Abstract
Abstract
At the start of the COVID-19 pandemic, the US faced nationwide shortages of nasopharyngeal swabs due to both overwhelmed supply chains and an increase in demand. To address this shortfall, multiple 3D printed swabs were ultimately produced and sold for COVID-19 testing. In this work, we present a framework for mechanical and functional bench-testing of nasopharyngeal swabs using standard and widely available material testing equipment. Using this framework, we offer a comprehensive, quantitative comparison of the 3D printed swabs to benchmark their performance against traditional flocked swabs. The test protocols were designed to emulate the clinical use of the nasopharyngeal swabs and to evaluate potential failure modes. Overall, the 3D printed swabs performed comparably to, or outperformed, the traditional swabs in all mechanical tests. While traditional swabs outperformed some of the new 3D printed swabs in terms of sample uptake and retention, similar amounts of RNA were recovered from both 3D printed and traditional swabs.
Graphic abstract
Keywords: COVID-19, 3D printing, Additive manufacturing, Mechanical, Absorption
Introduction
At the beginning of the COVID-19 pandemic, nationwide shortages surfaced in consumables used for testing. These shortages in the various components of viral testing kits, including nasopharyngeal swabs, were due to the simultaneous increase in demand and the decrease in supply, as temporary shutdowns of many production facilities reduced suppliers’ production capabilities. To bridge the gap between the demand for and supply of nasopharyngeal swabs, alternative manufacturing methods and sources were quickly turned to such as three-dimensional (3D) printing.1
Three-dimensional printing technologies have several advantages. First, 3D printing is geared toward fast prototyping for design iteration, enabling hundreds of different designs to be rapidly printed, tested, and then redesigned. Second, the wide variety of material and printing processes available for 3D printing means there are many options to create a swab. Last, 3D printers already in use in hospitals and clinics could easily be repurposed, allowing those entities to 3D print their own swabs to meet demand.
Absorbent tipped applicators, printed or traditional, are generally considered Class 1 medical devices under US Code of Federal Regulations 21 CFR 880.6025.2 Sterile swabs are exempt from premarket notification requirements3 and manufacturers are responsible for ensuring their product’s performance is appropriate for its clinical use. Ultimately, six different 3D printed nasopharyngeal swabs (Table I) were produced, tested clinically, and sold to various hospitals and clinics worldwide.4,5 These printed swabs utilized several different printing processes, materials, and various design methodologies for sample collection.
Table I.
Summary of the 3D printed and traditional nasopharyngeal swabs commercially available for COVID-19 testing.
| Printing process | Material | Design | |
|---|---|---|---|
| 3D printed swab | |||
| Abiogenix FAST Spiral NP Swab7 | HP Multi Jet Fusion | Polyamide 11 (Nylon) | Spiral |
| Formlabs–USF NP Swab8 | Formlabs’ Stereolithography | Formlabs Surgical Grade Resin | Bulbous |
| Formlabs–Northwell NP Swab9 | Formlabs’ Stereolithography | Formlabs Surgical Grade Resin | Bulbous with Holes |
|
EnvisionTEC NP Swab10 |
EnvisionTEC’s Continuous Digital Light Manufacturing | E-Guide Soft C-29C Resin | Lattice |
| Resolution Medical Lattice Swab12, 13 | Carbon’s Digital Light Synthesis | KeySplint Soft Clear Resin | Lattice |
| Traditional swabs | |||
|
Fosun Pharma NP Swab |
N/A | Nylon | Flocked |
| Copan FLOQSwab (510CS01) | N/A | Nylon | Flocked |
An additional 3D printed swab, made by Origin,6 was also produced and sold; however, the authors were unable to obtain these swabs for testing.
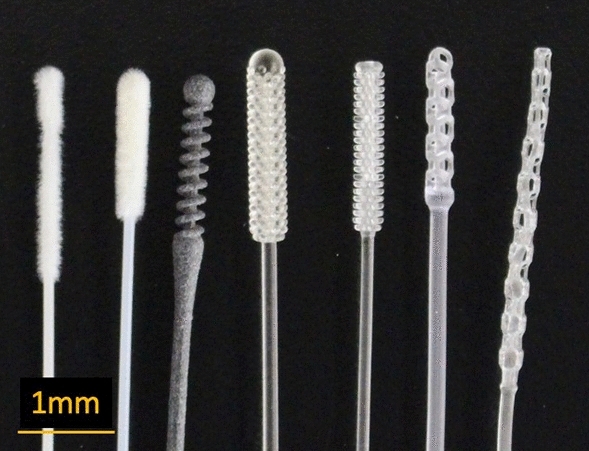
Traditional nasopharyngeal swabs are long and flexible, capable of being inserted into the nasopharyngeal space, with nylon flocking on the tip to enable sample collection. In appearance, the 3D printed swabs bear little resemblance to the traditional swabs (Figure 1). Some testing of the 3D printed swabs was conducted, most notably by the Veterans Health Administration (VHA)12 and Harvard University.1,13 The testing performed by the VHA compared the 3D printed and traditional swabs in actual use-case scenarios (e.g., ability to fit through the nasal cavity and reach the sampling location); however, some important usage scenarios were not evaluated (e.g., the ability of the swabs to be rotated at the sampling location). Additionally, several of the test protocols (i.e., mechanical testing) provide only qualitative assessments (e.g., asking users to assess prototypes’ mechanical properties such as breaking/bending rather than using a measurement tool) of their performance and allow for only a limited comparison among the different 3D printed (four prototypes) and traditional swabs.1,12,13 Additional quantitative assessments of some swabs (e.g., EnvisionTEC and Resolution Medical lattice swabs9,10) have been performed to compare their performance to traditional swabs; however, these studies do not allow for comparisons among other different 3D printed swabs. In this work, we present a framework for bench-testing nasopharyngeal swabs on mechanical and sample collection performance using standard and widely available material testing equipment. We aim to develop quantitative tests for nasopharyngeal swabs, covering the normal usage performance metrics, allowing for comparisons between the traditional and the newly manufactured, 3D printed swabs.
Figure 1.

Images of the traditional and 3D printed nasopharyngeal swabs. From top to bottom (top image) and left to right (bottom image): Fosun (traditional), Copan (traditional), Abiogenix (3D), Formlabs–USF (3D), Formlabs–Northwell (3D), EnvisionTEC (3D), and Resolution (3D).
Methods
Both 3D printed and traditional nasopharyngeal swabs were tested (Table I, Figure 1) on mechanical performance, sample collection efficiency (uptake/release and viral RNA recovery), and other preclinical metrics (i.e., PCR compatibility, physical abrasion). The mechanical test protocols developed were designed to emulate the clinical use of the nasopharyngeal swabs and to evaluate the potential failure modes. The mechanical analysis included testing of the swabs in tension to mimic swab catching when pulled out of the nasopharyngeal space; torsion to mimic catching on an obstruction when being rotated within the nasopharyngeal space, and flexure to mimic bending when inserted into a nasal cavity. The effects of sterilization method and shelf-aging on mechanical performance were also evaluated. The sample collection efficiency, and other preclinical metrics, were designed to ensure the swabs collected sufficient sample to test for the presence of the virus, without causing injury to the patient (e.g., epistaxis). Abiogenix swabs were provided by Abiogenix for testing. Formlabs swabs were printed by the authors using Formlabs Form 3B Printer and Formlabs Surgical Guide Resin. All other swabs were purchased from their respective suppliers. The number of replicates (i.e., swabs) used for each test is noted. All swabs were sterilized before testing either by the manufacturer or by the authors following the manufacturer’s recommendations.
These tests are summarized next.
- Mechanical Performance (Performed on a Universal Testing Machine (Instron), except for Torsion #2 and Combined Torsion and Flexure, which were performed on a Discovery Hybrid Rheometer [TA Instruments, DHR-1])
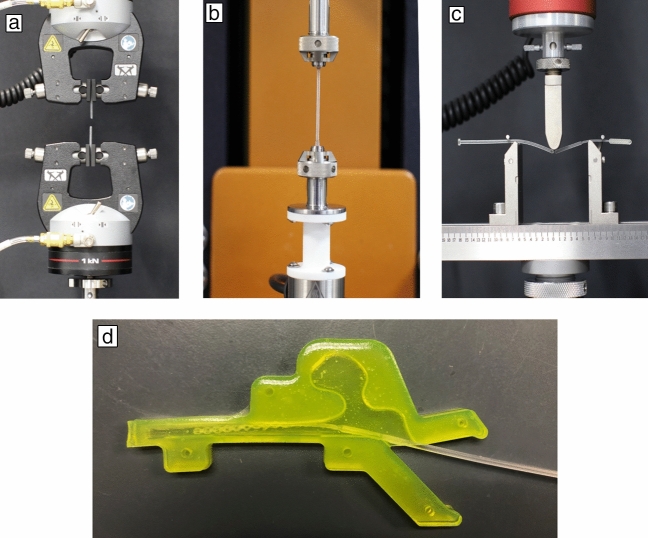
- Tensile #1 and #2–To mimic the worst-case scenario of the swab being caught on an obstruction when being pulled out of the nasopharyngeal space, the swab was pulled until it broke. For Tensile #1, the two ends of the swab were clamped between two pneumatic grips whereas in Tensile #2, the head and neck of the swab were clamped (Figure 2a). The two ends were pulled apart until the swab broke. Load required to break was recorded. Ten replicates were used.
- Torsion #1 and #2–To mimic the worst-case scenario of the swab being caught on an obstruction when being rotated within the nasopharyngeal space, the swab was twisted until it broke. For Torsion #1, the two ends of the swab were clamped between two grips whereas in Torsion #2, the head and the neck of the swab were clamped (Figure 2b). One end of the swab was held stationary while the other end was rotated counterclockwise until the swab broke. Torque required to break was recorded. For Torsion #1, 10 replicates were used; for Torsion #2, five replicates were used.
- Flexure–To mimic the use case of the swab bending when inserted into the nasal cavity, the flexibility of the joint between the head and the neck of the swab was measured by flexing it 90° at this joint (Figure 2c). The load required to flex the swab was recorded. Ten replicates were used.
- Combined Torsion and Flexure–To mimic the use case when swabbing, the swab was flexed and inserted into a 3D printed nasal cavity (filled with 1% wt/v locust gum as mucus mimic, Figure 2d) and then twisted for 25 rotations. Maximum torque required to rotate the swab was recorded. Five replicates were used.
- Break–Each swab handle has a breakpoint (Figure 1) that allows users to break off the end of the swab safely and easily into the transport vial. To evaluate the strength of the breakpoint of the swab, it was subjected to a standard three-point bend test at the breakpoint. Load required to break the swab was recorded. Three to ten replicates were used.
- Effect of sterilization and aging on mechanical performance–To evaluate the effects of the sterilization on the mechanical performance of the swabs, manufacturer-sterilized swabs, or in-house sterilized swabs (within 6 h of sterilization) were evaluated using Tensile #1. For aging tests, in-house sterilized only swabs were stored for up to 7 days in unopened sterilization pouches prior to testing. Swabs were sterilized in house, using protocols recommended by the manufacturer, as shown in Table II. Five to ten replicates were used.
- Sample Collection
- Uptake/Release–To quantify sample amount collected by the swabs, a simplified uptake/release test was performed. Swabs were first weighed prior to fully immersing the head of swab into a solution containing mucus mimic (1% wt/v locust gum in Hank’s buffered salt solution, HBSS) with 0.5% 150 kDa FITC labeled dextran as a tracer. Swabs were rotated clockwise for 15 s, quickly weighed, and then placed in 4 mL of HBSS. Swabs were kept in solution for 2.5 h and then vortexed vigorously. The amount of FITC-dextran released into the HBSS solution was measured using a Synergy H1 spectrometer. To quantify how much material adheres to the swab materials alone (i.e., in the absence of any 3D design), printed solid cylindrical swab shafts were used (four replicates). The amount collected was normalized by swab shaft surface area.
- PCR-Compatibility Testing–Swabs were first checked for any potential PCR-inhibitory material by allowing sterilized swabs (two replicates) to leach (swab-head down) into 3 mL HBSS for 4 days before PCR analysis.
- Viral RNA recovery–In a clinical setting, nasopharyngeal swabs are inserted until the swab contacts the nasopharynx. A collected sample therefore consists of both secretions and cells collected from the nasopharynx. To determine the ability of swabs to retain and release viral samples, swabs were used to collect samples from virus infected mucus mimic, as well as from virus infected cells. Swabs (three replicates) were immersed in 1.5 mL in mucus mimic spiked with 1 × 106 PFU murine coronavirus (mouse hepatitis virus, MHV) 14,15 and rotated for 15 s. A second set of swab samples (three replicates) were used to swab a monolayer (one well of a 96-well plate/replicate) of virus infected 17CL-1 mouse epithelial cells immersed in mucus mimic. Infected swabs were placed in a fresh tube containing 2 mL of media (Dulbecco’s Modified Eagle Medium (Thermo Scientific), 2% FBS, Pen/Strep antibiotic) and allowed to incubate at room temperature for 2.5 h for the mucus mimic or overnight at 4ºC for the infected cells. The samples were processed for RNA using QIAamp RNA Viral Mini kit (Qiagen) using a QIAcube Connect machine and the virus infected cells were pelleted and processed for RNA using RNeasy Mini Kit (Qiagen) and a QIAcube Connect machine. Viral RNA was eluted and quantitated in triplicate using a One Step PrimeScript RT-PCR Kit, (Takara) and MHV nsp2 specific primers and probes using cycling cycling conditions: 42 ºC/5 min, 95 ºC/30 sec, and 40 cycles of 95 ºC/5 sec, 60 ºC/1 min. The MHV nsp2 primers and probe used in the assay are described by Case et al.16
- Abrasion–To determine the abrasiveness of the swabs, 3D nasal cavity tissue mimics (Figure 2d) were molded using 6% agarose containing 1 mg/mL of FITC-dextran (70 kDa). Swabs (six replicates) were inserted into the 3D agarose nasal cavity and rotated clockwise for 15 s. Swabs were then immersed in HBSS and vigorously vortexed to allow for release of material scraped from nasal cavity. The amount of FITC-dextran released into the HBSS solution was measured using a Synergy H1 spectrometer.
Figure 2.
Images of the swabs in the different test setups: (a) Tensile, (b) Torsion, (c) Break, and (d) Abrasion. A similar nasal cavity mold as that shown in (d) was 3D printed and used for the Combined Torsion and Flexure Test (mold for mechanical testing was made out of surgical guide resin).
Table II.
Summary of the autoclave sterilization protocols.
| Autoclave Protocol No. | Description | Swabs Tested |
|---|---|---|
| Protocol #1 | 121°C for 30 min |
Abiogenix Formlabs–USF Formlabs–Northwell EnvisionTEC |
| Protocol #2 | 132°C for 4 min |
Abiogenix Formlabs–USF Formlabs–Northwell |
| Protocol #3 | 132°C for 4 min, Prevacuum Sterilization with four Preconditioning Pulses and a 30-min, 20-in. Hg vacuum dry time with three pulses | Resolution |
The data are expressed in tables as the mean ± standard deviation. Shapiro–Wilk normality test was used to determine if the data set was well-modeled by a normal distribution (i.e., Gaussian distribution). For non-Gaussian distribution, Kruskal–Wallis nonparametric test was used with Dunn’s test for multiple comparisons. Otherwise, ordinary one-way and two-way ANOVAs with Tukey post hoc tests were used for analysis between groups, where p < 0.05 was considered statistically significant.17
Results/discussion
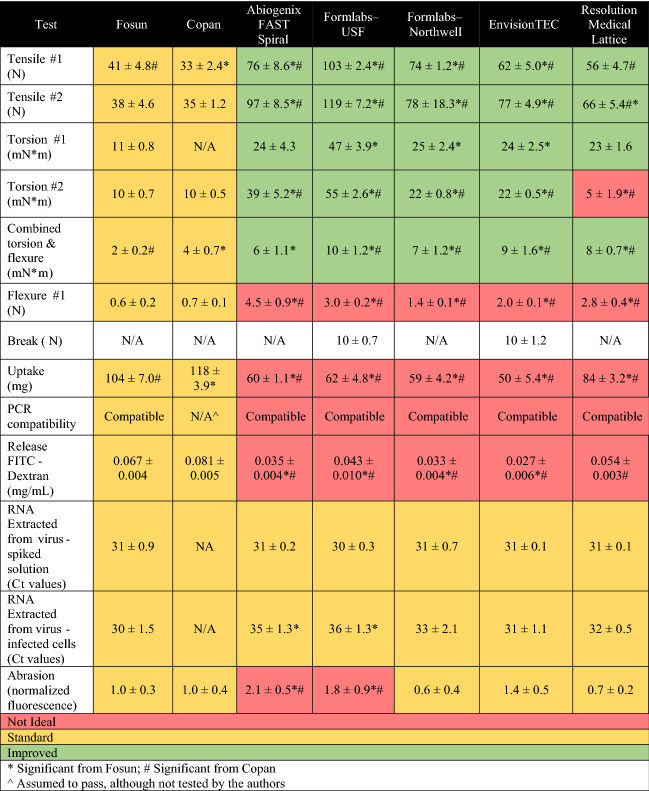
A summary of the results for all tests performed are shown in Tables III and IV.
Table III.
Summary of the testing results for the 3D printed nasopharyngeal swabs, with a comparison to the traditional swabs.
Table IV.
Summary of the tensile strengths measured using Tensile #1 for the 3D printed nasopharyngeal swabs before and after sterilization, for different sterilization protocols, and after aging.
| Pre-sterilization | Post-sterilization | After Aginga | |
|---|---|---|---|
| Abiogenix FAST spiral | 69 ± 2.2 |
AP#1: 67 ± 1.7 AP#2: 62 ± 2.7 |
AP#1: 65 ± 2.2 AP#2: 71 ± 2.8 |
| Formlabs–USF | Not tested |
AP#1: 106 ± 4.0 AP#2: 101 ± 2.8 |
AP#2: 122 ± 5.2 |
| Formlabs–Northwell | 94 ± 1.3 |
AP#1: 80 ± 2.3 AP#2: 66 ± 2.2 |
AP#1: 87 ± 5.3 |
| EnvisionTEC | 58 ± 2.5 |
AP#1: 51 ± 2.6 AP#1: 62 ± 5.0b |
AP#1: 59 ± 4.4 |
| Resolution Medical lattice | Not tested |
AP#3: 50 ± 2.3 AP#3: 56 ± 4.7b |
AP#3: 50 ± 3.0 |
Details on the Autoclave Protocols (AP) are summarized in Table II. All data are reported in Newtons. Bold font is used to indicate significant difference from pre-sterilized or unaged sterilized samples.
aSwabs were aged for 7 days, except for Resolution Medical, which were aged 5 days.
bSterilized by the manufacturer.
Mechanical performance
Understanding the mechanical performance is important for patient comfort as well as for safety (understanding break limits). During tensile testing, we directly tested loads required to break the swabs, mimicking the worst-case scenario of a swab being caught on an obstruction when being pulled out of the nasopharyngeal space. All the 3D printed swabs showed higher tensile strength (preferred) than the traditional swabs, in both tensile tests. For Tensile #1, only Abiogenix and EnvisionTEC swabs broke cleanly at the breakpoint of the swab (preferred); all the remaining swabs broke either in the neck region or at the head neck joint.
During swabbing, swabs are inserted into the nasal cavity and gently rotated. Simplified torsion testing was carried out to determine strength of swab to resist breaking when rotated, mimicking the worst-case scenario of a swab being caught on an obstruction. All the 3D printed swabs had higher torques to break (preferred) compared to traditional swabs in the two tests, except for Abiogenix and Resolution swabs, which did not perform significantly differently from traditional swabs in Torsion #1. It should be noted that although both Formlabs swab designs had higher torques to break compared to traditional swabs, they fragmented upon breaking which is not a desirable outcome. To better approximate the usage scenario of swabbing, swabs were rotated while bent inside a printed nasal cavity (Combined Torsion and Flexure). All the 3D printed swabs showed statistically significant higher torques compared to the traditional swabs. For the Combined Torsion and Flexure, three out of five Formlabs–USF and two of the five Abiogenix swabs broke at the breakpoint during this test; none of the other swabs broke.
For flexure testing, lower loads to flex are preferred as they correspond to a more flexible swab. Overall, the 3D printed swabs showed statistically significant higher loads to flex than the traditional swabs. Despite increased loads to flex, none of the swabs broke during this testing.
Finally, we evaluated the breakpoint of the swab. In this test, smaller loads are preferred to ensure the swab will easily break off when inserted into the testing vial. The Formlabs–USF and EnvisionTEC swabs broke cleanly at the breakpoint during the test; the other 3D printed and traditional swabs did not break, but rather slipped in the setup during testing, invalidating the results. Although this is a standard three-point bend test, used commonly for evaluating strength at break, the design of the swabs (both 3D printed and traditional) requires modifications to this test to allow for quantitative comparisons of performance.
While many of these 3D printed swabs can be purchased sterile, an attractive feature of 3D printing is the option to enable production of swabs at point-of-use. Therefore, it was important to look at how sterilization protocols and aging affect mechanical performance. (Note: these studies were not used to validate the effectiveness of the sterilization protocol.) Abiogenix, Formlabs and EnvisionTEC swabs showed statistically significant differences in tensile strength before and after autoclave sterilization, although for EnvisionTEC, there were significant differences only in swabs sterilized by the authors but not for those autoclaved sterilized by the manufacturer. The type of autoclave sterilization protocol (Protocol 1 versus Protocol 2) also resulted in significant differences for both Abiogenix and Formlabs swabs.
For all of the 3D printed swabs, except for the Resolution swabs, there were significant differences in tensile strength after aging. For Abiogenix, Formlabs, and EnvisionTEC the interaction between sterilization protocol and aging was also found to be significant (p < 0.001), indicating that the samples tended to be more susceptible to aging when autoclaved. We did not test beyond 1 week so it is unknown if the swab material properties will continue to change over a longer time scale. Nonetheless, shelf-life stability is indicated to play a role in tensile strength for post-sterilization.
Sample collection
For the uptake/release test, larger uptake and release amounts are preferred to ensure sufficient material is available for PCR viral testing. Given the difference in head designs and materials, we first tested the contribution of the printed material alone to attract the mucus mimic-FITC-dextran solution. No differences were noted between the different materials, suggesting fluid uptake in subsequent studies, could largely be attributed to the swab design. Although traditional swabs took up more material compared to any 3D printed swab, the total amount of FITC-dextran release from Fosun was not significantly different that the amount released by Resolution. While the materials of the 3D printed swabs are not absorbent compared to nylon bristles, 3D printing enables rapid prototyping for limitless design iterations to optimize swab parameters such as uptake efficiency. One such group11 demonstrated how their latticed swab head bulb geometries could be optimized for sample collection as well as patient comfort and after iterations resulted in a 3D printed swabs that was comparable or better that Copan swabs. Not only can 3D printed swabs head designs be varied but their dimensions can also be modified to create more patient-specific swabs such as 3D printed swabs for children younger than 3 years of age.18
Prior to quantifying viral RNA extracted, we first ensured that swab materials were compatible with PCR by allowing swabs to leach any potential PCR inhibitors into HBSS for 4 days. PCR compatibility tests showed no significant difference between printed and traditional swabs. In addition to potential leachates interfering with PCR measurements, another concern of leaching material could be in vivo toxicity. While this was beyond the scope of our bench testing, the 3D printed materials for the swabs used in these studies have been shown to be biocompatible by their manufacturers for this ex vivo application; however, the context for biocompatibility matters and use of these materials under different conditions (e.g., in vivo long-term exposure) requires further study.19
While uptake/release data in these tests provide a straightforward quantification across swab designs, more critical is how these results translate to virus collected and RNA extracted. Swabs were tested by either immersing in a virus-spiked mucus mimic or swabbing virus infected cells and then subjected to RNA isolation and quantitative RT-PCR. To measure viral load in RT-PCR, the cycle threshold (Ct) value is used to indicate the cycle number at which the signal exceeds background level. Each cycle represents one doubling and Ct levels are inversely proportional to the amount of nucleic acid in the sample where a lower Ct level indicates a higher viral load in the sample. Ct values showed no significant difference between amount of RNA recovered from 3D printed swabs immersed in virus spiked mucus mimic as compared to traditional swabs. Similarities in results between FITC-dextran spike mucus mimic versus virus spiked mucus mimic are likely due to the differences (e.g., stickiness, size) between the virus and FITC-dextran as well as differences between the two detection methods (i.e., fluorescence spectrometry versus PCR). Our results caution that uptake/release studies not be used to directly predict swab’s ability to capture virus for subsequent RNA extraction.
Interestingly, in the test case where swabs were used to swab a monolayer of virus infected cells, mimicking collection of cells from the nasopharynx during clinical swabbing, RNA amounts recovered from Formlabs–USF and Abiogenix differed significantly from traditional swabs. Clinical nasal swab specimens consist of both fluid and cells, so it was important to compare RNA recovery from virus infected cells in addition to virus-spiked mucus mimic. Despite differences observed in some of the 3D printed swab designs, ultimately the critical question is whether enough material is collected to qualify as a positive test. Ct values used to conclude a positive test vary among different tests since there is no standardization for Ct values across existing RT-PCR platforms, but typically high viral titers are associated with Ct values in low 30 s/high 20 s. It is likely that even the two printed swabs found to recover significantly different amounts of RNA compared to traditional swabs may still be suitable for viral capture and RNA extraction.
Although abrasive swabbing is likely to access deep layers of the mucosal barrier, excessive abrasiveness can cause injury to the patients (e.g., epistaxis) and must be considered when designing swabs. Except for Abiogenix and Formlabs–USF, 3D printed swabs were found to not be significantly different in abrasiveness compared to traditional swabs. These two printed swabs also had the widest heads (Abiogenix d = 3.2 mm, Formlabs-USF d = 4 mm), and it is likely that this increased the chances of contact and scraping with tissue mimic nasal cavity walls. In addition to head diameter and possibly the head design, flexibility of the swabs was noted to influence contact with the nasal cavity wall where less flexible swabs pushed the swab’s head up against the wall. The Abiogenix and Formlabs–USF were found to be less flexible in the flexure test than the other 3D printed and traditional swabs and this likely also contributed to the increased abrasiveness. Interestingly, for these two designs abrasiveness was inversely related to swab RNA recovery from swabbed monolayers of cells.
Conclusion
Our goal in this work was to provide a comprehensive, quantitative set of preclinical testing methods for nasopharyngeal swabs, covering the normal usage performance metrics, to glean a better understanding of where improvements are needed. We also used this framework for bench-testing 3D printed swabs with the traditional nasopharyngeal swabs. Critical to the function of swabs is that the swabs collect sufficient sample for RNA extraction, while causing the minimum of patient discomfort and without breaking during sample collection. The tensile, torsion, and flexure testing protocols outlined here can be used to assess whether the swabs will break during sample collection. Together the flexure and abrasion testing protocols can be used as metric toward predicting patient comfort, however, there are no metrics or standards at this time to suggest “x amount of flexibility” or “y amount of abrasiveness” are required for maximal patient comfort. Additional studies are needed to more closely link flexibility/abrasiveness with patient comfort. In summary, no single 3D printed swab performed exactly as traditional swabs and such results are to be expected based on the differences in both design and material. In most of the mechanical testing presented here, the 3D printed swabs demonstrated increased strength; however, this came at a cost to swab flexibility, which may influence patient comfort, as suggested by the abrasion results. However, regarding their primary function, collecting sufficient samples for RNA extraction, 3D printed swabs performed similarly to the traditional swabs.
Many of the 3D printed swabs tested here have already been sold worldwide and tested clinically.4,5 In some cases, 3D printed swabs were found to have fewer false positives compared to traditional swabs, further highlighting the need to improve sample collection for all types of swabs to improve diagnostic testing sensitivity.20 For swabs already in clinical use, our data presented here may serve to provide external validation of their results or provide additional data to consider for future iterations or improvements. Whether 3D printing swabs continues to prevail in a less urgent setting and in swab-scarce areas is still to be seen, but it is likely that 3D printed swabs could be used for other diagnostic testing beyond COVID-19 testing (e.g., crime scene swabbing). What this pandemic confirmed was the viability of additive manufacturing to rapidly respond to the critical need for production of nasopharyngeal swabs and highlights the potential of 3D printing as a parallel technology to conventional mass production.
Acknowledgments
This work was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344. Research was supported by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on response to COVID-19, with funding provided by the Coronavirus CARES Act. We kindly thank S. Weiss at the University of Pennsylvania Perelman School of Medicine for providing MHV strain A59. LLNL-JRNL-821044
Biographies
Angela Tooker
currently runs the Advanced Manufacturing Laboratory at Lawrence Livermore National Laboratory. She received her PhD degree in electrical engineering from the California Institute of Technology. For more than 15 years, she worked in microtechnology process and material development for development of in vivo and in vitro platforms for the study of neural networks and the treatment of a variety of neurological disorders. More recently, she has been researching mechanical properties of additively manufactured materials. Tooker can be reached by email at tooker1@llnl.gov.
Monica L. Moya
is a group leader for the Bioengineering and Advanced Fabrication Group in the Materials Engineering Division at Lawrence Livermore National Laboratory. She received her BS degree from Northwestern University and PhD degree in biomedical engineering from the Illinois Institute of Technology. Her current research interests include three-dimensional biofabrication strategies, biomaterials, vascularization, and microphysiological systems. Moya can be reached by email at moya3@llnl.gov.
Daniel N. Wang
is a graduate intern at Lawrence Livermore National Laboratory (LLNL), characterizing rheological and other mechanical properties of photopolymers for volumetric additive manufacturing. He received his MS degree in chemical engineering from Case Western Reserve University and BS degree in materials science and engineering from Cornell University. Prior to LLNL, he worked in industry developing thermoplastic sealing component materials for compressors used in high-pressure and temperature applications. Wang can be reached by email at wang122@llnl.gov.
Dennis Freeman
holds certificates in computer technology from Control Data Institute, Industrial Maintenance from Merritt College, and has completed course work in electromechanical technology from Chabot College. He has also completed numerous fracture mechanics and failure analysis courses at the University of California, Berkeley, University of California, Davis, and Texas A&M University. Freeman has worked at Lawrence Livermore National Laboratory (LLNL) for more than 40 years and has extensive experience in the field of mechanical testing of materials. He is considered a LLNL subject matter expert in the field of fatigue and fracture experimental methods and analysis. Freeman can be reached by email at freeman3@llnl.gov.
Monica Borucki
is a biomedical scientist at Lawrence Livermore National Laboratory. She received her PhD degree in microbiology from Colorado State University. Her research focuses on using molecular virology to understand the mechanisms of viral evolution and emergence used by different viral families during cross-species transmission. Additionally, she enjoys working as part of multidisciplinary teams of bioinformaticists, synthetic biologists, and bioengineers on a range of projects that address biodefense and pandemic preparedness. Borucki can be reached by email at borucki2@llnl.gov.
Elizabeth Wheeler
is the deputy director of the Center for Bioengineering and a deputy division leader in the Materials Engineering Division at Lawrence Livermore National Laboratory (LLNL). She received her PhD degree in chemical engineering from Stanford University. Her research focuses on integrating biology and engineering to yield new platforms or flexible devices for Homeland Security or medical applications. She has worked on numerous multidisciplinary teams that have field-tested technology developed at LLNL. Wheeler can be reached by email at wheeler16@llnl.gov.
Greg Larsen
is the supervisor of the Mechanics of Materials Group at Lawrence Livermore National Laboratory (LLNL). He received his BS degree in engineering technology from California Polytechnic State University, San Luis Obispo. He has been at LLNL for more than 30 years. Larsen can be reached by email at larsen4@llnl.gov.
Maxim Shusteff
is a staff engineer at Lawrence Livermore National Laboratory, affiliated with the Center for Engineered Materials and Manufacturing, and the Center for Bioengineering. He holds degrees in electrical engineering from Princeton University and the Massachusetts Institute of Technology. Shusteff has led a variety of additive manufacturing R&D efforts with an emphasis on advanced photopolymer materials, and optically driven three-dimensional printing. He also has experience in bioinstrumentation and microfluidics, for applications such as biological sample processing, pathogen detection, and novel cell culture methods. Shusteff can be reached by email at shusteff1@llnl.gov.
Eric B. Duoss
is the director of the Center for Engineered Materials and Manufacturing at Lawrence Livermore National Laboratory (LLNL), where he guides research activities and maps strategic directions in the areas of advanced materials and manufacturing. Duoss received his PhD degree in materials science and engineering from the University of Illinois. At LLNL, he leads teams that invent novel materials and manufacturing processes, with focus on creating designer architectures for chemical, mechanical, thermal, and functional properties for applications in the fields of defense, climate, transportation, energy, aerospace, human health, and others. Duoss can be reached by email at duoss1@llnl.gov.
Christopher M. Spadaccini
is currently the division leader for the Materials Engineering Division at the Lawrence Livermore National Laboratory (LLNL). He received his BS, MS, and PhD degrees from the Department of Aeronautics and Astronautics at the Massachusetts Institute of Technology. Spadaccini has been working in advanced additive manufacturing process development and architected materials for more than 13 years. He founded several new fabrication laboratories at LLNL for process development focused on micro- and nanoscale features and mixed-material printing. Spadaccini can be reached by email at spadaccini2@llnl.gov.
Data availability
The raw data required to reproduce these findings and the processed results are available on request from the corresponding author.
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Footnotes
Angela Tooker and Monica L. Moya contributed equally to this work.
Contributor Information
Angela Tooker, Email: tooker1@llnl.gov.
Monica L. Moya, Email: moya3@llnl.gov
Daniel N. Wang, Email: wang122@llnl.gov
Dennis Freeman, Email: freeman3@llnl.gov.
Monica Borucki, Email: borucki2@llnl.gov.
Elizabeth Wheeler, Email: wheeler16@llnl.gov.
Greg Larsen, Email: larsen4@llnl.gov.
Maxim Shusteff, Email: shusteff1@llnl.gov.
Eric B. Duoss, Email: duoss1@llnl.gov
Christopher M. Spadaccini, Email: spadaccini2@llnl.gov
References
- 1.Callahan C, Lee R, Zulauf KE, Tamburello L, Smith KP, Prefitera J, Cheng A, Green A, Azim AA, Yano A, Doraiswami N, Kirby JE, Arnaout RRA. Rapid open development and clinical validation of multiple new 3D-printed nasopharyngeal swabs in response to the COVID-19 pandemic. MedRxiv. 2020 doi: 10.1101/2020.04.14.20065094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.https://www.ecfr.gov
- 3.https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpcd/classification.cfm?id=2714
- 4.Cox J, Koepsell S. 3D-printing to address COVID-19 testing supply shortages. Lab. Med. 2020;51:e45. doi: 10.1093/labmed/lmaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ford J. A 3D-printed nasopharyngeal swab for COVID-19 diagnostic testing. 3D Print Med. 2020;6:1. doi: 10.1186/s41205-02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.https://www.origin.io/npswab/
- 7.https://www.abiogenix.com/
- 8.https://formlabs.com/covid-19-response/covid-test-swabs/
- 9.https://envisiontec.com/wp-content/uploads/2020/04/EnvisionTEC-NP-Swabs.pdf
- 10.https://www.resolutionmedical.com/latticeswabs
- 11.Bennet I. The rapid deployment of a 3D printed “latticed” nasopharyngeal swab for COVID-19 testing made using digital light synthesis. MedRxiv. 2020 doi: 10.1101/2020.05.25.20112201. [DOI] [Google Scholar]
- 12.https://3dprint.nih.gov/collections/covid-19-response/nasal-swabs/protocols
- 13.Callahan C. Open development and clinical validation of multiple 3D-printed sample collection swabs: Rapid resolution of a critical COVID-19 testing bottleneck. MedRxiv. 2020 doi: 10.1101/2020.04.14.20065094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed W. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191:110092. doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay JK. Design and multicenter clinical validation of a three-dimensionally printed nasopharyngeal swab for SARS-CoV-2 testing. JAMA Otolaryngol. Head Neck Surg. 2021 doi: 10.1001/jamaoto.2020.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case JB. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 2017;92(1):e01531–e1617. doi: 10.1128/JVI.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagano M, Gauvreau K. Principles of Biostatistics. 2. Boca Raton, FL: CRC Press; 2018. [Google Scholar]
- 18.Starosolski Z. Design of 3D-printed nasopharyngeal swabs for children is enabled by radiologic imaging. Am. J. Neurorad. 2020;41(12):2345. doi: 10.3174/ajnr.A6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers HB. Dental resins used in 3D printing technologies release ovo-toxic leachates. Chemosphere. 2020;270:129003. doi: 10.1016/j.chemosphere.2020.129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao AT. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J. Med. Virol. 2020;92(10):1755. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data required to reproduce these findings and the processed results are available on request from the corresponding author.