Abstract
In Saccharomyces cerevisiae, gene expression in the late G1 phase is activated by two transcription factors, SBF and MBF. SBF contains the Swi4 and Swi6 proteins and activates the transcription of G1 cyclin genes, cell wall biosynthesis genes, and the HO gene. MBF is composed of Mbp1 and Swi6 and activates the transcription of genes required for DNA synthesis. Mbp1 and Swi4 are the DNA binding subunits for MBF and SBF, while the common subunit, Swi6, is presumed to play a regulatory role in both complexes. We show that Stb1, a protein first identified in a two-hybrid screen with the transcriptional repressor Sin3, binds Swi6 in vitro. The STB1 transcript was cell cycle periodic and peaked in late G1 phase. In vivo accumulation of Stb1 phosphoforms was dependent on CLN1, CLN2, and CLN3, which encode G1-specific cyclins for the cyclin-dependent kinase Cdc28, and Stb1 was phosphorylated by Cln-Cdc28 kinases in vitro. Deletion of STB1 caused an exacerbated delay in G1 progression and the onset of Start transcription in a cln3Δ strain. Our results suggest a role for STB1 in controlling the timing of Start transcription that is revealed in the absence of the G1 regulator CLN3, and they implicate Stb1 as an in vivo target of G1-specific cyclin-dependent kinases.
In the budding yeast Saccharomyces cerevisiae, commitment to enter the mitotic cell cycle occurs during the late G1 phase; this commitment phase of the cell cycle is designated Start (reviewed in reference 33). Start is marked by the transcriptional induction of a subset of genes that catalyze entry into the mitotic cell cycle. Events that ensue once a yeast cell has passed through Start include initiation of DNA synthesis, budding, and spindle-pole body duplication.
The transcriptional activation of genes important for the Start transition is dependent upon two transcription factor complexes called SBF and MBF (reviewed in reference 6). The SBF (SCB-binding factor) complex contains the Swi4 and Swi6 proteins and activates transcription mainly through a cis-acting sequence element called the SCB (for “Swi4/6 cell cycle box”; the consensus sequence is CACGAAA). Genes activated by SBF include the G1 cyclins (CLN1, CLN2, PCL1, and PCL2), the HO endonuclease gene, the gene encoding the Swe1 protein kinase (42), and a number of genes required for cell wall biosynthesis (21). The Swi4 protein is the component of SBF responsible for specific binding to SCB sequences, while Swi6, on its own, does not bind DNA specifically. In the absence of Swi4 or Swi6, HO is not expressed, and G1 cyclin and cell wall biosynthetic gene expression is reduced. Swi6 also interacts with a second DNA binding protein, Mbp1, to form the transcription factor MBF (MCB-binding factor; also known as DSC1 [25]). MBF/DSC1 recognizes the so-called MCB element (for “MluI cell cycle box”; the consensus is ACGCGTNA) and activates G1-specific transcription of the S-phase cyclin genes CLB5 and CLB6, the SWI4 gene, and many genes needed for DNA synthesis such as CDC9 and POL1. In most strain backgrounds, SWI4, SWI6, and MBP1 are not essential genes. However, cells lacking both SWI4 and SWI6 or both SWI4 and MBP1 are not viable, arresting prior to DNA synthesis (25, 34). In both double mutants, the death results from inadequate expression of G1 cyclins, since ectopic expression of CLN2 can rescue the lethality.
Swi4, Swi6, and Mbp1 have both functional and sequence similarity to homologues in two other yeasts, Schizosaccharomyces pombe and Kluyveromyces lactis. Together, they form a family of closely related cell cycle-regulatory transcription factors (6, 25). In addition to the DNA binding domain (shared by the Swi4/Mbp1 homologues) and the C-terminal heterodimerization domain, all members of this family share a highly conserved central ankyrin repeat domain. The ankyrin repeat is a predominantly α-helical protein domain (17, 49) that is found in many functionally diverse proteins and has been implicated in mediating protein-protein interactions (reviewed in reference 30). Genetic studies suggest that the ankyrin repeats in Swi4, Swi6, and the Swi6 homologue Cdc10 in S. pombe play important roles (11a, 14; reviewed in reference 6). However, only the ankyrin repeats of Swi4 have been assigned a function; the ankyrin repeat region of Swi4 can bind the Cdc28 cyclin Clb2 (1, 44).
Both passage through Start and activation of SBF and MBF require the cyclin-dependent kinase (Cdk) gene CDC28 and one of the three G1 cyclin genes CLN1, CLN2, or CLN3. Although any one of the three G1 cyclins is, by itself, sufficient to drive Start, the Cln3-Cdc28 kinase probably functions upstream of SBF and MBF activation in the normal cell cycle (11, 48). CLN3 is essential for activation of CLN1 and CLN2 gene expression at the appropriate cell size. By contrast, CLN1 and CLN2 are not required for gene activation but are important for the proper execution of other Start-related events such as budding and DNA synthesis. Although genetic studies indicate a key role for Cln3 in activating SBF and MBF at Start, there is no evidence that Cln3-Cdc28 acts to directly phosphorylate or interact with components of SBF or MBF. Moreover, a cln3Δ strain is viable and still undergoes SBF- and MBF-dependent transcription, albeit at a larger cell size, indicating that alternative mechanisms must function to activate SBF/MBF (34) (see below). The BCK2 gene (for “bypass of C-kinase mutation”) appears to be involved in such an alternative pathway(s). Compared to bck2Δ and cln3Δ single mutants, bck2Δcln3Δ double mutants have a severe growth defect and show reduced levels of Start transcription (10, 13). The mitogen-activated protein kinase gene SLT2 (also called MPK1) may also be involved in regulating Start-dependent gene expression (21, 27).
In this paper, we identify another gene, STB1, that functions as an important regulator of the timing of Start transcription in the absence of CLN3. We identified Stb1 as a protein that bound to Swi6 in vitro. STB1 (for “Sin three-binding protein”) encodes a novel protein that was first identified in a two-hybrid screen with the general transcriptional repressor Sin3 (24), but the role of STB1 in transcriptional repression is unclear. Mutants with mutations in STB1 did not show significant defects in cell cycle progression or Start transcription. However, stb1Δcln3Δ double mutants exhibited a severe G1 progression defect and a longer delay in onset of Start transcription than was seen in the cln3Δ single mutant. Our data suggest that STB1 is important for the timing of Start transcription in the absence of CLN3 function and may define a parallel pathway for activation of gene expression at Start. Stb1 was phosphorylated by Cln-associated kinases in vitro, and Stb1 phosphoforms in vivo were dependent on CLN function. We discuss the significance of Stb1 phosphorylation by Cln kinases and the possibilities for CLN regulation of STB1.
MATERIALS AND METHODS
Yeast strains and plasmids.
Except where indicated, the genotype of the parental strain of the yeast strains used in this study was MATα TRP+ ura3-52 lys2-801a ade2-1070 his3Δ200 leu2-Δ1 (BY263, an S288C derivative [29]). The cln3ΔURA3 deletion strain (BY655) was constructed by transformation of strain BY263 with a cln3ΔURA3 disruption cassette (26). The stb1ΔTRP1 strain (BY806, MATα stb1ΔTRP1 trp1Δ63 ura3-52 lys2-801a ade2-1070 leu2-Δ1 his3Δ200) was constructed by using a PCR-based strategy that allowed targeted gene replacement of STB1 coding sequences with the URA3 gene. The resulting stb1ΔURA3 strain (BY805) was transformed with a URA3-TRP1 switcher plasmid (8) (details of strain construction are available on request). To construct a stb1Δ cln3Δ double mutant, strains BY806 (MATα stb1ΔTRP1) and BY655 (MATa cln3ΔURA3) were mated and BY822 (MATa stb1ΔTRP1 cln3ΔURA3) was recovered by dissecting tetrads. All gene disruptions were confirmed by Southern blot analysis.
For Western blot analysis of Stb1 phosphoforms, BY263 was transformed with either pRS425 (7) or pM2517 as the vector. Plasmid pM2517 contains a 3.1-kb genomic fragment of STB1 in pRS425 (24). For Western blot analysis of Stb1 phosphoforms in clnΔ strains, the wild-type strain used was BF305-15d (MATa ade1 arg5 his3 leu2 met14 trp1 ura3) (50). The clnΔsic1Δ strain was isogenic to BF305-15d except that it was cln1ΔHIS3 cln2ΔTRP1 cln3Δura3-GAL1-CLN3 sic1ΔURA3 (50). These strains were transformed with pM2517 for STB1 overexpression. For Northern blot experiments, the wild-type strain used was W303 (MATa ura3 trp1-1 ade2-1 his3-11,15 leu2-3,112 can1-100) and the stb1ΔTRP1 mutant strain was otherwise isogenic (24).
Yeast strains used for in vitro kinase assays were derived from a W303a parental strain. The HA-CLN1 (MT235), HA-CLN2 (MT244), and HA-CLB2 (MT537) strains were gifts from M. Tyers (strains as described in reference 51, except for W303 background).
To analyze suppression of the stb1Δcln3Δ slow-growth phenotype, the following plasmids were used to transform strain BY822: control vector YEp351 (LEU2); padh-CLN2, in which CLN2 is expressed from the constitutive S. pombe adh1 promoter (55); and 2μCLN1, which contains a genomic fragment including CLN1 in vector YEp351. The high-copy-number CLN1 plasmid was isolated from a yeast genomic library in a screen for plasmid suppressors of the growth defect of strain BY822 (7a). Control transformants included strain BY822 transformed with pM2517 (2μSTB1) and the isogenic wild-type strain (BY263) transformed with vector YEp351.
Protein affinity chromatography and microsequencing of p48.
Recombinant full-length Swi6 protein was expressed and purified from Escherichia coli as described previously (20, 43). The Swi6ΔM protein has 285 internal amino acids deleted, including the ankyrin repeats. The deletion was created by digestion of a full-length Swi6-containing plasmid with SacI followed by fill-in synthesis with Klenow polymerase, resulting in an in-frame 860-bp deletion.
Swi6 protein affinity chromatography was done as previously described (20), except that each column was washed with 10 volumes of SB buffer (20 mM HEPES [pH 2], 10% glycerol, 0.1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) plus 100 mM NaCl followed by 4 volumes of SB buffer plus 1 M NaCl before being eluted with 4 volumes of SB buffer containing 1% sodium dodecyl sulfate (SDS). Eluates determined by far-Western blotting (see below) to contain p48 were pooled, dialyzed to remove SDS, and concentrated by precipitation with methanol-acetone (1:1) before being loaded onto SDS-polyacrylamide gels. The p48 band could not be visualized by Coomassie blue or silver staining because it comigrated with a protein that bound to the AffiGel-10 resin. Gel slices at around 48 kDa were taken from both the Swi6 column and control column eluates for microsequencing.
Microsequencing of p48 was performed as described previously (54). The 48-kDa gel slices from both the Swi6 and control column eluates were digested with Achromobacter protease I, and the resulting peptides from each digest were resolved by high-pressure liquid chromatography (HPLC). The peptide maps were compared to find peptides unique to p48 that eluted only from the Swi6 column. The peptides were then sequenced with an automated sequencer (Applied Biosystems model 494).
Far-Western blot analysis.
To develop a Swi6 derivative useful for far-Western experiments, a 10× histidine tag and an HMK (heart muscle kinase) phosphorylation site were engineered at the N terminus of Swi6. The resulting plasmid, pET-HMKFL6, was transformed into E. coli BL21(DE3) pLysS (Novagen), and transformants were grown to an optical density at 600 nm of 0.8 in Luria broth LB supplemented with 70 μg of chloramphenicol per ml and 250 μg of ampicillin per ml. Protein expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, and the cultures were incubated at 25°C for 3 h. The His-HMK-Swi6 protein was then purified under native conditions by Ni2+-nitrilotriacetic acid (NTA) chromatography as recommended by the manufacturer (Qiagen).
Affinity column eluates were resolved on SDS-polyacrylamide gels and blotted onto nitrocellulose membrane (3). The resulting blot was renatured as described previously (53). The labelling of the Swi6 probe and the far-Western experiments were done essentially as described previously (4).
Synchronization of yeast cells and Northern blot analysis.
For synchronization by α-factor block and release, cells were arrested in late G1 by treatment with α-factor mating pheromone until more than 95% of the cells showed a shmoo morphology. The cells were then washed and released into fresh medium for collection of synchronous cell samples as previously described (5, 28).
For synchronization by centrifugal elutriation, cells were grown to an optical density at 600 nm of 0.6 to 0.8 in yeast extract-peptone (YP) medium containing sucrose. The cultures were concentrated to approximately 100 ml (final volume) and loaded at 12 to 14 ml/min into a Beckman JC-MI centrifugal elutriator (rotor type JE-5.0) running at 2,400 rpm and cooled to 5°C. The cells were elutriated at the loading speed with increases of 2 ml/min after each 100 to 150 ml of elutriated sample was recovered. The elutriated samples were checked by microscopy and by Coulter Channelizer analysis. The elutriated cells were quickly centrifuged and resuspended in fresh, prewarmed yeast extract-peptone-dextrose (YPD) medium for collection of synchronous samples at the time points specified. For both α-factor and elutriation synchronization experiments, 15 ml of cells was collected at the specified time points and RNA was prepared and analyzed by Northern blotting as described previously (23). Cells were analyzed for DNA content by fluorescence-activated cell sorting (FACS) as described previously (51), and the results were analyzed with LYSYS II software (Becton Dickinson). In elutriation synchrony experiments, cells at each time point were analyzed with a Coulter Channelizer and the mean cell size was determined.
The probes used for Northern blot analysis were a 600-bp EcoRI-HindIII fragment of the ACT1 gene (19), a 1.3-kb HindIII fragment containing the STB1 gene from pM2517, a 440-bp EcoRI fragment of CLB5 from the pMT895 vector (a gift from M. Tyers), an 864-bp PCR product containing the PCL1 coding sequence (29), a 1.7-kb BglII-EcoRI fragment of RNR1 (12), a 2.5-kb EcoRI fragment of CLN1 (18), and a 1.3-kb XhoI-NcoI fragment of CLN2 (19). Probes were labelled by random-primer synthesis with Klenow DNA polymerase in the presence of [α-32P]dATP. For RNA quantitation, Northern blots were exposed on a Molecular Dynamics screen and scanned with a Molecular Dynamics PhosphorImager and ImageQuant software.
Preparation of antibodies to Stb1 and Western blotting.
To construct a vector for preparation of recombinant Stb1 in bacteria, a 1.3-kb HindIII fragment containing STB1 was isolated from pM2517 and inserted into the HindIII site of pRSET-B (Invitrogen). The resulting plasmid, pRSET-StbH/H, expresses a derivative of Stb1 with an N-terminal His tag but lacking the first 85 N-terminal amino acids of full-length Stb1. pRSET-StbH/H was transformed into E. coli BL21(DE3), and the His-Stb1 fusion protein was purified under denaturing conditions by Ni2+-NTA affinity chromatography as recommended by the manufacturer (Qiagen). The purified protein was dialyzed gradually to 1 M urea and used to immunize rabbits (Faculty of Medicine, University of Toronto). Antisera were then affinity purified as described previously (2). For Western blotting, 10 μg of yeast extract was separated by SDS-polyacrylamide gel electrophoresis (PAGE) (9% polyacrylamide) and transferred to nitrocellulose with a semidry transfer apparatus (Bio-Rad). Immunoblots were probed with a 1:1,000 dilution of affinity-purified Stb1 antibody and subsequently with a 1:5,000 dilution of goat anti-rabbit immunoglobulin-peroxidase (Bio-Rad). Proteins were visualized by enhanced chemiluminescence (Renaissance detection system; New England Nuclear) followed by exposure to X-ray film (Xar-5; Kodak).
In vitro kinase assays with Cln-associated kinase.
Cln1-, Cln2-, and Clb2-associated kinases were immunoprecipitated from yeast strains bearing hemagglutinin-tagged (HA) forms of each cyclin, by using the 12CA5 antibody. The immunoprecipitation-kinase assay was performed as described previously (51). In each reaction, 0.5 μg of purified StbH/H protein (see the previous section) and 1.0 μg of histone H1 were added as exogenous substrates. For kinase reactions, the Stb1 protein was further dialyzed against kinase buffer prior to use.
Phosphatase analysis of Stb1 phosphoforms.
For phosphatase treatment of cell extracts, a log-phase yeast culture was lysed in modified lysis buffer (100 mM Tris [pH 8.0], 100 mM NaCl, 2 mM MnCl2, 10% glycerol, 1 mM dithiothreitol, and protease inhibitors as described in reference 20) by using agitation in the presence of glass beads. Approximately 150 μg of each extract was treated with 1,600 U of lambda protein phosphatase (New England Biolabs) for 0.5 h at 30°C. Where indicated, phosphatase inhibitors were present at 5 mM EDTA, 50 mM NaF, 50 mM β-glycerophosphate, and 1 mM sodium vanadate. The phosphatase reactions were stopped by addition of an equal volume of SDS sample buffer and electrophoresed on SDS–9% polyacrylamide gels for Western blot analysis.
Microscopy.
Cells were grown in YPD medium to log phase and observed at a magnification of ×630 with Nomarski optics and a charge-coupled device camera mounted on a Leica DM-LB microscope. Images from the camera were captured and analyzed by using the Northern Exposure Imaging system (Empix Imaging, Inc., Mississaugua, Ontario, Canada). Where indicated, the percentage of budded cells in each sample was determined by counting at least 300 cells per sample.
RESULTS
Interaction of Stb1 with Swi6.
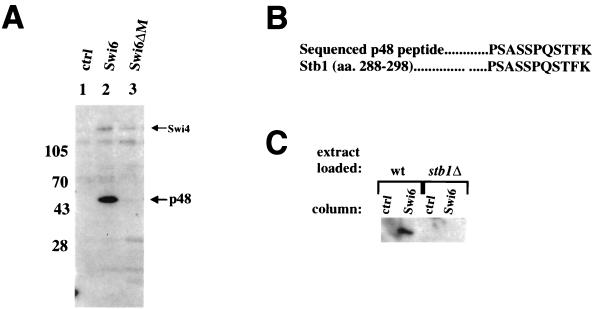
To investigate the function and regulation of Swi6, we used protein affinity chromatography to look for proteins in crude yeast extracts that physically associate with Swi6. Since some specific Swi6-binding proteins may be obscured by yeast proteins that bind nonspecifically to the column resin (Fig. 1 in reference 20), the Swi6 affinity column eluates were blotted onto nitrocellulose and probed with radiolabelled Swi6 protein. Using this far-Western protocol, we detected a 48-kDa protein (p48) in eluates from the Swi6 column (Fig. 1A, lane 2) but not in eluates from the control column (column resin with no coupled protein [lane 1]). The p48 band was absent in eluates from a column conjugated with a Swi6 protein derivative with the central ankyrin repeat region deleted (Swi6ΔM). Swi4 and Hrr25, two other Swi6-binding proteins, still bound to the Swi6ΔM ligand (lane 3) (20), indicating that the Swi6ΔM protein is sufficiently folded to maintain these protein interactions.
FIG. 1.
Binding of p48 (Stb1) to Swi6 protein affinity chromatography columns. (A) Yeast extracts were passed over a control column with resin alone (ctrl; lane 1), a column coupled with Swi6 protein (lane 2), and a column coupled with a Swi6 ligand with the central ankyrin region deleted (Swi6ΔM; lane 3). The columns were washed and eluted with 1% SDS. Eluates were resolved by SDS-PAGE, blotted onto nitrocellulose, and probed with a radioactively labelled Swi6 protein probe (see Materials and Methods). The positions of migration of p48 and Swi4 are shown to the right. Molecular size markers are indicated on the left (in kilodaltons). (B) Microsequencing of an 11-amino-acid peptide from the 48-kDa protein. Shown is an alignment of the sequence of a peptide derived from p48 and a predicted Stb1 peptide (amino acids [aa.] 288 to 298). (C) Swi6 protein affinity chromatography with extracts from stb1Δ cells. Extracts made from either wild-type cells (wt) or a stb1Δ strain were loaded onto either a control column (ctrl) or a Swi6-coupled column (Swi6) and eluted with 1% SDS. p48 (Stb1) was detected as described for panel A.
Microsequencing of p48 yielded an 11-amino-acid peptide (Fig. 1B) that matched perfectly with amino acids 288 to 298 of Stb1, a novel protein that was first identified in a two-hybrid screen with Sin3 (24). Other peptides obtained in the microsequencing analysis corresponded to a major protein contaminant that bound nonspecifically to the column control resin (Fig. 1 in reference 20). Due to the presence of this contaminant, the far-Western blot was necessary to visualize p48 in the column eluates. The p48 band was absent in eluates from Swi6 affinity columns loaded with an extract from a stb1Δ strain (Fig. 1C), confirming that p48 was indeed Stb1. The binding of Stb1 to Swi6 in the far-Western blot assay indicates that the Stb1-Swi6 interaction is direct. We conclude that Stb1 interacts directly with Swi6 in vitro and that this interaction requires the ankyrin repeats in Swi6.
Genetic interactions with stb1Δ.
Our finding that Stb1 interacts in vitro with Swi6, a regulator of Start transcription, prompted us to look for cell cycle defects in the stb1Δ mutant. We found no defects in the stb1Δ mutant in terms of growth rate, cell cycle progression, or transcript levels and cell cycle periodicity of a panel of Start-expressed genes (data not shown). Similarly, another study did not find a requirement for STB1 in expression of HO (24).
As mentioned above, certain combinations of mutations affecting the components of SBF and MBF exhibit lethal genetic interactions with one another (for example, swi4Δswi6Δ and swi4Δmbp1Δ strains are not viable). Because we found that Stb1 interacted physically with Swi6, we tested for genetic interactions between STB1 and SWI4, SWI6, and MBP1. No obvious additional defects were found in stb1Δswi4Δ, stb1Δswi6Δ, or stb1Δmbp1Δ strains in terms of growth rate or cell morphology (data not shown).
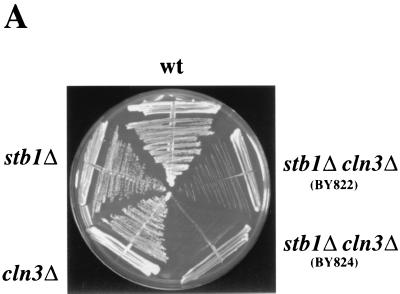
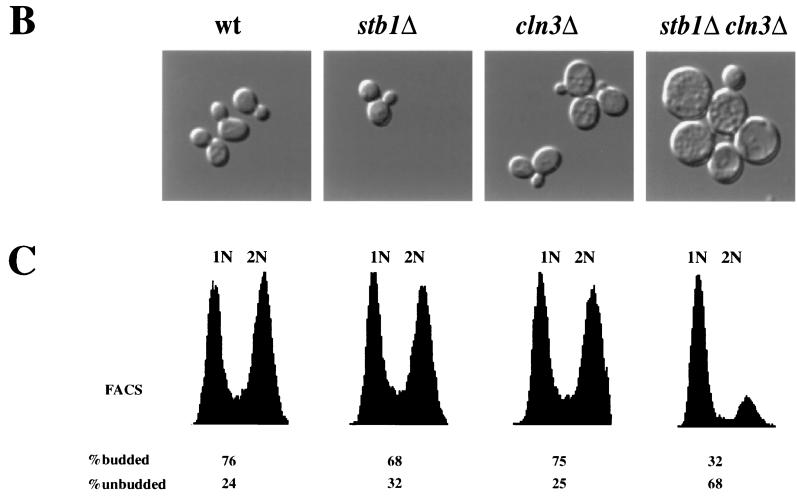
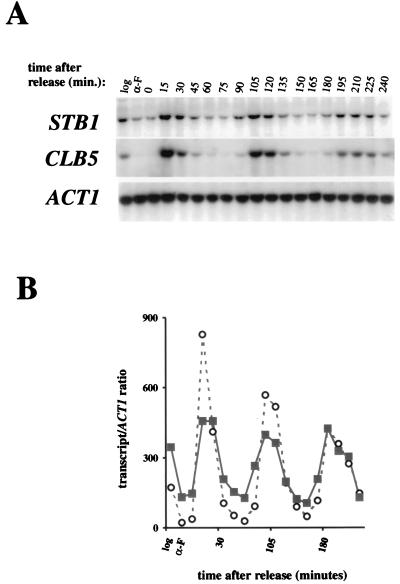
As noted previously, a number of genetic observations suggest that the Cln3-Cdc28 kinase is required for efficient activation of transcription at Start (11, 48). However, CLN3 is not absolutely required for activation of SBF- and MBF-dependent gene expression. In the absence of CLN3, activation of Start transcription is only delayed. These observations suggest that other regulators, such as BCK2, may activate SBF and MBF in the absence of CLN3 (10, 13). We constructed a stb1Δcln3Δ double mutant to test whether STB1 and CLN3 might be functioning in parallel to activate Start transcription. When cultured in rich or minimal media, both the cln3Δ strain and the stb1Δ mutant had growth rates comparable to the wild-type strain (Fig. 2A and data not shown). The stb1Δcln3Δ double mutant grew much more slowly than the wild-type strain, with an average doubling time in rich media of 4.5 h, compared with 1.5 h for the wild-type, cln3Δ, and stb1Δ strains (Fig. 2A and data not shown). Analysis of cell morphology and DNA content in log phase cultures revealed that the stb1Δcln3Δ double mutant cells accumulated in the G1 phase as predominantly large, unbudded cells (Fig. 2B and C). Of the Cdc28 G1 cyclins, only CLN3 was required for normal cell cycle progression in the absence of STB1; no additional growth defects were observed in stb1Δcln1Δcln2Δ mutants (data not shown). These observations support the notion that STB1 may be acting in parallel with CLN3 to regulate Start transcription. Consistent with a role for STB1 during the late G1 phase, STB1 transcript levels fluctuated during the cell cycle, with peak levels in late G1, coincident with maximal expression of other Start-regulated genes (Fig. 3 [CLB5 is shown]). Another group, using microarray hybridization, also found that STB1 was expressed at the same time as other Start-expressed genes (45). The promoter region 500 bp upstream of the STB1 start codon contains two matches to the MCB consensus and one potential SCB element (CGCGAAA); these sequences may be responsible for the G1-specific pattern of STB1 gene expression. Taken together, our results suggest that STB1 plays a role in G1 progression.
FIG. 2.
Growth characteristics of the stb1Δ cln3Δ double mutant. (A) Slow-growth phenotype of a stb1Δcln3Δ mutant strain. Wild-type (wt) (BY263), stb1Δ (BY805), and cln3Δ (BY655) strains and two isolates of the stb1Δcln3Δ double-mutant strain (BY822 and BY824) were streaked onto a YPD plate and incubated at 30°C. (B) Morphology of wild-type, stb1Δ, cln3Δ, and stb1Δcln3Δ strains. Wild-type (BY263), stb1Δ (BY806), cln3Δ (BY655), and stb1Δcln3Δ (BY822) strains were grown to mid-log phase in rich medium. The cells were viewed with Nomarski optics and photographed. (C) DNA content as measured by FACS analysis of samples shown in panel B). The positions of cells with G1 or G2 DNA contents are indicated by 1N and 2N, respectively. The percentages of budded and unbudded cells are shown below the FACS profile.
FIG. 3.
Analysis of STB1 gene expression through the cell cycle. (A) Cell cycle Northern blot analysis by a pheromone block-release method (see Materials and Methods). A wild-type strain (W303) was synchronized in the G1 phase by treatment with α-factor and released into YPD medium. RNA was prepared from samples taken every 15 min for Northern analysis with STB1, CLB5, and ACT1 (actin) probes. The lane labelled log shows RNA isolated from a log-phase culture, and the lane labelled α-F shows RNA isolated from the pheromone-arrested cells. (B) Quantitation of the Northern blot shown in panel A. The STB1 and CLB5 signals were quantitated by PhosphorImager analysis, and the values were normalized to the ACT1 loading control before plotting. Open circles, CLB5 transcript; solid squares, STB1 transcript.
STB1 is required for timely expression of Start genes in cln3Δ mutants.
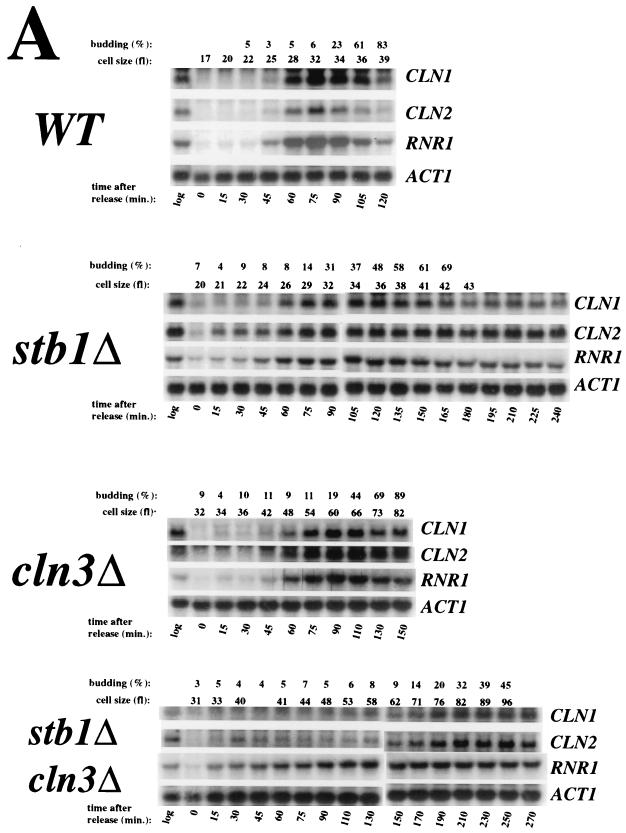
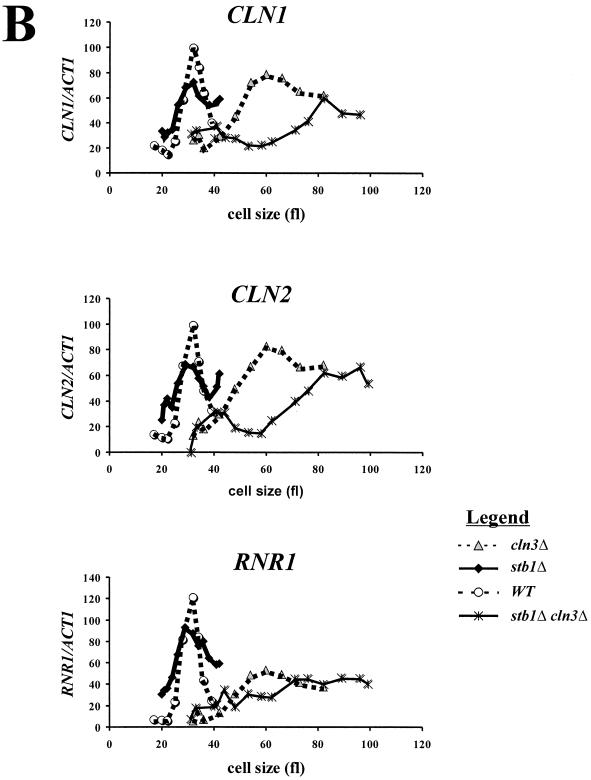
The phenotype of the stb1Δcln3Δ double mutant suggested that, like BCK2, STB1 may act in a parallel pathway with CLN3 for activating transcription at Start. To test this hypothesis, small G1 cells were isolated from wild-type, cln3Δ, stb1Δ, and stb1Δcln3Δ cultures by centrifugal elutriation. The elutriated cells were inoculated into fresh medium, and Start transcription, cell size, and budding index were monitored as the cells progressed synchronously through the cell cycle. Expression of the CLN1, CLN2, and RNR1 genes was analyzed. CLN1 transcription is regulated through MCB elements that are dependent upon SBF (Swi4/Swi6) (36), CLN2 is regulated through both SCB and MCB elements and also through a novel promoter element(s) that is dependent on SWI4 (9, 47), and RNR1 is controlled through MCB elements that are dependent on MBF (Mbp1/Swi6) (12). We found that CLN1, CLN2, and RNR1 transcripts began to accumulate in both the wild type and the stb1Δ mutant at an average cell volume of 25 fl (Fig. 4A and B). As previously reported (51), activation of these genes was significantly delayed in the cln3Δ mutant, with transcripts accumulating at a cell volume of roughly 40 fl (Fig. 4). The delay in transcriptional activation was even more pronounced in the stb1Δcln3Δ double mutant; the CLN1 and CLN2 transcripts began to accumulate at a cell volume of about 60 fl (Fig. 4). There was no clear induction of RNR1 in the stb1Δcln3Δ mutant. We conclude that although Start transcripts are induced with similar kinetics in wild-type and stb1Δ strains, STB1 is important for the induction of Start transcription in a cln3Δ strain.
FIG. 4.
Activation of Start transcription in stb1Δcln3Δ mutants. (A) Cell cycle Northern blot analysis by centrifugal elutriation. Wild type (WT, BY263), stb1Δ (BY805), cln3Δ (BY655), and stb1Δcln3Δ (BY822) cultures were synchronized by isolating small G1 daughter cells by centrifugal elutriation. The small daughter cells were inoculated into YPD medium. Aliquots were taken at 15- or 20-min intervals for total-RNA isolation, budding analysis, and FACS analysis of the DNA content (results not shown). Levels of CLN1, CLN2, RNR1, and ACT1 (actin) transcripts were determined by Northern blot analysis of the total RNA (relevant probe indicated to the right of each panel). The mean cell size for each fraction (in femtoliters), also shown at the top, was estimated by using a Coulter Channelizer. (B) Quantitation of the Northern blot data shown in panel A. CLN1, CLN2, and RNR1 transcript levels were measured by PhosphorImager analysis and normalized against ACT1 transcript levels as an internal control. The normalized expression levels were plotted against the mean cell size.
Figure 4B shows a small difference in the peak height of the transcript levels in the stb1Δ mutant relative to the wild-type strain. This difference between the stb1Δ mutant and the wild-type cells may reflect poorer synchronization of the stb1Δ strain by centrifugal elutriation, since stb1Δ cells synchronized by α-factor block and release exhibited Start transcription profiles identical to the wild type (data not shown).
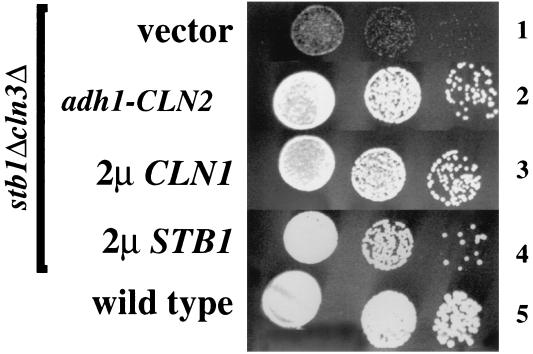
Overexpression of the G1 cyclins CLN1 and CLN2 can rescue the growth defects of strains mutated for known regulators of Start transcription (10, 13, 34, 35). This genetic suppression suggests that inadequate expression of CLN genes accounts for the cell cycle arrest or slow-growth phenotype of these strains. Likewise, the defect in timely activation of Start transcription in the stb1Δcln3Δ double mutant may account for the slow-growth phenotype in this strain. To test this possibility, we transformed the stb1Δcln3Δ double-mutant strain with a high-copy-number plasmid carrying the CLN1 gene or a plasmid expressing CLN2 from a constitutive promoter. Overexpression of either CLN1 or CLN2 suppressed the slow-growth phenotype of the stb1Δcln3Δ mutant, suggesting that the G1 delay in this strain is due to inadequate expression of CLNs (Fig. 5). Since Stb1 and Swi6 physically interact, Stb1 may affect Start transcription directly through SBF and/or MBF.
FIG. 5.
Suppression of the slow-growth defect of the stb1Δ cln3Δ mutant by overexpressed CLN genes. The following yeast strains were grown to log phase in minimal medium, serially diluted, and spotted onto YPD plates: BY822 (stb1Δcln3Δ) transformed with YEp351 (vector) (row 1), BY822 transformed with a plasmid expressing CLN2 from the low-level constitutive S. pombe adh1 promoter (adh1-CLN2) (row 2), BY822 transformed with a high-copy-number plasmid containing CLN1 (2μCLN1) (row 3), BY822 with STB1 on a high-copy-number plasmid (2μSTB1) (row 4), and a wild-type strain (BY263) transformed with vector (row 5). The plates were incubated at 30°C for 2 days and photographed.
Phosphorylation of Stb1 by Cln-associated kinases in vivo and in vitro.
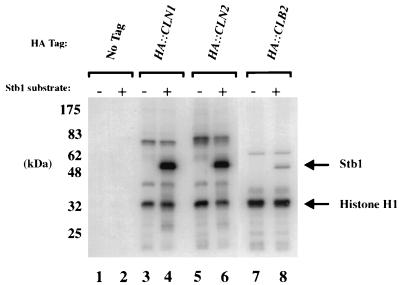
To further explore the function of Stb1, we investigated the relationship between Stb1 and Cln-Cdc28 kinase complexes. The Stb1 amino acid sequence contains five putative Cdc28 phosphorylation sites (S46, S111, S128, T191, and T351, using the consensus S/T, P, x, basic [31]). Epitope-tagged cyclins were used to immunoprecipitate Cln1-Cdc28 and Cln2-Cdc28 kinases from yeast extracts (51, 52). The immunoprecipitates were incubated with [γ-32P]ATP and tested for kinase activity toward purified recombinant Stb1. Histone H1, a substrate commonly used to assay Cdc28 kinase activity, was included as a control in the kinase reactions. Stb1 was an excellent substrate for both the Cln1-Cdc28 and Cln2-Cdc28 kinases in vitro and was a better in vitro substrate than histone H1 (Fig. 6, lanes 3 to 6). Stb1 was also phosphorylated by Cln3-Cdc28 kinase complexes purified from insect cells (53a). In contrast, Stb1 was less effectively phosphorylated by Cdc28 associated with the Clb2 mitotic cyclin (Fig. 6, lanes 7 and 8). All three forms of Cdc28 tested in the experiment in Fig. 6 were normalized with respect to phosphorylation of Histone H1.
FIG. 6.
Phosphorylation of Stb1 by Cdc28 kinases in vitro. Strains bearing HA (hemagglutinin)-tagged alleles of CLN1, CLN2, and CLB2 were lysed, and the tagged cyclin-Cdk complexes were immunoprecipitated with anti-HA antibodies. [γ-32P]ATP and exogenous substrates were added to the immunoprecipitates (1 μg of histone H1 to all reaction mixtures and 0.5 μg of Stb1 protein to reaction mixtures indicated above the lanes). Migrations of molecular size markers are shown to the left, and those of phosphorylated Stb1 and histone H1 are shown to the right.
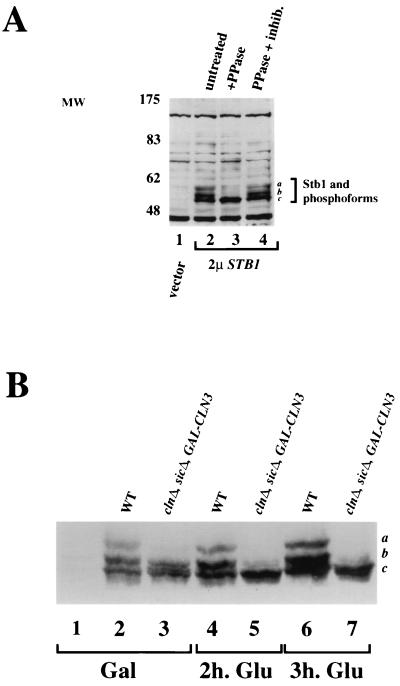
The ability of G1-specific forms of Cdc28 to phosphorylate Stb1 in vitro raised the possibility that Stb1 is a downstream target of Cln-Cdc28 kinases in vivo. To test this possibility, we first asked whether Stb1 was a phosphoprotein in vivo. Immunoblot analysis with affinity-purified antibodies to Stb1 recognized several isoforms of Stb1 in lysates from log-phase yeast cells (Fig. 7A, lane 2). The antibodies recognized endogenous Stb1 (data not shown), but the Stb1 isoforms were more easily detected in a strain bearing a high-copy-number plasmid expressing STB1 from its own promoter (see Materials and Methods) (Fig. 7A). Phosphatase treatment of the extract resulted in loss of the slower-migrating species (Fig. 7A, lane 3, bands a and b) and an increase in the fastest-migrating isoform (band c). The presence of phosphatase inhibitors in the phosphatase reaction prevented the conversion to the fastest-migrating isoform c (Fig. 7A, lane 4). We conclude that Stb1 is a phosphoprotein in vivo and that the slower-migrating species detected on SDS-PAGE correspond to hyperphosphorylated forms of Stb1.
FIG. 7.
Analysis of Stb1 phosphoforms by Western blot analysis. (A) Anti-Stb1 Western blot analysis of extracts prepared from log-phase cells transformed with either empty vector (vector, lane 1) or a high-copy-number STB1 plasmid (2μSTB1, lanes 2 to 4). Extracts were untreated (lane 2), treated with protein phosphatase (PPase) (lane 3), or treated with phosphatase and phosphatase inhibitors (lane 4) prior to Western blot analysis. (B) Anti-Stb1 Western blot on wild-type and cln1Δcln2Δcln3Δ extracts. Extracts were prepared from wild-type (WT) or cln1Δ cln2Δ cln3Δ sic1Δ (clnΔ, sicΔ, GAL-CLN3) mutant strains. The sic1Δ mutation permits growth of the clnΔ strain, which would otherwise die. The strain also carried a genomic GAL-CLN3 gene, which allowed CLN3 expression to be induced by culturing in galactose-containing medium. Extracts were prepared after growth in medium with galactose, where GAL-CLN3 is induced (Gal, lanes 2 and 3), and 2 h (lanes 4 and 5) and 3 h (lanes 6 and 7) after transfer to medium with glucose (Glu) to shut off CLN3 expression. Lane 1 shows an extract from a stb1Δ strain. The positions of migration of the various forms of Stb1 are indicated on the right by a, b, and c.
Because we saw in vitro phosphorylation of Stb1 by Cln-associated kinases, we tested whether the Stb1 phosphoforms detected in vivo are dependent on the CLN genes genetically. Although each CLN normally plays distinct roles in wild-type yeast cells, mutational analyses have shown that the CLN genes are genetically redundant. That is, any single CLN gene is sufficient to support cell viability and cell cycle progression. Given this genetic redundancy, we assayed Stb1 phosphoforms in yeast extracts from strains with all three CLN genes deleted. Since a strain lacking CLN1, CLN2, and CLN3 would normally arrest at Start, the cells were kept alive by deletion of the B-type Cdk inhibitor SIC1 (41, 50) and by expression of CLN3 from the inducible GAL promoter (GAL-CLN3) (52). The cln1,2,3Δ sic1ΔGAL-CLN3 strain was first grown on galactose-containing medium, to turn on CLN3 expression (Fig. 7B, lanes 1 to 3) and later transferred to glucose-containing medium to repress CLN3 expression, thereby depleting all Cln products from the yeast cell (lanes 4 to 7). We used anti-Stb1 antibody immunoblots on extracts from the Cln-depleted cells to show that the phosphorylated isoforms of Stb1 were dependent on CLN function (Fig. 7B). When Cln3 was the only source of Cln proteins, only the two lower phosphoforms of Stb1 were detected (Fig. 7B, lane 3, bands b and c). Relative to band c, band b was less intense in extracts from the clnΔGAL-CLN3 strain grown on glucose than in extracts from the wild-type strain. When GAL-CLN3 expression was shut off in the presence of glucose, the two upper bands (a and b) disappeared and band c increased in intensity. Therefore, Stb1 phosphoforms are dependent on CLN function in vivo. In conjunction with the specific phosphorylation of Stb1 by Cln kinases in vitro, this result strongly suggests that Stb1 is a direct target of Cln-associated kinases in vivo.
Cell cycle-dependent phosphorylation of Stb1.
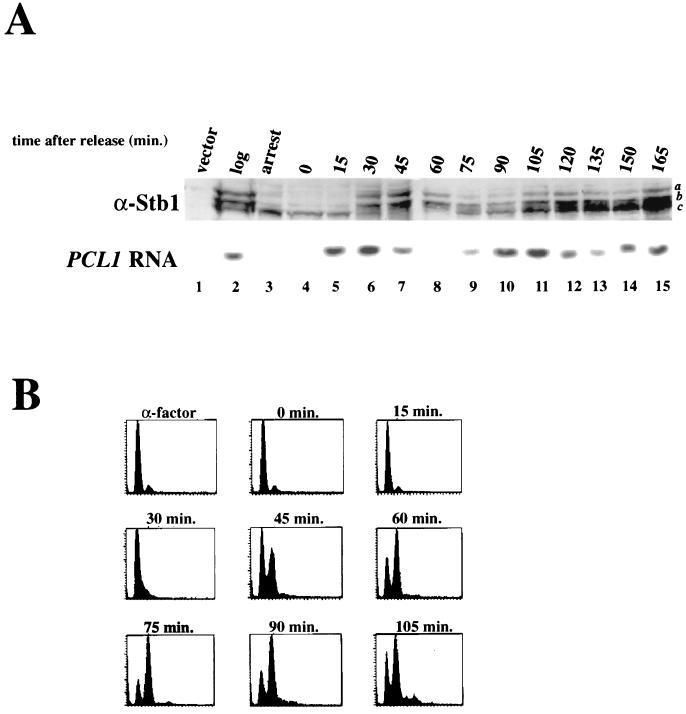
Because our analysis with asynchronous cultures showed that Stb1 was phosphorylated in a Cln-dependent manner, we next asked if phosphorylation of Stb1 was cell cycle periodic to reflect the periodic abundance of Cln proteins. To examine the phosphorylation of Stb1 throughout the cell cycle, α-factor mating pheromone was used to arrest cells in the G1 phase before Start, and the arrested cells were released into fresh medium. Extracts were prepared from samples as the culture progressed synchronously through the cell cycle (5). Figure 8 shows an immunoblot with anti-Stb1 antibodies on samples isolated from a synchronous culture. The slower-migrating, hyperphosphorylated forms of Stb1 (bands a and b) were absent in cells arrested by α-factor treatment (Fig. 8A, lane 3). The reduced phosphorylation of Stb1 in response to pheromone mirrored the inhibition of Cln1,2-Cdc28 kinase that is seen in α-factor-treated cells (39). The upper Stb1 phosphoforms then gradually reappeared as the cells progressed synchronously through the cell cycle (lanes 4 to 7). To monitor the timing of Stb1 phosphorylation during the cell cycle, the accumulation of the SBF-regulated PCL1 transcript was used as a marker for Start transcription (Fig. 8A), and FACS analysis of the cellular DNA content was used as an indicator for DNA synthesis and cell cycle position (Fig. 8B). We found that Stb1 phosphorylation reappeared after the appearance of PCL1 transcripts (Fig. 8A, lanes 5 and 6), but before DNA replication (Fig. 8B, 30 min). During the peak of Stb1 phosphorylation, only the two upper phosphoforms (a and b) were seen (Fig. 8A, lanes 6 to 8). The Stb1 phosphoforms then began to disappear in the G2 phase (Fig. 8A, lane 9; Fig. 8B, 75 min). Thus, Stb1 is phosphorylated in a cell cycle-dependent manner in vivo, with Stb1 phosphoforms beginning to accumulate early in the cell immediately after the appearance of Start-dependent transcripts. The timing of Stb1 phosphorylation is consistent with our data suggesting that Stb1 is a substrate for Cln-associated kinases.
FIG. 8.
Cell cycle analysis of Stb1 phosphoforms. (A) Western blot analysis of Stb1 phosphoforms in synchronized cell extracts. Extracts were prepared from log-phase (log, lane 2), α-factor-arrested (arrest, lane 3), or synchronized (lanes 4 to 15) cells. Cells were synchronized in the G1 phase by treatment with α-factor and released into fresh medium, and samples were taken from the synchronized cultures at the time points indicated above the lanes (15-min intervals). Various forms of Stb1 are denoted a, b, and c (on the right). Total RNA was isolated from the same samples, and PCL1 transcript levels were assessed by Northern blot analysis (below the Stb1 Western blot). The timing of PCL1 transcript appearance serves as a marker for Start transcription. (B) DNA content as measured by FACS analysis of α-factor-treated and synchronous cells used in panel A.
DISCUSSION
In this paper, we describe a series of experiments suggesting a role for Stb1 in the activation of transcription at Start; specifically, we found that STB1 was important for Start transcription when CLN3 is deleted. Compared with the cln3Δ mutant, the stb1Δcln3Δ mutant was slower growing, had larger cells, showed a more pronounced G1 delay, and underwent Start transcription at a larger cell volume. The slow-growth phenotype of the stb1Δcln3Δ mutant was suppressed by ectopic expression of CLN1 or CLN2, confirming that the slow-growth phenotype was due to the delayed timing of Start transcription. Stb1 may affect Start transcription through its interaction with Swi6. The Stb1 protein was phosphorylated by Cln-Cdc28 kinases in vitro and was phosphorylated in vivo in a CLN-dependent manner. We discuss the possible roles for Cln-dependent phosphorylation of Stb1 below.
Swi6-Stb1 interaction.
In this study, we describe a physical interaction between Swi6 and Stb1 in vitro. The Swi6 ankyrin repeats were required for binding of Stb1 to a Swi6 column, and Swi6 bound directly, in the absence of DNA or other proteins, to Stb1 in a far-Western blot assay. Consistent with our affinity chromatography results, a small amount of Swi6 can be coimmunoprecipitated with Stb1 from yeast extracts (7a). In choosing to study the biological role of STB1, we did not analyze in detail the regions of Stb1 or Swi6 required for their interaction. However, it will be interesting to assay Stb1 binding to Swi4 or Mbp1, which also contain ankyrin repeats. The close conservation of the ankyrin repeat region in cell cycle-regulatory transcription factors of three yeast species suggests that the ankyrin domains in these proteins may bind a similar or conserved protein (6, 25). Indeed, crystallographic studies of the Swi6 ankyrin repeat region predict that Swi6 family members have an arrangement of secondary-structure elements that is distinct from other ankyrin repeat proteins (15). However, despite the apparent conservation of the ankyrin repeat region in Swi6 family members, the Clb2 protein binds specifically to the Swi4 ankyrin repeats and not to those in Swi6 or Mbp1 (44). This result suggests that in Swi4, the ankyrin repeats mediate an interaction that is specific to the Swi4 motifs.
A substitution mutation in the Swi6 ankyrin repeats, G347D, has been suggested to cause a deficiency in protein-protein interactions (14, 15). Whether the binding of Swi6 to Stb1 requires the region of Swi6 defined by the G347D mutation remains to be tested. However, swi6 mutants defective for Stb1 binding may not have been isolated in the screen in which G347D was isolated; this screen selected for swi6 mutants that failed to express HO::lacZ, while stb1Δ mutants exhibit no defects in HO or SCB::lacZ expression (20a, 24).
Regulation of Start transcription by Stb1.
We found that Stb1 and Swi6 interact directly in vitro, suggesting that Stb1 may activate gene expression by interacting with Swi6, a common subunit in SBF and MBF. Stb1 was originally identified in a two-hybrid screen as a protein that bound the general transcriptional repressor, Sin3 (24). If Stb1 and Sin3 functionally interact, it is possible that Stb1, bound to promoters via Swi6, affects Start transcription through Sin3. However, no direct role for Stb1 has been reported in Sin3-dependent transcriptional repression. Moreover, HO is the only Start-expressed gene whose expression has been reported to be dependent on SIN3. Although SIN3 does affect HO transcription, a role for SIN3 in SBF-mediated transcription of HO has not been reported (46), and the cell cycle-regulated expression of HO or any other Start gene in a sin3Δ mutant has not been examined. Sin3 was not detected in the Swi6 affinity column eluates (20a), suggesting that Sin3 is not present in Swi6-Stb1 complexes.
Although a role for Stb1 in transcriptional activation or repression has not been ruled out, we suggest that Stb1 regulates the timing of Start transcription. Mutation of STB1 delays Start transcription in a cln3Δ mutant but does not significantly affect the overall levels of Start transcripts (Fig. 4B); also, the levels of Start transcripts in log-phase cultures of stb1Δ and stb1Δcln3Δ mutant cells were virtually identical to those in wild-type cells (data not shown). Moreover, a stb1Δ strain synchronized by a pheromone block-release protocol exhibited an identical transcriptional profile to a wild-type strain (data not shown; as noted in Results, the small difference between the stb1Δ and wild-type transcription profile in Fig. 4B may be due to differences in cell synchrony introduced by centrifugal elutriation). Therefore, Stb1 does not affect the transcriptional activation of Start genes per se but, rather, regulates the timing of Start transcription. The regulatory role of Stb1 is manifest in the stb1Δcln3Δ mutant. We suggest that Stb1, like Cln3, is a specific regulatory of the timing of Start transcription.
Role of CLN-dependent phosphorylation of Stb1.
Stb1 is phosphorylated in vitro by Cln-associated kinases, and Stb1 phosphoforms in vivo are dependent on the CLN genes. Although CLN3cln1Δcln2Δ cells showed Stb1 phosphoforms (Fig. 7B, lane 3, and data not shown), several observations suggest that Cln1-Cdc28 and Cln2-Cdc28 are the physiological kinases for Stb1 phosphorylation. First, we observed only low levels of Stb1 phosphorylation in a cln1Δcln2ΔGAL-CLN3 strain, despite overproduction of Cln3 (Fig. 7B, lane 3). Second, the Stb1 phosphoforms in a CLN1CLN2cln3Δ strain were comparable to those seen in a wild-type strain (data not shown). Third, Stb1 was phosphorylated after Start, at the time when Cln1 and Cln2 levels are induced. Finally, in the cln3Δ mutant, Cln1 and Cln2 are sufficient to phosphorylate Stb1 at the time of Start transcription (20a).
So far, our studies have not addressed the functional role of Stb1 phosphorylation. However, together with previous observations, our data suggest three possible models for the role of Cln1- and Cln2-dependent phosphorylation on Stb1. The first model invokes a role for Stb1 phosphorylation in the down-regulation of SBF-dependent transcription after Start. This model is supported by our observation that Stb1 is phosphorylated shortly after the burst of transcription at Start and by previous studies showing that CLN1 and CLN2 are required for the proper down-regulation of Start transcription (11, 48). Since CLN1 and CLN2 are important for budding and DNA synthesis, a second model is that Cln1 and Cln2 may phosphorylate Stb1 to mediate these post-Start functions. However, neither of these two models accounts for the delay in Start transcription that we observed in stb1Δcln3Δ cells.
A third model centers on the transcriptional defect of the stb1Δ cln3Δ mutant and posits that Cln1 and Cln2 kinases phosphorylate Stb1 to activate STB1-dependent transcription at Start. This hypothesis is consistent with several observations. First, Stb1 interacts directly with the Swi6 transcription factor in vitro. Second, CLN1/2 and STB1 are both important for Start transcription in the absence of CLN3 (cln1Δcln2Δcln3Δ mutants arrest before Start transcription, and stb1Δcln3Δ mutants are greatly delayed for Start transcription [reference 56 and this study]). Since both CLN1/2 and STB1 positively affect Start transcription, the simplest model for the function of Cln1- and Cln2-dependent phosphorylation on Stb1 is that Cln1 and Cln2 kinases phosphorylate Stb1 to activate Start transcription. This model is not consistent with the appearance of Stb1 phosphoforms after the peak in Start transcription in wild-type cells. However, in a cln3Δ mutant, where STB1 and CLN1/2 become genetically important for the timing of Start transcription, Stb1 phosphoforms appeared coincidentally with the appearance of Start transcripts (20a). This result suggests that Stb1 phosphoforms may become important in the cln3Δ mutant for activating Start transcription. Mutational analysis of the Stb1 phosphorylation sites will probably resolve this issue.
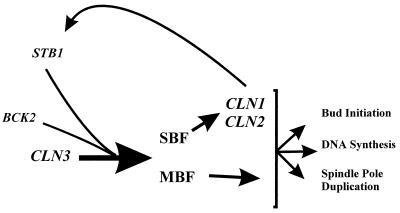
Parallel pathway(s) to CLN3 for activating start transcription.
Our data suggest that, like BCK2, STB1 functions parallel to CLN3 in the activation of Start transcription (Fig. 9). In any case, there are probably at least three pathways for activating Start transcription because stb1Δcln3Δ double mutants still undergo Start transcriptional activation, albeit at a much larger cell size. At this point, we do not understand the molecular details of how STB1, BCK2, or CLN3 activates transcription at Start, and our experiments do not specifically address whether STB1 and BCK2 function in the same or in parallel pathways. However, our data suggest that STB1 is a downstream component of at least one of these pathways, because it interacts directly with the Swi6 transcription factor.
FIG. 9.
Model for the role of STB1 in activation of transcription at Start. STB1 and BCK2 play important roles in activating Start transcription in the absence of CLN3 function and may define a parallel pathway(s) to CLN3 for activating gene expression at Start. Stb1 probably regulates transcriptional activation through interaction with the Swi6 subunit of the SBF and MBF transcription complexes. Stb1 is a substrate for Cln-Cdc28 kinases (most probably Cln1,2-Cdc28; see the text for a discussion of the possible roles for Stb1 phosphorylation).
What might be the physiological role of CLN3-independent pathways for activating Start transcription? In a wild-type cell, deletion of BCK2 or STB1 has little effect on the timing of Start (reference 10 and this study) whereas cln3Δ mutants undergo Start at a larger cell size (51). These data imply that CLN3 is the major activator of Start transcription and that the CLN3-independent pathways are important in the cln3Δ mutant only as a “back-up” pathway. However, most studies on CLN3, STB1, and BCK2 function used optimal growth conditions in defined or rich media. Under these growth conditions, CLN3 may indeed be the most significant activator of Start transcription. However, there is no evidence that deletion of CLN3 affects the timing of Start in cells grown under suboptimal conditions. Cln3 is downregulated, transcriptionally, translationally and by protein degradation, in response to poor nitrogen and carbon sources (16, 37, 38, 40). Paradoxically, cells grown in poor carbon sources, where Cln3 activity is low, undergo Start at a smaller cell size (22, 32). This result is unexpected since cln3Δ cells are large and cells overexpressing CLN3 are small (32, 52). These observation suggest that in poor carbon and/or nitrogen sources, CLN3 may play a less dominant role in activating Start transcription. The importance of the CLN3-independent pathways for activating SBF- and MBF-dependent transcription may become manifest in cells grown under suboptimal growth conditions.
ACKNOWLEDGMENTS
We thank A. Spence, L. Harrington, and M. Tyers for critical comments on the manuscript; M. Kasten and D. Stillman for unpublished reagents; C. Amille Walker for technical assistance.
Y.H. was supported in part by an Ontario Graduate Scholarship, M.C. was supported by a Connaught Scholarship and Open Fellowship from the University of Toronto, and R.K. was supported by NIH grant CA45508. This work was supported by a grant from the Medical Research Council (MRC) of Canada to B.A., who is an MRC Scientist.
REFERENCES
- 1.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 2.Andrews B J, Herskowitz I. Identification of a DNA binding factor involved in cell cycle-control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel R, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Blanar M, Rutter W. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 5.Breeden L. Alpha-factor synchronization of budding yeast. Methods Enzymol. 1997;283:332–341. doi: 10.1016/s0076-6879(97)83027-3. [DOI] [PubMed] [Google Scholar]
- 6.Breeden L. Start-specific transcription in yeast. Curr Top Microbiol Immunol. 1995;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- 7.Christianson T, Sikorski R, Dante M, Shero J, Heiter P. Multifunctional yeast high-copy shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 7a.Costanzo, M. Unpublished data.
- 8.Cross F. ’Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 9.Cross F, Hoek M, McKinney J D, Tinkelenberg A H. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiComo C J, Chang H, Arndt K T. Activation of CLN1 and CLN2 G1 cyclin expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirick L, Bohm T, Nasmyth K. Roles and regulation of the Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Donoviel, M., and Y. Ho. Unpublished data.
- 12.Elledge S J, Davis R W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 13.Epstein C, Cross F R. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewaskow S P, Sidorova J M, Hendle J, Emery J C, Lycan D E, Zhang K Y J, Breeden L L. Mutation and modeling analysis of the Saccharomyces cerevisiae Swi6 ankyrin repeats. Biochemistry. 1998;37:4437–4450. doi: 10.1021/bi972652e. [DOI] [PubMed] [Google Scholar]
- 15.Foord R, Taylor I, Sedgwick S, Smerdon S. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat Struct Biol. 1999;6:157–165. doi: 10.1038/5845. [DOI] [PubMed] [Google Scholar]
- 16.Gallego C, Gari E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorina S, Pavletich N. Structure of the p53 tumour suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 18.Hadwiger J A, Wittenberg C, Richardson H, De Barros Lopes M, Reed S I. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington L A, Andrews B J. Binding to the yeast Swi4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, HRR25, and the cell cycle regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Ho, Y. Unpublished data.
- 21.Igual J C, Johnson A L, Johnston L H. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 22.Jagadish M, Carter B. Genetic control of cell division in yeast cultured at different growth rates. Nature. 1977;269:145–147. doi: 10.1038/269145a0. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 24.Kasten M M, Stillman D J. Identification of the Saccharomyces cerevisiae genes STB1–STB5 encoding Sin3p binding proteins. Mol Gen Genet. 1997;256:376–386. doi: 10.1007/s004380050581. [DOI] [PubMed] [Google Scholar]
- 25.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 26.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 27.Madden K, Sheu Y-J, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- 28.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 29.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A, Andrews B. A family of cyclin-like proteins that interact with the cyclin-dependent kinase, Pho85. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaely P, Bennett V. The ANK repeat: a ubiquitous motif involved in macromolecular recognition. Trends Cell Biol. 1992;2:127–129. doi: 10.1016/0962-8924(92)90084-z. [DOI] [PubMed] [Google Scholar]
- 31.Morgan D. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 32.Nash R, Tokiwa G, Anand S, Erickson K, Futcher B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 34.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 35.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2 and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1 specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 36.Partridge J F, Mikesell G E, Breeden L L. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J Biol Chem. 1997;272:9071–9077. doi: 10.1074/jbc.272.14.9071. [DOI] [PubMed] [Google Scholar]
- 37.Parviz F, Heideman W. Growth-independent regulation of CLN3 mRNA levels by nutrients in Saccharomyces cerevisiae. J Bacteriol. 1998;180:225–230. doi: 10.1128/jb.180.2.225-230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parviz G, Hall D, Markwardt D, Heideman W. Transcriptional regulation of CLN3 expression by glucose in Saccharomyces cerevisiae. J Bacteriol. 1998;180:4508–4515. doi: 10.1128/jb.180.17.4508-4515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 40.Polymenis M, Schmidt E. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider B L, Yang Q-H, Futcher B. Linkage of replication to Start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 42.Sia R A L, Herald H A, Lew D J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidorova J, Breeden L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-phase specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegmund R F, Nasmyth K. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol Cell Biol. 1996;16:2647–2655. doi: 10.1128/mcb.16.6.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellman P T, Sherlock G, Zhang M, Iyer V, Anders K, Eisen M, Brown P, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sternberg P, Stern M J, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 47.Stuart D, Wittenberg C. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 49.Tevelev A, Byeon I, Selby T, Ericson K, Kim H, Kryanov V, Tsai M. Tumour suppressor p16INK4A: structural characterization of wild-type and mutant proteins by NMR and circular dichroism. Biochemistry. 1996;35:9475–9487. doi: 10.1021/bi960211+. [DOI] [PubMed] [Google Scholar]
- 50.Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for CLN G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinson C, LaMarco K, Johnson P, Landschulz W, McKnight S. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988;2:801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- 53a.Wang, J., and M. Tyers. Personal communication.
- 54.Wang R, Kobayashi R, Bishop J M. Cellular adherence elicits ligand-independent activation of the Met cell-surface receptor. Proc Natl Acad Sci USA. 1996;93:8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willems A, Lanker S, Patton E, Craig K, Nason T, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 56.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]