Abstract
c-myc has been shown to regulate G1/S transition, but a role for c-myc in other phases of the cell cycle has not been identified. Exposure of cells to colcemid activates the mitotic spindle checkpoint and arrests cells transiently in metaphase. After prolonged colcemid exposure, the cells withdraw from mitosis and enter a G1-like state. In contrast to cells in G1, colcemid-arrested cells have decreased G1 cyclin-dependent kinase activity and show hypophosphorylation of the retinoblastoma protein. We have found that overexpression of c-myc causes colcemid-treated human and rodent cells to become either apoptotic or polyploid by replicating DNA without chromosomal segregation. Although c-myc-induced polyploidy is not inhibited by wild-type p53 in immortalized murine fibroblasts, overexpression of c-myc in primary fibroblasts resulted in massive apoptosis of colcemid-treated cells. We surmise that additional genes are altered in immortalized cells to suppress the apoptotic pathway and allow c-myc-overexpressing cells to progress forward in the presence of colcemid. Our results also suggest that c-myc induces DNA rereplication in this G1-like state by activating CDK2 activity. These observations indicate that activation of c-myc may contribute to the genomic instability commonly found in human cancers.
Cell cycle checkpoints are complex systems of regulatory molecules that have evolved to maintain the fidelity of eukaryotic genomes during the G1 phase of the cell cycle, DNA replication, and chromosomal segregation (20, 42, 43, 48). The genome of normal cells is guarded by the coupling of DNA replication to the completion of a prior mitotic phase so that normal gene dosage is maintained in daughter cells. Exposure of normal cells to spindle inhibitors elicits the spindle assembly checkpoint, which does not allow cells to progress through the cell cycle until chromosomes are appropriately segregated. In Saccharomyces cerevisiae, several genes have been identified as participants in the spindle assembly checkpoint. Mutations in the BUB or MAD genes result in failure to arrest cell cycle progression in the presence of spindle inhibitors, subsequent premature DNA synthesis without cellular division, and eventually cell death (28, 36). Recent studies have identified homologous molecules in higher eukaryotes that function as components of the spindle checkpoint (11, 37, 47, 58, 59).
Defective surveillance of chromosome integrity leads to abnormalities in chromosomal structure and number, as well as other defects that have been implicated in tumorigenesis (26). Aneuploidy and chromosomal aberrations are hallmarks of most human cancers though, with the exception of p53 and the retinoblastoma (RB), the role of specific oncogene and tumor suppressor alterations in aneuploidy and chromosomal segregation defects are not well understood. The p53 tumor suppressor, which is commonly inactivated in human cancers, participates in the DNA damage and spindle assembly checkpoints (35). Loss of p53 function permits murine fibroblasts exposed to spindle inhibitors to undergo multiple rounds of DNA synthesis, becoming polyploid (12). Disruption of the RB tumor suppressor gene, which functions in G1/S control, also permits multiple rounds of DNA replication in the absence of mitosis (16, 29). Another hallmark of many common human cancers is the activation of the c-myc oncogene, whose product is a transcription factor that functions through dimerization with its partner protein, Max (2, 21, 25, 46). Overexpression of c-myc has been linked to genomic instability in cultured cells (39) and abnormal ploidy in in vivo animal models (54). We therefore sought to determine whether the c-myc oncogene is able to circumvent the cell cycle arrest induced by the spindle assembly checkpoint.
MATERIALS AND METHODS
Cell culture.
Rat1a, Rat1a-Myc, and Rat1a-Ras cells were grown as described previously (27). Rat1a-Ras cells were generated by cotransfecting Rat1a cells with EJ-ras and pBABE-puro (10:1 ratio) and selected by the addition of 0.75 μg of puromycin per ml for 2 weeks. H209, H209-Myc, H209-Ras lung cells (4), and A1N4 and A1N4-myc cells (60) were grown as described earlier. The 10(1)Val5 and VM10 cells were cultured as described previously (10), and those grown at 32°C were plated and cultured at 37.5°C until the cells were attached; the cells were then shifted to 32°C at least 8 h before any treatment was added to the medium and then grown at this temperature during the treatment. Cells were exposed to 25 ng of colcemid (Sigma) per ml for durations proportional to their doubling times.
To synchronize 10(1)Val5 and VM10 cells, the cells were incubated with mimosine (200 μM; Sigma) at 37.5°C for 22 h and then released into regular medium. Six hours after the release from mimosine, the cells were switched to 32°C and grown for 48 h with or without colcemid (25 ng/ml).
Cell cycle, apoptosis analysis, and DAPI staining.
DNA content flow cytometry and bromodeoxyuridine (BrdU) labeling was performed as described previously (55). The terminal deoxynucleotidyltransferase (TDT) end-labeling (TUNEL) assay was performed as described earlier (24). For DAPI (4′,6-diamidino-2-phenylindole) staining, cells grown as a monolayer on coverslips were washed once with phosphate-buffered saline (PBS) and fixed with 3.7% formaldehyde in PBS for 15 min at room temperature. After being washed once with PBS and permeabilized with 0.1% Nonidet P-40 (NP-40) in PBS for 10 min, the cells were stained with 0.5 μg of DAPI per ml at room temperature for 40 min and then washed with PBS vigorously before being mounted for fluorescent microscopy.
Videomicroscopy.
A1N4 and A1N4-Myc cells were plated the day before use into T25 flasks containing regular medium and then cultured for 24 h. Right before the filming, HEPES buffer (pH 7.55; 25 mM, final concentration) was added to the medium with or without colcemid (25 ng/ml), and the flasks were flushed with 5% CO2 in air for 15 s and then sealed tightly. Flasks were placed on a heated (37°C) microscope stage, and a time-lapse camera was used to make a recording every 15 s.
Western analysis.
Cell lysates were collected at different time points after exposure to colcemid, boiled in Laemmli buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to polyvinylidene difluoride membrane (Millipore). The membranes were prepared for immunoblotting as previously described (27). The antibodies for cyclins B, D, A, E, and MPM-2 were purchased from UBI. The antibodies for p34cdc2, CDK2, CDK4, and p53 (PAb240) were from Santa Cruz Biotechnology (Santa Cruz). An antibody against RB from Pharmingen was used to determine the phosphorylation status of RB. Antibody to actin was from Sigma. P21 antibody was purchased from Oncogene Research.
Immunoprecipitation of cyclin-dependent kinases and H1 kinase assay.
Cells were lysed by using immunoprecipitation buffer 1 (IPB1) containing 50 mM Tris-HCl (pH 7.4), 250 mM NaCl, 0.5 mM Na3VO5, 20 mM β-glycerophosphate, 15 mM phosphatase substrate p-nitrophenylphosphate, protease inhibitor Complete tablets (Boehringer Mannheim), and 0.1% NP-40. The protein concentration was measured by using the BCA kit (Pierce). Antibody against p34cdc2, CDK2, and CDK4 (all from Santa Cruz) were incubated with protein G beads in IPB1 at 4°C for 2 h and then washed once with IPB1. The washed protein G beads loaded with antibody were incubated with 300-μg lysate aliquots at 4°C for 2 h and washed three times with IPB1. Half of the beads with antibody-antigen complexes were used for the H1 kinase assay. Kinase assays were performed in 50 μl of kinase assay buffer containing 50 mM Tris (pH 7.4), 1 mM CaCl2, 5 mM MgCl2, 0.5 mM Na3VO5, 20 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 1 mM dithiothreitol, protease inhibitor Complete tablets, 50 μM ATP, 0.2 μg of purified histone H1 (Boehringer Mannheim) per ml, and 10 μCi of [γ-32P]ATP at 30°C for 30 min. The reaction was stopped by the addition of 50 μl of 2× Laemmli buffer and heating at 95°C for 5 min. The products were separated by SDS-PAGE, and autoradiography was then performed.
Metabolic labeling and immunoprecipitation of p53.
10(1)Val5 and VM10 cells were plated and grown at 37°C. Cells grown at 32°C were shifted to this temperature 12 h before being labeled. The cells were starved for methionine in Dulbecco modified Eagle medium with 2% fetal bovine serum for 1 h and then labeled with [35S]methionine at 20 μCi/ml for 4 h. The cells were lysed in IPB2 (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5 mM Na3VO5, 20 mM β-glycerophosphate, protease inhibitor Complete tablets, and 1% NP-40). Then, 0.3 g of lysate was incubated with Gamma-Bind beads (Pharmacia) at 4°C for an hour. The precleared lysates were incubated with monoclonal antibody PAb240 (Santa Cruz) overnight. The antigen-antibody complexes were incubated with fresh Gamma-Bind beads for 2 h and washed five times with IPB2. The immunoprecipitated products were separated by SDS-PAGE, and autoradiography was performed.
Luciferase reporter assay.
10(1)Val5 or VM10 cells (105) were plated into 10-cm dishes the day before transfection. Control luciferase reporter vector or luciferase reporter with wild-type or mutated p21 promoter was transfected by lipofection into these cells according to the manufacture’s protocol (GIBCO). The luciferase assay was performed 48 h after transfection according to the manufacture’s protocol (Promega).
RESULTS
Uncoupling of DNA replication from mitosis by overexpressed c-myc in rodent and human cells.
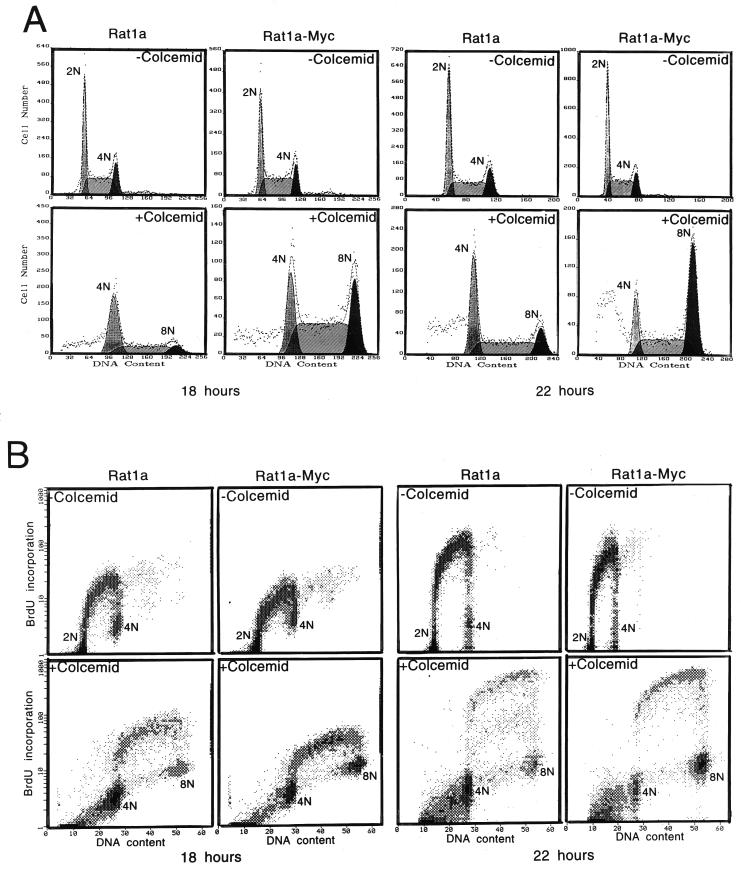
To determine whether c-myc is able to affect the spindle checkpoint, we assessed the ability of cells transformed by c-myc to arrest after exposure to the spindle inhibitor colcemid. The duration of exposure to colcemid for the different cell lines was varied to correspond to their doubling times. The distribution of cells in various phases of the cell cycle was analyzed by flow cytometry with the DNA stain propidium iodide. The DNA content in cells in G1, G2/M, and in cells that contained increased DNA content compared to cells in G2/M were designated as 2N, 4N, and 8N, respectively, for all cell lines. Rat1a fibroblasts are unique in that a single oncogenic event, such as overexpression of c-myc or ras alone, is sufficient to transform them (27, 56). Untransformed Rat1a fibroblasts exposed to colcemid for 18 h (ca. one doubling time) accumulated primarily with a 4N DNA content, and only a minority of cells entered the subsequent S phase prematurely (Fig. 1A). In contrast, the c-myc-transformed Rat1a-Myc cells displayed a >2-fold decrease in the percentage of arrested cells at 4N compared with Rat1a cells, and the majority entered subsequent S phase after treatment with colcemid; one-third of these became octoploid. After 22 h of colcemid exposure, >80% of the Rat1a-Myc cells contained DNA content greater than 4N, although this number is probably an underestimate due to a fraction of cells that underwent apoptosis, as will be described later.
FIG. 1.
Formation of polyploid population by overexpressed c-myc in the presence of colcemid in rodent and human cells. (A) DNA content (propidium iodide staining) flow cytometric histograms of untreated and colcemid-treated Rat1a and Rat1a-Myc cells. Cells with or without colcemid (25 ng/ml) were incubated for 18 and 22 h. Primary data are displayed as dots, and the computer best fit of data is shown as dashed lines and hatched contours. Without colcemid, the cells in the 2N, 2N-to-4N, and 4N populations account for 38.6, 43.8, and 17.6% for Rat1a and 35.4, 49.7, and 14.9% for Rat1a-Myc cells, respectively. With colcemid for 18 h, 53.8 and 23.0% of cells were arrested at 4N for Rat1a and Rat1a-Myc, respectively. Of the Rat1a cells, 36.5% were in the 4N-to-8N population, and 9.7% had 8N DNA content, while the percentages of the 4N-to-8N and 8N populations for Rat1a-Myc were 54.1 and 22.8%, respectively. After 22 h of colcemid incubation, 40.8 and 16.7% of the Rat1a and Rat1a-Myc cells were arrested at 4N, respectively. Of the Rat1a cells, 43.4% were in the S phase after 4N, and 15.7% accumulated at 8N, whereas 36.4% of Rat1a-Myc cells were in the S phase after 4N, and 46.9% were octaploid. (B) Two-dimensional BrdU incorporation (y axis) and DNA content (x axis) flow cytometric diagrams. The DNA contents of the cells are indicated in the diagrams. All experiments were replicated in separate studies. (C) Flow cytometric histograms of immortalized human breast epithelial A1N4 and A1N4-Myc cells with or without exposure to colcemid (25 ng/ml) for 24 and 48 h. Without colcemid, 57.1% of A1N4 cells were in G1, 28.7% were in S phase, and 14.2% were in G2/M phase. The percentages of A1N4-Myc cells in the G1, S, and G2/M phases were 49.0, 38.8, and 12.2%, respectively. With colcemid exposure for 24 h, 80.8% of A1N4 cells were arrested in G2/M, 15.5% were in the subsequent S phase, and 3.7% accumulated at 8N. At the same time point, 81.3% of the A1N4-Myc cells were arrested at 4N, 12.2% were in the interval from 4N to 8N, and 6.6% were at 8N. At 48 h after colcemid treatment, 68.8% of the A1N4 cells were arrested at 4N, 23.4% were in interval from 4N to 8N, and 7.8% were at 8N, whereas only 25.0% of the A1N4-Myc cells were arrested in G2/M, 31.8% of A1N4-Myc cells were within the interval from 4N to 8N, and 43.2% had a DNA content of 8N.
Whether colcemid-arrested cells were able to actively replicate DNA was assessed by measuring BrdU incorporation. The cells with DNA content between 4N and 8N incorporated BrdU (Fig. 1B), confirming that the 8N population arose from DNA rereplication of 4N cells. The absence of staining in the region below 4N suggests the failure of cellular division in both Rat1a and Rat1a-Myc cells after exposure to colcemid.
To determine whether wild-type c-Myc is required for the rereplication of DNA in the absence of cytokinesis, we studied transfected Rat1a cells that express a mutant c-Myc with a substitution of tryptophan to glutamate at amino acid position 135. The tryptophan-to-glutamate substitution generated at amino acid 135 in human c-Myc is identical to the transformation-inactive murine c-Myc mutant that is incapable of interacting with the novel Myc-associated protein TRRAP (33a, 40). The Rat1a cells overexpressing mutant Myc did not transform and failed to display premature DNA replication in the presence of colcemid. In addition, the phenotype we observed is not a drug-specific response since wild-type myc-overexpressing cells became polyploid when exposed to another microtubule inhibitor, vinblastine, a commonly used cancer therapy drug. In contrast, vinblastine-treated Rat1a cells were arrested with a 4N DNA content (data not shown).
We sought to determine whether this Myc-induced phenotype is restricted only to rodent cells, since it has been reported that human cells are different from rodent cells in their response to microtubule inhibitors (32). Our studies with an immortalized human breast epithelial cell line, A1N4, which can be transformed by overexpression of c-myc alone (60), suggested that overexpression of c-myc also is able to uncouple DNA replication from mitosis in human cells (Fig. 1C). As observed with the rat fibroblasts, deregulated c-myc expression in the A1N4-Myc cells caused premature DNA replication in cells treated with colcemid. At 24 h, >80% of both A1N4 and A1N4-Myc cells were arrested with a 4N DNA content, but during the next 24 h, three-quarters of the A1N4-Myc cells entered the subsequent S phase, whereas ca. 70% of the A1N4 cells remained arrested at 4N.
It is notable that the overexpression of c-myc does not significantly change the growth rate in these cells. The doubling times for Rat1a and Rat1a-Myc cells are 14.1 and 12.4 h, respectively, and those for A1N4 and A1N4-Myc cells are 18.1 and 17.3 h, respectively. These minor changes make it implausible that the phenotypic outcomes were due to differences in growth rates.
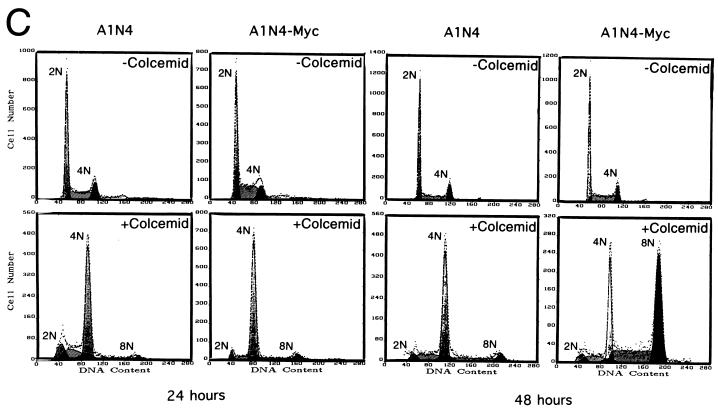
Transformation is insufficient to enhance DNA rereplication in the absence of mitosis.
We sought to determine whether the uncoupling of DNA replication from mitosis was associated with the oncogenic transformation of Rat1a cells or linked to deregulated c-myc expression. To do so, we assessed the ability of activated ras to induce premature DNA synthesis in colcemid-treated cells. Rat1a-Ras cells behaved like the Rat1a control cells when exposed to colcemid. Namely, cells with a 4N DNA content accumulated and a few entered a subsequent S phase (Fig. 2A). The Rat1a-Ras cells were, however, fully transformed and readily formed colonies in soft agar (data not shown), as previously reported (52). These observations indicate that the ras-induced transformed phenotype was insufficient to uncouple mitosis from DNA replication, suggesting that the phenotype seen in Rat1a-Myc cells is not simply a consequence of the transformed phenotype.
FIG. 2.
Overexpression of c-myc but not ras induces polyploidy. (A) DNA content flow cytometric histograms of Rat1a, Rat1a-Myc, and transformed Rat1a-Ras fibroblasts with or without exposure to colcemid (25 ng/ml) for 18 h. Culture conditions were as described in Fig. 1. The percentages of untreated cells in the G1, S, and G2/M phases were 39.7, 39.9, and 20.4%, respectively, for Rat1a; 39.7, 42.1, and 18.2%, respectively, for Rat1a-Myc; and 39.6, 38.4, and 22.0%, respectively, for Rat1a-Ras. With 18 h of treatment, the percentages of cells with 4N, 4N-to-8N, and 8N DNA contents were 40.9, 43.5, and 15.6%, respectively, for Rat1a; 25.6, 47.7, and 26.7%, respectively, for Rat1a-Myc cells; and 44.0, 36.6, and 19.4%, respectively, for Rat1a-Ras cells. (B) Flow cytometric histograms of human lung carcinoma NCI H209, H209-Myc, and H209-Ras cells with or without exposure to colcemid (25 ng/ml) for 72 h. The percentages of untreated cells in the G1, S, and G2/M phases were 46.8, 17.3, and 35.9%, respectively, for H209 cells; 48.6, 25.0, and 26.4%, respectively, for H209-Myc cells; and 50.0, 25.0, and 25.1%, respectively, for H209-Ras cells. After 72 h of exposure to colcemid, the percentages of cells with DNA contents of 4N, 4N to 8N, and 8N were 64.8, 16.0, and 19.2%, respectively, for H209 cells; 22.2, 24.4, and 53.5%, respectively, for H209-Myc cells; and 67.3, 8.9, and 23.8%, respectively, for H209-Ras cells.
To further confirm our observations, we studied an established human small cell lung carcinoma cell line, NCI H209 (4), which expresses very low levels of endogenous c-myc. The cell line was derived from a cancer and contains a mutated inactive RB (6). When exposed to colcemid, a small fraction of the parental H209 cells escaped 4N arrest and entered the subsequent S phase (Fig. 2B). This result is consistent with a previous study suggesting that the loss of RB function leads to premature DNA synthesis and polyploidy (16). Overexpression of c-myc in H209 cells markedly enhances polyploidy, in contrast to cells overexpressing activated ras. The doubling times for H209, H209-Myc, and H209-Ras cells are 43, 30, and 34 h, respectively, and are not proportional to the fraction of cells with >4N DNA content. In aggregate, these studies indicate that deregulated c-myc expression is able to accelerate the uncoupling of DNA replication from mitosis in both rodent and human cells.
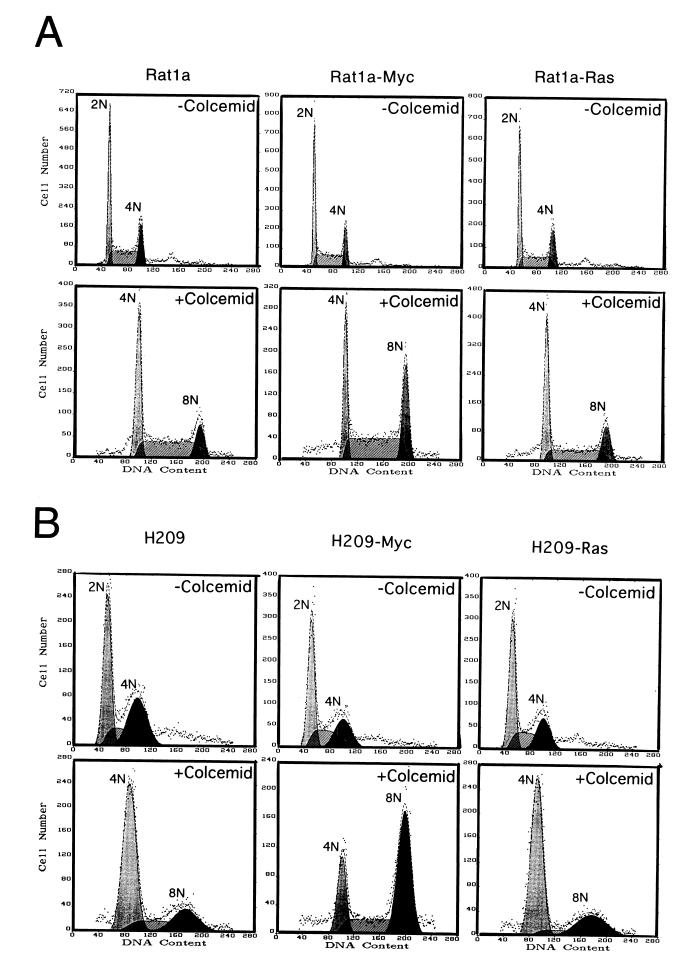
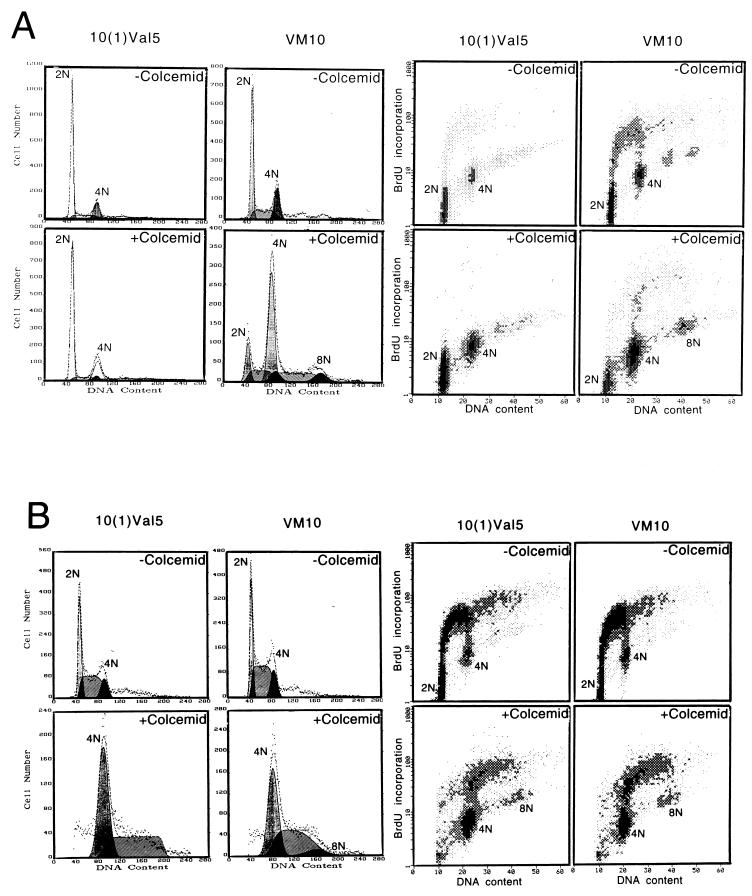
c-myc uncouples DNA replication from mitosis in the presence of p53.
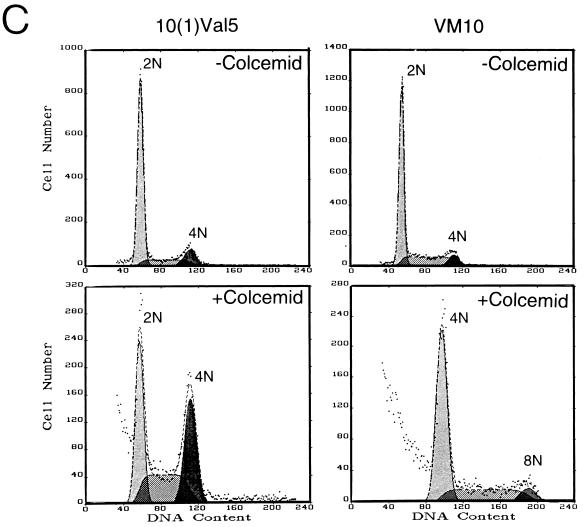
Because loss of p53 allows murine fibroblasts to overcome the arrest imposed by a spindle checkpoint, we determined whether p53 status affects the ability of c-myc overexpression to uncouple DNA replication from mitosis. p53 null murine fibroblasts that express either a temperature-sensitive p53 allele [10(1)Val5] or temperature-sensitive p53 together with murine c-myc (VM10) (10) were exposed to colcemid (Fig. 3A). At the permissive temperature of 32°C, overexpressed p53 in the wild-type conformation arrested the majority of 10(1)Val5 cells in G1, with only a small fraction of cells incorporating BrdU. With colcemid exposure for 48 h, most of these cells remained arrested in G1. In contrast, the VM10 cells that express c-myc ectopically showed a lower percentage in G1 without colcemid and underwent DNA synthesis and became octaploid with exposure to colcemid at 32°C.
FIG. 3.
Overexpression of c-myc induces polyploidy in the presence of overexpressed wild-type p53 in a murine fibroblast cell line. (A) Cells cultured at 32°C were left untreated or were treated with colcemid (25 ng/ml) for 48 h. Murine BALB/c fibroblasts line 10(1)Val5 expresses a temperature-sensitive p53 allele; VM10 is derived from 10(1)Val5 by transfection of a murine c-myc expression vector. The two left panel columns show DNA histograms, and the two right panel columns represent BrdU and propidium iodide two-dimensional flow cytometric diagrams. The percentages of untreated cells in 2N, 2N to 4N, and 4N were 74.6, 8.1, and 17.4%, respectively, for 10(1)Val5 cells and 49.6, 30.4, and 20.0%, respectively, for VM10 cells. With colcemid exposure, 57.9% of the 10(1)Val5 cells were in 2N, 13.6% were in 2N to 4N, and 28.6% were in 4N. Meanwhile, 9.3% of VM10 cells were in 2N, 11.4% were in 2N to 4N, 61.1% were in 4N, 12.9% were in 4N to 8N, and 5.7% were in 8N. (B) Cells grown at 37.5°C were left untreated or were treated with colcemid for 24 h. The two left panel columns show DNA histograms, and the two right panel columns represent BrdU and propidium iodide two-dimensional flow cytometric diagrams. Without colcemid, the percentages of cells in 2N, 2N to 4N and 4N were 36.7, 48.0, and 15.2%, respectively, for 10(1)Val5 and 32.0, 52.3, and 15.8%, respectively, for VM10 cells. With colcemid exposure, 52.2% of the 10(1)Val5 cells were in 4N and 47.8% were in 4N to 8N, whereas 45.9% of the VM10 cells were in 4N, 46.9% were in 4N to 8N, and 7.2% were in 8N. (C) The effect of colcemid on synchronized 10(1)Val5 and VM10 cells at 32°C. Cells were synchronized by incubating with mimosine (200 μM) at 37.5°C for 22 h and then released into regular medium. At 6 h after their release from mimosine, the cells were switched to 32°C and grown for 48 h with or without colcemid (25 ng/ml). Without colcemid, the percentages of cells in 2N, 2N to 4N, and 4N were 73.3, 14.6, and 12.0%, respectively, for 10(1)Val5 cells and 67.2, 25.7, and 7.1%, respectively, for VM10 cells. With colcemid exposure, 33.4% of the 10(1)Val5 cells were in 2N, 29.2% were in 2N to 4N, 30.7% were in 4N, 5.6% were in 4N to 8N, and 1.0% were in 8N. With colcemid exposure, most of the viable VM10 cells had a DNA content equal to or higher than 4N with 65.0% in 4N, 28.1% in 4N to 8N, and 6.9% at 8N.
When cultured at the nonpermissive temperature of 37.5°C, at which the temperature-sensitive p53 was inactive, the 10(1)Val5 cells displayed premature DNA synthesis and some octaploidy with colcemid exposure (Fig. 3B). This phenotype was also seen in p53 null parental 10(1)Val5 cells (data not shown) and is consistent with previously reported observations (12). c-myc overexpression in the absence of wild-type p53 in colcemid-exposed VM10 cells had no significant effect on premature BrdU incorporation and octoploidy.
We further studied the effects of wild-type p53 under conditions that lessened its ability to cause G1 arrest in the 10(1)Val5 cells and diminish the effects of colcemid at G2/M. The 10(1)Val5 and VM10 cell lines were first cultured at the nonpermissive temperature of 37.5°C, synchronized with mimosine at G1/S, released from mimosine into colcemid containing medium, and then cultured at the permissive temperature of 32°C. Under these conditions, the 10(1)Val5 cells were able to traverse into S and G2, but they were arrested by colcemid at 4N (Fig. 3C). In contrast, the VM10 cells continued to cycle, displaying 30% of cells with DNA content greater than 4N. These observations suggest that c-myc is able to bypass the spindle and G1/S checkpoints in the presence of wild-type p53 in immortalized murine fibroblasts.
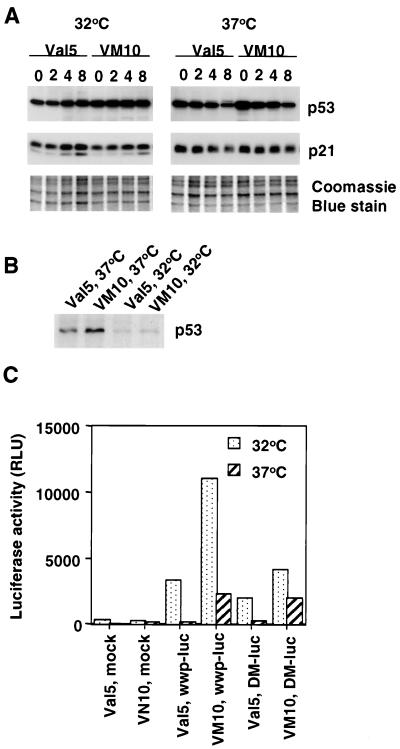
To verify the status of p53 in 10(1)Val5 and VM10 cells, we measured the induction of p21 by the chemotherapeutic agent etoposide (VP16) as a surrogate marker of p53 function (9) (Fig. 4A). p21 is a p53-responsive gene that binds to and inhibits cyclin-dependent kinases in G1 phase. In both 10(1)Val5 and VM10 cells, at the permissive temperature of 32°C, p21 was clearly induced by etoposide, while at the nonpermissive temperature of 37°C it remained unchanged after etoposide treatment, suggesting that p53 is functioning at 32°C but not at 37°C. We also noticed that both 10(1)Val5 and VM10 cells express a much higher level of p53 compared to primary rat embryonic fibroblasts or immortalized murine fibroblasts NIH 3T3 cells (data not shown). Overexpression of c-myc in VM10 cells does not change the total amount of p53 expressed at either temperature (Fig. 4A).
FIG. 4.
Status of p53 in (10)1Val5 (Val5) and VM10 cells. (A) Immunoblot of p21 induction by etoposide as a surrogate marker of p53 function. Val5 and VM10 cells were collected at different times (shown in hours at the top) after exposure to etoposide (40 μg/ml). A Coomassie blue-stained gel was used as loading control. (B) Immunoprecipitation of metabolically labeled p53 in Val5 and VM10 cells. (C) Luciferase reporter assay with p21 promoter. Cells were transfected with either a control promoterless vector with luciferase reporter gene (Mock), the wild-type 2.4-kb p21 promoter cloned into the reporter vector (wwp-luc), or a mutated p21 promoter with the p53 responsive element at 2.4 kb upstream deleted in the reporter vector (DM-luc) (19). Luciferase activity is shown as relative light units (RLU).
We further studied the status of p53 in these cells by immunoprecipitation with an antibody that recognizes the mutant but not wild-type p53 under nondenaturing conditions. PAb240 antibody precipitated a much larger amount of p53 from cells grown at 37°C than from those grown at 32°C (Fig. 4B) in both (10)1Val5 and VM10 cells, confirming that most of p53 is in mutant conformation at 37°C and wild-type at 32°C. Here again, overexpression of c-myc did not change the status of p53 in VM10 cells.
We also took a third approach to look at the p53 status. We performed a luciferase reporter assay with either a wild-type p21 promoter or a p21 promoter in which a p53-responsive element was deleted (19). At the permissive temperature of 32°C, the luciferase activity decreased twofold if the p53-responsive element is deleted in p21 promoter, while it remained the same at the nonpermissive temperature of 37°C in both (10)1Val5 and VM10 cells (Fig. 4C). Hence, our results from p21 induction by etoposide, immunoprecipitation by p53 mutant specific antibody and p21 promoter reporter assay all suggest that p53 is functional at the permissive temperature and inactive at the nonpermissive temperature in both (10)1Val5 and VM10 cells. c-myc overexpression does not seem to change p53 status in these cells.
To determine the status of p53 in other cells, we also performed reporter assays in Rat1a, Rat1a-Myc, A1N4, and A1N4-Myc cells. Deletion of the p53-responsive element in the p21 promoter also decreased the luciferase activities in these cells (data not shown), implying that p53 in these cells is also functional. In addition, it was previously reported that p53 is wild type in Rat1a cells (50). Sequence analysis revealed that p53 in A1N4 and A1N4-Myc cells are identical and do not bear hot spot coding sequence mutations (15).
Susceptibility of c-myc-overexpressing cells to apoptosis in the presence of colcemid.
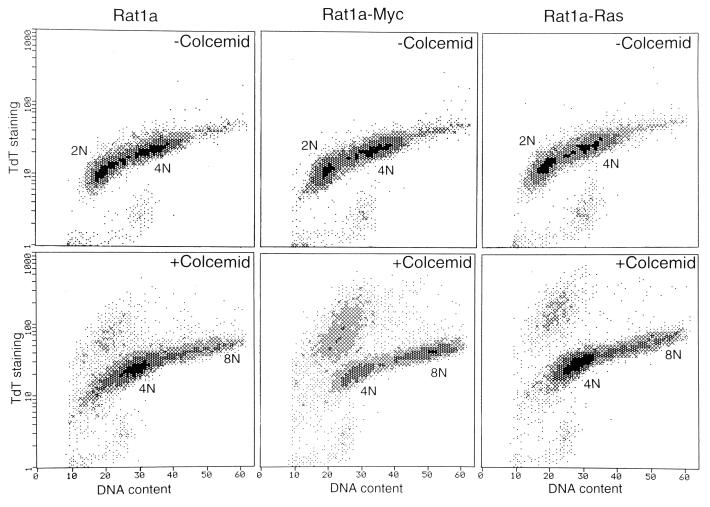
We determined whether c-myc potentiates the death of mammalian cells that fail to arrest in the presence of spindle-disrupting drugs. Our initial observations showed that polyploid cells were not recovered in long-term cultures. The extent of apoptosis was then determined by the TUNEL assay (23). Compared with Rat1a or Rat1a-Ras cells, Rat1a-Myc cells showed a much higher fraction of tetraploid cells that was stained by biotinylated-dUTP (Fig. 5), which is incorporated to nicked DNA by TDT, indicating that a large proportion of these cells underwent apoptosis.
FIG. 5.
Apoptosis of colcemid-treated Rat1a, Rat1a-Myc, and Rat1a-Ras cells. Cells were treated with colcemid for 18 h. DNA content determined by propidium iodide staining is shown on the x axis. A TDT end-labeling assay (i.e., a TUNEL assay) was performed as described by Gorczyca et al. (24). Without colcemid, the level of staining is lower than 3% for all three cell lines. With colcemid, 15.2% of Rat1a, 36.2% of Rat1a-Myc, and 13.3% of Rat1a-Ras cells were stained.
We also studied the effect of c-myc in a nonimmortalized cell culture, rat embryonic fibroblasts, which are similar to the precursors of the Rat1a cell line. The cells were infected with retroviruses expressing either human c-myc or a control virus with empty vector. When these cells were exposed to colcemid (25 ng/ml) at a concentration that is able to halt control cells at a 4N DNA content, the vast majority of c-myc-overexpressing cells underwent apoptosis (data not shown), which is consistent with our results with the Rat1a cell lines. Propidium iodide staining was unable to detect cells beyond 4N in the c-myc-overexpressing population, suggesting that additional genes are required to suppress the apoptotic pathway and push the cells forward to become octoploid (data not shown).
Adaptation of mitotic arrest after prolonged exposure to colcemid.
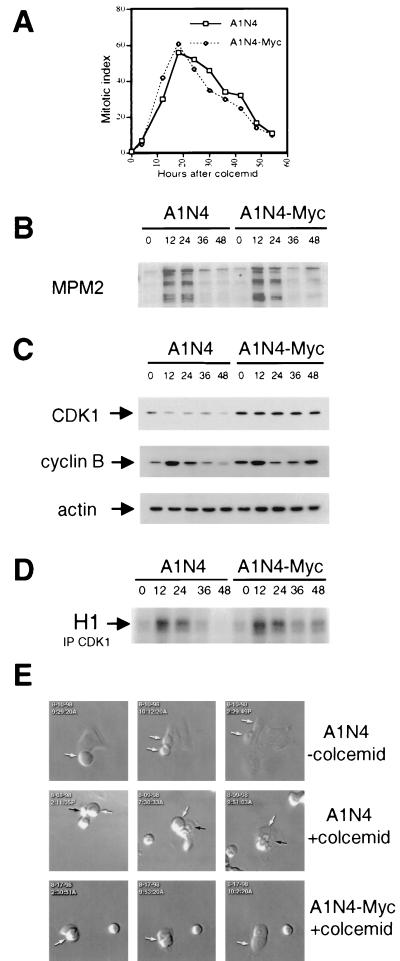
To delineate the means by which c-myc overexpression uncouples DNA replication and mitosis, we tested the hypothesis that either c-myc suppresses metaphase arrest or activates S-phase entry prematurely. We chose the human breast epithelial cells A1N4 for further studies, although comparable studies in Rat1a cells yielded similar results. We measured the mitotic index of A1N4 and A1N4-Myc cells by staining the cells with the DNA dye DAPI and assessing the percentage of mitotic cells with condensed chromosomes at different intervals after treatment with colcemid (Fig. 6A). In the first 20 h, both asynchronous A1N4 and A1N4-Myc cells showed an increased number of cells with condensed chromosomes and reached a maximal mitotic index at ca. 20 h, indicating a mitotic arrest. Thereafter, the number of mitotic cells declined, gradually dropping, although still maintaining a level higher than the untreated cells. This suggests that both A1N4 and A1N4-Myc cells withdraw from mitosis after prolonged exposure to colcemid.
FIG. 6.
Adaptation of human breast epithelial A1N4 and A1N4-Myc cells after prolonged exposure to colcemid. (A) Mitotic index of A1N4 and A1N4-Myc at different intervals of colcemid incubation. Asynchronous cells were treated with colcemid and fixed and stained with DAPI at different times. The mitotic index was measured by counting the percentage of cells with condensed chromosomes. (B) Immunoblot analysis of mitosis-specific phosphoepitope with MPM-2 antibody in lysates from asynchronous A1N4 or A1N4-Myc cells at various times (shown at the top in hours) after exposure to colcemid. (C) Protein levels of CDK1-cyclin B complex. The actin level was used as a loading control. (D) Autoradiogram of phosphorylated histone H1. Histone H1 kinase activities were measured from p34cdc2 immunoprecipitates (IP) of A1N4 or A1N4-Myc cell lysates obtained at different times after treatment with colcemid. (E) Videomi- croscopic photographs of A1N4 cells without (upper panel) or with (middle panel) colcemid exposure, as well as A1N4-Myc cells with colcemid treatment (lower panel). Arrows mark individual cells during time-lapse microscopy.
To further verify the exit of colcemid-treated A1N4 cells from mitosis, we examined total cellular extracts with the MPM-2 antibody, which recognizes mitosis-specific phosphoepitopes (13). Western analysis showed a correlation between MPM-2 reactivity and the mitotic index (Fig. 6B). This confirmed that both A1N4 and A1N4-Myc cells exit from mitosis after the transient block, after which A1N4 cells remain arrested in interphase. As a further confirmation of the progression of mitotic exit, we assayed the major enzyme p34cdc2 kinase (CDK1) that controls mitosis in eukaryotes. Activation of p34cdc2 promotes the initiation of mitosis, and its inactivation is normally required for exit from mitosis. While the protein level of p34cdc2 remained relatively constant, the level of its regulator, cyclin B, increased and then decreased in both A1N4 and A1N4-Myc cells during the first 24 h of colcemid exposure (Fig. 6C). Correspondingly, the histone H1 kinase activity of immunoprecipitated p34cdc2 rose and then declined in both cell lines within 24 h (Fig. 6D). Morphologically, colcemid-exposed A1N4 and A1N4-Myc cells both display aborted attempts to undergo mitosis with failed cytokinesis (Fig. 6E). Colcemid-treated cells observed by time-lapse videomicroscopy were rounded and demonstrated chromosomal condensation, followed by a lack of cytokinesis and subsequent flattening of the cells. Our results are consistent with previous studies suggesting that an adaptation or so-called “mitotic slippage” occurs in mammalian cells after prolonged exposure to microtubule inhibitors (3, 32, 33).
We also noted that, after the initial drop, there was a slight increase in MPM-2 reactivity and p34cdc2 H1 kinase activity only in A1N4-Myc cells, while those in A1N4 cells remained low (Fig. 6B, C, and D). The level of cyclin B showed an earlier and greater increase in A1N4-Myc cells after the decline, suggesting that, after withdrawing from mitosis, A1N4-Myc cells moved forward to cycle again, whereas A1N4 cells remained arrested in a G1-like state. Taken together, our measurements of mitotic index, MPM-2 reactivity, cyclin B level, and p34cdc2 kinase activity all suggest that A1N4 cells exit from mitosis and advance forward into a G1-like state, at which point they arrest. In contrast, the A1N4-Myc cells are able to overcome this G1-arrest state and continue cell cycle progression, suggesting that c-myc overrides the inhibition of DNA rereplication in the G1-arrested condition.
G1 cyclin-dependent kinase activities in colcemid-treated cells.
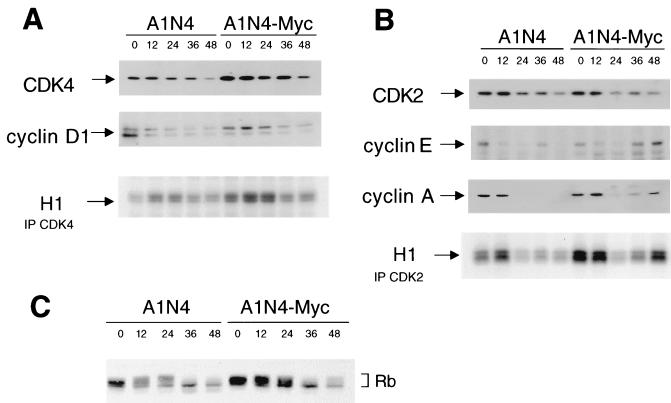
Since our results suggest an arrest of colcemid-treated A1N4 cells in G1-like phase and that A1N4-Myc cells are able to traverse through this G1 phase and initiate premature DNA replication, we sought to measure the activities of CDK4 and CDK2 kinases that regulate G1 and G1/S transition (53). CDK4 complexes, which are composed of the CDK4 catalytic subunit and cyclin D regulatory subunit family of proteins, promote S-phase entry by phosphorylating the retinoblastoma protein RB, releasing sequestered E2F transcription factors. Unleashed E2F transactivates downstream target genes, including those involved in the synthesis of DNA precursors, in DNA replication, and in the regulation of G1/S transition, such as cyclin E (14). Cyclin E and cyclin A are regulatory subunits of CDK2, whose activation triggers S-phase entry. CDK2 also phosphorylates RB to form an autoregulatory feedback loop which ensures the completion of G1/S transition. Our results showed that the levels of CDK4 and its regulator, cyclin D1, decreased slightly during exposure to colcemid, but the histone H1 kinase activity decreased in both A1N4 and A1N4-Myc cells over time (Fig. 7A). In contrast, while the protein level of CDK2 was unchanged during colcemid treatment, the levels of the regulatory subunits, cyclin E and cyclin A, showed a decrease first in both A1N4 and A1N4-Myc cells but rose again only in A1N4-Myc cells (Fig. 7B). Accordingly, the CDK2 histone H1 kinase activity paralleled the change in cyclin levels.
FIG. 7.
Activities of G1 kinases and phosphorylation status of RB protein in colcemid-treated A1N4 cells. (A) Protein abundance and kinase activity of CDK4-cyclin D1 complexes. Histone H1 kinase activities were measured from CDK4 immunoprecipitates (IP) of A1N4 or A1N4-Myc cell lysates obtained at various times after treatment with colcemid. (B) Protein abundance and kinase activity of CDK2-cyclin complexes. Histone H1 kinase activities were measured from CDK2 immunoprecipitates of A1N4 or A1N4-Myc cell lysates obtained at various times after treatment with colcemid. (C) Immunoblot analysis of RB protein in lysates obtained at various time points shows RB hypophosphorylation with colcemid exposure.
We studied the phosphorylation state of RB after treatment with colcemid, since hypophosphorylated RB arrests normal cells in G1 phase, and its phosphorylation relieves this inhibition (62, 63). Western analysis detected multiple phosphorylation forms of RB in asynchronous A1N4 and A1N4-Myc cells (Fig. 7C). In both cells, after constant exposure to colcemid, a hypophosphorylated form of RB appeared and remained the major species of RB after 36 h. This observation, together with the CDK data, suggests the possibility that the arrest in colcemid-treated A1N4 cells is imposed through a CDK4-RB axis. Furthermore, A1N4-Myc cells showed increased CDK2 activity and were able to overcome dephosphorylated RB.
DISCUSSION
The events of the cell cycle are ordered into pathways in which the initiation of late events is dependent on the completion of early events. This coordination and timing of the cell cycle is thought to be regulated by cell cycle checkpoints that ensure that critical events, such as DNA replication and chromosome segregation, are completed with high fidelity. In addition, checkpoints respond to damage by arresting the cell cycle to provide time for repair and by inducing transcription of genes that facilitate repair.
Adaptation of the mitotic checkpoint and the G1/S gatekeepers.
The mitotic checkpoint is tightly regulated and phylogenetically well conserved. While wild-type S. cerevisiae cells respond to microtubule inhibitors by arresting in metaphase, mutations in mitotic checkpoint genes fail to inhibit the metaphase-to-anaphase transition in the presence of inhibitors and thus allow the cells to progress through the cell cycle (20, 48). Because cytokinesis is still inhibited in the presence of microtubule inhibitors, rereplication of DNA in the subsequent S phase gives rise to polyploid cells.
In a survey of 14 rodent and human cell lines, Kung et al. (32) found that human cell lines respond to microtubule inhibitors differently than do rodent cells. The rodent cell lines tend to become polyploid with a fluctuation in the level of p34cdc2 activity, while most of the human cells remain arrested with a high level of cdc2 activity. In our study, the Rat1a fibroblast cell line did show a small population that escapes the 4N arrest. In contrast to the study by Kung et al., we showed that the human breast epithelial cell line did not stay permanently arrested in mitosis in the presence of spindle-disrupting agents but instead transited to the next G1-like stage. This adaptation, termed “mitotic slippage,” has been documented (3, 32, 41) when cells are exposed to damage for an extended period of time. As a consequence, the cells proceed through the cell cycle and enter the subsequent G1 phase, although the original perturbation persists.
Normal cells undergoing mitotic slippage must have developed multiple checkpoint pathways in response to intrinsic (mutations in spindle structure or kinetochore components) or extrinsic (microtubule inhibitors) signals to prevent premature entry into the next S phase. In mammalian cells, the tumor suppressor proteins, p53 and RB, as well as the cyclin-dependent kinase inhibitor p21, seem to be necessary for the G1 arrest of cells exposed to microtubule inhibitors (12, 16, 29, 33). When these proteins are inactivated, cells become polyploid when exposed to microtubule inhibitors. However, it appears that p53 is neither expressed in mitosis nor required for the metaphase arrest and is only expressed after the cells entered a G1-like stage (41). Recently, time-lapse videomicroscopy revealed that the p53 wild-type cells and the p53−/− rodent fibroblasts showed the same length of mitotic arrest by microtubule inhibitors before they entered a G1-like state (33). The lack of evidence of an immediate role of p53 in mitosis after exposure to these drugs favors the hypothesis that p53 only exerts its guarding effects in preventing an undivided cell from going into the next S phase rather than shortening the metaphase-anaphase interval. The roles of RB and p21 in G1 arrest after mitotic slippage are likewise implicated in regulating G1-to-S transition rather than in the mitotic checkpoint per se.
c-myc, genomic instability, and neoplastic progression.
By allowing repair to take place, checkpoint controls become crucial in maintaining genomic stability. Checkpoint loss often results in genomic instability and has been implicated in the evolution of normal cells into cancer cells that typically display aneuploidy and chromosomal abnormalities (26). In budding yeast, the mad and bub mutants show an increased frequency of spontaneous chromosome loss. Recently, mutations in BUB1 genes were found in human colorectal cancers (8), supporting the hypothesis that disrupted checkpoints contribute to tumor progression. Inactivation of the tumor suppressor gene encoding p53 eliminates cell cycle checkpoints and enhances the frequency of genomic rearrangements. Although both BUB1 and p53 genes participate in the cell cycle arrest imposed by spindle-disrupting agents, they function through distinct modes: BUB1-mutated cells escape the arrest in mitosis, but in p53 null cells the G1/S block is compromised, whereas the mitotic arrest checkpoint pathway seems to be intact.
Deregulation of c-myc is very frequently reported in human cancer cells that are often characterized by their altered genomes. Overexpression of inducible c-myc caused DNA amplifications in Rat1 fibroblasts (39), and ectopically overexpressed c-myc cooperates with transforming growth factor α to induce aneuploidy in the livers of transgenic mice (54). Although these observations are consistent with a link between c-myc and checkpoints guarding chromosome integrity, no direct evidence has been provided. In this study, we showed that the overexpression of c-myc disrupts the spindle checkpoint elicited by microtubule inhibitor and induces polyploid cells. Our results suggest that c-myc is likely to disrupt the G1-arrest checkpoint pathway involving p53 and RB, since the myc-overexpressing cells still undergo mitotic arrest, as do the parental cells, but rereplicate DNA after cells enter the G1 phase without cytokinesis. As such, this study indicates that deregulated c-myc may contribute to the genomic instability frequently found in tumor cells.
In one primary cell line, rat embryonic fibroblasts, overexpression of c-myc induced massive apoptosis in the presence of colcemid (data not shown). No population beyond 4N was detected in these cells treated with colcemid, suggesting that additional genes are required to suppress the apoptotic pathway and allow the cells to progress forward. This observation reflects the concept that multiple protective pathways are activated by spindle inhibitors to guard against premature cell cycle progression without cellular division. Recent work by Roussel, Sherr, and coworkers suggests that the overexpression of c-myc in murine embryonic fibroblasts induces p19/arf and p53 in turn, which might be responsible for increased apoptosis in low serum levels in these cells (65). In addition, cells that survived crisis and become immortalized by c-myc display a loss of p19 or p53. Therefore, loss of these genes might be required for c-myc to induce this phenotype in immortalized cells. A recent report showed that c-myc overexpression and p53 loss cooperate to promote polyploidy in a murine myeloid cell line (64). However, another study (41) indicates that the loss of p53 alone can cause polyploidy but that most of these cells will undergo apoptosis unless Bcl-xL is overexpressed. Hence, the loss of p53 does not necessarily protect against apoptosis when cells are exposed to spindle-damaging agents.
Although c-myc has been connected with cell cycle control in a variety of systems (2, 5), the precise cell cycle components at which c-myc exerts its effects remain to be determined. In our study, the fact that myc triggers S-phase entry in the presence of hypophosphorylated RB suggests that c-myc activates an alternative or downstream pathway independent of RB. Although RB/E2F seem to be the key regulators in the G1/S transition (62, 63), studies show that S-phase entry can occur without activating the RB/E2F pathway (34). In particular, cyclin E, which has been shown to be a target of E2F (7, 44) and a critical player in the G1/S transition (17, 18, 30, 31, 45), is able to induce the G1/S transition independently of the RB/E2F functions (34, 38, 45, 51). The observations that cyclin E is a potential target of c-myc (49) and that c-myc can bypass the RB/E2F pathway with effects indistinguishable from those of cyclin E (1) make cyclin E a good candidate target that couples c-myc with the cell cycle. In addition, c-myc may activate the cyclin E/CDK complex by inhibiting the function of a cyclin E/CDK inhibitor, p27 (57, 61). Although the involvement of cyclin E and p27 in Myc-mediated cell cycle phenotypes appears compelling, substantial further studies are needed before any firm conclusion can be reached. Cdc25A has been reported to be a direct target of c-myc (22); however, we did not find any effect of Cdc25A on premature DNA replication in colcemid-treated Rat1a cells. We also noted that the protein level of cdc2 and CDK4 is higher in A1N4-Myc cells than in A1N4 cells, although it remains the same during colcemid exposure. It is unclear whether either of these proteins contributes to the myc-induced phenotype.
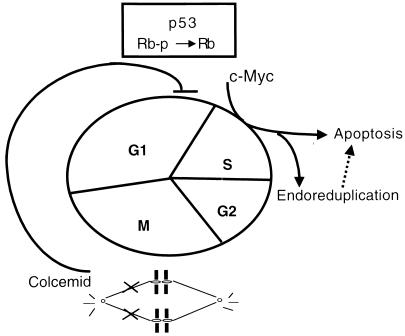
In aggregate, our observations, along with those of others, suggest a model of genome copy aberrations in neoplasms (Fig. 8). The mitotic checkpoint in mammalian cells, sensing the spindle status and chromosomal attachment and alignment, is activated by a disrupted spindle and arrests cells immediately in metaphase. Due to an intrinsic timing mechanism for mitotic exit or an adaptation of the inhibition, the cells may escape mitotic arrest after prolonged exposure to microtubule inhibitors and then proceed into the G1 phase. A signal, generated by detection of the initial defects, the failure of chromosomal segregation, or the absence of cytokinesis, however, is communicated to the cell cycle machinery in G1 phase to arrest progression beyond G1. An interaction between the detecting system and the cell cycle machinery can be predicted, although there is no clue to date as to its identity. It is possible that RB functions as the executor of this regulatory loop; therefore, the loss of RB eliminates the arrest. In support of this hypothesis, we observed that activation of the checkpoint by spindle inhibitors was associated with CDK4 and CDK2 inactivation and RB hypophosphorylation. Overexpression of c-myc overcomes this regulatory loop, probably via activating CDK2, and allows the cells to bypass the arrest in this G1-like state, which either activates the apoptotic pathway or enables premature DNA synthesis in the absence of mitosis, giving rise to polyploid cells.
FIG. 8.
Proposed model of the activity of c-myc in uncoupling DNA synthesis from mitosis. Spindle disruption is shown to cause hypophosphorylation of RB protein. c-myc is shown to bypass hypophosphorylated RB, inducing endoreduplication and apoptosis.
We discovered in this study that the ectopic expression of c-myc perturbs the coupling of DNA replication and mitosis, resulting in the formation of polyploid cells and increased cell death. Thus, the generation of chromosomal abnormalities and genomic instabilities very commonly found in human cancer cells may in part result from perturbed Myc and Max protein activities.
ACKNOWLEDGMENTS
We thank the members of our laboratory, E. Fearon, F. Spencer, and D. Wechsler, for their comments. We appreciate materials from L. Barr, S. Baylin, R. Dickson, and A. Levine, and we thank A. Partin and D. Coffey for the use of time-lapse micrography and J. Flook for technical assistance with flow cytometry.
This work was supported in part by NIH grant CA51497 (to C.V.D.).
REFERENCES
- 1.Alevizopoulos K, Vlach J, Hennecke S, Amati B. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati B, Alevizopoulos K, Vlach J. Myc and the cell cycle. Front Biosci. 1998;3:250–268. doi: 10.2741/a239. [DOI] [PubMed] [Google Scholar]
- 3.Andreassen P R, Margolis R L. Microtubule dependency of p34cdc2 inactivation and mitotic exit in mammalian cells. J Cell Biol. 1994;127:789–802. doi: 10.1083/jcb.127.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr L F, Campbell S E, Penno M B, Ball D W, Baylin S B. Cell-substratum interactions mediate oncogene-induced phenotype of lung cancer cells. Cell Growth Differ. 1996;7:1149–1156. [PubMed] [Google Scholar]
- 5.Berns K, Hijmans E M, Bernards R. Repression of c-Myc responsive genes in cycling cells causes G1 arrest through reduction of cyclin E/CDK2 kinase activity. Oncogene. 1997;15:1347–1356. doi: 10.1038/sj.onc.1201280. [DOI] [PubMed] [Google Scholar]
- 6.Bignon Y J, Shew J Y, Rappolee D, Naylor S L, Lee E Y, Schnier J, Lee W H. A single Cys706 to Phe substitution in the retinoblastoma protein causes the loss of binding to SV40 T antigen. Cell Growth Differ. 1990;1:647–651. [PubMed] [Google Scholar]
- 7.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Durr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahill D P, Lengauer C, Yu J, Riggins G J, Wilson J K, Markowitz S D, Kinzler K W, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 9.Canman C E, Wolff A C, Chen C Y, Fornace A J, Jr, Kastan M B. The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- 10.Chen J, Wu X, Lin J, Levine A J. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen R H, Waters J C, Salmon E D, Murray A W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 12.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 13.Davis F M, Tsao T Y, Fowler S K, Rao P N. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickson, R. Personal communication.
- 16.Di Leonardo A, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 17.Dulic V, Lees E, Reed S I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 18.Duronio R J, Brook A, Dyson N, O’Farrell P H. E2F-induced S phase requires cyclin E. Genes Dev. 1996;10:2505–2513. doi: 10.1101/gad.10.19.2505. [DOI] [PubMed] [Google Scholar]
- 19.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 20.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 21.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 22.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 23.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorczyca W, Gong J, Darzynkiewicz Z. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 1993;53:1945–1951. [PubMed] [Google Scholar]
- 25.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 26.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 27.Hoang A T, Lutterbach B, Lewis B C, Yano T, Chou T-Y, Barrett J F, Raffeld M, Hann S R, Dang C V. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoyt M A, Totis L, Roberts B T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 29.Khan S H, Wahl G M. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 1998;58:396–401. [PubMed] [Google Scholar]
- 30.Knoblich J A, Sauer K, Jones L, Richardson R, Saint R, Lehner C F. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 31.Koff A, Giordano A, Desai D, Yamashita K, Harper J W, Elledge S, Nishimoto T, Morgan D O, Franza B R, Roberts J M. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science. 1992;257:1689–1694. doi: 10.1126/science.1388288. [DOI] [PubMed] [Google Scholar]
- 32.Kung A L, Sherwood S W, Schimke R T. Cell line-specific differences in the control of cell cycle progression in the absence of mitosis. Proc Natl Acad Sci USA. 1990;87:9553–9557. doi: 10.1073/pnas.87.24.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanni J S, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Lee, L., and C. V. Dang. Unpublished data.
- 34.Leng X, Connell-Crowley L, Goodrich D, Harper J W. S-phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 35.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 38.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 39.Mai S, Hanley-Hyde J, Fluri M. c-Myc overexpression associated DHFR gene amplification in hamster, rat, mouse and human cell lines. Oncogene. 1996;12:277–288. [PubMed] [Google Scholar]
- 40.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 41.Minn A J, Boise L H, Thompson C B. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- 42.Nasmyth K. Viewpoint: putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- 43.Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 44.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Packham G, Cleveland J L. c-Myc and apoptosis. Biochim Biophys Acta. 1995;1242:11–28. doi: 10.1016/0304-419x(94)00015-t. [DOI] [PubMed] [Google Scholar]
- 47.Pangilinan F, Li Q, Weaver T, Lewis B C, Dang C V, Spencer F. Mammalian BUB1 protein kinases: map positions and in vivo expression. Genomics. 1997;46:379–388. doi: 10.1006/geno.1997.5068. [DOI] [PubMed] [Google Scholar]
- 48.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Roger I, Solomon D L, Sewing A, Land H. Myc activation of cyclin E/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene. 1997;14:2373–2381. doi: 10.1038/sj.onc.1201197. [DOI] [PubMed] [Google Scholar]
- 50.Qin X Q, Livingston D M, Kaelin W G, Jr, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Resnitzky D, Reed S I. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ricketts M H, Levinson A D. High-level expression of c-H-ras1 fails to fully transform Rat-1 cells. Mol Cell Biol. 1988;8:1460–1468. doi: 10.1128/mcb.8.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts J M, Koff A, Polyak K, Firpo E, Collins S, Ohtsubo M, Massague J. Cyclins, Cdks, and cyclin kinase inhibitors. Cold Spring Harbor Symp Quant Biol. 1994;59:31–38. doi: 10.1101/sqb.1994.059.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Sargent L M, Sanderson N D, Thorgeirsson S S. Ploidy and karyotypic alterations associated with early events in the development of hepatocarcinogenesis in transgenic mice harboring c-myc and transforming growth factor alpha transgenes. Cancer Res. 1996;56:2137–2142. [PubMed] [Google Scholar]
- 55.Shim H, Chun Y S, Lewis B C, Dang C V. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci USA. 1998;95:1511–1516. doi: 10.1073/pnas.95.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small M B, Hay N, Schwab M, Bishop J M. Neoplastic transformation by the human gene n-myc. Mol Cell Biol. 1987;7:1638–1645. doi: 10.1128/mcb.7.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steiner P, Philipp A, Lukas J, Godden-Kent D, Pagano M, Mittnacht S, Bartek J, Eilers M. Identification of a Myc-dependent step during the formation of active G1 cyclin-cdk complexes. EMBO J. 1995;14:4814–4826. doi: 10.1002/j.1460-2075.1995.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor S S, Ha E, McKeon F. The human homologue of bub3 is required for kinetochore localization of bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor S S, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 60.Thompson E W, Torri J, Sabol M, Sommers C L, Byers S, Valverius E M, Martin G R, Lippman M E, Stampfer M R, Dickson R B. Oncogene-induced basement membrane invasiveness in human mammary epithelial cells. Clin Exp Metastasis. 1994;12:181–194. doi: 10.1007/BF01753886. [DOI] [PubMed] [Google Scholar]
- 61.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 62.Weinberg R A. E2F and cell proliferation: a world turned upside down. Cell. 1996;85:457–459. doi: 10.1016/s0092-8674(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 63.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 64.Yin X Y, Grove L, Datta N S, Long M W, Prochownik E V. C-myc overexpression and p53 loss cooperate to promote genomic instability. Oncogene. 1999;18:1177–1184. doi: 10.1038/sj.onc.1202410. [DOI] [PubMed] [Google Scholar]
- 65.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]