Abstract
Background
Little is known about the role of DNA methylation (DNAm) epigenetic age acceleration in cognitive decline. Using a twin study design, we examined whether DNAm age acceleration is related to cognitive decline measured longitudinally in persons without a clinical diagnosis of dementia.
Methods
We studied 266 paired male twins (133 pairs) with a mean age of 56 years at baseline. Of these, 114 paired twins returned for a follow-up after an average of 11.5 years. We obtained 6 indices of DNAm age acceleration based on epigenome-wide data from peripheral blood lymphocytes. At both baseline and follow-up, we administered a battery of cognitive measures and constructed 2 composite scores, one for executive function and one for memory function. We fitted multivariable mixed regression models to examine the association of DNAm age acceleration markers with cognitive function within pairs.
Results
In cross-sectional analyses at baseline, there was no association between DNAm age acceleration and cognitive function scores. In longitudinal analyses, however, comparing twins within pairs, each additional year of age acceleration using the Horvath’s method was associated with a 3% decline (95% CI, 1%–5%) in the composite executive function score and a 2.5% decline (95% CI, 0.01%–4.9%) in the memory function score. These results did not attenuate after adjusting for education and other risk factors.
Conclusions
Middle-aged men who had older DNAm age relative to their brothers of the same demographic age showed a faster rate of cognitive decline in the subsequent 11.5 years. These results point to the role of epigenetic modifications in cognitive aging.
Keywords: Biomarkers, Cognitive aging, Epidemiology, Epigenetics, Twin studies
Background
Alzheimer’s disease-related dementias have a long preclinical phase during which typical pathologies develop (eg, amyloid plaques and neurofibrillary tangles) prior to the appearance of clinical symptoms. Thus, a key step for our ability to understand the pathophysiology of these conditions and to effectively prevent them is the identification of factors related to cognitive decline and dementia during the preclinical and early clinical stages. Several biomarkers have been associated with Alzheimer’s disease even decades before the onset of decline in cognitive function (1,2), and genetic variation is also known to contribute (3). However, age remains the strongest known risk factor for cognitive decline and dementias, unexplained by other risk factors or health conditions associated with aging (4). Understanding the molecular mechanisms through which the aging process affects susceptibility to cognitive impairment could provide important clues for the etiology of these neurodegenerative disorders and the identification of potentially modifiable risk factors.
Several studies have linked the aging process to epigenetic modifications across the genome (5,6). Epigenetic processes regulate gene expression and genome integrity through changes that are independent of DNA sequence (7) and may allow the identification of reversible molecular mechanisms through which exposures influence the expression of complex phenotypes. DNA methylation (DNAm), the addition of methyl groups at the 5′ position of cytosine rings in CpG dinucleotides to produce 5-methyl-cytosine, is the best known type of epigenetic modification (7).
Mathematical algorithms can estimate the age (in units of years) of cells, tissues, or organs, using DNAm (8,9). Using these DNAm age estimators, often referred to as “epigenetic clocks” (10), one can calculate DNAm age acceleration as the difference between DNAm age and chronological age (8–10). DNAm age acceleration represents a measure of the discrepancy between chronological and biological age and correlates with age-related physiological dysregulation in multiple tissues (9–11) as well as with premature mortality (12).
While various DNAm sites have been associated with cognitive performance (13) and neurodegenerative disorders including Alzheimer’s disease (14,15), less is known about the role of epigenetic clocks or epigenetic age acceleration in cognitive decline. Furthermore, very few studies have examined cognitive decline longitudinally. Thus, whether epigenetic dysregulation is an epiphenomenon or a causal factor in cognitive aging remains to be elucidated (16).
Epigenetic association studies are potentially confounded by environmental, demographic, and genetic factors. A twin study overcomes these weaknesses and thus is regarded as the most powerful design to study epigenetics (17). Twins are matched for age, the most important correlate of DNAm age. They also share in utero environment and early familial influences (eg, diet, socioeconomic, and parental factors), which contribute to epigenetic modifications and the expression of complex traits. Monozygotic (MZ) twins also carry identical genetic information from the primary sequence of DNA; dizygotic (DZ) twins, however, are also informative, as they are matched for the same characteristics as MZ except that they share half of their genes. Because epigenetic modifications are heritable, mostly through DNA variations at the CpG sites (18), a comparison of results between MZ and DZ twins provides estimates of the influence of genetic versus environmental influences on the associations. Using a twin study design, the purpose of the current investigation was to examine whether biological aging, estimated using a comprehensive set of DNAm age acceleration indices, is related to cognitive decline measured longitudinally among middle-aged and older twins without a clinical diagnosis of dementia.
Method
Study Cohort
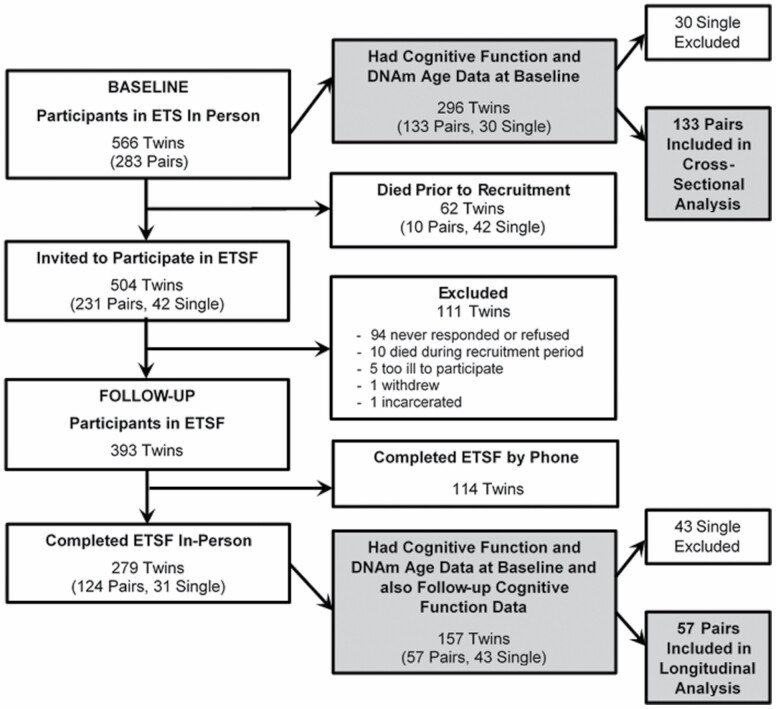
The participants in this study were recruited from the Vietnam Era Twin (VET) Registry, which is one of the largest national samples of adult male twin veterans who served on active duty during the Vietnam war era (1964–1975) (19). The present study is based on the Emory Twin Study (ETS) (20). For ETS, from VET Registry, we selected 283 MZ and DZ twin pairs (n = 566) born between 1946 and 1956 (representing 95% of all registry twins) who were discordant for depression or posttraumatic stress disorder (PTSD), as well as control pairs without these conditions. Twin pairs were excluded if either member of the twin pair had a history of cardiovascular disease at the baseline survey (21). Of the 283 ETS twin pairs, we invited 504 twins who were still alive (252 pairs) to participate in the Emory Twin Study Follow-up (ETSF) for a follow-up evaluation, in-person or over the phone (for those who were not able to travel), that was conducted on average 11.5 years after the initial ETS. A total of 393 twins have participated so far in ETSF (78%), and among them, 279 twins (including 124 pairs and 31 single twins) completed the in-person visit. Figure 1 shows the construction of the study population.
Figure 1.
Participant flow diagram. ETS = Emory Twin Study; ETSF = Emory Twin Study Follow-up; DNAm = DNA methylation.
At both in-person visits, all twin pairs were examined together at Emory University on the same day to minimize measurement error. Medical history, anthropometric measurements, behavioral and psychosocial measures, and cognitive performance were collected using identical protocols for the 2 twins. Zygosity data were obtained by DNA typing. We obtained written informed consent from all twins, and the Emory University institutional review board approved this study.
Genome-Wide DNA Methylation
At the baseline visit, we obtained specimens of whole blood for DNA extraction. We conducted genome-wide DNA methylation using the Illumina Infinium MethylationEPIC (850K) BeadChip, which builds upon the Infinium HumanMethylation450 BeadChip and measures more than 850 000 methylation sites in peripheral blood lymphocytes (22). The 850K chip includes 99% of the NCBI Reference Sequence (RefSeq) genes, with sites covering the promoter, untranslated regions, and gene body, more than 95% of CpG islands, and differentially methylated sites across tissue types.
For the Illumina Infinium I assay, fluorescent signals of each CpG site were measured from both site-specific methylated and unmethylated beads. For the Infinium II assay, a single bead type of each CpG site was measured for methylation status. We adopted a quantile normalization approach implemented in the R package “minfi” (23) for processing 850K data to correct for known technical shifts in methylation signals between the Illumina Infinium I and II probes and to generate adjusted β values for the association analyses. We set any results with a detection p value greater than .01 to missing and excluded 4 samples with more than 10% missing rate of methylation data. We also excluded CpG sites with more than a 5% missing rate. After all quality control procedures, a total of 846 459 CpG sites were kept for the analysis of DNAm age.
Estimation of DNA Methylation Age and Age Acceleration
We calculated 4 DNAm age estimators using the online calculator developed by Horvath’s group (https://dnamage.genetics.ucla.edu) (9). These included Horvath’s epigenetic clock (9), Hannum’s epigenetic clock (8), DNAm Pheno Age (11), and DNAm Grim Age (24). Horvath’s epigenetic clock is based on 353 CpGs and works well across different cell types and tissues. Hannum’s clock is based on 71 CpGs in leukocytes and is blood-specific. Both Horvath’s and Hannum’s DNAm biological age estimators were developed as predictors of chronological age as a surrogate of biological age. In contrast, DNAm Pheno Age, including 513 methylation sites, was developed as a predictor of aging-related conditions and overall mortality (denoting “phenotypic age”). After quality control, 334 out of 353 and 65 out of 71 CpG sites were available for the calculation of the Horvath and Hannum predicted methylation ages, respectively. All 513 CpG sites were available for the calculation of DNAm Pheno Age. The recent Grim Age estimator was developed using a 2-stage approach that first considered DNAm-based surrogate markers of smoking and other biomarkers of mortality, and, in a second stage, time to death. All these DNAm age biomarkers have been related to life span and age-related conditions (10,11,24).
From the DNAm age estimators above, we calculated DNAm age acceleration markers from the residuals resulting from regressing DNAm age estimators on chronological age in a linear model. All these DNAm age acceleration markers are independent of chronological age. We obtained Horvath’s DNAm age acceleration (Horvath AA), Hannum’s DNAm age acceleration (Hannum AA), Pheno age acceleration (Pheno AA), and Grim age acceleration (Grim AA). All these were calculated without adjustment for cell types. We also calculated 2 more indices that were adjusted for cell types: intrinsic epigenetic age acceleration (IEAA), from Horvath’s DNAm age, and extrinsic epigenetic age acceleration (EEAA), from Hannum’s DNAm age (12). In order to adjust for cell type, we calculated the proportions of blood cell subtypes (B cells, granulocytes, monocytes, natural killer cells, CD4, and CD8 T cells) using the method developed by Houseman et al. (25).
Measurements of Cognitive Function
At both the in-person baseline and in-person follow-up visits, we obtained measures of executive functioning and cognitive flexibility using the Trail Making Test (TMT). The TMT is a validated measure of working memory, which is administered in 2 parts (26). TMT-Part A is a timed visual-scanning task where participants are asked to connect 25 circles with lines as quickly as possible, while in TMT-Part B participants are asked to connect circles containing numbers or letters in an alternate numeric/alphabetical order (ie, 1-A, 2-B, 3-C, etc.). We recorded the time in seconds for participants to complete each task. Per standard procedures, TMT-A was discontinued at 150 seconds and TMT-B was discontinued at 300 seconds. Both tasks measure executive functioning, and TMT-B additionally measures cognitive flexibility, which reflects the ability to switch between different concepts simultaneously. Both TMT-A and TMT-B scores, in seconds, were log-transformed due to non-normality and then converted to Z scores and averaged together to obtain a composite normalized executive function score, following a common approach (27,28). To facilitate interpretation of results, the analysis was repeated by simply averaging the TMT-A and TMT-B scores. A higher score is indicative of worse executive function.
At both time points, we also assessed memory function using the Wechsler Memory Scale-Revised (WMS-R) (29). The WMS Logical Memory subtest measures immediate and 30-minute delayed recall of the elements in 2 different stories that were read aloud to each participant. The participant was then asked to retell each story as closely as possible to the original, either immediately (immediate recall) or after an interval of 30 minutes (delayed recall). A maximum score of 50 is attainable for both the immediate and the delayed recall task based on predefined criteria. Immediate recall reflects attention and memory, while delayed recall assesses memory retention. The ability to retain information from immediate to delayed recall assesses memory consolidation. Similar to the composite executive function score construction, the WMS immediate and delayed story recall scores were converted to Z scores and then averaged into a composite normalized memory function score. Again, to facilitate interpretation, we repeated the analysis using the average of the 2 raw scores. A higher score is indicative of better memory function.
Other Measurements
At the baseline visit, we performed a thorough assessment including medical history, sociodemographic information, health behaviors, blood pressure, and anthropometric data and drew blood samples to measure fasting blood glucose and lipid profile, as previously described (20). Physical activity was measured using the Baecke Questionnaire of Habitual Physical Activity, a 16-question instrument assessing physical activity levels at work, during sports and non-sports activities, rendering a global physical activity score (30). History of hypertension was defined as systolic blood pressure at least 140 mmHg or diastolic blood pressure at least 90 mmHg, or self-reported use of antihypertensive medications, following the Joint National Committee (JNC)-7 classification for Stage 1 hypertension which was the accepted staging at the time. History of coronary artery disease (CAD) that might have occurred from the time of the initial screen was defined as a previous diagnosis of myocardial infarction or angina pectoris or previous coronary revascularization procedures. Diabetes mellitus was defined as having a measured fasting glucose of more than 126 mg/dL or being treated with antidiabetic medications. Current use of beta-blockers, antidepressants, statins, and angiotensin-converting enzyme inhibitors was also recorded. A clinical diagnosis of major depression and PTSD (lifetime and current), as well as a diagnosis of alcohol abuse disorder, was obtained using the Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders, 4th Edition.
Prior to military induction, at an average age of 20 years, twins were administered the Armed Forces Qualification Test (AFQT Form 7A), a general test of cognitive ability (31). We used these scores, acquired from the military records and archived by the VET Registry, as an additional control factor in sensitivity analyses. The scores were not available in 16 twins (6%) in our analytical sample.
Statistical Analysis
We conducted descriptive analyses to summarize participants’ characteristics at the baseline visit, and describe DNA methylation age acceleration variables, and cognitive function at baseline. Continuous variables were described as mean and standard deviation (SD) and categorical variables as frequencies (percentage). Next, we evaluated the cross-sectional association between the within-pair difference in each of the DNAm age acceleration variables (as separate independent variables) and the within-pair difference in cognitive function measures at baseline (as separate dependent variables). We then examined the longitudinal association of the within-pair difference in DNAm age acceleration variables at baseline with the within-pair difference in cognitive function measures at follow-up. In a study of twins, within-pair differences intrinsically control for potential confounding by shared genetic and familial influences, in addition to environmental factors during the clinic visit, as twins were examined together. Because of the attrition between baseline and follow-up, we compared DNAm age acceleration, cognitive measures, and other key baseline variables between twins who participated in the in-person follow-up visit and those who did not participate. For both cross-sectional and longitudinal analyses, our primary outcome measures were the composite measures of executive and memory function. In secondary analyses, we examined individual cognitive function measures separately.
For all analyses, we fitted multivariable mixed-effects regression models and accounted for twin pairs as a random effect. In these models, the within-pair β coefficient describes the individual twin variation from the twin pair average (32); this formulation has the advantage of being independent of twin ordering and of including information on the individual twin value. The model equation is as follows:
Where represents the mean value of Y for the twin pair i, and represents the mean value of X for the twin pair i. The within-pair coefficient β w expresses the expected intrapair difference in Y (cognitive function scores) for a 1-year difference in DNAm age acceleration between the twin brothers. In order to better interpret effect sizes, we repeated the analyses by using averages of the raw cognitive testing subscores (both for executive function and memory function), which were log-transformed so that the modeling results could be interpreted as percent difference in cognitive status for each incremental year of DNAm age acceleration comparing the 2 brothers. To avoid model overfitting, we constructed a series of models to examine the impact of sets of a priori selected variables on the associations of interest. The base model, or Model 1, was unadjusted and included only the within-pair difference of the DNAm age acceleration variable. In Model 2, we adjusted for years of education, and in Model 3, we further adjusted for potential confounders, that is, variables that are likely related to both DNAm age acceleration and cognitive function (ever smoking, body mass index [BMI], and history of hypertension and alcohol abuse). In Models 4A and 4B, we further adjusted for diagnosis of major depression and PTSD, respectively. In a sensitivity analysis, we adjusted for AFQT raw score from military records. We used a similar modeling strategy for the longitudinal analysis, except that we additionally adjusted for baseline levels of cognitive function in all models.
To assess the potential shared genetic influence on DNAm age acceleration and cognitive function, we conducted stratified analyses in MZ and DZ twins separately and tested for the interaction by zygosity. Because MZ twins share 100% of their genes, while DZ twins only share 50% on average, if a larger association is found within DZ pairs than within MZ pairs, this suggests that genetic factors play a role. In a sensitivity analysis, we also adjusted for the history of CAD.
At baseline, 16 participants (5%) had missing TMT-A scores, and 1 participant had missing TMT-B scores; these missing scores were imputed using the average values in the rest of the sample. Missing data were rare (<5%) for all other variables, thus we used all available data without further imputation. We checked linearity assumptions for all continuous variables, as well as possible multicollinearity by variance inflation factors. A two-sided p value of less than .05 was used for statistical significance and 95% confidence intervals (CIs) were calculated from model parameters. All statistical analyses were performed using SAS, version 9.4 (SAS Institute, Cary, NC) and Stata 14.0 (StataCorp, College Station, TX).
Results
Participants Characteristics
Of the twin participants at the baseline visit, 296 individual twins had a sufficient amount of genomic DNA for epigenetic analysis, including 133 twin pairs (89 MZ pairs and 44 DZ pairs), and 30 single twins. The 266 individual twins from these 133 pairs represent our analytical sample for the within-pair cross-sectional analysis at baseline (Figure 1). Of the baseline sample, 62 twins died before the follow-up study (11%) and 393 participated in the follow-up study (78% of the survivors). Most of these (71%) participated in-person. Among them, 114 individual twins (57 pairs) had both baseline DNAm age and follow-up cognitive function data, thus they represent our analytical sample for the within-pair longitudinal analysis. Comparing twins who participated and those who did not participate in the in-person visit at follow-up, the 2 groups were similar (Supplementary Table 1). As expected, twins who did not return for follow-up tended to have more comorbidities, such as hypertension and alcohol abuse; they also smoked more and had lower levels of physical activity, but these differences were small and, except for physical activity, none were statistically significant. The 2 groups were also very similar for DNAm age acceleration and for cognitive function at baseline (Supplementary Table 1).
Of the 266 paired twins included in the analysis at baseline, 258 (97%) were White, with a mean age (SD) of 56 (3) years (Table 1). The sample had a substantial prevalence of behavioral and psychosocial risk factors. The average BMI was 30, 73% reported ever smoking, 53% met criteria for lifetime alcohol abuse, and 30% met criteria for lifetime major depression. Therefore, it is not surprising that, on average, the twins showed DNAm age acceleration, being between 0.08 years, that is, half a month (Pheno AA) to 0.22 years, or almost 3 months (Grim AA) “epigenetically older” than their demographic age. Virtually all the within-pair differences in DNAm age markers were correlated with each other (Supplementary Table 2).
Table 1.
Baseline Characteristics of 266 Twins (133 pairs)
| Characteristics, Mean (SD) or n (%) | Total (N = 266) |
|---|---|
| Sociodemographic factors | |
| Age, years | 56 (3) |
| White | 258 (97) |
| Education, years | 14 (2) |
| Employed | 203 (76) |
| Health factors | |
| BMI | 30 (5) |
| Ever smoker | 194 (73) |
| Baecke physical activity score | 7.2 (1.8) |
| Systolic blood pressure, mmHg | 124 (11) |
| Diastolic blood pressure, mmHg | 75 (8) |
| History of hypertension | 82 (31) |
| History of diabetes | 35 (13) |
| History of coronary heart disease | 32 (12) |
| History of alcohol abuse (lifetime) | 141 (53) |
| History of PTSD (lifetime) | 55 (21) |
| History of major depression (lifetime) | 81 (30) |
| Medication use | |
| β-Blockers | 21 (8) |
| Antidepressants | 41 (15) |
| Statins | 79 (30) |
| ACE inhibitors | 47 (18) |
| DNAm age acceleration (years) | |
| Horvath AA | 0.08 (3.84) |
| Hannum AA | 0.05 (3.81) |
| Grim AA | 0.22 (5.42) |
| Pheno AA | 0.04 (5.29) |
| IEAA | 0.07 (3.58) |
| EEAA | 0.10 (5.14) |
| Cognitive function | |
| TMT-A, seconds | 61 (46) |
| TMT-B, seconds | 145 (104) |
| Immediate recall score | 24 (7) |
| Delayed recall score | 20 (7) |
| Composite executive function score (raw) | 103 (61) |
| Composite memory function score (raw) | 21.7 (6.6) |
Note: AA = age acceleration; ACE = angiotensin-converting enzyme; BMI = body mass index; CAD = coronary artery disease; DNAm = DNA methylation; EEAA = extrinsic epigenetic age acceleration; IEAA = intrinsic epigenetic age acceleration; PTSD = posttraumatic stress disorder; SD = standard deviation; TMT = trail making test.
Association Between DNAm Age Acceleration and Cognitive Function
The mean within-pair difference in the raw composite executive function score was 55 points (SD, 50), and the mean within-pair difference in the raw composite memory function score was 5.5 points (SD, 4.4). All the DNAm age acceleration measures displayed nonsignificant cross-sectional associations with the composite executive and memory function scores at baseline, across all models (Table 2). For both composite scores, the unadjusted raw score difference per 1 year of DNA age acceleration was less than 2% for all DNAm age acceleration indices and close to zero for most of them.
Table 2.
Within-Pair Analysis of the Cross-Sectional Association Between Baseline DNA Methylation and Composite Cognitive Function
| Model 1 | Model 2 | Model 3 | Model 4A | Model 4B | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNAm variables | β (95% CI) | % Diff* | β (95% CI) | % Diff* | β (95% CI) | % Diff* | β (95% CI) | % Diff* | β (95% CI) | % Diff* |
| Composite Executive Function† | ||||||||||
| Horvath AA | −0.004 (−0.030 to 0.021) | −0.1 | −0.004 (−0.030 to 0.022) | −0.1 | −0.007 (−0.033 to 0.019) | −0.3 | −0.009 (−0.035 to 0.017) | −0.3 | −0.003 (−0.029 to 0.024) | 0.1 |
| Hannum AA | −0.021 (−0.050 to 0.008) | −1.9 | −0.02 (−0.050 to 0.009) | −1.7 | −0.021 (−0.052 to 0.009) | −1.7 | −0.022 (−0.053 to 0.008) | −1.7 | −0.017 (−0.048 to 0.014) | −1.4 |
| Grim AA | 0.003 (−0.021 to 0.027) | 0.7 | 0.002 (−0.022 to 0.026) | 0.5 | 0.004 (−0.023 to 0.032) | 0.8 | 0.003 (−0.025 to 0.031) | 0.9 | 0.004 (−0.024 to 0.031) | 0.8 |
| Pheno AA | −0.009 (−0.031 to 0.013) | 0.0 | −0.01 (−0.033 to 0.012) | −0.3 | −0.01 (−0.033 to 0.012) | −0.3 | −0.01 (−0.032 to 0.013) | −0.3 | −0.01 (−0.032 to 0.012) | −0.2 |
| IEAA | −0.012 (−0.039 to 0.015) | −0.8 | −0.011 (−0.038 to 0.016) | −0.7 | −0.014 (−0.041 to 0.013) | −0.8 | −0.017 (−0.044 to 0.011) | −0.8 | −0.01 (−0.037 to 0.017) | −0.5 |
| EEAA | −0.013 (−0.037 to 0.011) | −1.1 | −0.013 (−0.037 to 0.011) | −1.0 | −0.014 (−0.039 to 0.011) | −1.1 | −0.015 (−0.039 to 0.010) | −1.1 | −0.01 (−0.035 to 0.015) | −0.8 |
| Composite Memory Function‡ | ||||||||||
| Horvath AA | 0.01 (−0.017 to 0.037) | 0.5 | 0.01 (−0.017 to 0.037) | 0.5 | 0.006 (−0.021 to 0.033) | 0.3 | 0.011 (−0.017 to 0.038) | 0.5 | 0.006 (−0.021 to 0.033) | 0.3 |
| Hannum AA | −0.002 (−0.032 to 0.028) | −0.1 | −0.004 (−0.034 to 0.026) | −0.2 | −0.011 (−0.041 to 0.020) | −0.5 | −0.009 (−0.039 to 0.021) | −0.4 | −0.011 (−0.042 to 0.020) | −0.5 |
| Grim AA | −0.009 (−0.033 to 0.015) | −0.5 | −0.007 (−0.032 to 0.017) | −0.5 | −0.027 (−0.054 to 0.001) | −1.3 | −0.024 (−0.052 to 0.003) | −1.3 | −0.027 (−0.054 to 0.001) | −1.3 |
| Pheno AA | −0.014 (−0.036 to 0.009) | −0.4 | −0.011 (−0.034 to 0.012) | −0.3 | −0.014 (−0.037 to 0.009) | −0.5 | −0.015 (−0.038 to 0.008) | −0.5 | −0.014 (−0.037 to 0.009) | −0.5 |
| IEAA | 0.009 (−0.018 to 0.037) | 0.4 | 0.009 (−0.018 to 0.037) | 0.4 | 0.002 (−0.025 to 0.030) | 0.2 | 0.008 (−0.021 to 0.036) | 0.4 | 0.003 (−0.025 to 0.031) | 0.2 |
| EEAA | −0.001 (−0.024 to 0.024) | −0.1 | −0.001 (−0.025 to 0.023) | −0.1 | −0.005 (−0.030 to 0.020) | −0.3 | −0.003 (−0.028 to 0.022) | −0.2 | −0.005 (−0.030 to 0.020) | −0.2 |
Notes: AA = age acceleration; DNAm = DNA methylation; EEAA = extrinsic epigenetic age acceleration; IEAA = intrinsic epigenetic age acceleration. β values express within-pair differences in standardized composite cognitive scores, per 1-year increase in DNAm age acceleration within pairs. Percent differences, within pairs, are calculated from raw composite cognitive scores. Sample size N = 266 (or 133 pairs). Model 1: Unadjusted model; Model 2: + years of education; Model 3: Model 2 variables + ever smoking, BMI, and history of hypertension and alcohol abuse; Model 4A: Model 3 variables + history of major depression; Model 4B: Model 3 variables + history of PTSD.
*Within-pair difference in the raw composite score.
†A higher composite executive function score indicates worse cognitive function.
‡A higher composite memory function score indicates better cognitive function.
The mean length of follow-up was 11.5 years (SD, 2.2), with a range of 6.5–15.8 years. The mean within-person decline in the cognitive measures during follow-up was 33 points (SD, 65) for the composite executive function score and 6.8 points (SD, 9.7) for the composite memory function score. The mean within-pair difference in cognitive status changes during follow-up was 0.80 (SD, 0.73) for the executive function score and 0.93 (SD, 0.72) for the memory function score; the corresponding within-pair differences in raw score changes were 59 points (SD, 62) for executive function and 9.8 (SD, 7.3) for memory. The baseline and follow-up distribution of cognitive test scores is shown in Supplementary Table 3.
In longitudinal, within-pair analysis of baseline DNAm age acceleration and changes in cognitive function over the follow-up, Horvath’s AA and IEAA were associated with a decline in both the composite executive function and the memory function scores. Specifically, within-pair, a 1-year difference in Horvath’s AA was associated with a β of 0.09 (95% CI, 0.04–0.14) for the log-transformed normalized executive function score, while for IEAA, the β estimate was 0.08 (95% CI, 0.03–0.13). These changes corresponded to a 3% (95% CI, 1%–5%) greater decline in the composite raw executive function score for both DNAm acceleration measures. These differences did not substantially attenuate after adjusting for education and other potential confounding factors and mental health factors (Table 3). Horvath’s AA and IEAA were also associated with a decline in memory function. After adjusting for other factors, a 1-year within-pair difference in Horvath’s AA was associated with a β of −0.05 (95% CI, −0.10 to −0.01) for the normalized memory function score. For IEAA the β estimate was similar (β, −0.05, 95% CI, −0.09 to −0.01). These changes corresponded to a 2.5% (95% CI, 0.01%–4.9%) greater decline in the composite raw memory score for both measures. We also observed a weaker association of Pheno AA with the composite memory function score, which, however, was attenuated after adjustment for smoking and other risk factors.
Table 3.
Within-Pair Analysis of the Longitudinal Association Between Baseline DNA Methylation and Composite Cognitive Function at Follow-up
| Model 1 | Model 2 | Model 3 | Model 4A | Model 4B | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNAm variables | β (95% CI) | % Diff* | β (95% CI) | % Diff* | β (95% CI) | % Diff* | β (95% CI) | % Diff* | β (95% CI) | % Diff* |
| Composite Executive Function† | ||||||||||
| Horvath AA | 0.086 (0.038 to 0.135) | 3.3 | 0.084 (0.035 to 0.133) | 3.1 | 0.081 (0.031 to 0.131) | 3.4 | 0.069 (0.017 to 0.121) | 2.3 | 0.081 (0.031 to 0.131) | 3.2 |
| Hannum AA | 0.032 (−0.017 to 0.082) | 0.9 | 0.038 (−0.012 to 0.088) | 1.1 | 0.033 (−0.020 to 0.086) | 1.4 | 0.043 (−0.009 to 0.095) | 1.8 | 0.038 (−0.016 to 0.093) | 1.7 |
| Grim AA | 0.013 (−0.022 to 0.048) | −0.1 | 0.014 (−0.021 to 0.049) | −0.1 | 0.021 (−0.017 to 0.058) | −0.3 | 0.024 (−0.013 to 0.060) | −0.1 | 0.022 (−0.015 to 0.060) | −0.2 |
| Pheno AA | 0.027 (−0.008 to 0.062) | 0.0 | 0.025 (−0.009 to 0.060) | −0.3 | 0.022 (−0.012 to 0.057) | −0.2 | 0.022 (−0.011 to 0.055) | −0.4 | 0.022 (−0.013 to 0.056) | −0.4 |
| IEAA | 0.078 (0.030 to 0.125) | 3.2 | 0.077 (0.029 to 0.124) | 3.1 | 0.079 (0.030 to 0.128) | 3.4 | 0.067 (0.014 to 0.120) | 2.3 | 0.078 (0.029 to 0.127) | 3.3 |
| EEAA | 0.024 (−0.017 to 0.064) | 0.6 | 0.028 (−0.013 to 0.069) | 0.7 | 0.024 (−0.019 to 0.067) | 0.9 | 0.034 (−0.009 to 0.076) | 1.3 | 0.026 (−0.018 to 0.069) | 1.1 |
| Composite Memory Function‡ | ||||||||||
| Horvath AA | −0.057 (−0.105 to −0.009) | −2.5 | −0.052 (−0.098 to −0.005) | −2.3 | −0.056 (−0.101 to −0.012) | −2.5 | −0.055 (−0.101 to −0.010) | −2.5 | −0.053 (−0.098 to −0.009) | −2.4 |
| Hannum AA | −0.022 (−0.067 to 0.023) | −1.6 | −0.024 (−0.067 to 0.020) | −1.5 | 0.015 (−0.028 to 0.057) | 0.3 | 0.012 (−0.031 to 0.056) | 0.3 | 0.008 (−0.036 to 0.052) | 0.4 |
| Grim AA | −0.023 (−0.056 to 0.010) | −1.2 | −0.022 (−0.054 to 0.010) | −1.2 | −0.018 (−0.054 to 0.017) | −1.4 | −0.021 (−0.057 to 0.015) | −1.4 | −0.023 (−0.059 to 0.013) | −1.5 |
| Pheno AA | −0.044 (−0.078 to −0.010) | −1.6 | −0.038 (−0.072 to −0.004) | −1.4 | −0.028 (−0.060 to 0.003) | −1.0 | −0.030 (−0.061 to 0.002) | −1.0 | −0.027 (−0.059 to 0.005) | −1.0 |
| IEAA | −0.037 (−0.085 to 0.011) | −1.6 | −0.035 (−0.081 to 0.012) | −1.5 | −0.054 (−0.099 to −0.010) | −2.5 | −0.054 (−0.101 to −0.008) | −2.6 | −0.050 (−0.095 to −0.006) | −2.4 |
| EEAA | −0.017 (−0.054 to 0.019) | −1.0 | −0.018 (−0.053 to 0.018) | −0.9 | 0.014 (−0.020 to 0.049) | 0.6 | 0.012 (−0.023 to 0.047) | 0.6 | 0.010 (−0.024 to 0.045) | 0.5 |
Notes: AA = age acceleration; DNAm = DNA methylation; EEAA = extrinsic epigenetic age acceleration; IEAA = intrinsic epigenetic age acceleration. β values express within-pair difference in standardized composite cognitive scores, per 1-year increase in DNAm age acceleration within pairs. Percent differences, within pairs, are calculated from raw composite cognitive scores. Sample size N = 114 (or 57 pairs). Boldface font indicates significant association at p < .05. Model 1: Only adjusted for a respective composite cognitive score at baseline; Model 2: + years of education; Model 3: Model 2 variables + ever smoking, BMI, and history of hypertension and alcohol abuse; Model 4A: Model 3 variables + history of major depression; Model 4B: Model 3 variables + history of PTSD.
*Within-pair difference in the raw composite score.
†A higher composite executive function score indicates worse cognitive function.
‡A higher composite memory function score indicates better cognitive function.
Additional Analyses
Addition of the AFQT scores at age 20 did not substantially change the cross-sectional results for either executive function or memory. In stratified analysis by zygosity, the cross-sectional results showed mostly nonsignificant associations that were similar in MZ and in DZ, although for some measures, the coefficients tended to be larger in DZ than in MZ twins (Supplementary Tables 4A and 4B). Given the small number of DZ pairs in the longitudinal analysis (n = 17 pairs), this analysis could not be stratified by zygosity. Overall, the longitudinal association of DNAm age acceleration with individual cognitive function tests showed similar results as the composite scores, with Horvath’s AA and IEAA exhibiting the most consistent associations across all measures (Supplementary Tables 5A–5D). Finally, adjusting for the history of CAD did not change the results.
Discussion
In a study of veteran twins who were middle-aged at baseline, we found that those who were biologically older relative to their demographic age, indexed using DNAm age acceleration estimated with the Horvath’s method (Horvath AA and IEAA), were more likely to show a cognitive decline after an average of 11.5 years. Comparing twins, the “biologically older” twin experienced a 2.5%–3% greater cognitive decline for each incremental year of DNAm acceleration compared with his twin brother.
While we found that Horvath’s derived DNAm age acceleration measures were related to cognitive decline longitudinally, there was no association between DNAm acceleration and cognitive performance at baseline. The lack of association at baseline is not surprising given that the mean age of the twins at baseline was 56 years, potentially too young to show an effect of DNAm on cognitive aging. This is consistent with another study of twins of about the same age as our sample (28). However, despite their relatively younger age at baseline, after 11.5 years of mean follow-up, the association between DNAm acceleration and cognitive performance became apparent. The lack of an association at baseline, together with the demonstration of an association with longitudinal changes in cognitive function, eliminates the possibility of reverse causation between epigenetic age acceleration and cognitive function, that is, the possibility that lower cognitive performance is a cause rather than an effect of accelerated biological aging.
Epigenetic aging has been proposed as an important factor that can explain the aging process and its relationship with chronic conditions at the molecular level (10,16,33). Epigenetic aging estimated using DNAm age acceleration metrics, which are independent of chronological age, can be particularly useful in capturing biological alterations associated with cumulative environmental effects on the aging process over the lifetime, also known as “weathering” (9). Indeed, multiple studies of older individuals have linked accelerated DNAm age to earlier death, even after taking into account medical diagnoses and risk factors (12). DNAm age acceleration has also been associated with cognitive function in cross-sectional studies of predominantly older populations (34,35), especially in individuals with Alzheimer’s disease (27), as well as those with frailty (36), and Parkinson’s disease (37). Cross-sectional studies have also reported associations of DNAm age with histological markers of Alzheimer’s disease in postmortem brains (27) and with measures of degraded neural microstructural integrity (35).
Very few studies, however, have examined whether DNAm age acceleration is related to cognitive decline longitudinally. Marioni et al. (34) examined changes in cognitive measures in an older cohort followed up between the ages of 70 and 76 years and found no relationship between DNAm acceleration, measured with the Horvath algorithm, and cognitive decline, although a cross-sectional relationship was present at age 70. It is possible that the lack of longitudinal association was due to the relatively short follow-up time, when only modest changes in cognitive status occurred (34). A study of middle-aged male and female MZ twins also did not find a relationship of DNAm age acceleration, estimated using both the Horvath and the Hannum methods, with cognitive changes over 10 years (28). The twins, however, showed limited intrapair variation in cognitive measures and only small differences in cognitive function measured longitudinally. In our study, we found a mean within-pair difference in cognitive status change during follow-up of almost 1 SD of the normalized scores. Furthermore, our study included only men. Men have a faster DNAm age acceleration than women (38,39), and therefore, men may be more vulnerable to DNAm-related cognitive aging compared with women. Indeed, a recent study found an association between DNAm age acceleration and cognitive decline in men but not in women (38). Differences in the admixture of men and women in previous studies may play a role in the diverging results.
In our study, the Horvath’s derived metrics of age acceleration (Horvath AA and IEAA) showed the most robust associations with cognitive decline. An explanation for this could be inherent in the methodology for the construction of these age acceleration estimators. The Horvath’s epigenetic clock was constructed from epigenetic markers derived from multiple tissues, including the brain (9), while other age acceleration indices were derived from blood-related epigenetic markers only. The fact that we used peripheral blood cells for the measurement of DNAm age acceleration does not diminish this advantage, because of the demonstrated similarities of age-related DNA hypermethylation across tissues and cell types, including blood and brain (9,40,41). Furthermore, in contrast to Horvath’s clock, other DNAm age estimators were developed as predictors of phenotypic aging using blood-derived biomarkers of disease, such as white blood cell count, plasma proteins, inflammatory biomarkers, and glucose (10,11,24). Although these may be useful for the detection of age-related metabolic dysregulation and total mortality, they may be less relevant for cognition and brain disorders. Consistent with our findings, Horvath’s clock has been associated with Alzheimer’s disease, cognitive performance, and pathological biomarkers of brain aging and dementia in a number of studies (27,34,42–44). Although in our study Pheno AA was associated with memory function decline in unadjusted analysis, the association was weakened after adjusting for cardiovascular risk factors, highlighting the dependency of this aging biomarker on clinical indicators of cardiometabolic disorders. It should be noted that, in contrast to our results, a recent investigation reported that the EEAA metric was more consistently associated with cognitive decline than IEAA and other measures of DNAm acceleration in middle-aged men followed up for a mean of 4.7 years (38). However, in this study, there was substantial variability in associations across different cognitive tests and most of them showed a modest decline (38).
DNAm age acceleration may signal cognitive aging through a number of mechanisms. Epigenetic modifications are fundamental for gene regulation and the translation of environmental stimuli into gene expression. DNAm has been shown to contribute to learning processes, memory formation, and memory consolidation through epigenetic regulation of transcription in the central nervous system (45,46). There are a number of specific molecular pathways with potential long-term effects on learning and memory (10,47). These include synaptic plasticity and neurogenesis (47–49), stem cell function (50), immunosenescence (51), circadian rhythms (52), and the effects of cumulative stress through glucocorticoid signaling (53). Epigenetic clocks likely modulate these processes, and at least in part, these effects could be mediated by inflammatory and oxidative stress pathways resulting from cellular accumulation of reactive oxygen species (33). Because epigenetic processes are heritable, mostly due to genetic variations at the CpG sites (18), shared genetic pathways between epigenetic aging and cognitive decline could also be involved, as suggested by a previous negative study of MZ twins (28). However, in our study, we did not find marked differences in results based on the twin pairs’ zygosity, suggesting that genetic confounding should not be a dominant explanatory factor.
A limitation of our study is that our epigenetic markers were derived from blood cells and not brain tissue; however, age-related DNA hypermethylation is known to be conserved across tissues and cell types (9,40,41). Due to our relatively small sample size, especially for the longitudinal analysis, we may have had limited power to detect small differences, including differences in results by zygosity. The smaller sample at follow-up was in part dictated by the need for longitudinal data on both twins to conduct within-pair analyses. Although we cannot exclude that sample attrition between baseline and follow-up introduced bias in our analysis, this is unlikely given the similarity of characteristics comparing twins who participated at follow-up with those who did not. The original selection of participants in the ETS included samples of twin pairs who were discordant for depression or PTSD, in addition to normal pairs. This sampling strategy may have facilitated the detection of within-pair differences in age acceleration. Our study also has limited generalizability, because our twin sample was mostly White and all male. However, our co-twin control study design should have improved internal validity and precision by intrinsically adjusting for unknown or unmeasured familial confounders. In addition to the strengths of a matched twin design, ours is one of only a few investigations to examine longitudinal changes in cognitive function as a function of DNAm age acceleration, allowing the examination of the temporality of associations. Our study is further strengthened by a relatively long follow-up time and the use of a comprehensive set of DNAm acceleration indices derived using modern genome-wide DNA methylation methodology.
In conclusion, we found that DNAm age acceleration, measured with Horvath’s derived metrics of age acceleration (Horvath AA and IEAA), is associated with a faster rate of cognitive decline in the domains of executive function, cognitive flexibility, and memory. Our results contribute to a growing literature that supports the role of epigenetic modifications in cognitive aging and the risk of cognitive disorders. Because of their dynamic nature, epigenetic modifications are potentially reversible and thus may provide powerful means for prevention and intervention in cognitive decline.
Supplementary Material
Acknowledgments
The United States Department of Veterans Affairs has provided financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. Numerous organizations have provided invaluable assistance in the conduct of this study, including Department of Defense; National Personnel Records Center, National Archives and Records Administration; the Internal Revenue Service; National Institutes of Health; National Opinion Research Center; the National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University. Most importantly, the authors gratefully acknowledge the continued cooperation and participation of the members of the VET Registry and their families. Without their contribution, this research would not have been possible.
Funding
This work was supported by the National Institutes of Health (grants R01 HL68630, R01 AG026255, R01 HL125246, 2K24 HL077506, and R01 HL136205).
Conflict of Interest
None of the authors report conflicts of interest.
Author Contributions
V.V. conceived the study, obtained funding, interpreted the results, drafted the manuscript, and supervised all the research staff and the statistical analysis. M.H. performed the statistical analysis and contributed to manuscript writing. Z.W. analyzed the epigenetic data and constructed the age acceleration markers. Q.H. performed data management and data processing for the epigenetic data analysis. A.J.S., J.G., N.S., and J.D.B. contributed to study design and data collection and revised the manuscript for important intellectual content. B.K., N.M., O.M.L., L.S., and E.D. collected the data. Y.V.S. supervised the epigenetic analysis and revised the manuscript for important intellectual content.
References
- 1.Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiman EM, Quiroz YT, Fleisher AS, et al. Brain imaging and fluid biomarker analysis in young adults at genetic risk for autosomal dominant Alzheimer’s disease in the presenilin 1 E280A kindred: a case-control study. Lancet Neurol. 2012;11:1048–1056. doi: 10.1016/S1474-4422(12)70228-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 2020;16: 391–460. doi: 10.1002/alz.12068 [DOI] [PubMed] [Google Scholar]
- 5.Numata S, Ye T, Hyde TM, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet. 2012;90:260–272. doi: 10.1016/j.ajhg.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bocklandt S, Lin W, Sehl ME, et al. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230 [DOI] [PubMed] [Google Scholar]
- 8.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 11.Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10:573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8:1844–1865. doi: 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marioni RE, McRae AF, Bressler J, et al. Meta-analysis of epigenome-wide association studies of cognitive abilities. Mol Psychiatry. 2018;23:2133–2144. doi: 10.1038/s41380-017-0008-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein HU, Bennett DA, De Jager PL. The epigenome in Alzheimer’s disease: current state and approaches for a new path to gene discovery and understanding disease mechanism. Acta Neuropathol. 2016;132:503–514. doi: 10.1007/s00401-016-1612-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nativio R, Donahue G, Berson A, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci. 2018;21:497–505.doi: 10.1038/s41593-018-0101-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mather KA, Kwok JB, Armstrong N, Sachdev PS. The role of epigenetics in cognitive ageing. Int J Geriatr Psychiatry. 2014;29:1162–1171. doi: 10.1002/gps.4183 [DOI] [PubMed] [Google Scholar]
- 17.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McRae AF, Powell JE, Henders AK, et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014;15:R73. doi: 10.1186/gb-2014-15-5-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai M, Mori AM, Forsberg CW, et al. The Vietnam Era Twin Registry: a quarter century of progress. Twin Res Hum Genet. 2013;16:429–436. doi: 10.1017/thg.2012.122 [DOI] [PubMed] [Google Scholar]
- 20.Vaccarino V, Khan D, Votaw J, et al. Inflammation is related to coronary flow reserve detected by positron emission tomography in asymptomatic male twins. J Am Coll Cardiol. 2011;57:1271–1279. doi: 10.1016/j.jacc.2010.09.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherrer JF, Xian H, Bucholz KK, et al. A twin study of depression symptoms, hypertension, and heart disease in middle-aged men. Psychosom Med. 2003;65:548–557. doi: 10.1097/01.psy.0000077507.29863.cb [DOI] [PubMed] [Google Scholar]
- 22.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–399. doi: 10.2217/epi.15.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33:558–560. doi: 10.1093/bioinformatics/btw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Milano). 2019;11:303–327.doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudino EA, Geisler MW, Squires NK. Construct validity in the trail making test: what makes Part B harder? J Clin Exp Neuropsychol. 1995;17:529–535. doi: 10.1080/01688639508405143 [DOI] [PubMed] [Google Scholar]
- 27.Levine ME, Lu AT, Bennett DA, Horvath S. Epigenetic age of the pre-frontal cortex is associated with neuritic plaques, amyloid load, and Alzheimer’s disease related cognitive functioning. Aging (Albany NY). 2015;7:1198–1211. doi: 10.18632/aging.100864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starnawska A, Tan Q, Lenart A, et al. Blood DNA methylation age is not associated with cognitive functioning in middle-aged monozygotic twins. Neurobiol Aging. 2017;50:60–63. doi: 10.1016/j.neurobiolaging.2016.10.025 [DOI] [PubMed] [Google Scholar]
- 29.Wechsler D.WMS-R: Wechsler Memory Scale—Revised Manual. San Antonio, TX: The Psychological Corporation; 1987. [Google Scholar]
- 30.Richardson MT, Ainsworth BE, Wu HC, Jacobs DR Jr, Leon AS. Ability of the Atherosclerosis Risk in Communities (ARIC)/Baecke Questionnaire to assess leisure-time physical activity. Int J Epidemiol. 1995;24:685–693. doi: 10.1093/ije/24.4.685 [DOI] [PubMed] [Google Scholar]
- 31.Orme DR, Brehm W, Ree MJ. Armed forces qualification test as a measure of premorbid intelligence. Mil Psychol. 2001;13:187–197.doi: 10.1207/S15327876MP1304_1 [DOI] [Google Scholar]
- 32.Carlin JB, Gurrin LC, Sterne JA, Morley R, Dwyer T. Regression models for twin studies: a critical review. Int J Epidemiol. 2005;34:1089–1099. doi: 10.1093/ije/dyi153 [DOI] [PubMed] [Google Scholar]
- 33.Cencioni C, Spallotta F, Martelli F, et al. Oxidative stress and epigenetic regulation in ageing and age-related diseases. Int J Mol Sci. 2013;14:17643–17663. doi: 10.3390/ijms140917643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marioni RE, Shah S, McRae AF, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44:1388–1396.doi: 10.1093/ije/dyu277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf EJ, Logue MW, Hayes JP, et al. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–162. doi: 10.1016/j.psyneuen.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breitling LP, Saum KU, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany NY). 2015;7:1130–1142. doi: 10.18632/aging.100859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beydoun MA, Shaked D, Tajuddin SM, Weiss J, Evans MK, Zonderman AB. Accelerated epigenetic age and cognitive decline among urban-dwelling adults. Neurology. 2020;94:e613–e625. doi: 10.1212/WNL.0000000000008756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horvath S, Gurven M, Levine ME, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horvath S, Zhang Y, Langfelder P, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakyan VK, Down TA, Maslau S, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Degerman S, Josefsson M, Nordin Adolfsson A, et al. Maintained memory in aging is associated with young epigenetic age. Neurobiol Aging. 2017;55:167–171. doi: 10.1016/j.neurobiolaging.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 43.Raina A, Zhao X, Grove ML, et al. Cerebral white matter hyperintensities on MRI and acceleration of epigenetic aging: the atherosclerosis risk in communities study. Clin Epigenetics. 2017;9:21. doi: 10.1186/s13148-016-0302-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu AT, Hannon E, Levine ME, et al. Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat Commun. 2017;8:15353. doi: 10.1038/ncomms15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y, Langley B, Lubin FD, et al. Epigenetics in the nervous system. J Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu NK, Baek SH, Kaang BK. DNA methylation-mediated control of learning and memory. Mol Brain. 2011;4:5. doi: 10.1186/1756-6606-4-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, Jasper H, Rudolph KL. Aging-Induced stem cell mutations as drivers for disease and cancer. Cell Stem Cell. 2015;16:601–612. doi: 10.1016/j.stem.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grolleau-Julius A, Ray D, Yung RL. The role of epigenetics in aging and autoimmunity. Clin Rev Allergy Immunol. 2010;39:42–50. doi: 10.1007/s12016-009-8169-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh G, Ebrahimi S, Carlucci M, et al. Cytosine modifications exhibit circadian oscillations that are involved in epigenetic diversity and aging. Nat Commun. 2018;9:644. doi: 10.1038/s41467-018-03073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zannas AS, Arloth J, Carrillo-Roa T, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.