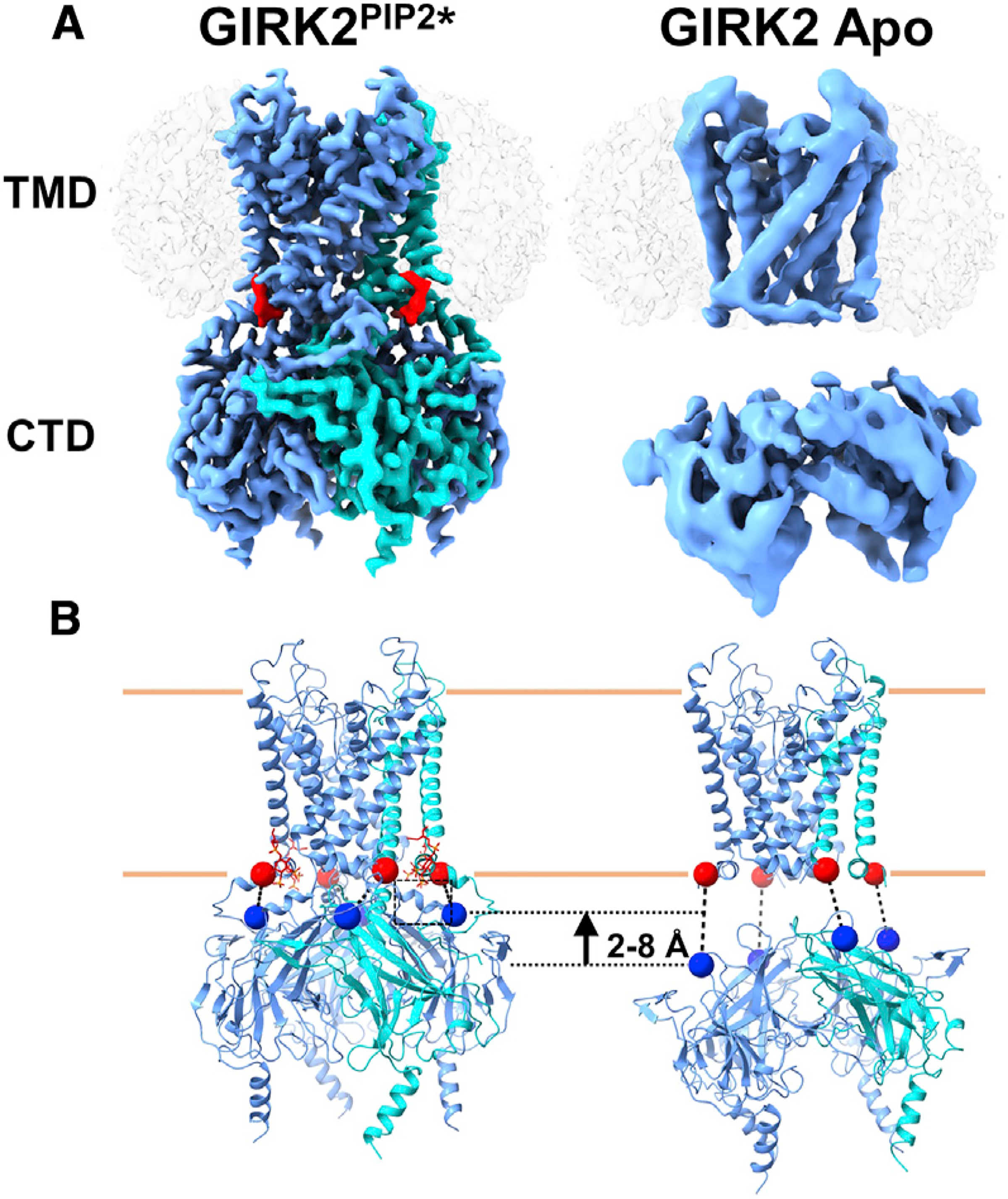
(A) CryoEM maps of the most prevalent GIRK2PIP2* and GIRK2 apo conformations; the CTD is disengaged from membrane in apo compared to the GIRK2PIP2* structure with the tether helix (residues 197–203) and N-terminal residues 67–78 disordered. PIP2 (red) is at the TMD-CTD interface; one subunit is highlighted in cyan and micelle is in transparency.
(B) Evaluation of CTD detachment in GIRK2PIP2* and GIRK2 apo based on the distance between Cα atoms of residues T80 (denoted by red sphere) and L229 (denoted by blue sphere). The ordered tether helix from one subunit of GIRK2PIP2* is shown with a dashed square.
See also Figures S1, S4, and S6 and Tables S1 and S2.
Figure 5. Different gating states for GIRK2 revealed by cryoEM.
Structures with different lipid modulators suggest varying conformations of CTD with respect to TMD. The structures reveal cholesterol (green) binding sites adjacent to PIP2 (red) pocket. These molecules stabilize the TMD-CTD interface, thereby aiding in anchoring CTD onto the membrane for further activation. The percentage fraction of total particles used for final round of 3D classification contributing to GIRK2 apo, GIRK2PIP2*, GIRK2PIP2**, GIRK2PIP2***, and GIRK2PIP2/CHS classes that were further used for refinement is 22%, 39%, 27%, 10%, and 62%, respectively.
See also Figures S2, S4, S5, and S6.
Functional effects of mutations in the CHS pockets on cholesterol potentiation of GIRK2
We used the GIRK2PIP2/CHS cryoEM structure with CHS modeled in site A and site B for mutagenesis, and then studied the effect on cholesterol potentiation of native GIRK2 channels expressed in HEK293 cells using electrophysiology. Whole-cell patch-clamp currents were recorded from HEK293 cells transiently transfected with GIRK2 cDNA that were exposed to either water-soluble cholesterol with methyl-β-cyclodextrin (MβCD) (+cholesterol) or MβCD alone (control) (Figure 6A). The cholesterol enrichment resulted in a significant (~2-fold) increase in the Ba2+-sensitive basal current for GIRK2 wild-type (WT) channels, with no change in inward rectification (Figures 6B and 6C) (cf. Bukiya et al., 2017).
Figure 6. Effect of mutations in CHS pockets on cholesterol potentiation of GIRK2 channels.
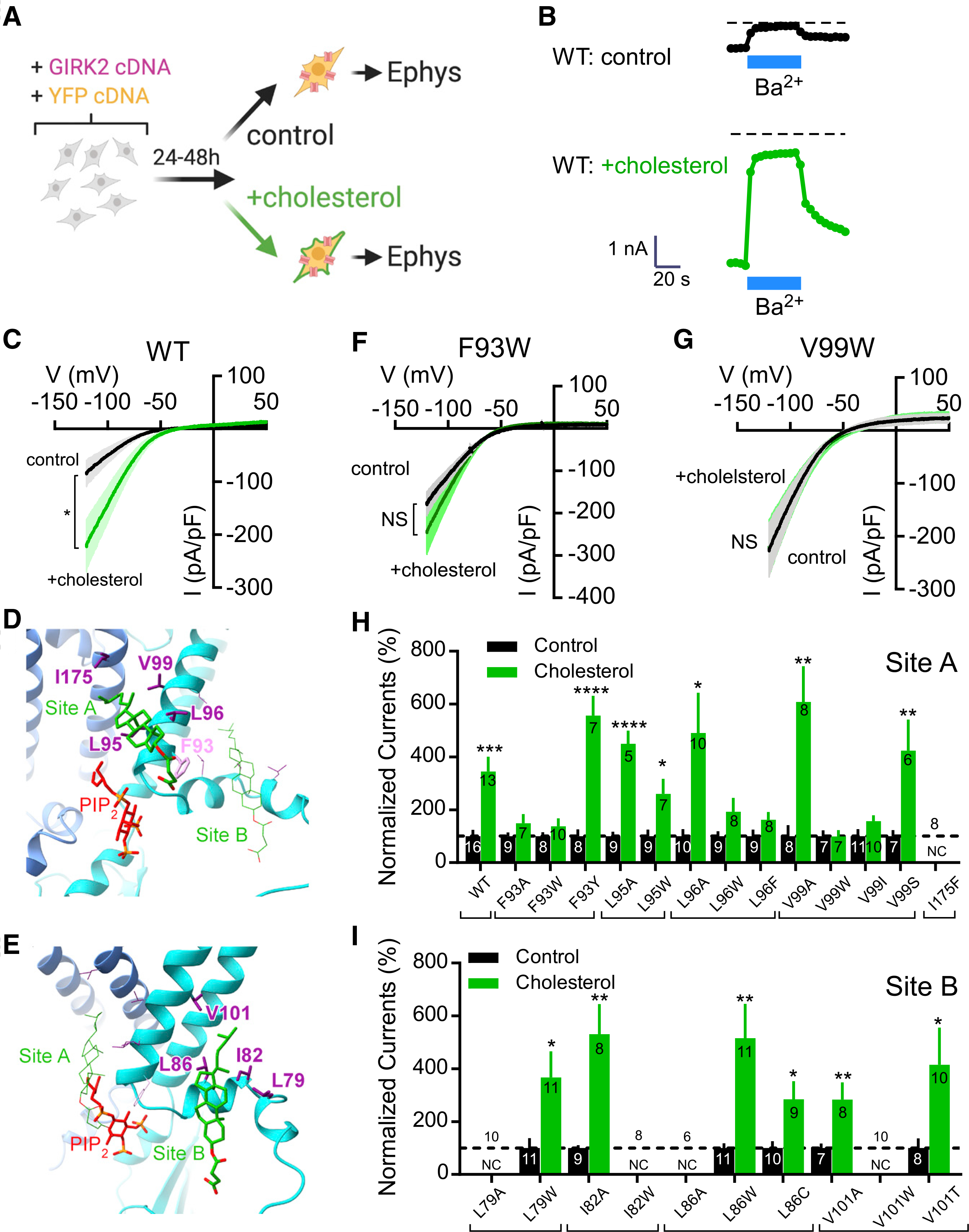
(A) Cartoon shows protocol used for cholesterol potentiation of GIRK2 channels expressed in HEK293 cells.
(B) The inward current (−120 mV) is plotted as a function of time, showing a large Ba2+-sensitive current in cholesterol-enriched HEK293 cells, as compared to control. Dashed line indicates zero current.
(C) Current-voltage plots show the mean Ba2+-sensitive current densities for control (black) and cholesterol-enriched (green) conditions for GIRK2 WT (n = 10 control; n = 10 +chol). A Student’s unpaired two-tailed t test was used for evaluation at −120 mV (*p < 0.05). SEM is shown in lighter color.
(D and E) Five residues surrounding the CHS in site A and four residues surrounding the CHS in site B were mutated and tested for cholesterol potentiation.
(F and G) Current-voltage plots show the mean Ba2+-sensitive current densities for control (black) and cholesterol-enriched (green) conditions for mutant F93W (F) (n = 8 control; n = 10 +chol) and V99W (g) (n = 7 control; n = 7 +chol) channels. NS, not significant.
(H and I) Bar graphs show the normalized % currents for GIRK2 WT compared to GIRK2 with a mutation in site A (H) or site B (I). For each, the Ba2+-sensitive basal current density for cholesterol enrichment (green bar) is normalized to each control condition (black bar). A Student’s unpaired two-tailed t test was used for evaluation at −120 mV (*p < 0.05, **p < 0.01, ****p < 0.001, control versus cholesterol treatment). N is indicated on the graph. Dashed line indicates 100% (i.e., no potentiation). NC, no current. Error bars represent SEM.
We next conducted an Ala/Trp scan of five residues in site A and four residues in site B (Figures 6D and 6E). In the site A CHS pocket, we found that increasing the side-chain volume at the L96 and V99 position (i.e., L96W, L96F, V99W, and V99I) eliminated cholesterol potentiation, whereas decreasing side chain volume (L96A, V99A) had little effect (Figures 6F–6H). Both L95A and L95W, alternatively, had no effect on cholesterol potentiation. Interestingly, both F93A and F93W disrupted cholesterol potentiation (Figure 6h). To determine whether the phenyl group was important, we examined the effect of F93Y mutation and observed normal cholesterol potentiation (Figure 6h). The effect of Ala/Trp mutations at L79, I82, L86, and V101 in site B did not eliminate cholesterol potentiation, or this effect created a non-functional channel (NC, Figure 6i). Taken together, these data provide evidence that the cholesterol interaction with site A is functionally relevant for potentiation of natively expressed GIRK2 channels.
DISCUSSION
Cholesterol binding sites on membrane proteins
Cholesterol binding sites on membrane proteins were initially characterized using a variety of techniques, including electron spin resonance, photoaffinity labeling, molecular modeling, and site-directed mutagenesis, which led to the identification of amino acid recognition motifs (Middlemas and Raftery, 1987; Jones and McNamee, 1988; Dreger et al., 1997; Hanson et al., 2008; Rosenhouse-Dantsker, 2019; Duncan et al., 2020a). Structures of membrane proteins (e.g., GPCRs and ion channels) determined in the presence of cholesterol or CHS have revealed interactions with “greasy hollows,” which refer to pockets in the transmembrane portion lined with hydrophobic residues (Lee, 2019; Duncan et al., 2020b). Previous work with GIRK2 has also supported the idea of a direct effect of cholesterol on the channel protein, as opposed to possible indirect effects on the bilayer (Bukiya et al., 2017; Glaaser and Slesinger, 2017). Furthermore, we now provide a structural view of a CHS/cholesterol pocket (site A) in GIRK2. The sterane rings and isooctyl tail of CHS are stacked against the TMD, between two adjacent subunits, and surrounded by hydrophobic residues. Generally, bulky substitutions at F93, L96, and V99 in site A eliminated cholesterol potentiation. One position (F93) was sensitive to both Ala/Trp mutations but not a Tyr mutation. This finding suggests that cholesterol binding may be favored by the phenyl group in this position. Some features of cholesterol binding in this pocket may be slightly different from CHS, since the head group in CHS is different from that of cholesterol (i.e., the CHS hemisuccinate is larger and more polar). For example, a salt bridge appears to form between the polar head group of CHS and R92, which also interacts with PIP2 (Tang et al., 2015). It is possible that cholesterol may interact with R92 as well. Interestingly, a R92Y mutation in GIRK2 eliminated cholesterol potentiation and greatly increased the Ba2+-sensitive basal current (data not shown). The putative interaction between R92, PIP2, and cholesterol requires further investigation to better understand the role of R92 in GIRK2 function.
Mutation of amino acids in the site B pocket, alternatively, appeared to have little effect on cholesterol potentiation. One possible explanation for this is that CHS is partially exposed to the intracellular side, and thus potentially less stable in that position and less sensitive to mutagenesis. Alternatively, site B may not be a cholesterol potentiation site, and instead another type of lipid may occupy this space. The higher percentage of non-functional channels could also indicate that this region of the channel is important for tetramer assembly.
Computational docking studies with cholesterol and a homology model of GIRK2 revealed two putative cholesterol-binding regions located at the center of the TMDs, near the TMD-CTD interface (Bukiya et al., 2017). These two regions are near to, but different from, the site A and site B pockets identified in the cryoEM GIRK2 structure (see Figure S8). Nonetheless, some of the amino acids previously identified using docking (e.g., V99 and V101) do overlap with site A and site B (Bukiya et al., 2017). It is possible that cholesterol may bind to several different regions of the channel in other lipid membrane environments, but it is more stable in site A under our experimental conditions.
Mechanism of PIP2 and cholesterol gating of GIRK2 channels
One of the main findings of our cryoEM studies is that the CTD is detached from the TMD in both the GIRK2 apo and a significant population of GIRK2PIP2 particles. While the CTD appears detached, it is actually connected via a stretched tethered linker that is unresolved. In contrast, the CTD is engaged with the TMD in the structure of GIRK2 apo solved by X-ray crystallography (Whorton and MacKinnon, 2011). One possibility for this difference could be due to lattice packing in X-ray crystal structures. Indeed, a recent cryoEM study of GIRK2 from MacKinnon and colleagues also demonstrated that the CTD is detached (i.e., extended) in the GIRK2 apo structure determined with cryoEM (Niu et al., 2020). A detached CTD in the absence of PIP2 and other auxiliary proteins (e.g., G proteins, SUR) may be a common feature of inward rectifiers (Whorton and MacKinnon, 2013; Martin et al., 2019), since the CTD is also detached in the Kir2.2 apo X-ray crystal structure (Hansen et al., 2011).
Based on the observation of several different conformations of GIRK2 in the absence and presence of PIP2, we suggest a plausible model for gating transitions from closed to open in GIRK2 channels. First, without PIP2 the CTD is uncoupled (i.e., detached) from the TMD and the channel cannot open. This is consistent with the functional studies showing that PIP2 is necessary for channel activation (Huang et al., 1998; Sui et al., 1998; Glaaser and Slesinger, 2017). Second, the inter-domain region and a wobbling CTD are stabilized by the binding of four PIP2 molecules at the TMD-CTD interface (see Video S1), thus repositioning the stretched tether helix into a helical conformation, rotating the CTD by ~31°–38° clockwise (when viewed top-down), and reducing the distance between Cα atoms of T80 and L229 by 2–8 Å (Figure 4). Similarly, the CTD rotates ~35° from GIRK2 apo to GIRK2-PIP2 cryoEM structures (Niu et al., 2020). In contrast, the CTD rotates in the opposite direction by ~4° with Gβγ in the X-ray structure of GIRK2-Gβγ-PIP2 (Whorton and MacKinnon, 2013). Taken together, these studies suggest that the PIP2 binding induces a major conformational change in GIRK2, involving a “twisting engagement” of the CTD with the TMD, and then a “locking rotation” of the CTD with the TMD after engagement that is required for the two gates to open. This observation potentially explains why GIRK channels are not gated open by Gβγ, ethanol, or cholesterol in the absence of PIP2
The cryoEM structures also reveal subtle differences in the coordination of PIP2 between GIRK2PIP2/CHS and GIRK2PIP2*, and the X-ray crystal structure of GIRK2-PIP2 (PDB: 3SYA). The 1′ PO4 of PIP2 in the cryoEM structures interacts with the side chain of R92, whereas in the X-ray crystal structure PIP2 is coordinated by the amide backbone of R92 and the side chain is not resolved (Figures 3A; Figure S7C). In addition, the 4′ PO4 interaction with K64 in the 3SYA structure is not evident in the cryoEM structures. The phosphate atom of 1′ PO4 of PIP2 is moved from the membrane toward the cytoplasmic side by 3 Å along with an ~3° rotation of the CTD around the 4-fold axis, resulting in a slightly wider pore opening at the inner-helix gate in the GIRK2PIP2* cryoEM structure compared to the crystal structure of GIRK2 (PDB: 3SYA; Figure S7; Figure 3B). These differences could indicate a possible unique transition state, but additional studies are needed to rule out an effect of experimental conditions, such as the concentration of PIP2.
What is the role of cholesterol/CHS in GIRK2 channel activation? In the presence of PIP2 and CHS, our analysis revealed predominantly a conformation of GIRK2PIP2/CHS that is remarkably similar to the GIRK2PIP2* structure. In the absence of CHS, there is a decrease in the fraction of particles assuming the fully engaged conformation relative to the disengaged states, suggesting that CHS may stabilize the four PIP2-bound GIRK2 with an engaged CTD. By comparison, the presence of cholesterol increases the thermal stability of GPCRs and often lowers the energy barrier for agonist-induced conformational changes (Gimpl, 2016). The binding of CHS in peripheral hydrophobic sites with no highly specific interactions is in agreement with the lack of large conformational changes on GIRK2 as a result of this engagement (i.e., the similar structure of GIRK2PIP2* and GIRK2PIP2/CHS), implying the importance of CHS/cholesterol for primarily enhancing PIP2 binding. The increase in PIP2 affinity, as promoted by cholesterol (Glaaser and Slesinger, 2017), might increase the probability of the channel entering the open state. Similarly, Niu et al. (2020) found that increasing the PIP2 concentration shifts the equilibrium of GIRK2 between a mix of engaged and disengaged states, toward the engaged form. However, one noticeable difference is that GIRK2 with PIP2 in the absence of Gβγ has a wider pore opening at the inner-helix gate and, importantly, is gated open by PIP2 and potentiated further by cholesterol even in the absence of Gβγ.
Physiological significance of cholesterol gating of GIRK2 channels
The potential connection between elevated plasma cholesterol and increased risk for cardiac disease is well known (Rosenson et al., 2018). Less well understood is the role of cholesterol in the brain. Nearly all cholesterol in the CNS is derived from de novo synthesis, and it is therefore not influenced by dietary changes in cholesterol (Dietschy and Turley, 2001). Nevertheless, changes in brain cholesterol have been implicated in several neurodegenerative diseases, such as Alzheimer’s disease, Huntington’s disease, Parkinson’s disease, and Niemann-Pick disease (Vance, 2012). Hypercholesterolemia causes impairment of dopamine signaling and psychomotor dysfunction in mice (Engel et al., 2016; Paul et al., 2017a, 2018). Increases in cholesterol levels have been shown to elevate β-amyloid precursor protein levels in cholesterol-enriched lipid rafts (Cho et al., 2019) and are associated with increased risk for AD (Zarrouk et al., 2018). Accordingly, there is a growing need to understand how cholesterol affects brain function.
GIRK2 is relatively unique among members of the Kir family because it is potentiated in response to cholesterol rather than inhibited, as occurs with most other members of the Kir family (Rosenhouse-Dantsker, 2019). Acute elevation of brain cholesterol in hippocampal neurons increases both basal and GPCR-dependent activity of GIRK channels (Bukiya et al., 2017), suggesting that changes in brain cholesterol in vivo could affect both the basal and neurotransmitter-activated GIRK currents. Our cryoEM study of GIRK2 provides important information on the structural mechanism of cholesterol potentiation and reveals a cholesterol pocket that may be suitable for targeting with drugs. The structure of the alcohol pocket in GIRK2 was recently used in a virtual screen for GIRK modulators and identified a GIRK1-activating compound, GiGA1 (Zhao et al., 2020). Now, a similar screening can be conducted using the structure of the CHS site A in GIRK2, which may lead to the discovery of a new family of GIRK modulators.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| GFP Mouse Anti-tag, clone: GF28R | Invitrogen | Cat#: MA515256 |
|
| ||
| Biological samples | ||
|
| ||
| Yeast strain Pichia Pastoris SMD 1163H | Invitrogen | SMD1163H |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 1-Propanol | Sigma-Aldrich | Catalog #: 402893 |
| Barium Chloride | Sigma-Aldrich | Catalog #: 342920 |
| Cholesterol-methyl-β-cyclodextrin | Sigma-Aldrich | Catalog #: C4951 |
| Salts for electrophysiology | Sigma-Aldrich | |
| ACMA (9-Amino-6-Chloro-2-Methoxyacridine) | Life Technologies | Catalog #: A1324 |
| CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) | Sigma-Aldrich | Catalog #: C2759 |
| Valinomycin | Life Technologies | Catalog #:V1644 |
| MTS-HE (2-hydroxyethyl methanethiosulfonate) | Toronto Research Chemicals | Catalog #: H942250 |
| Cholesterol, ovine wool | Avanti Polar Lipids | Catalog #: 700000 |
| CHS (Cholesteryl Hemisuccinate Tris Salt) | Anatrace | Catalog #: CH210 |
| Lipids-PE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine) | Avanti Polar Lipids | Catalog #: 850757 |
| Lipids-PG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) | Avanti Polar Lipids | Catalog #: 840457 |
| PIP2 (PI(4,5)P2-diC8) | Echelon | Cat #: P-4508 |
| DDM (n-Dodecyl-β-D-Maltopyranoside) | Anatrace | Catalog #: D310 |
| GIRK2 recombinant protein | This study | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| QuikChange II XL | Agilent Technology | N/A |
|
| ||
| Deposited data | ||
|
| ||
| GIRK2PIP2/CHS map | EMDB | EMD-22154 |
| GIRK2PIP2* map | EMDB | EMD-22150 |
| GIRK2PIP2** map | EMDB | EMD-22151 |
| GIRK2PIP2*** map | EMDB | EMD-22152 |
| GIRK2PIP2****map | EMDB | EMD-22153 |
| GIRK2Apo map | EMDB | EMD-22155 |
| GIRK2PIP2/CHS model | PDB | 6XEV |
| GIRK2PIP2* model | PDB | 6XEU |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T cells | ATCC | Catalog #: CRL-3216 |
|
| ||
| Oligonucleotides | ||
|
| ||
| DNA primers for mutagenesis | N/A | |
| GIRK2 L79A F GAG ACG TAC CGA TAC GCG ACG GAC ATC TTC ACC | IDT DNA technologies | N/A |
| GIRK2 L79W F GAG ACG TAC CGA TAC TGG ACG GAC ATC TTC ACC | IDT DNA technologies | N/A |
| GIRK2 I82A F CGA TAC CTG ACG GAC GCC TTC ACC ACC CTG G | IDT DNA technologies | N/A |
| GIRK2 I82W F CCG ATA CCT GAC GGA CTG GTT CAC CAC CCT GGT G | IDT DNA technologies | N/A |
| GIRK2 L86A F GAC ATC TTC ACC ACC GCG GTG GAC CTG AAG TGG | IDT DNA technologies | N/A |
| GIRK2 L86C F GAC ATC TTC ACC ACC TGC GTG GAC CTG AAG TGG | IDT DNA technologies | N/A |
| GIRK2 L86W F GAC ATC TTC ACC ACC TGG GTG GAC CTG AAG TGG | IDT DNA technologies | N/A |
| GIRK2 F93A F GTG GAC CTG AAG TGG AGA GCC AAC CTG TTG ATC TTT G | IDT DNA technologies | N/A |
| GIRK2 F93Y F GTG GAC CTG AAG TGG AGA TAC AAC CTG TTG ATC TTT G | IDT DNA technologies | N/A |
| GIRK2 F93W F GGA CCT GAA GTG GAG ATG GAA CCT GTT GAT CTT TG | IDT DNA technologies | N/A |
| GIRK2 L95A F CCT GAA GTG GAG ATT CAA CGC GTT GAT CTT TGT CAT GTC | IDT DNA technologies | N/A |
| GIRK2 L95W F CCT GAA GTG GAG ATT CAA CTG GTT GAT CTT TGT CAT GTC | IDT DNA technologies | N/A |
| GIRK2 L96A F CTG AAG TGG AGA TTC AAC CTG GCG ATC TTT GTC ATG GTC | IDT DNA technologies | N/A |
| GIRK2 L96F F CTG AAG TGG AGA TTC AAC CTG TTC ATC TTT GTC ATG GTC | IDT DNA technologies | N/A |
| GIRK2 L96W F CTG AAG TGG AGA TTC AAC CTG TGG ATC TTT GTC ATG GTC | IDT DNA technologies | N/A |
| GIRK2 V99A F CAA CCT GTT GAT CTT TGC CAT GGT CTA CAC AGT GAC G | IDT DNA technologies | N/A |
| GIRK2 V99I F CAA CCT GTT GAT CTT TAT CAT GGT CTA CAC AGT G | IDT DNA technologies | N/A |
| GIRK2 V99S F GAT TCA ACC TGT TGA TCT TTT CCA TGG TCT ACA CAG TGA CG | IDT DNA technologies | N/A |
| GIRK2 V99W F GAT TCA ACC TGT TGA TCT TTT GGA TGG TCT ACA CAG TGA CGT G | IDT DNA technologies | N/A |
| GIRK2 V101A F GAT CTT TGT CAT GGC CTA CAC AGT GAC GTG | IDT DNA technologies | N/A |
| GIRK2 V101T F GTT GAT CTT TGT CAT GAC CTA CAC AGT GAC GTG GC | IDT DNA technologies | N/A |
| GIRK2 V101W F CCT GTT GAT CTT TGT CAT GTG GTA CAC AGT GAC GTG GC | IDT DNA technologies | N/A |
| GIRK2 I175F F GAT TAT TCT CCT CTT ATT CCA GTC CGT GTT GG | IDT DNA technologies | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| mouse GIRK2c cDNA | Horvath et al., 2018 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/ |
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-software |
| CTFFIND4 | Rohou and Grigorieff, 2015 | https://grigoriefflab.janelia.org/ctffind4 |
| Relion | Scheres, 2012, 2015, 2016; Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Download_%26_install |
| Phenix | Afonine et al., 2018 | https://phenix-online.org/ |
| 3DVA, cryoSPARC | Punjani and Fleet, 2021 | https://cryosparc.com/ |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| HOLE program | Smart et al., 1996 | http://www.holeprogram.org/ |
| Prism 9.0 | Graphpad | RRID:SCR_002798 |
| BioRender | https://biorender.com | |
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Paul Slesinger (paul.slesinger@mssm.edu).
Materials availability
Plasmids generated in this study are available from the lead contact upon request.
Data and code availability
CryoEM maps of GIRK2PIP2/CHS, GIRK2PIP2*, GIRK2PIP2**, GIRK2PIP2***, GIRK2PIP2**** and GIRK2 apo are deposited in Electron Microscopy Data Bank under accession codes EMD-22154, EMD-22150, EMD-22151, EMD-22152, EMD-22153 and EMD-22155, respectively. The coordinates of GIRK2PIP2/CHS and GIRK2PIP2* models are deposited in Protein Data Bank under accession codes 6XEV and 6XEU, respectively.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Molecular biology and cell culture
A truncated Mus musculus Girk2 (Kcnj6) cDNA (containing amino acids 52–380) linked in-frame with an HRV 3C protease site, green fluorescent protein (GFP) and a decahistidine (HIS10) tag (a generous gift from R. MacKinnon, The Rockefeller University, New York, NY) in pPICZ (ThermoFisher) was transformed into Pichia pastoris using electroporation for protein expression (according to manufacturer protocols). Transformants were screened based upon Zeocin resistance (> 1 mg/ml) and GFP emission. Highest expressing clones were selected for large-scale purification.
Mutations were introduced to mouse GIRK2c using site-directed mutagenesis (QuickChange II XL, Agilent Technology) and confirmed by DNA sequencing. Human Embryonic Kidney 293T (HEK293T) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% (v/v) Fetal Bovine Serum (FBS), 100 U/ml penicillin, 100 mg/ml streptomycin and 1X Glutamax (ThermoFisher) in a humidified 37C incubator with 5% CO2. Cells were plated 12-well plates and transiently transfected with cDNA (GIRK2c WT or mutant: 0.5 μg; eYFP: 0.02 μg, to identify transfected cells) using Lipofectamine 2000 (ThermoFisher). 48h after transfection, cells were plated onto poly-D-lysine (100 mg/ml; SigmaAldrich) coated 12-mm glass coverslips in 24-well plates for whole-cell patch-clamp recording.
METHOD DETAILS
Protein purification and reconstitution
All GIRK channels were expressed and purified in P. pastoris as described previously (Glaaser and Slesinger, 2017). Briefly, the highest-expressing clone was grown in BMGY medium and induced in BMM medium containing 1% methanol. Cells were harvested, resuspended in buffer (50 mM HEPES, pH 7.4; 150 mM KCl; 1 mM TCEP; 1 mM AEBSF and Complete EDTA-free protease inhibitor tablets (Roche)), flash frozen in liquid nitrogen, and stored at −80 °C. Frozen cells were lysed in a Mixer Mill (Retsch) 5-times for 3 minutes at 25 Hz and stored as powder at −80°C until needed. The cell powder was solubilized in buffer containing 50 mM HEPES, pH 7.35; 150 mM KCl; 1 mM TCEP; 1 mM AEBSF; Complete ULTRA EDTA-free protease inhibitor tablets (Roche) and either 2% (w/v) n-Dodecyl-β-D-maltoside (DDM; Anatrace) or 2% (w/v) DDM supplemented with 0.2% (w/v) Cholesteryl Hemisuccinate Tris Salt (Anatrace) with gentle stirring at 4 °C. Unsolubilized material was separated by centrifugation at 40,000 × g for 40 min at 4 °C and filtered. The supernatant was incubated with HISPur Cobalt charged resin (ThermoFisher) equilibrated in wash buffer (50 mM HEPES, pH 7.0; 150 mM KCl; 0.2% DDM or 0.2%DDM/0.02% CHS; 20 mM imidazole). The resin was subsequently washed in 10 column volumes (CV) of wash buffer, 5 CV containing 40 mM Imidazole, and eluted in buffer containing 300 mM imidazole. The eluate was pooled, exchanged into imidazole-free buffer and digested overnight at 4 °C with HRV 3 C protease, purified as described (Shaya et al., 2011) (a generous gift of D. Minor, UCSF, San Francisco, CA). The protein was subsequently concentrated and run on a Superdex-200 gel filtration column in buffer containing 20 mM TRIS-HCl pH 7.5, 150 mM KCl, 20 mM DTT, 3 mM TCEP, and 1 mM EDTA, 0.025% (w/v) DDM (anagrade) alone or with 0.0025% CHS. Fractions eluting at a volume consistent with the GIRK channel tetramer were pooled, concentrated and examined by SDS-PAGE and Coomassie blue staining.
Purified GIRK2 channels were reconstituted into lipid vesicles as described previously (Glaaser and Slesinger, 2017). Briefly, a lipid mixture containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho(1’-rac-glycerol) (POPG), and L-α-phosphatidylinositol-4,5-bisphosphate (Brain, PI(4,5)P2 (Porcine)) at mass ratios of 3:1:0.04 (POPE:POPG:PIP2) or 3:1 (POPE:POPG) was prepared, reconstituted in vesicle buffer (20 mM K-HEPES, pH 7.4; 150 mM KCl; 0.5 mM EDTA containing 35 mM CHAPS) and incubated with protein in detergent at a 1:200 protein: lipid ratio unless otherwise indicated. Where indicated, cholesterol (Ovine wool; Anatrace) was added to vesicles at a mole percentage of 5% or Cholesteryl Hemisuccinate (CHS; Anatrace) was incorporated at 2.5% mol percentage. Detergent was removed through sequential addition of Bio-beads SM-2 (Bio-rad). All phospholipids, cholesterol, and Brain PIP2 were purchased from Avanti Polar Lipids, Inc. Water-soluble PIP2 (diC8-PIP2) was purchased from Echelon Biosciences.
Flux assay
The fluorescence-based flux assay for GIRK channel activity was performed as described previously (Whorton and MacKinnon, 2013; Glaaser and Slesinger, 2017). Briefly, Liposomes were diluted 1:20 into flux buffer (20 mM Na-HEPES, pH 7.4; 150 mM NaCl; 0.5 mM EDTA) containing 5 μM of the H+ sensitive dye 9-Amino-6-chloro-2-methoxyacridine (ACMA)(Invitrogen). Fluorescence was measured using a Flexstation 3 microplate reader (Molecular Devices) with the following parameters: 410 nm excitation, 480 nm emission, 455 nm cutoff, medium PMT sensitivity, and sampling at 2 seconds. After a stable baseline fluorescence (150 s) was obtained, the H+ ionophore m-chlorophenyl hydrazone (CCCP)(Sigma) was automatically added (1 μM final), then a second addition of vehicle or methanethiosulfonate hydroxyethyl (100 μM final; MTS-HE, Toronto Research Chemicals) was added 150 s later, followed 900 s later by a third addition with the K+ ionophore Valinomycin (100 nM final; Invitrogen), to determine the maximal K+ flux. GIRK2 channels are likely arranged in both orientations in the liposomes. However, we expect the channels oriented insideout to support high K+ flux because of high Na+ in the flux buffer and high K+ in the liposome (Glaaser and Slesinger, 2017). The percentage relative K+ flux (or relative fluorescence intensity) was calculated by measuring the extent of quenching 10 s before VM addition (F), and dividing by the total quenching capacity of the liposomes normalized to the relative fluorescence units (RFU) 10 s before the addition of vehicle or MTS-HE (F0). Liposome flux assay illustration created with Biorender.com
CryoEM sample preparation, data collection, and processing
The quality of purified samples was initially screened by negative stain EM using established protocols (Peisley and Skiniotis, 2015). For cryoEM, 3.5 μl of GIRK2 at a concentration of 7–10 mg/ml was applied to glow-discharged Quantifoil Au1.2/1.3, 200 mesh grids, blotted and then plunge-frozen in liquid ethane using FEI Vitrobot. All cryoEM data were collected at 300 kV on a Titan Krios equipped with a Gatan K3 direct detection camera. Raw images were collected as movies, recorded at a magnification of 58,824 X corresponding to 0.85 pixel per Å at the specimen level. Each movie was recorded for 3 s at 0.05 s/frame with a total dose of 60 electrons/pixel and defocus values ranging from −0.8 to −2.2 μm.
The movies were motion-corrected and dose-weighted using Motioncor2 (Zheng et al., 2017) and defocus values were determined by CTFFIND4 (Rohou and Grigorieff, 2015). Template-based particle picking, 2D/3D classification, and 3D refinements were performed using the Relion pipeline (Scheres, 2012, 2015, 2016; Zivanov et al., 2018). The 3D classes with both domains distinguishable were refined with C4 or C1 symmetry as shown in Figures S2, S4, and S6, post processed with a mask encompassing both the domains, corrected for modulation transfer function (MTF) of K3 direct detection camera at 300 kV and sharpened with suitable B factor (Tables S1 and S2). The resolution of the maps reported here was estimated according to the 0.143 “gold-standard” Fourier Shell Correlation (FSC) criterion. The resolution of GIRK2PIP2/CHS, GIRK2PIP2*, GIRK2PIP2**, GIRK2PIP2***, GIRK2PIP2****, and GIRK2 apo was indicated globally as 3.5, 3.2, 4.8, 7.7, 7.7 and 4.8 Å, respectively. Local resolution was estimated using Relion. In GIRK2 apo, two 3D class with significant disengagement and similar relative orientation of domains were combined for the final 3D refinement (Figure S6). The input particle stack for 3D classification was also used for cryoSPARC 3D variability analysis (Punjani and Fleet, 2021) (Video S1).
Modeling of protein and lipids in the cryoEM maps
The GIRK2 backbone and PIP2 at the TMD-CTD interface as determined in the X-ray crystal structure of GIRK2 (PDB: 3SYA) was initially fit as rigid-body into the EM maps using UCSF Chimera (Pettersen et al., 2004) and subsequently interactively adjusted using COOT (Emsley and Cowtan, 2004). The GemSpot pipeline (Robertson et al., 2020) was used for modeling CHS into EM map followed by refinement in COOT (Emsley and Cowtan, 2004). The models in the target EM maps were refined further using Phenix real-space refinement (Afonine et al., 2018). Pore dimension in the model was analyzed using the HOLE program (Smart et al., 1996). The figures in the manuscript were prepared using UCSF ChimeraX (Goddard et al., 2018).
Electrophysiological studies
Mutant and wild-type GIRK2 channels were transiently expressed in HEK293T cells and whole-cell patch-clamp recordings were made as described previously (Zhao et al., 2020). All mutants were confirmed by DNA sequencing. Borosilicate glass electrodes (Warner Instruments) of 3–6 MΩ were filled with an intracellular solution containing 130 mM KCl, 20 mM NaCl, 5 mM EGTA, 5.46 mM MgCl2, 2.56 mM K2ATP, 0.3 mM Li2GTP and 10 mM HEPES (pH 7.4, ~320 mOsm). The extracellular ‘20K’ solution contained 20 mM KCl, 140 mM NaCl, 0.5 mM CaCl2, 2 mM MgCl2, and 10 mM HEPES (pH 7.4; ~310 mOsm). Currents were elicited at 0.5 Hz with a voltage step to −120 mV from a holding potential of −40 mV, followed by a ramp voltage protocol (−120 mV to +50 mV, EK = −50 mV with 20 mM Kout). K+ currents were adjusted for series resistance, and measured at −120 mV with either 20K solution or 20K + Ba2+ (1 mM) solution. The basal current was defined as the Ba2+-sensitive current. Cholesterol-mediated potentiation of the basal current was normalized as a percentage of the control (vehicle) basal current (Normalized Current %). All currents are expressed as current density (current / cell capacitance; pA/pF).
Cholesterol enrichment
We used water-soluble cholesterol with methyl-β-cyclodextrin (MβCD) (40mg cholesterol per gram, Sigma-Aldrich; C4951) for cholesterol enrichment in HEK293T cells. MβCD is a well-known cholesterol donor (Zidovetzki and Levitan, 2007). On the day of the recording, transfected HEK293T cells were incubated in DMEM containing 0.625 mM cholesterol modified from previously described on HEK293 cells (Wu et al., 2013). We kept the molar ratio of cholesterol: MβCD ~1:8. Controls were incubated with serum-free DMEM. After 1 h of incubation, which was shown to potentiate GIRK currents in hippocampal pyramidal neurons (Bukiya et al., 2017), HEK293T cells were washed once with FBS, and subjected to whole-cell patch-clamp recording. The data that support the findings of this study are available from the corresponding author upon reasonable request.
QUANTIFICATION AND STATISTICAL ANALYSIS
For three or more groups, a one-way ANOVA followed with a Dunnett’s multiple comparisons post hoc test was used for evaluating significant differences (*p < 0.05, **p < 0.01, **** p < 0.001) (Figure 1). For groups of two, a Student’s unpaired two-tailed t test was used for evaluating significant differences (*p < 0.05, **p < 0.01, **** p < 0.001) (Figure 6). All statistical tests were conducted with Prism 9 (Graphpad). The mean ± SEM are shown. The specific statistical details of experiments can be found in the figure legends.
Supplementary Material
Highlights.
CryoEM structure identifies cholesterol binding site in brain GIRK2 channel
Mutational and functional studies reveal key residues in cholesterol potentiation
Elucidating cholesterol binding pockets can aid in developing new modulators
ACKNOWLEDGMENTS
An earlier version of this manuscript was uploaded on bioRxiv (Mathiharan et al., 2020). This work was supported in part by the National Institute on Alcohol Abuse and Alcoholism (AA018734) to P.A.S. The cryoEM data for this study were collected at the Stanford-SLAC cryoEM facility. We thank the Slesinger and Skiniotis laboratories for discussions on the experiments, and E. Montabana for help with cryoEM data collection. Some illustrations were created with BioRender.com
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109619.
A video abstract is available at https://doi.org/10.1016/j.celrep.2021.109619#mmc4.
REFERENCES
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsteiner H, Zangerl-Plessl EM, Chen X, and Stary-Weinzinger A (2019). Conduction through a narrow inward-rectifier K+ channel pore. J. Gen. Physiol 151, 1231–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KA, He S, Jin R, Miller DM, Bolla JR, Clarke OB, Johnson P, Windley M, Burns CJ, Hill AP, et al. (2020). A constricted opening in Kir channels does not impede potassium conduction. Nat. Commun 11, 3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukiya AN, and Rosenhouse-Dantsker A (2017). Synergistic activation of G protein-gated inwardly rectifying potassium channels by cholesterol and PI(4,5)P2. Biochim. Biophys. Acta Biomembr. 1859, 1233–1241. [DOI] [PubMed] [Google Scholar]
- Bukiya AN, Durdagi S, Noskov S, and Rosenhouse-Dantsker A (2017). Cholesterol up-regulates neuronal G protein-gated inwardly rectifying potassium (GIRK) channel activity in the hippocampus. J. Biol. Chem 292, 6135–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YY, Kwon OH, Park MK, Kim TW, and Chung S (2019). Elevated cellular cholesterol in familial Alzheimer’s presenilin 1 mutation is associated with lipid raft localization of β-amyloid precursor protein. PLoS ONE 14, e0210535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Laucht M, Ridinger M, Wodarz N, Rietschel M, Maier W, Lathrop M, Lourdusamy A, Zimmermann US, Desrivieres S, and Schumann G (2011). KCNJ6 is associated with adult alcohol dependence and involved in gene 3 early life stress interactions in adolescent alcohol drinking. Neuropsychopharmacology 36, 1142–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, Wischmeyer E, Dityatev A, Sytnyk V, Veh RW, Karschin A, and Schachner M (2002). The neural cell adhesion molecule regulates cell-surface delivery of G-protein-activated inwardly rectifying potassium channels via lipid rafts. J. Neurosci 22, 7154–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietschy JM, and Turley SD (2001). Cholesterol metabolism in the brain. Curr. Opin. Lipidol 12, 105–112. [DOI] [PubMed] [Google Scholar]
- Dreger M, Krauss M, Herrmann A, and Hucho F (1997). Interactions of the nicotinic acetylcholine receptor transmembrane segments with the lipid bilayer in native receptor-rich membranes. Biochemistry 36, 839–847. [DOI] [PubMed] [Google Scholar]
- Duncan AL, Corey RA, and Sansom MSP (2020a). Defining how multiple lipid species interact with inward rectifier potassium (Kir2) channels. Proc. Natl. Acad. Sci. USA 117, 7803–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AL, Song W, and Sansom MSP (2020b). Lipid-dependent regulation of ion channels and G protein-coupled receptors: Insights from structures and simulations. Annu. Rev. Pharmacol. Toxicol 60, 31–50. [DOI] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Engel DF, de Oliveira J, Lopes JB, Santos DB, Moreira ELG, Farina M, Rodrigues ALS, de Souza Brocardo P, and de Bem AF (2016). Is there an association between hypercholesterolemia and depression? Behavioral evidence from the LDLr−/− mouse experimental model. Behav. Brain Res. 311, 31–38. [DOI] [PubMed] [Google Scholar]
- Epshtein Y, Chopra AP, Rosenhouse-Dantsker A, Kowalsky GB, Logothetis DE, and Levitan I (2009). Identification of a C-terminus domain critical for the sensitivity of Kir2.1 to cholesterol. Proc. Natl. Acad. Sci. USA 106, 8055–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpl G (2016). Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids 199, 61–73. [DOI] [PubMed] [Google Scholar]
- Glaaser IW, and Slesinger PA (2017). Dual activation of neuronal G protein-gated inwardly rectifying potassium (GIRK) channels by cholesterol and alcohol. Sci. Rep 7, 4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SB, Tao X, and MacKinnon R (2011). Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, and Stevens RC (2008). A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KG, Alva H, Blednov YA, and Cunningham CL (2003). Reduced ethanol-induced conditioned taste aversion and conditioned place preference in GIRK2 null mutant mice. Psychopharmacology (Berl.) 169, 108–114. [DOI] [PubMed] [Google Scholar]
- Horvath GA, Zhao Y, Tarailo-Graovac M, Boelman C, Gill H, Shyr C, Lee J, Blydt-Hansen I, Drögemöller BI, Moreland J, et al. (2018). Gain-of-function KCNJ6 Mutation in a Severe Hyperkinetic Movement Disorder Phenotype. Neuroscience 384, 152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Feng S, and Hilgemann DW (1998). Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gβγ. Nature 391, 803–806. [DOI] [PubMed] [Google Scholar]
- Jin U, Park SJ, and Park SM (2019). Cholesterol metabolism in the brain and its association with Parkinson’s disease. Exp. Neurobiol 28, 554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones OT, and McNamee MG (1988). Annular and nonannular binding sites for cholesterol associated with the nicotinic acetylcholine receptor. Biochemistry 27, 2364–2374. [DOI] [PubMed] [Google Scholar]
- Lee AG (2019). Interfacial binding sites for cholesterol on G protein-coupled receptors. Biophys. J 116, 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis DE, Kurachi Y, Galper J, Neer EJ, and Clapham DE (1987). The βγ subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature 325, 321–326. [DOI] [PubMed] [Google Scholar]
- Lüscher C, and Slesinger PA (2010). Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat. Rev. Neurosci 11, 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM, Sung MW, Yang Z, Innes LM, Kandasamy B, David LL, Yoshioka C, and Shyng SL (2019). Mechanism of pharmacochaperoning in a mammalian KATP channel revealed by cryo-EM. eLife 8, e46417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mathiharan YK, Glaaser IW, Zhao Y, Robertson MJ, Skiniotis G, and Slesinger PA (2020). Structural basis of GIRK2 channel modulation by cholesterol and PIP2. bioRxiv. 10.1101/2020.06.04.134544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middlemas DS, and Raftery MA (1987). Identification of subunits of acetylcholine receptor that interact with a cholesterol photoaffinity probe. Biochemistry 26, 1219–1223. [DOI] [PubMed] [Google Scholar]
- Munoz MB, and Slesinger PA (2014). Sorting nexin 27 regulation of G protein-gated inwardly rectifying K+ channels attenuates in vivo cocaine response. Neuron 82, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Tao X, Touhara KK, and MacKinnon R (2020). Cryo-EM analysis of PIP2 regulation in mammalian GIRK channels. eLife 9, e60552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Choudhury A, Chandra Boruah D, Devi R, Bhattacharya P, Choudhury MD, and Borah A (2017a). Hypercholesterolemia causes psychomotor abnormalities in mice and alterations in cortico-striatal biogenic amine neurotransmitters: Relevance to Parkinson’s disease. Neurochem. Int 108, 15–26. [DOI] [PubMed] [Google Scholar]
- Paul R, Choudhury A, Kumar S, Giri A, Sandhir R, and Borah A (2017b). Cholesterol contributes to dopamine-neuronal loss in MPTP mouse model of Parkinson’s disease: Involvement of mitochondrial dysfunctions and oxidative stress. PLoS ONE 12, e0171285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Dutta A, Phukan BC, Mazumder MK, Justin-Thenmozhi A, Manivasagam T, Bhattacharya P, and Borah A (2018). Accumulation of cholesterol and homocysteine in the nigrostriatal pathway of brain contributes to the dopaminergic neurodegeneration in mice. Neuroscience 388, 347–356. [DOI] [PubMed] [Google Scholar]
- Peisley A, and Skiniotis G (2015). 2D projection analysis of GPCR complexes by negative stain electron microscopy. Methods Mol. Biol. 1335, 29–38. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Punjani A, and Fleet DJ (2021). 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol 213, 107702. [DOI] [PubMed] [Google Scholar]
- Reuveny E, Slesinger PA, Inglese J, Morales JM, Iñiguez-Lluhi JA, Lefkowitz RJ, Bourne HR, Jan YN, and Jan LY (1994). Activation of the cloned muscarinic potassium channel by G protein βγ subunits. Nature 370, 143–146. [DOI] [PubMed] [Google Scholar]
- Rifkin RA, Huyghe D, Li X, Parakala M, Aisenberg E, Moss SJ, and Slesinger PA (2018). GIRK currents in VTA dopamine neurons control the sensitivity of mice to cocaine-induced locomotor sensitization. Proc. Natl. Acad. Sci. USA 115, E9479–E9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MJ, van Zundert GCP, Borrelli K, and Skiniotis G (2020). GemSpot: A pipeline for robust modeling of ligands into Cryo-EM maps. Structure 28, 707–716.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenhouse-Dantsker A (2019). Cholesterol binding sites in inwardly rectifying potassium channels. Adv. Exp. Med. Biol 1135, 119–138. [DOI] [PubMed] [Google Scholar]
- Rosenhouse-Dantsker A, Leal-Pinto E, Logothetis DE, and Levitan I (2010). Comparative analysis of cholesterol sensitivity of Kir channels: Role of the CD loop. Channels (Austin) 4, 63–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenhouse-Dantsker A, Noskov S, Durdagi S, Logothetis DE, and Levitan I (2013). Identification of novel cholesterol-binding regions in Kir2 channels. J. Biol. Chem 288, 31154–31164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenson RS, Brewer HB Jr., Barter PJ, Björkegren JLM, Chapman MJ, Gaudet D, Kim DS, Niesor E, Rye KA, Sacks FM, et al. (2018). HDL and atherosclerotic cardiovascular disease: Genetic insights into complex biology. Nat. Rev. Cardiol 15, 9–19. [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2015). Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol 189, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2016). Processing of structurally heterogeneous Cryo-EM data in RELION. Methods Enzymol. 579, 125–157. [DOI] [PubMed] [Google Scholar]
- Shaya D, Kreir M, Robbins RA, Wong S, Hammon J, Brüggemann A, and Minor DL Jr. (2011). Voltage-gated sodium channel (NaV) protein dissection creates a set of functional pore-only proteins. Proc. Natl. Acad. Sci. USA 108, 12313–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorini S, Liao YJ, Duncan SA, Jan LY, and Stoffel M (1997). Normal cerebellar development but susceptibility to seizures in mice lacking G protein-coupled, inwardly rectifying K+ channel GIRK2. Proc. Natl. Acad. Sci. USA 94, 923–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376. [DOI] [PubMed] [Google Scholar]
- Sui JL, Petit-Jacques J, and Logothetis DE (1998). Activation of the atrial KACh channel by the βγ subunits of G proteins or intracellular Na+ ions depends on the presence of phosphatidylinositol phosphates. Proc. Natl. Acad. Sci. USA 95, 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QY, Larry T, Hendra K, Yamamoto E, Bell J, Cui M, Logothetis DE, and Boland LM (2015). Mutations in nature conferred a high affinity phosphatidylinositol 4,5-bisphosphate-binding site in vertebrate inwardly rectifying potassium channels. J. Biol. Chem 290, 16517–16529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE (2012). Dysregulation of cholesterol balance in the brain: Contribution to neurodegenerative diseases. Dis. Model. Mech 5, 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, and MacKinnon R (2011). Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, and MacKinnon R (2013). X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature 498, 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickman KD, Iñiguez-Lluhl JA, Davenport PA, Taussig R, Krapivinsky GB, Linder ME, Gilman AG, and Clapham DE (1994). Recombinant G-protein β γ-subunits activate the muscarinic-gated atrial potassium channel. Nature 368, 255–257. [DOI] [PubMed] [Google Scholar]
- Wu W, Wang Y, Deng XL, Sun HY, and Li GR (2013). Cholesterol down-regulates BK channels stably expressed in HEK 293 cells. PLoS ONE 8, e79952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrouk A, Debbabi M, Bezine M, Karym EM, Badreddine A, Rouaud O, Moreau T, Cherkaoui-Malki M, El Ayeb M, Nasser B, et al. (2018). Lipid biomarkers in Alzheimer’s disease. Curr. Alzheimer Res. 15, 303–312. [DOI] [PubMed] [Google Scholar]
- Zhang J, and Liu Q (2015). Cholesterol metabolism and homeostasis in the brain. Protein Cell 6, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ung PM, Zahoránszky-Kohalmi G, Zakharov AV, Martinez NJ, Simeonov A, Glaaser IW, Rai G, Schlessinger A, Marugan JJ, and Slesinger PA (2020). Identification of a G-protein-independent activator of GIRK channels. Cell Rep. 31, 107770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovetzki R, and Levitan I (2007). Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim. Biophys. Acta 1768, 1311–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CryoEM maps of GIRK2PIP2/CHS, GIRK2PIP2*, GIRK2PIP2**, GIRK2PIP2***, GIRK2PIP2**** and GIRK2 apo are deposited in Electron Microscopy Data Bank under accession codes EMD-22154, EMD-22150, EMD-22151, EMD-22152, EMD-22153 and EMD-22155, respectively. The coordinates of GIRK2PIP2/CHS and GIRK2PIP2* models are deposited in Protein Data Bank under accession codes 6XEV and 6XEU, respectively.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the Lead Contact upon request.