Objectives:
Unexpected ICU readmission is associated with longer length of stay and increased mortality. To prevent ICU readmission and death after ICU discharge, our team of intensivists and data scientists aimed to use AmsterdamUMCdb to develop an explainable machine learning–based real-time bedside decision support tool.
Derivation Cohort:
Data from patients admitted to a mixed surgical-medical academic medical center ICU from 2004 to 2016.
Validation Cohort:
Data from 2016 to 2019 from the same center.
Prediction Model:
Patient characteristics, clinical observations, physiologic measurements, laboratory studies, and treatment data were considered as model features. Different supervised learning algorithms were trained to predict ICU readmission and/or death, both within 7 days from ICU discharge, using 10-fold cross-validation. Feature importance was determined using SHapley Additive exPlanations, and readmission probability-time curves were constructed to identify subgroups. Explainability was established by presenting individualized risk trends and feature importance.
Results:
Our final derivation dataset included 14,105 admissions. The combined readmission/mortality rate within 7 days of ICU discharge was 5.3%. Using Gradient Boosting, the model achieved an area under the receiver operating characteristic curve of 0.78 (95% CI, 0.75–0.81) and an area under the precision-recall curve of 0.19 on the validation cohort (n = 3,929). The most predictive features included common physiologic parameters but also less apparent variables like nutritional support. At a 6% risk threshold, the model showed a sensitivity (recall) of 0.72, specificity of 0.70, and a positive predictive value (precision) of 0.15. Impact analysis using probability-time curves and the 6% risk threshold identified specific patient groups at risk and the potential of a change in discharge management to reduce relative risk by 14%.
Conclusions:
We developed an explainable machine learning model that may aid in identifying patients at high risk for readmission and mortality after ICU discharge using the first freely available European critical care database, AmsterdamUMCdb. Impact analysis showed that a relative risk reduction of 14% could be achievable, which might have significant impact on patients and society. ICU data sharing facilitates collaboration between intensivists and data scientists to accelerate model development.
Keywords: decision support techniques, information dissemination, machine learning, mortality, patient discharge, patient readmission
Real-time bedside computerized decision support tools leveraging the power of machine learning models and the vast amount of routinely collected data in the ICU hold great promise to improve patient outcomes. However, large-scale implementation is currently hampered by model performance and lack of generalizability across centers and countries (1–14).
A significant hurdle is lack of access to additional training and validation data, preferably from other hospitals. Due to technical, legal, ethical, and privacy concerns, ICU data are currently not shared on a large scale. However, the Joint Data Science Task Force of the Society of Critical Care Medicine (SCCM) and the European Society of Intensive Care Medicine (ESICM) recently launched their Data Sharing initiative to ameliorate this situation (15). AmsterdamUMCdb is the first freely accessible European critical care database released under this initiative (16).
To demonstrate the usefulness of its release for bringing data science closer to the bedside, we set out to use AmsterdamUMCdb to develop a model to prevent ICU readmission or death after discharge from the ICU. The transition of care to a less monitored environment may lead to preventable errors and adverse events. In addition, unexpected ICU readmission is associated with longer length of stay and an increase in mortality (17, 18). However, unnecessarily prolonging ICU stays is wasteful especially considering limited ICU capacity and may be detrimental to the patient (19, 20).
In view of clinical acceptability and compliance, we aimed to develop a model that will present trends for its predictions and insight in those predictions. To facilitate implementability at the bedside, we sought to only use clinically relevant features that are readily available through interfaces with existing electronic health records. Given these considerations, we united a team of experienced intensivists and data scientists.
MATERIALS AND METHODS
AmsterdamUMCdb contains data from a 32-bed mixed surgical-medical academic ICU and a 12-bed high-dependency unit (Medium Care Unit [MCU]). Pseudonymized data were extracted from its main source (MetaVision, iMDsoft, Tel Aviv, Israel). Data from patients older than 18 years and admitted to the ICU between 2004 and March 2016 were included in the analysis. ICU admissions longer than 30 days were excluded, as these patients typically follow a discharge workflow closely coordinated with the receiving ward. The local Medical Ethics Committee reviewed the study protocol and considered it to be outside the scope of the Law on Scientific Research on Humans. In addition, they approved a waiver for the need for written informed consent for the use of pseudonymized data based on the very large number of included patients in accordance with the General Data Protection Regulation.
End Points
The primary outcome was ICU readmission and/or death, both within 7 days of ICU discharge. Both are likely to influence decisions on ICU discharge and represent competing risks. ICU readmission was defined as a transfer from the ICU to the general ward and back to the ICU or MCU during the same hospital stay. Palliative care patients and patients with do-not-resuscitate or do-not-intubate orders were excluded from the analysis. Patients transferred to other hospitals were also excluded.
Feature Engineering
Patient demographics (e.g., age and sex), clinical observations (e.g., nursing scores and Glasgow Coma Scale score), automated physiologic measurements from devices (e.g., patient monitor, ventilator, and continuous renal replacement therapy), laboratory studies, medication (e.g., sedatives and vasopressors), and other support (e.g., enteral feeding and intermittent hemodialysis) were considered as input for the model. Extensive discussions in expert sessions with intensivists and visual analysis using heat maps on all available variables in the dataset were used to determine potential features. Demographics and characteristics of the admission (e.g., length of stay or time spent at the hospital before the ICU admission) were directly used as features. For variables that were measured or documented multiple times during the admission, extensive preprocessing was performed in order to extract informative features (7, 12, 13) (see Feature Engineering section, Tables S1 and S2, Supplemental Digital Content 1, http://links.lww.com/CCX/A783).
Model Development
After feature engineering, the total number of features was initially 5,466 per patient. Model development and analysis were performed using scikit-learn on Python (21). Using scikit-learn Pipelines, feature selection and model training were performed sequentially with stratified 10-fold cross-validation. In addition, the output of automatic feature selection was manually reviewed to remove features that have no biological plausibility or were expected to generalize poorly (e.g., the number of laboratory measurements per day). To prevent information leakage, feature selection was done exclusively on the training set of each fold.
The frequency of our combined endpoint was relatively low compared with an uneventful (good) outcome (5.3% vs 94.7%), also known as an imbalanced dataset. We applied both logistic regression and more advanced machine learning algorithms to study which method would lead to the best performance for this type of prediction problem. Algorithms were trained to predict the outcome using grid search to optimize hyperparameters (see the sections Model Training, Hyperparameter Tuning, and Tables S3 and S4, Supplemental Digital Content 1, http://links.lww.com/CCX/A783).
Performance
Model performance was validated on data from patients admitted to the same ICU between March 2016 and 2019. Data were extracted from the hospitalwide electronic health records (EpicCare Inpatient, Epic Systems, Verona, WI) that replaced the previous system on the ICU. Since the primary outcome was imbalanced, we constructed both the receiver operating characteristic (ROC) curve as well as the precision-recall curve (PRC), and calculated the areas under these curves (area under the receiver operating characteristic curve [AUROC] and area under the precision-recall curve [AUPRC], respectively) (22–24) (for definitions, see Fig. S1, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). Calibration of the model was evaluated by constructing the probability calibration curve. In addition, we evaluated the effect of feature engineering on model performance.
Feature Importance
To determine feature importance, we used SHapley Additive exPlanations (SHAP) (25, 26). SHAP determines for each patient individually the contribution of all features to that patient’s prediction and thus can be used to interpret the model’s prediction for an individual patient for complex machine learning algorithms.
Sensitivity Analysis
A number of critical care societies, including SCCM and ESICM, advocate using ICU readmission within 2 days of discharge as a quality indicator (27). To evaluate the robustness of our model, we retrained the model on the composite outcome after 2 days and compared performance with the model developed for 7 days.
Clinical Relevance and Impact
We used decision curve analysis to quantify the usefulness of our model based on the net benefit, defined as the difference between the true positives (actual readmissions/deaths) and the false positives (incorrectly identified patients that could have been discharged), corrected by a factor determined by a threshold the clinician chooses to accept: the readmission probability (Fig. S4, Supplemental Digital Content 1, http://links.lww.com/CCX/A783) (28, 29).
To explore potential clinical impact on the validation cohort, we first divided patients into: 1) short-stay patients (ICU stay < 2 days, 65%) and 2) long-stay patients (≥ 2 days, 35%). Using decision curve analysis, a clinically reasonable risk threshold of 6% was chosen to further divide the “short-stay” patients into “high-risk” and “low-risk” patients. For “long-stay” patients, we assessed potential clinical impact based on analysis of readmission probability-time curves. These were obtained by using our prediction model to calculate the probability of the primary outcome for each day of ICU stay, thus describing its trend throughout the admission. After expert panel sessions with intensivists together with user interface software designers, a prototype of the software displaying the readmission probability-time curve and feature importance was developed. Finally, we explored the impact of our model on readmission rates and length of stay using two scenarios. In the first scenario, we assume the readmission rates of the groups that will be kept in the ICU longer to drop by 15% and in the second scenario to drop by 30%.
RESULTS
After excluding patients that did not survive their first ICU admission and ICU admissions longer than 30 days, our training data set included 14,105 admissions (Table 1). Most patients were ventilated and received vasoactive drugs during their admission: 86.3% and 68.2%, respectively. The combined readmission/mortality rate within 7 days of ICU discharge was 5.3%, with readmission and mortality rates of 4.3% and 1.2%, respectively. As expected, patients that were readmitted or died within 7 days of ICU discharge were more often emergency patients (65% vs 43%) and had a longer initial length of stay (6.4 vs 5.1 d). In addition, patients that died within 7 days of ICU discharge were older (71.5 vs 63.2 yr). The validation cohort contained 3,929 patients that suffered from a slightly higher combined outcome of 6.7% (Table S5, Supplemental Digital Content 1, http://links.lww.com/CCX/A783).
TABLE 1.
Derivation Cohort
| Characteristic | Total | No Events | Readmission and/or Death | Readmission Only | Death Only |
|---|---|---|---|---|---|
| ICU admissions, n (%) | 14,105 (100) | 13,354 (94.7) | 751 (5.3) | 610 (4.3) | 173 (1.2) |
| Demographics | |||||
| Age, yr, mean (sd) | 63.3 (15.3) | 63.2 (15.3) | 65.0 (15.5) | 63.5 (15.4) | 71.5 (13.5) |
| Female, n (%) | 4,439 (32.1) | 4,180 (32.0) | 259 (34.9) | 206 (34.2) | 71 (41.5) |
| Body mass index, kg/m2, mean (sd) | 26.3 (4.9) | 26.4 (4.9) | 25.1 (4.8) | 25.1 (4.8) | 24.7 (4.9) |
| Admission type | |||||
| Surgical, n (%) | 3,691 (66.7) | 3,529 (67.9) | 162 (48.5) | 150 (53.8) | 22 (29.7) |
| Emergency admission, n (%) | 6,247 (44.3) | 5,761 (43.1) | 486 (64.7) | 368 (60.3) | 139 (80.3) |
| Length of stay, d, mean (sd) | 3.5 (5.2) | 3.3 (5.1) | 5.5 (6.4) | 5.0 (6.1) | 6.8 (7.1) |
| Supportive care | |||||
| Received mechanical ventilation, n (%) | 12,172 (86.3) | 11,592 (86.8) | 580 (77.2) | 474 (77.7) | 130 (75.1) |
| Received vasopressors/inotropes, n (%) | 9,623 (68.2) | 9,082 (68.0) | 541 (72.0) | 440 (72.1) | 122 (70.5) |
| Risk scores | |||||
| Sequential Organ Failure Assessment maximum, during admission, mean (sd) | 4.4 (4.2) | 4.3 (4.1) | 5.7 (4.7) | 5.6 (4.6) | 6.2 (5.0) |
| Modified Early Warning Score at admission, mean (sd) | 3.0 (1.9) | 3.0 (1.9) | 3.6 (2.1) | 3.5 (2.1) | 4.0 (2.2) |
| Stability and Workload Index for Transfer score at discharge, mean (sd) | 6.4 (5.0) | 6.4 (4.9) | 5.9 (5.9) | 5.9 (5.9) | 5.6 (6.1) |
Patients are grouped by outcome events after ICU discharge: readmission and/or death within 7 d of discharge. Sequential Organ Failure Assessment score ranges from 0 to 24; higher ranges indicate greater severity of illness. Modified Early Warning Score ranges from 0 to 14; higher ranges indicate more abnormal physiologic variables (30). Stability and Workload Index for Transfer score ranges from 0 to 64; higher ranges indicate higher readmission risk (31). Note: classification in Surgical/Medical admissions was only available for patients admitted in 2010 and later.
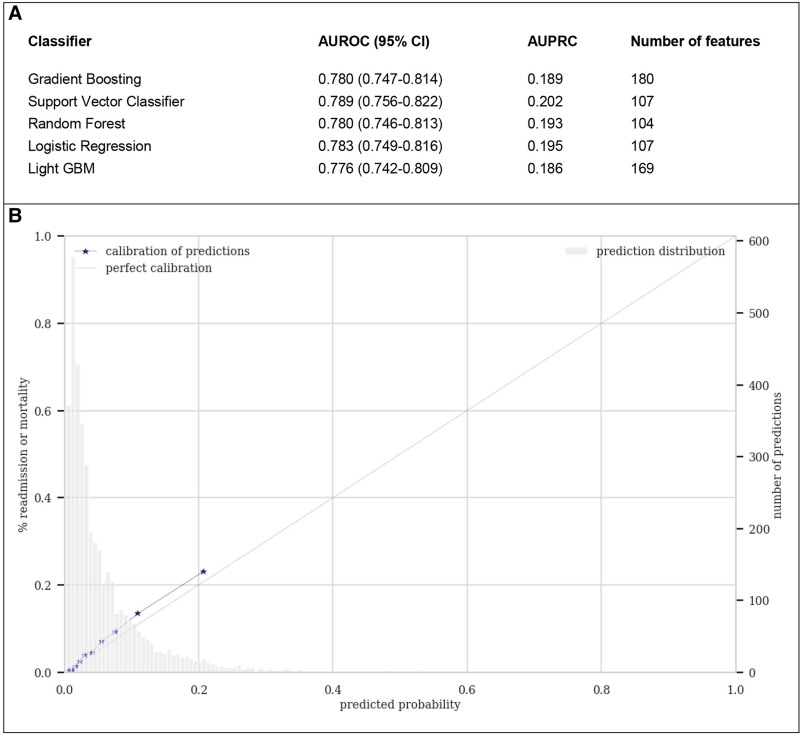
Figure 1 shows both model discrimination and calibration performance on the validation cohort. Figure S1, Supplemental Digital Content 1 (http://links.lww.com/CCX/A783) displays the constructed ROC curve and PRC for all tested algorithms. All algorithms showed similar good discrimination (AUROC range, 0.78–0.79; AUPRC range, 0.19–0.20), with nonsignificant differences in performance (Table S6, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). Please note that, for a random, noninformative, model, the baseline value for AUROC is always to 0.5, whereas the baseline value for AUPRC corresponds to the proportion of “positive” cases, in our case the combined readmission or mortality rate within 7 days (0.067). Based on cross-validation performance (Table S7, Supplemental Digital Content 1, http://links.lww.com/CCX/A783) and prior experience with the Gradient Boosting (XGBoost) algorithm as the current state-of-the-art classification algorithm, this model was selected to be included in further model analysis and development (32). Discrimination did not improve significantly by combining the prediction of separate models for readmission and mortality (Table S6, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). The model was capable of separating readmission and/or mortality versus good outcome, but overlap remains in the low probability range (Fig. S3, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). The value of additional feature engineering, such as squaring the features, was nonsignificant (logistic regression, AUROC without squared features: 0.78 vs with squared features: 0.78; XGBoost without squared features: 0.78 vs with squared features: 0.78).
Figure 1.
Discrimination and calibration model performance on the validation set. A, Discrimination by different algorithms using area under the receiver operating characteristic curve (AUROC) and area under the precision-recall curve (AUPRC) as metrics. B, Probability calibration curve for the Gradient Boosting model. The predicted probabilities are very similar to the true outcome of patients in the validation cohort. Please note that the calibration curve does not extend beyond ~0.2 model since the model rarely outputs values greater than 0.3. GBM = gradient boosting machine.
As a typical real-world model, it did not show perfect discrimination but showed good calibration with predicted probabilities very similar to the true outcomes (Fig. 1B). For the chosen 6% threshold, the model has a less-than ideal sensitivity (recall) of 0.72, specificity of 0.70, and a positive predictive value (PPV) of 0.15 (Fig. S2, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). Though a PPV of 0.15 may appear relatively low, this does suggest that the model identifies those patients with a two-to-three time higher than baseline risk (0.067), which ultimately is the goal of an adverse outcome prediction model (24).
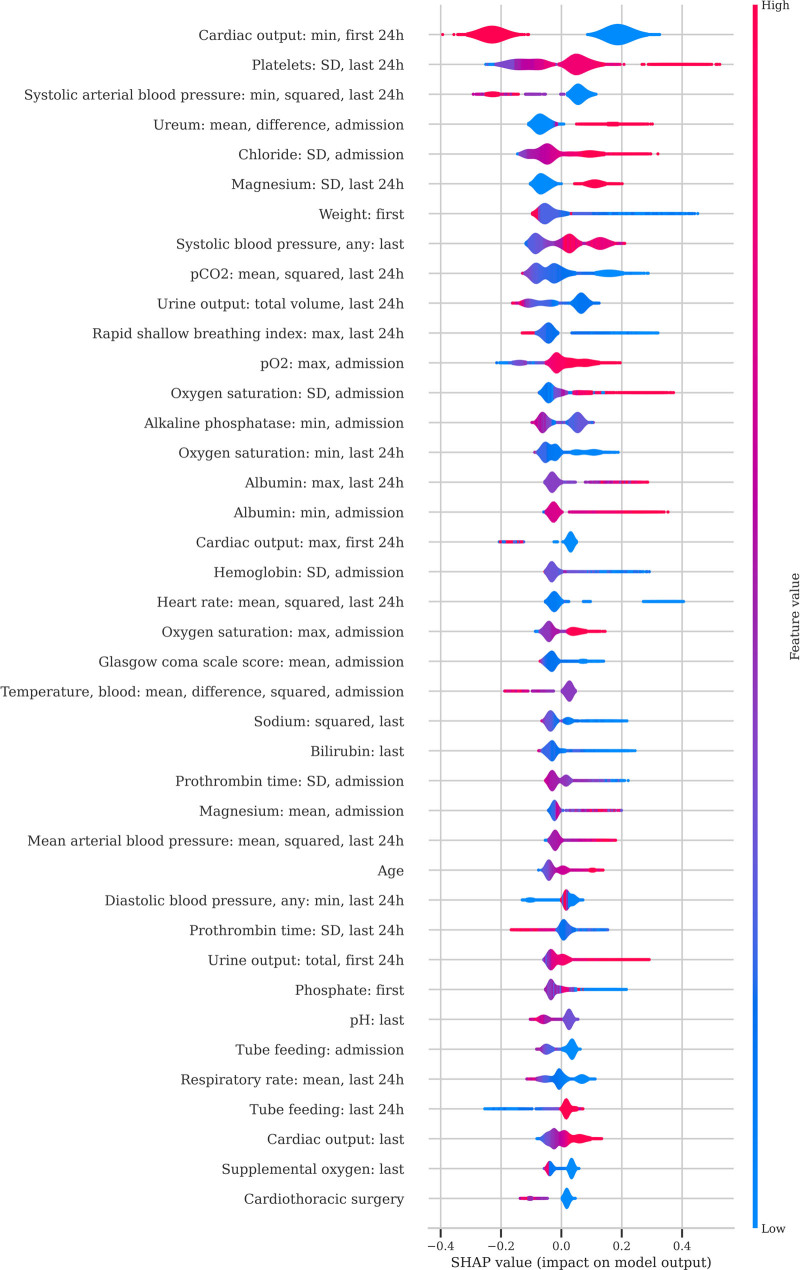
The features used as input for the XGBoost model after feature selection are shown in Table 2. The distribution of SHAP values of the most predictive features is given in Figure 2. Most features are common clinical parameters, such as type of patient (elective surgery) and physiologic variables (respiratory rate, mean arterial pressure, urine output, and oxygen requirements), but also less-apparent features such as the need for tube feeding have a significant effect on the predicted risk. The most predictive features take into consideration the severity of illness at admission (e.g., cardiac output and urine output in the first 24 hr) as well as the condition of the patient before discharge (Pco2 and respiratory rate of the last 24 hr).
TABLE 2.
Features Used as Input for the Final Gradient Boosting Model
| Feature Category | Feature Name | Number of Features |
|---|---|---|
| General information | ||
| Patient characteristics | Age, gender, and weight at admission | 3 |
| Admission information | Origin department | 3 |
| Laboratory results | ||
| Blood gas analysis | pH, Paco2, Pao2, actual bicarbonate, base excess, and arterial oxygen saturation | 15 |
| Hematology | Hemoglobin, WBC count, platelet count, activated partial thromboplastin time, and prothrombin time | 16 |
| Routine chemistry | Sodium, potassium, creatinine, ureum, creatinine/ureum ratio, chloride, ionized calcium, magnesium, phosphate, lactate dehydrogenase, glucose, lactate, C-reactive protein, and albumin | 43 |
| Cardiac enzymes | Creatinine kinase and troponin-T | 5 |
| Liver and pancreas tests | Bilirubin, alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, Gamma-glutamyltransferase, and amylase | 11 |
| Vital signs and device data | ||
| Circulation | Heart rate, arterial blood pressure (systolic/diastolic/mean), noninvasive blood pressure (systolic/diastolic), cardiac output, temperature, and central venous pressure | 34 |
| Respiration | Fio2, positive end-expiratory pressure, tidal volume, respiratory rate, peripheral oxygen saturation, and rapid shallow breathing index | 18 |
| Clinical observations and scores | ||
| Neurology | Glasgow Coma Scale score, Richmond Agitation-Sedation Scale, pupil response, and pupil diameter | 9 |
| Respiration | Bronchial suctioning, coughing reflex, and Pao2/Fio2 | 10 |
| Nephrology | Urine output | 2 |
| Diagnostics and therapeutics | ||
| Lines, drains and tubes | Endotracheal tube and urine catheter | 3 |
| Interventions | Supplemental oxygen, continuous renal replacement therapy, and tube feeding | 8 |
| Total | 180 | |
After manual selection, logistic regression with an L1 penalty, and training using 10-fold cross-validation, these features were used as input for the final Gradient Boosting model. The number of features includes aggregations of primary features.
Figure 2.
Feature importance. For the Gradient Boosting model, feature performance is plotted for the first 40 features using SHapley Additive explanations (SHAP). For each feature, each red or blue dot represents the impact of that feature on the prediction for that patient. Red dots represent patients with a high value for the specific feature, whereas blue dots represent patients with a low value for the specific feature. Red dots on to the right side of the distribution indicate that high values of the feature are associated with a high risk of readmission or mortality. Conversely, blue dots on the right side of the distribution indicate that low values of the feature are associated with a high risk.
Mortality has been shown to be more easily predictable than readmission (7). By retraining the model to predict ICU readmission and/or mortality within the first 2 days, performance decreased (AUROC, 0.73), which is expected since our 7-day model postdischarge included relatively more patients that died (2-d mortality is 0.3% vs 1.8% readmission and 7-d mortality 1.2% vs 4.3% readmission).
Decision curve analysis identifies the net benefit of using the model for a range of thresholds. These are determined by the importance assigned by intensivists to untimely discharging patients (false negatives) compared with unnecessarily keeping patients in the ICU (false positives). For example, using a threshold of 5%, we consider an untimely discharge 19 times more important than an unnecessarily prolonged stay. For clinically relevant thresholds (~3 to ~30%), net benefit using our model is higher than the default strategies (Fig. S5, Supplement Digital Content 1, http://links.lww.com/CCX/A783).
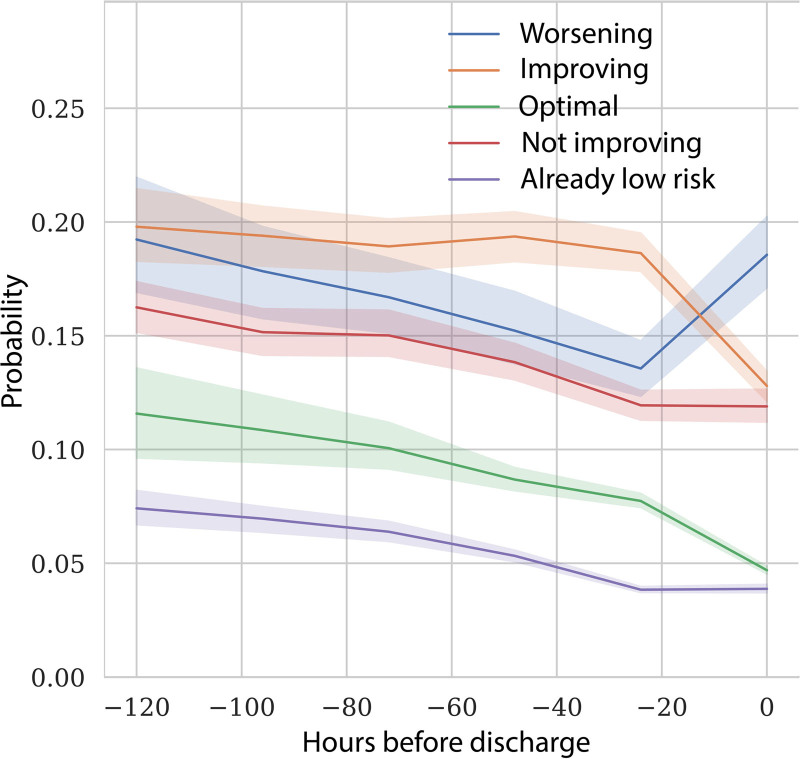
For “long-stay” patients, on average, predicted readmission probability decreased as patients get closer to ICU discharge. However, there was large variation between the patients allowing us to identify five subgroups. Figure 3 shows readmission probabilities for those subgroups at different time points of ICU admission. Table S8 and the Impact Analysis section (Supplemental Digital Content 1, http://links.lww.com/CCX/A783) give detailed definitions for these groups, but in short, “improving” represents patients that have a high risk but were improving by at least 2% points, “not improving” have a high risk but were improving by less than 2%, “already low risk” are low-risk patients that already had a low risk the day before, “optimal” patients recently improved toward a low risk, and “worsening” are patients that showed a high risk that increased by more than 2%.
Figure 3.
Mean predicted readmission probability at different moments of ICU admission. The colored lines are the average (with 95% CI) for five groups of patients that differ in the way the predicted risk changes toward the moment of discharge. See the Impact Analysis section (Supplemental Digital Content 1,http://links.lww.com/CCX/A783) for the definition of these groups.
In our impact analysis, we suggest discharge strategies based on these groups: postponing discharge, discharging as planned, or discharging a day earlier (Table S9, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). Using two possible scenarios, we show that integrating the readmission probability-time curve in a discharge workflow and changing management for high-risk short-stay patients and long-stay groups “not improving,” “already low risk,” and “worsening” could lead to a decrease of up to 14% in readmission rate with an increase of only about 1.6% in average length of stay (Table S10 and Impact Analysis section, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). A preproduction software version of the user interface, demonstrating an overview of the admitted patients and individual prediction trends and features, is shown in Figure S6, Supplemental Digital Content 1 (http://links.lww.com/CCX/A783).
DISCUSSION
For the first time, we describe the development of an explainable machine learning model for clinical decision support to optimize timing of ICU discharge using AmsterdamUMCdb, the first freely available European critical care database. The model demonstrated good performance on a dataset from more recent patients, and by using feature importance techniques and displaying risk prediction for readmission and mortality throughout the admission, application of our model as a bedside decision support tool seems feasible.
Several attempts to develop prediction models to prevent ICU readmission and/or death after discharge from the ICU for general adult critical care patients have been made previously (3, 5–8, 10–14, 33, 34). Earlier models have used logistic regression for prediction focusing on very few parameters, whereas the newer models use more advanced machine learning algorithms (Table S11, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). Our XGBoost machine learning model represents an improvement over the models reported in the literature in terms of AUROC. However, the improvement in performance is modest, and by choosing a time window of 7-day postdischarge, more patients have been included that died after discharge, which is a more predictable task (7). Nevertheless, given the large and increasing number of ICU admissions worldwide, this modest reduction may have significant impact for patients and society.
Our article has several strengths. First, compared with the current literature, we performed more extensive feature engineering. Unsurprisingly, this allowed the logistic regression model to achieve a comparable performance to more advanced machine learning algorithms.
Second, our model reduces the wide gap between model development and bedside implementation (35). From the start, we developed our model, pipeline, and software design with clinical implementation in mind, based on our previous experience implementing bedside decision support (36, 37). This required close collaboration between intensivists and data scientists for feature engineering with a focus on features that are available in real time, innovative approaches with respect to interpretability, actionable insights, and feature importance, as well as performance evaluations and impact analyses.
Finally, we chose to report both discrimination and calibration, with both ROC curve and PRC, and probability calibration curves, respectively. Predicting relatively uncommon occurrences like adverse events after ICU discharge is still a major challenge. Evaluation of predictive performance requires more than analysis using ROC curves that tend to give an overly optimistic view of model performance in imbalanced datasets (22–24). Though the AUPRC of 0.189 (Fig. 1C) is significantly higher than the baseline of the combined outcome of 6.7% (0.067) in the validation cohort, there is still room for improvement, even for the complex models we describe.
Our article also has several limitations. First, although model performance was measured using a separate validation dataset, our study is still a single-center study. Unfortunately, validating the model on data of other hospitals is a relatively slow process, both due to legal issues and the required harmonization (mapping) of the data between different hospitals.
Second, in our Dutch setting where ICU capacity is strained, we specifically chose to target readmissions and mortality until 7 days after ICU discharge to include patients that might suffer from complications that typically occur later, for example, respiratory failure or sepsis and not the quality indicator of ICU readmission after 2 days. Data suggest that the majority of ICU admissions within 48 hours after ICU discharge are not preventable (38). In addition, an analysis of early (< 72 hr) versus late (> 72 hr) IC readmissions showed that patients with late ICU readmissions were more often discharged “after hours,” had developed acute renal failure, and showed a trend toward more severe comorbidities, suggestion a group of patients that are not captured when limiting the scope to 2 days. In a prospective study validating an algorithm predicting death or readmission within 14 days after ICU discharge, the median time to readmission was 4 days (interquartile range [IQR], 2–9 d) and 7 days (IQR, 3–19 d) for death on the ward after ICU discharge, suggesting that limiting predictions to only the first 2 days may be too restrictive (12). However, since the performance of our retrained model for the outcome after 2 days dropped significantly, this does show that predicting early readmissions is a more difficult task, which is in line with the literature where models (mainly) predicting mortality after ICU discharge typically have higher AUROCs (Table S11, Supplemental Digital Content 1, http://links.lww.com/CCX/A783).
Third, predicting and possibly preventing readmissions may not influence outcomes in some healthcare systems. In fact, even unexpected ICU readmissions may not unequivocally lead to an increase in hospital mortality as some authors have shown in a prospective study (39).
Furthermore, since the model was trained on features of discharged patients, a low predicted probability in a readmission probability-time curve does not necessarily imply that a patient is ready for discharge, since other conditions, such as the need for mechanical ventilation or vasoactive drugs, may prevent this. In fact, these counterfactual predictions (i.e., estimating outcome probability assuming the patient had been discharged) may possibly lead to incorrect conclusions. However, this is an inherent problem when implementing predictive models, because using the model may expedite the timing of decision-making, thus leading to less available data for the model to base its predictions on. To mitigate incorrect conclusions by using the software, the predicted outcomes are accompanied by symbols denoting possible situations preventing discharge, such as need for mechanical ventilation, amount of supplemental oxygen, and vasoactive drugs (Figure S6, Supplemental Digital Content 1, http://links.lww.com/CCX/A783). In addition, end-user training with these limitations in mind will be part of the implementation strategy.
Finally, it should be realized that ICU discharge and readmission decisions captured in AmsterdamUMCdb are subject to the preferences and biases of the intensivists practicing at our center and that the model learns this human subjectivity (40).
Clinical model adoption depends on ease of use and the trade-off between the cost associated with a readmission (mortality and length of stay) and the cost of an unnecessary prolonged stay (length of stay, canceled elective surgery, or denied admissions). Our explorative readmission probability-time curves suggest that it is feasible to prevent readmissions and deaths from premature ICU discharge with only a small increase in total length of stay. Furthermore, if patients that do not improve over the last days of admission (“not improving” group in Fig. 3) would be discharged earlier as suggested by the model, not only readmission rate but also total length of stay might be reduced. By using the predicted readmission risk and the change in risk compared with the previous day, together with clinical intuition, we hope intensivists can make better informed decisions. These promising results have prompted us to proceed with multicenter validation and clinical implementation at the bedside in Amsterdam UMC.
CONCLUSIONS
Our findings showed that the vast amount of data stored in AmsterdamUMCdb, the first freely available European critical care database, can be used to develop an explainable model that may aid in identifying patients with high risk of readmission and mortality after ICU discharge. By using readmission probability-time curves, our analysis showed that the model may decrease the readmission rate with a relative risk reduction of up to 14% while only minimally increasing the average length of stay. Model performance can and should be further improved with data from other hospitals following the SCCM/ESICM Joint Data Sharing initiative and similarly releasing their critical care patient data responsibly. In addition, by joining forces with data scientists, intensivists can advance the process from clinical model to bedside decision support tool.
ACKNOWLEDGMENTS
We thank the ICU and MCU nurses, residents, fellows, and staff for their efforts in documenting and continuously improving the collected data in the patient data management system. We also thank Jan Peppink for database administration and support of the patient data management system.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Drs. Thoral and Elbers wrote the study protocol. Mr. Driessen designed the data extraction process. Dr. Thoral, Dr. Fornasa, Mr. de Bruin, Mr. Tonutti, and Mr. Hovenkamp analyzed and interpreted the data. Dr. Thoral, Dr. Fornasa, and Mr. de Bruin drafted the article. All authors read, commented, and approved the final article.
The local Medical Ethics Committee reviewed the study protocol and considered it to be outside the scope of the Law on Scientific Research on Humans. In addition, they approved a waiver for the need for written informed consent for the use of pseudonymized data based on the very large number of included patients in accordance with the General Data Protection Regulation.
Amsterdam University Medical Centers is entitled to royalties from Pacmed. Mr. Hovenkamp is cofounder of Pacmed, a Dutch technology company in health care. The remaining authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Daly K, Beale R, Chang RW: Reduction in mortality after inappropriate early discharge from intensive care unit: Logistic regression triage model. BMJ. 2001; 322:1274–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez R, Baigorri F, Navarro G, et al. : A modified McCabe score for stratification of patients after intensive care unit discharge: The Sabadell score. Crit Care. 2006; 10:R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gajic O, Malinchoc M, Comfere TB, et al. : The Stability and Workload Index for Transfer score predicts unplanned intensive care unit patient readmission: Initial development and validation. Crit Care Med. 2008; 36:676–682 [DOI] [PubMed] [Google Scholar]
- 4.Fernandez R, Serrano JM, Umaran I, et al. ; Sabadell Score Study Group: Ward mortality after ICU discharge: A multicenter validation of the Sabadell score. Intensive Care Med. 2010; 36:1196–1201 [DOI] [PubMed] [Google Scholar]
- 5.Frost SA, Tam V, Alexandrou E, et al. : Readmission to intensive care: Development of a nomogram for individualising risk. Crit Care Resusc. 2010; 12:83–89 [PubMed] [Google Scholar]
- 6.Ouanes I, Schwebel C, Français A, et al. ; Outcomerea Study Group: A model to predict short-term death or readmission after intensive care unit discharge. J Crit Care. 2012; 27:422.e1–422.e9 [DOI] [PubMed] [Google Scholar]
- 7.Badawi O, Breslow MJ: Readmissions and death after ICU discharge: Development and validation of two predictive models. PLoS One. 2012; 7:e48758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jo YS, Lee YJ, Park JS, et al. : Readmission to medical intensive care units: Risk factors and prediction. Yonsei Med J. 2015; 56:543–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo YF, Rumshisky A: Interpretable topic features for post-ICU mortality prediction. AMIA Annu Symp Proc. 2016; 2016:827–836 [PMC free article] [PubMed] [Google Scholar]
- 10.Desautels T, Das R, Calvert J, et al. : Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: A cross-sectional machine learning approach. BMJ Open. 2017; 7:e017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venugopalan J, Chanani N, Maher K, et al. : Combination of static and temporal data analysis to predict mortality and readmission in the intensive care. Annu Int Conf IEEE Eng Med Biol Soc. 2017; 2017:2570–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabes J, Seligman W, Barrett C, et al. : Does the implementation of a novel intensive care discharge risk score and nurse-led inpatient review tool improve outcome? A prospective cohort study in two intensive care units in the UK. BMJ Open. 2017; 7:e018322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojas JC, Carey KA, Edelson DP, et al. : Predicting intensive care unit readmission with machine learning using electronic health record data. Ann Am Thorac Soc. 2018; 15:846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue Y, Klabjan D, Luo Y: Predicting ICU readmission using grouped physiological and medication trends. Artif Intell Med. 2019; 95:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Society of Intensive Care Medicine: Data Science Section Data Science. ESICM. Available at: https://www.esicm.org/groups/data-science/. Accessed January 20, 2020 [Google Scholar]
- 16.Thoral PJ, Peppink JM, Driessen RH, et al. ; Amsterdam University Medical Centers Database (AmsterdamUMCdb) Collaborators and the SCCM/ESICM Joint Data Science Task Force: Sharing ICU patient data responsibly under the Society of Critical Care Medicine/European Society of Intensive Care Medicine Joint Data Science Collaboration: The Amsterdam University Medical Centers Database (AmsterdamUMCdb) Example. Crit Care Med. 2021; 49:e563–e577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen LM, Martin CM, Keenan SP, et al. : Patients readmitted to the intensive care unit during the same hospitalization: Clinical features and outcomes. Crit Care Med. 1998; 26:1834–1841 [DOI] [PubMed] [Google Scholar]
- 18.Alban RF, Nisim AA, Ho J, et al. : Readmission to surgical intensive care increases severity-adjusted patient mortality. J Trauma. 2006; 60:1027–1031 [DOI] [PubMed] [Google Scholar]
- 19.Bose S, Johnson AEW, Moskowitz A, et al. : Impact of intensive care unit discharge delays on patient outcomes: A retrospective cohort study. J Intensive Care Med. 2019; 34:924–929 [DOI] [PubMed] [Google Scholar]
- 20.Williams TA, Ho KM, Dobb GJ, et al. ; Royal Perth Hospital ICU Data Linkage Group: Effect of length of stay in intensive care unit on hospital and long-term mortality of critically ill adult patients. Br J Anaesth. 2010; 104:459–464 [DOI] [PubMed] [Google Scholar]
- 21.Pedregosa F, Varoquaux G, Gramfort A, et al. : Scikit-learn: Machine learning in python. J Mach Learn Res. 2012; 12:2825–2830 [Google Scholar]
- 22.Saito T, Rehmsmeier M: The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015; 10:e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis J, Goadrich M: The relationship between precision-recall and ROC curves. Proceedings of the 23rd International Conference on Machine learning - ICML ’06, Pittsburgh, PA, June 25–29, 2006. ACM Press, New York, NY, 2006, pp 233–240 [Google Scholar]
- 24.Leisman DE: Rare events in the ICU: An emerging challenge in classification and prediction. Crit Care Med. 2018; 46:418–424 [DOI] [PubMed] [Google Scholar]
- 25.Lundberg SM, Erion GG, Lee S-I: Consistent individualized feature attribution for tree ensembles. arXiv:1802.03888 [cs.LG] [Google Scholar]
- 26.Lundberg SM, Nair B, Vavilala MS, et al. : Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018; 2:749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maharaj R, Terblanche M, Vlachos S: The utility of ICU readmission as a quality indicator and the effect of selection. Crit Care Med. 2018; 46:749–756 [DOI] [PubMed] [Google Scholar]
- 28.Vickers AJ, Elkin EB: Decision curve analysis: A novel method for evaluating prediction models. Med Decis Making. 2006; 26:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Calster B, Wynants L, Verbeek JFM, et al. : Reporting and interpreting decision curve analysis: A guide for investigators. Eur Urol. 2018; 74:796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subbe CP, Kruger M, Rutherford P, et al. : Validation of a modified Early Warning Score in medical admissions. QJM. 2001; 94:521–526 [DOI] [PubMed] [Google Scholar]
- 31.Kareliusson F, De Geer L, Tibblin AO:Risk prediction of ICU readmission in a mixed surgical and medical population. J Intensive Care. 2015; 3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen T, Guestrin C: XGBoost: A scalable tree boosting system. KDD '16: The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, August 13–17, 2016, pp 785–794 [Google Scholar]
- 33.Ng YH, Pilcher DV, Bailey M, et al. : Predicting medical emergency team calls, cardiac arrest calls and re-admission after intensive care discharge: Creation of a tool to identify at-risk patients. Anaesth Intensive Care. 2018; 46:88–96 [DOI] [PubMed] [Google Scholar]
- 34.McWilliams CJ, Lawson DJ, Santos-Rodriguez R, et al. : Towards a decision support tool for intensive care discharge: Machine learning algorithm development using electronic healthcare data from MIMIC-III and Bristol, UK. BMJ Open. 2019; 9:e025925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosein FS, Bobrovitz N, Berthelot S, et al. : A systematic review of tools for predicting severe adverse events following patient discharge from intensive care units. Crit Care. 2013; 17:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weijs PJ, Stapel SN, de Groot SD, et al. : Optimal protein and energy nutrition decreases mortality in mechanically ventilated, critically ill patients: A prospective observational cohort study. JPEN J Parenter Enteral Nutr. 2012; 36:60–68 [DOI] [PubMed] [Google Scholar]
- 37.Elbers PW, Girbes A, Malbrain ML, et al. : Right dose, right now: Using big data to optimize antibiotic dosing in the critically ill. Anaesthesiol Intensive Ther. 2015; 47:457–463 [DOI] [PubMed] [Google Scholar]
- 38.Al-Jaghbeer MJ, Tekwani SS, Gunn SR, et al. : Incidence and etiology of potentially preventable ICU readmissions. Crit Care Med. 2016; 44:1704–1709 [DOI] [PubMed] [Google Scholar]
- 39.Santamaria JD, Duke GJ, Pilcher DV, et al. ; Discharge and Readmission Evaluation (DARE) Study Group: Readmissions to intensive care: A prospective multicenter study in Australia and New Zealand. Crit Care Med. 2017; 45:290–297 [DOI] [PubMed] [Google Scholar]
- 40.McLennan S, Lee MM, Fiske A, et al. : AI ethics is not a panacea. Am J Bioeth. 2020; 20:20–22 [DOI] [PMC free article] [PubMed] [Google Scholar]