Abstract
The estrogen receptor (ER) regulates the expression of target genes in a ligand-dependent manner. The ligand-dependent activation function AF-2 of the ER is located in the ligand binding domain (LBD), while the N-terminal A/B domain (AF-1) functions in a ligand-independent manner when isolated from the LBD. AF-1 and AF-2 exhibit cell type and promoter context specificity. Furthermore, the AF-1 activity of the human ERα (hERα) is enhanced through phosphorylation of the Ser118 residue by mitogen-activated protein kinase (MAPK). From MCF-7 cells, we purified and cloned a 68-kDa protein (p68) which interacted with the A/B domain but not with the LBD of hERα. Phosphorylation of hERα Ser118 potentiated the interaction with p68. We demonstrate that p68 enhanced the activity of AF-1 but not AF-2 and the estrogen-induced as well as the anti-estrogen-induced transcriptional activity of the full-length ERα in a cell-type-specific manner. However, it did not potentiate AF-1 or AF-2 of ERβ, androgen receptor, retinoic acid receptor alpha, or mineralocorticoid receptor. We also show that the RNA helicase activity previously ascribed to p68 is dispensable for the ERα AF-1 coactivator activity and that p68 binds to CBP in vitro. Furthermore, the interaction region for p68 in the ERα A/B domain was essential for the full activity of hERα AF-1. Taken together, these findings show that p68 acts as a coactivator specific for the ERα AF-1 and strongly suggest that the interaction between p68 and the hERα A/B domain is regulated by MAPK-induced phosphorylation of Ser118.
Fat-soluble ligands such as steroid and thyroid hormones, retinoids, and vitamin D regulate many biological processes, including cell proliferation and differentiation, through transcriptional control of target gene expression mediated by their cognate receptors (5, 10, 32). These nuclear receptors belong to a large family of ligand-inducible transcription factors, and studies of their molecular structure have identified five or six functional domains (designated A to F). The C domain, located in the middle of the protein, is responsible for specific binding to DNA-responsive elements. The ligand binding domain (LBD) is localized in the C-terminal E/F domain and contains a ligand-dependent transcriptional activation function (AF-2) (30, 46), whereas another activation function (AF-1), which is constitutive in the absence of the LBD, is located in the A/B region (46). The activities of both AF-1 and AF-2 are promoter context and cell type specific and can synergize (45). The role of AF-1 in the ligand-induced transactivation function of estrogen receptor α (ERα) is prominent in the actions of estrogen (17β-estradiol [E2]) and partial agonists such as tamoxifen. Tamoxifen blocks the AF-2 but not the AF-1 activity of ERα (6), exerting agonistic or antagonistic activities in a tissue-specific way. Moreover, we have shown that the human ERα (hERα) AF-1 activity is potentiated through phosphorylation of the Ser118 residue by mitogen-activated protein kinase (MAPK), which is activated by growth factors such as insulin, insulin-like growth factors and tumor necrosis factor alpha (26). To achieve such complex AF activity, the presence of common transcriptional mediators or cofactors mediating AF activity to basic transcription machinery was suggested. The observations that AF-1 and AF-2 of steroid receptors are transcriptionally squelched or interfere with each other further supported this idea (37, 45). Some components of the basic transcription machinery such as TATA-associated factors (22, 35, 36) and TFIIB (4) associate with nuclear receptors in a ligand-independent manner. Indeed, putative transcriptional mediators and cofactors interacting with and activating the AF-2 activities of nuclear receptors in a ligand-dependent fashion have been recently identified (17). They include the TIF2/SRC-1 (2, 9, 11, 39, 47) and CBP/p300 families (24), TIF1 (29), ARA70 (52), and many others (40). Some of these factors (CBP/p300 and SRC-1) exhibit autonomous histone acetyltransferase activity (11, 42) and appear to associate with P/CAF (51), and therefore some of the effects at the chromatin level might be mediated by histone acetylation. Despite the information on coactivators for AF-2, little is known about possible coactivator(s) for AF-1.
To study the AF-1 of the ERα with respect to the MAPK-induced potentiation of this activity, we sought to identify a coactivator for AF-1 of the hERα. Here we report the purification and cloning of a 68-kDa protein (p68) and show that it interacts with the hERα A/B domain but not with its LBD and that this interaction is enhanced by the MAPK-mediated phosphorylation of the hERα at Ser118. Furthermore, the transcriptional activity of ERα AF-1, but not AF-2, was potentiated by p68, whereas overexpression of p68 had no effect on either AF-1 or AF-2 of ERβ, androgen receptor, retinoic acid receptor alpha, or mineralocorticoid receptor. The RNA helicase activity previously ascribed to p68 (12, 16) was dispensable for the coactivator activity. p68 bound to CBP in vitro. Thus, these findings demonstrate that p68 acts as a coactivator specific for the ERα AF-1 and indicate that phosphorylation of the hERα at Ser118 by MAPK potentiates this interaction.
MATERIALS AND METHODS
Vectors.
The GAL4 chimeric proteins with hERα deletion mutants [pM-A/B(1–180), pM-AB-Ms, and pM-LBD(295–595)], with human ERβ deletion mutants [pM-A/B(1–95) and pM-E/F(213–477)], human androgen receptor A/B domain [pM-AR(1–557)], human mineralocorticoid receptor A/B domain [pM-MR(1–279)], and human retinoic acid receptor alpha A/B domain [pM-RARα(1–99)] were generated by subcloning cDNA fragments into the appropriate sites of the multicloning region of the pM vector (Clontech). The regions of the nuclear receptors subcloned are given as the numbers of the amino acid sequences in parentheses. VP16-hERα and glutathione S-transferase-tagged hERα (GST-hERα) fusion protein expression vectors were prepared in the same way, by subcloning into pVP16 (Clontech) and pGEX-2T/4T (Amersham Pharmacia Biotech), respectively. p68 cDNA was isolated by reverse transcription-PCR from an MCF-7 cDNA library, and the PCR-amplified cDNA was subcloned into the appropriate multicloning sites of the expression vectors pET-17b (Novagen), pM, and pSG5. SRC-1 and human ERβ cDNAs were isolated from the HeLa cDNA library, and the expression vectors were constructed by introducing the cDNAs, which were verified (27, 39), into pcDNA3 (Invitrogen) (44, 50). Amino acid substitution of p68 (p68K144R) was performed by PCR-based mutagenesis with primers 5′-CAGACTGGATCTGGGAGAACATTGT-CTTATTTGC-3′ and 5′-GGAAGCAAA-TAAGACAATGTTCTCCCAGATCCAG-3′.
GST pull-down and in vitro binding assays.
GST-ER/p68 fusion proteins were expressed in bacteria and bound to glutathione-Sepharose 4B (Pharmacia) beads as described previously (20). Metabolic labeling of MCF-7 was performed by culturing the cells for 4 h in medium containing [35S]methionine (Amersham Pharmacia Biotech) (9). In vitro translation for the linearized expression vectors was performed with [35S]methionine by using the TNT kit (Promega). For the GST pull-down assay, a 50% suspension of GST-protein beads (50 μl), which contained up to 1.0 μg of protein, was resuspended in the same volume of binding buffer (20 mM Tris-HCl [pH 7.5], 0.12 M NaCl, 10% [vol/vol] glycerol, 0.055% 2-mercaptoethanol, 1 mM EDTA, 0.1 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5% Nonidet P-40) (50). The nuclear extract of 35S-labelled MCF-7 (3 × 106 cells) in 300 μl of binding buffer or an aliquot (15 μl) of the in vitro translation reaction mixture was mixed with GST-protein beads and suspended for 1 h at 4°C. The beads were then washed four times with washing buffer (replacing 0.12 M NaCl in the binding buffer with 0.1 M NaCl) and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. After electrophoresis, radiolabeled proteins were visualized with an image analyzer (BAS2000; Fuji Film, Tokyo, Japan).
In vitro phosphorylation.
The GST-HE15 and GST-HE15/457 proteins (5 μg) purified on glutathione-Sepharose 4B beads were phosphorylated in vitro with MAPK as described previously (26). The activated MAPK was a gift from E. Nishida (Kyoto University, Kyoto, Japan). Thus, phosphorylated GST fusion proteins were separated from the free ATP and used as probes in the GST pull-down experiment.
Purification and microsequencing of the 68-kDa protein.
The nuclear extract of MCF-7 cells was incubated for 1 h at 4°C with 30 mg of GST-HE15 protein immobilized on 750 μl of glutathione-Sepharose. After the beads were washed three times in 1.5 ml of washing buffer (replacing 0.15 M NaCl in binding buffer with 0.1 M NaCl), proteins were eluted in 750 μl of elution buffer 1 (replacing 0.15 M NaCl in binding buffer with 0.2 M NaCl). After mixing for 5 min, the supernatant was removed again and the beads were resuspended in 750 μl of elution buffer 2 (replacing 0.15 M NaCl in binding buffer with 0.3 M NaCl). After mixing for 5 min, the supernatant was collected and concentrated by trichloroacetic acid precipitation. The precipitated proteins were resuspended in SDS-PAGE sample buffer, electrophoresed on a 10% polyacrylamide gel, and electroblotted on a polyvinylidene difluoride (PVDF) membrane (ProBlott; Perkin-Elmer, Norwalk, Conn.) at 7 V/cm at 4°C for 15 h in 10 mM cyclohexyl aminopropene sulfonic acid-NaOH (pH 11)–10% methanol. After blotting, protein bands were visualized by Coomassie brilliant blue staining, and the band corresponding to the p68 protein was excised. Cysteine residues of the protein on the PVDF membrane were carboxymethylated by reductive alkylation as described by Iwamatsu (21). S-carboxymethylated proteins were digested in situ with lysylendopeptidase, and the resulting peptides were separated by micro-fast-performance liquid chromatography (FPLC) with the SMART system (Amersham Pharmacia Biotech) (21). Microsequencing was performed with a 473A protein sequencer (Perkin-Elmer). Peptide fragment sequences were searched in the nucleotide/protein database by using the BLAST search method.
Northern analysis.
MTN blots were obtained from Clontech. We also extracted and analyzed poly(A)+ RNA from various human cell lines derived from estrogen target tissues, as follows: MCF-7, breast cancer cell line; T47D, breast cancer cell line; LNCaP, prostate carcinoma cell line; HOS, osteosarcoma cell line; HTOA, ovarian cancer cell line; Ishikawa, uterine body cancer cell line. The cDNA probe of p68 RNA helicase was labeled with [α-32P]dCTP by the random-primer method. Prehybridizations and hybridizations were done as described previously (43). A human glyceraldehyde-3-phosphate dehydrogenase probe from Clontech was used for control experiments.
Transient transfection, reporter assay, and mammalian two-hybrid assay.
For transfection, COS-1 cells were seeded in 100-mm dishes containing phenol red-free Dulbecco’s minimal essential medium (GIBCO BRL) supplemented with 10% charcoal dextran-treated fetal bovine serum. At 40 to 60% confluency, the cells were transfected with 2 μg of ERE-G-CAT or 17M2G-CAT reporter plasmid, 0.4 μg of ER expression vector, 0.15 to 0.6 μg of p68 expression vector, and 3 μg of pCH110 β-galactosidase reporter, and Bluescribe M13+ was used as the carrier DNA to adjust the total amount of DNA (26). After 20 to 24 h, the medium was replaced with fresh medium with or without 10−7 M E2, 10−7 M 4-hydroxytamoxifen (OHT) or 10−7 M ICI164,384 (ICI). After 24 to 48 h, the cells were lysed to determine the β-galactosidase and chloramphenicol acetyltransferase activity (25).
Immunoblot analysis.
The cell lysates of COS-1 transfected both with hERα and the p68 expression vector were subjected to SDS-PAGE and transferred to a nitrocellulose membrane (Hybond-ECL; Amersham Pharmacia Biotech). The membrane was probed with the monoclonal anti-hERα antibody F3 (1) and then with the peroxidase-labeled second antibody. Antibody staining was visualized with an enhanced chemiluminescence system (Amersham Pharmacia Biotech).
RESULTS
A 68-kDa protein interacts with the hERα A/B domain, but not the E/F domain.
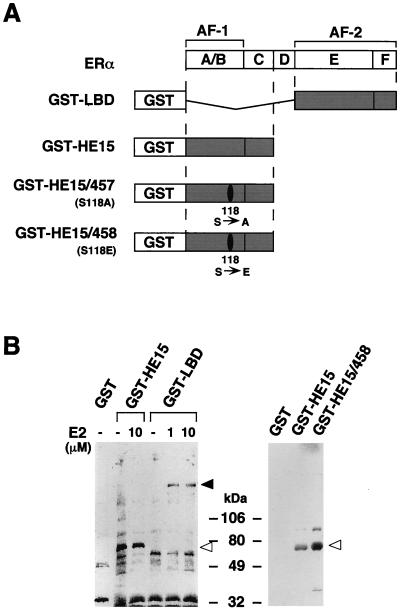
To isolate a hERα AF-1-specific coactivator, a 35S-labeled MCF-7 nuclear extract was loaded on a GST-tagged hERα N-terminal A–C region (GST-HE15) (Fig. 1A) column. After extensive washing, a 68-kDa protein (p68) interacting with GST-HE15 was detected (Fig 1B). Under similar conditions, no 68-kDa protein interacted with GST alone or with the ERα LBD (GST-LBD [Fig. 1]) in either the absence or presence of 17β-estradiol (E2) whereas a 160-kDa protein (9, 14) specifically interacted with GST-LBD in the presence of E2 (Fig. 1B). Interestingly, the interaction of p68 with the hERα N-terminal region was enhanced by replacing the hERα Ser118 residue with a glutamate residue (S118E; GST-HE15/458 [Fig. 1B, right panel]). Since this amino acid substitution mimics the phosphorylation of the Ser118 residue in terms of transactivation properties (1), these results indicated that the interaction between p68 and the hERα A/B region might be increased by phosphorylation of Ser118.
FIG. 1.
Detection of binding proteins to the hERα A/B domain in MCF-7 cells. (A) The hERα (with regions A to F shown) and the GST-ER fusion proteins used. (B) Binding proteins to the hERα A/B domain in MCF-7 cells. Aliquots of the 35S-labeled MCF-7 nuclear extract were incubated with glutathione-Sepharose beads loaded with GST alone, GST-HE15, GST-HE15/458, GST-HE15/457, or GST-LBD in the absence or presence of E2 at 1 and 10 μM. The bound proteins were subjected to SDS-PAGE (5 to 20% polyacrylamide gradient gel) followed by autoradiography. Open arrowheads indicate the position of a protein of 68 kDa. Size markers are indicated in kilodaltons. The solid arrowhead indicates the position of the SRC-1/TIF2 160-kDa family proteins (9, 14).
From these findings, we speculated that p68 is a putative coactivator specific for ERα AF-1 and is involved in the enhancement of AF-1 activity by the MAPK-mediated phosphorylation of the Ser118 residue in the hERα A/B region.
Peptide microsequencing identifies the purified 68-kDa protein as p68 RNA helicase.
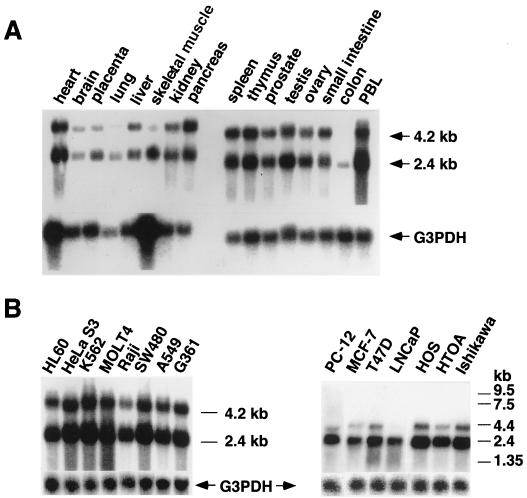
To purify p68 from the MCF-7 cell nuclear extracts, we used a GST pull-down assay to concentrate the protein. The p68 protein absorbed to a GST-chimeric hERα A/B fusion protein (GST-HE15) on the beads was dissociated in a 0.25 to 0.3 M NaCl fraction in a stepwise elution, subjected to SDS-PAGE, and electroblotted on a PVDF membrane. The 68-kDa band was cut out of the Coomassie brilliant blue (G)-stained membrane, S-carboxymethylated, and digested with lysylendopeptidase. The five digested peptides were isolated by reversed-phase semi-micro-high-performance liquid chromatography. The peptide sequences of five peptides perfectly matched the sequences of the known p68 RNA helicase (31), as underlined in Fig. 2. We cloned the p68 RNA helicase cDNA by reverse transcription-PCR from an MCF-7 cDNA library and verified the DNA sequence. With this cloned cDNA, we studied the tissue distribution of the p68 transcripts and their expression in cell lines by Northern blotting. Two transcripts of mouse p68 of 2.2 and 3.5 kb were found in human tissue (31). We detected human p68 transcripts of 2.4 and 4.2 kb (Fig. 3), both of which were expressed ubiquitously in all tissues except the colon. These findings reveal no significant tissue specificity in p68 gene expression. Relatively high-level expression of p68 was seen in all cell lines; however, even in steroid hormone-dependent cell lines like MCF-7 and LNCaP, cell type specificity in p68 gene expression was not detected (Fig. 3B, right panel). The levels of the two transcripts differed among tissues and cell lines, and the smaller transcript (2.4 kb) was expressed at higher levels than the 4.2-kb transcript. However, the biological significance of the difference in p68 transcript size remains to be elucidated.
FIG. 2.
Amino acid sequence of human p68 RNA helicase protein. The five sequences determined by microsequencing are underlined and completely matched to the reported p68 RNA helicase protein (GenBank accession no. X15729 and X52104). The nuclear receptor recognition motif (LXXLL motif [15]) is doubly underlined, and the DEAD box motif is boxed.
FIG. 3.
Expression patterns of p68 RNA helicase transcripts in normal human tissues (A) and cancer cell lines (B). Northern blot analysis was performed as described previously (43). PBL, peripheral blood leukocyte. The cancer cell lines are as follows: HL60, promyelocytic leukemia cell line; HeLa S3, cervical carcinoma cell line; K562, chronic myelogenous leukemia cell line; Raji, Burkitt’s lymphoma cell line; SW480, colorectal adenocarcinoma cell line; A549, lung carcinoma cell line; G361, melanoma cell line; MCF-7, breast cancer cell line; T47D, breast cancer cell line; LNCaP, prostate carcinoma cell line; HOS, osteosarcoma cell line; HTOA, ovarian cancer cell line; Ishikawa, uterine body cancer cell line; PC12, rat pheochromocytoma cell line. The glyceraldehyde-3-phosphate dehydrogenase (G3PDH) transcript was used as an internal control. The relative positions of RNA markers (in kilobases) are shown on the right of panel B.
Direct interaction of p68 with the hERα A/B domain in vitro and in vivo.
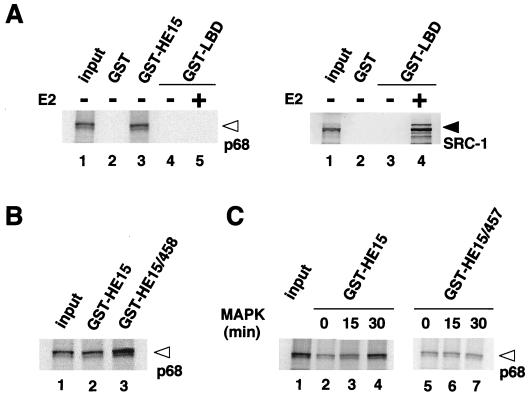
A possible p68 interaction with the hERα A/B domain was studied in vitro with a recombinant p68 protein produced by in vitro translation in rabbit reticulocyte with the cloned p68 cDNA. The interaction of p68 with the hERα A/B domain was tested by the GST pull-down assay (Fig. 1A), used to purify endogenous p68 protein from MCF-7 cells. The recombinant p68 protein interacted with GST-HE15 but not with GST-LBD in both the presence and absence of E2 (Fig. 4A) and the E2 antagonists OHT and ICI (data not shown), consistent with previous data (Fig. 1). Lack of association of p68 with the LBD was further supported by the fact that in vitro-translated SRC-1 binds strongly to this GST-LBD in an E2-dependent manner (Fig. 4A, right panel). The interaction of p68 with the A/B region was further potentiated when the Ser118 residue was replaced with Glu (S118E) (Fig. 4B), suggesting that the MAPK-mediated phosphorylation of Ser118 increases the binding of p68 to the A/B domain. To directly test this idea, GST-HE15 was phosphorylated by MAPK in vitro and subjected to a GST pull-down assay. The binding of p68 to the A/B region appeared to be dependent to some extent on the phosphorylation of the A/B region by MAPK (Fig. 4C). In addition, the binding preference that was increased by the phosphorylation of Ser118 was abrogated by replacement of Ser118 with Ala in the A/B region (S118A) (GST-HE15/457; Fig. 4C).
FIG. 4.
The recombinant p68 RNA helicase specifically binds to the hERα A/B domain in vitro. (A) p68 interacts only with the hERα A/B domain but not with the LBD, irrespective of the presence of E2. In vitro-translated p68 RNA helicase recombinant protein (left panel, lane 1) was analyzed by SDS-PAGE (5 to 20% polyacrylamide gradient gel). GST-ER fusion proteins, immobilized on beads, were mixed with 15 μl of in vitro translation reaction mixtures of p68; 2 μl of translation reaction mixture was loaded on the input lane. SRC-1 interacts with the hERα LBD only in the presence of E2 (right panel; lane 4) as previously reported (39). The open arrowhead indicates the position of a p68 RNA helicase protein, and the solid arrowhead indicates the position of the SRC-1 protein. (B) The p68 interaction is enhanced by replacing the Ser118 residue with Glu (S118E) in the bacterially expressed GST fusion protein [GST-HE15/458]. (C) Phosphorylation of the hERα A/B domain by MAPK increases its binding to p68. GST-HE15 or GST-HE15/457 that was incubated with activated MAPK for 0, 15, or 30 min at 30°C was used as the probe for an in vitro pull-down assay with 35S-labeled p68 protein.
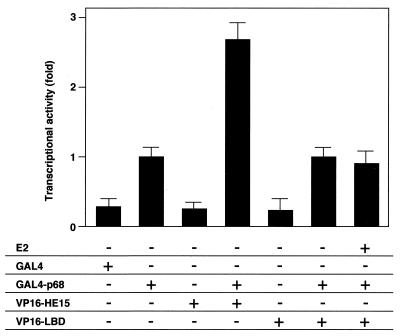
To further study the p68 interaction in vivo, a mammalian two-hybrid assay in COS-1 cells was performed with a p68 chimeric protein fused to a GAL4 DNA binding domain (DBD) (GAL4-p68) and two hERα deletion mutants fused to a VP16 activation domain (VP16-HE15 and VP16-LBD). Interaction of p68 with the A/B domain was detected, whereas the LBD did not interact with p68 even in the presence of E2 (Fig. 5). We also detected an intrinsic transactivation function of p68 by comparing GAL4-p68 with GAL4 DBD only (Fig. 5).
FIG. 5.
p68 interacts with the hERα A/B domain but not with the LBD in vivo. p68 interacts with the hERα A/B domain in the mammalian two-hybrid system. A mammalian two-hybrid system with GAL4-p68 fusion protein and VP16-ER fusion proteins (VP16-HE15 and VP16-LBD) was used in COS-1 cells. COS-1 cells were cotransfected with 1 μg of either GAL4-DBD, GAL4-p68, VP16-HE15, or VP16-LBD in the presence or absence of E2 (10−7 M), along with 2 μg of 17M2G-CAT reporter plasmid. Significant interaction was detected only between p68 and the hERα A/B domain.
p68 acts as a specific coactivator for hERα AF-1.
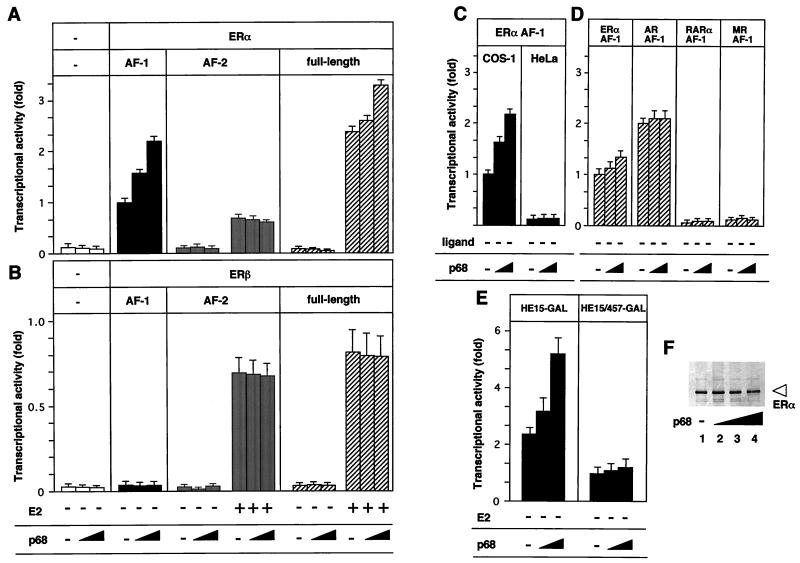
From in vivo and in vitro interaction studies, it appeared that p68 RNA helicase acts as a coactivator for the hERα AF-1. To address this extra function, the effect of p68 in the transactivation functions of the hERα and hERβ (33) was investigated with ER fusion proteins of the GAL4 DBD. Forced expression of p68 significantly enhanced the AF-1 activity but not the AF-2 activity of the hERα in the absence and presence of E2 (Fig. 6A). The enhanced transactivation function of p68 was detected with the full length of hERα but was not as marked as that seen with AF-1 alone. However, neither hERβ AF-1 nor AF-2 was activated by p68 (Fig. 6B). Moreover, the AF-1 activities of other nuclear receptors tested (Fig. 6D), as well as their AF-2 activities (data not shown), were not enhanced by p68. However, overexpression of p68 could not potentiate the activity of the hERα AF-1 in HeLa cells, in which the hERα AF-1 is known to be very low (Fig. 6C), suggesting that the p68 action is cell type specific. Note that overexpression of p68 does not affect the expression levels of hERα (Fig. 6F) or of chimeric receptors (data not shown).
FIG. 6.
p68 potentiates the ligand-induced transactivation function of hERα through AF-1. (A) p68 potentiates AF-1 but not AF-2 of hERα. COS-1 cells were cotransfected with 0.40 μg of HE15-GAL(AF-1), LBD-GAL(AF-2), or HEGO (AF-1 plus AF-2) (26) and with either 0, 0.3, or 0.6 μg of pSG5-p68 in the presence or absence of E2 (10−7 M), along with 2 μg of 17M2G-CAT or 2 μg of ERE-G-CAT (for HEGO only). p68 potentiated the ligand-induced transactivation of the full-length hERα and AF-1 but not AF-2 activated by E2. (B) p68 has no effect in the transactivation function (AF-1 and AF-2) of hERβ. (C) p68 potentiates the hERα AF-1 activity in COS-1 cells but not in HeLa cells. (D) p68 has no effect on the AF-1 activities of the other nuclear receptors. (E) p68 potentiates the hERα AF-1 phosphorylated at the Ser118 residue. COS-1 cells were cotransfected with HE15-GAL or HE15/457-GAL along with either 0, 0.3, or 0.6 μg of pSG5-p68 and 2 μg of 17M2G-CAT. (F) p68 does not affect the expression levels of the hERα A/B domain. COS-1 cells were transfected with 0.40 μg of HE15-GAL and with either 0, 0.15, 0.3, or 0.6 μg of pSG5-p68. A Western blot analysis shows that the amount of the expressed chimeric protein is not affected by p68 expression. The open arrowhead indicates the position of the protein.
Phosphorylation of Ser118 by MAPK is essential for enhancement of hERα AF-1 activity by p68.
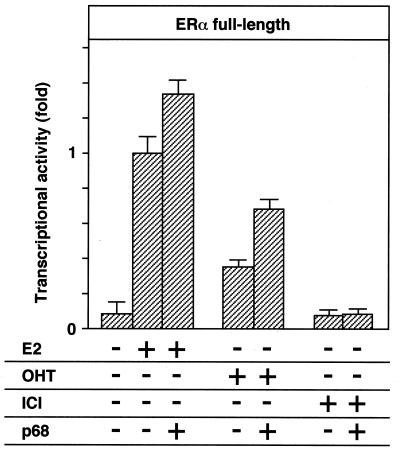
Previous studies showed that several serine residues of the ERα A/B domain are phosphorylated in cells treated with growth factors, which can potentiate the actions of E2 (1, 3, 19). We further demonstrated that MAPK, which is activated by growth factors, specifically phosphorylates Ser118 of the ERα A/B domain and that this phosphorylation enhances the AF-1 activity (26). Taken together with the binding preference of p68 for the A/B domain phosphorylated by MAPK at the Ser118 residue (Fig. 1 and 4), our result indicates that p68 acts as a coactivator for the hERα AF-1 in a MAPK-mediated phosphorylation-dependent way. To address this point, the effects of p68 on the AF-1 activity of the phosphorylated A/B domain were examined by comparing the wild-type A/B domain (HE15-GAL) and the A/B domain point mutant (HE15/457-GAL), in which the Ser118 residue is replaced by alanine and is unable to be phosphorylated by MAPK (1, 26). As shown in Fig. 6E, the AF-1 activity enhanced by p68 was abrogated by Ser118 replacement (HE15/457-GAL). Thus, these results demonstrate that the phosphorylation of Ser118 by MAPK is indispensable for the enhanced activity of ERα AF-1 by p68. To further clarify the role of p68 in the ERα AF-1 activity, the action of p68 in the full-length ERα activated by OHT and ICI was examined. Although ICI was shown to suppress ERα transactivation function as a pure antagonist, OHT is considered a partial antagonist, blocking only AF-2 function and not AF-1 function. p68 could not induce the transactivation function of hERα-bound ICI; however, it potentiated the function of hERα-bound OHT (Fig. 7), again supporting the idea that p68 acts as a coactivator specific for hERα AF-1.
FIG. 7.
p68 potentiates the transactivation function of hERα induced by OHT but not ICI. COS-1 cells were cotransfected with 0.40 μg of HEGO and 2 μg of ERE-G-CAT, along with either 0, 0.3, or 0.6 μg of pSG5-p68, and treated with 100 nM E2, OHT, or ICI.
p68 coactivator activity requires the hERα A/B domain but does not require intrinsic RNA helicase activity.
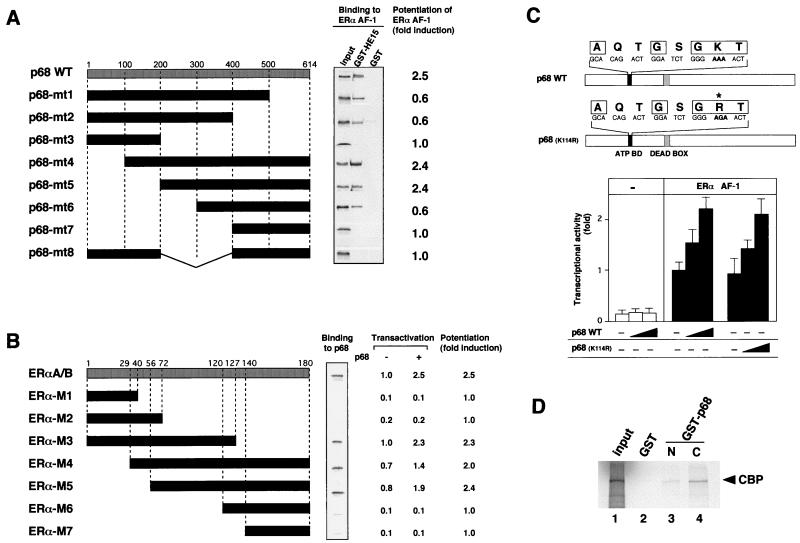
To delineate the region of p68 responsible for the interaction with ERα A/B domain, a series of p68 deletion mutants were examined by the GST pull-down assay, with the GST-HE15 protein as a probe, and by the transient-expression assay to establish their roles in the ERα AF-1 activity (Fig. 8). Truncations of the N-terminal region up to amino acid (aa) 300 (p68-mt4, p68-mt5, and p68-mt6) did not reduce the p68 interaction in vitro, but a further 100-aa deletion caused complete loss of the interaction (p68-mt7) (Fig. 8A, middle panel). Consistent with the N-terminal truncations, the region from aa 300 to 400 was required in the C-terminal truncation mutants (p68-mt1, p68-mt2, and p68-mt3). In the transactivation assay, p68-mt4 and p68-mt5 were as potent as the wild-type p68 but p68-mt1, p68-mt2, and p68-mt6 suppressed the ERα AF-1 activity (Fig. 8A, right panel). From these results, it is likely that the regions surrounding the interaction domain are required for the full coactivator activity of p68. Next, to determine the interaction regions for p68 in the A/B domain of ERα, a series of truncated mutants of the A/B domain fused to GST were generated and examined for binding to in vitro-translated p68 in GST pull-down assays. As shown in Fig. 8B, p68 directly interacted with the middle region (aa 56 to 127) in the A/B domain, which contains the Ser118 residue and was also essential for the AF-1 activity. Transactivation assays showed that p68 was able to potentiate the transcriptional activities of only the GAL4-fused deletion mutants of the A/B domain which contain the interaction region for p68. From results obtained from in vitro binding assays and in vivo transactivation assays, we concluded that the interaction regions identified in GST pull-down assays were also functional in vivo and that direct binding between p68 and the A/B domain in vivo was absolutely necessary for the potentiation of AF-1 activity by p68.
FIG. 8.
The RNA helicase activity of p68 is not required for the coactivator activity for the hERα AF-1. (A) The interaction domain of p68 for the hERα A/B domain is essential for the p68 coactivator activity for hERα AF-1. Representations of the p68 deletion mutants used for in vitro GST pull-down assay (middle panel) and the transient-expression assay (right panel) are shown. 35S-labeled p68 mutants were assayed with the GST-HE15 protein as a probe, showing that the region (aa 300 to 400) of the ATP binding domain and the DEAD box motif is required for direct interaction with the hERα A/B domain. The coactivator activities of p68 deletion mutants were determined in the transient-expression assay. COS-1 cells were cotransfected with 0.40 μg of HE15-GAL, 2 μg of 17M2G-CAT, and 0.6 μg of the expression vector of the p68 mutant. The mean fold induction of the AF-1 activity by the p68 mutant from three independent experiments is shown. (B) The interaction region of the hERα A/B domain for p68 is necessary for the potentiation of AF-1 activity by p68. Representations of the hERα A/B domain deletion mutants used for in vitro GST pull-down assay (middle panel) and the transient-expression assay (right panel) are shown. 35S-labeled p68 was assayed with the GST-fused A/B domain deletion mutant proteins as a probe, showing that the region (aa 56 to 127) in the A/B domain is required for direct p68 binding. The coactivator activities of p68 for the A/B domain deletion mutants were determined in the transient-expression assay. COS-1 cells were cotransfected with either 0.40 μg of A/B deletion mutants-plus-GAL (pM-A/B-Ms) with 2 μg of 17M2G-CAT and 0.6 μg of the expression vector of p68. The mean fold induction of the AF-1 activity by p68 from three independent experiments is shown. (C) A p68 mutant with a mutation in the ATP binding domain essential for the RNA helicase activity still potentiates the hERα AF-1. The ATP binding domain and DEAD box motif are indicated by solid and shaded boxes, respectively. The amino acid residues (AXXGXGKT), which are highly conserved among helicases, are boxed. The asterisk shows the replaced amino acid. COS-1 cells were cotransfected with 0.40 μg of pM-HE15 and 2 μg of 17M2G-CAT and with 0, 0.3, or 0.6 μg of the expression vector for the p68 mutant. The activity of hERα AF-1 was enhanced by both the wild-type p68 and the K144R mutant of p68. (D) p68 binds to CBP. To test the binding between p68 and CBP, GST control protein, GST-p68N (aa 1 to 387) fusion protein, or GST-p68C (aa 388 to 614) fusion protein was immobilized on beads and mixed with 15 μl of in vitro CBP translation reaction mixtures. Binding proteins were analyzed by SDS-PAGE. In vitro-translated CBP binds to both GST-fused p68N and p68C but not to the GST control protein.
Previous reports demonstrating that the p68 protein is an ATP-dependent RNA helicase raised the question of whether RNA helicase activity (16) was required for the p68 coactivator activity for the hERα AF-1. To address this issue, a point mutation (Lys144 to Arg) in the p68 ATP binding site was introduced to abolish RNA helicase activity (12, 13) (Fig. 8C, upper panel). This mutation had no effect on the p68 coactivator activity, leading us to conclude that p68 helicase activity is not required for its hERα coactivator function.
Since p68 itself exhibited only weak intrinsic activity in transactivation (Fig. 5), we explored a possible interaction of p68 with known coactivators. For this study, we used two major classes of coactivators, the SRC-1/TIF2 family proteins and the CBP/p300 class (44). All SRC-1/TIF2 family proteins showed no interaction with p68 in a GST pull-down assay (data not shown). However, both the N-terminal (aa 1 to 387) and C-terminal (aa 388 to 614) domains of p68 fused to GST bound to in vitro-translated CBP (Fig. 8D). These results demonstrate that the coactivator activity of p68 for the hERα AF-1 is mediated at least by CBP.
The p68 interaction region in the hERα A/B domain is essential for full activity of the hERα AF-1.
To assess if the p68 interaction with the hERα A/B domain in vivo reflects the p68 coactivator activity for the hERα AF-1, we studied the relationship between p68 interaction and the enhanced AF-1 activity mediated by p68 with a series of the hERα A/B domain deletion mutants. In vitro GST pull-down assays demonstrated that the central region (from aa 56 to 127) in the hERα A/B domain is required for the in vitro p68 interaction (Fig. 8B). In agreement with this interaction, p68 potentiated the transactivation functions of the hERα A/B domain deletion mutants which retain the central region. Interestingly, this interaction region was also essential for the full activity of the hERα AF-1 (Fig. 8B). Thus, from these findings, it is likely that the full activity of hERα AF-1 requires p68 interaction through a central region in the hERα A/B domain.
DISCUSSION
Identification of ERα AF-1 specific coactivator, p68.
Several classes of putative nuclear receptor coactivators such as the SRC-1/TIF2 (2, 9, 11, 39, 47) and CBP/p300 (24) family proteins, TIF1 (29), ARA70 (52), and many others (22, 35, 36, 40) have been investigated in terms of transcriptional mediation between nuclear receptors and the basal transcriptional machinery (5, 17). Significant enhancement of ligand-induced transactivation of many nuclear receptors is observed in the SRC-1/TIF2 family proteins when they are expressed in mammalian cells. Moreover, overexpression of the SRC-1/TIF2 family proteins can reduce transcriptional interference among nuclear receptors and nuclear receptor autosquelching (11, 39, 47). By biochemical approaches, direct interactions of the SRC-1/TIF2 family proteins with the LBD have been further demonstrated to occur in a ligand-dependent manner (2, 9, 11, 47), and ligand-induced interactions of ERα with the SRC-1/TIF2 family proteins are induced by agonists but not by antagonists (9, 14). Analysis of the interacting domains of coactivators led to the identification of consensus motifs (LXXLL) for the direct interaction with nuclear receptors (15). From these observations, an SRC-1/TIF2 family of proteins was recognized as containing the best-characterized coactivators for AF-2 of various nuclear receptors, including ERα and ERβ. Like the SRC-1/TIF2 family proteins, the reported coactivators were found while searching for a coactivator for AF-2 but not for AF-1. To our knowledge, there is no report of a coactivator specific for AF-1 of nuclear receptors. However, since the ratios between the AF-1 and AF-2 activities, at least in ERα, are cell type specific (6, 46), the existence of a coactivator that specifically interacts with and activates AF-1 is a distinct possibility (37, 45). More recently, the SRC-1/TIF2 family proteins have been reported to stimulate the ERα AF-1 activity of nuclear receptors synergistically with the AF-2 (34, 49), but these coactivators seem only to partially support the AF-1 activity of ERα (49). Thus, even if this family of proteins are coactivators for AF-1, an additional coactivator directly interacting with the ERα A/B domain is believed to exist. When recombinant A/B and LBD domains of the hERα were used as probes to purify interactants from the nuclear extracts of various cell lines, no interactant for the 160-kDa protein of the ERα A/B domain was found in the present study, while a ligand-dependent interaction with the LBD was seen in the interactant for the 160-kDa protein, presumably SRC-1/TIF2 family proteins (9, 14). In contrast to the 160-kDa interactant, we found that p68 interacts with the A/B domain but not the LBD of ERα even in the presence of E2 and E2 antagonists. Purification and cloning identified this p68 protein as a previously reported p68 RNA helicase protein. The recombinant p68 protein interacted in vivo and in vitro specifically with the ERα A/B domain. The overexpression of p68 potentiated the hERα AF-1 activity but not other AF-1 and AF-2 activities of tested nuclear receptors, including ERβ. Interestingly, deletion of the hERα A/B domain showed that p68 interacts with the central region, which is essential for the hERα AF-1 activity (Fig. 8B). Although the p68 coactivator activity was less potent in the full length of hERα bound to E2 than in AF-1 alone, p68 could potentiate the ligand-induced transactivation function of the full-length hERα by OHT, which is considered to be an ERα AF-1 agonist and an AF-2 antagonist (1, 6). Taken together, these results indicate that p68 is a coactivator that specifically interacts with and activates the AF-1 of ERα.
The p68 RNA helicase acts as a coactivator.
The p68 RNA helicase protein was first reported to have immunological cross-reactivity with an antibody against the simian virus 40 large T antigen (12). p68 is a nuclear protein, and its localization in the nucleus varies during the cell cycle. At telophase, it translocates from the nucleoplasm to the nucleoli (18). p68 is a member of the DEAD-box protein family of putative RNA helicases (16) and contains an ATPase A motif that is responsible for RNA helicase activity (12). p68 is proposed to be important in diverse cellular processes, including RNA processing, transcription, translation, cell growth, and division. Moreover, p68 binds calmodulin in a Ca2+-dependent manner and is phosphorylated by protein kinase C through the IQ domain in the C-terminal region (7). Since the p68 ATPase activity is inhibited by both calmodulin binding and protein kinase C phosphorylation, the RNA-unwinding activity of p68 is supposed to be modulated by dual Ca2+-signaling pathways (7). In the present study, we first found that this p68 also functions as a transcriptional coactivator but that the coactivator activity of p68 does not require the RNA helicase activity. Since p68 itself possesses weak intrinsic transactivation activity, a region other than that containing the RNA-unwinding activity appears to regulate the p68 coactivator function of the ERα AF-1. Since CBP exhibited in vitro affinity to the N-terminal and C-terminal domains of p68 (Fig. 8D), p68 may serve as an adapter protein to associate with AF-2 coactivators. Since the ERα AF-1 activity is cell type specific and p68 is ubiquitiously expressed (Fig. 3), it is possible to speculate that upon binding ERα AF-1, p68 recruits an unknown coactivator(s) which acts in a cell-type-specific manner. This idea was further supported by the present observation that p68 overexpression cannot potentiate the ERα AF-1 activity in HeLa cells (Fig. 6C).
p68 potentiates ERα AF-1 phosphorylated by MAPK.
It was first reported in 1993 that ligand-dependent phosphorylation occurs on the Ser118 residue of ERα (1) and this phosphorylation supports the full activity of ERα AF-1. We further demonstrated that MAPK, which is activated by growth factors, undergoes this phosphorylation at Ser118 and potentiates the ERα AF-1 activity (26). In the present study, we found that the p68 binding affinity for the the hERα A/B region is increased by MAPK-mediated phosphorylation. Moreover, p68 does not potentiate the ERα AF-1 mutant, in which Ser118 is replaced by alanine and cannot be phosphorylated. Thus, it is most likely that when the Ser118 residue is phosphorylated, p68 associates tightly with the ERα A/B region to potentiate the ERα AF-1 function.
Like the ERα AF-2, the ERα AF-1 function is also induced by ligand binding to the ERα LBD (6, 46). During this process, physical and functional interactions between the AF-1 and AF-2 of ERα induced by ligand binding are believed to occur. More recently, it was reported that SRC-1 mediates the ligand-dependent interaction of AF-1 and AF-2 (34, 49). Since p68 potentiated the ligand-bound ERα but not unbound ERα, the possibility exists that p68 binds to the A/B domain only when the AF-2 is activated by ligand binding. Alternatively, it is speculated that like the CBP/p300 proteins, a coactivator(s) bound to the LBD in a ligand-dependent manner recruits p68 to associate with the A/B region. To test these hypothesis, further studies of p68 relating to the ligand-induced interaction of the A/B domain and LBD of hERα are required.
ACKNOWLEDGMENTS
We thank Pierre Chambon for helpful discussions throughout the study, Hiroaki Fuse for discussions and technical assistance, and Eisuke Nishida for the kind gift of purified MAPK.
This work was supported by funds from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Medicale, and the Human Frontier Science Program and by a grant-in-aid for priority areas from the Ministry of Education, Science, Sports and Culture of Japan (S.K.)
REFERENCES
- 1.Ali S, Metzger D, Bornert J M, Chambon P. Modulation of transcriptional activation by ligand-dependent phosphorylation of the human oestrogen receptor A/B region. EMBO J. 1993;12:1153–1160. doi: 10.1002/j.1460-2075.1993.tb05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AlB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Aronica S M, Katzenellenbogen B S. Stimulation of estrogen receptor-mediated transcription and alteration in the phosphorylation state of the rat uterine estrogen receptor by estrogen, cyclic adenosine monophosphate, and insulin-like growth factor-I. Mol Endocrinol. 1993;7:743–752. doi: 10.1210/mend.7.6.7689695. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad A, Ha L, Reinberg D, Tsai S, Tsai M-J, O’Malley B. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 6.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buelt M K, Glidden B J, Storm D R. Regulation of p68 RNA helicase by calmodulin and protein kinase C. J Biol Chem. 1994;269:29367–29370. [PubMed] [Google Scholar]
- 8.Bunone G, Briand P A, Miksicek R J, Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 9.Cavailles V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 11.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 12.Ford M J, Anton I A, Lane D P. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988;332:736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- 13.Gross C H, Shuman S. The nucleoside triphosphatase and helicase activities of vaccinia virus NPH-II are essential for virus replication. J Virol. 1998;72:4729–4736. doi: 10.1128/jvi.72.6.4729-4736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 15.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Hirling H, Scheffner M, Restle T, Stahl H. RNA helicase activity associated with the human p68 protein. Nature. 1989;339:562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 18.Iggo R D, Lane D P. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ignar-Trowbridge D, Teng C T, Ross K A, Parker M G, Korach K S, McLachlan J A. Peptide growth factors elicit estrogen receptor-dependent transcriptional activation of an estrogen-responsive element. Mol Endocrinol. 1993;7:992–998. doi: 10.1210/mend.7.8.8232319. [DOI] [PubMed] [Google Scholar]
- 20.Imai T, Matsuda K, Shimojima T, Hashimoto T, Masuhiro Y, Kitamoto T, Sugita A, Suzuki K, Matsumoto H, Masushige S, Nogi Y, Muramatsu M, Handa H, Kato S. ERC-55, a binding protein for the papilloma virus E6 oncoprotein, specifically interacts with vitamin D receptor among nuclear receptors. Biochem Biophys Res Commun. 1997;233:765–769. doi: 10.1006/bbrc.1997.6531. [DOI] [PubMed] [Google Scholar]
- 21.Iwamatsu A. S-Carboxymethylation of proteins transferred onto polyvinylidene difluoride membranes followed by in situ protease digestion and amino acid microsequencing. Electrophoresis. 1992;13:142–147. doi: 10.1002/elps.1150130129. [DOI] [PubMed] [Google Scholar]
- 22.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 23.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 25.Kato S, Tora L, Yamauchi J, Masushige S, Bellard M, Chambon P. A far upstream estrogen response element of the ovalbumin gene contains several half-palindromic 5′-TGACC-3′ motifs acting synergistically. Cell. 1992;68:731–742. doi: 10.1016/0092-8674(92)90148-6. [DOI] [PubMed] [Google Scholar]
- 26.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 27.Kuiper G G J M, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J. Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LeDouarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 29.LeDouarin B, Zechel C, Garnier J M, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lees J A, Fawell S E, Parker M G. Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res. 1989;17:5477–5488. doi: 10.1093/nar/17.14.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire L, Heinlein U A. High-level expression in male germ cells of murine P68 RNA helicase mRNA. Life Sci. 1993;52:917–926. doi: 10.1016/0024-3205(93)90526-9. [DOI] [PubMed] [Google Scholar]
- 32.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maruyama K, Endoh H, Sasaki I H, Kanou H, Shimaya E, Hashimoto S, Kato S, Kawashima H. A novel isoform of rat estrogen receptor beta with 18 amino acid insertion in the ligand binding domain as a putative dominant negative regular of estrogen action. Biochem Biophys Res Commun. 1998;246:142–147. doi: 10.1006/bbrc.1998.8590. [DOI] [PubMed] [Google Scholar]
- 34.McInerney E M, Tsai M J, O’Malley B W, Katzenellenbogen B S. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone receptor coactivator. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mengus G, May M, Carre L, Chambon P, Davidson I. Human TAF(II)135 potentiates transcriptional activation by the AF-2s of the retinoic acid, vitamin D3, and thyroid hormone receptors in mammalian cells. Genes Dev. 1997;11:1381–1395. doi: 10.1101/gad.11.11.1381. [DOI] [PubMed] [Google Scholar]
- 37.Meyer M E, Gronemeyer H, Turcotte B, Bocquel M T, Tasset D, Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989;57:433–442. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- 38.Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 39.Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 40.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 41.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang G E, Cavailles V, Parker M G, Kushner P J. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 43.Takeyama K, Kitanaka S, Sato T, Kobori M, Yanagisawa J, Kato S. 25-Hydroxyvitamin D3 1alpha-hydroxylase and vitamin D synthesis. Science. 1997;277:1827–1830. doi: 10.1126/science.277.5333.1827. [DOI] [PubMed] [Google Scholar]
- 44.Takeyama K, Masuhiro Y, Fuse H, Endoh H, Murayama A, Kitanaka S, Suzawa M, Yanagisawa J, Kato S. Selective interaction of vitamin D receptor with transcriptional coactivators by a vitamin D analolog. Mol Cell Biol. 1999;19:1049–1055. doi: 10.1128/mcb.19.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 46.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 47.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 48.vom Baur E, Zechel C, Heery D, Heine M J, Garnier J M, Vivat V, Le D B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 49.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen M P, Chen D, Huang S M, Subramanian S, McKinerney E, Katzenellenbogen B S, Stallcup M R, Kushner P J. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 50.Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Toriyabe T, Kashiwagi K, Watanabe M, Kawabata M, Miyazono K, Kato S. Convergence of TGFβ and vitamin D signaling pathways on SMAD proteins acting as common transcriptional co-activators. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- 51.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 52.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]