ABSTRACT.
Reliable data on antimicrobial resistance (AMR) transmission dynamics in Uganda remains scarce; hence, we studied this area. Eighty-six index patients and “others” were recruited. Index patients were those who had been admitted to the orthopedic ward of Mulago National Referral Hospital during the study period; “others” included medical and non-medical caretakers of the index patients, and index patients’ immediate admitted hospital neighbors. Others were recruited only when index patients became positive for carrying antimicrobial-resistant bacteria (ARB) during their hospital stay. A total of 149 samples, including those from the inanimate environment, were analyzed microbiologically for ARB, and ARB were analyzed for their antimicrobial susceptibility profiles and mechanisms underlying observed resistances. We describe the diagnostic accuracy of the extended-spectrum β-lactamase (ESBL) production screening method, and AMR acquisition and transmission dynamics. Index patients were mostly carriers of ESBL-producing Enterobacteriaceae (PE) on admission, whereas non-ESBL-PE carriers on admission (61%) became carriers after 48 hours of admission (9%). The majority of ESBL-PE carriers on admission (56%) were referrals or transfers from other health-care facilities. Only 1 of 46 samples from the environment isolated an ESBL-PE. Marked resistance (> 90%) to β-lactams and folate-pathway inhibitors were observed. The ESBL screening method’s sensitivity, specificity, positive predictive value, and negative predictive value were 100%, 50%, 90%, and 100%, respectively. AMR acquisition and transmission occurs via human–human interfaces within and outside of health-care facilities compared with human–inanimate environment interfaces. However, this remains subject to further research.
INTRODUCTION
For many decades, antibiotics have contributed greatly to the reduction in morbidity and mortality, particularly in the treatment and prevention of infections.1–3 However, antibiotics are currently threatened by the emergence and spread of antimicrobial resistance (AMR) within a wide range of infectious agents.4–6 The rise in AMR likely contributes to the post-antibiotic era in which minor infections become fatal as a result of the ineffectiveness of currently available antibiotics, and such infections are on the increase globally.1,2,7 The post-antibiotic era first described as an apocalyptic fantasy before the 21st century has now become a very real possibility.1,2,7
It is now estimated that, each year, at least 700,000 people worldwide die of infections caused by antimicrobial-resistant bacteria (ARB). By 2050, infections involving ARB are predicted to cause the death of an estimated 10 million lives per year, with Africa alone contributing about 4.15 million deaths annually.8,9 It is also predicted8 that the economic cost in lost global production caused by AMR will amount to approximately $100 trillion between now and 2050 if the AMR problem is not tackled.
In East Africa, accurate and reliable data expressly about AMR acquisition and transmission dynamics remain scarce, and yet these data are essential for formulating and monitoring effective responses to AMR.3,9,10 Hence, this study aimed to leverage a source perspective to elaborate the acquisition and transmission dynamics of AMR and the risk factors influencing them in the orthopedic ward of Mulago National Referral Hospital, Uganda, to contribute ultimately to a deeper understanding of these AMR-related topics.
MATERIALS AND METHODS
Study design and duration.
This was a laboratory-based unicentral longitudinal study that was part of and foundational to a larger study titled “Understanding Transmission Dynamics and Acquisition of AMR at Referral Hospitals and Community Settings in East Africa” (GCA-AMR-2019). Our study was conducted between September 2019 and March 2020, and aimed to leverage a source perspective to understand transmission dynamics and acquisition of AMR in the orthopedic ward of Mulago National Referral Hospital, Uganda. The specific aims of this study included determining the carriage of ARB by study participants, antimicrobial susceptibility profiles of the isolated ARB, common phenotypic mechanisms mediating resistance in the ARB, and based on these results, transmission dynamics and acquisition of AMR in the orthopedic ward.
Study population and sampling strategy.
The sample sizes were determined based on the admitted index patients positive for ARB during their hospital stay. Briefly, purposive consecutive sampling was used to recruit index patients admitted into the orthopedic ward of Mulago National Referral Hospital. Samples were collected from index patients on admission or within 24 hours of their stay in the orthopedic ward. In the event the index patients were positive for carrying ARB on admission or within 24 hours of their stay, sampling of their home environment was done. However, in the event index patients were negative for carrying ARB on admission or within 24 hours of their stay, similar additional samples were obtained subsequently from these patients 48 and 72 hours after their stay in an attempt to isolate ARB. When index patients became positive for carrying ARB 48 or 72 hours after their stay, similar samples were obtained from the other study participants (medical caretakers or health-care workers attending to the index patients, non-medical caretakers of the index patients, the immediate admitted hospital neighbors of the index patients) and the index patients’ immediate inanimate environment (bed headboards and bed handrails) in an attempt to isolate ARB. In addition, environmental sampling was done twice (at the start of the study and 3 months after the start of the study). During this sampling, samples were collected using 0.9% saline premoistened sterile cotton swabs and included those from the inanimate ward environment (electrical switches, health-care worker handwashing sinks, health-care worker reception tables, and ward door handles). Because of the low number of admissions per week (n = 4–5), the purposive consecutive sampling of the index patients was based on five, with a consent rate of 100%. In other words, all five patients admitted each week were included in the study. Also, because of the limited number of medical caretakers or health-care workers attending to the index patients during the day shift, some were included more than once. However, during analysis, their data were considered once.
Study samples, sample collection, and transportation.
Study samples included rectal swabs and swabs from the inanimate environment: index patient bed headboard, index patient bed handrails, and inanimate ward environment (electrical switches, health-care worker handwashing sinks, health-care worker reception tables, and ward door handles). The samples were collected aseptically and were then labeled properly and transported to the clinical microbiology laboratory of Mulago National Referral Hospital for testing within 2 hours after collection.
Microbial culture and antimicrobial susceptibility profiling.
Isolation of AMR Gram-negative and Gram-positive bacteria was done using conventional microbiological culture. Briefly, isolation of the Gram-negative bacteria was done by inoculating the collected samples on MacConkey media, a selective and differential culture media containing 4 µg/mL cefotaxime, a third-generation cephalosporin, or 16 µg/mL ceftazidime, also a third-generation cephalosporin. Determination of the concentrations of the antibiotics cefotaxime and ceftazidime was done per the Clinical Laboratory and Standard Institute (CLSI) 2019 guidelines.11
Similarly, isolation of the Gram-positive bacteria was done by inoculating the samples on Mannitol salt agar, a selective and differential culture media containing 6 µg/mL oxacillin. Determination of the concentration of oxacillin was done per the CLSI 2019 guidelines.11 After inoculation of the samples on the respective culture media, incubation was done at 35 ± 2°C for 16 to 18 hours in ambient air.11
After incubation, identification of the bacteria was done using the organisms’ phenotypic characteristics (respective morphological, cultural, and biochemical characteristics).12,13 The morphological characteristics were used in the identification were based on the Gram-staining properties of the respective bacteria. Cultural characteristics included the colony morphology characteristics of respective colonies of the different bacteria on culture media. Biochemical characteristics were those derived from different tests—namely, catalase, triple sugar iron (TSI), sulfur indole and motility, citrate, urease, and oxidase.12,13
In particular, Escherichia coli colonies were identified by their lactose fermentation characteristic; entire, fixed margins; and their steady growth pattern that creates concentric growth rings in the colonies when viewed using a microscope.12,13 Also, positive catalase, TSI, motility, indole, and negative citrate, urease, and oxidase were used to identify E. coli.12,13
Klebsiella pneumoniae colonies were identified by their characteristic Gram-negative reaction; short, plump, and straight rod appearance when viewed using a microscope; lactose fermenting mucoid dome-shaped characteristic; positive catalase, TSI, citrate, and urease; and negative indole, motility, and oxidase.12–14
Antimicrobial susceptibility profiling was performed with the Kirby Bauer disk diffusion method on Mueller Hinton Agar (Biolab, Budapest, Hungary) as recommended by the CLSI 2019 guidelines.11 Briefly, two to three colonies were emulsified in sterile saline. The turbidity of the resulting suspension was then adjusted to the 0.5 McFarland standard. Antibiotic disks (Biolab) used included ampicillin, 10 µg; amoxicillin–clavulanic acid, 20/10 µg; cefotaxime, 30 µg; ceftazidime, 30 µg; meropenem, 30 µg; trimethoprim/sulfamethoxazole, 1.25 mg/23.75 mg; chloramphenicol, 30 µg; gentamycin, 10 µg; ciprofloxacin, 5 µg; cefepime, 30 µg; piperacillin–tazobactam, 100 mg/10 mg; tetracycline, 30 µg; amikacin, 30 µg; colistin, 10 µg; aztreonam, 30 µg; and cefoxitin, 30 µg. The results were interpreted according to CLSI 2019 guidelines.11 Escherichia coli ATCC 25992/ATCC 35218 and K. pneumoniae ATCC 700603 were used to control the antimicrobial susceptibility profiling for quality.
Testing for extended-spectrum and class C β-lactamases (AmpC) production.
Extended-spectrum β-lactamase (ESBL) production was screened for using the modified double-disk synergy method.15,16 During testing, disks of 30 μg cefotaxime, 30 μg ceftazidime, and 30 μg cefepime were placed at 30 mm and 20 mm (for cefotaxime) from 20/10 μg amoxicillin–clavulanic acid or 100/10 μg piperacillin–tazobactam.15,16 Cefepime was placed at 25 mm when 100/10 μg piperacillin–tazobactam was used in place of amoxicillin–clavulanic acid.15,16 The results of this test were reported to two groups after incubation at 35 ± 2°C for 16 to 18 hours in ambient air as follows: group 1 (ESBL producers), which included ESBL producers sensitive, intermediate, or resistant to cefotaxime and ceftazidime; and group 2 (derepressed mutants), which include non-ESBL producers resistant to cefotaxime and ceftazidime.15,16 AmpC b-lactamase producers are resistant to b-lactam–b-lactam inhibitor combinations and give positive and negative tests during ESBL production screening and confirmation,15 it is on these features that screening and confirmation of AmpC b-lactamase production in this study was based. Briefly, the target isolates were first screened for AmpC production using 30 µg cefoxitin. Isolates with inhibition zones ≤ 18 mm were considered positive for AmpC β-lactamase production. Confirmation of AmpC β-lactamase production was done by using a combination of 30 µg cefoxitin and 200 µg cloxacillin, and by comparing the resulting inhibition zones against those of 30 µg cefoxitin without 200 µg cloxacillin. Differences in inhibition zones ≥ 4 mm in the cefoxitin and cloxacillin combination compared with cefoxitin alone were considered positive for AmpC β-lactamase production.
Data management and analysis.
Data obtained from microbial culture, antimicrobial susceptibility profiling, and interviewer-administered questionnaire responses were checked for completeness. The data were also double-entered for validation using EpiData (v. 3.1; EpiData Association, Odense, Denmark). The data were then exported to STATA (v.13; Stata Corp., College Station, TX) for analysis. Pearson χ2 analysis was used to assess for any differences between ESBL production and non-ESBL production regarding demographic characteristics. Means of continuous variables (particularly age and inhibition zone) were compared using the independent t-test. Crude logistic regression analysis was used to explore laboratory characteristics of ESBL producers for comparison with non-ESBL producers. The differences were considered significant at P ≤ 0.05. Multi-drug resistance was defined as resistance to three or more classes of antibiotics.17 The types of antibiotics included amikacin and gentamycin (aminoglycosides); amoxicillin–clavulanic acid and piperacillin–tazobactam (β-lactam inhibitor combinations); meropenem (a carbapenem); ceftazidime, cefotaxime, and cefepime (extended-spectrum cephalosporins/third-generation cephalosporins); cefoxitin (a cephamycin); ciprofloxacin (a fluoroquinolone); sulfamethoxazole/trimethoprim (a folate pathway inhibitor); ampicillin (a penicillin); chloramphenicol (a phenicol); aztreonam (a monobactam); colistin (a polymyxin); and tetracycline (a tetracycline). The sensitivity, specificity, and positive and negative predictive values of the screening test were determined as described elsewhere.18 In these computations, the modified double-disk synergy testing method was considered the gold standard. Microbial culture and antimicrobial susceptibility data are described elsewhere19,20 and were used to determine the acquisition and transmission dynamics of ARB in the orthopedic ward.
Ethics approval and consent to participate.
Ethical approval was obtained from the School of Biomedical Sciences–Research and Ethics Committee, College of Health Sciences, Makerere University (approval no. SBS-650); Mulago National Referral Hospital-Research and Ethics Committee (approval no. MHREC-1702); and the Uganda National Council for Science and Technology (approval no. HS411ES). Permission to access the ward was also sought from the orthopedic department of Mulago National Referral Hospital and the orthopedic ward in-charge. Written informed consent was obtained from each index patient, non-medical caretakers of the index patients, medical caretakers or health-care workers attending to the index patients, and the index patients’ immediate hospital neighbors before sample collection. Also, consent for children was provided by their parents or guardians.
RESULTS
Demographic characteristics of the study participants.
A total of 103 participants were enrolled. Of these, 84% were index patients and 16% were “others.” Males constituted 57% of the study participants whereas females constituted 43% of the study participants. Although the study participants were from different regions of Uganda (i.e., western, northern, southern, eastern, and central), the majority (64%) were from Kampala District, which is the capital city of Uganda, and Wakiso District, which partly encircles Kampala, both found in the central region. Several study participants (35%) stated they were employed informally in several skilled and unskilled trades of the informal sector, including farming, carpentry, bricklaying, welding, building and construction, public transport, and business. The unemployed category consisted of pupils (those attending primary school), students (those attending secondary school, university, tertiary or vocational institutions), and housewives. The formally employed category included public servants. The majority of the study participants (93%) had attained a minimum of primary education (Table 1).
Table 1.
Demographic characteristics of the study participants (N = 103)
| Characteristic | Value, n (%) |
|---|---|
| Gender | |
| Male | 59 (57) |
| Female | 44 (43) |
| Age, y | |
| 0–16 | 32 (31) |
| 17–35 | 43 (42) |
| 36–52 | 20 (19) |
| ≥ 53 | 8 (8) |
| Participant definition | |
| Index | 86 (84) |
| Others | 17 (16) |
| Participant address | |
| Kampala | 34 (33) |
| Wakiso | 32 (31 |
| Other | 37 (36) |
| Nature of address | |
| Urban | 80 (78) |
| Suburban | 7 (7) |
| Rural | 16 (15) |
| Occupation | |
| Formal | 18 (17) |
| Informal | 36 (35) |
| Unemployed | 49 (48) |
| Religion | |
| Catholic | 45 (44) |
| Anglican | 16 (16) |
| Muslim | 14 (14) |
| Pentecostal | 13 (12) |
| Other | 15 (14) |
| Education level | |
| No formal education | 7 (7) |
| Primary school | 37 (36) |
| Secondary school | 34 (33) |
| Technical/vocational | 17 (16) |
| University | 8 (8) |
Time to positivity of the index patients after their admission to the orthopedic ward.
The majority of the index patients (70%) was positive for carrying ARB within 48 hours of their admission to the orthopedic ward. Noticeably, the majority (61%) of the index patients was positive for carrying ARB within 24 hours of admission to the orthopedic ward. No index patients were positive for carrying ARB after 72 hours of their stay in the orthopedic ward. With regard to the origin of the index patients positive for carrying ARB on admission or within 24 hours of their stay in the orthopedic ward, the majority (56%) were not from home, but were referrals or transfers from other wards of Mulago National Referral Hospital and other regional referral hospitals as well as local clinics and health centers. Index patients negative for carrying ARB on admission or within 24 hours of their stay in the orthopedic ward were mostly from home (77%). Of the seven positive “others,” four were health-care workers and three were non-medical caretakers of the index patients (Table 2).
Table 2.
Time to positivity and origin of the index patients (N = 86)
| Characteristic | Value, n (%) |
|---|---|
| Time to positivity | |
| 24 h | 52 (61) |
| 48 h | 8 (9) |
| 72 h | 0 (0) |
| Negative after 72 h | 26 (30) |
| Others positive, n = 24* | 7/24 |
| Origin of index patients positive during 24 h, n = 52 | |
| Home | 23 (44) |
| Local clinics and health centers | 19 (36) |
| Wards other than orthopedic | 7 (14) |
| Other regional referral hospitals | 3 (6) |
| Origin of index patients negative during 24 h, n = 34 | |
| Home | 26 (77) |
| Local clinics and health centers | 8 (24) |
| Wards other than orthopedic | 0 (0) |
| Other regional referral hospitals | 0 (0) |
Nonmedical caretakers of the index patients, medical caretakers or health-care workers attending to the index patients, and index patients’ immediate admitted hospital neighbors. “Others” was linked specifically to patients who became positive 48 to 72 hours after admission.
Bacterial species isolated from the study participant samples.
The majority of samples obtained from the index patients (90%) had bacterial species belonging to the family Enterobacteriaceae—namely, E. coli and K. pneumoniae. The other bacterial species isolated were defined as unidentified Gram-negative bacteria (Table 3). Samples that had been obtained from inanimate environments immediate to the index patients (index patients’ bed headboard and hand rails) did not yield any ARB. Of the other samples obtained from the inanimate environment, only 1 of 30 collected at the start of the study yielded E. coli. This sample was from the health-care worker handwashing sink (Table 3).
Table 3.
Bacterial species isolated from the participant samples (N = 80)
| Escherichia coli, n (%) | Klebsiella pneumonia, n (%) | UGNB, n (%) | Bacterial isolates from inanimate environment, n (%)* | ||
|---|---|---|---|---|---|
| 1 | 2 | 3 | |||
| 57 (71) | 15 (19) | 8 (10) | 0/8 (0) | 0/8 (0) | 1/30 (3.3) |
UGNB = unidentified Gram-negative bacteria
1, index patients’ bed’s headboards; 2, index patients’ bed’s handrails; 3, samples from inanimate ward environment (electrical switches; 4, health-care workers’ handwashing sinks; 5, health-care workers’ reception table; 6, ward door handles)
Antimicrobial susceptibility profiles of the isolated bacterial species.
Resistance to cefotaxime, ampicillin, and cotrimoxazole was more pronounced, with 96% of the bacteria isolates showing resistance to each of the antibiotics. Worth noting, a greater percentage of the bacterial isolates showed resistance to cefotaxime (96%) compared with ceftazidime (55%). Although analysis using binomial logistic regression revealed that ESBL production was associated significantly with resistance to cefotaxime, but not with resistance to ceftazidime as well as the bacterial species. Analysis using the independent sample t-test revealed that inhibition zones of cefotaxime and ceftazidime were associated statistically and significantly with ESBL production. Bacterial isolates were predominantly sensitive to meropenem (94%), colistin (93%), and amikacin (91%).
Upon initial phenotypic screening of the bacterial isolates for the production of ESBLs using ceftazidime or cefotaxime containing media, all (100%) were producers of ESBLs. However, upon confirmatory phenotypic testing using the modified double-disk synergy test, 90% were true producers of ESBLs whereas the other bacterial isolates were derepressed mutants (i.e., non-ESBLs resistant to ceftazidime and cefotaxime). Of the eight isolated positive for ESBL production upon screening, and negative for ESBL production upon confirmation, eight (100%) were producers of AmpC β-lactamases. In addition, the majority of the bacterial isolates (n = 75, 94%) were multi-drug resistant. Use of the modified double-disk synergy test showed sensitivity, specificity, and positive and negative predictive values of 100%, 50%, 90%, and 100%, respectively. Analysis using Pearson’s χ2 and the independent t-test revealed no statistically significant association between the phenotype of the bacterial isolates (i.e., ESBLs or non-ESBLs) and participant age, gender, address, nature of address, occupation, religion, education level, or Enterobacteriaceae species (Table 4).
Table 4.
Phenotypic characterization of antimicrobial resistance in the isolated bacterial species, sensitivity, specificity, and positive and negative predictive values of the screening test (N = 80)
| Characteristic | Value |
|---|---|
| ESBL positive, initial screening, n (%) | 80 (100) |
| ESBL positive, confirmatory testing, n (%) | 72 (90) |
| Derepressed mutants, n (%) | 8 (10) |
| AmpC production testing, n (%) | 8/8 (100) |
| AmpC inclusive, n (%) | 0/0 (0) |
| Multidrug-resistant carbapenem- susceptible bacteria species, n (%) | 75/80 (94) |
| Sensitivity of screening method, % | 100 |
| Specificity of screening method, % | 50 |
| Positive predictive value of screening method, % | 90 |
| Negative predictive value of screening method, % | 100 |
AmpC = Class C β-lactamases; ESBL = extended-spectrum β-lactamase.
Antimicrobial resistance acquisition and transmission dynamics in the orthopedic ward: Index patients positive for carrying ARB on or 24 hours after admission to the ward.
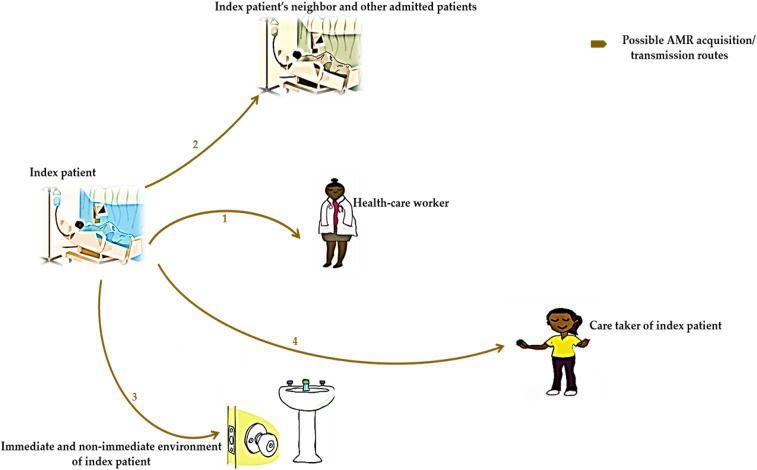
Our results show that when index patients were positive for carrying ARB on admission or 24 hours after admission to the orthopedic ward, the spread of these bacteria was from index patients to others (i.e., medical caretakers or health-care workers attending to the index patients, non-medical caretakers of the index patients, and immediate admitted hospital neighbors of the index patients) via routes 1, 2, and 4. The findings, however, show no ARB spread from index patients to their inanimate environment via route 3. Broadly, the findings also showed that, in this case, index patients are reservoirs of ARB (Figure 1).
Figure 1.
Schematic showing antimicrobial resistance (AMR) acquisition and transmission dynamics in the orthopedic ward of index patients positive for carrying antimicrobial-resistant bacteria 24 hours after admission. 1 = AMR transmission from the index patient to the health-care worker; 2 = AMR transmission from the index patient to the index patient’s neighbor and other admitted patients; 3 = AMR transmission from the index patient to the immediate and non-immediate environment of the index patient; 4 = AMR transmission from the index patient to their non-medical caretaker. This figure appears in color at www.ajtmh.org.
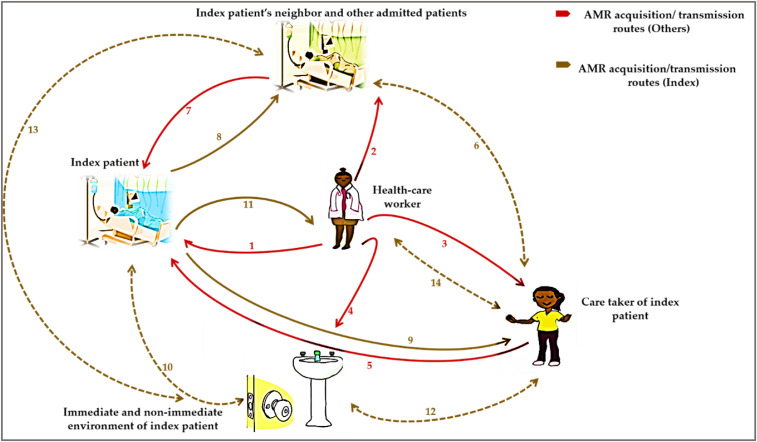
Antimicrobial resistance acquisition and transmission dynamics in the orthopedic ward: Index patients positive for carrying ARB 48 hours after admission to the ward.
Our results show that the practical interfaces for AMR acquisition and spread in the orthopedic ward were predominantly human–human interaction compared with human–inanimate environment interaction. After 48 hours in the orthopedic ward, index patients acquired ARB primarily from their medical caretakers or health-care workers on the ward and not by any other source (via route 1). During this time, the medical caretakers or health-care workers also spread the ARB to either the index patients’ non-medical caretakers, immediate admitted hospital neighbor, and other admitted patients as well as the inanimate environment (via routes 2, 3, and 4).
Index patients acquired ARB from their non-medical caretakers as well as other admitted hospital neighbors (i.e., immediate and other admitted patients) after 48 hours (via routes 5 and 7). It is unlikely that index patients acquired and transmitted ARB from or to the inanimate environment during this time (via route 10) (Figure 2).
Figure 2.
Schematic showing antimicrobial resistance (AMR) acquisition and transmission dynamics in the orthopedic ward of index patients positive for carrying antimicrobial-resistant bacteria 48 hours after hospital stay. 1 = AMR transmission from health-care worker to the index patient; 2 = AMR transmission from health-care worker to index patient’s neighbor and other admitted patients; 3 = AMR transmission from health-care worker to the non-medical caretaker of the index patient; 4 = AMR transmission from health-care worker to the immediate and non-immediate environment of the index patient; 5 = AMR transmission from the non-medical caretaker of the index patient to the index patient; 6 = AMR transmission/acquisition among the non-medical caretaker and index patient’s neighbor and other admitted patients; 7 = AMR transmission from the index patient’s neighbor and other admitted patients to the index patient; 8 = AMR transmission from index patient to index patient’s neighbor and other admitted patients; 9 = AMR transmission from the index patient to the non-medical caretaker of the index patient; 10 = AMR transmission/acquisition between the index patient and the immediate and non-immediate environment of the index patient; 11 = AMR transmission from the index patient to the health-care worker; 12 = AMR transmission/acquisition between the non-medical caretaker of the index patient and the immediate and non-immediate environment of the index patient; 13 = AMR transmission/acquisition among the index patient’s neighbor and other admitted patients and the immediate and non-immediate environment of the index patient; 14 = AMR transmission/acquisition between the health-care worker and the non-medical caretaker of the index patient. This figure appears in color at www.ajtmh.org.
Similar to the events that took place during the 24-hour period, index patients positive for carrying ARB at 48 hours spread these bacteria to their medical caretakers or health-care workers, non-medical caretakers, and immediate hospital neighbors (via routes 8, 9, and 11), but not to their inanimate environment (via route 10) (Figure 2).
Interactions between the non-medical caretakers of the index patients, medical caretakers, or health-care workers attending to the index patients and immediate admitted neighbors of the index patients as well as the inanimate environment were not routes for AMR acquisition and transmission during the index patients’ hospital stay (routes 6, 12, 13, and 14) (Figure 2). In the orthopedic ward, medical caretakers or health-care workers attending to index patients are reservoirs of ARB.
DISCUSSION
We explored AMR acquisition and transmission dynamics in health-care settings based on the orthopedic ward of Mulago National Referral Hospital in Uganda. Our results reveal thought-provoking findings concerning AMR acquisition and transmission dynamics and we make several recommendations to inform the need as well as the nature of AMR surveillance.
Regarding the time to positivity and origin of the index patients, the majority of our index patients were positive for carrying ARB on admission or within 24 hours of their stay on the orthopedic ward. For index patients who were negative on or 24 hours after admission to the ward, carriage of ARB was reported markedly after 48 hours. Some of the index patients remained negative for carrying ARB after 72 hours. With regard to the origin of the index patients, the majority—particularly those who were positive for carrying ARB on or 24 hours after admission—did not come from home but were referrals or transfers from several units/wards within Mulago National Referral Hospital and other regional referral hospitals, or had been admitted to several local clinics or health-care centers prior their transfer to Mulago National Referral Hospital. Similar to other related studies,21–23 our results demonstrate that AMR in the community and health-care settings is increasing progressively. Also, similar to another related study,24 we showed, to some extent, how larger health-care settings and smaller health-care settings can be contributing to the AMR problem, as well as the potential subsequent spillover of healthcare-associated pathogens to the community. Because of the substantial public health consequences, including the numerous challenges in managing community- and hospital-acquired bacterial infections—particularly those involving ARB—there is a need to evaluate accurately the interactions between the community and health-care settings to identify sources and/or reservoirs and sinks of AMR to understand further the transmission dynamics and acquisition of AMR; to initiate monitoring and surveillance of AMR originating from the community and health-care settings; to identify, recommend, and spearhead the implementation of strategies that prevent increases in reservoirs and sinks of AMR in the community and health-care settings; to leverage unique antimicrobial use behaviors and practices of individuals who remain negative for ARB after their hospital stay in efforts to combat AMR; and to ascertain the role of other Mulago National Referral Hospital units/wards, regional referral hospitals, and smaller health-care settings (local clinics and health-care centers) in AMR.
Our results evidence an alarming prevalence of ESBL-PE (90%) from fecal samples of our study participants—several-fold greater than the 5.3% reported in a related study conducted in 2006 and 2007 at Mulago National Referral Hospital,21 and greater than the 62% reported in another related study at Mulago National Referral Hospital in 2014,25 but only slightly greater than the 89% reported in a study conducted in 2015 and 201626 at Kasese regional referral hospital, southwestern Uganda. The ESBLs-PE isolated in this study were E. coli and K. pneumoniae. Similar to other related studies,21–23 ours provides data suggestive of persistent intestinal shedding of ESBL-PE by both healthy individuals and hospitalized patients. This lessens treatment options for both community- and hospital-acquired infections involving ESBL-PE.27 The source of ESBL-PE in participant samples from the community could have been a consequence of participants visiting the health-care setting or interacting with health-care workers in that setting. This was also suggested as a possibility in other related studies.27,28 However, some studies suggested that population exposure to antibiotics could explain this observation,21,29 but others demonstrated ESBL-PE in individuals with no history of having taken antibiotics;30,31 others highlighted food products from animals as potential sources of the bacteria.30,31 These findings underscore the need to spearhead the introduction of monitoring and surveillance of AMR originating in the community and health-care settings to guide both clinical care and the review of national clinical guidelines for the management of infections involving ESBL-PE, agitate for a multi-layered behavioral-based approach aimed at (but not limited to) reducing the volume of antimicrobial use wherever and whenever possible. Also, we need to investigate further the potential of using ESBL-PE from sewage as AMR detection markers that could be used in monitoring and surveillance, and develop a scheme (including the use of double-selected culture media in a single step to detect ESBL-PE, as used in our study) to reduce the turnaround time and cost of phenotypic ESBL-PE screening while eliminating the need for molecular approaches.
The screening approach using double-selected culture media in a single step to detect ESBL-PE in our study had a high diagnostic accuracy with regard to its ability to identify ARB (specifically, ESBL producers) directly from patient/non-patient samples. This screening method could potentially revolutionize AMR monitoring and surveillance, especially in limited-resource settings, because the inability to diagnose several diseases quickly has been documented as a significant cause of the disproportionate number of deaths resulting from communicable diseases in limited-resource settings compared with high-income settings.32,33
The potential of using ESBL-PE from sewage as AMR detection markers that can then be used in monitoring and surveillance—particularly E. coli and K. pneumoniae—is supported by the fact that these are documented priority pathogens as a result of their widespread levels of AMR, distribution in the gastrointestinal tract of humans as well as animals, and involvement in the sharing of AMR gene carrying mobile genetic elements with other bacteria that are found in similar environments.34,35
Regarding the abundance of ARB in the hospital environment (ward environment). Environmental sampling in our study revealed that hospital environments were mostly free of ARB except for the health-care worker handwashing sink, where these bacteria were isolated. Unlike related studies that have indicated the presence of ARB from inanimate surfaces,36,37 our results reveal otherwise. However, our findings are similar to those of other related studies, all of which point toward the transmission of AMR through human–human interface compared with human–inanimate environment interface, emphasizing the potential role of health-care workers and patient attendants in the cycle of AMR transmission.38,39 Patient exchange among hospital units/wards and regional referral hospitals driven primarily by limited space,40 specialization of care,24 and other factors (e.g., the availability of free care in these facilities24) could be exacerbating human–human transmission and acquisition of ARB. Also, patient exchange suggests a high patient turnover and a probable absence of ARB transmission and acquisition to and from inanimate surfaces, hence a reduction in the ARB that results in rapid decays and eventual pathogen extinction due to limited adaptation time. By contrast, humans can potentially act as reservoirs,38,39 orchestrating continuous transmission and acquisition of ARB. In light of this, there remains a need to evaluate accurately the interactions among humans in the community and health-care settings to understand AMR transmission dynamics and acquisition further at the human–human interface. However, this finding could be the result of common hygienic and sanitation practices on the target ward that could have also limited the isolation of ARB from the inanimate environment. Poor hygienic and sanitary practices have been shown previously to drive the spread of resistant bacteria in health environment surfaces.41,42 Hence, the influence of common hygienic and sanitation practices should be investigated further in the context of this work.
We report high rates of resistance to antibiotics in common usage–namely, penicillins with ESBL-resistant penicillins, sulfonamides in combination with trimethoprim, third-generation cephalosporins, and combinations of penicillins including b-lactamase inhibitors. Also, we report high multi-drug resistance (i.e., resistance to three or more classes of antibiotics) prevalence in ESBL-PE isolates. Similar to other related studies,21,43 we report high rates of multi-drug-resistant ESBL-PE in both community samples of healthy and diseased individuals, and those of hospitalized patients. Like other related studies,21,43–46 we attribute these findings to the empirical use of antibiotics in health-care settings; community exposure to antibiotics; acquisition/transmission of the bacteria from environmental sources exacerbated by several factors that include, among others, community hygiene levels and peoples’ antibiotic use behaviors; and the lack of an AMR monitoring and surveillance system for the community and health-care settings. Multi-drug-resistant ESBL-PE remains a major threat to patients because infections involving these organisms render possible treatments impractical and call for the use of more expensive treatment alternatives. Hence, these findings underscore the need to boost microbiology laboratory capacity to spearhead culture-guided antibiotic therapy, strengthen monitoring and surveillance of AMR in community and health-care settings, and pursue infection control interactive community and health-care setting-based education sessions aimed at evaluating and exploring the acceptability of habit-based infection control behavior change interventions.
Our study had limitations. First, despite our initial hope of recruiting more index patients into the study, we discovered that only a limited number of admissions were possible each week because of space-related challenges on the target ward and the emergence of the coronavirus disease 2019 pandemic, which triggered a countrywide lockdown and the cessation of all research activities. Second, we were unable to sample index patients’ home environment as initially conceptualized because we learned the majority of our index patients were not from home, but were referrals or transfers from several units/wards within Mulago National Referral Hospital or other regional referral hospitals, or had been admitted to several local clinics or health-care centers prior their transfer to Mulago National Referral Hospital. This could have limited our comprehensive understanding of the transmission dynamics and acquisition of AMR not only at the community level, but also at health-care setting level. In addition, we encountered unanticipated resources (especially funding) required to explore the additional sites, including obtaining administrative and ethical clearance at the respective sites, materials and consumables, as well as other expenses. Third, although genomic data have been described as being better at providing closer insight into AMR transmission dynamics and acquisition,34,47 we do not present these data, which could have affected our interpretation of AMR transmission dynamics and acquisition data.
Despite these limitations, we are able to provide invaluable insight into AMR transmission dynamics in health-care settings using our unique approach of basing our study on the orthopedic ward of Mulago National Referral Hospital in Uganda. Our findings are crucial and have the potential to spur further monitoring and surveillance of AMR. They could also promote the development and deployment of effective interventions and innovations to reduce AMR incidence, especially in limited-resource settings.
CONCLUSION
AMR transmission and acquisition occur primarily via human–human interface within and outside of health-care facilities, compared with human–inanimate environment interface. However, this remains subject to additional research. A better understanding of human-human and human-environment interactions could aid in the development of evidence-based and effective interventions that could ultimately reduce the burden of AMR in humans. Also, whole-genome sequencing approaches, when explored, could provide a greater understanding of AMR transmission dynamics and acquisition in health-care and community settings.
ACKNOWLEDGMENTS
We thank Jackie Nabukenya, from the School of Nursing and Midwifery, Mulago National Referral Hospital, who helped with participant recruitment and data collection; Emmanuel Nyei, from the Clinical Microbiology Laboratory, Mulago National Referral Hospital, Uganda, who helped with data analysis; and Harriet Nakayiza, from the Department of Immunology and Molecular Biology, School of Biomedical Sciences, College of Health Sciences, Makerere University, for administrative assistance. The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
References
- 1.Nathan C, Cars O, 2014. Antibiotic resistance: problems, progress, and prospects. N Engl J Med 371: 1761–1763. [DOI] [PubMed] [Google Scholar]
- 2.Rossolini GM, Arena F, Pecile P, Pollini S, 2014. Update on the antibiotic resistance crisis. Curr Opin Pharmacol 18: 56–60. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization , 2014. Antimicrobial Resistance: Global Report on Surveillance. Geneva, Switzerland: WHO. [Google Scholar]
- 4.Boerlin P, Reid-Smith RJ, 2008. Antimicrobial resistance: its emergence and transmission. Anim Health Res Rev 9: 115. [DOI] [PubMed] [Google Scholar]
- 5.Goldmann DAet al. , 1996. Strategies to prevent and control the emergence and spread of antimicrobial-resistant microorganisms in hospitals: a challenge to hospital leadership. JAMA 275: 234–240. [PubMed] [Google Scholar]
- 6.Baker S, Thomson N, Weill FX, Holt KE, 2018. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 360: 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pärnänen K, Karkman A, Tamminen M, Lyra C, Hultman J, Paulin L, Virta M, 2016. Evaluating the mobility potential of antibiotic resistance genes in environmental resistomes without metagenomics. Sci Rep 6: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasovský D, Littmann J, Zorzet A, Cars O, 2016. Antimicrobial resistance: a threat to the world’s sustainable development. Ups J Med Sci 121: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill J, 2014. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations: December 2014. London, UK: Review on Antimicrobial Resistance.
- 10.Ampaire Let al. , 2016. A review of antimicrobial resistance in East Africa. Afr J Lab Med 5: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical Laboratory and Standard Institute , 2019. Performance Standards for Antimicrobial Susceptibility Testing, 29th edition. CLSI supplement M100. Wayne, PA: Clinical Laboratory Standards Institute. [Google Scholar]
- 12.WC W, 2006. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology, 6th edition. Philadelphia, PA: Lippincott Williams & Wilkins, 845–853. [Google Scholar]
- 13.Cheesbrough M, 2006. District Laboratory Practice in Tropical Countries Part 2. New York, NY: Cambridge University Press, 300–301. [Google Scholar]
- 14.Patel SSet al. , 2017. Isolation and identification of Klebsiella pneumoniae from sheep: case report. Int J Curr Microbiol Appl Sci 6: 331–334. [Google Scholar]
- 15.Aruhomukama D, 2020. Review of phenotypic assays for detection of extended-spectrum β-lactamases and carbapenemases: a microbiology laboratory bench guide. Afr Health Sci 20: 1090–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drieux L, Brossier F, Sougakoff W, Jarlier V, 2008. Phenotypic detection of extended‐spectrum β‐lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 14: 90–103. [DOI] [PubMed] [Google Scholar]
- 17.Kateete DP, Nakanjako R, Namugenyi J, Erume J, Joloba ML, Najjuka CF, 2016. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago Hospital in Kampala, Uganda (2007–2009). Springerplus 5: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh R, Mathai A, Parikh S, Sekhar GC, Thomas R, 2008. Understanding and using sensitivity, specificity and predictive values. Indian J Ophthalmol 56: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panta K, Ghimire P, Rai SK, Mukhiya RK, Singh RN, Rai G, 2013. Antibiogram typing of Gram negative isolates in different clinical samples of a tertiary hospital. Asian J Pharm Clin Res 6: 153–156. [Google Scholar]
- 20.Flournoy DJ, 1982. Quantitative antibiogram as a potential tool for epidemiological typing. Infect Control Hosp Epidemiol 3: 384–387. [DOI] [PubMed] [Google Scholar]
- 21.Najjuka CF, Kateete DP, Kajumbula HM, Joloba ML, Essack SY, 2016. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes 9: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birgy Aet al. , 2012. Community faecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in French children. BMC Infect Dis 12: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calva JJ, Sifuentes-Osornio J, Céron C, 1996. Antimicrobial resistance in fecal flora: longitudinal community-based surveillance of children from urban Mexico. Antimicrob Agents Chemother 40: 1699–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouyos RD, Zur Wiesch PA, Bonhoeffer S, 2011. On being the right size: the impact of population size and stochastic effects on the evolution of drug resistance in hospitals and the community. PLoS Pathog 7: e1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kateregga JN, Kantume R, Atuhaire C, Lubowa MN, Ndukui JG, 2015. Phenotypic expression and prevalence of ESBL-producing Enterobacteriaceae in samples collected from patients in various wards of Mulago Hospital, Uganda. BMC Pharmacol Toxicol 16: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrew B, Kagirita A, Bazira J, 2017. Prevalence of extended-spectrum beta-lactamases-producing microorganisms in patients admitted at KRRH, southwestern Uganda. Int J Microbiol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naseer U, Haldorsen B, Simonsen GS, Sundsfjord A, 2010. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect 16: 171–178. [DOI] [PubMed] [Google Scholar]
- 28.Young BE, Lye DC, Krishnan P, Chan SP, Leo YS, 2014. A prospective observational study of the prevalence and risk factors for colonization by antibiotic resistant bacteria in patients at admission to hospital in Singapore. BMC Infect Dis 14: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woerther PLet al. , 2013. Characterization of fecal extended-spectrum-β-lactamase-producing Escherichia coli in a remote community during a long time period. Antimicrob Agents Chemother 57: 5060–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey JK, Pinyon JL, Anantham S, Hall RM, 2010. Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J Med Microbiol 59: 1331–1339. [DOI] [PubMed] [Google Scholar]
- 31.Overdevest Iet al. , 2011. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis 17: 1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma S, Zapatero-Rodríguez J, Estrela P, O’Kennedy R, 2015. Point-of-care diagnostics in low resource settings: present status and future role of microfluidics. Biosensors (Basel) 5: 577–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz Granados CA, Cardo DM, McGowan JE, Jr., 2008. Antimicrobial resistance: international control strategies, with a focus on limited-resource settings. Int J Antimicrob Agents 32: 1–9. [DOI] [PubMed] [Google Scholar]
- 34.Wee BA, Muloi DM, van Bunnik BA, 2020. Quantifying the transmission of antimicrobial resistance at the human and livestock interface with genomics. Clin Microbiol Infect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.European Food Safety Authority, European Centre for Disease Prevention and Control , 2015. EU summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J 13: 4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sserwadda I, Lukenge M, Mwambi B, Mboowa G, Walusimbi A, Segujja F, 2018. Microbial contaminants isolated from items and work surfaces in the post-operative ward at Kawolo General Hospital, Uganda. BMC Infect Dis 18: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber DJ, Anderson D, Rutala WA, 2013. The role of the surface environment in healthcare-associated infections. Curr Opin Infect Dis 26: 338–344. [DOI] [PubMed] [Google Scholar]
- 38.Allegranzi B, Pittet D, 2009. Role of hand hygiene in healthcare-associated infection prevention. J Hosp Infect 73: 305–315. [DOI] [PubMed] [Google Scholar]
- 39.van den Dool C, Haenen A, Leenstra T, Wallinga J, 2016. The role of nursing homes in the spread of antimicrobial resistance over the healthcare network. Infect Control Hosp Epidemiol 37: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kigera JWM, 2010. Patterns of spine surgeries at Mulago Hospital. East Afr Orthop J 4. [Google Scholar]
- 41.Akinyemi KO, Atapu AD, Adetona OO, Coker AO, 2009. The potential role of mobile phones in the spread of bacterial infections. J Infect Dev Countries 3: 628–632. [DOI] [PubMed] [Google Scholar]
- 42.Shobo COet al. , 2020. Bacterial diversity and functional profile of microbial populations on surfaces in public hospital environments in South Africa: a high throughput metagenomic analysis. Sci Total Environ 719: 137360. [DOI] [PubMed] [Google Scholar]
- 43.Seni Jet al. , 2013. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Uganda. BMC Res Notes 6: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pena Cet al. , 2006. Risk-factors for acquisition of extended-spectrum β-lactamase-producing Escherichia coli among hospitalised patients. Clin Microbiol Infect 12: 279–284. [DOI] [PubMed] [Google Scholar]
- 45.George EA, Sankar S, Jesudasan MV, Sudandiradoss C, Nandagopal B, 2014. Incidence of extended spectrum beta lactamase producing Escherichia coli among patients, healthy individuals and in the environment. Indian J Med Microbiol 32. [DOI] [PubMed] [Google Scholar]
- 46.Babu R, Kumar A, Karim S, Warrier S, Nair SG, Singh SK, Biswas R, 2016. Faecal carriage rate of extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalised patients and healthy asymptomatic individuals coming for health check-up. J Glob Antimicrob Resist 6: 150–153. [DOI] [PubMed] [Google Scholar]
- 47.Aruhomukama D, Sserwadda I, Mboowa G, 2019. Investigating colistin drug resistance: the role of high-throughput sequencing and bioinformatics. F1000 Res 8. [DOI] [PMC free article] [PubMed] [Google Scholar]