Abstract
Translation elongation factor 1β (EF-1β) is a member of the family of guanine nucleotide exchange factors, proteins whose activities are important for the regulation of G proteins critical to many cellular processes. EF-1β is a highly conserved protein that catalyzes the exchange of bound GDP for GTP on EF-1α, a required step to ensure continued protein synthesis. In this work, we demonstrate that the highly conserved C-terminal region of Saccharomyces cerevisiae EF-1β is sufficient for normal cell growth. This region of yeast and metazoan EF-1β and the metazoan EF-1β-like protein EF-1δ is highly conserved. Human EF-1β, but not human EF-1δ, is functional in place of yeast EF-1β, even though both EF-1β and EF-1δ have previously been shown to have guanine nucleotide exchange activity in vitro. Based on the sequence and functional homology, mutagenesis of two C-terminal residues identical in all EF-1β protein sequences was performed, resulting in mutants with growth defects and sensitivity to translation inhibitors. These mutants also enhance translational fidelity at nonsense codons, which correlates with a reduction in total protein synthesis. These results indicate the critical function of EF-1β in regulating EF-1α activity, cell growth, translation rates, and translational fidelity.
Translation elongation requires the function of soluble protein factors. In eukaryotic organisms, the translation elongation factor 1 (EF-1) delivers aminoacyl-tRNAs (aa-tRNAs) to the A site of an elongating ribosome (6). EF-2 functions after peptide bond formation to translocate the peptidyl-tRNA to the P site (30). EF-3 is a third fungus-specific elongation factor (3). All three factors have a requirement for energy from either GTP or ATP. Only EF-1, however, requires a nucleotide exchange factor to maintain the pool of active nucleotide triphosphate-bound protein.
EF-1 is composed of four subunits in metazoans (α, β, γ, and δ) (6) but only three subunits in the yeast Saccharomyces cerevisiae (α, β, and γ) (31). The α subunit is a classic G protein that binds aa-tRNAs in a GTP-dependent manner and delivers them to the A site of the elongating ribosome. After GTP hydrolysis, the resulting GDP remains bound to EF-1α, and the protein is unable to reenter the elongation cycle until GTP is rebound. The EF-1β subunit functions as a guanine nucleotide exchange factor in vitro (38). While EF-1β is found associated with the γ subunit, the role of EF-1γ remains unknown, although it modestly stimulates the activity of EF-1β in vitro (41). The δ subunit of metazoans also functions as a guanine nucleotide exchange factor in vitro but may function in vivo in the assembly of higher-order complexes of EF-1 with aa-tRNA synthetases (2). There is no biochemical or genetic evidence that yeasts contain an EF-1δ subunit.
The β subunit of yeast EF-1 (yEF-1) is encoded by the single-copy essential gene TEF5 (14). The requirement for yEF-1β can be relieved by the presence of an extra copy of the gene encoding yEF-1α (20). The resulting yEF-1β-deficient strains, however, show dramatically slowed growth and reduced translational fidelity. Reduced translational fidelity is also seen for dominant and recessive mutations in yEF-1α (9, 11, 33). Other mutations in components of the translational apparatus, such as ribosomal proteins and rRNAs, also affect translation fidelity (13, 23). Thus, many factors play discrete roles in maintaining efficient and accurate protein synthesis.
In this work, we demonstrate that the C-terminal region of yEF-1β is functional as the only form of the protein in vivo. Similar C-terminal proteolytic fragments of Artemia salina and human EF-1β (hEF-1β) retain guanine nucleotide exchange activity in vitro (27, 41). This region contains a nearly identical cluster of amino acids found in all EF-1β and EF-1δ proteins sequenced to date. Mutational analysis directed to the first two residues of this cluster results in a large series of substitutions that confer conditional growth defects and severe sensitivity to translation inhibitors. The site of these mutations is predicted, based on the nuclear magnetic resonance structure of the C terminus of hEF-1β and the crystal structure of the prokaryotic homologs EF-Tu and EF-Ts, to lie at the end of a β-strand opposite the proposed functional loops for nucleotide exchange (18, 28). While expression of the hEF-1β protein can replace the essential yeast protein in vivo, the hEF-1δ cannot. This indicates that, while this conserved cluster defines an important region of yEF-1β, more is required for the function of this class of proteins. Strains with mutations in this C-terminal motif show reduced total translation and enhanced translational fidelity at all three nonsense codons, thus indicating the important role of EF-1β in maintaining efficient and accurate translation.
MATERIALS AND METHODS
Strains and media.
Escherichia coli NM522 and DH5α were used for plasmid preparation. S. cerevisiae strains used in these studies are listed in Table 1. JM749 was the diploid parent of all tef5::TRP1 strains (24). Standard yeast genetic methods were employed (25, 35). Yeast cells were grown in either YEPD (1% Bacto yeast extract, 2% peptone, 2% dextrose) or defined synthetic complete medium (C or C-) supplemented with 2% dextrose as a carbon source unless noted otherwise, where 2% galactose was used. Yeast cells were transformed by the lithium acetate method (15). Strain JWY4231 was prepared by mating JWY4201 with MC1160 (33). Diploids were sporulated and dissected to identify Trp+ Ura+ colonies (tef5::TRP1 pTEF5 URA3) containing the met2-1 and his4-713 +1 insertion alleles and the lys2-801 UGA nonsense allele.
TABLE 1.
Parental S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JWY4134 | MATa ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 his3-Δ200 | 19 |
| JWY4175 | MATa/α ura3-52/ura3-52 trp1-Δ101/trp1-Δ101 lys2-801/lys2-801 leu2-Δ1/leu2-Δ1 his3-Δ200/his3-Δ200 TEF5/tef5::TRP1 | 20 |
| JWY4200 | MATa ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 his3-Δ200 tef5::TRP1 pTEF5 URA3 | 20 |
| JWY4229 | MATa ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 his3-Δ200 tef5::TRP1 pTEF5 LEU2 | 20 |
| MC1160 | MATa ura3-52 trp1-Δ101 lys2-801 leu2-3,112 met2-1 his4-713 | 33 |
| JWY4231 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pTEF5 URA3 | This work |
| JWY4247 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pTEF5 LEU2 | This work |
| JWY4298 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pGAL1TEF5Δ60 URA3 | This work |
| JWY4299 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pGAL1TEF5Δ85 URA3 | This work |
| TKY169 | MATa ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 his3-Δ200 tef5::TRP1 pGAL1TEF5-HA URA3 | This work |
| TKY235 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pTEF5 LEU2 | This work |
| TKY236 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-4 LEU2 | This work |
| TKY237 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-10 LEU2 | This work |
| TKY238 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-1 LEU2 | This work |
| TKY239 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-8 LEU2 | This work |
| TKY240 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-2 LEU2 | This work |
| TKY241 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-9 LEU2 | This work |
| TKY242 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-5 LEU2 | This work |
| TKY243 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-7 LEU2 | This work |
| TKY244 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-3 LEU2 | This work |
| TKY251 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 ptef5-6 LEU2 | This work |
| TKY256 | MATa ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 his3-Δ200 tef5::TRP1 pGAL1hEF-1β URA3 | This work |
| TKY257 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pGAL1TEF5Δ96 URA3 | This work |
| TKY258 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pTEF5-HA URA3 | This work |
| TKY266 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pTEF5Δ96 URA3 | This work |
| TKY285 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pGAL1TEF5-HA URA3 | This work |
| TKY286 | MATα ura3-52 trp1-Δ101 lys2-801 leu2-Δ1 met2-1 his4-713 tef5::TRP1 pGAL1hEF-1β URA3 | This work |
DNA manipulations.
Recombinant DNA techniques were performed as described previously (32). Restriction endonucleases and DNA-modifying enzymes were obtained from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Plasmids designed to be used in both E. coli and yeast were the URA3 low-copy CEN plasmids YCp50 and pRS316 (17, 36) and the high-copy 2μm plasmid YEp24 (5). The TEF2 gene is cloned in the low-copy YCp50 (YCpMS29) and high-copy YEp24 (YCpMS42) vectors (33). The TEF3 (pJWB2853) and TEF4 (pJWB2824) genes are cloned on the low-copy YCp50 vector (19).
Preparation of truncations of the TEF5 gene.
A series of GAL1 promoter constructs with N-terminal hemagglutinin (HA) tags (pRD series, prepared by R. Deshaies and provided by B. Stillman) were used to produce C-terminal fragments of yEF-1β, encoded by TEF5. A XhoI site was introduced 5′ of the AUG of the full-length TEF5 gene (lacking the intron, pTKB104) by PCR with oligonucleotide TEF5XhoI (5′-GAATATATACACTCGAGAATGGCATC-3′). The fragment was digested with XhoI and cloned into plasmid pRD58 to produce plasmid pTKB170, which encodes a full-length form of yEF-1β with an N-terminal HA tag (yEF-1β-HA). Plasmid pJWB3013 was cut at the EcoRI site at amino acid 61 and the ClaI site of the vector polylinker and cloned into vector pRD54. The resulting plasmid, pJWB3062, expresses an HA-tagged yEF-1β from amino acid 61 to 206 (yEF-1βΔ60-HA). A NarI site was introduced at amino acid 86 by site-directed mutagenesis with oligonucleotide TEF5Ala86 (5′-CGATTTATTCGGCGCCGACGATGAAGAAGC-3′) (21). This results in the substitution of alanine for serine 86. The resulting plasmid, pJWB3028, was digested with NarI and ClaI, and the fragment was cloned into vector pRD54 to produce plasmid pJWB3064 (yEF-1βΔ85-HA). A HindIII site at amino acid 97 was introduced by PCR mutagenesis with oligonucleotide TEF5Hind (5′-CTGAAAAGCTTGAAGGC-3′). The resulting fragment was digested with HindIII and XhoI and cloned into pRD58, resulting in plasmid pTKB268 (yEF-1βΔ96-HA). The vectors were transformed into yeast strain JWY4247, and the TEF5 LEU2 helper plasmid was lost by dilution following growth in C-Ura-galactose, producing strains TKY285 (yEF-1β-HA), JWY4298 (yEF-1βΔ60-HA), JWY4299 (yEF-1βΔ85-HA), and TKY257 (yEF-1βΔ96-HA). The N-terminal yEF-1β truncation was prepared by cloning the 0.7-kb XhoI fragment of pTKB298 into pRD58, producing an HA-tagged N-terminal 96-amino-acid fragment of yEF-1β. The construct was transformed into yeast strains JWY4229 and JWY4175.
The full-length yEF-1β-HA and the yEF-1βΔ96-HA truncation were cloned under the control of the authentic TEF5 promoter. The TEF5 promoter was prepared with plasmid pJWB3013 as a template and oligonucleotides TEF5EagI (5′-CAATACCGGCCGCTTTTGACATA-3′) and TEF5BamHI (5′-CGGTGGATCCTTATGTGTGTAT-3′). The resulting fragment was digested with both enzymes and cloned into plasmids pTKB170 and pTKB268, producing full-length yEF-1β-HA (pTKB269) and yEF-1βΔ96-HA (pTKB278). This replaces the GAL1 promoter with the TEF5 promoter. Both constructs were transformed into JWY4247, and the TEF5 LEU2 helper plasmid was spontaneously lost following growth in C-Ura, producing strains TKY258 (yEF-1β-HA) and TKY266 (yEF-1βΔ96-HA).
The production of stable proteins was confirmed by Western blot analysis with antibodies against the HA tag. The relative expression levels of the different proteins were standardized with an anti-phosphatidylglycerol kinase 1 antibody as an internal control in the Western blots.
Cloning and expression of hEF-1β and hEF-1δ cDNAs.
Human cDNAs encoding EF-1β and EF-1δ were generously provided by W. Moller and J. Dijk (University of Leiden, Leiden, The Netherlands). The hEF-1β cDNA was subcloned into pBluescript (pTKB121) and further subcloned under the control of the yeast GAL1 promoter by using a derivative of pRD54 (pTKB154) containing the PGK1 transcriptional terminator. A SalI site was introduced into the hEF-1β coding sequence in frame with the HA tag by PCR mutagenesis with oligonucleotide hEF1Beta-1 (5′-GATACAGTCGACACC-3′), producing pTKB250 (hEF-1β-HA). A SalI site was introduced into the hEF-1δ coding sequence in frame with the HA tag by PCR mutagenesis with oligonucleotide hEF1DeltaSalI (5′-GGCGTCGACAAATGGCTAC-3′), producing pTKB151 (hEF-1δ-HA). The truncated form of hEF-1δ lacking amino acids 1 to 178 was prepared by introducing a SalI site at residue 177 with PCR mutagenesis and oligonucleotide TruncDelta (5′-GCGGACAAGGAGTCGACCAGCTGCGGG-3′) to produce pTKB301 (hEF-1δΔ172-HA).
The three plasmids were introduced into strains JWY4229 and JWY4247, and expression of the HA-tagged proteins was confirmed by Western blot analysis. The TEF5 LEU2 plasmid was lost by strains containing pTKB250 (hEF-1β) by spontaneous growth in C-Ura-galactose, producing strains TKY256 and TKY286, respectively. Plasmids pTKB151 and pTKB301 were unable to support loss of the TEF5 LEU2 plasmid from JWY4229. Plasmids pTKB151 and pTKB301 expressing hEF-1δ-HA and hEF-1δΔ172-HA were transformed into the heterozygous diploid with the tef5::TRP1 null allele and sporulated, and spores were monitored by tetrad dissection and random spore analysis. Both hEF-1δ constructs were introduced into strains TKY238 and TKY243 to monitor the ability to suppress the conditional defects conferred by the tef5-1 or tef5-7 mutant allele.
Coimmunoprecipitations of EF-1 subunits.
Yeast strains were grown in C-Ura-galactose to an A600 of approximately 1.0. Cell pellets were resuspended in immunoprecipitation buffer (50 mM Tris [pH 7.5], 50 mM NaCl, 0.02% sodium azide, 1 mM phenylmethylsulfonyl fluoride, 1 mg of aprotinin per ml, 1% Triton X-100), lysed with glass beads, and centrifuged at 4°C for 30 min. A 250-μl reaction mixture containing 50 μg of precleared extracts was mixed with 3 μl of either anti-yEF-1α polyclonal antibody or anti-Rpa1p polyclonal antibody (kindly provided by Steven Brill, Rutgers University) or without antibody and mixed at 4°C for 1 h. Forty microliters of recombinant protein A-Sepharose beads (Repligen Corp.) was added and mixed at 4°C for 1 h. Pellets were washed four times with 4 volumes of immunoprecipitation buffer prepared with 75 mM NaCl and without 1% Triton X-100. Loading dye was added to pellets, which were boiled for 5 min and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein gels were transferred to nitrocellulose, probed with monoclonal antibodies against the HA epitope, and detected with the ECL kit (Amersham).
Isolation of conditional mutant alleles of TEF5.
A 1.8-kb fragment containing the TEF5 gene encoding yEF-1β cloned into pBluescript (pJWB2962) was used as the template for PCR mutagenesis. An oligonucleotide containing a mixture of all four deoxynucleotides (X in the oligonucleotide sequence) for the first two bases of the Lys-120 and Ser-121 codons of the TEF5 gene (5′-AAGCCAGCTGCTXXGXXCATTGTCACTCTAG-3′) was used in combination with the pBluescript reverse primer to amplify the 3′ end of the TEF5 gene. The fragment was gel purified and used as the 3′ primer in combination with the pBluescript universal primer to amplify the entire TEF5 gene. The resulting fragment was gel purified and transformed into strain JWY4200 along with a fragment of pJWB3013 (TEF5 LEU2) produced by digestion with NdeI and NcoI to remove 75% of the TEF5 coding sequence. Transformants resulting from homologous recombination in vivo to reconstitute the intact TEF5 LEU2 plasmid were selected on C-Leu medium. Analysis of growth on 5-fluoroorotic acid (5-FOA) indicated that 500 colonies were unable to lose the pTEF5 URA3 plasmid and 2,200 colonies contained viable forms of the TEF5 gene (4).
The 2,200 colonies were replica plated from 5-FOA to YEPD medium at 13, 30, and 37°C to identify colonies with conditional growth phenotypes. The TEF5 LEU2 plasmids were isolated from yeast, shuttled through E. coli, and retransformed into JWY4200 to confirm that the conditional defects were conferred by the plasmid-borne gene. The tef5 mutant alleles on the plasmids were sequenced. Strains TKY235-244 and TKY251 were prepared by transforming wild-type and mutant TEF5 LEU2 plasmids into JWY4231 and monitoring loss of pTEF5 URA3 on 5-FOA (4).
Temperature sensitivity, translational fidelity, and growth of tef5::TRP1 strains.
Temperature sensitivity was assayed by spotting 5 μl of a suspension of each of the tef5 strains at A600 = 1.0 onto YEPD plates, followed by incubation at 13, 23, 30, and 37°C for 3 to 7 days. Phenotypic suppression of the lys2-801 (UGA) mutation was determined by spotting 10 μl of strains containing wild-type TEF5 or a tef5 allele onto complete medium (C) or complete medium lacking lysine (C-Lys) and incubating for 2 to 5 days at 30°C. Paromomycin-induced misreading was similarly assayed on the same media containing 0.1, 0.2, or 0.5 mg of paromomycin per ml. Doubling times were determined by measuring the growth in liquid culture of at least two independent isolates for each mutant. Cultures grown for 1 day in YEPD at 30°C were diluted to an A600 of approximately 0.1 in fresh YEPD and grown at 30°C with vigorous shaking. Optical density (A600) was assayed approximately every 2 h. Cultures were diluted into fresh YEPD when the A600 reached mid-log phase (0.4 to 0.6 U) to allow continued monitoring.
Drug sensitivity.
Two-milliliter cultures of each strain were grown at 30°C in YEPD to mid-log phase. At least two independent colonies were assayed for each mutant allele tested. For each culture, 0.3 ml was spread plated onto YEPD plates and 10 μl of each drug was pipetted onto sterile BBL 1/4-in.-diameter paper discs. The concentrations of drugs used were 2.5 mM cycloheximide, 5 mM hygromycin B, 48 mM paromomycin, 5 mM streptomycin, and 5 mM kanamycin. A maximum of two filters were placed on each plate, and the plates were incubated for 2 to 3 days at 30°C. Sensitivity to each drug was measured by the radius of inhibition around each disc.
Suppression of conditional growth defects of strains containing tef5 alleles.
tef5::TRP1 strains containing either one of the 20 tef5 alleles or wild-type TEF5 on a LEU2 CEN plasmid were transformed with the CEN vectors pRS316 (URA3), YCpMS29 (TEF2 URA3), pJWB2853 (TEF3 URA3), and pJWB2824 (TEF4 URA3) and the 2μm vector YCpMS42 (TEF2 URA3). Transformants were selected and maintained on C-Ura. Colonies were grown overnight in C-Ura liquid medium, diluted to equal cell numbers, and spotted as a dilution series on C-Ura solid medium. Plates were incubated at 13, 23, 30, and 37°C for 3 to 7 days.
Nonsense suppression assays.
Nonsense suppression assays were performed on strains containing the URA3 wild-type lacZ control plasmid pUKC815tail (lacZ under the PGK1 promoter with the PGK1 transcriptional terminator) or a plasmid with an in-frame nonsense codon in lacZ: pUKC819tail (UGA), pUKC817tail (UAA), or pUKC818tail (UAG). The mutant and wild-type strains containing each plasmid were grown overnight at 30°C in C-Ura to mid-log phase. At least four samples for each strain and plasmid were analyzed in duplicate by the o-nitrophenyl-β-d-galactopyranoside assay as previously described (9).
In vivo [35S]methionine incorporation.
Strains TKY235, TKY238, and TKY243 were converted to MET2 by transformation of a PCR fragment containing the wild-type MET2 gene and homologous recombination. Liquid cultures (100 ml) of cells were grown in C-Met at 30°C to an A600 of 0.5 to 0.7. At the zero time point, 50 μM cold methionine and 1 μCi of [35S]methionine (7.9 mCi/ml, 293.0 MBq/ml; NEN) per ml were added to each culture. At 10-min intervals, the optical density (A600) of the cultures was determined, 1-ml aliquots of the cultures were removed, and labeled methionine incorporation was monitored by cold trichloroacetic acid (TCA) precipitation. Ice-cold 50% TCA (0.2 ml) was added to each aliquot, and then aliquots were incubated on ice for 10 min, heated to 70°C for 20 min, and filtered through Whatman GF/C filters. Filters were washed with 10 ml of 5% TCA (4°C) and 10 ml of 95% ethanol, dried, and counted in a scintillation counter. All time points were analyzed in triplicate, and all errors were less than 4%.
RESULTS
The C-terminal half of yEF-1β is sufficient for normal growth of yeast.
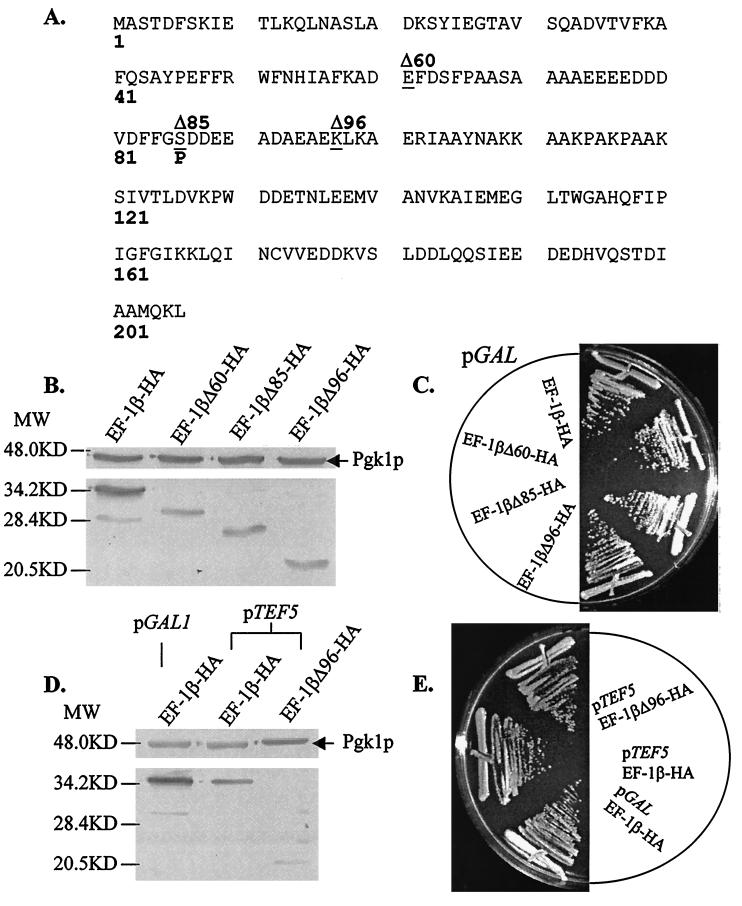
To address the functional significance of the highly conserved C terminus of EF-1β, the N terminus was removed and the C terminus was expressed under the control of a galactose-inducible promoter with an N-terminal HA epitope tag (Fig. 1A). Full-length yEF-1β-HA and three truncations that remove 60 (yEF-1βΔ60-HA), 85 (yEF-1βΔ85-HA), and 96 (yEF-1βΔ96-HA) amino acids from the N terminus were all stably expressed in yeast (Fig. 1B). All four forms of the protein were able to function as the only form of EF-1β and showed no growth defects (Fig. 1C) or sensitivity to translation inhibitors (data not shown). The N-terminal 96 amino acids of yEF-1β were separately expressed as an HA-tagged peptide and stably produced; however, it did not support growth of a tef5Δ strain (data not shown).
FIG. 1.
Only the C terminus of EF-1β is essential in vivo. (A) Sequence of S. cerevisiae EF-1β indicating sites of truncations (underlined). (B) Western blot analysis of extracts of strains expressing the HA epitope-tagged full-length and truncated yEF-1β. Lanes from left to right are TKY285 (yEF-1β-HA), JWY4298 (yEF-1βΔ60-HA), JWY4299 (yEF-1βΔ85-HA), and TKY257 (yEF-1βΔ96-HA) grown in galactose and expressed from the GAL1 promoter. (C) Growth of strains expressing full-length and truncated yEF-1β. Strains from the top are TKY285 (yEF-1β-HA), JWY4298 (yEF-1βΔ60-HA), JWY4299 (yEF-1βΔ85-HA), and TKY257 (yEF-1βΔ96-HA) grown on YEP-galactose at 30°C for 4 days. (D) Comparison of expression from the GAL1 and TEF5 promoters. Lanes from left to right are TKY285 (GAL1 promoter, yEF-1β-HA) and the TEF5 promoter expressing full-length yEF-1β-HA (TKY258) and truncated yEF-1βΔ96-HA (TKY266). (E) Growth of strains expressing (from top to bottom) TKY285 (GAL1 promoter, yEF-1β-HA) and TEF5 promoter expressing full-length yEF-1β-HA (TKY258) and truncated yEF-1βΔ96-HA (TKY266) grown on YEP-galactose at 30°C for 4 days. All blots were probed with anti-HA and anti-phosphatidylglycerol kinase antibodies. For panels B and D, MW indicates molecular mass in kilodaltons.
One possible explanation for the ability of the truncated proteins to function in place of full-length yEF-1β was that expression from the GAL1 promoter resulted in artificially high levels of protein. To rule out this possibility, full-length yEF-1β and yEF-1βΔ96 were expressed as HA-tagged proteins under the control of the authentic TEF5 promoter. Quantitative Western blot analysis of the full-length proteins indicated that the TEF5 and GAL1 promoters result in levels of protein expression that differ by approximately 50% (Fig. 1D). Both full-length yEF-1β and yEF-1βΔ96 expressed from the authentic TEF5 promoter can function as the only form of the protein. Analysis of growth rates, drug sensitivity, and the ability of these strains to grow at low and high temperatures indicated that the truncations did not dramatically alter the function of yEF-1β in vivo (Fig. 1E and data not shown). Polyribosome profile analysis of strains TKY258 (yEF-1β-HA) and TKY266 (yEF-1βΔ96-HA) did not indicate any change in the polyribosome content or distribution (data not shown).
The C terminus of EF-1β contains the most highly conserved region between species.
Comparison of the sequences of the EF-1β proteins from S. cerevisiae, the fission yeast (Schizosaccharomyces pombe), Xenopus laevis, Bombyx mori (silkworm), rice, A. salina, wheat, Trypanosoma cruzi, rabbits, and humans indicates that the proteins have an overall identity of 16.2% and conservative substitutions of 32.9% among all 10 species. The majority of the sequence conservation is limited to the C terminus, where the identity is 25.3% and conservation is 48.5%. This region contains a highly conserved cluster of amino acids, K120SIVTLDVKPWDD132 (Fig. 2). This region is also nearly identical in the metazoan EF-1β-like protein, EF-1δ (34). In humans, the sequences of hEF-1β and hEF-1δ are 100% identical in this region (Fig. 2). At the end of this region lies the ETNL motif, proposed by comparison to the structure of the prokaryotic homolog EF-Ts to play a critical role in nucleotide exchange (18).
FIG. 2.
Conserved cluster of residues in S. cerevisiae, S. pombe (fission yeast), X. laevis, B. mori (silkworm), Oryza sativa (rice), A. salina, wheat germ, T. cruzi, rabbit, and human EF-1β proteins and hEF-1δ. Hyphens indicate residues identical to S. cerevisiae EF-1β.
The human homolog of EF-1β is functional in yeast.
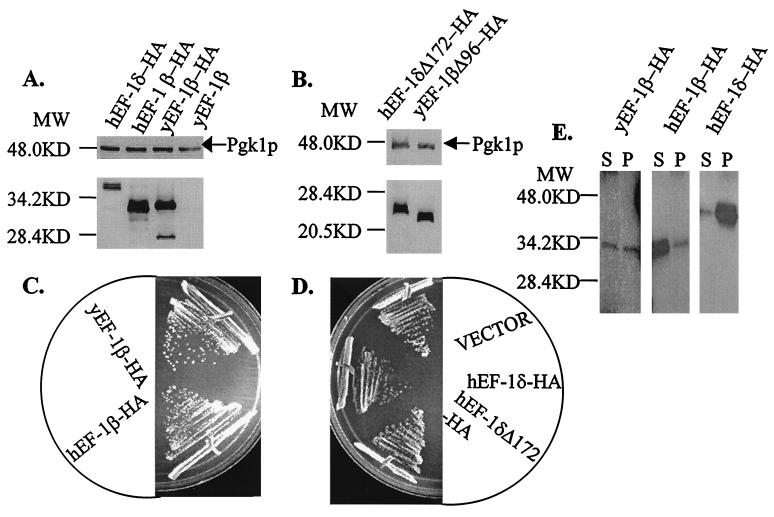
Based on the highly identical sequence cluster in EF-1βs and EF-1δs (Fig. 2), we determined if the human homologs were functional in vivo in yeast. The human cDNAs encoding hEF-1β and hEF-1δ were both stably expressed as HA fusion proteins with the yeast GAL1 promoter (Fig. 3A). hEF-1β functions as the only form of the protein in vivo, and the resulting strains showed no change in growth (Fig. 3C) or sensitivity to translational inhibitors (data not shown). hEF-1δ could neither complement a tef5::TRP1 null allele nor suppress the growth defects of strain TKY238 or TKY243 containing the tef5-1 or tef5-7 allele. Expression of hEF-1δ resulted in slight growth defects of tef5 mutant strains (data not shown) and the isogenic wild-type strain TKY235 (Fig. 3D). To determine if the unique 52-amino-acid N-terminal extension of hEF-1δ was interfering with the function of the protein, a C-terminal fragment of hEF-1δ corresponding to the smallest functional form of yEF-1β was constructed and assayed. The truncated hEF-1δ protein (hEF-1δΔ172-HA) was made (Fig. 3B); however, it was unable to function in place of yEF-1β or partially function to suppress the growth defects of a strain containing a conditional allele of TEF5, TKY238 (tef5-1) or TKY243 (tef5-7). One difference, however, is that hEF-1δΔ172-HA did not adversely affect the growth of mutant strains or an isogenic wild-type strain (Fig. 3D).
FIG. 3.
The human homolog of yEF-1β, but not the hEF-1β-like protein EF-1δ, is functional in yeast. The cDNAs encoding hEF-1β and hEF-1δ were fused to the HA epitope tag and expressed under the control of the yeast GAL1 promoter. (A) Western blot analysis indicates that HA-tagged full-length forms of hEF-1β and hEF-1δ are expressed under the GAL1 promoter. Lanes from left to right are hEF-1δ-HA (JWY4229 with pTKB301), hEF-1β-HA (TKY256), yEF-1β-HA (TKY169), and a strain containing an untagged form of yEF-1β (JWY4229). (B) Truncated hEF-1δ is stably expressed under the control of the GAL1 promoter in yeast. Lanes from left to right are hEF-1δΔ172-HA (JWY4229 with pTKB158) and yEF-1βΔ96-HA (TKY257). (C) Growth of strains containing HA-tagged full-length forms of yEF-1β-HA (TKY169, top) and hEF-1β-HA (TKY256, bottom) on C-Ura-galactose at 30°C for 4 days. (D) Growth of strain JWY4229 expressing full-length and truncated hEF-1δ. The strain contains the chomosomal wild-type TEF5 gene and (from top to bottom) pRS316 (empty vector), hEF-1δ-HA, or hEF-1δΔ172-HA and was grown on C-Ura-galactose at 30°C for 4 days. (E) yEF-1β, hEF-1β, and hEF-1δ coimmunoprecipitate with yEF-1α. Shown are Western blots with anti-HA antibodies of a portion of the supernatants (S) and the entire pellets (P) of an immunoprecipitation of yeast extracts containing yEF-1β-HA (TKY169), hEF-1β-HA (TKY286), and hEF-1δ-HA (TKY4231 plus pTKB151) precipitated with an anti-yEF-1α polyclonal antibody. For panels A, B, and E, MW indicates molecular mass in kilodaltons.
hEF-1β and hEF-1δ physically interact with yEF-1α.
To determine if the inability of hEF-1δ to complement an yEF-1β deficiency was a result of the protein’s inability to interact with yEF-1α, we examined the interaction of these proteins by coimmunoprecipitation. As expected, both yEF-1β and hEF-1β were detected in complexes precipitated by anti-yEF-1α polyclonal antibodies (Fig. 3E, pellet) but were not detected when either a nonspecific polyclonal antibody (against Rpa1p) or no antibody was utilized (data not shown). The association of yEF-1β with yEF-1α was weak, as evidenced by unbound protein in the supernatant. This is not surprising given that this association is transient and that the high content of GTP in the extract will favor EF-1α–GTP complexes. Interestingly, full-length hEF-1δ was also present in the yEF-1α-immunoprecipitated complexes (Fig. 3E). EF-1δ showed a strong association with yEF-1α, as determined by the amount of EF-1δ in the pellet relative to protein in the supernatant.
Mutant alleles of tef5 confer conditional growth defects.
Based on the truncation results and the sequence and functional homology of yEF-1β and hEF-1β, mutagenesis was targeted to the first two identical residues of the conserved cluster in the essential C terminus. By substituting the first two bases of each codon for Lys-120 and Ser-121, 13 or 14 single substitutions are possible at each residue, respectively. Furthermore, many double mutations are possible. PCR mutagenesis and in vivo recombination produced 2,700 colonies. Approximately 500 contained lethal mutations in the TEF5 gene; however, sequence analysis of 12 of these mutations indicated that all were −1 or −2 frameshift mutations (data not shown). Of the remaining 2,200 colonies, 20 conferred cold-sensitive (Cs−) or both temperature-sensitive (Ts−) and Cs− defects. No mutants conferred only a Ts− defect. The sequence substitutions of the tef5-1 through tef5-20 mutant alleles and the phenotypes conferred are shown in Tables 2 (Cs− mutations) and 3 (Ts− mutations).
TABLE 2.
Suppression of the Cs− (13°C) growth defects of tef5 mutant strains by excess copies of the gene encoding the EF-1α subunita
| Allele | Mutation | Phenotype | EF-1α CEN | EF-1α 2μm |
|---|---|---|---|---|
| TEF5 | None | Cs+ | ⇓ | ⇓⇓ |
| tef5-1 | S121L | Cs− | ⇑ | ⇑⇑ |
| tef5-2 | S121I | Cs− | ⇑⇑ | ⇑ |
| tef5-3 | S121N | Cs− | ⇑⇑ | |
| tef5-4 | K120R S121L | Cs− | ⇑ | ⇑⇑ |
| tef5-5 | K120R S121I | Cs− | ⇓ | |
| tef5-6 | K120R S121N | Cs− | (⇑) | |
| tef5-7 | K120R S121Δ I122Δ | Cs− | ⇑⇑ | ⇑ |
| tef5-8 | K120L S121D | Cs− | ⇑ | ⇑⇑ |
| tef5-9 | K120E S121V | Cs− | ⇑⇑ | ⇑⇑ |
| tef5-10 | K120Q S121N | Cs− | ⇑⇑ | ⇑⇑ |
| tef5-11 | K120P S121L | Cs− | ⇑⇑ | ⇑ |
| tef5-12 | K120P S121A | Cs− | (⇑) | |
| tef5-13 | K120M S121G | Cs− | ⇓ | |
| tef5-14 | K120S S121D | Cs− | ⇑⇑ | ⇑ |
| tef5-15 | A119G K120F | Cs− | ⇑ | ⇑ |
| tef5-16 | K120M S121N I122F | Cs− | ⇓ | |
| tef5-17 | K120L S121V I122S | Cs− | ||
| tef5-19 | A119V K120I S121Δ | Cs− | ⇑⇑ | |
| tef5-20 | A118V K120V S121Δ | Cs− | ⇓ |
Growth is scored relative to the same strain containing the URA3 plasmid pRS316 as follows: ⇑⇑, significantly increased growth; ⇑, increased growth; (⇑), slightly increased growth; (⇓), slightly reduced growth; ⇓, reduced growth; and ⇓⇓, significantly reduced growth.
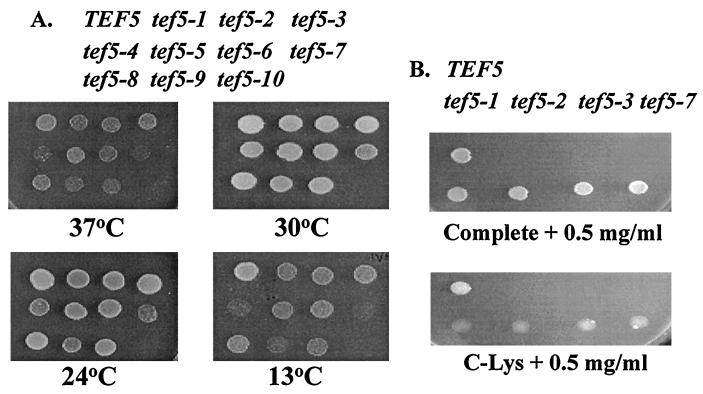
Three single mutations were isolated at S121, resulting in substitution for Leu, Ile, and Asn. Furthermore, three double mutations that contain these three individual substitutions plus the conservative K120R substitution were obtained. The other mutations included a series of double substitutions and two weak Ts− and Cs− triple substitutions (tef5-16 and tef5-17). Several mutations contained one (tef5-18, -19, -20) or two (tef5-7) residue deletions. Further analysis focused on the tef5-1 through tef5-10 alleles. Strains containing these alleles had the tightest Ts− or Cs− phenotypes (Fig. 4A). A slight growth defect was observed for some tef5 mutant strains even at the permissive temperature of 30°C (Fig. 4A), and the doubling time increased slightly from 2.05 ± 0.13 h for a TEF5 strain to 2.26 ± 0.28 h for a tef5-1 strain and statistically significantly for tef5-2, tef5-3, and tef5-7 strains (2.54 ± 0.14, 2.49 ± 0.19, and 3.04 ± 0.23 h, respectively).
FIG. 4.
Growth defects of strains containing the tef5 mutant alleles. (A) Strains containing the lys2-801 allele (UAG) and either the wild-type TEF5 gene or one of the tef5-1 to tef5-10 alleles on a LEU2 CEN plasmid were grown at 30°C, and equal numbers of cells were spotted on YEPD medium. From top left are shown TKY235 (TEF5), TKY238 (tef5-1), TKY240 (tef5-2), TKY244 (tef5-3), TKY236 (tef5-4), TKY242 (tef5-5), TKY251 (tef5-6), TKY243 (tef5-7), TKY239 (tef5-8), TKY241 (tef5-9), and TKY237 (tef5-10). Growth was monitored following 3 to 7 days at 37, 30, 24, or 13°C. (B) Growth was monitored on complete medium–0.5 mg of paromomycin per ml (top) and C-Lys–0.5 mg of paromomycin per ml (bottom) for strains (from top left) TKY235 (TEF5), TKY238 (tef5-1), TKY240 (tef5-2), TKY244 (tef5-3), and TKY243 (tef5-7) following 7 days at 30°C.
Strains containing mutant alleles of TEF5 show sensitivity to translation elongation inhibitors.
The tef5-1 through tef5-10 alleles were transferred by plasmid shuffling to a strain background containing three reporter alleles for translational fidelity (met2-1 and his4-713 for a +1 frameshift and lys2-801 for nonsense suppression) for all further studies. These strains were analyzed for effects on elongation by assaying sensitivity to the aminoglycosides paromomycin and hygromycin B, the elongation inhibitor cycloheximide, and the prokaryotic translation inhibitors streptomycin and kanamycin. All 10 mutant alleles showed significant increases in sensitivity to cycloheximide, hygromycin B, and paromomycin (Table 4). The S121L and S121I single mutations showed a further increased sensitivity to all three drugs when combined with the K120R substitution. Interestingly, the S121N mutation was less sensitive to translation inhibitors in combination with the K120R substitution. All effects were specific to eukaryotic elongation inhibitors, since no strains showed sensitivity to streptomycin or kanamycin (data not shown).
TABLE 4.
Drug sensitivity of strains containing tef5 allelesa
| Allele | Mutation | Paros | Hygros | Cyclos |
|---|---|---|---|---|
| TEF5 | None | 2.25 | 4.25 | 10 |
| tef5-1 | S121L | 4.5 | 5.5 | 12.25 |
| tef5-2 | S121I | 4 | 5.5 | 14 |
| tef5-3 | S121N | 4 | 6 | 13.25 |
| tef5-4 | K120R S121L | 4.5 | 7.25 | 14 |
| tef5-5 | K120R S121I | 4.5 | 6.5 | 16 |
| tef5-6 | K120R S121N | 3.75 | 5.75 | 10.75 |
| tef5-7 | K120R S121Δ I122Δ | 6.25 | 6 | 14.5 |
| tef5-8 | K120L S121D | 4.25 | 5.75 | 11.5 |
| tef5-9 | K120E S121V | 5.25 | 7.75 | 15.5 |
| tef5-10 | K120Q S121N | 4.5 | 7 | 14.25 |
Sensitivity is determined by measuring the radius of inhibition of growth of a lawn of cells (in millimeters) around a filter disc containing 10 μl of 48 mM paromomycin (Paro), 5 mM hygromycin B (Hygro), or 2.5 mM cycloheximide (Cyclo).
Strains containing tef5 mutant alleles are suppressed by excess yEF-1α but not yEF-1γ.
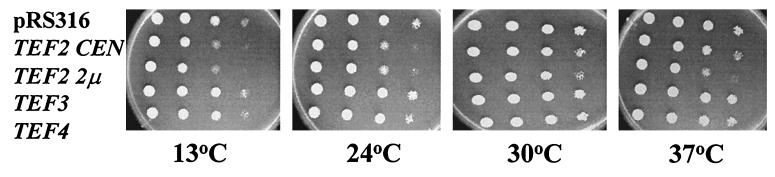
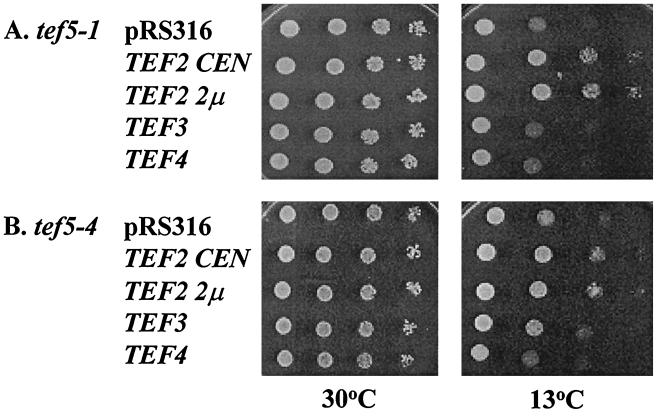
In order to understand the effects of yEF-1β mutations on the interaction with the substrate for the guanine nucleotide exchange reaction, yEF-1α, we determined if excess yEF-1α would suppress the conditional growth defects of a tef5 mutant strain. Both low (CEN)- and high (2μm)-copy plasmids containing the TEF2 gene encoding yEF-1α were transformed into derivative strains of JWY4200 containing 1 of the 20 tef5 mutant alleles. As a control, excess yEF-1α was placed in the TEF5 wild-type strain. Interestingly, excess yEF-1α resulted in conditional growth defects at 13°C and to a lesser extent 37°C (Fig. 5). Thus, any enhanced growth of the tef5 mutant strains at the nonpermissive temperature of the mutant alleles indicated both suppression of the conditional growth defects conferred by the tef5 mutation and a lack of negative consequences of the presence of excess yEF-1α.
FIG. 5.
Excess copies of the gene encoding yEF-1α but not of that encoding yEF-1γ (TEF3 and TEF4, respectively) result in a conditional growth defect in a wild-type strain. Strain TKY235 (TEF5 LEU2) was transformed with URA3 plasmids containing no TEF gene (pRS316), TEF2 CEN, TEF2 2μm, TEF3 CEN, and TEF4 CEN. The strains were grown at 30°C in liquid C-Ura, and equal numbers of cells for each were spotted in 1/10 serial dilutions and grown on C-Ura at 13, 24, 30, and 37°C.
Tables 2 and 3 show the results of overexpression of yEF-1α on these strains. None of the mutant strains with a Ts− growth defect was suppressed by excess yEF-1α (Table 3), although a few showed less severe growth defects associated with EF-1α overexpression compared to the TEF5 wild-type strain (tef5-1, -5, -8, and -18). Strains containing several tef5 mutant alleles were less sensitive to the Cs− defects caused by overexpression of yEF-1α (tef5-5, -13, -16, -17, and -20 [Table 2]). Additionally, some strains grew well at 13°C and thus did not show the growth defects conferred by excess yEF-1α and suppressed the conditional defect conferred by the tef5 allele (tef5-1-4, 7-12, -14, and -19). In some cases, suppression was dosage dependent, better with high- than with low-copy yEF-1α (strains containing tef5-1, tef5-4, and tef5-8 [Fig. 6 and data not shown]). Strains with tef5-9 and tef5-10 showed a high level of suppression with either the low- or the high-copy TEF2 plasmid. Overall, strains with 1 of the 20 different mutations in EF-1β are less sensitive to the negative effects of overexpressing EF-1α. As opposed to excess yEF-1α, there was no effect of an extra copy of either the TEF3 or the TEF4 gene encoding yEF-1γ proteins on a wild-type strain (Fig. 5) or any tef5 mutant strain (Fig. 6 and data not shown).
TABLE 3.
Suppression of the Ts− (37°C) growth defects of tef5 mutant strains by excess copies of the gene encoding the EF-1α subunita
| Allele | Mutation | Phenotype | EF-1α CEN | EF-1α 2μm |
|---|---|---|---|---|
| TEF5 | None | Ts+ | ⇓ | ⇓⇓ |
| tef5-1 | S121L | Ts− | ||
| tef5-2 | S121I | (Ts−) | ⇓ | ⇓⇓ |
| tef5-3 | S121N | (Ts−) | ⇓ | ⇓ |
| tef5-4 | K120R S121L | Ts− | ⇓ | ⇓⇓ |
| tef5-5 | K120R S121I | (Ts−) | ||
| tef5-6 | K120R S121N | (Ts−) | (⇓) | |
| tef5-7 | K120R S121Δ I122Δ | Ts− | ⇓ | ⇓⇓ |
| tef5-8 | K120L S121D | (Ts−) | ||
| tef5-9 | K120E S121V | (Ts−) | ⇓ | ⇓ |
| tef5-10 | K120Q S121N | Ts− | (⇓) | (⇓) |
| tef5-15 | A119G K120F | (Ts−) | ⇓ | |
| tef5-16 | K120M S121N I122F | Ts− | ⇓ | ⇓⇓ |
| tef5-17 | K120L S121V I122S | Ts− | ⇓⇓ | |
| tef5-18 | A119Δ K120W S121Y | Ts− |
Growth is scored relative to the same strain containing the URA3 plasmid pRS316 as follows: (⇓), slightly reduced growth; ⇓, reduced growth; and ⇓⇓, significantly reduced growth. (Ts−), slight Ts− growth defect.
FIG. 6.
Suppression of the Cs− defects of mutant strains containing tef5-1 (TKY238 S121L) (A) and tef5-4 (TKY236 K120R S121L) (B) by excess yEF-1α. Plasmids bearing the URA3 marker and no TEF gene (pRS316), TEF2 CEN, TEF2 2μm, TEF3 CEN, and TEF4 CEN were transformed into the strain and grown in C-Ura. Equal numbers of cells for each strain were spotted in 1/10 serial dilutions and grown on C-Ura at 30 and 13°C.
Mutations in yEF-1β enhance the fidelity of decoding nonsense codons.
One possible explanation for the slow growth of the yEF-1β mutant strains is a general slowing of translation. This is also consistent with the model that guanine nucleotide exchange is the rate-limiting step in elongation. Based on models of translational fidelity and the elongation rate, this would result in enhanced translational fidelity at the A site of the ribosome (22). We used two assays to monitor nonsense suppression. Strains containing wild-type TEF5 or the tef5-1, -2, -3, or -7 allele were grown on C-Lys and C-Lys–0.5 mg of paromomycin per ml to monitor suppression of the lys2-801 (UGA) allele. Slightly better growth was seen for the wild-type strain than for the mutant strains on C-Lys. The paromomycin-induced reduced fidelity allowed the wild-type TEF5 strain, but not the mutant strains, to grow on C-Lys–0.5 mg of paromomycin per ml (Fig. 4B). This effect was not due to the paromomycin sensitivity of these strains, since no growth defect was observed on complete medium containing 0.5 mg of paromomycin per ml. We further quantitatively assayed the ability of these strains to read a UAG, UAA, or UGA stop codon with lacZ reporter constructs. Strains containing the wild-type TEF5 and tef5-1, -2, -3, and -7 alleles were assayed for the production of β-Gal from lacZ constructs containing an in-frame UAA, UAG, or UGA codon (Table 5). Compared to a strain with wild-type TEF5, all mutants showed less readthrough of the three nonsense codons, or enhanced fidelity. The largest effect, a 10-fold reduction in reading of a UAG stop codon, was seen for a strain containing the tef5-7 allele. This effect was not due to any changes in the level of the lacZ mRNA (data not shown).
TABLE 5.
Reading of nonsense codons in strains containing tef5 alleles
| Allele | Mutation | % Misreadinga
|
||
|---|---|---|---|---|
| UAA | UAG | UGA | ||
| TEF5 | None | 0.88 ± 0.29 | 1.3 ± 0.36 | 1.2 ± 0.66 |
| tef5-1 | S121L | 0.28 ± 0.12 | 0.55 ± 0.08 | 0.33 ± 0.17 |
| tef5-2 | S121I | 0.32 ± 0.22 | 0.31 ± 0.06 | 0.36 ± 0.16 |
| tef5-3 | S121N | 0.29 ± 0.11 | 0.24 ± 0.06 | 0.48 ± 0.30 |
| tef5-7 | K120R S121Δ I122Δ | 0.21 ± 0.08 | 0.18 ± 0.07 | 0.35 ± 0.17 |
Misreading was monitored by determining the β-galactosidase activity in cells containing either a wild-type lacZ reporter gene or a variant containing the indicated stop codon in frame with lacZ. Percent misreading is the β-galactosidase activity for the indicated nonsense reporter divided by the activity for the wild-type lacZ reporter.
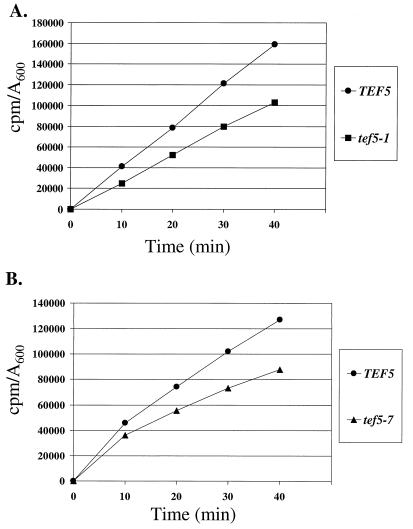
Based on this phenotype, total translation was monitored by measuring total methionine incorporation in strains containing either wild-type TEF5 or the tef5-1 or tef5-7 mutant alleles. Incorporation assays were performed at the permissive temperature of 30°C for comparison to the conditions used to monitor translational fidelity. Both the tef5-1 and tef5-7 mutant strains showed an approximately 30% decrease in the incorporation of [35S]methionine over 40 min of growth at the permissive temperature (Fig. 7). Thus, mutations in the highly conserved motif of EF-1β dramatically affect total translation in the cell.
FIG. 7.
Total methionine incorporation in strains containing wild-type TEF5 (TKY235, circles), the tef5-1 allele (TKY238, squares) (A), or the tef5-7 allele (TKY243, triangles) (B). Strains were grown to mid-log phase in C-Met and labeled for varying times in [35S]methionine. Incorporation (in counts per minute) is expressed per A600 unit of cells.
DISCUSSION
The β subunit of the EF-1 complex has demonstrated activity as the guanine nucleotide exchange factor for the G protein EF-1α (6, 38). For EF-1β as a guanine nucleotide exchange factor, models of G protein regulation allow several predictions of the effects of changes in EF-1β activity. First, although active EF-1α–GTP can be regenerated by the spontaneous release of GDP, EF-1β enhances the rate of this reaction. Thus, EF-1β contributes to a larger pool of active EF-1–GTP, making possible increased rates of translation elongation. Deletion of EF-1β should thus slow elongation, likely below the threshold for viability, consistent with the essential nature of the TEF5 gene encoding yEF-1β (14). Second, as the substrate for EF-1β, excess EF-1α should be able to at least partially compensate for the loss of EF-1β activity, which we have previously demonstrated for yeast (20).
A third prediction is that a critical function catalyzed by EF-1β should be conserved, which we find by both sequence and function conservation. Analysis of the sequence identity among 10 different EF-1β proteins from many different species clearly supports the importance of the C terminus of this protein in its function in vivo. We demonstrate that an EF-1β fragment containing the C terminus is able to function as the only form of the protein in vivo, with no associated growth defects. While previous studies have indicated that a C-terminal protease fragment of A. salina EF-1β containing residues 106 to 206 maintains guanine nucleotide exchange activity in vitro (40, 41), these experiments demonstrate that this region is sufficient for normal growth and results in no sensitivity to translation inhibitors or changes in polyribosome content or distribution.
The human homolog of EF-1β is also functional in vivo in yeast. Expression of the hEF-1β-like protein hEF-1δ is unable to complement the lack of yEF-1β in vivo, which may not be surprising since the level of full-length and especially truncated hEF-1δ expressed was lower than that of hEF-1β. It is more surprising that the hEF-1δ was unable to even partially suppress the conditional growth defects of strains containing mutant forms of yEF-1β and actually showed a slight negative effect on cell growth. Since hEF-1δ does strongly physically interact with yEF-1α, as shown by coimmunoprecipitation (Fig. 3E), this negative growth effect is likely caused by dominantly interfering with the function of yEF-1β. Future analysis of any potential dependence of this association on EF-1β and EF-1γ, with strains deficient in these subunits, will provide insight into the EF-1 complex.
Removal of the more divergent N-terminal region of hEF-1δ negates this effect, likely by reducing the affinity for yEF-1α and relieving inhibition of yEF-1β function. Association of neither hEF-1δΔ172-HA nor yEF-1βΔ96-HA with yEF-1α could be detected by coimmunoprecipitation (data not shown). Thus, the cell tolerates both reduced levels of EF-1β protein, as seen for the truncated forms of yEF-1β or the constructs expressed from the GAL1 promoter (Fig. 1), and reduced association with EF-1α, as seen for the truncations (Fig. 3), while still allowing normal growth. While hEF-1δ may possess guanine nucleotide exchange activity in vitro and the conservation of sequence allows association with yEF-1α, the small number of changes between hEF-1β and hEF-1δ, which are 85.3% identical in the conserved C-terminal 109 amino acids, are clearly important for EF-1β function. It appears that EF-1δ may be specific to metazoans, perhaps by functioning in the assembly of the higher-order aa-tRNA synthetase complexes found in these organisms (2), but provides an important tool for analyzing the functional differences between EF-1β and EF-1δ.
Mutations in the conserved C terminus of EF-1β affect both the efficiency and the accuracy of translation elongation. Strains with mutations in residues K120 and S121 of yEF-1β show severe growth defects and sensitivities to elongation inhibitors relative to a wild-type strain. These results indicate that mutations in yEF-1β alter translation elongation, either directly or through the activity of yEF-1α. According to the nuclear magnetic resonance structure of the C terminus of hEF-1β, residues K120 and S121 lie at the end of a β-sheet opposite the loops predicted to play critical roles in guanine nucleotide exchange (28). Thus, the in vivo analysis of yEF-1β has yielded new insight into residues not predicted to play a critical role in the function of this protein. It is of particular interest that the Cs− defects of strains containing many of the yEF-1β mutations are suppressed by excess yEF-1α. No effects are seen on the Ts− mutant phenotype. Cs− defects are often associated with reduced complex formation; thus, these mutations may reduce the interaction between EF-1α and EF-1β. Alternatively, if EF-1β activity is limiting, the presence of excess EF-1α may allow for growth by a mechanism similar to that seen when EF-1α is overexpressed in cells completely lacking EF-1β (20). These effects are different from the growth seen when EF-1β activity is totally bypassed by expression of a third copy of an EF-1α gene (20). EF-1β-deficient strains with three copies of EF-1α are extremely slow growing, while examples such as the tef5-1 allele shown in Fig. 6 are suppressed to wild-type levels of growth.
The critical role of EF-1β in maintaining a pool of active EF-1α–GTP supports the prediction that these mutations would alter translational fidelity. An additional phenotype of strains containing a mutant allele of TEF5 is enhanced sensitivity to paromomycin, which is often predictive of effects on translational fidelity (26, 37). The lack of readthrough of nonsense mutations in a lacZ reporter construct and the lys2-801 (UGA) allele clearly demonstrates that mutations in the highly identical K120 and S121 residues enhance the fidelity of nonsense recognition in yeast. Many mutations in tRNA and ribosomal protein genes that reduce translational fidelity and suppress nonsense codons have been isolated (13). Most mutations that enhance fidelity were isolated and analyzed as antisuppressors of strains bearing suppressor mutations (13). The effect of the EF-1β mutants is seen for all three nonsense codons, indicating that these mutants show omnipotent hyperaccuracy. This phenotype is similar to the antisuppressor phenotypes that result from overexpression of Sup35p (eRF3) and Sup45p (eRF1) (39) and some rRNA mutations (8) in yeast. Thus, a potential mechanism for the increased fidelity at stop codons may be more efficient competition for recognition of the stop codon by release factors due to an increased ratio of the less abundant release factors to active EF-1α.
It is interesting to compare these mutants to the restrictive mutants of bacteria and corresponding mutations in yeast (1, 12). Restrictive mutants enhance translational fidelity by lowering the stability of the ribosome–aa-tRNA interaction, resulting in an increased aa-tRNA discard rate (22). The prediction of such mutations is that any increase in translational fidelity would require reduced growth rates and a lower translational efficiency (22). E. coli ribosome mutants with altered processivity also show increased accuracy (10). These predictions are supported by the growth phenotypes and the reduced total translation of strains containing the tef5-1 or tef5-7 mutant allele. Thus, EF-1β activity is likely limiting for translation elongation, such that mutations that alter EF-1β activity slow total translation and consequently enhance translational fidelity. Clearly, EF-1β plays an important role in the efficiency and accuracy of the translation elongation process. Further genetic and biochemical analysis of these mutants will provide insight into factors that may regulate translation elongation, such as kinases (7, 16, 29), or interactions with other components of the translational apparatus such as the ribosome or release factors.
ACKNOWLEDGMENTS
We thank Mark Sandbaken and Mike Culbertson for providing the TEF2 plasmids, Stuart Peltz and Jonathan Dinman for providing reporter plasmids and comments on the manuscript, and John L. Woolford for initial support for this project. All sequencing was performed in the UMDNJ DNA Synthesis and Sequencing Laboratory, with thanks to Regina Felder and Sheila Mazar.
This work was funded by grants to T.G.K. from the New Jersey Affiliate of the American Heart Association and the National American Heart Association and a predoctoral fellowship to A.C.-S. from the New Jersey Affiliate of the American Heart Association. L.V. and T.W. were supported by NIH training grant no. 5R25GM55145.
REFERENCES
- 1.Alksne L E, Anthony R A, Liebman S W, Warner J R. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci USA. 1993;90:9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bec G, Kerjan P, Waller J-P. Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1βγδ complex. J Biol Chem. 1994;269:2086–2092. [PubMed] [Google Scholar]
- 3.Belfield G P, Tuite M F. Translation elongation factor 3: a fungal-specific translation factor? Mol Microbiol. 1993;9:411–418. doi: 10.1111/j.1365-2958.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 5.Botstein D, Falco S C, Stewart S E, Brennan M, Scherer S, Stinchcomb D T, Struhl K, Davis R W. Sterile host yeast (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho M G, Carvalho J F, Merrick W C. Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch Biochem Biophys. 1984;234:603–611. doi: 10.1016/0003-9861(84)90310-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen C-J, Traugh J A. Expression of recombinant elongation factor 1 beta from rabbit in Escherichia coli. Phosphorylation by casein kinase II. Biochim Biophys Acta. 1995;1264:303–311. doi: 10.1016/0167-4781(95)00166-2. [DOI] [PubMed] [Google Scholar]
- 8.Chernoff Y O, Vincent A, Liebman S W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinman J D, Kinzy T G. Translational misreading: mutations in translation elongation factor 1α differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA. 1997;3:870–881. [PMC free article] [PubMed] [Google Scholar]
- 10.Dong H, Kurland C G. Ribosome mutants with altered accuracy translate with reduced processivity. J Mol Biol. 1995;248:551–561. doi: 10.1006/jmbi.1995.0242. [DOI] [PubMed] [Google Scholar]
- 11.Farabaugh P J, Vimaladithan A. Effect of frameshift-inducing mutations of elongation factor 1α on programmed +1 frameshifting in yeast. RNA. 1998;4:38–46. [PMC free article] [PubMed] [Google Scholar]
- 12.Gorini L. Streptomycin and misreading of the genetic code. In: Nomura M, Tissieres A, Lengyel P, editors. Ribosomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. pp. 791–803. [Google Scholar]
- 13.Hinnebusch A, Liebman S W. Protein synthesis and translational control in Saccharomyces cerevisiae. In: Broach J R, Jones E W, Pringle J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Genome dynamics, protein synthesis and energetics. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 627–735. [Google Scholar]
- 14.Hiraga K, Suzuki K, Tsuchiya E, Miyakawa T. Cloning and characterization of the elongation factor EF-1β homologue of Saccharomyces cerevisiae. EF-1β is essential for growth. FEBS Lett. 1993;316:165–169. doi: 10.1016/0014-5793(93)81208-h. [DOI] [PubMed] [Google Scholar]
- 15.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen G M C, Maessen G D F, Amons R, Moller W. Phosphorylation of elongation factor 1β by an exogenous kinase affects its catalytic nucleotide exchange activity. J Biol Chem. 1988;263:11063–11066. [PubMed] [Google Scholar]
- 17.Johnston M, Davis R. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R. The structure of the Escherichia coli EF-Tu · EF-Ts complex at 2.5Å resolution. Nature. 1996;379:511–518. doi: 10.1038/379511a0. [DOI] [PubMed] [Google Scholar]
- 19.Kinzy T G, Ripmaster T R, Woolford J L., Jr Multiple genes encode the translation elongation factor EF-1γ in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:2703–2707. doi: 10.1093/nar/22.13.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinzy T G, Woolford J L., Jr Increased expression of Saccharomyces cerevisiae translation elongation factor EF-1α bypasses the lethality of a TEF5 null allele encoding EF-1β. Genetics. 1995;141:481–489. doi: 10.1093/genetics/141.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 22.Kurland C G, Jorgensen F, Richter A, Ehrenberg M, Bilgin N, Rojas A-M. Through the accuracy window. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome. Structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 513–526. [Google Scholar]
- 23.Liebman S W, Chernoff Y O, Liu R. The accuracy center of a eukaryotic ribosome. Biochem Cell Biol. 1995;73:1141–1149. doi: 10.1139/o95-123. [DOI] [PubMed] [Google Scholar]
- 24.Maddock J R, Weidenhammer E M, Adams C C, Lunz R L, Woolford J J L. Extragenic suppressors of Saccharomyces cerevisiae prp4 mutations identify a negative regulator of PRP genes. Genetics. 1994;136:833–847. doi: 10.1093/genetics/136.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortimer R K, Hawthorne D C. Genetic mapping in Saccharomyces. Genetics. 1966;53:165–173. doi: 10.1093/genetics/53.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer E, Wilhelm J M, Sherman F. Phenotypic suppression of nonsense mutants in yeast by aminoglycoside antibiotics. Nature. 1979;277:148–150. doi: 10.1038/277148a0. [DOI] [PubMed] [Google Scholar]
- 27.Perez J M J, Kriek J, Dijk J, Canters G W, Moller W. Expression, purification and spectroscopic studies of the guanine nucleotide exchange domain of human elongation factor, EF-1β. Protein Expr Purif. 1998;13:259–267. doi: 10.1006/prep.1998.0895. [DOI] [PubMed] [Google Scholar]
- 28.Perez J M J, Siegal G, Kriek J, Dijk J, Canters G W, Moller W. The solution structure of the guanine nucleotide exchange domain of human elongation factor 1β reveals a striking resemblance to that of EF-Ts from Escherichia coli. Structure. 1999;7:217–226. doi: 10.1016/s0969-2126(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 29.Peters H I, Chang Y-W E, Traugh J A. Phosphorylation of elongation factor 1 (EF-1) by protein kinase C stimulates GDP/GTP-exchange activity. Eur J Biochem. 1995;234:550–556. doi: 10.1111/j.1432-1033.1995.550_b.x. [DOI] [PubMed] [Google Scholar]
- 30.Riis B, Rattan S I S, Clark B F C, Merrick W C. Eukaryotic protein elongation factors. Trends Biol Chem. 1990;15:420–424. doi: 10.1016/0968-0004(90)90279-k. [DOI] [PubMed] [Google Scholar]
- 31.Saha S K, Chakraburtty K. Protein synthesis in yeast. Isolation of variant forms of elongation factor 1 from the yeast Saccharomyces cerevisiae. J Biol Chem. 1986;261:12599–12603. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sandbaken M G, Culbertson M R. Mutations in elongation factor EF-1α affect the frequency of frameshifting and amino acid misincorporation in Saccharomyces cerevisiae. Genetics. 1988;120:923–934. doi: 10.1093/genetics/120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders J, Raggiaschi R, Morales J, Moller W. The human leucine zipper-containing guanine-nucleotide exchange protein elongation factor-1δ. Biochim Biophys Acta. 1993;1174:87–90. doi: 10.1016/0167-4781(93)90097-w. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 36.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in S. cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh A, Ursic D, Davies J. Phenotypic suppression and misreading in Saccharomyces cerevisiae. Nature. 1979;277:146–148. doi: 10.1038/277146a0. [DOI] [PubMed] [Google Scholar]
- 38.Slobin L I, Moller W. Purification and properties of an elongation factor functionally analogous to bacterial elongation factor Ts from embryos of Artemia salina. Eur J Biochem. 1978;84:69–77. doi: 10.1111/j.1432-1033.1978.tb12142.x. [DOI] [PubMed] [Google Scholar]
- 39.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Damme H, Amons R, Janssen G, Moller W. Mapping the functional domains of the eukaryotic elongation factor 1βγ. Eur J Biochem. 1991;197:505–511. doi: 10.1111/j.1432-1033.1991.tb15938.x. [DOI] [PubMed] [Google Scholar]
- 41.van Damme H T F, Karssies R, Timmers C J, Janssen G M C, Moller W. Elongation factor 1β of artemia: localization of functional sites and homology to elongation factor 1δ. Biochim Biophys Acta. 1990;1050:241–247. doi: 10.1016/0167-4781(90)90174-z. [DOI] [PubMed] [Google Scholar]