Abstract
Myb-related cdc5p is required for G2/M progression in the yeast Schizosaccharomyces pombe. We report here that all detectable cdc5p is stably associated with a multiprotein 40S complex. Immunoaffinity purification has allowed the identification of 10 cwf (complexed with cdc5p) proteins. Two (cwf6p and cwf10p) are members of the U5 snRNP; one (cwf9p) is a core snRNP protein. cwf8p is the apparent ortholog of the Saccharomyces cerevisiae splicing factor Prp19p. cwf1+ is allelic to the prp5+ gene defined by the S. pombe splicing mutant, prp5-1, and there is a strong negative genetic interaction between cdc5-120 and prp5-1. Five cwfs have not been recognized previously as important for either pre-mRNA splicing or cell cycle control. Further characterization of cwf1p, cwf2p, cwf3p, and cwf4p demonstrates that they are encoded by essential genes, cosediment with cdc5p at 40S, and coimmunoprecipitate with cdc5p. We further show that cdc5p associates with the U2, U5, and U6 snRNAs and that cells lacking cdc5+ function are defective in pre-mRNA splicing. These data raise the possibility that the cdc5p complex is an intermediate in the assembly or disassembly of an active S. pombe spliceosome.
The Schizosaccharomyces pombe cdc5+ gene was identified in the original screen for fission yeast mutants defective for cell cycle progression (51). At the restrictive temperature, cells harboring the temperature-sensitive cdc5-120 mutation become arrested with a 2N content of DNA in the G2 phase of the cycle (51, 56). The cloning and initial characterization of cdc5+ showed that its function is essential for viability and that its predicted protein product shares significant homology to the DNA binding domain of the vertebrate proto-oncoprotein c-Myb (56). The DNA binding domain of c-Myb consists of three imperfect repeats that contain evenly spaced hydrophobic residues, typically tryptophan but in some cases phenylalanine, isoleucine, or tyrosine (36). These residues have been shown by nuclear magnetic resonance structural analysis to make up the backbone of the DNA binding motif (53, 54). cdc5p contains two Myb repeats and an additional repeat that is similar to a Myb repeat but lacks certain residues characteristic of a canonical Myb repeat (55). Sequence similarity to a family of known DNA binding proteins led us to suggest initially that cdc5p might be required for entry into mitosis via regulation of transcription (56).
Since the initial characterization of cdc5+, work in other organisms and database searches have revealed proteins that are closely related to cdc5p. To date, apparent cdc5p orthologs have been identified in Arabidopsis thaliana, Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Xenopus laevis, and human (7, 21, 25, 55, 69). This family of proteins was shown to be functionally conserved since DNAs encoding full-length versions of the Drosophila and Arabidopsis proteins and a truncated version of the human protein will rescue the growth defect of the S. pombe temperature-sensitive mutant, cdc5-120 (25, 55). Further the S. cerevisiae ortholog, CEF1 (S. cerevisiae homolog of cdc5), is essential for mitotic progression; cells lacking Cef1p arrest as large budded cells with aberrant spindle morphologies (55). Other links to cell cycle progression for cdc5 family members were reports that mammalian cells overexpressing hCdc5 showed a shortened G2 and reduced cell size, and cells expressing a carboxy-terminally truncated version of hCdc5 slowed G2 progression (6).
Although it is highly conserved, the exact biochemical function of cdc5p/Cef1p has not been elucidated. Evidence supporting a role for this protein in DNA binding has been described. The bacterially expressed Myb repeats of the Arabidopsis thaliana ortholog bound to the double-stranded DNA sequence CTCAGCG in vitro (25). Also, the human ortholog was shown to be a nuclear protein (7) and a chimeric molecule consisting of the carboxy terminus of human Cdc5p coupled to the DNA binding domain of GAL4 transactivated a reporter gene in COS-7 cells (6).
To address the function of cdc5p, we have examined its biochemical properties in S. pombe. As expected for a potential DNA binding protein, cdc5p was found exclusively within the nucleus. Unexpectedly, we also found that all detectable cdc5p is contained within a discrete 40S complex. Because the function of this complex seemed likely to be intimately entwined with that of cdc5p, we purified the complex and have identified 10 of its subunits. All are previously undescribed S. pombe open reading frames. Nine of the 10 components have obvious orthologs in other organisms, many of which are involved in pre-mRNA splicing. However, the functions of several orthologs have not been described. cdc5p also copurifies with the U2, U5, and U6 snRNAs demonstrating an association with S. pombe snRNPS. Consistent with this association, cells lacking cdc5+ function are defective in pre-mRNA splicing. Together, these data raise the possibility that the cdc5p complex is a stable intermediate in the assembly or disassembly of the S. pombe spliceosome. We discuss potential mechanisms whereby defects in pre-mRNA splicing give rise to cell cycle blocks.
MATERIALS AND METHODS
Strains, growth media, cell synchronization, and genetic methods.
S. pombe strains used in this study (Table 1) were grown in yeast extract medium or minimal medium with appropriate supplements (44). Transformations were performed by electroporation (61) or by the dimethyl sulfoxide-enhanced lithium acetate method (32). Genomic DNA was isolated as described earlier (44). For flow cytometric analysis, cells were treated as previously detailed (67), except that Sytox Green (final concentration, 1 μM; Molecular Probes, Eugene, Oreg.) was used to stain the DNA.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| KGY28 | 972 h+ | P. Nurse |
| KGY162 | cdc5-120 h+ | P. Nurse |
| KGY450 | cdc5+/cdc5::ura4 ade6+/ade6-704 ura4-D18/ura4-D18 leu1-32/leu1-32 h+/h− | Lab stock |
| KGY792 | cdc5HA h− | This study |
| KGY1218 | ura4-D18/ura4-D18 leu1-32/leu1-32 his3-D1/his3-D1 ade6-M210/ade6-M216 h+/h− | Lab stock |
| KGY1403 | cdc10-V50 h+ | P. Nurse |
| KGY1421 | cwf3myc leu1-32 ura4-D18 his3-D1 ade6-M210 h− | This study |
| KGY1423 | cwf4myc leu1-32 ura4-D18 his3-D1 ade6-M210 h+ | This study |
| KGY1425 | cwf1myc leu1-32 ura4-D18 his3-D1 ade6-M210 h− | This study |
| KGY1429 | cwf2myc leu1-32 ura4-D18 his3-D1 ade6-M216 h+ | This study |
| KGY1432 | cwf3HA leu1-32 ura4-D18 his3-D1 ade6-M210 h− | This study |
| KGY1434 | cwf4HA leu1-32 ura4-D18 his3-D1 ade6-M216 h− | This study |
| KGY1437 | cwf1HA leu1-32 ura4-D18 his3-D1 ade6-M216 h− | This study |
| KGY2177 | cwf10myc leu1-32 ura4-D18 ade6-M210 h− | This study |
| KGY1565 | cwf1+/cwf1::KanR ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 his3-D1/his3-D1 h+/h− | This study |
| KGY1567 | cwf4+/cwf4::KanR ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 his3-D1/his3-D1 h+/h− | This study |
| KGY1607 | cdc5myc leu1-32 ura4-D18 ade6-M210 h− | This study |
| KGY1671 | cwf2+/cwf2::KanR ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 his3-D1/his3-D1 h+/h− | This study |
| KGY1672 | cwf3+/cwf3::KanR ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 his3-D1/his3-D1 h+/h− | This study |
| KGY1490 | cdc5::ura4+ pREP41cdc5 leu1-32 ura4-D18 ade6-704 | This study |
| KGY1876 | (DK623) prp5-1 leu1-32 h− | Potashkin et al. (59) |
| KGY1141 | prp2-1 h− | Potashkin et al. (58) |
| KGY1937 | prp5-1 h+ | This study |
| KGY352 | nuc-663 h− | P. Nurse |
| KGY3 | cdc25-22 leu1-32 h− | P. Nurse |
Plasmids and molecular biology techniques.
Plasmid manipulations and bacterial transformations were performed by standard techniques (66). Sequencing reactions were performed by using either Sequenase 2.0 (USB, Cleveland, Ohio) or a Thermosequenase radiolabeled terminator cycle sequencing kit (Amersham, Cleveland, Ohio) according to manufacturer’s instructions. PCR amplifications were performed with Taq polymerase and Gene Amp reagents (Perkin-Elmer, Norwalk, Conn.), Pfu polymerase, BioExact (ISC Bioexpress, Kaysville, Utah), or TaqPlus Precision (Stratagene, La Jolla, Calif.) according to the manufacturer’s instructions. Amplifications were accomplished by using a PTC-100 programmable thermal controller or a PTC-150 Mini-Cycler (PTC-100 or PTC-150; MJ Research, Watertown, Mass.). Sequences of all oligonucleotides are available upon request.
HA tagging and gene replacement of cdc5+.
A NotI site was added to the 3′ end of the cdc5+ coding region by amplifying a 280-bp fragment by using the following primers: cdc55′bgl and cdc5NRV. This fragment was subcloned into pBS-SK+ (Stratagene) and sequenced to check for PCR induced mutations. A NotI fragment encoding three copies of the hemagglutinin (HA) epitope (provided by Bruce Futcher) recognized by the monoclonal antibody 12CA5 (Boehringer Mannheim, Indianapolis, Ind.) was subcloned into the engineered NotI site. A BglII to EcoRV fragment encoding the HA-tagged carboxy terminus of cdc5p was cloned into a pIRT2 plasmid containing a genomic copy of cdc5+ (pKG321). The functionality of this construct (pKG562) was confirmed by transforming it into diploid cells (KGY450), sporulating the diploids, and selecting for haploid cells that could grow in the absence of leucine and uracil. A gene replacement construct was made by amplifying 1,240 bp of the 3′ genomic region of cdc5+ by using the primers cdc53′kpn and PucMCSRI and subcloning the amplification product into pKG562. A 4.1-kb fragment containing the 0.6-kb 5′ genomic region, the entire cdc5+ coding region (including the HA tags), and 1.2 kb of the 3′ flank region was transformed into temperature-sensitive cdc5-120 mutant cells. A stable integrant (KGY792) was selected at 36°C and confirmed as a gene replacement by Southern blot analysis.
Generation of anti-cdc5p antibodies.
Full-length cdc5+ cDNA was amplified by PCR from pKG307 (pTZ19R [Pharmacia, Piscataway, N.J.] which contains full-length cdc5+ cDNA) by using the primers cdc5BamST and cdc5STOP, which introduce BamHI restriction sites at the 5′ and 3′ ends of the cDNA. The resulting fragment was treated with the Klenow fragment of Escherichia coli DNA polymerase I, subcloned into the SmaI site of pTZ19R, and sequenced in its entirety to ensure the absence of PCR-induced mutations (pKG469). Recombinant cdc5p was produced by using the Xpress System (Invitrogen, Carlsbad, Calif.) by cloning the BamHI cdc5+ cDNA fragment into the BamHI site of pRSETA. Insoluble cdc5p was purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electroeluted, and used to immunize two rabbits, Pearl and Jam. While antibodies from both rabbits recognize both bacterially expressed and endogenous cdc5p, Jam antisera were used exclusively for this study.
Indirect immunofluorescence.
For visualization by indirect immunofluorescence, cells were fixed with 10% methanol–3.7% formaldehyde for 30 min at room temperature (18), washed with phosphate-buffered saline, and processed as described earlier (3). Polyclonal antisera against cdc5p or preimmune sera were incubated at a 1/50 dilution followed by a 1/100 dilution of Texas red-conjugated goat anti-rabbit secondary antibody (Molecular Probes). Visualization of HA-tagged cdc5p was performed with 12CA5 antibodies (Boehringer Mannheim) at a concentration of 20 μg/ml followed by the addition of a 1/100 dilution of Texas red-conjugated goat anti-mouse secondary antibody (Molecular Probes). All fluorescence microscopy was performed on a Zeiss microscope (Axioskope; Zeiss, Inc., Thornwood, N.Y.) with appropriate filters. Images were captured by using a cooled charge-coupled device camera (ZVS47DEC; Optronics, Goleta, Calif.).
Immunoprecipitations and immunoblots.
Protein lysates were made by glass bead disruption of the cell walls in a minimal volume of Nonidet P-40 (NP-40) buffer. For “denatured” lysates, lysed cells were heated to 95°C in SDS lysis buffer (10 mM NaPO4, pH 7.4; 1.0% SDS; 1 mM dithiothreitol; 1 mM EDTA; 50 mM NaF; 100 μM Na3VO4; 4 μg of leupeptin per ml) for 2 min and extracted with NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1.0% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 100 μM Na3VO4, 4 μg of leupeptin per ml) and protease inhibitors. For “native” lysates, heating in SDS lysis buffer was omitted. 35S-labeled lysates were prepared in an identical manner except that cells were grown overnight in minimal medium and then grown for 4 h in the presence of 1 mCi of Tran[35S]-label (ICN Pharmaceuticals, Costa Mesa, Calif.) prior to lysis. In vitro cdc5p was produced by using the cdc5+ cDNA (pKG469) to program an in vitro transcription-translation reaction (TNT-coupled T7 reticulocyte lysate; Promega, Madison, Wis.).
For immunoblots, a 1/5 volume of 5× sample buffer was added to the extracts. Immunoprecipitations were performed by incubating 5 μl of polyclonal antibodies with the extracts for 1 h on ice, followed by a 30-min incubation with 50 μl of a 1:1 slurry of protein A-Sepharose (Pharmacia). Immunoprecipitates were washed six times with NP-40 buffer and then resuspended in sample buffer. Anti-myc immunoprecipitations were performed by using 5 μg of 9E10 antibody and 5 μg of rabbit anti-mouse (Cappel, Organon Teknika Corp., West Chester, Pa.). Unless otherwise noted, 12CA5 immunoprecipitations were performed by using 20 μg of 12CA5 which had been coupled to protein A-Sepharose by using DMP (Sigma, St. Louis, Mo.) (22). After 1.5 h of incubation, the immunoprecipitates were washed six times in RIPA buffer (NP-40 buffer plus 1.0% deoxycholate and 0.1% SDS) and then resuspended in sample buffer.
Proteins were resolved on SDS–6 to 20% polyacrylamide gels. For immunoblotting, proteins were then transferred by electroblotting to a polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore Corp., Bedford, Mass.). cdc5p immunoblots were performed by incubation with anti-cdc5p serum JAM (1:10,000 dilution in a 5% milk–Tris-buffered saline [TBS] blocking solution), followed by the addition of horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibodies (Sigma). Similar procedures were used to probe for anti-fatty acid synthetase (FAS) antibodies at 1:2,000. Epitope-tagged proteins were detected with 12CA5 (HA tag) or 9E10 (to detect the myc tag) antibodies at 2 μg/ml in TBS followed by treatment with horseradish peroxidase-conjugated goat anti-mouse polyclonal antibodies (Sigma). Immunoblots were visualized by using enhanced chemiluminescence (Amersham). For visualization of 35S-labeled proteins, the protein gels were fixed, treated for fluorography (Amplify; Amersham), dried, and exposed to film.
Glycerol and sucrose gradient analysis.
Two to four milligrams of total protein was layered onto a 10 to 30% glycerol or sucrose gradient which was subsequently spun at 28,000 rpm for 15 h in an SW50ti rotor (Beckman, Palo Alto, Calif.). Fractions from these gradients were collected, mixed with sample buffer, and resolved by SDS-PAGE. After transfer, the blots were probed with anti-cdc5p antibodies. For size standards, parallel gradients were run on samples containing thyroglobulin (19S) and catalase (11.3S) (HWM Standards; Pharmacia) or 2 mg of lysate from S. cerevisiae which was subsequently probed with antibodies recognizing FAS as a 40S marker (a generous gift from S. J. Wakil).
Complex purification.
Cells were lysed in NP-40 buffer by using a Bead-Beater (Biospec Products, Bartlesville, Okla.), and the initial lysate was cleared of cellular debris with a 10-min spin at 2,000 × g. A second clearing spin was performed for 1 h at 38,000 rpm (100,000 × g) in a 70ti rotor (Beckman). The lysate was then dialyzed extensively into Tris-KCl buffer (50 mM Tris-HCl, pH 7.4; 150 mM KCl; 2 mM EDTA). Proteins were precipitated with 55% ammonium sulfate; the precipitate was then resuspended and dialyzed into NP-40 buffer. This lysate was then applied to a 12CA5 column which was made by coupling purified 12CA5 antibody to protein A-Sepharose by using the cross-linking agent DMP (22). The column was washed with 80 column volumes of NP-40 buffer followed by 10 column volumes of NP-40 buffer containing only 0.1% NP-40 rather than 1.0% NP-40. The proteins were eluted at room temperature with 2 volumes of this buffer that also contained 1.0% SDS. The eluent was concentrated by using an Ultrafree centrifugal filter device (Millipore Corp.), suspended in sample buffer, and resolved by SDS-PAGE. The proteins were visualized by silver staining (Plusone; Pharmacia) according to the manufacturer’s instructions. To prepare proteins for microsequencing, the resolved proteins were transferred to a PVDF membrane (Immobilon P; Millipore Corp.) in lieu of silver staining. Immobilized proteins were visualized by using ponceau-S (Sigma), and bands of interest were excised for microsequencing.
In vivo tagging.
myc-tagged and HA-tagged strains were constructed by using a system described previously (79) with cassettes and techniques described by Bahler et al. (2). The following primers were used to amplify either the myc13-kan or HA3-kan cassette: for cwf1+, SP#10AFORtag and SP#10AREVtag; for cwf2+, SP#10BFORtag and SP#10BREVtag; for cwf3+, SP#7FORtag and SP#7REVtag; for cwf4+, SP#9FORtag and SP#9REVtag; and for cwf10+, cwf10FORtag and cwf10REVtag. These fragments were transformed into diploid cells (KGY1218), allowed to recover for 12 h on yeast extract (YE) plates, and then replica plated onto YE plates containing 100 μg of G418 (Geneticin; Gibco BRL, Grand Island, N.Y.) per ml. G418-resistant colonies were screened for homologous recombinants by Southern blotting; the presence of the epitope was confirmed by immunoblotting with either 9E10 or 12CA5 antibodies. Sporulation and tetrad analysis showed 2:2 segregation of the G418 resistance, confirming that the haploid strains cwf1HA (KGY1437), cwf1myc (KGY1425), cwf2myc (KGY1429), cwf3HA (KGY1432), cwf3myc (KGY1421), cwf4HA (KGY1434), cwf4myc (KGY1423), and cwf10myc (KGY2177) were viable.
Cloning of prp5+.
The prp5+ gene was cloned by functional complementation of the temperature-sensitive (Ts−) growth defect of the S. pombe pre-mRNA splicing mutant prp5-1 (59). We transformed S. pombe DK623 by the lithium acetate procedure (44) with an S. pombe genomic library (provided by Paul Young, Queen’s University, Kingston, Ontario, Canada) generated by partial digestion of wild-type DNA with HindIII and insertion into the shuttle vector pWH5 (81). Approximately 8,000 leu+ transformants recovered on minimal medium at the permissive temperature (23°C) were screened for temperature-resistant (Ts+) clones by replica plating at 37°C. Two colonies were isolated that grew well at 37°C and did not exhibit the elongated cellular morphology characteristic of the cell division cycle defect (cdc−) in the prp5-1 mutant (59). Northern blot analysis of RNA from the Ts+ clones indicated that the prp5-1 pre-mRNA splicing defect was also rescued. The two Ts+ clones carried plasmids that shared a 7-kb HindIII fragment, which when inserted into the S. pombe vector pSP1 (15) or pIRT31 (64) was able to fully rescue the Ts− growth and cdc− defects upon reintroduction into the original prp5-1 strain. Integration of the 7-kb HindIII fragment into the genome of a prp5-1 strain resulted in stable rescue of the Ts− and cdc− phenotypes, and genetic mapping indicated that the integrated fragment was tightly linked to the prp5 locus. Thus, the 7-kb fragment very likely contained the prp5+ gene. DNA sequence determination of the minimum complementing prp5+ clone revealed an open reading frame identical to cwf1+.
Gene disruption of cwf1+, cwf2+, cwf3+, and cwf4+.
Genes were deleted using the HA-kan cassette (2). The entire open reading frames of cwf genes were targeted for deletion by using the following primers to amplify by PCR the HA-kan cassette: for cwf1+, SP#10AKOFOR and SP#10AREVtag; for cwf2+, SP#10BKOFOR and SP#10BREVtag; for cwf3+, SP#7KOFOR and SP#7REVtag; and for cwf4+, SP#9KOFOR and SP#9REVtag. As in the epitope-tagging procedure, the resulting PCR products were transformed into diploid cells (KGY1218), allowed to recover for 12 h on YE plates, and then replica plated onto YE-G418. Homologous targeting, confirmed by Southern blotting, resulted in the following deletion strains: cwf1::kanR/cwf1+ (KGY1565), cwf2::kanR/cwf2+ (KGY1671), cwf3::kanR/cwf3+ (KGY1672), and cwf4::kanR/cwf4+ (KGY1567). Sporulation and tetrad analysis of the resulting diploid strains showed two wild-type spores per tetrad, and no G418-resistant haploid colonies were recovered, indicating that these genes are essential in S. pombe.
Generation of conditional expression of cdc5+.
A repressible form of cdc5+ was constructed by subcloning the cdc5+ cDNA into the pREP41 vector (41). This construct was transformed into the cdc5::ura4+/cdc5+ diploid strain. Sporulation on medium lacking uracil and leucine selected for cells in which the only copy of cdc5+ was the plasmid-borne copy (pREP41 cdc5+). Repression of cdc5+ expression was obtained by growing the cells in medium containing thiamine at 5 μg/ml (41, 42).
RNA and Northern blots.
Total RNA from cells was prepared as described by Moreno et al. (44). To isolate RNA from the immunoprecipitates, the immunoprecipitates were resuspended in PK buffer (200 mM Tris, pH 8.0; 25 mM EDTA; 300 mM NaCl; 2% SDS), digested for 30 min with 100 μg of proteinase K (Boehringer Mannheim), and extracted once with phenol-chloroform; the nucleic acids were then precipitated by using a 1/10 volume of 3 M sodium acetate, 30 μg of glycogen, and 2.5 volumes of ethanol.
To detect mRNAs, total RNA was resolved by using formaldehyde-agarose gels and then capillary was blotted to GeneScreen+ (Dupont-NEN, Boston, Mass.) or Duralon-UV (Stratagene). To detect snRNAs, samples were resolved on an 8% polyacrylamide–8 M urea gel and transferred to Duralon-UV membrane by using a semidry blotter at 250 mA for 1 h in 0.1× TAE (10 mM Tris-acetate, pH 7.8; 5 mM sodium acetate; 0.5 mM EDTA). U6 snRNA was detected by using 32P-labeled oligonucleotides complementary to either the intron (U6 I) or the mature sequence (U6 E). tf2d RNA was detected by using 32P-labeled oligonucleotides complementary to both intronic (TFIID I) and exonic (TFIID E) sequences under the conditions described by Potashkin et al. (37, 58). Other snRNAs were detected by using 32P-labeled oligonucleotides complementary to S. pombe U1 (SPU1), U2 (U2B), U4 (SPU4), and U5 (YU5). Blots were exposed to PhosphorImager screens and visualized on a PhosphorImager with MD ImageQuant version 3.3 (Molecular Dynamics).
RESULTS
Detection and localization of cdc5p.
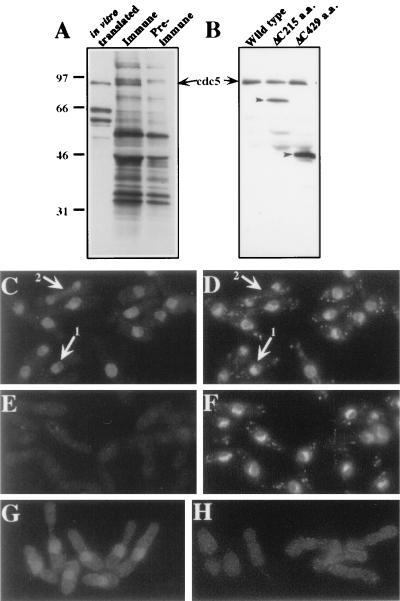
In order to begin investigating the biochemical properties of cdc5p, we raised polyclonal antibodies against a bacterially produced cdc5 fusion protein. From denatured lysates of 35S-labeled wild-type cells, anti-cdc5p serum precipitated a protein of approximately 95 kDa which comigrated with in vitro-translated cdc5p protein (Fig. 1A). These antibodies also recognized a single band of approximately 95 kDa in an immunoblot analysis of immunoprecipitates (Fig. 1B). In immunoprecipitations from cells expressing genomic clones of cdc5+ truncated at the 3′ end, smaller proteins of the appropriate sizes were recognized by the antibodies (Fig. 1B). These results established that the antibodies recognized the cdc5+ gene product.
FIG. 1.
cdc5p is a 97 kDa nuclear protein. Anti-cdc5p antibodies recognize a protein of 97 kDa from Tran[35S]-labeled S. pombe lysates (A and B) and from in vitro-produced cdc5p (A). (A) Autoradiograph of in vitro-produced cdc5p and immunoprecipitates from cells metabolically labeled with Tran[35S]-label by using immune and preimmune sera. (B) Immunoblot of anti-cdc5p immunoprecipitations from cells containing no plasmid, a plasmid encoding a 215-amino-acid truncation of cdc5p, and a plasmid encoding a 429-amino-acid truncation of the carboxy terminus of cdc5p. (C to H) Cdc5p is localized to the nucleus by indirect immunofluorescence. (C) Indirect immunofluorescence performed on wild-type S. pombe cells fixed with methanol-formaldehyde and probed with anti-cdc5p polyclonal antibodies. (D) DAPI staining of field in panel C. (E) Indirect immunofluorescence with preimmune sera. (F) DAPI staining of field in panel E. (G) Indirect immunofluorescence with 12CA5 in the cdc5HA strain. (H) 12CA5 staining of wild-type cells. (Panels G and H were exposed for the same length of time.)
By using these antibodies, cdc5p was localized to the nucleus by indirect immunofluorescence (Fig. 1C); preimmune serum produced only diffuse background staining (Fig. 1E). cdc5p staining was excluded from the non-DAPI (4′,6-diamidino-2-phenylindole) staining portion of the nucleus (Fig. 1C and D, arrow 1). cdc5p did not colocalize exclusively with chromatin since during mitosis cdc5p showed a diffuse nucleoplasmic localization (Fig. 1C and D, arrow 2). Further confirmation of the nuclear localization of cdc5p came from examining cells in which the endogenous cdc5+ gene had been engineered to express an epitope-tagged version, cdc5p-HA. This version encodes cdc5p tagged with three copies of the HA epitope at its carboxy terminus. When these cells (KGY792) were probed with 12CA5 antibody, staining also was restricted to the nucleus (Fig. 1G). No specific 12CA5-dependent staining was detected in wild-type cells that did not contain an epitope-tagged protein (Fig. 1H).
cdc5p is part of a multiprotein high-molecular-weight complex.
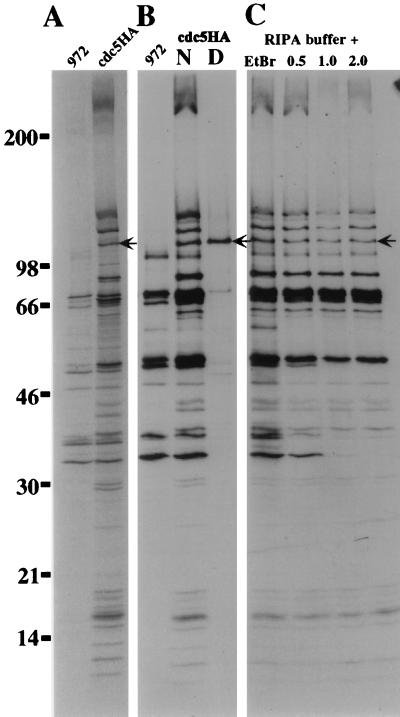
To examine whether other proteins bound stably to cdc5p, the cdc5HA strain was labeled with Tran[35S]-label, and immunoprecipitates were analyzed by SDS-PAGE. Numerous polypeptides coimmunoprecipitated with cdc5p-HA. These proteins were not seen in immunoprecipitates from wild-type cultures prepared in parallel (Fig. 2A), or when the 12CA5 antibodies were preincubated with 12CA5 peptide prior to immunoprecipitation (data not shown). Further, these proteins did not coimmunoprecipitate when the immunoprecipitations were performed from protein lysates which had been prepared under denaturing conditions (Fig. 2B).
FIG. 2.
Multiple proteins coimmunoprecipitate with cdc5p in a salt-resistant and DNA-independent manner. (A) Autoradiogram of immunoprecipitates from cells metabolically labeled with Tran[35S]-label. Immunoprecipitation was done with 12CA5 antibody from strain 972 h− or strain cdc5HA. (B) Immunoprecipitations were done as in panel A except that those done with cdc5HA were done under either native (N) or denatured (D) conditions. (C) Association of specific proteins with cdc5HA is neither DNA dependent nor salt sensitive. Immunoprecipitations were performed as in panel A except that ethidium bromide at 100 μg/ml was maintained throughout the immunoprecipitation or immunopellets were washed with radioimmunoprecipitation assay buffer containing increasing concentrations of NaCl (0.5, 1.0, and 2.0 M). The arrow indicates cdc5p-HA.
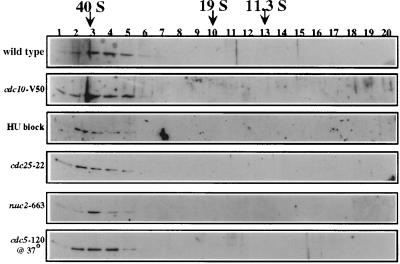
In order to determine the size of the cdc5p-containing complex, cell lysates were subjected to glycerol gradient sedimentation. As judged by immunoblot analysis of fractions from a 10 to 30% glycerol gradient, cdc5p sedimented at approximately 40S (Fig. 3). No monomeric cdc5p was observed even upon longer exposure of the immunoblot (data not shown). We also tested the possibility that the defect in cdc5-120 was a result of cdc5p falling out of the 40S complex. This was not the case, as we failed to detect any appreciable change in the sedimentation profile of cdc5p in this mutant grown at the restrictive temperature (Fig. 3). That cdc5p was a member of a high-molecular-weight complex was confirmed by gel filtration chromatography on a Sepharose CL-2B column. Endogenous cdc5p peaked early in the “postaggregate” fractions, and no peaks of lower molecular weight were detected (data not shown).
FIG. 3.
cdc5p is a component of a 40S complex. Shown is an immunoblot of fractions collected from 10 to 30% glycerol gradients which had been centrifuged for 15 h at 28,000 rpm. Strains from which lysates were prepared were as follows: 972 h− (wild type), cdc10-V50 block (G1 block), hydroxyurea block (HU) (S-phase block), cdc25-22 block (G2 block), nuc2-663 block (M-phase block), or cdc5-120 block. Blots were probed with anti-cdc5p. The migrations of FAS (40S), thyroglobulin (19S), and catalase (11.3S) collected from parallel gradients are indicated.
The cdc5p-associated complex is not DNA dependent and is resistant to salt.
Because of the similarity of cdc5p to a family of known DNA binding proteins, it was possible that the apparent association of other proteins with cdc5p could be a result of multiple proteins binding independently to a stretch of DNA. To address this possibility, we preincubated Tran[35S]-labeled cdc5HA extract with 100 μg of ethidium bromide per ml and maintained this concentration throughout the immunoprecipitation and washes. Lai and Herr (33) have shown that ethidium bromide can be used to distinguish between genuine protein-protein interactions and ones that are a result of two or more proteins binding independently to a stretch of DNA, presumably as a result of the intercalation of ethidium into the DNA, causing conformational changes that no longer allow protein-DNA interaction (33). Ethidium bromide treatment had no effect on the ability of the members of the cdc5p-associated complex to be coimmunoprecipitated with cdc5p-HA (Fig. 2C).
In order to assay the relative stability of the cdc5p-associated complex, we subjected the immunoprecipitations to stringent salt washes. Increasing the salt concentration in the washes to up to 2.0 M NaCl removed only a few of the specifically associated components and reduced the levels of certain background proteins (Fig. 2C). This indicated that the complex is quite stable even at high ionic concentrations.
cdc5p remains in a high-molecular-weight complex through the cell cycle.
In asynchronously growing cultures, all detectable cdc5 protein in the cell was associated with a 40S complex (Fig. 3). However, it was possible that cdc5p was not present in this complex at all stages of the cell cycle. To test this possibility, cells that had been blocked at various stages of the cell cycle by using either temperature-sensitive mutants or drugs were collected and lysed, and the lysates were subjected to glycerol gradient analysis. To block cells in G1, the temperature-sensitive cdc10-V50 mutant was used (39); hydroxyurea was used to block cells in S phase (46), and the cdc25-22 mutant was used to block cells in G2 (65); and the nuc2-663 mutant was used to block cells in mitosis (24). At all of these block points, cdc5p remained in the 40S complex. These data do not exclude the possibility that cdc5p leaves the complex at some portion of the cell cycle not represented by these block points. They also do not exclude the possibility that certain members of the complex are entering and exiting the complex through the cell cycle without having a dramatic effect on its overall size. However, the fact that cdc5p remains in a complex throughout the cell cycle and that we cannot detect any uncomplexed cdc5p within the cell strongly suggests that cdc5p is performing its essential role as part of this complex.
Purification of cdc5p-interacting proteins.
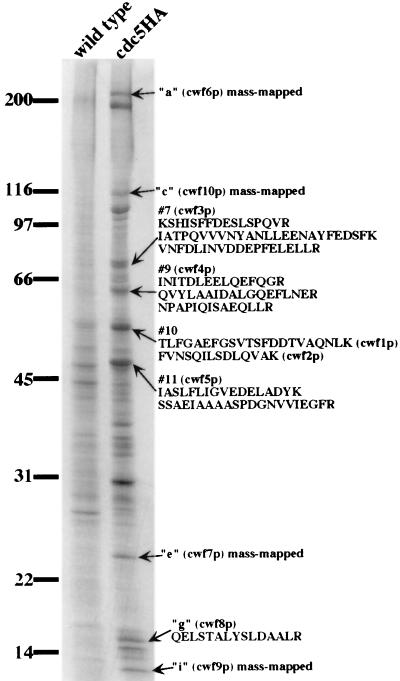
To better understand the role that this complex plays in cdc5p function, we set out to identify its subunits. We purified the complex utilizing a 12CA5 immunoaffinity column. The purification procedure was performed in parallel on cdc5HA and wild-type cells. A portion of these samples was resolved by SDS-PAGE and visualized by silver staining. Comparison of the silver-stained eluents showed that a number of proteins specifically associated with cdc5p-HA (Fig. 4). Another portion of the sample was resolved by SDS-PAGE and transferred to a PVDF membrane, and the proteins were visualized by ponceau-S staining. Bands of interest were excised; four were identified by mass mapping (27), and five others required microsequencing for positive identification (Fig. 4) . Database searches showed that these nine protein bands corresponded to ten previously undescribed open reading frames (band 10 contained two proteins), which we have designated cwf1 to cwf10 (complexed with cdc5) (Fig. 4 and Table 2).
FIG. 4.
Purification and identification of cdc5p-associated proteins. A silver-stained gel shows that eluted protein from the immunoaffinity purification of the cdc5p-HA-associated complex compared to the eluted protein from an identical purification from an untagged strain. Arrows indicate protein bands from which microsequence data were obtained. Peptide sequences corresponding to each band are shown to the right of the arrow.
TABLE 2.
Identities of cwf proteins
| cwf protein | Predicted molecular size (kDa) | S. cerevisiae ortholog | Putative orthologs of described function in other species | Protein motifs |
|---|---|---|---|---|
| prp5p/cwf1p | 52.4 | YPL151c | A. thaliana PRL1 (Q42384); Homo sapiens (AF044333) | WD/40, beta transducin |
| cwf2p | 46.4 | YDL209c | ||
| cwf3p | 94.6 | Syf1p-YDR416w | TPR | |
| cwf4p | 80.8 | Syf3p-YLR117c | D. melanogaster crn (AL009171) | TPR |
| cwf5p | 39.6 | Ecm2p-YBR065c | RRMc | |
| cwf6p/prp15p | 274.5 | Prp8p-P33334 | H. sapiens (2463577) | |
| cwf7p | 12.6a | |||
| cwf8p | 54.2b | Prp19p-P32523 | ||
| cwf9p | 13.1 | SMD2-YLR275w | H. sapiens SM D (P43330) | |
| cwf10p | 111.2 | Snu114p-YKL173w | Mus musculus U5-116 (2105430) | GTP binding |
Identity of cwf proteins.
All of the cdc5p complex subunits identified above, with the exception of cwf7p, have apparent orthologs in S. cerevisiae and higher eukaryotic organisms (see Table 2). The cwf6+ gene encodes the S. pombe ortholog of S. cerevisiae Prp8p and human p220, a subunit of the U5 snRNP (1). This highly conserved protein has been shown to be integral to the splicing reaction and undergoes extensive contacts with the substrate RNA at both the 5′ exon and 3′ splice site regions (5, 63, 74–76). As it did not correspond to any previously published S. pombe splicing mutant (59, 60, 77), and it is almost certain that cwf6+ will perform the same function in S. pombe, we have redesignated it prp15+. cwf10+ encodes the S. pombe ortholog of another component of the U5 snRNP: human U5-115 kDa/S. cerevisiae Snu114p (19, 20). cwf9p is most similar to the core snRNP protein, D2 (23). Band “g” in the purification, cwf8p, is a degradation product of the S. pombe ortholog of the essential S. cerevisiae splicing factor, Prp19p. S. cerevisiae Prp19p also has been shown to be a component of a high-molecular-weight complex (72, 73) that likely represents a homologous complex in S. cerevisiae.
The orthologs of cwf1p also have been implicated in pre-mRNA splicing. The cwf1p protein contains four canonical WD-40 repeats, and human cwf1p was recently identified as a protein that copurifies with the spliceosome from HeLa cell extracts (49). The A. thaliana ortholog of cwf1p, PRL1 (pleiotropic regulatory locus 1) is reported to be involved in a variety of cellular processes surrounding glucose signaling. While this protein was shown to associate in vitro with the protein kinase C-βII isoform and demonstrated a two-hybrid interaction with α-importin, no clear biochemical role has been described for this protein (47). Interestingly, this A. thaliana protein was identified earlier as a cDNA that caused an aberrant morphology when overexpressed in S. pombe (82).
cwf3p and cwf4p are members of a subfamily of tetratricopeptide repeat (TPR) proteins most similar to D. melanogaster crooked neck (crn) (78, 83). The D. melanogaster crn gene has also been implicated in cell cycle control based on its embryonic lethal phenotype (83). The pre-mRNA splicing factors, Prp39p and Prp42p, while also members of this family of TPR repeat proteins, do not represent the S. cerevisiae orthologs of cwf3p and cwf4p (Table 2). cwf5p contains RNA recognition motifs, which suggests that RNA interactions might be important for the function of this protein. The S. cerevisiae ortholog of this protein was implicated in cell surface assembly as a mutant that conferred hypersensitivity to calcofluor white (38). The remaining proteins, cwf2p and cwf7p, contain no obvious protein motifs nor do they have orthologs of ascribed function in other organisms (Table 2).
cwf1p, cwf2p, cwf3p, cwf4p, and cwf10p are bona fide members of the cdc5p-associated complex.
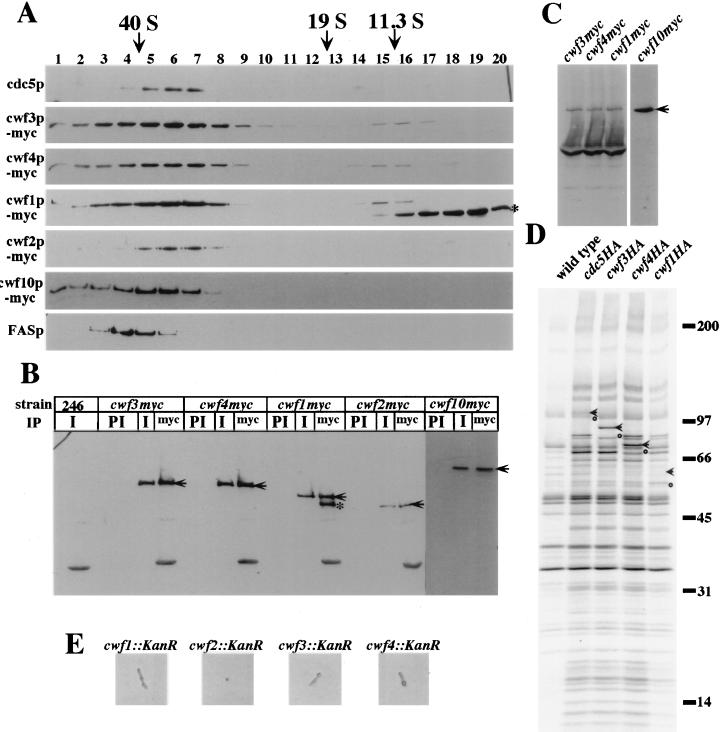
As described above, apparent orthologs of several of the cwf proteins were identified as components of the spliceosome. To determine what proportion of each was associated with cdc5p, the endogenous copies of cwf1+, cwf2+, cwf3+, cwf4+, and cwf10+ were tagged at the 3′ end of their open reading frames with sequences encoding either 3 copies of the HA epitope or 13 copies of the myc epitope. Sucrose gradient analysis of lysates prepared from the myc-tagged strains showed that the majority of the endogenous cwf proteins cosedimented with cdc5p at 40S (Fig. 5A). A lower-molecular-weight form of cwf1p-myc, presumably N-terminally truncated since it retained the C-terminal tag, was found in fractions consistent with the size of a monomer.
FIG. 5.
S. pombe cwf1p, cwf2p, cwf3p, cwf4p, and cwf10p associate with the cdc5p complex in vivo. (A) Immunoblots on fractions from 10 to 30% sucrose gradients probed with anti-cdc5 (cdc5p), anti-myc epitope, 9E10 (cwf3p-myc, cwf4p-myc, cwf1p-myc, cwf2p-myc, and cwf10p-myc, or anti-FAS (FAS-40S marker, FASp). The small asterisk on the right side of the panel indicates an apparent amino-terminal truncation of cwf1p-myc. The migration of thyroglobulin (19S) and catalase (11.3S) collected from parallel gradients is indicated. (B) Endogenously myc-tagged versions of cwf1p, cwf2p, cwf3p, cwf4p, and cwf10p coimmunoprecipitate with cdc5p in vivo. An anti-myc immunoblot of immunoprecipitates from wild-type (246), cwf3myc, cwf4myc, cwf1myc, cwf2myc, or cwf10myc strains is shown. Immunoprecipitations were performed with anti-cdc5p immune sera (I), preimmune sera (PI), or anti-myc antibodies (9E10, myc). Arrows indicate full-length tagged proteins. The small asterisk indicates an apparent amino-terminal truncation of cwf1p-myc. (C) Anti-cdc5p immunoblot of immunoprecipitates from cwf3myc, cwf4myc, cwf1myc, or cwf10myc strains with 9E10. The arrow indicates endogenous cdc5p. (D) Autoradiograph of immunoprecipitates from strains metabolically labeled with Tran[35S]-label as follows: wild-type, cdc5HA, cwf3HA, cwf4HA, and cwf1HA. The arrows indicate HA-tagged proteins in each respective strain. The circles indicate the untagged versions of the respective proteins. (E) cwf1+, cwf2+, cwf3+, and cwf4+ are essential genes. Shown are results for representative phenotypes of cwf::KanR spores.
To confirm that the cwf proteins were associated with cdc5p, we performed immunoprecipitations from the myc-tagged strains and immunoblotting with anti-cdc5p antibodies. In each case, cdc5p coimmunoprecipitated with the myc-tagged proteins (Fig. 5C). In reciprocal experiments, anti-cdc5p antibodies precipitated each of the myc-tagged proteins (Fig. 5B), whereas preimmune serum failed to do so (Fig. 5B). Identical results were obtained with the HA-tagged versions of the proteins (data not shown). Additionally, the same pattern of proteins coimmunoprecipitated with cwf1p-HA, cwf3p-HA, and cwf4p-HA as with cdc5p-HA from 35S-labeled cell lysates (Fig. 5D).
cwf1+, cwf2+, cwf3+, and cwf4+ are essential genes.
We wished to test whether other members of the cdc5p-associated complex, like cdc5+, were encoded by essential genes. In a diploid background, strains were constructed in which one copy of the entire open reading frames of cwf1+, cwf2+, cwf3+, or cwf4+ was replaced with a marker conferring resistance to G418. Analysis of these strains revealed two viable G418-sensitive colonies per tetrad, which indicated that cwf1+, cwf2+, cwf3+, and cwf4+ encode essential proteins (data not shown and Fig. 5E).
prp5-1 (cwf1) shows a synthetic phenotype with cdc5-120.
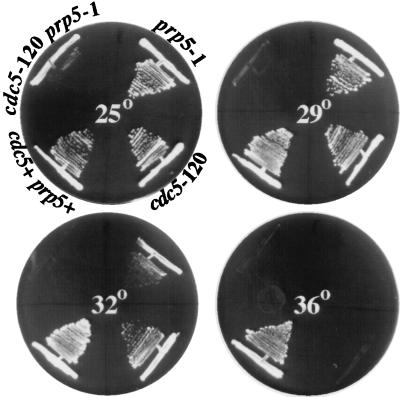
The prp5+ gene was cloned by complementation of the prp5-1 mutant and found to be identical to cwf1+. Further confirmation of this identity was the inability to recover temperature-sensitive, G418-resistant colonies from a mating of cwf1myc and prp5-1 (data not shown). Given the physical association between cdc5p and prp5p/cwf1p, we tested for genetic interactions between cdc5-120 and prp5-1. The double-mutant strain showed a reduced restrictive temperature. It failed to grow at 29°C, a temperature at which either single mutant grew quite well (Fig. 6). The clear negative genetic interaction between cdc5+ and prp5+/cwf1+, along with the interaction between their products, suggests overlapping functions for these two proteins.
FIG. 6.
cdc5-120 and prp5-1 show a reduced restrictive temperature. The wild type and prp5-1, cdc5-120, and cdc5-120 prp5-1 mutants were streaked onto agar medium at 25, 29, 32, and 36°C. The double-mutant strain was capable of growth at 25°C but not at 29°C. Each single mutant was capable of growth at both 29 and 32°C.
cdc5p is associated with U2, U5, and U6 snRNAs.
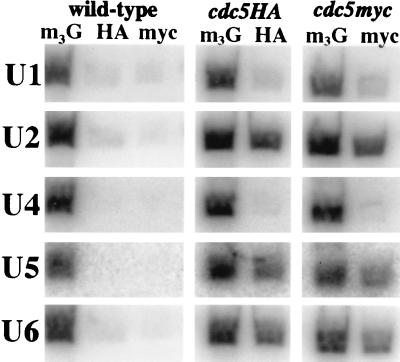
Due to the interaction of cdc5p with known snRNP proteins, we wished to test which, if any, of the S. pombe snRNAs associated with cdc5p. Utilizing oligonucleotides complementary to S. pombe U1, U2, U4, U5, and U6 snRNAs, we found that cdc5p immunoprecipitates contained the U2, U5, and U6 snRNAs but did not contain the U1 or U4 snRNA (Fig. 7). Also, cdc5 immunoprecipitates did not contain detectable amounts of the snoRNA, U3 (data not shown).
FIG. 7.
cdc5p associates with U2, U5, and U6 snRNAs. RNA was isolated from anti-m3G cap, anti-myc, or anti-HA immunoprecipitations from the wild-type, cdc5HA, or cdc5myc strain and subsequently probed with 32P-labeled oligonucleotides complementary to the U1, U2, U4, U5, or U6 snRNA.
cdc5+ function is required for pre-mRNA splicing.
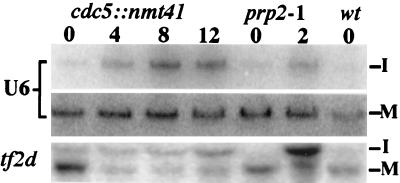
Because of the physical association of cdc5p with multiple splicing factors and snRNAs, we tested whether S. pombe cells require cdc5+ function for pre-mRNA splicing. In this experiment, we utilized cells containing the null mutant of cdc5 and a plasmid carrying the cdc5 cDNA under control of the thiamine-regulatable nmt1-T41 promoter (4). This strain grows in the absence of thiamine, but when cdc5+ expression is repressed by the addition of thiamine, the cells cease division after 12 to 14 h (data not shown). We looked for accumulation of the precursor form of two intron-containing genes (U6 and tf2d) by Northern blot analysis over a 12-h time course of cdc5 repression using wild-type and prp2-1 cells as controls. The S. pombe U6 RNA is synthesized from a precursor RNA that contains a single pre-mRNA-type intron (60, 71). The tf2d gene contains three. Like prp2-1 cells, cells lacking cdc5 function accumulated unspliced RNAs (Fig. 8), indicating that cdc5+ function is required for pre-mRNA splicing.
FIG. 8.
cdc5+ function is required for pre-mRNA splicing. RNA was prepared from KGY1490 cells grown in the presence of thiamine for 0, 4, 8, or 12 h, prp2-1 cells grown at 37°C for 0, 2, or 4 h, or wild-type (wt) cells and then hybridized to oligonucleotides complementary to the U6 intron, mature U6 RNA, and both intron and exon sequences within the tf2d gene.
DISCUSSION
S. pombe cdc5+ was identified in a screen for cell division cycle mutants on the basis of a single mutant allele, cdc5-120 (51). Further characterization of the cell cycle defects of cdc5-120 showed that its 87-kDa Myb-related gene product was essential specifically for G2/M progression (51, 56). Subsequent cloning and characterization of cdc5p orthologs from other organisms have indicated that cdc5p’s role in cell cycle progression is likely to be conserved; expression of cdc5 relatives from A. thaliana, D. melanogaster, and human rescues the growth defect of cdc5-120 (25, 55). The fact that the S. cerevisiae ortholog, Cef1p, also is essential for G2/M progression argues strongly that cdc5p function is conserved throughout evolution (55). In this report, we show that this highly conserved protein is a subunit of a discrete 40S complex that contains known protein and snRNA components of the pre-mRNA splicing machinery. Further, cdc5+ function is required for pre-mRNA splicing.
The discovery that cdc5p is a member of a stable high-molecular-weight complex was unexpected since c-Myb and most other Myb-related proteins function either as monomers or as heterodimers (see, for example, references 17, 30, 40, and 52). However, there are precedents for Myb-related proteins being components of larger protein complexes. The Myb-related SNAP190 protein is found in the SNAPc complex that is involved in the basal transcription of snRNAs (80). Two other Myb-related proteins, Swi3p (57) and Rsc8p (11), are subunits of the even larger, megadalton-size SWI/SNF and RSC complexes that are important for chromatin remodeling in vivo (10–12, 26). As yet, it is not clear what the functions of the Swi3p and Rsc8p proteins are within these large complexes. It is possible that the Myb repeats of these proteins are not important for binding to DNA since the Myb repeats of several proteins have been shown to be involved in protein-protein interactions (16, 30, 31).
To gain insight into the function of cdc5p, the 40S complex was purified, and 10 subunits were identified, each representing previously undescribed S. pombe open reading frames. These identifications raised the unanticipated possibility that cdc5p is involved in pre-mRNA splicing, as five cwfs have orthologs involved in this process. cwf9p is the S. pombe core snRNP protein D2 and thus is predicted to be a subunit of all spliceosomal snRNPs: U1, U2, U4/6, and U5 (34, 62, 70). Since cwf6p/prp15p is the apparent ortholog of S. cerevisiae and human U5 snRNP proteins Prp8p/p220, it is likely that cwf6p/prp15p is a subunit of the S. pombe U5 snRNP (1). cwf10p also should be a component of the S. pombe U5 snRNP since it is highly related to human U5-116 and S. cerevisiae Snu114p (19, 20). cwf8p is the S. pombe ortholog of S. cerevisiae Prp19p, a protein known to have an essential role in pre-mRNA processing (13, 14). Consistent with cdc5p’s association with these known spliceosomal proteins, the human homolog of cdc5p, as well as cwf1p and cwf8p, recently has been reported to be a component of an in vitro-assembled mammalian spliceosome (48). It is of note that we have identified several highly conserved proteins as a part of this complex that previously have not been identified as important for pre-mRNA splicing in any organism or involved in cell cycle progression. It will be important to determine whether these novel proteins are indeed required for one or both processes. In addition to these proteins, we found that cdc5p copurified with a specific set of snRNAs, which confirmed that cdc5p is indeed snRNP associated.
There are two likely possibilities to explain the presence of predicted components of the S. pombe spliceosome within the cdc5p complex. One is that this complex represents an active S. pombe spliceosome, a possibility supported by the presence of those snRNAs which would be predicted to be present within a catalytically active spliceosome (U2, U5, and U6) (references 43, 45, and 50 and references therein), and that the size of this complex is reminiscent of that of an in vitro-assembled S. cerevisiae spliceosome (35). While CDC5 family members are capable of associating with an in vitro-assembled spliceosome (9, 49), proof that this complex represents an in vivo spliceosome would require a demonstration that splicing intermediates are present within this complex. A second possibility is that the cdc5p-associated complex represents a remnant of a disassembled spliceosome. If this were the case, then the complex might contain excised introns or else completely lack intermediates or products of the splicing reaction. The fact that cells lacking cdc5+ function accumulate unspliced RNAs supports either hypothesis, since a factor necessary for recycling spliceosomal components for subsequent rounds of pre-mRNA splicing would indirectly be needed for pre-mRNA processing. Further analysis of the RNA composition of cdc5p complexes should allow us to distinguish between these possibilities.
A major question raised by this and other studies is why mutants in components of the pre-mRNA splicing machinery generate specific cell cycle phenotypes (8). For example, S. cerevisiae Prp8p has not only been shown to be an essential splicing factor but also seems to be important for S-phase progression (68). Interestingly, it is possible to isolate mutants that are defective for only one of these two processes; prp8 mutants have a splicing defect but no apparent cell cycle defects, and dbf3 mutants are defective for S-phase progression but show no defects in macromolecular RNA synthesis (28, 29, 68). In S. pombe, most prp mutants exhibit cell cycle defects, but they do not all show the same cell cycle defects (37, 59, 77). cwf1+/prp5+ itself has been shown not only to display a defect in pre-mRNA splicing but also blocks cell cycle progression in G2 (59). One better-characterized example is the cdc28+/prp8+ gene, which, like cdc5+ and cwf1+/prp5+, appears to be required for both cell cycle progression at G2/M and pre-mRNA splicing (37). In contrast to the strong negative genetic interaction between cdc5-120 and prp5-1, there is no genetic interaction between cdc5-120 and a cdc28 mutant and no evidence that cdc28p is in the cdc5p complex (data not shown).
The most straightforward explanation for the cell cycle arrest phenotypes of pre-mRNA splicing mutants is that an intron-containing transcript(s) becomes rate limiting for a particular cell cycle transition. Since we have found that cells lacking CEF1 function also are defective in pre-mRNA splicing (9), this possibility can be more easily addressed in S. cerevisiae, in which just 4% of the genes are predicted to contain introns. Another possibility to explain why pre-mRNA splicing mutants arrest at the G2/M transition in two widely disparate yeasts is that defects in pre-mRNA splicing lead to changes in the organization of the nucleus such that cell cycle progression through mitosis is prevented. This possibility is more difficult to address experimentally, unless the G2 arrest is mediated by a checkpoint. If it is, it should be possible to isolate extragenic mutants that allow cdc5-120 cells to enter mitosis or mutants within cdc5+ which prevent cell cycle progression but do not affect pre-mRNA splicing. Analysis of such mutants would allow us to more clearly dissect this relationship between cell cycle progression and pre-mRNA splicing.
ACKNOWLEDGMENTS
We thank J. Bahler and J. R. Pringle for providing us with the myc13-kan and HA3-kan tagging cassettes. We are grateful to J. Leszyk (Worcester Foundation) for the expert peptide sequencing of isolated proteins and R. Carnahan for his assistance in the epitope tagging of cwf10p-myc. We thank all members of the Gould lab, including D. McCollum, M. K. Balasubramanian, and L. D. Berry, for their valuable discussions and technical advice. N.S. and D.F. also thank J. Niemiec for dedicated technical assistance.
This work was supported by NIH grants GM47728 to K.L.G. and GM38242 to D.F. W.H.M. was supported by NCI grant T32 CA09592. D.F. was the recipient of an Irma T. Hirschl Career Scientist Award. K.L.G. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Anderson G J, Bach M, Luhrmann R, Beggs J D. Conservation between yeast and man of a protein associated with U5 small nuclear ribonucleoprotein. Nature. 1989;342:819–821. doi: 10.1038/342819a0. [DOI] [PubMed] [Google Scholar]
- 2.Bahler J, Wu J Q, Longtine M S, Shah N G, McKenzie III A, Steever A B, Wach A, Philippsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian M K, McCollum D, Gould K L. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- 4.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point of thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 5.Beggs J D, Teigelkamp S, Newman A J. The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J Cell Sci Suppl. 1995;19:101–105. doi: 10.1242/jcs.1995.supplement_19.15. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein H S, Coughlin S R. A mammalian homolog of fission yeast Cdc5 regulates G2 progression and mitotic entry. J Biol Chem. 1998;273:4666–4671. doi: 10.1074/jbc.273.8.4666. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein H S, Coughlin S R. Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J Biol Chem. 1997;272:5833–5837. doi: 10.1074/jbc.272.9.5833. [DOI] [PubMed] [Google Scholar]
- 8.Burns, C. G., and K. L. Gould. Connections between pre-mRNA processing and regulation of the eukaryotic cell cycle. In S. L. Chew (ed.), Posttranscriptional regulation of gene expression and its importance to the endocrine system, in press. Karger, Basel, Switzerland.
- 9.Burns, C. G., R. Ohi, A. Krainer, and K. L. Gould. Myb-related CDC5 proteins are required for pre-messenger RNA splicing. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 10.Burns L G, Peterson C L. The yeast SWI-SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 12.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng S C. Formation of the yeast splicing complex A1 and association of the splicing factor PRP19 with the pre-mRNA are independent of the 3′ region of the intron. Nucleic Acids Res. 1994;22:1548–1554. doi: 10.1093/nar/22.9.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng S C, Tarn W Y, Tsao T Y, Abelson J. PRP19: a novel spliceosomal component. Mol Cell Biol. 1993;13:1876–1882. doi: 10.1128/mcb.13.3.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cottarel G, Beach D, Deuschle U. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr Genet. 1993;23:547–548. doi: 10.1007/BF00312650. [DOI] [PubMed] [Google Scholar]
- 16.Cutler G, Perry K M, Tjian R. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol Cell Biol. 1998;18:2252–2261. doi: 10.1128/mcb.18.4.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 18.Demeter J, Morphew M, Sazer S. A mutation in the RCC1-related protein pim1 results in nuclear envelope fragmentation in fission yeast. Proc Natl Acad Sci USA. 1995;92:1436–1440. doi: 10.1073/pnas.92.5.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabrizio P, Esser S, Kastner B, Luhrmann R. Isolation of S. cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science. 1994;264:261–265. doi: 10.1126/science.8146658. [DOI] [PubMed] [Google Scholar]
- 20.Fabrizio P, Laggerbauer B, Lauber J, Lane W S, Luhrmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groenen P M, Vanderlinden G, Devriendt K, Fryns J P, van de Ven W J. Rearrangement of the human CDC5L gene by a t(6;19)(p21;q13.1) in a patient with multicystic renal dysplasia. Genomics. 1998;49:218–229. doi: 10.1006/geno.1998.5254. [DOI] [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 23.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Luhrmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirano T, Hiraoka Y, Yanagida M. A temperature-sensitive mutation of the Schizosaccharomyces pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirayama T, Shinozaki K. A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:13371–13376. doi: 10.1073/pnas.93.23.13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 27.James P, Quadroni M, Carafoli E, Gonnet G. Protein identification in DNA databases by peptide mass fingerprinting. Protein Sci. 1994;3:1347–1350. doi: 10.1002/pro.5560030822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston L H, Thomas A P. A further two mutants defective in initiation of the S phase in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:445–448. doi: 10.1007/BF00729467. [DOI] [PubMed] [Google Scholar]
- 29.Johnston L H, Thomas A P. The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:439–444. doi: 10.1007/BF00729466. [DOI] [PubMed] [Google Scholar]
- 30.Kanei-Ishii C, Tanikawa J, Nakai A, Morimoto R I, Ishii S. Activation of heat shock transcription factor 3 by c-Myb in the absence of cellular stress. Science. 1997;277:246–248. doi: 10.1126/science.277.5323.246. [DOI] [PubMed] [Google Scholar]
- 31.Kanei-Ishii C, Yasukawa T, Morimoto R I, Ishii S. c-Myb-induced trans-activation mediated by heat shock elements without sequence-specific DNA binding of c-Myb. J Biol Chem. 1994;269:15768–15775. [PubMed] [Google Scholar]
- 32.Keeney J B, Boeke J D. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehmeier T, Raker V, Hermann H, Luhrmann R. cDNA cloning of the Sm proteins D2 and D3 from human small nuclear ribonucleoproteins: evidence for a direct D1-D2 interaction. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin R J, Lustig A J, Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987;1:7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- 36.Lipsick J S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 37.Lundgren K, Allan S, Urushiyama S, Tani T, Ohshima Y, Frendewey D, Beach D. A connection between pre-mRNA splicing and the cell cycle in fission yeast: cdc28+ is allelic with prp8+ and encodes an RNA-dependent ATPase/helicase. Mol Biol Cell. 1996;7:1083–1094. doi: 10.1091/mbc.7.7.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lussier M, White A M, Sheraton J, di Paolo T, Treadwell J, Southard S B, Horenstein C I, Chen-Weiner J, Ram A F, Kapteyn J C, Roemer T W, Vo D H, Bondoc D C, Hall J, Zhong W W, Sdicu A M, Davies J, Klis F M, Robbins P W, Bussey H. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marks J, Fankhauser C, Reymond A, Simanis V. Cytoskeletal and DNA structure abnormalities result from bypass of requirement for the cdc10 start gene in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1992;101:517–528. doi: 10.1242/jcs.101.3.517. [DOI] [PubMed] [Google Scholar]
- 40.Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 41.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 42.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 43.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. pp. 303–358. [Google Scholar]
- 44.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 45.Murray H L, Jarrell K A. Flipping the switch to an active spliceosome. Cell. 1999;96:599–602. doi: 10.1016/s0092-8674(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 46.Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- 47.Nemeth K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kalman Z, Stankovic-Stangeland B, Bako L, Mathur J, Okresz L, Stabel S, Geigenberger P, Stitt M, Redei G P, Schell J, Koncz C. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 1998;12:3059–3073. doi: 10.1101/gad.12.19.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Luhrmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 50.Nilsen T W. RNA-RNA interactions in the spliceosome: unraveling the ties that bind. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 51.Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 52.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb- specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 53.Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A, et al. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat Struct Biol. 1995;2:309–320. doi: 10.1038/nsb0495-309. [DOI] [PubMed] [Google Scholar]
- 54.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 55.Ohi R, Feoktistova A, McCann S, Valentine V, Look A T, Lipsick J S, Gould K L. Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol Cell Biol. 1998;18:4097–4108. doi: 10.1128/mcb.18.7.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohi R, McCollum D, Hirani B, Den Haese G J, Zhang X, Burke J D, Turner K, Gould K L. The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J. 1994;13:471–483. doi: 10.1002/j.1460-2075.1994.tb06282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 58.Potashkin J, Frendewey D. Splicing of the U6 RNA precursor is impaired in fission yeast pre-mRNA splicing mutants. Nucleic Acids Res. 1989;17:7821–7831. doi: 10.1093/nar/17.19.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Potashkin J, Kim D, Fons M, Humphrey T, Frendewey D. Cell-division-cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr Genet. 1998;34:153–163. doi: 10.1007/s002940050381. [DOI] [PubMed] [Google Scholar]
- 60.Potashkin J, Li R, Frendewey D. Pre-mRNA splicing mutants of Schizosaccharomyces pombe. EMBO J. 1989;8:551–559. doi: 10.1002/j.1460-2075.1989.tb03409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prentice H L. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raker V A, Plessel G, Luhrmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 63.Reyes J L, Kois P, Konforti B B, Konarska M M. The canonical GU dinucleotide at the 5′ splice site is recognized by p220 of the U5 snRNP within the spliceosome. RNA. 1996;2:213–225. [PMC free article] [PubMed] [Google Scholar]
- 64.Rotondo G, Gillespie M, Frendewey D. Rescue of the fission yeast snRNA synthesis mutant snm1 by overexpression of the double-strand-specific Pac1 ribonuclease. Mol Gen Genet. 1995;247:698–708. doi: 10.1007/BF00290401. [DOI] [PubMed] [Google Scholar]
- 65.Russell P, Nurse P. Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell. 1986;45:781–782. doi: 10.1016/0092-8674(86)90550-7. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 67.Sazer S, Sherwood S W. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 68.Shea J E, Toyn J H, Johnston L H. The budding yeast U5 snRNP Prp8 is a highly conserved protein which links RNA splicing with cell cycle progression. Nucleic Acids Res. 1994;22:5555–5564. doi: 10.1093/nar/22.25.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stukenberg P T, Lustig K D, McGarry T J, King R W, Kuang J, Kirschner M W. Systematic identification of mitotic phosphoproteins. Curr Biol. 1997;7:338–348. doi: 10.1016/s0960-9822(06)00157-6. [DOI] [PubMed] [Google Scholar]
- 70.Sumpter V, Kahrs A, Fischer U, Kornstadt U, Luhrmann R. In vitro reconstitution of U1 and U2 snRNPs from isolated proteins and snRNA. Mol Biol Rep. 1992;16:229–240. doi: 10.1007/BF00419662. [DOI] [PubMed] [Google Scholar]
- 71.Tani T, Ohshima Y. The gene for the U6 small nuclear RNA in fission yeast has an intron. Nature. 1989;337:87–90. doi: 10.1038/337087a0. [DOI] [PubMed] [Google Scholar]
- 72.Tarn W Y, Hsu C H, Huang K T, Chen H R, Kao H Y, Lee K R, Cheng S C. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tarn W Y, Lee K R, Cheng S C. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc Natl Acad Sci USA. 1993;90:10821–10825. doi: 10.1073/pnas.90.22.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teigelkamp S, Newman A J, Beggs J D. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teigelkamp S, Whittaker E, Beggs J D. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 1995;23:320–326. doi: 10.1093/nar/23.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Umen J G, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Urushiyama S, Tani T, Ohshima Y. Isolation of novel pre-mRNA splicing mutants of Schizosaccharomyces pombe. Mol Gen Genet. 1996;253:118–127. doi: 10.1007/s004380050304. [DOI] [PubMed] [Google Scholar]
- 78.Verhasselt P, Volckaert G. Sequence analysis of a 37.6 kbp cosmid clone from the right arm of Saccharomyces cerevisiae chromosome XII, carrying YAP3, HOG1, SNR6, tRNA-Arg3 and 23 new open reading frames, among which several homologies to proteins involved in cell division control and to mammalian growth factors and other animal proteins are found. Yeast. 1997;13:241–250. doi: 10.1002/(SICI)1097-0061(19970315)13:3<241::AID-YEA61>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 79.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 80.Wong M W, Henry R W, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–377. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wright A, Maundrell K, Heyer W D, Beach D, Nurse P. Vectors for the construction of gene banks and the integration of cloned genes in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Plasmid. 1986;15:156–158. doi: 10.1016/0147-619x(86)90051-x. [DOI] [PubMed] [Google Scholar]
- 82.Xia G, Ramachandran S, Hong Y, Chan Y S, Simanis V, Chua N H. Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 1996;10:761–769. doi: 10.1046/j.1365-313x.1996.10040761.x. [DOI] [PubMed] [Google Scholar]
- 83.Zhang K, Smouse D, Perrimon N. The crooked neck gene of Drosophila contains a motif found in a family of yeast cell cycle genes. Genes Dev. 1991;5:1080–1091. doi: 10.1101/gad.5.6.1080. [DOI] [PubMed] [Google Scholar]