Abstract
As a severe public health problem, methamphetamine (METH) abuse places a heavy burden on families and society. A growing amount of evidence has indicated communication between gut microbiota and the CNS in drug addiction, with associations to neural, endocrine and immune pathways. Thus, we searched for alterations in the gut microbiota and their potential effects in METH users through 16S rRNA gene sequencing. A decreased Shannon index indicated lower bacterial diversity in the METH users than in the age-matched control group. The gut microbial community composition in the METH users was also altered, including reductions in Deltaproteobacteria and Bacteroidaceae abundances and increases in Sphingomonadales, Xanthomonadales, Romboutsia and Lachnospiraceae abundances. Moreover, the Fusobacteria abundance was correlated with the duration of METH use. Enterobacteriaceae, Ruminococcaceae, Bacteroides, and Faecalibacterium had statistically significant correlations with items related to the positive and negative symptoms of schizophrenia and to general psychopathology in the METH users, and all have previously been reported to be altered in individuals with psychotic syndromes, especially depression. Abstraction, one of the items of the cognitive assessment, was positively related to Blautia. These findings revealed alterations in the gut microbiota of METH users, and these alterations may play a role in psychotic syndrome and cognitive impairment. Although the mechanisms behind the links between these disorders and METH abuse are unknown, the relationships may indicate similarities in the pathogenesis of psychosis induced by METH abuse and other causes, providing a new paradigm for addiction and METH use disorder treatment.
Subject terms: Microbiology, Diseases
Introduction
Methamphetamine (METH) abuse has been a severe public health concern in recent decades1,2. METH is the most popular addictive drug in China, and there were 118.6 million individuals with METH use disorder in 2019, as reported by the Office of the National Narcotics Control Commission. The release of the neurotransmitter dopamine and decrease in dopamine transporters caused by METH might be the main factors responsible for dose escalation3. Chronic METH use causes a selective pattern of cerebral deterioration, including cerebral metabolic abnormalities and structural brain alterations4. A meta-analysis showed that impairments of many cognitive domains, such as attention, executive functions, visual memory and working memory, occurred in individuals with METH use disorders5. Anxiety or depression, violent behavior (often risk-taking), insomnia, repetitive movements (stereotypy), and psychosis have also been observed with chronic METH use6. Individuals with methamphetamine-related disorders also have a higher risk of schizophrenia7. However, the mechanisms underlying the cognitive impairment and behavioral abnormalities are still unknown.
Studies of the microbiota–gut–brain axis have increased rapidly in the past few years, and the commonly recognized gut microbiota play important roles in the regulation of anxiety8, depression9, cognition10 and pain11,12. Short-term intragastric treatment of adult mice with an antibiotic mixture impaired their novel object recognition memory by disrupting the bacterial community in the colon13. Exposure to a social disruption stressor changed the structure of the colonic mucosa-associated microbiota in mice and resulted in anxiety-like behavior14. In addition, comorbid gastrointestinal (GI) inflammation accompanies humoral immunity to food antigens early during the course of schizophrenia15, and the compromised endothelial barrier16 and permeability of the blood–brain barrier lead to translocation of food-derived peptides and resident gut microbiota17.
METH-mediated release of neurotransmitters might cause enteric neuron damage and death and subsequent GI dysfunction by leading to the generation of oxidative stress molecules, including reactive oxygen (ROS) and nitrogen (RNS) species18–20. METH use may also cause GI vasoconstriction and bowel ischemia, ultimately leading to abdominal or stomach cramping, severe constipation and/or diarrhea and tissue dehydration21–23. METH use is associated with ischemic colitis24–26 and vasculitis of the distal colon27, which may reflect inflammation caused by disruption of the intestinal barrier. METH also alters two major tight junction proteins, claudin-1 and ZO-1, which regulate gut epithelial paracellular permeability to alter colon barrier integrity28. Thus, the gut microbiome may also be influenced in the METH-treated animals or abusers because of these alterations in the intestine. Among men who use METH and also have sex with men, the overall composition of the gastrointestinal microbiome was altered29. It was reported that METH increased microbial diversity30, elevated the pathogenic bacteria31 and altered the composition of gut bacteria32 and metabolites in animals33. However, the fecal microbiota composition in heterosexual or Asian METH users remains unknown.
Changes in intestinal microbiota may influence behavior and alleviate neuropsychiatric conditions. Germ-free mice exhibited anxiolytic-like behavior in an elevated plus-maze accompanied by changes in the mRNA and protein levels of genes implicated in anxiety and stress reactivity, such as NR2B, BDNF and 5HT1A34. The human milk oligosaccharides 3′-sialylactose (3'SL) or 6′-sialylactose (6′SL) could attenuate stressor-induced anxiety-like behavior by supporting normal microbial communities14, and probiotic treatment could reverse the reduction in swim behavior and increase in immobility in maternal separation rats in a forced swim test35. These findings suggest that the intestinal microflora plays an important role in the regulation of cognition and mood. The therapeutic application of probiotic bacteria for the neuropsychiatric conditions caused by METH use still has broad prospects.
Thus, in this study, 16 subjects with a clinical diagnosis of METH addiction from Wuhan Mental Health Center and 14 age- and gender-matched healthy subjects were selected, and high-throughput 16S rRNA sequencing and bioinformatics analysis were performed to analyze the variations in gut microbiota between the two groups, including the differences in the diversity of the bacterial community, the microbiota composition and the taxa that best characterized each group. In addition, we analyzed the correlation between the initial age or duration of METH use and the composition of gut microbiota to determine whether the dosage of METH influenced the gut microbiota. Furthermore, the correlations between the neuropsychiatric conditions and cognitive impairments caused by METH use and the changes in gut microbiota were also a major focus of this study.
Materials and method
Subject selection
The study protocol was approved by the Ethics Committee of Wuhan Mental Health Center (Wuhan, Hubei, China). All participants provided written informed consent prior to enrollment after receiving a written description of the study.
We recruited 16 subjects with a clinical diagnosis of methamphetamine use disorder (average age, 36 ± 7.8; age range 27–55), defined as the METH group hereinafter, and compared them with 14 age-matched healthy subjects who had never used METH or any other addictive drug (average age, 36 ± 9.4; age range 22–54), defined as the Ctr group hereinafter. The MUD is diagnosed based on the Diagnostic and Statistical Manual of Mental Disorders 5 (DSM-5) by the consensus of two psychiatrists (Supplementary). The two groups were named METH and Ctr, respectively. All of the subjects were male. Neither their marital status nor their level of education were significantly different. All of the subjects belonged to the same nationality (Han Chinese) and lived in Wuhan city. No antibiotics, probiotics, or prebiotics were taken in the 3 months prior to sample collection. None of the subjects were on anti-inflammatory or antioxidant drugs.
Information on METH use was also collected. The average age of initial METH use was 27.5 ± 5.82 years, with a range of 22–40 years, and the average duration of METH use was 8 ± 3.40 years, with a range of 4–15 years. The average frequency of METH use was 4 ± 0.81 days per week. The subjects in the METH group had been hospitalized 1–6 times (Table 1).
Table 1.
Characteristics of the study population.
| METH | Ctr | |
|---|---|---|
| Subjects (n) | 16 | 14 |
| Proportion of males, no. (%) | 16 (100%) | 14 (100%) |
| Age (years; means ± SD) (range) | 36 (7.8) (27–55) | 36 (9.4) (22–54) |
| Employed, no. (%) | 3 (18.75%) | 7 (50.00%) |
| Education | ||
| Primary school, no. (%) | 5 (31.25%) | 4 (28.57%) |
| Middle school, no. (%) | 3 (18.75%) | 5 (35.71%) |
| High school, no. (%) | 1 (6.25%) | 2 (14.29%) |
| University or higher, No. (%) | 7 (43.75%) | 3 (21.43%) |
| Married, no. (%) | 8 (50.00%) | 9 (64.29%) |
| Age of initial METH use (years; mean ± SD) (range) | 27.5 (5.82) (22–40) | NA |
| Duration of METH use (years; mean ± SD) (range) | 8 (3.40) (4–15) | NA |
| Frequency of METH use (days per week; mean ± SD) (range) | 4 (0.81) (2–5) | NA |
| Number of hospitalizations (number; mean ± SD) (range) | 2 (1.45) (1–6) | NA |
Assessment of cognition and symptoms of schizophrenia
One experienced psychiatrist from Wuhan Mental Health Center performed the screening examinations. Mild cognitive impairment (MCI) and dementia were assessed using the Montreal Cognitive Assessment (MoCA). The positive and negative syndrome scale (PANSS) was used to assess schizophrenia and psychosis spectrum disorders. All patients provided written informed consent.
Fecal sample collection and DNA isolation
Fecal samples were collected from the patients who abuse METH and the matched controls in the morning and stored at − 80 °C prior to analyses. DNA was extracted using the Magnetic Soil and Stool DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Then, the purity and concentration of DNA were determined by 1% agarose gel electrophoresis. Sample DNA was diluted to 1 ng/μL with sterile water.
Polymerase chain reaction (PCR)
16S rRNA genes were amplified using the specific primers with barcodes (Primer: 16S V4: 515F-806R). All PCRs were carried out in 30 μL with 15 μL of Phusion High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM forward and reverse primers, and approximately 10 ng of template DNA. Thermal cycling consisted of initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s and final extension at 72 °C for 5 min. A Bio-Rad T100 gradient PCR instrument was used.
The same volume of 1× loading buffer (containing SYBR green) was mixed with the PCR products, and electrophoresis was performed on a 2% agarose gel for detection. Samples with bright main bands between 400–450 bp were chosen for further experiments.
The PCR products were mixed in equidensity ratios. Then, the mixed PCR products were purified with a GeneJET Gel Extraction Kit (Thermo Scientific).
Library preparation and sequencing
Sequencing libraries were generated using an Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA) with index codes added following the manufacturer’s recommendations. The library quality was assessed on a Qubit 2.0 fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina NovaSeq platform, and 250 bp paired-end reads were generated.
Data analysis
The raw data from the Illumina NovaSeq platform were spliced to obtain clean tags with quality control. Paired-end reads from the original DNA fragments were merged by FLASH and were assigned to each sample according to the unique barcodes36. Sequences were analyzed using the QIIME (Quantitative Insights Into Microbial Ecology) software package, and in-house Perl scripts were used to analyze alpha (within samples) and beta (among samples) diversity. First, reads were filtered by QIIME quality filters. Then, we used pick_de_novo_otus.py to pick operational taxonomic units (OTUs) by making an OTU table. Sequences with ≥ 97% similarity were assigned to the same OTU. We selected representative sequences for each OTU and used the RDP classifier to annotate taxonomic information for each representative sequence. Species richness is the number of bacterial species assigned on the basis of the OTUs detected in the samples. Alpha diversity was used to analyze the within-community microbial community diversity diversity37. To compute alpha diversity under the 97% consistency threshold for different samples, we rarified the OTU table and calculated three metrics: Chao1, which estimates the species abundance; the observed species, which estimates the number of unique OTUs found in each sample; and the Shannon index (Supplementary Table 1). Rarefaction curves were generated based on these three metrics. A Venn diagram was used to show the numbers of shared and unique OTUs between the two groups (R Software, http://www.r-project.org/, Version 3.0.3).
QIIME calculates both weighted and unweighted UniFrac distances (Qiime Software, http://qiime.org/, Version 1.9.1), which are phylogenetic measures of beta diversity. We used both weighted and unweighted UniFrac distances for principal coordinate analysis (PCoA), obtaining principal coordinates and visualizing them from complex, multidimensional data. We also calculated the Bray–Curtis UniFrac distances and visualize them by nonmetric multidimensional scaling (NMDS) (R Software, Version 2.15.3). To mine deeper into the microbial diversity data to explore the differences between the samples, significance tests were conducted with statistical analysis methods, including MetaStat (R Software, Version 2.15.3), LEfSe (LEfSe Software, Version 1.0) and ADONIS (R Software, Version 2.15.3). Correlations between variables were calculated using Spearman’s rank-correlation analysis (R Software, Version 2.15.3). All tests for significance were two sided, and p < 0.05 was considered significant.
Ethics approval and consent to participate
The study was conducted after the approval by Wuhan Mental Health Center Ethics Committee at January 2019, and all written informed consents were obtained from the enrolled subjects, in compliance with national legislation and the Code of Ethical Principles for Medical Research Involving Human Subjects of the World Medical Association (Declaration of Helsinki). This program belongs to the Clinical cohort study on addiction-related disorders in China and registered in Chinese Clinical Trial Registry (ChiCTR2000032198).
Results
Diversity and community composition of gut microbiota
To characterize the gut microbiota associated with METH use disorder, the alpha diversity index was analyzed. The trend in species richness in patients with METH use disorder was similar to that in the control individuals, based on the rarefaction analysis estimates (Supplementary Fig. 1a). There were no significant differences between the METH and Ctr groups either in community richness or the number of observed species. However, the significant difference in the Shannon index showed that the Ctr group had greater species diversity (Wilcoxon rank-sum test, p = 0.025) (Supplementary Fig. 1b). There were 611 shared OTUs, 118 OTUs in the METH group only and 126 OTUs in the Ctr group only (Supplementary Fig. 1c).
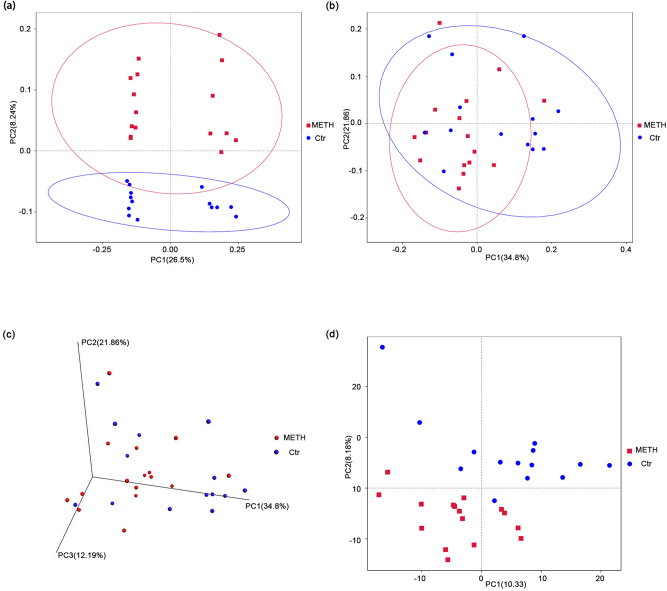
Beta diversity is used to compare the microbial community composition of different samples38. The beta diversity calculated on the basis of the weighted UniFrac distances, the unweighted UniFrac distances and the Bray–Curtis distances revealed a strong effect of METH use disorder on the gut microbiota39,40. The METH samples clustered distinctly from the controls in PCoA space and differed significantly in composition (Supplementary Table 2, with ADONIS R = 0.074, p = 0.013, unweighted UniFrac; ADONIS R = 0.078, p = 0.041, weighted UniFrac), indicating robust differences in the membership of gut bacteria between the METH and Ctr groups (Fig. 1a–c). The Bray–Curtis distances also showed significant differences between the METH and Ctr groups (Supplementary Table 2, with ADONIS R = 0.093, p = 0.006). Principal component analysis (PCA) based on Euclidean distances, which extracts the most important elements and structures in the data and shows the difference in multidimensional data on a two-dimensional coordinate chart41, also describes the similarity of the samples in the same group and the disparity between the two groups (Fig. 1d).
Figure 1.
Subjects who abuse METH harbor an altered bacterial gut microbiota. PCoA of the bacterial beta diversity based on the (a) unweighted and (b) weighted UniFrac distances. (c) Three-dimensional PCoA of bacterial beta diversity based on the weighted UniFrac distances. (d) PCA of bacterial beta diversity based on the Euclidean distances. Subjects who abuse METH and Ctr subjects are colored in red and blue, respectively.
Composition of gut microbiota in the METH and control groups
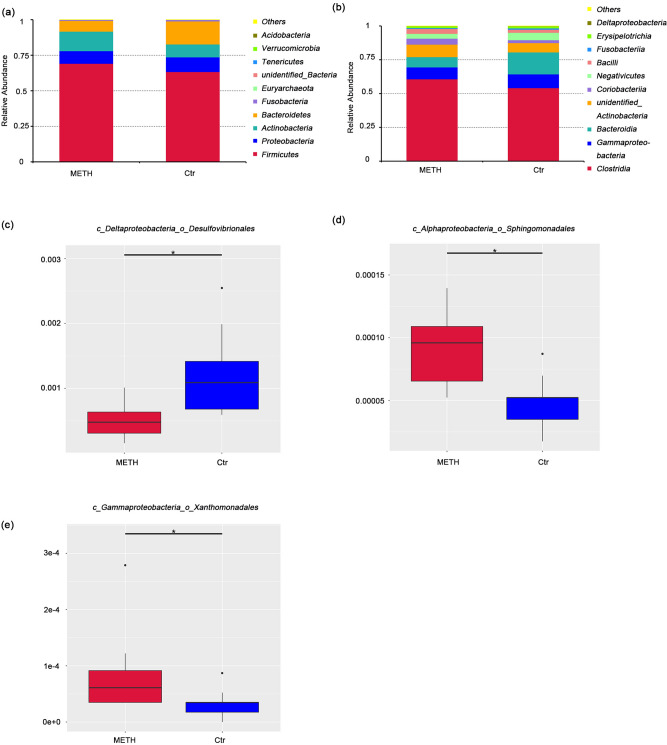
The overall microbial compositions of the METH and Ctr groups were examined at different taxonomic levels. The top five most abundant phyla were Firmicutes, Proteobacteria, Actinobacteria, Bacteroidetes and Fusobacteria (Fig. 2a, Supplementary Table 2). The top five most abundant classes were Clostridia, Gammaproteobacteria, Bacteroidia, unidentified Actinobacteria and Coriobacteriia (Fig. 2b, Supplementary Table 2). The results showed no phyla or classes taxa with significant differences between the two groups. At the order level, Desulfovibrionales was significantly decreased (MetaStat, p = 0.001, q = 0.021; Fig. 2c, Supplementary Table 3) and Sphingomonadales (MetaStat, p = 0.001, q = 0.021; Fig. 2d, Supplementary Table 3) and Xanthomonadales (MetaStat, p = 0.001, q = 0.021; Fig. 2e, Supplementary Table 3) were significantly increased in the METH group, but there were no significant differences in the dominant bacteria (top 10 most abundant) between the two groups. Clostridiales, Enterobacteriales, Bacteroidales and Bifidobacteriales were the top four most abundant orders (Supplementary Table 3). We found 8 statistically significant differences between the METH and Ctr groups at the family level (Supplementary Table 3), including an increase in the relative abundances of Rikenellaceae, an unidentified Clostridiales and Desulfovibrionaceae and a reduction in the relative abundances of Tannerellaceae, Lactobacillaceae, Xanthomonadaceae, Sphingomonadaceae and Peptococcaceae in the METH group. Among the top ten most abundant families, only Clostridiales significantly was increased in the METH group (MetaStat, p = 0.001, q = 0.019; Supplementary Table 3). We also found 12 significant differences in the METH group compared with the Ctr group at the genus level (Supplementary Table 3), but the abundances of all the species were low.
Figure 2.
Composition of gut microbiota in the METH and control groups. Relative abundances of (a) phylum-level and (b) class-level gut microbial taxa. At the order level, (c) Desulfovibrionales was significantly decreased, and (d) Sphingomonadales and (e) Xanthomonadales were significantly increased in the METH group. *q < 0.05 (MetaStat).
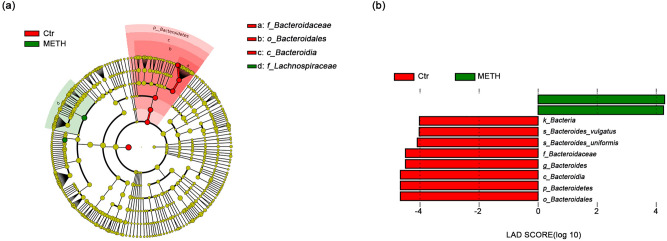
We further analyzed the bacterial community structure by using the linear discriminant effect size (LEfSe), an algorithm for high-dimensional biomarker discovery that uses linear discriminant analysis (LDA) to estimate the effect size of each taxon that is differentially represented in METH and Ctr groups33. LEfSe analysis revealed significant increases in the relative abundances of several bacterial taxa, including Romboutsia and Lachnospiraceae, and a significant reduction in the relative abundance of Bacteroidaceae in the METH group compared with the Ctr group (Fig. 3a,b).
Figure 3.
Taxonomic representation of significant differences in relative abundances between the METH and Ctr groups. (a) Cladograms generated by LEfSe indicating differences in the bacterial taxa between the subjects with METH use disorder (METH) and the control (Ctr) subjects. Nodes in red indicate taxa that were enriched in the Ctr compared to the METH group, while nodes in green indicate taxa that were enriched in the Ctr compared to the METH group. (b) LDA scores for the bacterial taxa with differential abundances between the METH and Ctr groups. Positive and negative LDA scores indicate bacterial taxa enriched in the METH and Ctr groups, respectively. Only the taxa with p < 0.01 (Wilcoxon rank-sum test) and LDA > 4.0 are shown in the figure legend.
Gut microbiota may be associated with age and the duration of METH use
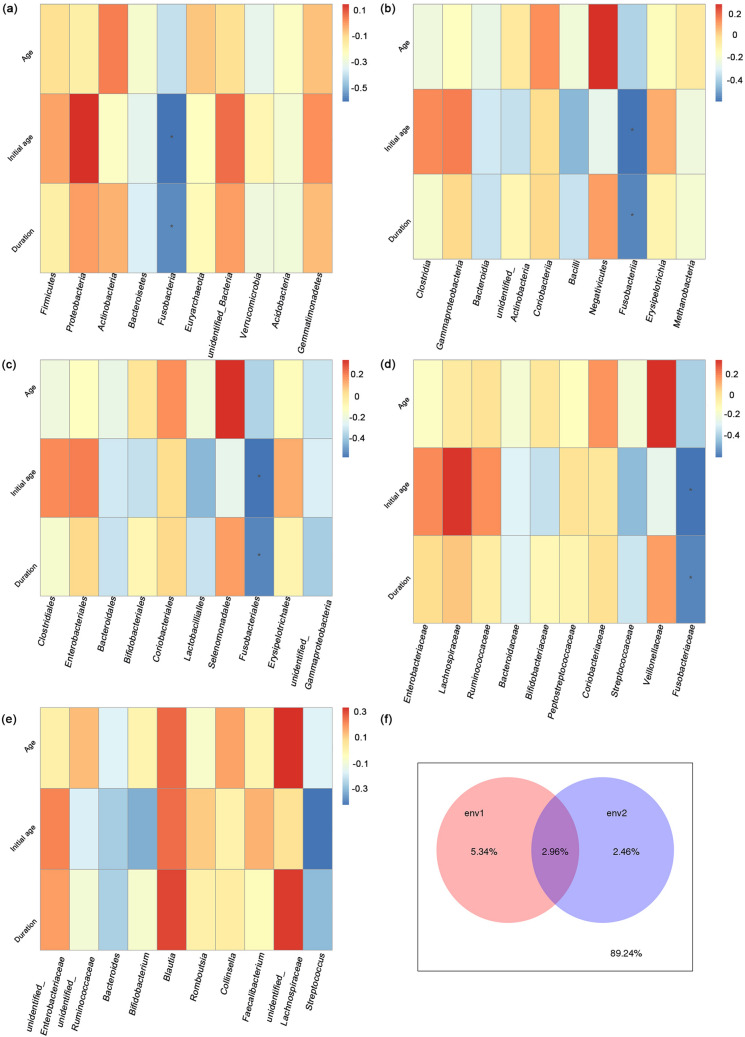
The above results show the influence of METH use disorder on gut microbiota but do not reveal whether the dosage of METH or the duration of METH use influences the gut microbiota. Thus, we analyzed the correlation between different taxa of the gut microbiota and age, the initial age of METH use and the duration of METH use. At the phylum level, the duration (Spearman rank correlation, R = − 0.589, p = 0.016) and age (Spearman rank correlation, R = − 0.545, p = 0.029) were both negatively related to Fusobacteria (Fig. 4a, Supplementary Table 4). Similarly, at the class, order and family levels, the duration (Spearman rank correlation, R = − 0.589, p = 0.016) and age (Spearman rank correlation, R = − 0.545, p = 0.029) were both negatively related to Fusobacteriia, Fusobacteriales, and Fusobacteriaceae, respectively (Fig. 4b–d; Supplementary Table 4). The initial age of METH use was not significantly related to the gut microbiota. The duration was negatively related to Streptococcus (Spearman rank correlation, R = − 0.440, p = 0.088) at the genus level (Fig. 4e). Considering the interactions between microbiota and physiological aging processes42, we sought to determine which factor affected the microbial community distribution more, age or the duration of METH use. Variance partitioning canonical correspondence analysis (VPA) showed that the duration of METH use contributed more to the differences in the microbial community distribution than the age of the individuals did (Supplementary Fig 1, Fig. 4f.).
Figure 4.
Gut microbiota may be associated with age and duration of METH use. The correlation between gut microbiota of different taxa and age, the initial age of METH use and duration of METH use (years) at the (a) phylum, (b) class, (c) order, (d) family, and e genus levels. The ordinate represents age (Age), initial age of METH use (Initial age) and duration of METH use (Duration). The abscissa represents the species information. Spearman rank correlation, *p < 0.05. (f) VPA showed that age (env2) contributed less to the microbial community than did the duration of METH use (env1). The intersecting part of the circle is the common explanatory quantity of env1 and env2, while the outside of the circle is the unexplainable quantity.
Positive and negative symptoms of schizophrenia and general psychopathology and cognition were related to gut microbiota
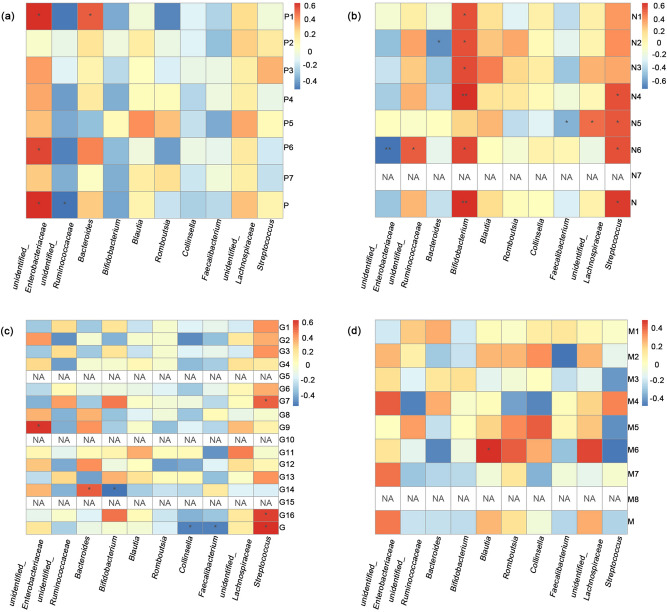
The PANSS was first developed in 1987 to assess both positive and negative symptoms of schizophrenia as well as general psychopathology43. METH-associated psychosis (MAP) is common and includes hallucinations and delusions44–46. Thus, we discuss whether phenotypes such as schizophrenia or other psychopathologies caused by METH use disorder are related to changes in gut microbiota. The positive scale showed that except for grandiosity, every index was abnormal to different degrees (Supplementary Table 5). At the genus level, an unidentified Enterobacteriaceae was positively related to delusions (Spearman rank correlation, R = 0.616, p = 0.011), suspiciousness (Spearman rank correlation, R = 0.568, p = 0.022) and the total positive scale (Spearman rank correlation, R = 0.620, p = 0.010) (Supplementary Table 6, Fig. 5a). The total positive scale was also negatively related to an unidentified Ruminococcaceae (Spearman rank correlation, R = − 0.507, p = 0.045) (Supplementary Fig 2). Delusions were positively correlated with Bacteroides (Spearman rank correlation, R = 0.523, p = 0.038) (Supplementary Table 6, Fig. 5a). Conceptual disorganization, hallucinatory behavior, excitement, grandiosity and hostility were not significantly correlated with the top ten most abundant phyla. All the individuals were absent of stereotyped thinking, and most of them received a minimal or mild level of criteria among the negative items (Supplementary Table 5). Bifidobacterium was positively related to blunted affect (Spearman rank correlation, R = 0.570, p = 0.021), emotional withdrawal (Spearman rank correlation, R = 0.561, p = 0.024), poor rapport (Spearman rank correlation, R = 0.582, p = 0.018), passive-apathetic social withdrawal (Spearman rank correlation, R = 0.652, p = 0.006), lack of spontaneity and flow of conversation (Spearman rank correlation, R = 0.594, p = 0.015) and the total negative scale (Spearman rank correlation, R = 0.650, p = 0.006) (Supplementary Table 6, Fig. 5b). Streptococcus was negatively related to passive-apathetic social withdrawal (Spearman rank correlation, R = 0.568, p = 0.022), difficulty in abstract thinking (Spearman rank correlation, R = 0.548, p = 0.028), lack of spontaneity and flow of conversation (Spearman rank correlation, R = 0.565, p = 0.023) and the total negative scale (Spearman rank correlation, R = 0.616, p = 0.011) (Supplementary Table 6, Supplementary Fig 3, Fig. 5b). There was a negative correlation between emotional withdrawal and Bacteroides (Spearman rank correlation, R = − 0.619, p = 0.011). Difficulty in abstract thinking was negatively related to Faecalibacterium (Spearman rank correlation, R = − 0.510, p = 0.044) and positively related to an unidentified Lachnospiraceae (Spearman rank correlation, R = 0.501, p = 0.048) (Supplementary Table 6, Fig. 5b). Lack of spontaneity and flow of conversation was negatively related to an unidentified Enterobacteriaceae (Spearman rank correlation, R = − 0.714, p = 0.002) and positively related to an unidentified Ruminococcaceae (Spearman rank correlation, R = 0.537, p = 0.032) (Supplementary Table 6, Supplementary Fig. 4, Fig. 5b). For the general psychopathology scale, the results for mannerisms and posturing, for disorientation and for preoccupation all showed no abnormality (Supplementary Table 5). Fusobacteria was positively related to motor retardation (Spearman rank correlation, R = 0.510, p = 0.044), active social avoidance (Spearman rank correlation, R = 0.573, p = 0.020) and the total general psychopathology scale (Spearman rank correlation, R = 0.617, p = 0.011) (Supplementary Table 6, Fig. 5c). Unusual thought content was positively related to an unidentified Enterobacteriaceae (Spearman rank correlation, R = 0.606, p = 0.013) (Supplementary Table 6, Fig. 5c). Poor impulse control was negatively related to Bifidobacterium (Spearman rank correlation, R = − 0.545, p = 0.029) and positively related to Bacteroides (Spearman rank correlation, R = 0.522, p = 0.038) (Supplementary Table 6, Fig. 5c). The total general psychopathology scale was negatively related to Collinsella (Spearman rank correlation, R = − 0.504, p = 0.047) and Faecalibacterium (Spearman rank correlation, R = − 0.502, p = 0.047) (Supplementary Table 6, Fig. 5c) (Supplementary Fig 5).
Figure 5.
Positive and negative symptoms of schizophrenia as well as general psychopathology and cognition were related to gut microbiota. The correlations between gut microbiota at the genus level for different taxa and (a) the positive symptoms of schizophrenia (P1 = delusions, P2 = conceptual disorganization, P3 = hallucinatory behavior, P4 = excitement, P5 = grandiosity, P6 = suspiciousness, P7 = hostility, P = the total positive scale), (b) the negative symptoms of schizophrenia (N1 = blunted affect, N2 = emotional withdrawal, N3 = poor rapport, N4 = passive-apathetic social withdrawal, N5 = difficulty in abstract thinking, N6 = lack of spontaneity and flow of conversation, N7 = stereotyped thinking, N = the total negative scale) and (c) general psychopathology (G1 = somatic concern, G2 = anxiety, G3 = guilty feelings, G4 = tension, G5 = mannerisms and posturing, G6 = depression, G7 = motor retardation, G8 = uncooperativeness, G9 = unusual thought content, G10 = disorientation, G11 = poor attention, G12 = lack of judgment and insight, G13 = disturbance of volition, G14 = poor impulse control, G15 = preoccupation, G16 = active social avoidance, G = general psychopathology scale) from PANSS and (d) the MoCA (M1 = visuospatial executive, M2 = naming, M3 = attention, M4 = calculation, M5 = language, M6 = abstract, M7 = delayed recall, M8 = orientation, M = the total MoCA scale). Spearman rank correlation, *p < 0.05, **p < 0.001.
The MoCA, developed by Nasreddine et al. in 2005, is widely used to facilitate the detection of MCI39. We also analyzed the correlation between the scales of MoCA and the gut microbiota at the genus level. Blautia was positively related to abstraction (Spearman rank correlation, R = 0.502, p = 0.047) (Supplementary Table 6, Fig. 5d) (Supplementary Fig 5).
Discussion
This study of 16 individuals with METH use disorder and 14 age-matched subjects found that METH use significantly impacted gut microbial composition, and the impacts were related to the age of the individual and the duration of METH use. We found differences in the Shannon index, which indicated greater bacterial diversity in the Ctr group. However, greater fecal microbial diversity (estimated using the Shannon and Simpson indices) in METH CPP rats has been reported before30. The strict control over the animal-related variables, including the living environment, age, sex, etc., may have contributed greatly to these results. The nonnegligible differences in the duration of drug use also affected the gut microbiota. The average duration of METH use in the study subjects was 8 years, far longer than that in the METH CCP rats. In addition, many studies have shown that in adults, higher microbiota diversity correlates with increased overall healt42.
In this study, changes in gut microbial community composition were identified from the beta diversity differences between the two groups assessed via ADONIS analysis of the weighted UniFrac, unweighted UniFrac and Bray–Curtis distances. A cohort study from the University of California also showed significant variation in the overall gut microbiota composition of METH users29. In addition, gut dysbiosis was characterized by significant taxonomical differences between one of the major classes and three of the orders in METH users versus control subjects. Deltaproteobacteria abundance was reduced in the METH group and has also been reported to be reduced with chronic inflammation of the gastrointestinal (GI) tract and Crohn’s disease47, and there were also cases of chronic METH use with presentations of the signs and symptoms of Crohn's disease48. Furthermore, we discovered that the relative abundances of the orders Sphingomonadales and Xanthomonadales were significantly increased in the gut microbiota of the subjects in the METH group compared to that of the control subjects, while the relative abundance of the order Desulfovibrionales was significantly reduced in the individuals in the METH group.
Cases of people who chronically use METH and suffer from gastrointestinal diseases, such as nonocclusive mesenteric ischemia48 and intestinal ischemia49, may help explain these changes in the gut microbiota composition. Our results showed that several gut microorganisms, including Fusobacteria, Fusobacteriia, Fusobacteriales and Fusobacteriaceae, were negatively correlated with the duration of METH use. Fusobacteria are anaerobic gram-negative bacilli that are natural constituents of the microbiota in the mouth and other mucosal sites in humans50. However, several Fusobacteria taxa were significantly more abundant in METH CPP rats30, indicating the different influence between chronic and short-term METH use on gut microbiota. Fusobacteria and Fusobacteriia were both reported to increase in the stool microbiome of individuals with colorectal cancer compared with healthy individuals51,52. Patients with Crohn's disease showed an increased abundance of Fusobacteriaceae53. The role of those bacteria in the pathogenesis of gastrointestinal tract disease or gastrointestinal tract impairment related to METH use is unknown. The positive correlations between Fusobacteria and motor retardation, active social avoidance and the total general psychopathology scale may also indicate its important role in METH use disorders. Although age was negatively related to Fusobacteria, Fusobacteriia, Fusobacteriales, Fusobacteriaceae and Fusobacterium at different taxonomic levels in both the METH and Ctr groups, there are no studies about the relationship between those bacteria and aging (Supplementary Table 5). Therefore, these are potential new correlations of microbiota with aging among Asians in both METH users and non-drug users. VPA showed that the duration of METH use contributed more to the differences in microbial community distribution than did the age of the individuals, so the role of METH could not be ignored.
METH can induce psychosis and even a persistent psychotic syndrome that shows similarities to schizophrenia among recreational and chronic users54. Chronic METH users have higher rates of deficits in memory, executive functioning, anxiety, depression, and most notably psychosis48. The METH users in our study also showed abnormalities in the PANSS and MoCA tests (Supplementary Table 5). A higher abundance of the Enterobacteriaceae family triggers colonic inflammatory conditions and is distinctly increased in the colon of patients with non-tremor-dominant Parkinson disease (PD)55,56. P. mirabilis, belonging to the Enterobacteriaceae family, worsened motor symptoms and dopaminergic neuronal damage in mice at the premotor phase of PD57, which may be related to the positive symptoms of schizophrenia induced by METH use in the same way (Supplementary Table 6). The abundance of Ruminococcaceae was reported to be correlated with a lower severity of negative symptoms58 and was found to be negatively related to the total positive scale for the METH users in our study (Supplementary Table 6). Although the relative abundances of fecal Bacteroides were reported to be negatively correlated with brain signatures associated with depression59, we found that Bacteroides was positively correlated with delusions and poor impulse control but negatively correlated with emotional withdrawal (Supplementary Table 6). GABA could be produced by some Bacteroides59, and its uptake and release were reduced in cortical synaptosomes prepared from schizophrenic brains60, which may be related to the positive correlation we found in the present study (Supplementary Table 6). Bifidobacterium longum 1714™ has been shown to reduce stress-related behaviors in preclinical studies and improve stress responses61,62 and cognitive function in healthy volunteers63, but Bifidobacterium was positively related to several items of the negative scale and poor impulse control in our study (Supplementary Table 6). Faecalibacterium was reported to be negatively correlated with the severity of depressive symptoms64 and negatively related to difficulty in abstract thinking in this study, which may be influenced by its butyrate-producing function65. Although opposite trends of Enterobacteriaceae and Faecalibacterium abundance were found in patients with major depressive disorder59, these bacteria were all negatively correlated with the items of the negative scale in METH users (Supplementary Table 6). Enterobacteriaceae and Faecalibacterium were also correlated with the items of the general psychopathology scale in METH users (Supplementary Table 6). Furthermore, Lachnospiraceae and Ruminococcaceae, which were positively related to the items of the negative scale (Supplementary Table 6), were all found to be decreased in the responding individuals with major depressive disorder59. Blautia, which was positively related to abstraction in the cognition assessment (Supplementary Table 6), was also found to be increased in patients with major depressive disorder64,66.
Our results showed alterations in gut microbiota composition and significantly different taxa at various levels in METH users and age-matched individuals. However, the absence of diet data and the unique ethnicity, gender and sexual orientation may limit the generalizability of our findings to other groups. Fortunately, this work could supplement the research reporting the gut microbiota of male METH users who had sex with men29. Although not all of the altered genera were the same in their research and ours, the reduction in Bacteroides was similar and significant29. Considering the differences in ethnicity and sexual behavior patterns, the variation in the results was acceptable.
Here, we observed an altered intestinal microbial community, including richer bacterial diversity and significant changes in the relative abundance of several genera, in individuals who abused METH. These features may be associated with impairment of the gastrointestinal tract in chronic METH users. In addition, the genera that were significantly related to the duration of METH use highlight the chronic effect of this drug. There is currently no accepted pharmaceutical treatment for psychosis induced by METH. Thus, further research on gut microbiota alterations in people who abuse METH, especially those with symptoms of schizophrenia or cognitive impairment, may inform therapeutic approaches. The common altered microbes in psychosis induced by METH and other factors also revealed potential relationships with pathogenesis.
Supplementary Information
Acknowledgements
The authors would like to thank the staff of Wuhan Mental Health Center for sample collection and performing the screening examinations. We thank Novogene Co. (Beijing, China) for 16S rDNA sequencing and technical support. We thank Nellie M., Senior Editor, of Springer Nature Author Services, for medical editing assistance with an earlier version of the manuscript, funded by National Nature Science Foundation. All the authors are grateful to the enrolled subjects.
Author contributions
Y.D.Y. collected the samples and clinical data. X.Y. performed the data analysis and original draft writing. X.B.L. and G.Y.L. searched for the the enrolled subjects and performed data curation. K.Z. and G.W. came up with conception and design of the study. All authors reviewed the manuscript.
Funding
The authors are supported by the National Key R&D Program of China (2017YFC1310400), Youth Program of Hubei Province Nature Science Foundation (2018CFB334), Youth Program of Wuhan Municipal Health Commission Foundation (WX18Q41).
Data availability
All datasets generated for this study are included in the manuscript/supplementary files. The data have already been uploaded to SRA(ID: PRJNA679237).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yongde Yang and Xuan Yu.
Contributor Information
Kuan Zeng, Email: 773847352@qq.com.
Gang Wang, Email: 532614206@qq.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97548-1.
References
- 1.Jones CM, Compton WM, Mustaquim D. Patterns and characteristics of methamphetamine use among adults—United States, 2015–2018. MMWR Morb. Mortal. Wkly. Rep. 2020;69:317–323. doi: 10.15585/mmwr.mm6912a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song XB, Shao XT, Liu SY, Tan DQ, Wang Z, Wang DG. Assessment of metformin, nicotine, caffeine, and methamphetamine use during Chinese public holidays. Chemosphere. 2020;258:127354. doi: 10.1016/j.chemosphere.2020.127354. [DOI] [PubMed] [Google Scholar]
- 3.Wilson JM, Kalasinsky KS, Levey AI, Bergeron C, Reiber G, Anthony RM, Schmunk GA, Shannak K, Haycock JW, Kish SJ. Striatal dopamine nerve terminal markers in human, chronic methamphetamine users. Nat. Med. 1996;2:699–703. doi: 10.1038/nm0696-699. [DOI] [PubMed] [Google Scholar]
- 4.Thompson PM, Hayashi KM, Simon SL, Geaga JA, Hong MS, Sui Y, Lee JY, Toga AW, Ling W, London ED. Structural abnormalities in the brains of human subjects who use methamphetamine. J. Neurosci. 2004;24:6028–6036. doi: 10.1523/JNEUROSCI.0713-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potvin S, Pelletier J, Grot S, Hebert C, Barr AM, Lecomte T. Cognitive deficits in individuals with methamphetamine use disorder: A meta-analysis. Addict. Behav. 2018;80:154–160. doi: 10.1016/j.addbeh.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 6.Moszczynska A, Callan SP. Molecular, behavioral, and physiological consequences of methamphetamine neurotoxicity: Implications for treatment. J. Pharmacol. Exp. Ther. 2017;362:474–488. doi: 10.1124/jpet.116.238501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer BA, Richardson EJ, Heesacker M, DePue MK. Public stigma and the label of gambling disorder: Does it make a difference? J. Gambl. Stud. 2018;34:1281–1291. doi: 10.1007/s10899-017-9735-x. [DOI] [PubMed] [Google Scholar]
- 8.Jiang HY, Zhang X, Yu ZH, Zhang Z, Deng M, Zhao JH, Ruan B. Altered gut microbiota profile in patients with generalized anxiety disorder. J. Psychiatr. Res. 2018;104:130–136. doi: 10.1016/j.jpsychires.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Rogers GB, Keating DJ, Young RL, Wong ML, Licinio J, Wesselingh S. From gut dysbiosis to altered brain function and mental illness: Mechanisms and pathways. Mol. Psychiatry. 2016;21:738–748. doi: 10.1038/mp.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Sun G, Feng T, Zhang J, Huang X, Wang T, Xie Z, Chu X, Yang J, Wang H, Chang S, Gong Y, Ruan L, Zhang G, Yan S, Lian W, Du C, Yang D, Zhang Q, Lin F, Liu J, Zhang H, Ge C, Xiao S, Ding J, Geng M. Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer's disease progression. Cell Res. 2019;29:787–803. doi: 10.1038/s41422-019-0216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo R, Chen LH, Xing C, Liu T. Pain regulation by gut microbiota: Molecular mechanisms and therapeutic potential. Br. J. Anaesth. 2019;123:637–654. doi: 10.1016/j.bja.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 12.Cryan JF, Dinan TG. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 13.Frohlich EE, Farzi A, Mayerhofer R, Reichmann F, Jacan A, Wagner B, Zinser E, Bordag N, Magnes C, Frohlich E, Kashofer K, Gorkiewicz G, Holzer P. Cognitive impairment by antibiotic-induced gut dysbiosis: Analysis of gut microbiota-brain communication. Brain Behav. Immun. 2016;56:140–155. doi: 10.1016/j.bbi.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarr AJ, Galley JD, Fisher SE, Chichlowski M, Berg BM, Bailey MT. The prebiotics 3'Sialyllactose and 6'Sialyllactose diminish stressor-induced anxiety-like behavior and colonic microbiota alterations: Evidence for effects on the gut-brain axis. Brain Behav. Immun. 2015;50:166–177. doi: 10.1016/j.bbi.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alander T, Svardsudd K, Johansson SE, Agreus L. Psychological illness is commonly associated with functional gastrointestinal disorders and is important to consider during patient consultation: A population-based study. BMC Med. 2005;3:8. doi: 10.1186/1741-7015-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei J, Hemmings GP. Gene, gut and schizophrenia: The meeting point for the gene-environment interaction in developing schizophrenia. Med. Hypotheses. 2005;64:547–552. doi: 10.1016/j.mehy.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Severance EG, Yolken RH, Eaton WW. Autoimmune diseases, gastrointestinal disorders and the microbiome in schizophrenia: More than a gut feeling. Schizophr. Res. 2016;176:23–35. doi: 10.1016/j.schres.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dive A, Foret F, Jamart J, Bulpa P, Installe E. Effect of dopamine on gastrointestinal motility during critical illness. Intensive Care Med. 2000;26:901–907. doi: 10.1007/s001340051279. [DOI] [PubMed] [Google Scholar]
- 19.Dunser MW, Hasibeder WR. Sympathetic overstimulation during critical illness: Adverse effects of adrenergic stress. J. Intensive Care Med. 2009;24:293–316. doi: 10.1177/0885066609340519. [DOI] [PubMed] [Google Scholar]
- 20.Zizzo MG, Mule F, Mastropaolo M, Serio R. D1 receptors play a major role in the dopamine modulation of mouse ileum contractility. Pharmacol. Res. 2010;61:371–378. doi: 10.1016/j.phrs.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Herr RD, Caravati EM. Acute transient ischemic colitis after oral methamphetamine ingestion. Am. J. Emerg. Med. 1991;9:406–409. doi: 10.1016/0735-6757(91)90073-S. [DOI] [PubMed] [Google Scholar]
- 22.Brannan TA, Soundararajan S, Houghton BL. Methamphetamine-associated shock with intestinal infarction. MedGenMed. 2004;6:6. [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson TL, Plackett TP, Gagliano RA, Jr, Smith RR. Methamphetamine-induced paralytic ileus. Hawaii J. Med. Public Health. 2012;71:44–45. [PMC free article] [PubMed] [Google Scholar]
- 24.Beyer KL, Bickel JT, Butt JH. Ischemic colitis associated with dextroamphetamine use. J. Clin. Gastroenterol. 1991;13:198–201. doi: 10.1097/00004836-199104000-00016. [DOI] [PubMed] [Google Scholar]
- 25.Dirkx CA, Gerscovich EO. Sonographic findings in methamphetamine-induced ischemic colitis. J. Clin. Ultrasound. 1998;26:479–482. doi: 10.1002/(SICI)1097-0096(199811/12)26:9<479::AID-JCU9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Holubar SD, Hassinger JP, Dozois EJ, Masuoka HC. Methamphetamine colitis: a rare case of ischemic colitis in a young patient. Arch. Surg. 2009;144:780–782. doi: 10.1001/archsurg.2009.139. [DOI] [PubMed] [Google Scholar]
- 27.Link DP, Chi YW. Massive hematochezia: A complication of methamphetamine-induced vasculitis treated by transcatheter hemostasis. Case Rep. Radiol. 2011;2011:919236. doi: 10.1155/2011/919236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persons AL, Bradaric BD, Dodiya HB, Ohene-Nyako M, Forsyth CB, Keshavarzian A, Shaikh M, Napier TC. Colon dysregulation in methamphetamine self-administering HIV-1 transgenic rats. PLoS ONE. 2018;13:e0190078. doi: 10.1371/journal.pone.0190078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook RR, Fulcher JA, Tobin NH, Li F, Lee DJ, Woodward C, Javanbakht M, Brookmeyer R, Shoptaw S, Bolan R, Aldrovandi GM, Gorbach PM. Alterations to the gastrointestinal microbiome associated with methamphetamine use among young men who have sex with men. Sci. Rep. 2019;9:14840. doi: 10.1038/s41598-019-51142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ning T, Gong X, Xie L, Ma B. Gut microbiota analysis in rats with methamphetamine-induced conditioned place preference. Front. Microbiol. 2017;8:1620. doi: 10.3389/fmicb.2017.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen LJ, Zhi X, Zhang KK, Wang LB, Li JH, Liu JL, Xu LL, Yoshida JS, Xie XL, Wang Q. Escalating dose-multiple binge methamphetamine treatment elicits neurotoxicity, altering gut microbiota and fecal metabolites in mice. Food Chem. Toxicol. 2021;148:111946. doi: 10.1016/j.fct.2020.111946. [DOI] [PubMed] [Google Scholar]
- 32.Forouzan S, Hoffman KL, Kosten TA. Methamphetamine exposure and its cessation alter gut microbiota and induce depressive-like behavioral effects on rats. Psychopharmacology. 2021;238:281–292. doi: 10.1007/s00213-020-05681-y. [DOI] [PubMed] [Google Scholar]
- 33.Angoa-Perez M, Zagorac B, Winters AD, Greenberg JM, Ahmad M, Theis KR, Kuhn DM. Differential effects of synthetic psychoactive cathinones and amphetamine stimulants on the gut microbiome in mice. PLoS ONE. 2020;15:e0227774. doi: 10.1371/journal.pone.0227774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil. 2011;23:255–264. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 35.Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–1188. doi: 10.1016/j.neuroscience.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Magoc T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Zhang X, Guo F, Wu W, Zhang T. Characterization of tetracycline resistant bacterial community in saline activated sludge using batch stress incubation with high-throughput sequencing analysis. Water Res. 2013;47:4207–4216. doi: 10.1016/j.watres.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avershina E, Frisli T, Rudi KD. Novo semi-alignment of 16S rRNA gene sequences for deep phylogenetic characterization of next generation sequencing data. Microbes Environ. 2013;28:211–216. doi: 10.1264/jsme2.ME12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Toole PW, Jeffery IB. Gut microbiota and aging. Science. 2015;350:1214–1215. doi: 10.1126/science.aac8469. [DOI] [PubMed] [Google Scholar]
- 43.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 1987;13:261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 44.Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, Charlson FJ, Norman RE, Flaxman AD, Johns N, Burstein R, Murray CJ, Vos T. Global burden of disease attributable to mental and substance use disorders: Findings from the Global Burden of Disease Study 2010. Lancet. 2013;382:1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- 45.Swanson SM, Sise CB, Sise MJ, Sack DI, Holbrook TL, Paci GM. The scourge of methamphetamine: Impact on a level I trauma center. J. Trauma. 2007;63:531–537. doi: 10.1097/TA.0b013e318074d3ac. [DOI] [PubMed] [Google Scholar]
- 46.McKetin R, McLaren J, Lubman DI, Hides L. Hostility among methamphetamine users experiencing psychotic symptoms. Am. J. Addict. 2008;17:235–240. doi: 10.1080/10550490802019816. [DOI] [PubMed] [Google Scholar]
- 47.Magro DO, Santos A, Guadagnini D, de Godoy FM, Silva SHM, Lemos WJF, Vitulo N, Torriani S, Pinheiro LV, Martinez CAR, Saad MJA, Coy CSR. Remission in Crohn's disease is accompanied by alterations in the gut microbiota and mucins production. Sci. Rep. 2019;9:13263. doi: 10.1038/s41598-019-49893-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson JE, Brown IE, Olson KA, Iverson K, Cocanour CS, Galante JM. Nonocclusive mesenteric ischemia in patients with methamphetamine use. J .Trauma Acute Care Surg. 2018;84:885–892. doi: 10.1097/TA.0000000000001855. [DOI] [PubMed] [Google Scholar]
- 49.Choi K, Imrie A, Lourie R, Cross T. Intestinal ischemia due to methamphetamine use: A case report. Int. J. Surg. Case Rep. 2019;58:11–13. doi: 10.1016/j.ijscr.2019.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brennan CA, Garrett WS. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allali I, Boukhatem N, Bouguenouch L, Hardi H, Boudouaya HA, Cadenas MB, Ouldim K, Amzazi S, Azcarate-Peril MA, Ghazal H. Gut microbiome of Moroccan colorectal cancer patients. Med. Microbiol. Immunol. 2018;207:211–225. doi: 10.1007/s00430-018-0542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wearne TA, Cornish JL. A comparison of methamphetamine-induced psychosis and schizophrenia: A review of positive, negative, and cognitive symptomatology. Front. Psychiatry. 2018;9:491. doi: 10.3389/fpsyt.2018.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov. Disord. 2015;30:350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 56.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi JG, Kim N, Ju IG, Eo H, Lim SM, Jang SE, Kim DH, Oh MS. Oral administration of Proteus mirabilis damages dopaminergic neurons and motor functions in mice. Sci. Rep. 2018;8:1275. doi: 10.1038/s41598-018-19646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen TT, Kosciolek T, Maldonado Y, Daly RE, Martin AS, McDonald D, Knight R, Jeste DV. Differences in gut microbiome composition between persons with chronic schizophrenia and healthy comparison subjects. Schizophr. Res. 2019;204:23–29. doi: 10.1016/j.schres.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, Lewis K. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019;4:396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savignac HM, Kiely B, Dinan TG, Cryan JF. Bifidobacteria exert strain-specific effects on stress-related behavior and physiology in BALB/c mice. Neurogastroenterol. Motil. 2014;26:1615–1627. doi: 10.1111/nmo.12427. [DOI] [PubMed] [Google Scholar]
- 62.Savignac HM, Tramullas M, Kiely B, Dinan TG, Cryan JF. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behav. Brain Res. 2015;287:59–72. doi: 10.1016/j.bbr.2015.02.044. [DOI] [PubMed] [Google Scholar]
- 63.Allen AP, Hutch W, Borre YE, Kennedy PJ, Temko A, Boylan G, Murphy E, Cryan JF, Dinan TG, Clarke G. Bifidobacterium longum 1714 as a translational psychobiotic: Modulation of stress, electrophysiology and neurocognition in healthy volunteers. Transl. Psychiatry. 2016;6:939. doi: 10.1038/tp.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, Wang W, Tang W, Tan Z, Shi J, Li L, Ruan B. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 65.Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019;4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 66.Chung YE, Chen HC, Chou HL, Chen IM, Lee MS, Chuang LC, Liu YW, Lu ML, Chen CH, Wu CS, Huang MC, Liao SC, Ni YH, Lai MS, Shih WL, Kuo PH. Exploration of microbiota targets for major depressive disorder and mood related traits. J. Psychiatr. Res. 2019;111:74–82. doi: 10.1016/j.jpsychires.2019.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the manuscript/supplementary files. The data have already been uploaded to SRA(ID: PRJNA679237).