ABSTRACT
Merkel cell polyomavirus (MCPyV) infects most of the human population asymptomatically, but in rare cases it leads to a highly aggressive skin cancer called Merkel cell carcinoma (MCC). MCC incidence is much higher in aging and immunocompromised populations. The epidemiology of MCC suggests that dysbiosis between the host immune response and the MCPyV infectious cycle could contribute to the development of MCPyV-associated MCC. Insufficient restriction of MCPyV by normal cellular processes, for example, could promote the incidental oncogenic MCPyV integration events and/or entry into the original cell of MCC. Progress toward understanding MCPyV biology has been hindered by its narrow cellular tropism. Our discovery that primary human dermal fibroblasts (HDFs) support MCPyV infection has made it possible to closely model cellular responses to different stages of the infectious cycle. The present study reveals that the onset of MCPyV replication and early gene expression induces an inflammatory cytokine and interferon-stimulated gene (ISG) response. The cGAS-STING pathway, in coordination with NF-κB, mediates induction of this innate immune gene expression program. Further, silencing of cGAS or NF-κB pathway factors led to elevated MCPyV replication. We also discovered that the PYHIN protein IFI16 localizes to MCPyV replication centers but does not contribute to the induction of ISGs. Instead, IFI16 upregulates inflammatory cytokines in response to MCPyV infection by an alternative mechanism. The work described herein establishes a foundation for exploring how changes to the skin microenvironment induced by aging or immunodeficiency might alter the fate of MCPyV and its host cell to encourage carcinogenesis.
IMPORTANCE MCC has a high rate of mortality and an increasing incidence. Immune-checkpoint therapies have improved the prognosis of patients with metastatic MCC. Still, a significant proportion of the patients fail to respond to immune-checkpoint therapies or have a medical need for iatrogenic immune-suppression. A greater understanding of MCPyV biology could inform targeted therapies for MCPyV-associated MCC. Moreover, cellular events preceding MCC oncogenesis remain largely unknown. The present study aims to explore how MCPyV interfaces with innate immunity during its infectious cycle. We describe how MCPyV replication and/or transcription elicit an innate immune response via cGAS-STING, NF-κB, and IFI16. We also explore the effects of this response on MCPyV replication. Our findings illustrate how healthy cellular conditions may allow low-level infection that evades immune destruction until highly active replication is restricted by host responses. Conversely, pathological conditions could result in unbridled MCPyV replication that licenses MCC tumorigenesis.
KEYWORDS: Merkel cell polyomavirus, tumor virus, DNA viruses, polyomavirus, viral oncogenesis, innate immunity
INTRODUCTION
Merkel cell polyomavirus (MCPyV) infection can be detected on the skin of most healthy adults (1), yet details of its virology and infectious cycle remain sparse. Evidence from serological studies suggests that MCPyV infects most people during early childhood and prevalence of exposure to the virus increases as populations age (2–4). A vast majority of MCPyV infections are asymptomatic (5), but some result in an aggressive skin cancer called Merkel cell carcinoma (MCC) (6–9). Over 80% of MCC tumors can be traced to a viral etiology by the presence of integrated MCPyV genomic sequence in the cellular chromatin (7). Though MCC cases are rare, the incidence of MCC has tripled over the last 2 decades (10–12). MCC has a high rate of mortality with 5-year overall survival around 51% for patients presenting with local disease at the time of diagnosis and worse prognoses for those with more advanced stages of disease (13). Chemotherapies have thus far failed to produce durable responses in patients with metastatic disease (14, 15). A recent burgeoning of anti-PD1 and anti-PDL1 treatments for MCC have shown promise as first-line therapies, though a significant proportion of patients do not respond and the durability of responses varies (16–18). The pursuit of more targeted MCC therapies necessitates a better understanding of the oncogenic underpinnings of MCC and the role of MCPyV in this process.
MCPyV has an ∼5.4 kb circular double-stranded DNA (dsDNA) genome. As a small DNA virus, MCPyV encodes an efficient repertoire of viral proteins. The viral genome is divided into early and late regions by a noncoding control region (NCCR) containing the viral origin of replication and bidirectional promoters that drive early and late gene transcription (19). The early region expresses large tumor antigen (LT) and small tumor antigen (sT), which support replication, as well as 57kT and an alternate LT open reading frame (ALTO), with functions that are less defined (20, 21). Major and minor capsid proteins, VP1 and VP2, respectively, are expressed from the MCPyV late region along with a microRNA (miRNA) that modulates early gene expression (22). In documented cases of virus-associated MCC, however, MCPyV DNA is integrated into the tumor cell genome such that expression of native sT and a truncated LT (LTT) is invariably preserved (23–25). Expression of these viral oncoproteins drives oncogenesis in virus-positive (v+) MCC tumors and is required for their survival (24, 26, 27). The mechanism and prerequisite conditions through which MCPyV integrates, however, are unknown.
Factors driving MCPyV integration and oncogenesis are difficult to probe in a laboratory setting because the virus has a narrow tropism, and the original cell of MCC is still the subject of speculation. Some evidence suggests that the absence of healthy immune surveillance and suppression of MCPyV in the skin enable development of v+MCC. For example, several factors that can dampen antiviral immunity such as advanced age, chronic UV exposure, and immunosuppression increase the risk of MCC development (28). Most MCC patients have experienced chronic, high levels of UV exposure, maintain lower melanin content in their skin, and are over the age of 60 (10). In addition, those at the greatest risk for MCC relative to others in the same age range include persons who are immunocompromised as a result of HIV/AIDs, chronic lymphocytic leukemia, and treatment for autoimmunity or organ transplantation (29–33).
While immune escape of nascently transformed cells likely contributes to increased incidence of MCC in these at-risk populations, there is additional evidence that these groups control MCPyV infection poorly. A study in Japan found that MCPyV DNA prevalence and viral load on sun-exposed skin increased sharply in individuals over the age of 40 and remained high for the oldest groups (34). HIV-positive men more frequently have detectable MCPyV DNA on their skin, and those with poorly controlled HIV infection have higher MCPyV DNA loads than do those with better-controlled infections (35). In kidney transplant recipients, MCPyV DNA was more readily detected in the urine of those with BKPyV-DNAemia and with histologically verified polyomavirus-associated nephropathy (36). Though it is impossible to determine whether v+MCC patients had high MCPyV titers prior to disease onset, they produce MCPyV VP1-specific circulating antibodies more frequently and at higher titers, markers that positively correlate with high MCPyV DNA and capsid load on the skin (37–40). More recently, it was also found that higher MCPyV DNA load correlates with worse survival outcomes in MCC patients (41).
Collectively, these epidemiological data afford the possibility that altered immune status in the skin enables unimpeded propagation of MCPyV and a favorable environment for oncogenesis. Rampant MCPyV infection may promote pro-oncogenic conditions such as more frequent replication errors that could result in MCPyV integration and mutation and/or entry into the original MCC cell from its reservoir. Studies of the host responses that keep MCPyV infection in check have been limited until recently due to a lack of tools and systems with which to study infection. Using ex vivo skin sections and in vitro cultures of cells isolated from human foreskins, we found that human dermal fibroblasts (HDFs) are uniquely capable of supporting MCPyV infection (42). Establishing this model infection system has made it possible to explore cellular responses to MCPyV infection and to determine what impact those responses have on the course of MCPyV proliferation.
In the present study, we discovered that MCPyV induces expression of interferon-stimulated genes (ISGs) and inflammatory cytokines in primary HDFs during later stages of infection. Using a CRISPR knockout approach, we found that this response to infection is mediated by the cGAS-STING and NF-κB pathways. While others have explored how expressed MCPyV genes might modulate host immune factors in different cellular contexts (43–46), to our knowledge these are the first observations of host cellular responses in the context of MCPyV infection. The present study therefore represents an important step in broadening our understanding of this enigmatic virus and its role in initiating human cancer.
RESULTS
MCPyV infection induces an ISG/inflammatory cytokine response concomitant with peak viral replication and transcription.
Since discovering that HDFs support MCPyV infection, we have been able to observe distinct stages of infection using this model system (Fig. 1A). We previously described that MCPyV entry is enhanced in the absence of fetal bovine serum (FBS) and that addition of FBS enables MCPyV gene expression, replication, and transcriptional changes in cellular genes (42). Because further MCPyV spread is limited in the presence of FBS (42), we are able to parse the cellular response to a single round of the infectious cycle (Fig. 1A).
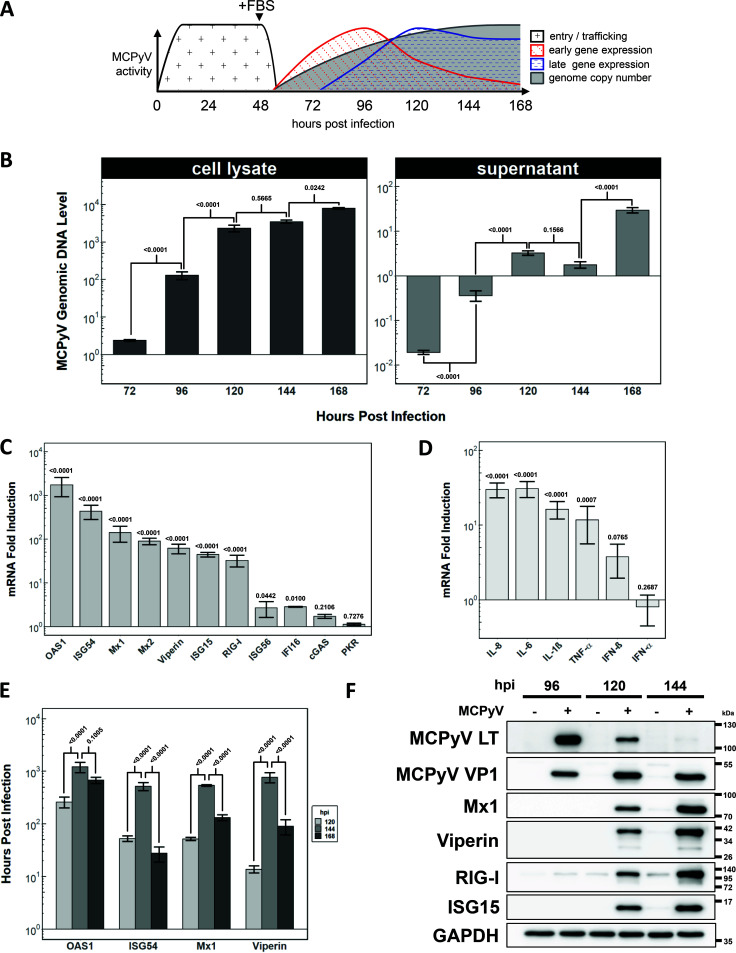
FIG 1.
ISGs and innate inflammatory cytokines are induced in coordination with MCPyV early gene expression and viral replication. (A) Schematic visualization of MCPyV infection events in HDFs. The time frame and magnitude of each event are estimated by the data shown in our previous studies (42, 62, 63) and the current study. (B) MCPyV genome abundance measured by qPCR of whole-cell lysates and supernatants at the indicated time points relative to HDFs treated with heat-inactivated MCPyV. The y axes in both panels are on a log10 scale with a horizontal line (y = 1) representing the control condition. Supernatant genome values in MCPyV infection are initially lower than that of the control condition, likely because the heat-inactivated viruses added to the control group have lower tendency to enter the cells and therefore remained in the supernatant. For the live virus treated group, as output MCPyV exceeds input virus in the supernatant at later time points, the values become many fold greater than those of control. (C and D) RT-qPCR analysis of the transcript level of ISGs (C) and innate immune cytokines (D) at 144 h.p.i. in HDFs infected with MCPyV relative to those in HDFs treated with heat-inactivated MCPyV. Error bars represent the standard error of the mean. (E) RT-qPCR analysis of select ISGs from MCPyV infection relative to those from heat-inactivated MCPyV-treated condition harvested at 120, 144, and 168 h.p.i. Error bars represent the standard error of the mean. (F) Western blot time course of MCPyV early gene, LT, late gene, and VP1, as well as ISGs, for 96, 120, and 144 h.p.i. Lanes loaded with samples from heat-inactivated MCPyV-treated HDFs are labeled (−) and those infected with MCPyV are labeled (+). Cellular GAPDH from all samples was blotted as a loading control.
In this study, we infected HDFs using our established conditions as described in reference 42 to examine both viral activities and the host response to viral infection. In order to control for the impact of noninfectious viral particles, loose nucleic acid, and other components of the virus preparation, we heat-inactivated half of the MCPyV stock and used it in the control experiments. By comparing total DNA extracted from MCPyV-infected HDFs and those treated with the same volumes of heat-inactivated-MCPyV (control), we found that the rate of increase of MCPyV replication is the greatest between 72 and 120 h postinfection (h.p.i.) (Fig. 1B). MCPyV replication continues up to at least 168 h.p.i at a lower rate. In a similar manner, we performed quantitative PCR (qPCR) on supernatants from MCPyV and control conditions to measure relative release of output viral genomes into the extracellular environment (Fig. 1B). With control supernatant containing a relatively constant level of MCPyV DNA, MCPyV-infected supernatants show the greatest increases between 72 and 96 h and 144 to 168 h (Fig. 1B). These spikes in relative MCPyV genome abundance suggest that MCPyV is being released from infected cells continually after replication begins.
After delineating a time frame in which MCPyV replication is elevated, we sought to determine if there was evidence of an innate immune response to MCPyV activity. We performed reverse transcriptase quantitative PCR (RT-qPCR) with a set of primers targeting ISGs and inflammatory cytokines with documented antiviral functions (Fig. 1C and D). This experiment revealed that relative to the control condition, MCPyV infection of HDFs induced up to 1,000-fold stimulation of several ISGs, including OAS1, ISG54, Mx1, Mx2, Viperin, ISG15, and RIG-I at 144 h.p.i. (Fig. 1C). In addition to the robust induction of ISGs, MCPyV infection also caused 10- to 30-fold induction of inflammatory cytokines, including interleukin 8 (IL-8), IL-6, IL-1β, and tumor necrosis factor alpha (TNF-α), whereas the level of interferons (IFNs) was not significantly stimulated by MCPyV (less than 3-fold) (Fig. 1D). We have consistently observed similar responses to MCPyV infection using low-passage primary HDFs isolated from more than 10 donors (see Materials and Methods), thus ruling out donor-to-donor variability. A time course RT-qPCR of MCPyV infection revealed that peak ISG transcriptional activity occurred around 144 h.p.i. (Fig. 1E).
To establish whether the ISG response to MCPyV infection was present at the protein level, we harvested whole-cell lysates for protein extraction from MCPyV-infected and control conditions at 96, 120, and 144 h.p.i. (Fig. 1F). Immunoblotting for MCPyV LT and VP1, early and late genes, respectively, served as a reference for infection stage. As expected, MCPyV LT, which directly supports MCPyV replication, declined at the protein level coinciding with slowing MCPyV replication that occurs between 120 and 144 h.p.i (Fig. 1B and F). MCPyV VP1, however, increases in abundance from 96 h.p.i and remains high by 144 h.p.i. Crucially, ISGs that were induced at the RNA level by MCPyV infection were also upregulated at the protein level after 120 h.p.i. (Fig. 1 C and F). From these studies, it is clear that the ISGs/cytokines are induced after infected cells have reached peak viral replication and early transcription, suggesting that these viral activities produce pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) to trigger the cellular response.
IFI16 localizes to MCPyV replication centers.
Canonical immune responses to viral infection begin when PAMPs or DAMPs, like viral DNA, are sensed by pattern recognition receptors (PRRs). PRR agonist binding stimulates conserved intracellular signaling cascades that activate transcription factors belonging to the IFN regulatory factor (IRF) and nuclear-factor kappa B (NF-κB) families (47–49). These activated transcription factors can in turn stimulate production of IFNs and cytokines that are secreted from the cell to function via autocrine and paracrine modalities. Cytokines binding cell surface receptors activate downstream signaling, including the Jak-STAT pathways (47–49). Cytokine-mediated signaling events can also spread and intensify ISG expression and the antiviral status by further stimulating NF-κB and IRF transcriptional activity (47–49).
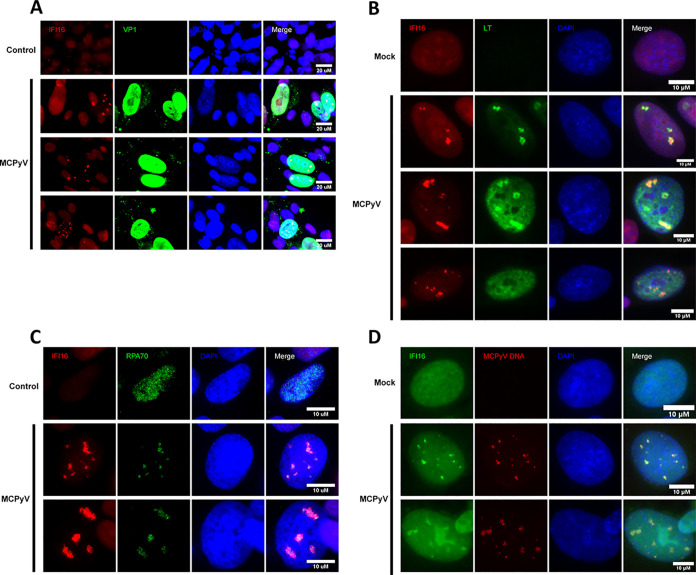
Given the evidence that peak MCPyV replication and gene expression precede a marked ISG response, we wanted to identify the sensor responsible for detecting the putative MCPyV-associated molecular pattern. We performed immunofluorescence microscopy (IF) on MCPyV-infected and control HDFs using antibodies targeting known sensors of viral nucleic acid to determine if they are differentially localized in MCPyV-infected cells. This approach established interferon-γ inducible protein 16 (IFI16), an upstream component for stimulator of interferon genes (STING) (50–52), as a candidate sensor for MCPyV. Immunostaining for IFI16 revealed that a small percentage of nuclei in the MCPyV infection setting developed a distinct punctate or track-like IFI16 pattern (Fig. 2). On the other hand, all cells in the control condition and the vast majority of cells in MCPyV infection exhibited diffuse, pan-nuclear IFI16 staining that was sometimes enriched in nucleoli. Costaining experiments revealed that the intense punctate IFI16 signal only occurred in nuclei that were VP1 or LT positive (Fig. 2A and B). When IFI16 punctate signal was present in nuclei containing LT puncta, markers of MCPyV replication centers, the intense IFI16 and LT signals generally overlapped (Fig. 2B). Replication centers, or foci, are subnuclear regions in which active viral DNA synthesis takes place and where most MCPyV DNA is present in the nucleus (42). Because IFI16 formed puncta in a small subset of MCPyV-positive nuclei and those puncta overlapped with LT-containing replication foci, we hypothesized that IFI16 was localizing to MCPyV DNA at brief periods during the infectious cycle independently of LT.
FIG 2.
IFI16 localizes to MCPyV replication centers exclusively in MCPyV-positive nuclei. (A) IF staining for IFI16 and VP1 in MCPyV-infected nuclei. MCPyV-infected and heat-inactivated-MCPyV-treated HDFs were stained using IFI16 and VP1 antibodies and counterstained with DAPI. (B) IF staining for IFI16 and LT in MCPyV-infected nuclei. MCPyV-infected and mock-infected HDFs were stained using IFI16 and LT antibodies and counterstained with DAPI. (C) IF staining for IFI16 and RPA70 in MCPyV-infected nuclei. MCPyV-infected and heat-inactivated-MCPyV-treated HDFs were stained using IFI16 and RPA70 antibodies and counterstained with DAPI. (D) Immuno-FISH experiment staining for IFI16 and probing for MCPyV genomic DNA. Mock- or MCPyV-infected HDFs were harvested at 120 h postinfection. The cells were subjected to sequential immunofluorescence staining using IFI16 antibody (Sigma, catalog number HPA002134) and FISH with an MCPyV probe (Biosearch technologies). Bar, 10 μm. Pan-nuclear IFI16 staining and slight enrichment in nucleoli were evident in control conditions and in most cells in the MCPyV-infected condition. A subset of those cells with MCPyV DNA foci revealed adjacent or overlapping signal with IFI16 puncta.
To test whether IFI16 localizes to centers of actively synthesizing MCPyV genomes, we performed IFI16/RPA70 costaining (Fig. 2C). We have shown previously that RPA70, which binds single-stranded DNA, forms intense puncta at MCPyV replication centers likely undergoing DNA synthesis (53). The experiment showed that concentrations of ssDNA represented by nuclear RPA70 foci that were unique to MCPyV infection specifically overlapped with IFI16 foci (Fig. 2C). To confirm that IFI16 puncta were forming at regions of MCPyV DNA replication foci, we performed immunofluorescent in situ hybridization (immuno-FISH) staining for IFI16 and probing for MCPyV-specific DNA sequence (Fig. 2D). As was true with other markers of MCPyV nuclear presence, IFI16 puncta occurred in a small population of MCPyV DNA foci containing nuclei, yet this signal was unique to MCPyV-positive cells and correlated with the pattern of MCPyV DNA-containing foci (Fig. 2D). That IFI16 formed puncta in a small proportion of MCPyV-positive cells led us to suspect that this is either a rare or a temporally brief event taking place during MCPyV infection. We also concluded that while IFI16 and MCPyV replication foci did not coincide 1:1 in the fashion of true colocalization, the signals always overlap or are adjacent when present in the same nucleus. Because IFI16 foci are unique to MCPyV-infected cells and appear to localize to sites of active MCPyV DNA synthesis, we hypothesized that IFI16 detects MCPyV genomes in the nucleus and elicits downstream ISG and innate inflammatory cytokine induction.
MCPyV infection activates the STING/TBK1/IRF3 and NF-κB pathways.
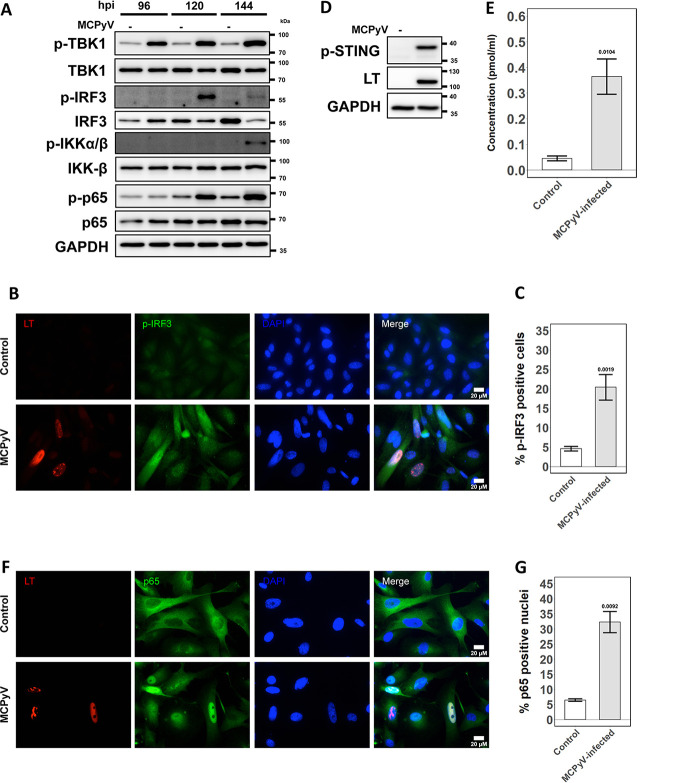
IFI16 is a DNA-binding protein reported to stimulate type I IFN and ISG induction through interaction with the cyclic GMP-AMP synthase (cGAS)-STING pathway in response to infection with other DNA viruses (51, 54, 55). Since IFI16 is localized to replication centers and ISGs are upregulated during MCPyV infection, we set out to understand whether cGAS-STING pathway factors were being activated. Again, we performed time course Western blotting comparing the key downstream molecules in MCPyV-infected and control whole-cell lysates (Fig. 3A). Downstream effectors of STING, tank-binding kinase 1 (TBK1), and IFN regulatory factor 3 (IRF3) appear to be more abundant in their active, phosphorylated state in infection conditions than in control conditions. The frequency of active, phosphorylated IRF3 (p-IRF3) signal in cells was also elevated during MCPyV infection as observed by immunofluorescence (IF) (Fig. 3B and C). Given that cGAS-STING effector proteins were likely activated by infection, we also confirmed that the activated form of STING was more abundant in MCPyV-infected cells by Western blotting (Fig. 3D).
FIG 3.
STING-TBK1-IRF3 and NF-κB pathways are activated during MCPyV infection in HDFs. (A) Whole-cell immunoblot time course of the STING-TBK1-IRF3 pathway and the NF-κB pathway, respectively, for 96, 120, and 144 h.p.i. Lanes loaded with samples from heat-inactivated MCPyV-treated HDFs are labeled (−) and those infected with MCPyV are labeled (+). Cellular GAPDH from all samples was blotted as a loading control. (B) IF costaining for phosphorylated IRF3 (p-IRF3) and LT in HDFs infected with MCPyV or treated with heat-inactivated MCPyV 6 days postinfection/treatment. (C) Quantification for panel B. (D) HDFs were infected with active (+) or heat-inactivated (−) MCPyV. At 120 h.p.i., the whole lysates were blotted with the indicated antibodies. (E) ELISA quantification of cGAMP present in whole-cell lysates of mock-infected and MCPyV-infected cells at 120 h.p.i. One-tailed Student’s t test produced a P value of 0.0052 (<0.01). (F) IF costaining for MCPyV LT and NF-κB p65 subunit in HDFs infected with MCPyV or treated with heat-inactivated MCPyV. All cells were fixed at 144 h.p.i. (G) Quantification for panel F. Error bars represent standard error of the mean.
Canonical activation of STING in response to viral pathogens involves cGAS binding foreign DNA, which activates its catalysis of the STING agonist cyclic GMP-AMP (cGAMP) (56). Although discovered as a cytoplasmic sensor, cGAS has been found to localize in the nucleus (57). It has been suggested that the cGAMP produced by activated nuclear cGAS could diffuse through nuclear pores to activate STING in the cytoplasm (57). Our candidate sensor of MCPyV DNA, IFI16, has also been reported to cooperate with cGAS in some instances to initiate STING signaling (54, 58). Therefore, we examined the possible involvement of cGAS in activating the STING-TBK1-IRF3 pathway. We performed an enzyme-linked immunosorbent assay (ELISA) to quantify cGAMP production in whole-cell lysates from mock-infected and MCPyV-infected HDFs. cGAMP concentration was significantly higher in MCPyV-infected samples than in control samples (Fig. 3E). Thus, the catalytic activity of cGAS is likely upregulated by MCPyV infection and may contribute to the host cell innate response.
NF-κB can also be regulated by STING and is capable of upregulating the transcription of inflammatory cytokines such as those we observed (Fig. 1D). Therefore, we assessed the activation status of the NF-κB pathway during MCPyV infection. When the NF-κB pathway is activated, the inhibitor of NF-κB kinase (IKK) and the p65 subunit of NF-κB are phosphorylated, allowing p65 to be phosphorylated and to translocate into the nucleus (59). Immunoblotting for phosphorylated epitopes on IKK-β and p65 revealed elevated levels in MCPyV infection relative to those in the control (Fig. 3A). IF costaining for MCPyV LT and p65 also made evident an increased nuclear localization of p65 in MCPyV-infected conditions (Fig. 3F and G). Collectively, these data suggest that the cGAS-STING-TBK1-IRF3 and NF-κB pathways are stimulated by later events in MCPyV infection and may contribute to an antiviral response with induced cytokine/ISG expression.
CRISPR knockout of the cGAS-STING-TBK1-IRF3 pathway, but not IFI16, ablates ISG response to MCPyV infection.
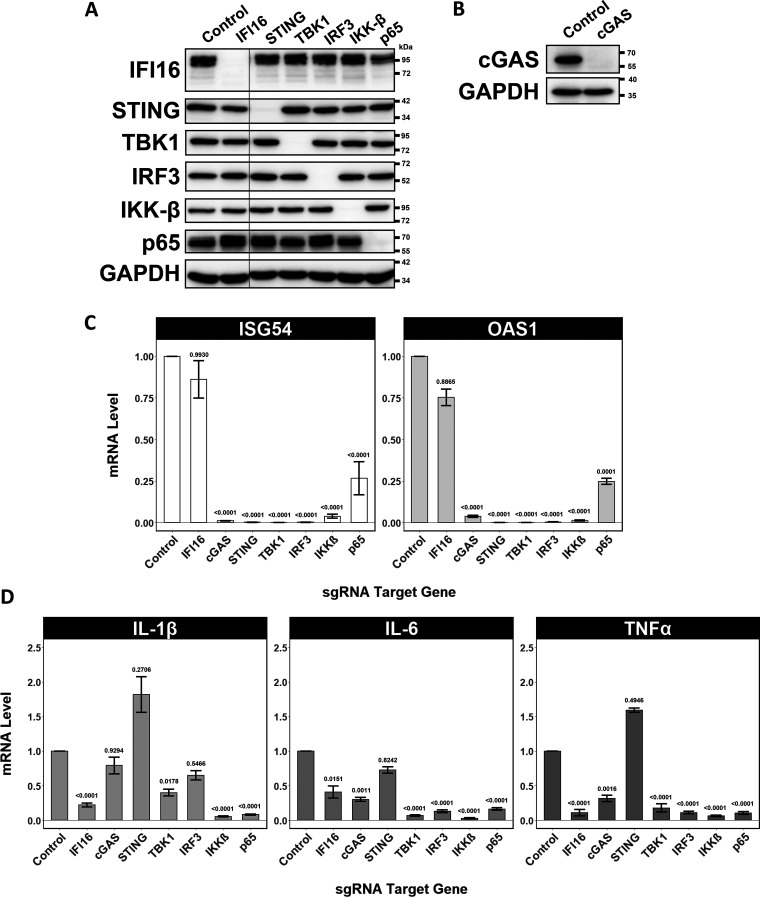
Next, we aimed to discern how these immune regulatory factors affect the course of the innate response to MCPyV and its proliferation. We used a lentiviral CRISPR-Cas9 system to knock out several regulatory genes in the cGAS-STING-TBK1-IRF3 and NF-κB pathways in primary HDFs. Since these primary cells cannot be grown from a single clone, we selected for successful expression of the given single guide RNA (sgRNA) and validated knockout efficiency by Western blotting (Fig. 4A and B). We verified that each of these cell lines, along with a control cell line expressing an sgRNA targeting the nonmammalian gene, was capable of supporting MCPyV infection by IF (data not shown). For RT-qPCR analysis, we harvested cell lysates at 144 h.p.i., which is the peak time point for ISG induction as determined by our time course (Fig. 1E). We found that individual knockout of cGAS, STING, TBK1, IRF3, IKK-β, or p65 ablated normal ISG induction during MCPyV infection (Fig. 4C). This result implied that each of these factors is required for inducing ISGs in response to MCPyV. Surprisingly, knockout of IFI16 did not significantly reduce induction of ISGs relative to that in the control.
FIG 4.
CRISPR knockout of STING-TBK1-IRF3 and NF-κB pathways ablates the ISG response to MCPyV infection in HDFs. (A) Immunoblot validation of HDF CRISPR knockout cell lines. Cell lysates were harvested at the time of seeding for infection. Lanes are labeled with the genes targeted by the stably expressed sgRNA. HDFs stably expressing Cas9 and an sgRNA targeting the nonmammalian luciferase gene were used as a negative control. A lane was cropped out between IFI16 and STING lanes. (B) Immunoblot validation of cGAS cell line used in shown experiments was blotted separately. (C and D) RT-qPCR analysis of marker ISGs (C) and inflammatory cytokines (D) in MCPyV-infected HDF CRISPR knockout cell lines targeted by the indicated sgRNAs. Cells were analyzed at 144 h.p.i. For each gene of interest, expression levels for each cell line are displayed relative to the control cell line stably expressing Cas9 and a luciferase-targeted sgRNA. Error bars represent standard error of the mean.
We then assessed the transcriptional upregulation of IL-1β, IL-6, and TNF-α by RT-qPCR in each knockout setting during MCPyV infection. It is well documented that these cytokines are transcriptionally regulated by the NF-κB pathway (49). This fact was substantiated with regard to MCPyV infection by the result that knockout of either IKK-β or p65 dramatically reduced IL-1β, IL-6, and TNF-α RNA levels (Fig. 4D). IFI16 knockout also significantly reduced the expression of each of these cytokines, suggesting that it may influence the cytokine response to MCPyV independently of the mechanism driving ISG induction. On the other hand, cGAS and STING knockout does not appear to have a significant impact on cytokine expression, while knockout of TBK1 and IRF3 moderately inhibits IL-6 and TNF-α induction by MCPyV. These observations indicate that both the NF-κB and IRF3 axes participate in the transcriptional induction of IL-1β, IL-6, and TNF-α triggered by active MCPyV infection.
ISG and inflammatory cytokine induction has a modest, direct impact on MCPyV proliferation.
After establishing that these immunological pathways participate in a host response to MCPyV, we assayed whether the response has measurable consequences for the propagation of MCPyV genomes in HDFs. To accomplish this, we compared MCPyV genome levels in lysates and supernatants harvested at 168 h.p.i. across the HDF knockout cell lines. Knockout of cGAS, IKK-β, or p65 significantly increased the replication of MCPyV genomes in whole-cell lysates (Fig. 5A). However, none of the knockout cell lines showed a significantly increased level of MCPyV genomes in the supernatant (Fig. 5A).
FIG 5.
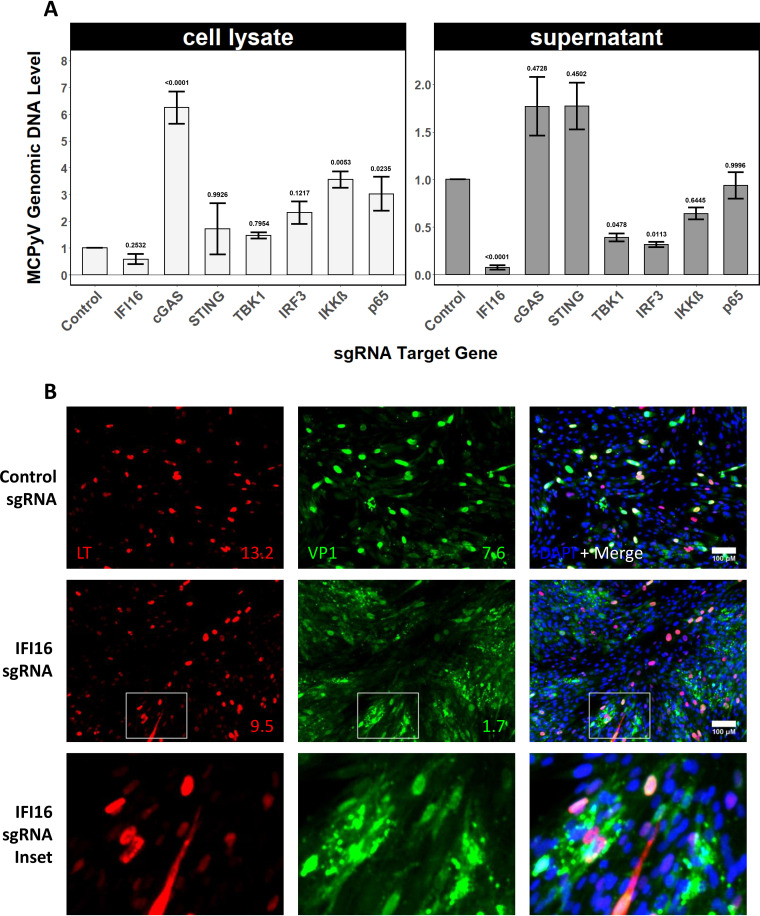
CRISPR knockout of some STING-TBK1-IRF3 and NF-κB pathway components stimulates MCPyV replication in HDFs. (A) qPCR-measured abundance of MCPyV genome equivalents in cell lysates and supernatants of HDF CRISPR knockout cell lines targeted by the indicated sgRNAs. Shown are the MCPyV DNA levels in MCPyV-infected HDF CRISPR knockout cells relative to those in the same cells treated with heat-inactivated MCPyV at 168 h.p.i. Error bars represent the standard error of the mean. (B) Representative images from IF costaining for LT and VP1 in MCPyV-infected CRISPR knockout HDF cell lines fixed around 160 h.p.i. Number values in lower right corner indicate the percentage of intact, positive nuclei in that condition for that antigen. Final row is a magnification of the inset outlined by the white box in the second row.
Unexpectedly, elimination of some factors, especially IFI16, resulted in substantially lower MCPyV genomes detected in the supernatant. This finding was of particular interest because it coincides with the observation that high-titer infection of IFI16 knockout HDFs results in apparent fragmentation of VP1-positive nuclei (Fig. 5B). Unlike the control cells that show the typical pan-nuclear VP1 signal, most of the IFI16 knockout HDFs showed scattered VP1 signal that often spreads outside of the infected cells, indicating that severe cell lysis is occurring in the IFI16 knockout samples (Fig. 5B). Quantification of the IF results reveals that, compared to that of the control cell line, IFI16 knockout cell lines showed a markedly lower number of intact VP1-positive nuclei (Fig. 5B value insets). Therefore, it is possible that disruption of MCPyV-containing cell membranes and early cell death in IFI16 knockout cells either prevents the completion of MCPyV genome replication in these cells or alters the mode of egress of MCPyV into the extracellular space. Considered together, these results imply that the presence or absence of innate immune effectors during highly active MCPyV infection could alter the rate of MCPyV replication and, ultimately, host cell fate.
DISCUSSION
MCPyV maintains asymptomatic persistent infection in more than 80% of the general population (1, 60, 61) but tends to cause MCC in elderly and immunocompromised individuals (28). Epidemiological studies have revealed a strong correlation between immunosuppression, elevated MCPyV genome loads, and increased risk for MCC (28, 35). These observations suggest that the virus strikes a balance in healthy hosts where it is restricted by immune mechanisms but avoids eradication. Furthermore, settings where immune restriction is reduced increase the likelihood of MCPyV-induced tumorigenesis. However, little is known about the immune response elicited by MCPyV. By extension, neither is much understood about the immune evasion strategies that enable MCPyV to establish persistent or poorly controlled infection in different settings. Such insights could explain the duality of MCPyV infection outcomes involving either long-term asymptomatic infection or integration and rapid oncogenesis.
In the current study, we employed the MCPyV infection model established in our recent study (42, 62, 63) to examine the host innate immune response to MCPyV infection. We discovered that MCPyV infection of low-passage primary normal HDFs induces robust expression of key antiviral ISGs and inflammatory cytokines at 144 h.p.i., while no significant induction of these genes was observed at the early time points (Fig. 1E and F). We found that the ISG response reliably follows peak MCPyV infection activities (Fig. 1F), suggesting that it is most likely induced by MCPyV DNA replication and/or transcription. In addition to the robust induction of ISGs, MCPyV infection also caused 10- to 30-fold induction of inflammatory cytokines, including IL-1β, IL-6, TNF-α, and IL-8, (Fig. 1C and D).
Our observation that MCPyV infection results in ISG induction builds upon existing literature that examines cellular responses to other polyomaviruses. It has been demonstrated that overexpression of SV40, JCPyV, or BKPyV T antigens in mouse embryonic fibroblasts leads to upregulation of ISGs and an antiviral state (64, 65). In addition, JCPyV infection of human renal proximal tube epithelial cells results in IFN secretion and subsequent ISG induction in a manner that partially controls the extent of infection (66). Infection of the same cells with BKPyV nonetheless fails to produce the same ISG response and protection. In microvascular endothelial cells found in the lung and bladder, however, BKPyV infection seems to be blunted by IFN-mediated innate immune gene induction that coincides with later stages of the infectious cycle (67). Intriguingly, like with BKPyV, the induction of ISGs by MCPyV occurs relatively late in the viral life cycle. Perhaps further investigation of the cell-type-specific innate immune responses to polyomaviruses may offer an explanation as to why certain cell types are more or less permissible to MCPyV. Antiviral mechanisms unique to the as yet unidentified cell of origin of MCC, for example, could increase the frequency of MCPyV integration events.
We also found that IFI16 localizes to the subnuclear sites of MCPyV replication in a small population of MCPyV-positive cells (Fig. 2). The rarity of the IFI16 punctate phenotype and its tendency to localize to MCPyV replication foci led us to conclude that it is likely enriched at MCPyV genomes at a brief and specific stage of the infectious cycle. Despite being a promising candidate as the initiating factor of the ISG response to infection for its ability to shuttle between nucleus and cytoplasm (68), IFI16 was not necessary for MCPyV-associated ISG induction (Fig. 4C). Instead, cGAS proved to be likely activated by MCPyV infection, essential for ISG induction, and involved in repressing MCPyV replication (Fig. 3E, Fig. 4C, and Fig. 5A). The manner in which cGAS senses MCPyV is still in question. It remains to be determined whether cGAS localizes and detects MCPyV DNA in the nucleus or senses the viral genomes that may leak into the cytoplasm during highly active replication or egress (Fig. 6).
FIG 6.
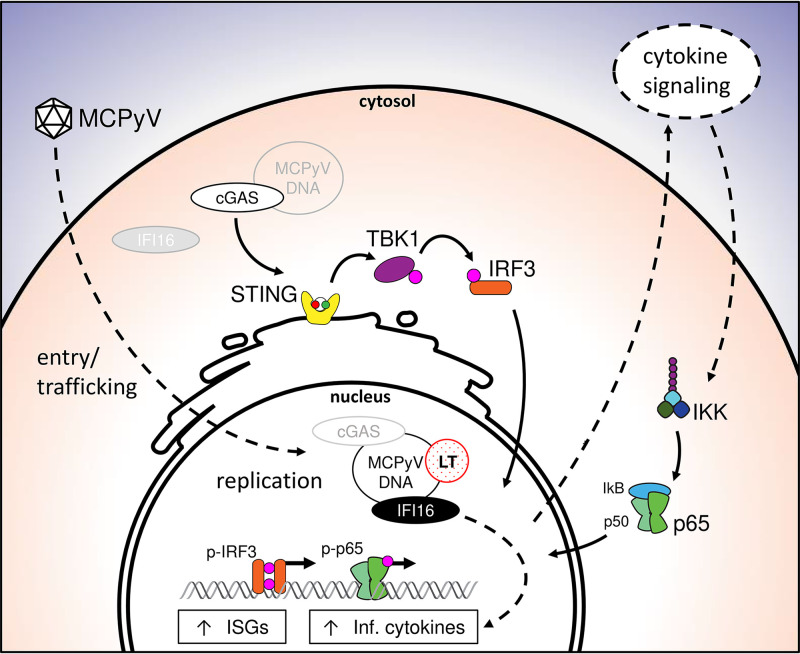
Schematic working model of cellular responses to MCPyV infection. Solid black arrows represent functional findings in the present paper and expected relationships based on established canonical pathways. Dotted line arrows indicate suspected mechanism based on indirect evidence and literature. Light gray items represent possible alternative localizations of that factor. MCPyV undergoes attachment, entry, and trafficking to the nucleus without eliciting ISG or inflammatory cytokine induction. Naked MCPyV DNA in the nucleus enables viral gene transcription and replication. cGAS likely detects MCPyV DNA in either the cytoplasm or the nucleus to stimulate signaling through the STING-TBK1-IRF3 axis. Upregulation of cytokines is suspected to then function in an autocrine and paracrine fashion to turn on other innate immune pathways such as NF-κB. ISGs and inflammatory cytokines are ultimately upregulated as a result of MCPyV activity. IFI16 contributes to the induction of cytokines, but not of ISGs, independently of cGAS-STING in a manner that is yet to be determined. (Inf. cytokines, inflammatory cytokines: IL-1β, TNF-α, IL-6).
Due to the role of STING signaling in mediating cGAS- and IFI16-induced responses to viral DNA (50–52), we assessed the activation status of STING downstream effectors during MCPyV infection and their functional relevance to the antiviral status of the host cell. We found that STING, TBK1, IRF3, IKK-β, and the NF-κB subunit p65 are activated by phosphorylation during MCPyV infection (Fig. 3). In addition, CRISPR knockout of these key immune mediators represses MCPyV-induced ISG and cytokine expression (Fig. 4), allowing increased MCPyV replication (Fig. 5). Our data therefore support a model in which MCPyV infection generates PAMPs or DAMPs that activate STING- and NF-κB-mediated induction of cytokines and ISGs, which in turn negatively regulate MCPyV infection (Fig. 6).
The cytokines upregulated by MCPyV, such as IL-1β and TNF-α, are capable of further positive regulation of ISG expression via IRF1, IRF3, and NF-κB (69–78). Therefore, we suspect that these cytokines function in both an autocrine and a paracrine manner during MCPyV infection to generate and amplify the antiviral status of infected cells and neighboring uninfected cells alike (Fig. 6). Unlike the inflammatory cytokines, the IFN-β mRNA level was not significantly induced in MCPyV-infected cells compared to that in the cells treated with heat-inactivated MCPyV (Fig. 1D). Whether this level of induction is biologically relevant to the cascade of events regulating ISG expression remains to be investigated.
As this is the first study to examine innate intrinsic responses to MCPyV infection, the findings herein pose questions that should be addressed in the future. For example, while cGAS is likely the predominant MCPyV PRR responsible for the induction of ISGs (Fig. 4), the role of IFI16 throughout MCPyV infection remains to be fully detailed. We found that IFI16 localizes to sites of MCPyV replication and contributes to the induction of inflammatory cytokines but not of ISGs (Fig. 2 and Fig. 4C and D). This suggests that it may support transcriptional upregulation of cytokines by means other than the cGAS-STING and NF-κB pathways.
One mechanism by which IFI16 could induce inflammatory cytokine induction is that MCPyV DNA-bound IFI16 could shuttle to the cytoplasm where it may engage in inflammasome activation and pyroptosis as has been reported in herpesvirus infection (79, 80). The VP1-positive nuclei fragments found in IFI16 knockout conditions during MCPyV infection could indicate that the absence of IFI16 disrupts typical cell death responses to infection. If IFI16 does not promote inflammasome-related pyroptosis, it may promote other cell fates such as senescence. For example, it has been reported that IFI16 is accumulated in aging fibroblasts (81) and that the loss of IFI16 in fibroblasts allows a bypass of cellular senescence (82). Recently, another group showed that MCPyV genome transfected into HDFs causes a secretory senescence phenotype (83). Together, these findings hint that IFI16 may promote a secretory senescence phenotype in MCPyV-infected fibroblasts in tandem with innate signaling from cGAS-STING and NF-κB. Changes in IFI16 activity and abundance in aging skin could also alter the course of MCPyV propagation and host cell fate.
Knockout of IKK-β, p65, or cGAS led to elevated MCPyV replication as measured by qPCR analysis of whole-cell lysates (Fig. 5). The most significant of these changes was that of cGAS knockout, which resulted in ∼6× higher levels of MCPyV DNA in cell lysates (Fig. 5). One explanation for the dramatic impact of cGAS presence on MCPyV replication compared to that of the other factors is that, as a principal sensor of MCPyV DNA, cGAS impacts all downstream aspects of the host cellular response. Another possibility is that cGAS binding to MCPyV DNA can directly inhibit MCPyV replication in addition to initiating host signaling events. That knockout of effectors downstream of cGAS was not sufficient to limit MCPyV replication could imply that MCPyV has evolved strategies to antagonize the STING-mediated antiviral immune response. Alternatively, the varied impacts of knocking out the downstream components on MCPyV replication may be due to unexpected influences outside of canonical pathways. For example, one group found that knockout of innate immune regulators like IFI16, STING, and IRF3 had pleiotropic effects on herpesvirus replication (84). It is possible that other antiviral immune signaling pathways not explored here are involved in controlling MCPyV infection. Therefore, how MCPyV evades the host immune responses to achieve persistent infection remains an important question to address in the future.
Our current data suggest that ISGs and inflammatory cytokines induced by MCPyV infection could dampen viral spread even in the absence of intervention by innate and adaptive immune cells. The extent to which this antiviral immune response can prevent uncontrolled viral propagation within the skin microenvironment will need to be tested in an in vivo model once one becomes available. Until then, exploring the mechanisms by which MCPyV counteracts the innate host cell response will yield insights regarding MCPyV persistence in healthy hosts. Such a mechanism would clarify how the lack of antiviral pressure in at-risk populations encourages MCC tumorigenesis.
MATERIALS AND METHODS
Isolation and culture of fibroblasts from human foreskin.
Human neonatal foreskins were obtained from the Penn Skin Biology and Diseases Resource-based Research Center. Protocols for isolation and culture of HDFs have been described and visualized previously (42, 63) and are outlined briefly here for convenience. Fat and subcutaneous tissue were trimmed from the foreskin sample. The remaining tissue sample was cut into four smaller pieces which were then incubated in 5 ml of 10 mg/ml Dispase II (Roche, 04942078001) supplemented with Antibiotic-Antimycotic (Life technologies, 15240-062) at 4°C overnight. After this digestion, the dermal layer was transferred to a 15-ml tube containing 5 ml 1.5 mg/ml collagenase type IV (Life technologies, 17104-019) supplemented with Antibiotic-Antimycotic. The samples were incubated at 37°C in 5% CO2 for 4 to 6 h with periodic shaking until macroscopic tissue was no longer visible. The samples were centrifuged at 180 × g for 5 min. The dissociated cells were pelleted and plated in Dulbecco modified Eagle medium (DMEM) (Life technologies, 11965084) supplemented with 10% FBS (HyClone, SH30071.03), 1× nonessential amino acids (Gibco), and 1× glutamine (Gibco). The same medium was used to expand and passage dermal fibroblasts in 10 cm2 dishes for use in subsequent experiments. With each passage, fibroblasts were allowed to grow to confluence before they were split 1 to 4. HDFs isolated from more than 10 donors were used in this study and similar observations were made.
MCPyV infection of dermal fibroblasts.
HDFs and lentiCRISPRv2 sgRNA stable HDFs were infected with MCPyV using the method described previously (62, 63). The pR17b MCPyV plasmid and MCPyV T-antigen expression plasmids pMtB and pADL used in the production of MCPyV virions were kindly provided by Christopher Buck. MCPyV viral fractions were normalized by quantifying viral genome equivalents using qPCR analysis and making dilutions with Dulbecco’s phosphate-buffered saline (DPBS) to the nearest 1 × 107 genomes/μl. All MCPyV preparations used in the experiments were between 3 × 108 to 1 × 109 genomes/μl. To extract DNA from preparations of infectious MCPyV particles, 1 μl of a given MCPyV fraction was added to 50 μl of Lucigen DNA extraction buffer. The extraction steps were as follows: vortex 15 sec, incubate 15 min at 65°C, vortex 15 sec, incubate 6 min at 95°C, and store at −80°C. The DNA extraction samples were then used as the template in qPCR analysis using Applied Biosystems SYBR green FAST master mix with NCCR-targeted primers. Within any technical replicate, the same viral preparation was mixed well by pipetting up-and-down and used for all conditions. Treatment with control heat-inactivated MCPyV for each experiment was performed by sealing the tube containing the same virus preparation used for infection with a plastic lock, boiling at 100°C for 10 min, cooling on ice, spinning down, and mixing before adding the same volume as that used in the MCPyV infection to control wells.
qPCR and RT-qPCR.
qPCRs were made using Applied Biosystems SYBR green Fast Mix on an Applied Biosystems QuantStudio3 quantitative thermal cycler. qPCR primers used for quantification of MCPyV genome equivalents relative to cellular DNA are as listed in Table 1. The pR17b plasmid (containing the MCPyV genome) and cellular DNA from control conditions were used as negative controls to check for possible background amplification. Primers used to quantify gene expression in RT-qPCR experiments are shown in Table 2. RNA was isolated for RT-qPCR using Takarabio Nucleospin RNA XS Kits and immediately subjected to reverse transcription for downstream analysis and long-term storage at −80°C. An identical set of RNA isolate samples that underwent the same procedure without reverse transcriptase present were used as control for background nucleic acid contamination. For 96-well plates, lysates harvested from 3 to 5 wells of cells were used for a given replicate. Fold change in expression of cellular genes such as ISGs and inflammatory cytokines was calculated by RT-qPCR 2–ΔΔCt as done previously (85), where the control gene is glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the control condition is treatment with the same MCPyV preparation that has been heat-inactivated. In RT-qPCR analysis of CRISPR sgRNA knockout HDFs, the control condition was MCPyV infection of HDFs stably expressing Cas9 and an sgRNA for the nonmammalian gene luciferase.
TABLE 1.
The following primers were used for qPCR analysis; all oligonucleotides were ordered from Integrated DNA Technologies (IDT)
| Primer target | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| MCPyV NCCR | TAGGCAGCCAAGTTGTGGTTA | CGTCTCCCTCCCAAACAGAAA |
| Genomic GAPDH | GGCCCTGACAACTCTTTTCATCTT | CAACTGTGAGGAGGGGAGATTC |
TABLE 2.
The following primers were used for RT-qPCR analysis; all oligonucleotides were ordered from Integrated DNA Technologies (IDT)
| Primer target | Forward primer sequence (5′→3′) | Reverse primer sequence (5′→3′) |
|---|---|---|
| ISG54 | GCACTGCAACCATGAGTGAGA | GCTTGCCTCAGAGGGTCAAT |
| Viperin | TGGGTGCTTACACCTGCTG | TGAAGTGATAGTTGACGCTGGT |
| OAS1 | AGAAGGCAGCTCACGAAACC | CCACCACCCAAGTTTCCTGTA |
| RIG-I | TTGCCACCTCAGTTGCTGAT | ACTGCTTTGGCTTGGGATGT |
| Mx1 | CAGCCTGCTGACATTGGGTA | CCACATTACTGGGGACCACC |
| Mx2 | ATGATTTCTCCATCCTGAACGTG | CCTGAAGCTCTAGCTCGGTG |
| ISG15 | GCGCAGATCACCCAGAAGAT | GTTCGTCGCATTTGTCCACC |
| ISG56 | GCTTACACCATTGGCTGCTG | CCATTTGTACTCATGGTTGCTGT |
| IFI16 | AGAGCCATCTTCGGACTCCT | CAGTCTTGGTTTCAACGTGGT |
| cGAS | ACGTGCTGTGAAAACAAAGAAG | GTCCCACTGACTGTCTTGAGG |
| PKR | AGAGTAACCGTTGGTGACATAACCT | GCAGCCTCTGCAGCTCTATGTT |
| GAPDH | GGTGGTCTCCTCTGACTTCAACA | GTTGCTGTAGCCAAATTCGTTGT |
| IL-8 | CACCGGAAGGAACCATCTCA | GGCAAAACTGCACCTTCACA |
| IL-6 | AGGAGACTTGCCTGGTGAAA | CAGGGGTGGTTATTGCATCT |
| IL-1β | CTGAGCTCGCCAGTGAAATG | TGTCCATGGCCACAACAACT |
| TNF-α | GCTGCACTTTGGAGTGATCG | TCACTCGGGGTTCGAGAAGA |
| IFN-β | GCCGCATTGACCATCTATGAGA | GAGATCTTCAGTTTCGGAGGTAAC |
| IFN-α | TTTCTCCTGCCTGAAGAACAG | GCTCATGATTTCTGCTCTGACA |
Immunofluorescent staining.
Cells were fixed with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min. Immunofluorescent (IF) staining was performed as described previously (86). The following primary antibodies were used in 3% bovine serum albumin (BSA), 0.5% Triton X-100: CM2B4 (MCPyV LT) (1:500, sc-136172, Santa Cruz), anti-MCPyV VP1 (1:2,000, Christopher Buck Laboratory), anti-RPA 70 (1:100, 2267S, Cell Signaling Technologies), anti-NF-κB p65 (1:1,500, 8242S, Cell Signaling Technologies), anti-IRF3 (1:200, sc-33641, Santa Cruz), and anti-IFI16 (1:200, ab55328, Abcam) antibodies. The secondary antibodies used were Alexa Fluor 594 goat anti-mouse IgG (1:1,000, A11032, ThermoFisher Scientific) and Alexa Fluor 488 goat anti-rabbit IgG (1:500, A11034, ThermoFisher Scientific). The IFI16 antibody (Sigma, catalog number HPA002134) and an MCPyV probe (Biosearch technologies) were used for the sequential immunofluorescence staining of IFI16 and MCPyV genome. DAPI (4′,6-diamidino-2-phenylindole, D9542, Sigma) staining at 1:8,000 was included in the secondary antibody incubation mixture. All IF images were collected using an inverted fluorescence microscope (IX81; Olympus) connected to a high-resolution charge-coupled-device camera (FAST1394; QImaging). Images were analyzed and presented using SlideBook (version 5.0) software (Intelligent Imaging Innovations, Inc.). The scale bars were added using ImageJ software.
Western blotting.
To prepare whole-cell lysates, cells were lysed in lysis buffer (10 mM HEPES [pH 7.9], 500 mM NaCl, 3 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5% Triton X-100 supplemented with protease inhibitors). Phosphatase inhibitors were added to the lysis buffer when probing for levels of phosphorylated epitopes. In order to obtain sufficient lysate volume from 96-well plates, cells collected from 10 to 15 wells with the same experimental conditions were combined in lysis buffer for a single replicate. After 30 to 60 min incubation on ice with vortexing, whole-cell lysates were centrifuged at 15,000 × g for 10 min at 4°C to remove debris. Protein concentration of whole-cell lysates was determined using the Bradford assay. The protein samples were resolved on SDS-PAGE gels, transferred onto polyvinylidene difluoride (PVDF) membranes, and immunoblotted with specific primary antibodies as indicated in the figure legends. The primary antibodies used in this study include anti-IFI16 (1:1,000, ab55328, Abcam), anti-cGAS (1:1,000, D1D3G, Cell Signaling Technology), anti-STING (1:1,000, 13647S, Cell Signaling Technology), anti-TBK1/NAK (1:1,000, D1B4, Cell Signaling Technology), anti-IRF3 (1:250, sc-33641, Santa Cruz Biotechnology), anti-p65 (1:400, sc-8008, Santa Cruz), anti-IKKβ (1:1,000, 2684S, Cell Signaling Technology), and anti-GAPDH (1:2,000, 5174S, Cell Signaling Technology). Primary antibodies used in this study recognizing phosphorylated epitopes include anti-p-TBK1 S172 (1:1,000, 5483S, Cell Signaling Technology), anti-p-IRF3 S396 (1:250, 4947S, Cell Signaling Technology), anti-p-p65 S536 (1:250, 3033S, Cell Signaling Technology), and anti-p-IKKα/β S176/180 (1:250, 2697S, Cell Signaling Technology). The secondary antibodies used were HRP-linked anti-rabbit IgG (1:3,000, 7074S, Cell Signaling Technology) and HRP-linked anti-mouse IgG (1:3,000, 7076S, Cell Signaling Technology). Western blots were developed using Western Lightning ECL solution (PerkinElmer) and images were captured using an Amersham Imager 600 device (GE/Cytiva).
Immunofluorescent-smFISH (single molecule fluorescence in situ hybridization).
HDF cells were fixed with 3.7% formaldehyde in PBS for 10 min, and then the immunofluorescent staining was performed using IFI16 antibody (Sigma, catalog number HPA002134) with 1:500 dilution as previously described (86). After immunofluorescent staining, the cells were fixed with 3.7% formaldehyde in PBS for 10 min and then washed twice with PBS and once with wash buffer A (Biosearch Technologies) following the manufacturer’s instructions. Hybridization was performed with the coverslips upside down on a drop of MCPyV LT probes (synthesized by Biosearch Technologies, 1:50 dilution) in the hybridization buffer. After being sealed with rubber cement, the coverslips (containing the target virus DNA and probes) were heated/denatured at 94°C for 3 min and then incubated at 37°C overnight in a humidified hybridization chamber as previously described (42). Following hybridization, the samples were incubated at 37°C for 30 min with wash buffer A and then incubated at 37°C for 30 min with 100 ng/ml DAPI in wash buffer A. After being washed twice with wash buffer B (Biosearch Technologies), cells were mounted and observed using an inverted fluorescence microscope (IX81; Olympus).
cGAMP ELISA.
ELISA for cGAMP was performed using the 2′,3′-cyclic GAMP ELISA kit (Arbor Assays, catalog number K067-H1). Mock-infected and MCPyV-infected HDFs were grown in a 6-well plate. At 120 h.p.i., cells were washed once with PBS and followed by 150 μl of M-per buffer plus 1 mM EDTA mixture being added to each well. The plate was hand-shaken for 5 min at room temperature. Wells were observed with a tissue culture microscope to ensure complete coverage and lysis across the well. A standard curve was made using a stock cGAMP dilution of 0.0328 pmol/ml. All other steps were performed following the manufacturer’s instructions.
Generation of CRISPR/Cas9 knockout HDF stable cells.
sgRNAs were cloned into the LentiCRISPR v2 (pXPR_023) plasmid (Addgene) following protocols from the Genetic Perturbation Program and the Feng Zhang Lab at the Broad Institute that have been adapted by the Elizabeth White Laboratory at the University of Pennsylvania. Oligonucleotides for the generation of sgRNA cassettes targeting genes of interest were selected from the Brunello library from the Broad Institute as well as successful CRISPR knockout exhibited in work by others (Table 3). Reconstituted oligonucleotides were annealed in a Bio-Rad PCR thermal cycler using T4 ligation buffer and T4 PNK enzyme (New England Biolabs). The LentiCRISPR v2 plasmid was cut and dephosphorylated at 37°C for 30 min in a reaction mixture containing FastDigest BsmBI (Esp3I) (Thermo/Fermentas) restriction enzyme, FastDigest Buffer, and 0.1 mM DTT. The 12.8-kb linearized vector was gel purified and ligated to 1:250 diluted sgRNA cassettes. The resulting constructs were transformed into competent Escherichia coli (DH5α), which were then plated on ampicillin-LB plates and incubated overnight at 37°C. Picked colonies were miniprepped and validated by sequencing using a U6 promoter-targeted primer.
TABLE 3.
The following sgRNAs were used to generate cassettes that were cloned into the LentiCRISPR v2 plasmid; all oligonucleotides were ordered from Integrated DNA Technologies (IDT)
| sgRNA target | Oligonucleotide 1 | Oligonucleotide 2 | Source |
|---|---|---|---|
| IFI16 | CACCGGAATGAAGTCTTCCGAGTGA | AAACTCACTCGGAAGACTTCATTCC | 87 |
| cGAS | CACCGATGATATCTCCACGGCGGCG | AAACCGCCGCCGTGGAGATATCATC | 88 |
| STING | CACCGCCCGTGTCCCAGGGGTCACG | AAACCGTGACCCCTGGGACACGGGC | 89 |
| TBK1 | CACCGAAGGATGTTTGCAAGAACAG | AAACCTGTTCTTGCAAACATCCTTC | 90 |
| IRF3 | CACCGGAGGTGACAGCCTTCTACCG | AAACCGGTAGAAGGCTGTCACCTCC | 89, 88 |
| IKK-β | CACCGTCAGCCCCCGGAACCGAGAG | AAACCTCTCGGTTCCGGGGGCTGAC | 91 |
| p65 | CACCGGGAAGATCTCATCCCCACCG | AAACCGGTGGGGATGAGATCTTCCC | 88 |
| Luciferase | CACCGCTTCGAAATGTCCGTTCGGT | AAACACCGAACGGACATTTCGAAGC | 92 |
For the production of lentivirus harboring the LentiCRISPRv2 construct stably expressing desired sgRNA, HEK293T cells were cultured in 10-cm dishes to 90% confluence. LentiCRISPRv2 plasmids were transfected into HEK 293T cells together with psPAX2 and pMD.2G using Lipofectamine 2000 (Invitrogen). At 6 h posttransfection, the culture medium was changed to fresh medium. Twenty-four hours later, lentiviruses were harvested from the supernatant and filtered through a 0.45-μm filter. Passage 2 HDFs were transduced with the purified lentiviruses supplemented with Polybrene. Starting on day 2 after transduction, cells were selected using 0.5 to 2 μg/ml puromycin where the lower range of concentration would be used at lower cell densities. Throughout selection, nontransduced HDFs were exposed to the same selection conditions in parallel to ensure complete killing of cells lacking the resistance gene. HDFs remained under selection with these parameters while being expanded to passage 4, at which point stocks were frozen. Prior to infection, thawed cells were expanded to confluence under puromycin selection and replated the following day. Knockout efficiency was reconfirmed by Western blotting at the point of MCPyV infection.
Statistical analyses.
Statistical analysis for RT-qPCR and qPCR experiments was performed with Prism software (version 8.3.0). Fold induction presented on the graphs for each of these experiments was calculated by the expression 2−(Δ − Δ), where Δ − Δ is the difference in Ct values of the gene of interest and a housekeeping gene of the experimental condition from that of the control condition. Experimental analysis for these data were carried out at the level of Δ − Δ. The specific statistical analyses for each data set were as follows.
Fig. 1B: Two-way analysis of variance (ANOVA): P < 0.0001; with Sidak’s multiple comparison’s test.
Fig. 1C and D: Multiple t tests; P values controlled for multiple comparisons using Holm-Sidak method.
Fig. 1E: Two-way ANOVA: P < 0.0001; with Dunnett’s multiple comparison’s test.
Fig. 4B and C: Two-way ANOVA: P < 0.0001; with Dunnett’s multiple comparison’s test.
Fig. 5A: Two-way ANOVA: P < 0.0001; with Dunnett’s multiple comparison’s test.
ACKNOWLEDGMENTS
We acknowledge all members of the Jianxin You laboratory not listed as authors for their feedback and support throughout the course of this project. We thank the members of the Elizabeth White Laboratory for providing guidance throughout the project, including the production of lentiCRISPRv2 sgRNA stable cells. We also thank the Erle Robertson Laboratory for the use of their Applied Biosystems Quantstudio3 thermal cycler and Sunny Shin for helpful discussion. This work was supported by National Institutes of Health (NIH) grants R01CA187718, R21AR074073, and R21AI149761, NCI Cancer Center Support grant NCI P30 CA016520, and Penn CFAR pilot award P30 AI 045008.
Contributor Information
Jianxin You, Email: jianyou@pennmedicine.upenn.edu.
Lawrence Banks, International Centre for Genetic Engineering and Biotechnology.
REFERENCES
- 1.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, Hedman L, Mattila PS, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. 2011. Serological evidence of Merkel cell polyomavirus primary infections in childhood. J Clin Virol 50:125–129. 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 3.Martel-Jantin C, Pedergnana V, Nicol JT, Leblond V, Tregouet DA, Tortevoye P, Plancoulaine S, Coursaget P, Touze A, Abel L, Gessain A. 2013. Merkel cell polyomavirus infection occurs during early childhood and is transmitted between siblings. J Clin Virol 58:288–291. 10.1016/j.jcv.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Viscidi RP, Rollison DE, Sondak VK, Silver B, Messina JL, Giuliano AR, Fulp W, Ajidahun A, Rivanera D. 2011. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin Vaccine Immunol 18:1737–1743. 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. 2011. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg Infect Dis 17:1371–1380. 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schadendorf D, Lebbe C, Zur Hausen A, Avril MF, Hariharan S, Bharmal M, Becker JC. 2017. Merkel cell carcinoma: epidemiology, prognosis, therapy and unmet medical needs. Eur J Cancer 71:53–69. 10.1016/j.ejca.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gjoerup O, Chang Y. 2010. Chapter 1 - Update on Human Polyomaviruses and Cancer, p 1–51. In George FVW, George K (ed), Advances in Cancer Research, vol 106. Academic Press. [DOI] [PubMed] [Google Scholar]
- 9.Harms PW. 2017. Update on Merkel cell carcinoma. Clin Lab Med 37:485–501. 10.1016/j.cll.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Stang A, Becker JC, Nghiem P, Ferlay J. 2018. The association between geographic location and incidence of Merkel cell carcinoma in comparison to melanoma: an international assessment. Eur J Cancer 94:47–60. 10.1016/j.ejca.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulson KG, Park SY, Vandeven NA, Lachance K, Thomas H, Chapuis AG, Harms KL, Thompson JA, Bhatia S, Stang A, Nghiem P. 2018. Merkel cell carcinoma: current US incidence and projected increases based on changing demographics. J Am Acad Dermatol 78:457–463.e452. 10.1016/j.jaad.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald TL, Dennis S, Kachare SD, Vohra NA, Wong JH, Zervos EE. 2015. Dramatic increase in the incidence and mortality from Merkel cell carcinoma in the United States. Am Surg 81:802–806. 10.1177/000313481508100819. [DOI] [PubMed] [Google Scholar]
- 13.Harms KL, Healy MA, Nghiem P, Sober AJ, Johnson TM, Bichakjian CK, Wong SL. 2016. Analysis of prognostic factors from 9387 Merkel cell carcinoma cases forms the basis for the new 8th edition AJCC staging system. Ann Surg Oncol 23:3564–3571. 10.1245/s10434-016-5266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer JG, Blom A, Doumani R, Lewis C, Tarabadkar ES, Anderson A, Ma C, Bestick A, Parvathaneni U, Bhatia S, Nghiem P. 2016. Response rates and durability of chemotherapy among 62 patients with metastatic Merkel cell carcinoma. Cancer Med 5:2294–2301. 10.1002/cam4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cowey CL, Mahnke L, Espirito J, Helwig C, Oksen D, Bharmal M. 2017. Real-world treatment outcomes in patients with metastatic Merkel cell carcinoma treated with chemotherapy in the USA. Future Oncol 13:1699–1710. 10.2217/fon-2017-0187. [DOI] [PubMed] [Google Scholar]
- 16.Nghiem P, Bhatia S, Lipson EJ, Sharfman WH, Kudchadkar RR, Brohl AS, Friedlander PA, Daud A, Kluger HM, Reddy SA, Boulmay BC, Riker AI, Burgess MA, Hanks BA, Olencki T, Margolin K, Lundgren LM, Soni A, Ramchurren N, Church C, Park SY, Shinohara MM, Salim B, Taube JM, Bird SR, Ibrahim N, Fling SP, Homet Moreno B, Sharon E, Cheever MA, Topalian SL. 2019. Durable tumor regression and overall survival in patients with advanced Merkel cell carcinoma receiving pembrolizumab as first-line therapy. J Clin Oncol 37:693–702. 10.1200/JCO.18.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Angelo SP, Russell J, Lebbé C, Chmielowski B, Gambichler T, Grob J-J, Kiecker F, Rabinowits G, Terheyden P, Zwiener I, Bajars M, Hennessy M, Kaufman HL. 2018. Efficacy and safety of first-line avelumab treatment in patients with stage iv metastatic Merkel cell carcinoma: a preplanned interim analysis of a clinical trial. JAMA Oncol 4:e180077. 10.1001/jamaoncol.2018.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker JC, Stang A, Hausen AZ, Fischer N, DeCaprio JA, Tothill RW, Lyngaa R, Hansen UK, Ritter C, Nghiem P, Bichakjian CK, Ugurel S, Schrama D. 2018. Epidemiology, biology and therapy of Merkel cell carcinoma: conclusions from the EU project IMMOMEC. Cancer Immunol Immunother 67:341–351. 10.1007/s00262-017-2099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. 2011. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J Mol Biol 409:529–542. 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter JJ, Daugherty MD, Qi X, Bheda-Malge A, Wipf GC, Robinson K, Roman A, Malik HS, Galloway DA. 2013. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc Natl Acad Sci U S A 110:12744–12749. 10.1073/pnas.1303526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. 2009. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol 83:12118–12128. 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo GJ, Chen CJ, Sullivan CS. 2009. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383:183–187. 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. 2013. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol 87:6118–6126. 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest 121:3623–3634. 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A 105:16272–16277. 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. 2010. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol 84:7064–7072. 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, Becker JC, Schrama D. 2012. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer 130:847–856. 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 28.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. 2008. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol 58:375–381. 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. 2002. Merkel cell carcinoma and HIV infection. Lancet 359:497–498. 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- 30.Koljonen V, Kukko H, Pukkala E, Sankila R, Bohling T, Tukiainen E, Sihto H, Joensuu H. 2009. Chronic lymphocytic leukaemia patients have a high risk of Merkel-cell polyomavirus DNA-positive Merkel-cell carcinoma. Br J Cancer 101:1444–1447. 10.1038/sj.bjc.6605306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemminki K, Liu X, Ji J, Sundquist J, Sundquist K. 2012. Kaposi sarcoma and Merkel cell carcinoma after autoimmune disease. Int J Cancer 131:E326–328. 10.1002/ijc.27376. [DOI] [PubMed] [Google Scholar]
- 32.Penn I, First MR. 1999. Merkel’s cell carcinoma in organ recipients: report of 41 cases. Transplantation 68:1717–1721. 10.1097/00007890-199912150-00015. [DOI] [PubMed] [Google Scholar]
- 33.Buell JF, Trofe J, Hanaway MJ, Beebe TM, Gross TG, Alloway RR, First MR, Woodle ES. 2002. Immunosuppression and Merkel cell cancer. Transplant Proc 34:1780–1781. 10.1016/S0041-1345(02)03065-8. [DOI] [PubMed] [Google Scholar]
- 34.Hashida Y, Kamioka M, Tanaka M, Hosokawa S, Murakami M, Nakajima K, Kikuchi H, Fujieda M, Sano S, Daibata M. 2016. Ecology of Merkel cell polyomavirus in healthy skin among individuals in an Asian cohort. J Infect Dis 213:1708–1716. 10.1093/infdis/jiw040. [DOI] [PubMed] [Google Scholar]
- 35.Wieland U, Silling S, Scola N, Potthoff A, Gambichler T, Brockmeyer NH, Pfister H, Kreuter A. 2011. Merkel cell polyomavirus infection in HIV-positive men. Arch Dermatol 147:401–406. 10.1001/archdermatol.2011.42. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Strassl R, Helantera I, Aberle SW, Bond G, Hedman K, Weseslindtner L. 2019. Multiplex analysis of Human Polyomavirus diversity in kidney transplant recipients with BK virus replication. J Clin Virol 120:6–11. 10.1016/j.jcv.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Touze A, Le Bidre E, Laude H, Fleury MJ, Cazal R, Arnold F, Carlotti A, Maubec E, Aubin F, Avril MF, Rozenberg F, Tognon M, Maruani A, Guyetant S, Lorette G, Coursaget P. 2011. High levels of antibodies against Merkel cell polyomavirus identify a subset of patients with Merkel cell carcinoma with better clinical outcome. JCO 29:1612–1619. 10.1200/JCO.2010.31.1704. [DOI] [PubMed] [Google Scholar]
- 38.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. 2009. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog 5:e1000578. 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faust H, Pastrana DV, Buck CB, Dillner J, Ekstrom J. 2011. Antibodies to Merkel cell polyomavirus correlate to presence of viral DNA in the skin. J Infect Dis 203:1096–1100. 10.1093/infdis/jiq173. [DOI] [PubMed] [Google Scholar]
- 40.Pastrana DV, Wieland U, Silling S, Buck CB, Pfister H. 2012. Positive correlation between Merkel cell polyomavirus viral load and capsid-specific antibody titer. Med Microbiol Immunol 201:17–23. 10.1007/s00430-011-0200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von der Grun J, Winkelmann R, Meissner M, Wieland U, Silling S, Martin D, Fokas E, Rodel C, Rodel F, Balermpas P. 2019. Merkel cell polyoma viral load and intratumoral CD8+ lymphocyte infiltration predict overall survival in patients with Merkel cell carcinoma. Front Oncol 9:20. 10.3389/fonc.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Yang R, Payne AS, Schowalter RM, Spurgeon ME, Lambert PF, Xu X, Buck CB, You J. 2016. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe 19:775–787. 10.1016/j.chom.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffiths DA, Abdul-Sada H, Knight LM, Jackson BR, Richards K, Prescott EL, Peach AH, Blair GE, Macdonald A, Whitehouse A. 2013. Merkel cell polyomavirus small T antigen targets the NEMO adaptor protein to disrupt inflammatory signaling. J Virol 87:13853–13867. 10.1128/JVI.02159-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akhbari P, Tobin D, Poterlowicz K, Roberts W, Boyne JR. 2018. MCV-miR-M1 targets the host-cell immune response resulting in the attenuation of neutrophil chemotaxis. J Invest Dermatol 10.1016/j.jid.2018.03.1527. [DOI] [PubMed] [Google Scholar]
- 45.Shahzad N, Shuda M, Gheit T, Kwun HJ, Cornet I, Saidj D, Zannetti C, Hasan U, Chang Y, Moore PS, Accardi R, Tommasino M. 2013. The T antigen locus of Merkel cell polyomavirus downregulates human Toll-like receptor 9 expression. J Virol 87:13009–13019. 10.1128/JVI.01786-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdul-Sada H, Muller M, Mehta R, Toth R, Arthur JSC, Whitehouse A, Macdonald A. 2017. The PP4R1 sub-unit of protein phosphatase PP4 is essential for inhibition of NF-kappaB by Merkel polyomavirus small tumour antigen. Oncotarget 8:25418–25432. 10.18632/oncotarget.15836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Yarilina A, Ivashkiv LB. 2010. Type I interferon: a new player in TNF signaling. Curr Dir Autoimmun 11:94–104. 10.1159/000289199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonizzi G, Karin M. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 25:280–288. 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. 2013. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc Natl Acad Sci U S A 110:2969–2974. 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, Jakobsen MR, Nevels MM, Bowie AG, Unterholzner L. 2018. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-kappaB signaling after nuclear DNA damage. Mol Cell 71:745–760.e745. 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Li J, Schowalter RM, Jiao J, Buck CB, You J. 2012. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog 8:e1003021. 10.1371/journal.ppat.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM. 2015. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci U S A 112:E1773–1781. 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diner BA, Lum KK, Toettcher JE, Cristea IM. 2016. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. mBio 7. 10.1128/mBio.01553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ni G, Ma Z, Damania B. 2018. cGAS and STING: at the intersection of DNA and RNA virus-sensing networks. PLoS Pathog 14:e1007148. 10.1371/journal.ppat.1007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gentili M, Lahaye X, Nadalin F, Nader GPF, Puig Lombardi E, Herve S, De Silva NS, Rookhuizen DC, Zueva E, Goudot C, Maurin M, Bochnakian A, Amigorena S, Piel M, Fachinetti D, Londoño-Vallejo A, Manel N. 2019. The N-Terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep 26:2377–2393.e2313. 10.1016/j.celrep.2019.01.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Almine JF, O’Hare CAJ, Dunphy G, Haga IR, Naik RJ, Atrih A, Connolly DJ, Taylor J, Kelsall IR, Bowie AG, Beard PM, Unterholzner L. 2017. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat Commun 8:14392. 10.1038/ncomms14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christian F, Smith EL, Carmody RJ. 2016. The regulation of NF-κB subunits by phosphorylation. Cells 5:12. 10.3390/cells5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. 2009. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer 125:1250–1256. 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguière A, Manuguerra J-C, Caro V, Eloit M. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7:e38499. 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Krump NA, MacDonald M, You J. 2018. Merkel cell polyomavirus infection of animal dermal fibroblasts. J Virol 92 10.1128/JVI.00476-18. 10.1128/JVI.00476-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu W, Krump NA, Buck CB, You J. 2019. Merkel cell polyomavirus infection and detection. J Vis Exp 10.3791/58950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rathi AV, Cantalupo PG, Sarkar SN, Pipas JM. 2010. Induction of interferon-stimulated genes by Simian virus 40 T antigens. Virology 406:202–211. 10.1016/j.virol.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giacobbi NS, Gupta T, Coxon AT, Pipas JM. 2015. Polyomavirus T antigens activate an antiviral state. Virology 476:377–385. 10.1016/j.virol.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Assetta B, De Cecco M, O’Hara B, Atwood WJ. 2016. JC Polyomavirus infection of primary human renal epithelial cells is controlled by a type I IFN-induced response. mBio 7 10.1128/mBio.00903-16. 10.1128/mBio.00903-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.An P, Saenz Robles MT, Duray AM, Cantalupo PG, Pipas JM. 2019. Human polyomavirus BKV infection of endothelial cells results in interferon pathway induction and persistence. PLoS Pathog 15:e1007505. 10.1371/journal.ppat.1007505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schowalter RM, Reinhold WC, Buck CB. 2012. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS One 7:e42181. 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aarreberg LD, Wilkins C, Ramos HJ, Green R, Davis MA, Chow K, Gale M. Jr, 2018. Interleukin-1beta Signaling in dendritic cells induces antiviral interferon responses. mBio 9 10.1128/mBio.00342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rivieccio MA, John GR, Song X, Suh HS, Zhao Y, Lee SC, Brosnan CF. 2005. The cytokine IL-1beta activates IFN response factor 3 in human fetal astrocytes in culture. J Immunol 174:3719–3726. 10.4049/jimmunol.174.6.3719. [DOI] [PubMed] [Google Scholar]
- 71.Mayer-Barber KD, Yan B. 2017. Clash of the cytokine titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell Mol Immunol 14:22–35. 10.1038/cmi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orzalli MH, Smith A, Jurado KA, Iwasaki A, Garlick JA, Kagan JC. 2018. An antiviral branch of the IL-1 signaling pathway restricts immune-evasive virus replication. Mol Cell 71:825–840.e826. 10.1016/j.molcel.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aarreberg LD, Esser-Nobis K, Driscoll C, Shuvarikov A, Roby JA, Gale M. Jr, 2019. Interleukin-1β induces mtDNA release to activate innate immune signaling via cGAS-STING. Mol Cell 74:801–815.e806. 10.1016/j.molcel.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W, Xu L, Brandsma JH, Wang Y, Hakim MS, Zhou X, Yin Y, Fuhler GM, van der Laan LJ, van der Woude CJ, Sprengers D, Metselaar HJ, Smits R, Poot RA, Peppelenbosch MP, Pan Q. 2016. Convergent transcription of interferon-stimulated genes by TNF-α and IFN-α augments antiviral activity against HCV and HEV. Sci Rep 6:25482. 10.1038/srep25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W, Xu L, Su J, Peppelenbosch MP, Pan Q. 2017. Transcriptional regulation of antiviral interferon-stimulated genes. Trends Microbiol 25:573–584. 10.1016/j.tim.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bartee E, Mohamed MR, Lopez MC, Baker HV, McFadden G. 2009. The addition of tumor necrosis factor plus beta interferon induces a novel synergistic antiviral state against poxviruses in primary human fibroblasts. J Virol 83:498–511. 10.1128/JVI.01376-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Venkatesh D, Ernandez T, Rosetti F, Batal I, Cullere X, Luscinskas FW, Zhang Y, Stavrakis G, García-Cardeña G, Horwitz BH, Mayadas TN. 2013. Endothelial TNF receptor 2 induces IRF1 transcription factor-dependent interferon-β autocrine signaling to promote monocyte recruitment. Immunity 38:1025–1037. 10.1016/j.immuni.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yarilina A, Park-Min K-H, Antoniv T, Hu X, Ivashkiv LB. 2008. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon–response genes. Nat Immunol 9:378–387. 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 79.Johnson KE, Chikoti L, Chandran B. 2013. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol 87:5005–5018. 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iqbal J, Ansari MA, Kumar B, Dutta D, Roy A, Chikoti L, Pisano G, Dutta S, Vahedi S, Veettil MV, Chandran B. 2016. Histone H2B-IFI16 recognition of nuclear herpesviral genome induces cytoplasmic interferon-beta responses. PLoS Pathog 12:e1005967. 10.1371/journal.ppat.1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duan X, Ponomareva L, Veeranki S, Panchanathan R, Dickerson E, Choubey D. 2011. Differential roles for the interferon-inducible IFI16 and AIM2 innate immune sensors for cytosolic DNA in cellular senescence of human fibroblasts. Mol Cancer Res 9:589–602. 10.1158/1541-7786.MCR-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xin H, Pereira-Smith OM, Choubey D. 2004. Role of IFI 16 in cellular senescence of human fibroblasts. Oncogene 23:6209–6217. 10.1038/sj.onc.1207836. [DOI] [PubMed] [Google Scholar]
- 83.Siebels S, Czech-Sioli M, Spohn M, Schmidt C, Theiss J, Indenbirken D, Gunther T, Grundhoff A, Fischer N. 2020. Merkel cell polyomavirus DNA replication induces senescence in human dermal fibroblasts in a Kap1/Trim28-dependent manner. mBio 11 10.1128/mBio.00142-20. 10.1128/mBio.00142-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y, Liu Y, Wu J, Roizman B, Zhou GG. 2018. Innate responses to gene knockouts impact overlapping gene networks and vary with respect to resistance to viral infection. Proc Natl Acad Sci U S A 115:E3230–E3237. 10.1073/pnas.1720464115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 86.Liu W, Stein P, Cheng X, Yang W, Shao NY, Morrisey EE, Schultz RM, You J. 2014. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ 21:1950–1960. 10.1038/cdd.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, Stetson DB. 2016. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity 45:255–266. 10.1016/j.immuni.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sali TM, Pryke KM, Abraham J, Liu A, Archer I, Broeckel R, Staverosky JA, Smith JL, Al-Shammari A, Amsler L, Sheridan K, Nilsen A, Streblow DN, DeFilippis VR. 2015. Characterization of a novel human-specific STING agonist that elicits antiviral activity against emerging alphaviruses. PLoS Pathog 11:e1005324. 10.1371/journal.ppat.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodwin JM, Dowdle WE, DeJesus R, Wang Z, Bergman P, Kobylarz M, Lindeman A, Xavier RJ, McAllister G, Nyfeler B, Hoffman G, Murphy LO. 2017. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Rep 20:2341–2356. 10.1016/j.celrep.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang R, Jiang Q, Zhou X, Wang C, Guan Y, Tao J, Xi J, Feng JM, Jiang Z. 2017. MAVS activates TBK1 and IKKepsilon through TRAFs in NEMO dependent and independent manner. PLoS Pathog 13:e1006720. 10.1371/journal.ppat.1006720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vysochan A, Sengupta A, Weljie AM, Alwine JC, Yu Y. 2017. ACSS2-mediated acetyl-CoA synthesis from acetate is necessary for human cytomegalovirus infection. Proc Natl Acad Sci U S A 114:E1528–E1535. 10.1073/pnas.1614268114. [DOI] [PMC free article] [PubMed] [Google Scholar]