Abstract
Objective
Provide a reference to elucidate the mechanism of circRNAs regulating osteoarthritis (OA) and the clinical treatment.
Methods
Herein, articles about circRNAs (hsa-circ) and osteoarthritis in the recent 5 years have been reviewed and the differential expression and regulatory effect of circRNAs in OA deduced. Based on these conclusions and Protein-Protein Interaction (PPI), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the acquired circRNAs, the potential functions and interactions of circRNAs in OA and the involved signaling pathways are discussed.
Results
A total of 33 studies meeting the inclusion criteria were included in this study, and 27 circRNAs were upregulated and 8 circRNAs were downregulated in OA. A total of 31 circRNAs were finally included in the PPI, GO, and KEGG analyses. From PPI, 12 map nodes and 7 map edges were interrelated. VWF had the biggest node and edge size. From GO, VWF showed a majority of the functions. From KEGG, circRNAs are enriched in PI3K/AKT, human papillomavirus infection (HPI), and focal adhesion (FA) pathways, and VWF was involved in major pathways.
Conclusion
We found that most articles about circRNAs regulating OA in the recent 5 years focused on the mechanism, especially the absorption effect of circ-miRNA as sponges in the recent 2 years, while most of the articles about their functions addressed ECM and PI3K, AKT, and mTOR signaling pathways. Future studies might focus on the functions of circRNAs, and circRNA VWF, with preferable functions, interactions, and involvement, can be used as a biological indicator to detect OA in clinical practice.
1. Introduction
Osteoarthritis (OA) is a common clinical disease that has a long process from early inflammation in the joint to the wear and tear of the cartilage layer and the formation of subchondral osteophytes, eventually leading to the failure of the joint to carry out daily movements and perform daily functions [1, 2]. In clinical practice, OA can only be relieved and improved but cannot be cured fully [3, 4]. The mechanism of OA has not yet been defined in existing studies; however, some studies have shown that circular RNAs (circRNAs) play a role in the occurrence and development of OA, but the functions and mechanism of circRNAs in OA were still not very clear [5, 6]. The present study reviewed the articles about circRNAs and OA in the recent 5 years to provide some reference to elucidate the mechanism of circRNAs in OA. Also, based on PPI, GO, and KEGG analysis of the acquired circRNAs, the potential functions of circRNAs in OA and the involved signaling pathways are also discussed in this article. This review can provide some reference for the fundamental research of the prevention and treatment of OA.
2. Material and Methods
2.1. Data Source
“Circular RNA (circRNA)” and “osteoarthritis (OA)” were used as keywords to search relevant articles from January 1, 2016, to 2021 in China National Knowledge Infrastructure (CNKI), PubMed, and Web of Science. There are no ethics committee approval and informed consent in this article.
2.2. Criteria
2.2.1. Inclusion Criteria
Inclusion criteria are as follows: (1) experimental articles with the keywords in the databases and (2) articles with the circRNA ID starting with hsa (human gene).
2.2.2. Exclusion Criteria
Exclusion criteria are as follows: (1) overviews in the databases, (2) repeated articles, (3) articles of poor quality, (4) articles with the circRNA ID starting with mmu (mouse gene), and (5) articles about rheumatoid arthritis (RA).
2.3. PPI, GO, and KEGG Analyses
Included circRNAs were retrieved from circBase (http://www.circbase.org/) to check the accuracy of information, and the circRNAs that had no relevant information were deleted.
The PPI network (https://string-db.org/) was mapped. Choose multiple proteins-gene symbol, and the minimum required interaction score was 0.400.
GO analysis (DAVID, https://david.ncifcrf.gov/summary.jsp) and KEGG analysis (KOBAS, http://kobas.cbi.pku.edu.cn/kobas3) were performed to discuss the potential functions and the participating signaling pathways (species, Homo sapiens; input type, gene symbol; p < 0.05). As shown in Figure 1.
Figure 1.
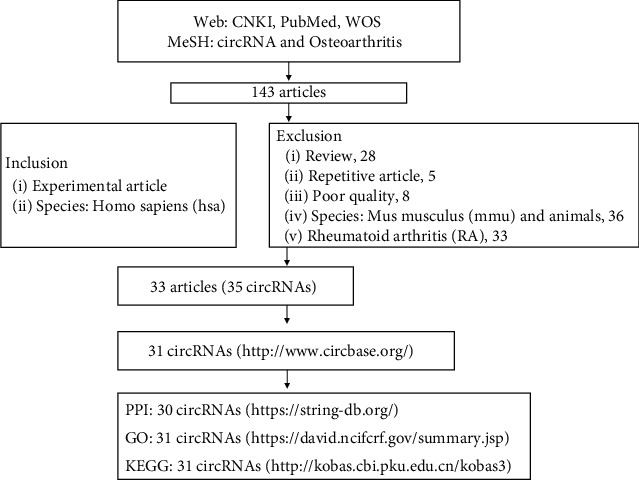
Flowchart.
2.4. Data Processing and Analysis
The acquired data were analyzed and mapped using Cytoscape (3.7.2) and R language (R x64 4.0.2).
3. Results
3.1. Current Studies on the Correlation between circRNAs and OA
A total of 33 studies meeting the inclusion criteria were included in this study, and 35 circRNAs were sorted out by circRNA ID, gene symbol, regulation, miRNA, target gene/signal pathway, reference, and year (Table 1). As shown in Table 1, 81.82% of the articles about circRNAs in the recent 5 years discussed the circ-miRNA axis. As shown in Table 2, 27 circRNAs had upregulated expression and 8 had downregulated expression. As shown in Figure 2, most articles (61%) about circRNAs were published in 2020, and 15% of the articles were published in 2021.
Table 1.
Articles about the correlation between circRNAs and OA in the recent 5 years.
| circRNA ID | Gene symbol | Regulation | miRNAs | Target gene/pathway | Reference | Year | |
|---|---|---|---|---|---|---|---|
| 1 | hsa_circ_0141827 | SERPINE2 | Down | miR-1271 | ERG pathway, SOX, COL2 | Shen et al. [7] | 2019 |
| 2 | hsa_circ_0092516 | NT5C2 | Down | miR-337-3p | MMP-1, COL2 | Huang et al. [8] | 2020 |
| 3 | hsa_circ_0020014 | DUSP5 | NA | NA | NA | Wang et al. [9] | 2020 |
| 4 | hsa_circ_0041552 | UBE2G1 | Down | miR-373 | HIF-1a | Chen et al. [10] | 2020 |
| 5 | hsa_circ_0000448 | GCN1L1 | Down | miR-330-3p | TNF-α, ADAMTS4 | Zhu et al. [11] | 2020 |
| 6 | hsa_circ_0021592 | HIPK3 | Down | miR-124 | SOX-8 | Wu et al. [12] | 2020 |
| 7 | hsa_circ_0129854 | VCAN | NA | NA | NF-κB pathway | Ma et al. [13] | 2020 |
| 8 | hsa_circ_0080978 | CDK14 | Down | miR-125a-5p | SOX-9, Smad-2 | Shen et al. [14] | 2020 |
| 9 | hsa_circ_0136474 | ASH2L | Down | miR-127-5p | MMP-13 | Li et al. [15] | 2019 |
| 10 | hsa_circ_0129214 | PDE4D | Down | miR-103a-3p | FGF18 | Wu et al. [16] | 2021 |
| 11 | hsa_circ_0055722 | ANKRD36 | Down | miR-599 | Casz1 | Zhou et al. [17] | 2021 |
| 12 | hsa_circ_0005105 | SEC24A | Down | miR-26a | NAMPT | Wu et al. [18] | 2017 |
| 13 | hsa_circ_0032131 | PRKCH | Down | miR-1182 | NA | Wang et al. [19, 20] | 2019 |
| 14 | hsa_circ_0026176 | TMBIM6 | Down | miR-27a | MMP-13 | Bai et al. [21] | 2020 |
| 15 | hsa_circ_9119# | NA | Down | miR-26a | PTEN | Chen et al. [22] | 2020 |
| 16 | hsa_circ_0025119 hsa_circ_0025113 hsa_circ_0009897 hsa_circ_0002447 |
VWF
VWF PLOD1 COL6A3 |
NA | NA | NA | Wang et al. [23] | 2020 |
| 17 | hsa_circ_7# | NA | Down | miR-7 | PI3K/AKT/mTOR | Zhou et al. [24, 25] | 2020 |
| 18 | hsa_circ_0045714 hsa_circ_0002485 hsa_circ_0005567 |
UNK
ATP9B EPS15 |
NA | NA | NA | Xiao et al. [26] | 2019 |
| 19 | NA# | PSM3 | Down | miR-296-5p | Ni et al. [27] | 2020 | |
| 20 | hsa_circ_100226# | MSR | Down | miR-875 | TNF-α | Liu et al. [28] | 2017 |
| 21 | hsa_circ_0001946 | CDR1 | Down | miR-641 | COL2, IL-6 | Zhang et al. [29] | 2020 |
| 22 | hsa_circ_0040639 | CDH13 | Down | miR-296-3p | PTEN | Zhou et al. [30] | 2020 |
| 23 | hsa_circ_0023404 | RNF121 | Down | miR-665 | MYD88 | Wang et al. [31] | 2020 |
| 24 | hsa_circ_0141827 | SERPINE2 | Down | miR-495 | TGFBR2 | Zhang et al. [32] | 2020 |
| 25 | hsa_circ_0035826 | CSNK1G1 | Down | miR-4428 | FUT2 | Xiao et al. [33] | 2020 |
| 26 | hsa_circ_0005567 | EPS15 | Down | miR-495 | ATG14 | Zhang et al. [34] | 2020 |
| 27 | hsa_circ_0010014 | DHRS3 | Down | miR-183-5p | GREM1 | Jiang et al. [35] | 2020 |
| 28 | hsa_circ_0072655 | ADAMTS6 | Down | miR-431-5p | IL-β | Fu et al. [36] | 2020 |
| 29 | hsa_circ_0045714 | UNK | Down | miR-193b | IGF1R | Li et al. [37] | 2017 |
| 30 | hsa_circ_0114876 | PTPRA | Down | miR-671 | TRAF2 | Wang et al. [38] | 2021 |
| 31 | hsa_circ_0104873 hsa_circ_0104595 hsa_circ_0101251 |
IQGAP1
SCAPER RP11-909M7.3 |
NA | NA | NA | Yu et al. [39] | 2018 |
| 32 | hsa_circ_0017855 | RSU1 | Down | miR-93-5p | MAP3K8 | Yang et al. [40] | 2021 |
| 33 | hsa_circ_0045714 | UNK | Down | miR-218-5p | HRAS | Jiang et al. [41] | 2021 |
Note: “NA” means not available, and “#” means no relevant information in circBase.
Table 2.
Expression of circRNAs in OA.
| Expression of 35 circRNAs in OA | |
|---|---|
| Upregulation (27) | Downregulation (8) |
| NT5C2 [8], DUSP5 [9], UBE2G1 [10], GCN1L1 [11], HIPK3 [12], VCAN [13], ASH2L [15], SEC24A [18], PRKCH [19, 20], TMBIM6 [21], VWF [23], PLOD1 [23], COL6A3 [23], hsa_circ_7 [24, 25], ATP9B [26], PSM3 [27], MSR [28], CDR1 [29], CDH13 [30], RNF121 [31], CSNK1G1 [33], DHRS3 [35], PTPRA [38], IQGAP1 [39], SCAPER [39], RP11-909M7.3 [39], RSU1 [40] | SERPINE2 [7, 32], CDK14 [14], PDE4D [16], ANKRD36 [17], hsa_circ_9119 [22], EPS15 [26, 34], ADAMTS6 [36], UNK [37, 41] |
Figure 2.

Year of issue of the 33 articles.
3.2. PPI, GO, and KEGG Analyses of circRNAs
Repeated circRNAs (SERPINE2, VWF, EPS15, and UNK) and those having no information in circBase (hsa_circ_9119, hsa_circ_7, PSM3, and hsa_circ_100226) were excluded, and a total of 31 circRNAs were finally included in the PPI, GO, and KEGG analyses. The final result showed p < 0.05.
Figure 3 of PPI analysis shows a network of 30 circRNAs (RP11-909M7.3 not found in STRING); of these, 12 map nodes and 7 map edges were interrelated. VWF and DUSP5 had the biggest map node size (degree 2); IQGAP1-VWF-SERPINE2 and PLOD1-COL6A3 had the bigger map edge size (0.906, 0.928, and 0.913).
Figure 3.
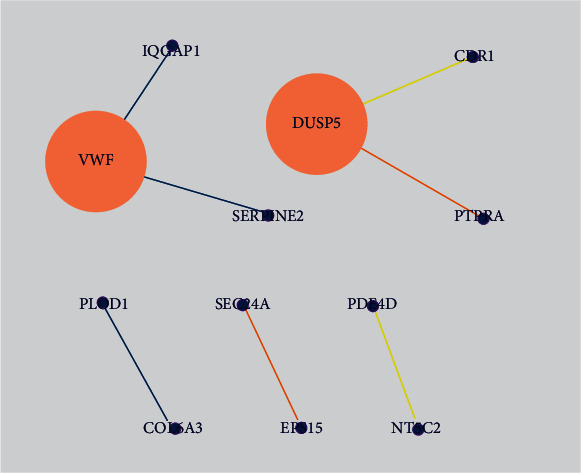
PPI network of circRNAs. Note: map node size to degree and map edge size to combined score, low values to small sizes and bright colors.
GO enrichment analysis usually covers molecular function (MF), cellular component (CC), and biological process (BP). The results of this study showed that the functions of 31 circRNAs were mainly focused on MF, including protein kinase activity and glycosaminoglycan binding; CC were proteinaceous ECM, platelet α granule, extracellular matrix (ECM), cellular exosome, and endoplasmic reticulum membrane; BP included peptidyl-serine phosphorylation, ECM organization, and cell adhesion (Figure 4). Among them, VWF showed a majority of the functions (6/10, Table 3).
Figure 4.
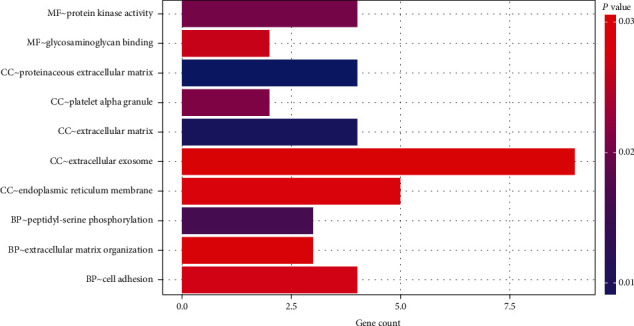
GO analysis of the 31 circRNAs.
Table 3.
Functions of circRNAs.
| Term | Genes |
|---|---|
| MF: protein kinase activity | PRKCH, CDK14, HIPK3, CSNK1G1 |
| MF: glycosaminoglycan binding | VCAN, SERPINE2 |
| CC: proteinaceous COL6A3 extracellular matrix | VCAN, VWF, COL6A3, ADAMTS6 |
| CC: platelet alpha granule | SERPINE2, VWF |
| CC: extracellular matrix | VCAN, SERPINE2, VWF |
| CC: extracellular exosome | PRKCH, VWF, PTPRA, CDH13, COL6A3, UBE2G1, PLOD1, IQGAP1, RSU1 |
| CC: endoplasmic reticulum membrane | SEC24A, TMBIM6, PLOD1, DHRS3, RNF121 |
| BP: peptidyl-serine phosphorylation | PRKCH, HIPK3, CSNK1G1 |
| BP: extracellular matrix organization | VCAN, VWF, COL6A3 |
| BP: cell adhesion | VCAN, VWF, CDH13, COL6A3 |
KEGG signaling pathway analysis showed that circRNAs are enriched in PI3K/AKT, human papillomavirus infection (HPI), focal adhesion (FA), and other seven pathways (Figure 5), and VWF and COL6A3 were involved in 4/7 pathways (Table 4).
Figure 5.
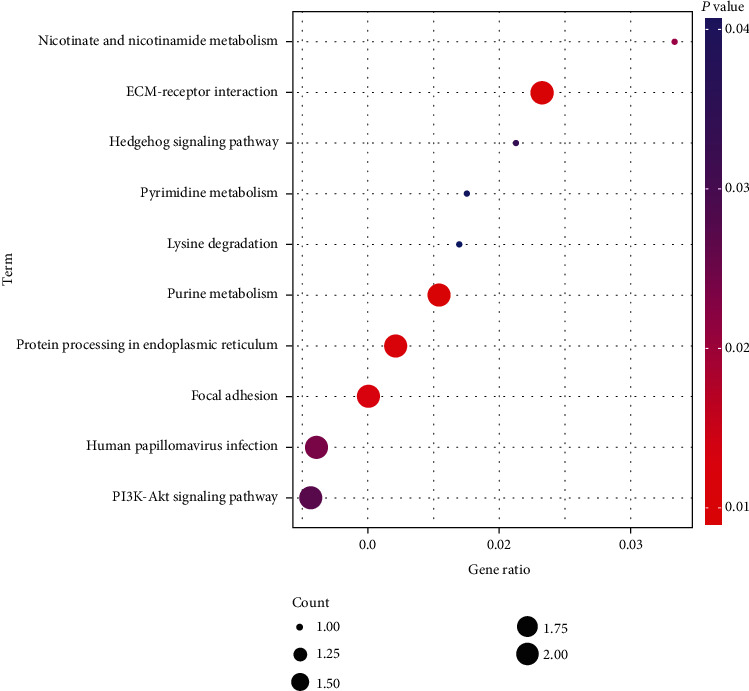
KEGG analysis of the 31 circRNAs.
Table 4.
Signaling pathways involving circRNAs.
| Term | Input |
|---|---|
| ECM-receptor interaction | VWF|COL6A3 |
| Purine metabolism | PDE4D|NT5C2 |
| Protein processing in the endoplasmic reticulum | SEC24A|UBE2G1 |
| Focal adhesion | VWF|COL6A3 |
| Nicotinate and nicotinamide metabolism | NT5C2 |
| Human papillomavirus infection | VWF|COL6A3 |
| PI3K-Akt signaling pathway | VWF|COL6A3 |
| Hedgehog signaling pathway | CSNK1G1 |
| Pyrimidine metabolism | NT5C2 |
| Lysine degradation | PLOD1 |
4. Discussion
4.1. Brief Information and Functions of circRNAs
circRNAs are a type of noncoding RNAs mainly found in the cytoplasm of mammalian cells. circRNAs consist of the 3′- and 5′-phosphodiester bonds covalently linked to form a circular structure, which is stable and resistant to RNA exonuclease-mediated degradation, and hence are termed as circRNAs. Three types of circRNAs, exon circRNAs (exon circular RNA (ecircRNAs)), intron circRNAs (intron circular RNA (ciRNAs)), and exon-intron circRNAs (exon-intron circular RNA (EIcircRNAs)) [42–44], especially ciRNAs, are conserved across evolution and have a half-life of >48 h, which also confirms their high stability. In addition, circRNAs are highly stable and sensitive in body fluids and used for biochemical tests [45–47].
Current studies showed that the functions of circRNAs are as follows. (1) They adsorb microRNAs (miRNAs), bind to miRNAs as sponges, affect the corresponding message RNAs (mRNAs), and eventually regulate the expression of target genes. (2) They regulate the activity of RNA-binding proteins (RBPs) and transport them or act as their scaffold to facilitate the formation of new complexes. Additionally, circRNAs can also interact with proteins, selectively cut or transcribe parent genes (binding RNA polymerases), and encode the proteins [48–52], as shown in Figure 6.
Figure 6.
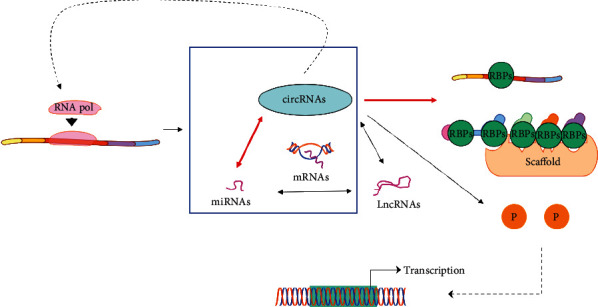
Functions of circRNAs.
4.2. Studies on the Mechanism of circRNAs in OA and circRNAs as Biological Indicators of OA
This study showed that the articles on circRNAs in OA in the recent 5 years mainly focused on the mechanism while they also discussed circRNAs as clinical, biological indicators of OA.
According to the statistical results of this study, 61% of the relevant articles in the recent 5 years were published in 2020 and 15% in 2021. The majority of these articles focused on the mechanism of circ-miRNA with respect to the absorption effect of circRNAs as sponges on miRNAs in OA. Kulcheski et al. [53] proposed that circRNAs are sponges of miRNAs and can serve as the novel type of biomarkers. circRNA 0092516 regulates chondrocyte differentiation and apoptosis via miRNA-337-3p/PTEN (phosphatase and tensin homolog), according to Huang et al. [8], while circRNA UBE2G1 regulates lipopolysaccharide- (LPS-) induced OA chondrocytes via miR-373/hypoxic inducible factor 1 alpha (HIF-1α), according to Chen et al. [9]. Wu et al. [12] demonstrated that lowly expressed circRNA HIPK3 regulates SRY-related high-mobility group box gene 8 (SOX-8), a critical marker of chondrocyte development as the sponge of miR-124, thus promoting the apoptosis of osteoarthritis chondrocytes. Ma et al. [13] found that circRNA VCAN promotes the apoptosis of OA chondrocytes by blocking the NF-κB signaling pathway. Wu et al. [16] showed that circRNA PDE4D protected OA by binding to miR-103a-3p and regulating the fibroblast growth factor 18 (FGF18), and Zhou et al. [17] found that circRNA ANKRD36 regulated Casz1 (miR-599 target gene) and prevented the apoptosis and inflammation of OA chondrocytes by targeting miR-599.
Additionally, some studies also discussed circRNAs as biological indicators to detect and evaluate OA. In the study by Wang et al. [9], patients with Kashin–Beck disease (KBD) and OA were subjected to circRNA sequencing to observe differential expression; the result of which showed that circRNA 0020014 could serve as the potential marker of OA to evaluate the progression of OA. Wang et al. [23] analyzed the gene expression profile, wherein VWF (hsa_circ_0025119) and other three genes served as OA markers. Xiao et al. [26] demonstrated that, on the Illumina HiSeq platform, circRNA 0045714 was expressed differentially in OA. Xiang et al. [54] revealed the expression profile of circRNAs in OA through RNA sequencing and identified 122 circRNAs of differential expression. Based on these studies, VWF (hsa_circ_0025119) had the highest value (Figure 3, Tables 3 and 4), indicating a significant interaction between VWF and other circRNAs; also, additional functions and signaling pathways were detected in the BP. Therefore, we speculated that VWF (hsa_circ_0025119) is more feasible to be used as a biological indicator compared to other circRNAs, to detect OA in clinical practice.
4.3. Studies on the Potential Functions of circRNAs in OA and Involved Signaling Pathways
The current study showed that circRNAs play a critical role in ECM. Shen et al. [7] showed that the overexpression of circRNA SERPINE2 downregulates the miR-1271-ERG (E26 transformation-specific-related gene) pathway to reduce HCS (human chondrocyte) apoptosis and promote ECM anabolism, thus slowing down OA development. Zhu et al. [11] found that circRNA GCN1L1 regulates miR-330-3p and TNF-α to promote OA synovial cells and reduce ECM anabolism. Wu et al. [18] demonstrated that circRNA 0005105 upregulates the expression of NAMPT (miR-26a target gene) and promotes ECM degradation in chondrocytes by absorbing miR-26a as sponges. In addition, circRNA TMBIM6 promotes ECM degradation of OA-induced chondrocytes via the miR-27a/matrix metalloproteinase-13 (MMP-13) axis, according to Bai et al. [21]. circRNA SERPINE2 reduces IL-1β-induced apoptosis and ECM degradation of chondrocytes by regulating the miR-495/transforming growth factor-beta receptor 2 (TGFBR2) axis [32]. Furthermore, the functions of circRNAs also include protein kinase activity, glycosaminoglycan binding, endoplasmic reticulum membrane, and peptidyl-serine phosphorylation, which can be the focus of future studies on the mechanism of OA.
In this study, VWF and COL6A3 are involved in the PI3K/AKT signaling pathway (Table 4). According to Zhou et al. [24], circRNA7 regulates PI3K/AKT/mTOR by absorbing miR-7, thus aggravating OA and indicating that the PI3K/AKT signaling pathway may play a critical role in circRNAs regulating the development of OA. The PI3K/AKT/mTOR signaling pathway functions in cartilage degeneration, subchondral bone dysfunction, and synovial inflammation [55–57]. Therefore, in future studies on the mechanism of circRNA-regulated OA chondrocytes and synovial cells, the correlation between the circ-PI3K/AKT/mTOR axes can be observed, and the role of PI3K/AKT/mTOR is discussed. Multiple collagen factors were also detected in the ECM-receptor interaction pathway in Figure 5. Collagen is a vital component of cartilage composition and plays a crucial role in protecting cartilage tissues [58–60]. This finding suggested that the ECM-receptor interaction signaling pathway may also play a critical role in the mechanism underlying circRNA-regulated OA (Figure 7).
Figure 7.
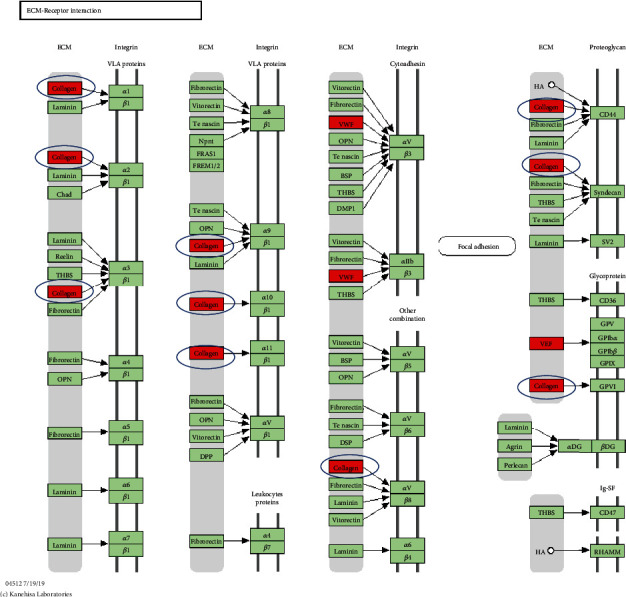
ECM-receptor interaction signaling pathway.
We also found that the expression of most circRNAs was upregulated, while a few were downregulated in OA. According to Wang et al. [31], circRNA RNF121 aggravated the progression of OA via the miR-665/MYD88 axis (MYD88 is the canonical adaptor for inflammatory pathway), and according to Xiao et al. [33], circRNA CSNK1G1 promotes the progression of OAs by targeting the miR-4428/FUT2 (fucosyltransferase) axis. Jiang et al. [35] demonstrated that circRNA DHRS3 accelerates OA progression via miR-183-5p/GREM1 (Gremlin, the miR-183-5p target gene). Wang et al. [38] found that circRNA 0114876 aggravates OA via the miR-671/TRAF2 (TNF receptor-associated factor 2) axis. Yang et al. [40] found that circRNA RSU1 aggravates OA via the miR-93-5p/MAP3K8 (mitogen-activated protein kinase 8) axis, and Shen et al. [14] showed that circRNA CDK14 protects OA via the sponge tissue miR-125a-5p and enhances the expression of Smad2 (gene of TGF-β family). Moreover, in the study by Chen et al. [22], circRNA 9119 was shown to prevent apoptosis of IL-1β-treated OA chondrocytes by blocking the miR-26a/PTEN axis, and circRNA ADAMTS6 protects OA by absorbing miR-431-5p [36]. Another study showed that circRNA 0045714 exerted a protective effect on OA via the miR-193b/insulin-like growth factor 1 receptor (IGF1R) axis [37]. In summary, 77.78% of the circRNAs were upregulated and 22.23% were downregulated, and the overexpression of the majority of the circRNAs aggravates the occurrence and development of OA.
Herein, the studies on the correlation between circRNAs and OA in the recent 5 years and the circRNAs with differential expression and reliable mechanism of action in OA were reviewed. We found that most articles about circRNAs regulating OA in the recent 5 years focused on the mechanism, especially the absorption effect of circ-miRNA as sponges in the recent 2 years, while most of the articles about their functions addressed ECM and PI3K, AKT, and mTOR signaling pathways. Based on the GO and KEGG analysis results, future studies might focus on the functions of circRNAs, such as protein kinase activity, glycosaminoglycan binding, endoplasmic reticulum membrane, and peptidyl-serine phosphorylation, as well as ECM-receptor interaction-related signaling pathways. circRNA VWF, with preferable functions, interactions, and involvement, can be used as a biological indicator to detect OA in clinical practice. However, although the absorption effect of circ-miRNA as sponges in the mechanism of OA has been under intensive focus in the recent 2 years, studies are still rare. Therefore, further studies would focus on the database of the circ-miRNA axis in OA in order to provide a reference for the clinical treatment based on the mechanism of OA.
Acknowledgments
This study was funded by Shanghai Key Laboratory of Human Sport Competence Development and Maintenance (Shanghai University of Sport) (No. 11DZ2261100) and the National Natural Science Foundation of China (81871835).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
The first author took responsibility for the conception and design of the study, or acquisition of data, or analysis and interpretation of data and drafted the article or revised it critically for important intellectual content. The corresponding author took responsibility for the final approval of the version to be submitted. Wei Wu is the first author.
References
- 1.van den Bosch M. H. J. Osteoarthritis year in review 2020: biology. Osteoarthritis and Cartilage. 2021;29(2):143–150. doi: 10.1016/j.joca.2020.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Billesberger L. M., Fisher K. M., Qadri Y. J., Boortz-Marx R. L. Procedural treatments for knee osteoarthritis: a review of current injectable therapies. Pain Research & Management. 2020;2020, article 3873098:1–11. doi: 10.1155/2020/3873098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunt M. A., Charlton J. M., Esculier J. F. Osteoarthritis year in review 2019: mechanics. Osteoarthritis and Cartilage. 2020;28(3):267–274. doi: 10.1016/j.joca.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Abramoff B., Caldera F. E. Osteoarthritis: pathology, diagnosis, and treatment options. The Medical Clinics of North America. 2020;104(2):293–311. doi: 10.1016/j.mcna.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Yu C. X., Sun S. An emerging role for circular RNAs in osteoarthritis. Yonsei Medical Journal. 2018;59(3):349–355. doi: 10.3349/ymj.2018.59.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H. Z., Lin Z., Xu X. H., Lin N., Lu H. D. The potential roles of circRNAs in osteoarthritis: a coming journey to find a treasure. Bioscience Reports. 2018;38(5) doi: 10.1042/BSR20180542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen S., Wu Y., Chen J., et al. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Annals of the Rheumatic Diseases. 2019;78(6):826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z., Ma W., Xiao J., Dai X., Ling W. CircRNA_0092516 regulates chondrocyte proliferation and apoptosis in osteoarthritis through the miR-337-3p/PTEN axis. The Journal of Biochemistry. 2021;169(4):467–475. doi: 10.1093/jb/mvaa119. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Wu C., Zhang Y., et al. Screening for differentially expressed circRNA between Kashin-Beck disease and osteoarthritis patients based on circRNA chips. Clinica Chimica Acta. 2020;501:92–101. doi: 10.1016/j.cca.2019.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Chen G., Liu T., Yu B., Wang B., Peng Q. CircRNA-UBE2G1 regulates LPS-induced osteoarthritis through miR-373/HIF-1a axis. Cell Cycle. 2020;19(13):1696–1705. doi: 10.1080/15384101.2020.1772545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H., Hu Y., Wang C., Zhang X., He D. CircGCN1L1 promotes synoviocyte proliferation and chondrocyte apoptosis by targeting miR-330-3p and TNF-α in TMJ osteoarthritis. Cell Death & Disease. 2020;11(4):p. 284. doi: 10.1038/s41419-020-2447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Q., Yuan Z. H., Ma X. B., Tang X. H. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. European Review for Medical and Pharmacological Sciences. 2020;24(15):7937–7945. doi: 10.26355/eurrev_202008_22476. [DOI] [PubMed] [Google Scholar]
- 13.Ma H. R., Mu W. B., Zhang K. Y., Zhou H. K., Jiang R. D., Cao L. CircVCAN regulates the proliferation and apoptosis of osteoarthritis chondrocyte through NF-κB signaling pathway. European Review for Medical and Pharmacological Sciences. 2020;24(12):6517–6525. doi: 10.26355/eurrev_202006_21635. [DOI] [PubMed] [Google Scholar]
- 14.Shen P., Yang Y., Liu G., et al. CircCDK14 protects against osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics. 2020;10(20):9113–9131. doi: 10.7150/thno.45993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Yuan B., Pei Z., et al. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. Journal of Cellular and Molecular Medicine. 2019;23(10):6554–6564. doi: 10.1111/jcmm.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Hong Z., Xu W., et al. Circular RNA circPDE4D protects against osteoarthritis by binding to miR-103a-3p and regulating FGF18. Molecular Therapy. 2021;29(1):308–323. doi: 10.1016/j.ymthe.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J. L., Deng S., Fang H. S., du X. J., Peng H., Hu Q. J. Circular RNA circANKRD36 regulates Casz1 by targeting miR-599 to prevent osteoarthritis chondrocyte apoptosis and inflammation. Journal of Cellular and Molecular Medicine. 2021;25(1):120–131. doi: 10.1111/jcmm.15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y., Zhang Y., Zhang Y., Wang J. J. CircRNA hsa_circ_0005105 upregulates NAMPT expression and promotes chondrocyte extracellular matrix degradation by sponging miR-26a. Cell Biology International. 2017;41(12):1283–1289. doi: 10.1002/cbin.10761. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Wu C., Zhang F., et al. Screening for differentially expressed circular RNAs in the cartilage of osteoarthritis patients for their diagnostic value. Genetic Testing and Molecular Biomarkers. 2019;23(10):706–716. doi: 10.1089/gtmb.2019.0108. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Wu C., Yang Y., Ren Z., Lammi M. J., Guo X. Preliminary exploration of hsa_circ_0032131 levels in peripheral blood as a potential diagnostic biomarker of osteoarthritis. Genetic Testing and Molecular Biomarkers. 2019;23(10):717–721. doi: 10.1089/gtmb.2019.0036. [DOI] [PubMed] [Google Scholar]
- 21.Bai Z. M., Kang M. M., Zhou X. F., Wang D. CircTMBIM6 promotes osteoarthritis-induced chondrocyte extracellular matrix degradation via miR-27a/MMP13 axis. European Review for Medical and Pharmacological Sciences. 2020;24(15):7927–7936. doi: 10.26355/eurrev_202008_22475. [DOI] [PubMed] [Google Scholar]
- 22.Chen C., Yin P., Hu S., Sun X., Li B. Circular RNA-9119 protects IL-1β-treated chondrocytes from apoptosis in an osteoarthritis cell model by intercepting the microRNA-26a/PTEN axis. Life Sciences. 2020;256, article 117924 doi: 10.1016/j.lfs.2020.117924. [DOI] [PubMed] [Google Scholar]
- 23.Wang B., Zhong J. L., Xu X. H., et al. Gene expression profiling analysis to identify key genes and underlying mechanisms in meniscus of osteoarthritis patients. Combinatorial Chemistry & High Throughput Screening. 2021;24(8):1151–1167. doi: 10.2174/1386207323666200902140656. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X., Li J., Zhou Y., et al. Down-regulated ciRS-7/up-regulated miR-7 axis aggravated cartilage degradation and autophagy defection by PI3K/AKT/mTOR activation mediated by IL-17A in osteoarthritis. Aging. 2020;12(20):20163–20183. doi: 10.18632/aging.103731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X., Jiang L., Fan G., et al. Role of the ciRS-7/miR-7 axis in the regulation of proliferation, apoptosis and inflammation of chondrocytes induced by IL-1β. International Immunopharmacology. 2019;71:233–240. doi: 10.1016/j.intimp.2019.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Xiao K., Xia Z., Feng B., et al. Circular RNA expression profile of knee condyle in osteoarthritis by illumina HiSeq platform. Journal of Cellular Biochemistry. 2019;120(10):17500–17511. doi: 10.1002/jcb.29014. [DOI] [PubMed] [Google Scholar]
- 27.Ni J. L., Dang X. Q., Shi Z. B. CircPSM3 inhibits the proliferation and differentiation of OA chondrocytes by targeting miRNA-296-5p. European Review for Medical and Pharmacological Sciences. 2020;24(7):3467–3475. doi: 10.26355/eurrev_202004_20805. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q., Zhang X., Hu X., et al. Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Molecular Therapy - Nucleic Acids. 2017;7:223–230. doi: 10.1016/j.omtn.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W., Zhang C., Hu C., Luo C., Zhong B., Yu X. Circular RNA-CDR1as acts as the sponge of microRNA-641 to promote osteoarthritis progression. Journal of Inflammation. 2020;17(1) doi: 10.1186/s12950-020-0234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z., Ma J., Lu J., Chen A., Zhu L. Circular RNA CircCDH13 contributes to the pathogenesis of osteoarthritis via CircCDH13/miR-296-3p/PTEN axis. Journal of Cellular Physiology. 2021;236(5):3521–3535. doi: 10.1002/jcp.30091. [DOI] [PubMed] [Google Scholar]
- 31.Wang T., Hao Z., Liu C., et al. LEF1 mediates osteoarthritis progression through circRNF121/miR-665/MYD88 axis via NF-кB signaling pathway. Cell Death & Disease. 2020;11(7):p. 598. doi: 10.1038/s41419-020-02769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q., Qiao X., Xia W. CircSERPINE2 weakens IL-1β-caused apoptosis and extracellular matrix degradation of chondrocytes by regulating miR-495/TGFBR2 axis. Bioscience Reports. 2020;40(11, article BSR20201601) doi: 10.1042/BSR20201601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao J., Wang R., Zhou W., Cai X., Ye Z. Circular RNA CSNK1G1 promotes the progression of osteoarthritis by targeting the miR‑4428/FUT2 axis. International Journal of Molecular Medicine. 2020;47(1):232–242. doi: 10.3892/ijmm.2020.4772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Zhang J., Cheng F., Rong G., Tang Z., Gui B. Hsa_circ_0005567 activates autophagy and suppresses IL-1β-induced chondrocyte apoptosis by regulating miR-495. Frontiers in Molecular Biosciences. 2020;7:p. 216. doi: 10.3389/fmolb.2020.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang R., Gao H., Cong F., Zhang W., Song T., Yu Z. Circ_DHRS3 positively regulates GREM1 expression by competitively targeting miR-183-5p to modulate IL-1β-administered chondrocyte proliferation, apoptosis and ECM degradation. International Immunopharmacology. 2020;91, article 107293 doi: 10.1016/j.intimp.2020.107293. [DOI] [PubMed] [Google Scholar]
- 36.Fu Q., Li L., Wang B., et al. CircADAMTS6/miR-431-5p axis regulate interleukin-1β induced chondrocyte apoptosis. The Journal of Gene Medicine. 2021;23 doi: 10.1002/jgm.3304. [DOI] [PubMed] [Google Scholar]
- 37.Li B. F., Zhang Y., Xiao J., et al. Hsa_circ_0045714 regulates chondrocyte proliferation, apoptosis and extracellular matrix synthesis by promoting the expression of miR-193b target gene IGF1R. Human Cell. 2017;30(4):311–318. doi: 10.1007/s13577-017-0177-7. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q., Luo S., Yang J., et al. Circ_0114876 promoted IL-1β-induced chondrocyte injury by targeting miR-671/TRAF2 axis. Biotechnology Letters. 2021;43(4):791–802. doi: 10.1007/s10529-020-03070-1. [DOI] [PubMed] [Google Scholar]
- 39.Yu F., Xie C., Sun J., Feng H., Huang X. Circular RNA expression profiles in synovial fluid: a promising new class of diagnostic biomarkers for osteoarthritis. International Journal of Clinical and Experimental Pathology. 2018;11(3):1338–1346. [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y., Shen P., Yao T., et al. Novel role of circRSU1 in the progression of osteoarthritis by adjusting oxidative stress. Theranostics. 2021;11(4):1877–1900. doi: 10.7150/thno.53307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang H., Dai J., Zhang C., Sun H., Tang X. Circ_0045714 alleviates TNF-α-induced chondrocyte injury and extracellular matrix degradation through miR-218-5p/HRAS axis. Journal of Bioenergetics and Biomembranes. 2021;53(1):97–107. doi: 10.1007/s10863-020-09868-y. [DOI] [PubMed] [Google Scholar]
- 42.Li R., Jiang J., Shi H., Qian H., Zhang X., Xu W. CircRNA: a rising star in gastric cancer. Cellular and Molecular Life Sciences. 2020;77(9):1661–1680. doi: 10.1007/s00018-019-03345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zang J., Lu D., Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. Journal of Neuroscience Research. 2020;98(1):87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y., Jia X., Xu J. The new function of circRNA: translation. Clinical & Translational Oncology. 2020;22(12):2162–2169. doi: 10.1007/s12094-020-02371-1. [DOI] [PubMed] [Google Scholar]
- 45.Wesselhoeft R. A., Kowalski P. S., Anderson D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nature Communications. 2018;9(1):p. 2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L. L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nature Reviews Molecular Cell Biology. 2020;21(8):475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 47.Shi X., Wang B., Feng X., Xu Y., Lu K., Sun M. circRNAs and exosomes: a mysterious frontier for human cancer. Molecular Therapy - Nucleic Acids. 2020;19:384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10(8):3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao M. S., Ai Y., Wilusz J. E. Biogenesis and functions of circular RNAs come into focus. Trends in Cell Biology. 2020;30(3):226–240. doi: 10.1016/j.tcb.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W. Y., Cai Z. R., Liu J., Wang D. S., Ju H. Q., Xu R. H. Circular RNA: metabolism, functions and interactions with proteins. Molecular Cancer. 2020;19(1):p. 172. doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Q., Li F., He A., Yang B. B. Circular RNAs: expression, localization, and therapeutic potentials. Molecular Therapy. 2021;29(5):1683–1702. doi: 10.1016/j.ymthe.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W., Qi L., Chen R., et al. Circular RNAs in osteoarthritis: indispensable regulators and novel strategies in clinical implications. Arthritis Research & Therapy. 2021;23(1):p. 23. doi: 10.1186/s13075-021-02420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulcheski F. R., Christoff A. P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Journal of Biotechnology. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 54.Xiang S., Li Z., Bian Y., Weng X. RNA sequencing reveals the circular RNA expression profiles of osteoarthritic synovium. Journal of Cellular Biochemistry. 2019;120(10):18031–18040. doi: 10.1002/jcb.29106. [DOI] [PubMed] [Google Scholar]
- 55.Sun K., Luo J., Guo J., Yao X., Jing X., Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis and Cartilage. 2020;28(4):400–409. doi: 10.1016/j.joca.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y., Cai W., Han G., et al. Panax notoginseng saponins prevent senescence and inhibit apoptosis by regulating the PI3K‑AKT‑mTOR pathway in osteoarthritic chondrocytes. International Journal of Molecular Medicine. 2020;45(4):1225–1236. doi: 10.3892/ijmm.2020.4491. [DOI] [PubMed] [Google Scholar]
- 57.Xue J. F., Shi Z. M., Zou J., Li X. L. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomedicine & Pharmacotherapy. 2017;89:1252–1261. doi: 10.1016/j.biopha.2017.01.130. [DOI] [PubMed] [Google Scholar]
- 58.Honvo G., Lengelé L., Charles A., Reginster J. Y., Bruyère O. Role of collagen derivatives in osteoarthritis and cartilage repair: a systematic scoping review with evidence mapping. Rheumatology and Therapy. 2020;7(4):703–740. doi: 10.1007/s40744-020-00240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Etich J., Rehberg M., Eckes B., Sengle G., Semler O., Zaucke F. Signaling pathways affected by mutations causing osteogenesis imperfecta. Cellular Signalling. 2020;76, article 109789 doi: 10.1016/j.cellsig.2020.109789. [DOI] [PubMed] [Google Scholar]
- 60.Gencoglu H., Orhan C., Sahin E., Sahin K. Undenatured type II collagen (UC-II) in joint health and disease: a review on the current knowledge of companion animals. Animals. 2020;10(4):p. 697. doi: 10.3390/ani10040697. [DOI] [PMC free article] [PubMed] [Google Scholar]


