Abstract
Fen1/Rad27 nuclease activity, which is important in DNA metabolism, is stimulated by proliferating cell nuclear antigen (PCNA) in vitro. The in vivo role of the PCNA interaction was investigated in the yeast Rad27. A nuclease-defective rad27 mutation had a dominant-negative effect that was suppressed by a mutation in the PCNA binding site, thereby demonstrating the importance of the Rad27-PCNA interaction. The PCNA-binding defect alone had little effect on mutation, recombination, and the methyl methanesulfonate (MMS) response in repair-competent cells, but it greatly amplified the MMS sensitivity of a rad51 mutant. Furthermore, the PCNA binding mutation resulted in lethality when combined with a homozygous or even a heterozygous pol3-01 mutation in the 3′→5′ exonuclease domain of DNA polymerase δ. These results suggest that phenotypically mild polymorphisms in DNA metabolic proteins can have dramatic consequences when combined.
Polymorphisms occur throughout the human genome. In some cases, they may affect biological function and may be responsible for poorly understood differences in individual susceptibilities to diseases. Because genetic instability is a major causative factor in tumorigenesis, the consequences of subtle functional alterations in proteins that act on DNA are of particular interest. The Fen1 nuclease, termed Rad27 in the yeast Saccharomyces cerevisiae, plays an important role in DNA metabolism (reviewed in reference 24). It seems plausible that even subtle changes in Fen1 could have significant deleterious effects because deletion of the Fen1/Rad27 gene in yeast cells causes severe effects, including temperature sensitivity and hypersensitivity to the DNA-alkylating agent methyl methanesulfonate (MMS). The rad27-Δ (a deletion null allele) strain exhibits instabilities in short DNA repeats, microsatellites and minisatellites, expansions of the trinucleotide repeat CTG/CAG, duplications between separated short direct random repeats, intrachromosomal and interchromosomal mitotic recombination, and chromosome loss (8, 15, 20, 38, 41, 44, 48). RAD27 becomes essential for viability in strains lacking double-strand break (DSB) recombinational repair (42, 44) or the 5′→3′ exonuclease EXO1 (43) or in strains carrying mutation pol3-01 in the 3′→5′ exonuclease proofreading domain of DNA polymerase δ (Pol δ) (20), and in strains with a temperature-sensitive mutation in DNA2 (DNA helicase/3′→5′ exonuclease) (1, 3).
The Fen1 nuclease recognizes specific types of DNA structures (reviewed in reference 24). In vitro, it is highly active toward 5′-flap DNA, a branched structure that can result from strand displacement during DNA synthesis. Fen1 acts as an endonuclease to cleave the displaced flap strand at the single-strand/double-strand junction. It also acts as a 5′→3′ exonuclease at nicks in duplex DNA. Significantly, Fen1 can remove a 5′-terminal ribonucleotide as well. During lagging strand DNA synthesis, RNA primers are removed by RNase H1; however, this enzyme cannot excise the final 5′-terminal ribonucleotide at the RNA-DNA junction. The completion of RNA primer removal by Fen1 is essential for Okazaki fragment processing in reconstituted replication assays (14, 35, 47, 49). At the restrictive temperature (37°C), a rad27-Δ mutant accumulates short DNA fragments of the size range expected for unprocessed Okazaki fragments (27). Fen1 can cleave oligonucleotide substrates with 5′-terminal abasic sites that mimic reaction intermediates that arise during base excision repair (BER) (6, 32). In reconstituted long-patch BER assays, Fen1 is essential for the excision step (18, 19). The relevance of these in vitro observations to BER in vivo is supported by the MMS sensitivity of rad27-Δ mutants.
In addition to its structure-specific nuclease activity, Fen1 exhibits a protein-protein interaction with proliferating cell nuclear antigen (PCNA). PCNA is essential for DNA synthesis during replication and repair (reviewed in references 17 and 50). It is a homotrimeric ring-shaped protein that serves as an accessory factor for Pol δ and Pol ɛ. DNA-bound PCNA forms a sliding clamp that tethers Pol δ or Pol ɛ to template DNA and thus promotes processive DNA synthesis. It also binds to several other proteins involved in DNA metabolism, including DNA ligase I (22) and DNA-(cytosine-5) methyltransferase (4). PCNA binds to Fen1 and stimulates nuclease activity on oligonucleotide-based 5′ flaps and nicked duplex DNA substrates (23, 54). These properties suggest that Fen1 and PCNA interact during the course of DNA replication, DNA repair, or both. Support for the importance of Fen1-PCNA binding in BER has come from studies with a reconstituted long-patch BER system, in which Fen1-PCNA interaction was shown to contribute to excision efficiency (9).
The possible involvement of the Fen1-PCNA interaction in DNA replication has been proposed (23), but it has not been investigated specifically. Recently, a hypothetical model of the Fen1-PCNA complex was presented that illustrated PCNA binding and nuclease activities working in tandem during replication (13). We have selectively inactivated nuclease and PCNA binding activities of yeast Fen1/Rad27 to dissect the contributions of each to overall function in vivo. Unexpectedly, we found that PCNA binding in Rad27/Fen1 has a function beyond mere stimulation of nuclease activity. A mutation of Rad27 eliminating PCNA binding had very little effect by itself but had major consequences via intragenic or intergenic interactions. The PCNA nonbinding rad27 allele greatly enhanced the repair deficiency of a rad51-null strain and caused inviability of yeast cells with a minor Pol δ defect. Our results show that while subtle defects in specific DNA metabolic proteins may have little impact on their own, they can result in severe phenotypic effects in combination. This principle, when applied to naturally occurring polymorphisms, can have profound implications for the inheritance of susceptibility to disease.
MATERIALS AND METHODS
Plasmids.
An EcoRI-StuI fragment of S. cerevisiae genomic DNA from plasmid yEP24-3a (3) containing the entire RAD27 open reading frame and flanking transcriptional control elements was subcloned into the CEN plasmid pRS416 or yIP pRS406 to produce pRG8X and pRG101A, respectively. These wild-type RAD27 plasmids were used as templates for mutagenesis to create CEN and yIP plasmids (pRG103B and pRG104A, respectively) containing the mutation D179A (rad27-n) or CEN and yIP plasmids (pRG95E and pRG102D, respectively) containing the mutation F346A/F347A (rad27-p). The D179A mutation was created by using the primer pair 5′-AGCAAGTGAAGATATGGCCACACTCTGTTATAGAACACCCT-3′ and 5′-AGGGTGTTCTATAACAGAGTGTGGCCATATCTTCACTTGCT-3′, which created an MscI site to facilitate screening, and the F346A/F347A mutation was created by using the primer pair 5′-CATTCAGGGTAGGTTAGATGGCGCCGCCCAAGTGGTGCCTAAGACAAAG-3′ and 5′-CTTTGTCTTAGGCACCACTTGGGCGGCGCCATCTAACCTACCCTGAATG-3′, which created an EheI (SfoI) site. The RAD27 TRP1+ CEN plasmid pLC80B was constructed by subcloning the NotI-XhoI fragment from pRG101A into pRS314. Plasmids pLC76A, pLC78A, and pLC77A for the bacterial expression of C-terminal His6-tagged wild-type Rad27 and mutant Rad27-n and Rad27-p proteins, respectively, were made by PCR amplification with pRG101A, pRG104A, and pRG102D as templates with primers 5′-AGCACCATGGGTATTAAAGGTTTGAA-3′ and 5′-TCGCTCGAGTCTTCTTCCCTTTGTGACTTTA-3′ and then ligating them into pET28b. Plasmids pRG106A, pRG105A, pRG107A, and pRG108A are URA3+ 2-μm multicopy plasmids derived from the vector yEP195-SPGAL (5) for galactose-inducible overexpression in yeast cells of wild-type Rad27 or mutant D179A (Rad27-n), F346A/F347A (Rad27-p), and D179A/F346A/F347A (Rad27-n,p) proteins, respectively, by using the GAL1 promoter. These were constructed by three-fragment ligation of the XbaI-BstXI N-terminal fragment from pLC76A (for pRG106A and pRG107A) or pLC78A (for pRG105A and pRG108A), the BstXI-HindIII C-terminal fragment from pRG101A (for pRG106A and pRG105A) or pRG102D (for pRG107A and pRG108A), and the XbaI-HindIII-digested vector yEP195-SPGAL (for all four constructs). The yeast PCNA bacterial expression plasmid pLC93C was made by PCR by using yeast open reading frame YBR088C (Research Genetics) as template with primers 5′-AAGAACATGTTAGAAGCAAAATTTGAAGAAGC-3′ and 5′-ACGGAAGCTTATTCTTCGTCATTAAATTTAGG-3′ and then ligating the AflIII-HindIII-digested PCR product with NcoI-HindIII-digested pET28b. Rad27 and PCNA in plasmids were verified by DNA sequencing. pLC93C contains synonymous codons at PCNA residues C81 (TGC) and I147 (ATC) that differ from those reported in GenBank (accession numbers X16676 and Z35957) but do not affect the amino acid sequence of the encoded protein.
Expression, purification, and characterization of Rad27 proteins.
His6-tagged wild-type and mutant Rad27 proteins were expressed in bacteria, purified by Ni2+-metal chelate affinity chromatography, quantitated, and characterized enzymatically as previously described for human Fen1 (9).
Yeast genetic procedures and strains.
Standard yeast media and procedures of yeast genetics were used (36). Haploid strains used to study the effects of rad27 mutations were isogenic to CG379 (S1 in our collection) MATα ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52 (28). Rates of forward mutations were measured in the CAN1 gene. Strains with reporter lys2 alleles used to detect frameshift mutations in homonucleotide runs A12 (+1 frameshifts) and A14 (−1 frameshifts) have been described (45, 46). Haploid strains ALE100 and ALE101 were constructed for measuring the rate of interchromosomal recombination between two lys2 sequences located in nonhomologous chromosomes II and III, similar to previously described strains (25). lys2::HS-D in the chromosome II contains the 658-bp insert of a direct repeat of two human-specific Alu consensus sequences cloned from plasmid pPD39 (2) in the BamHI site of LYS2. The plasmid pRS305L3 with the 5′-truncated lys2-Δ5′ (1,691-bp XhoI-HindIII fragment in the polylinker of pRS305) was integrated in the vicinity of LEU2 in chromosome III. Because lys2-Δ5′ and lys2::HS-D overlap for 382 bp from 5′ of the insert and for 1,309 bp from 3′ of the insert, they can produce Lys+ recombinants via gene conversion or/and crossing over. The latter can lead to translocations (25).
Derivatives of CG379 (S1) carrying pol3-01, pol2-4, and exo1 mutations were described previously (45, 46). Deletion replacement constructs with hisG-URA3-hisG gene blaster were used to obtain null mutations in rad27, pR2.10 (41) and rad51, pΔRAD51 (40). Plasmids containing rad27-p (pRG102D) or rad27-n (pRG104A) mutant alleles (Fig. 1) in the polylinker of the pRS406 (URA3) integrating plasmid were used for two-step replacements of the wild-type RAD27 with rad27 mutant alleles. The resulting strains carried mutant rad27 in its normal genomic environment. Because each mutation introduced a restriction site, genotyping was done by restriction digestion of a PCR-amplified region overlapping the mutation. For the rad27-n mutation, the oligonucleotides RGYKL541 (5′-CCGGCTGGTAAGTTATGATA-3′) and RGYKL1487 (5′-CAAGTCGAGTCCTCTCAAAACTA-3′) were used. MscI digestion of the 987-bp PCR product containing rad27-n produced 113- and 874-bp products. For genotyping of rad27-p, the oligonucleotides RGYKLSEQ3 (5′-TCTTCTTCCCTTTGTGACTTTATTC-3′) and RGYKLSEQ5U (5′-GACTGGCCTTACAAACAAGCA-3′) were used. SfoI digestion of the 324-bp PCR fragment containing the rad27-p resulted in 210- and 114-bp products.
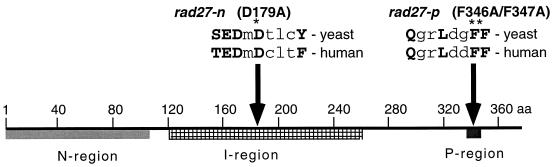
FIG. 1.
Mutations in Rad27. The N region (lightly shaded) and I region (striped) are catalytically important regions common to Fen1 and XPG-family exo- and endonucleases. The rad27-n mutation (asterisk) lies within the sequence Ser/Thr-Asp/Glu-Asp-X-Asp-X-X-X-Phe/Tyr (conserved residues are in uppercase letters) that is present in all eukaryote Fen1 and XPG homologs examined to date. The P region (darkly shaded) is the PCNA binding motif. The rad27-p mutation (double asterisk) changes two residues involved in hydrophobic interaction with PCNA. The sequence Gln-X-X-Leu/Ile/Met-X-X-Phe-Phe/Tyr (critical residues are in uppercase letters) is present in several PCNA binding proteins.
In order to obtain rad27-n/RAD27 heterozygous diploids, we first obtained the chromosomally integrated rad27-n plasmid and then crossed integrants containing both wild-type and rad27-n alleles with strain E67 (MATa ade2-1 arg4-8 leu2-3,112 lys2-BX thr1-4 trp1-1 ura3-52 cup1-1) and then identified rad27-n/RAD27 heterozygotes among independent popouts of the URA3 marker.
In order to assess the viability of double mutants carrying either rad27-p or rad27-Δ combined with exo1-null, rad51-null, or pol3-01, we first transformed the strain carrying only rad27 with the ARS-CEN plasmid LC80B (TRP1 RAD27) and then obtained a deletion or replacement in the second gene. Double mutants carrying LC80B were replica plated twice on complete yeast extract-peptone-dextrose (YPD) medium at 2-day intervals, suspended in water, plated at low density to YPD, and then incubated for either 3 or 6 to 7 days (to allow the appearance of slow-growing variants). The frequency of loss of the TRP1 marker was determined among 200 to 600 colonies by replica plating them to medium without tryptophan. In such an assay, pLC80B is lost with a frequency of as much as 80%. The absence or very infrequent (<1%) loss of TRP1 was considered to be evidence of synthetic lethality of the double mutation.
Heterozygous pol3-01/POL3 diploids carrying various combinations of rad27-Δ, rad27-p, and RAD27 (wild-type) alleles were obtained by crossing MATα (S1 background) rad27-p pol3-01 or rad27-Δ pol3-01 strains carrying the pLC80B (TRP1 RAD27) plasmid with MATa strains carrying either the rad27-Δ or RAD27 (wild-type) allele. For each cross, we used two different MATa strains, TE01 (MATa lys2::HIS3 trp1-del1 leu2-2 his3-15,11 ura3-x ade2-Δ) and VL6-48-a (MATa his3-200 trp1-Δ1 met14 ura3-52 ade2-101 lys2-801).
PCNA binding assay.
Yeast PCNA and His6-tagged wild-type Rad27, Rad27-p, and Rad27-n proteins were expressed in BL21(DE3). Cells from 100-ml cultures were lysed in 5 ml of 20 mM Tris-HCl (pH 7.9)–500 mM NaCl–5 mM imidazole–0.2 mg of lysozyme per ml with protease inhibitors and then sonicated, and the lysate was next clarified by centrifugation. Binding assay mixtures consisted of 100 μl of 50% NiSO4-charged iminodiacetic acid resin (HisBind; Novagen), 1.5 ml of lysate from cells expressing His6-tagged wild-type or mutant Rad27, and 300 μl of lysate from cells expressing untagged wild-type PCNA. In control experiments with either the Rad27 or the PCNA omitted, lysate was replaced by an equivalent volume (1.5 ml or 300 μl) of lysis buffer. Mixtures were incubated for 5 h at 4°C, and then the resin was washed with 10 mM Tris-HCl (pH 7.9)–250 mM NaCl–30 mM imidazole. Protein complexes were eluted with Laemmli buffer and analyzed on 12% gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
Rad27 immunoblot.
A wild-type yeast strain was transformed with the yEP195-SPGAL or yEP195-SPGAL derivatives pRG106A, pRG105A, pRG107A, and pRG108A to obtain galactose-inducible expression of wild-type Rad27, Rad27-n, Rad27-p, or Rad27-n,p. Cells were grown in glucose medium without uracil and then transferred to galactose for 12 h. Cells were suspended in 50 mM Tris-HCl (pH 8.0)–5% glycerol–1 mM dithiothreitol–phenylmethylsulfonyl fluoride–0.5 mM EDTA and lysed by vortexing them with glass beads. Lysate was resolved on 10% gels by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride, and the membrane was incubated with a 1:1,600 dilution of rabbit polyclonal antibody (immunoglobulin G [IgG] fraction, 5.2 mg/ml) raised against human Fen1 (cross-reactivity with Rad27 was confirmed by prior immunoblotting against Rad27 expressed in bacteria) and 1:5,000 mixture of peroxidase-conjugated goat anti-rabbit IgG; Rad27 bands were then detected by enhanced chemiluminescence.
RESULTS
Mutations in Rad27.
Eukaryotic Fen1 homologs are strongly conserved. Human Fen1 and S. cerevisiae Rad27 are 380 and 382 amino acids long, respectively, and 58% identical in amino acid sequence. Fen1/Rad27 has discrete nuclease and PCNA binding domains (Fig. 1). The N and I regions (12, 37) comprise the catalytic domain that is responsible for exo- and endonuclease activities. The D181A mutation in human Fen1 abolishes catalytic activity but does not affect binding to DNA flap substrates (39). We made the corresponding mutation in Rad27 (D179A, rad27-n) to likewise inactivate nuclease activity. The PCNA binding activity of human Fen1 has been localized to a short region near the C terminus (10, 52), and the F343A/F344A mutation within this region abolishes PCNA binding without affecting nuclease activity (9). We therefore made the corresponding mutation (F346A/F347A, rad27-p) in Rad27 to specifically inactivate its PCNA binding.
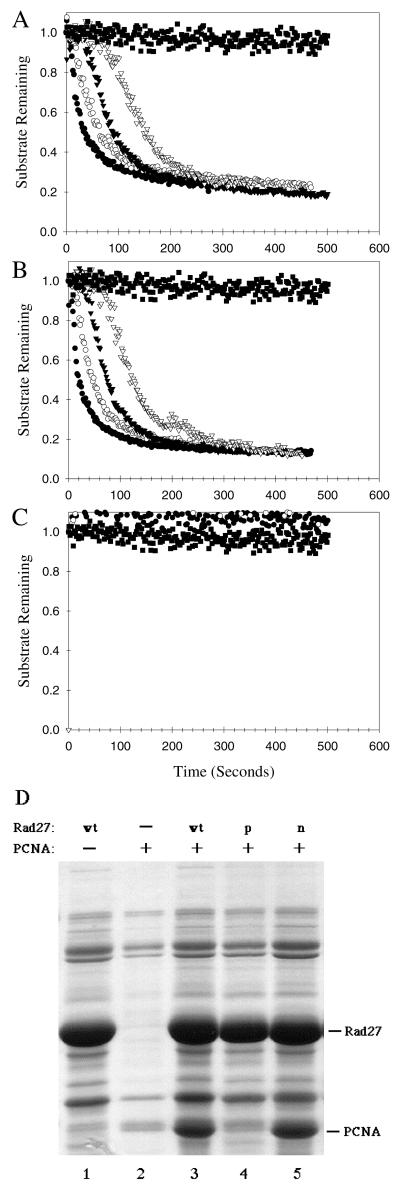
The endonuclease activities of wild-type Rad27, Rad27-n, and Rad27-p proteins with C-terminal hexahistidine (His6) tags were measured in an assay based on flow cytometry (31). The His6 tag does not alter the catalytic activity of Fen1 (31). The DNA cleavage kinetics of wild-type Rad27 and Rad27-p were essentially identical (Fig. 2A and B). In contrast, Rad27-n exhibited no detectable cleavage activity at any concentration tested (Fig. 2C), confirming that nuclease activity was abolished by the D179A mutation. The abilities of these proteins to bind to yeast PCNA were also evaluated (Fig. 2D). Lysates containing His6-tagged wild-type or mutant Rad27 proteins were mixed with yeast PCNA, and metal chelate resin was used to recover the complexes. Although several bacterial proteins had strong affinity for the resin, including some with a mobility similar to that of yeast PCNA (Fig. 2D, lane 1), nonspecific binding by PCNA in the absence of Rad27 was minimal (Fig. 2D, lane 2). PCNA bound well to wild-type Rad27 and Rad27-n (Fig. 2D, lanes 3 and 5), but specific binding to Rad27-p was negligible (Fig. 2D, lane 4). Thus, the D179A and F346A/F347A mutations in Rad27 specifically inactivated catalytic and PCNA binding activities, respectively.
FIG. 2.
Characterization of wild-type and mutant Rad27 proteins. Wild-type Rad27, Rad27-p (F346A/F347A), and Rad27-n (D179A) were expressed in bacteria and purified. The cleavage of a fluoresceinated 5′-flap DNA substrate was monitored by flow cytometry to measure the nuclease activities of wild-type Rad27 (A), Rad27-p (B), and Rad27-n (C). Bead-associated fluorescence (normalized to an initial value of 1.0), which indicates uncleaved substrate, and the reaction time (in seconds) are plotted. Reactions were conducted with wild-type or mutant Rad27 protein at 200 nM (●), 100 nM (○), 50 nM (▴), 25 nM (▵), or no enzyme (■). The PCNA binding activities of wild-type and mutant Rad27 proteins were determined using a bead pulldown assay (D). Ni2+-charged metal chelate resin was incubated with bacterial lysate from cells expressing His6-tagged wild-type Rad27 (lanes 1 and 3), Rad27-p (lane 4), or Rad27-n (lane 5) or an equal volume of lysis buffer without protein (lane 2). Lysis buffer alone (lane 1) or bacterial lysate from cells expressing untagged wild-type yeast PCNA (lanes 2 to 5) was added. Complexes were analyzed by SDS-PAGE and Coomassie blue staining. The migration positions of Rad27 and PCNA are indicated.
Functional interaction of nuclease activity and PCNA binding in Rad27.
In order to explore the effects of a change that leaves the PCNA binding and the DNA substrate recognition properties intact, the nuclease activity of Rad27 was eliminated. We expected that yeast strains carrying this enzymatically inactive allele would exhibit a phenotype similar to that of rad27-Δ, and therefore we used MMS sensitivity when screening for replacements of RAD27 by rad27-n. All MMS-sensitive isolates formed small colonies. Based on the following observations, the slow growth and MMS sensitivity were due to the rad27-n mutation. Among 55 isolates from five different strains, all carried the rad27-n mutation. Among six complete tetrads from a rad27-n/RAD27 diploid, there were always two normal-size colonies and two microcolonies (overall spore viability in 16 tetrads, 75%). The 12 normal colonies carried the wild-type RAD27 allele, and the 12 microcolonies carried rad27-n.
Because growth was inhibited even in comparison with the rad27-Δ strain (Fig. 3A), the Rad27-n protein appeared to be toxic. Overexpression of the rad27-n mutant allele in the presence of RAD27 in haploid strains also inhibited growth (Fig. 3B). Growth inhibition was much greater in the rad51 repair-defective mutant than in the wild type, suggesting that Rad27-n might cause DNA damage. Unlike rad51 mutations, neither exo1 nor pol3-01 (a mutation that eliminates Pol δ 3′→5′ exonuclease) affected the sensitivity of yeast to overexpressed rad27-n (Fig. 3C), even though all three are synthetically lethal with rad27-Δ.
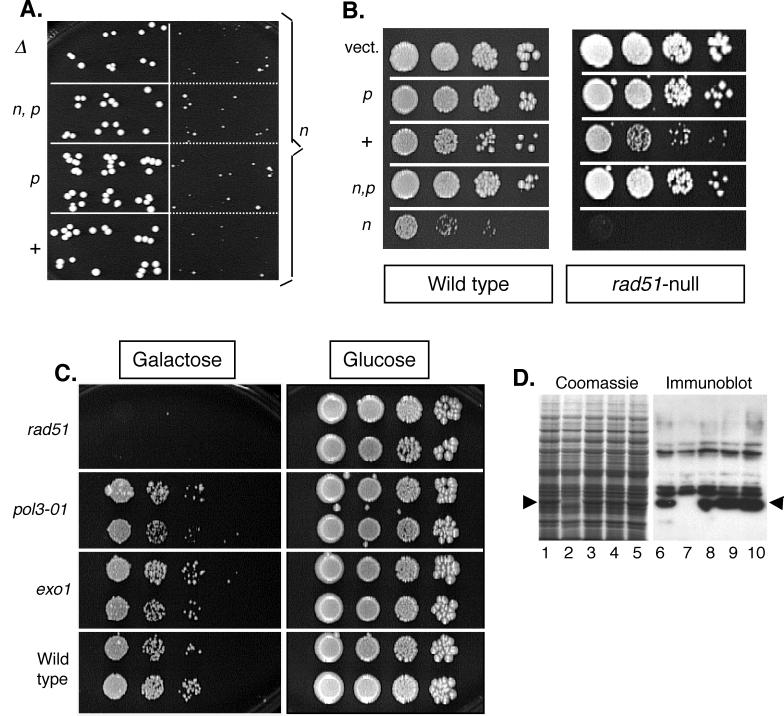
FIG. 3.
Effect of rad27 alleles on yeast growth. (A) Single-copy genomic alleles rad27-Δ (Δ), rad27-n,p (n,p), rad27-p (p), RAD27 wild type (+), and rad27-n (n). Drops of yeast suspensions containing approximately 3 to 5 CFU per drop were placed onto complete YPD medium (all compared strains on the same plate) and incubated for 3 days. Four independent isolates in areas divided by dashed lines are presented for the rad27-n. (B) Effect of galactose-induced overexpressing various rad27 alleles on growth of yeast strains carrying a wild-type RAD27 and either wild-type RAD51 or rad51-null allele. Drops of serial 10-fold dilutions of yeast suspensions were placed on synthetic medium lacking uracil (to select for the presence of the plasmid) with either glucose or galactose. Yeast density starts from approximately 5 × 103 cells/drop and decreases in 10-fold steps from left to right. Plates were incubated for 6 days to allow growth for slow-growing strains. Only galactose plates are shown. There was no detectable growth delay of any strain on glucose plates (parallel glucose plate is not shown; see example presented in panel C). (C) Effect of galactose-induced overexpression of the plasmid rad27-n allele on growth of yeast strains carrying a wild-type RAD27 and other mutations as indicated. Plating was the same as that described for panel B. (D) Inducible Rad27 protein expression was confirmed by immunoblot analysis. A wild-type yeast strain was transformed with an episomal plasmid that used the GAL-1/10 promoter to express wild-type Rad27 (lanes 1 and 6), the nuclease-defective D179A mutant Rad27-n (lanes 3 and 8), the PCNA nonbinding mutant F346A/F347A Rad27-p (lanes 4 and 9), or the combined mutant D179A/F346A/F347A Rad27-n,p (lanes 5 and 10). Cells transformed with the parental plasmid lacking the RAD27 insert served as a negative control (lanes 2 and 7). Induced cell lysates were analyzed by SDS-PAGE and Coomassie blue staining (lanes 1 to 5) or by immunoblotting with an antibody that recognizes Rad27 (lanes 6 to 10). Arrowheads indicate the migration position of Rad27.
The rad27-p allele, which eliminates PCNA binding, did not affect yeast growth by itself. However, this mutation completely eliminated the toxicity of single-copy or overexpressed rad27-n when both mutations were combined in the same allele (rad27-n,p; Fig. 3A and B). The toxic effect was eliminated even in the rad51 mutant strain, which was especially sensitive to rad27-n overexpression (Fig. 3B, right panel).
In order to exclude the possibility that rad27-p suppressed the toxicity of rad27-n by affecting protein stability, we confirmed the presence of wild-type and mutant Rad27 proteins by immunoblot analysis (Fig. 3D). The levels of the Rad27-n,p and Rad27-n proteins were similar; therefore, intragenic suppression was not due to the elimination of the toxic protein.
Elimination of PCNA binding suppressed only the negative effect of rad27-n on growth, but it did not restore other deficiencies caused by rad27-n. The rad27-n,p double mutant was as sensitive to 2 mM MMS as rad27-n and rad27-Δ (not shown), whereas both wild-type and rad27-p strains grew even on 4 mM MMS (Fig. 4A). We examined the impact of the rad27 mutations on the rates of frameshift mutations in long homonucleotide runs, on forward CAN1 mutations, and on interchromosomal recombination. The rates of all of these events were increased by rad27-Δ, in agreement with other reports (20, 33, 41, 48) (Table 1). Comparable increases were also found with the rad27-n and rad27-n,p mutants. Thus, based on MMS sensitivity and genetic instability measurements, the Rad27-n protein is nonfunctional in repair and in mutation prevention, and a second mutation, rad27-p, can suppress the toxic effects of rad27-n but cannot restore functionality to the protein.
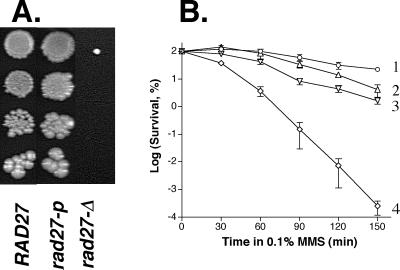
FIG. 4.
Effect of rad27-p on MMS sensitivity. (A) Effect of various rad27 alleles on growth in presence of 4 mM (0.034% [vol/vol]) MMS. Plating was on MMS-containing YPD medium was as described for Fig. 3B. Plates were incubated at 30°C for 6 days. (B) Synergistic effect of rad27-p and rad51-null on MMS sensitivity. Graph shows survival of yeast strains after different times of exposure to 11.8 mM (0.1% [vol/vol]) MMS in 0.1 M KPO4 buffer (pH 7.0). Wild type (1, ○), rad27-p (2, ▵), rad51-null (3, ▿), and rad27-p rad51-null (4, ◊) yeast strains were exposed to MMS as described in Materials and Methods. Means and standard errors (represented by vertical bars, which sometime overlap with symbols) reflect the findings for three independent measurements.
TABLE 1.
Rates of mutation and interchromosomal recombination in various rad27 mutants and RAD27 wild-type strains
| Genotype | Mutation and interchromosomal recombination rates (107)a of:
|
|||
|---|---|---|---|---|
| lys2-A12 (+1 bp mutations) | lys2-A14 (−1 bp mutations) | CAN1 (forward mutations) | Interchromosomal recombination | |
| Wild type | 0.72 (0.48–1.5) | 2.3 (1.1–3.4) | 3.2 (2.7–5.2) | 1.96 (1.06–2.58) |
| rad27-Δ | 19 (15–29) | 140 (93–196) | 77 (63–153) | 15 (8–95) |
| rad27-n | 52 (25–61) | 193 (92–426) | 123 (90–216) | ND |
| rad27-n,p | 26 (17–46) | 145 (97–302) | 129 (101–279) | 28 (17–49) |
| rad27-p | 1.3 (0.81–2.0) | 4.7 (2.6–12) | 6.9 (5.1–8.0) | 2.3 (1.9–7.7) |
| pol2-4 | 1.7 (0.41–2.8) | 4.7 (1.7–6.6) | 6.0 (3.4–8.7) | ND |
| exo1 | 1.8 (1.3–3.7) | 41 (21–205) | 16 (12–29) | ND |
| rad27-Δ pol2-4 | 300 (190–520) | 350 (210–570) | 359 (285–574) | ND |
| rad27-p pol2-4 | 5.3 (3.4–11) | 8.1 (5.6–9.1) | 16.8 (11–22) | ND |
| rad27-p exo1 | 6.3 (3.8–7.8) | 87 (52–150) | 24 (18–36) | ND |
95% confidence intervals are given in parentheses. ND, not determined.
PCNA binding by Rad27 is not required for normal genetic stability.
Despite complete elimination of both in vitro PCNA binding and the strong intragenic interaction with rad27-n, the rad27-p mutation did not cause genetic instability characteristic of rad27-Δ. rad27-p did not cause statistically significant increases in the rates of +1 and −1 frameshifts in long homonucleotide runs, in the rates of forward mutations inactivating CAN1, or in the rates of interchromosomal recombination, whereas rad27-Δ caused 8- to 61-fold increases in these reporter systems (Table 1).
Because rad27-Δ is nonviable in combination with several defects in DNA replication and/or repair (3, 20, 42, 44; see also our data below), we looked for possible synergistic effects between rad27-Δ or rad27-p and other defects in DNA metabolism. We found that the mutation eliminating the 3′→5′ exonuclease activity of DNA polymerase ɛ (Pol ɛ), pol2-4 (28), amplified the effect of rad27-Δ in three mutation reporter systems. Unlike the case of the rad27-Δ pol2-4 double mutant, the rad27-p pol2-4 had rates of change that were very close to those expected from an additive interaction between these two mutations. Double mutants carrying rad27-Δ and exo1 are nonviable (reference 43 and our data below); therefore, exo1-null strains might be sensitized to subtle defects in the Rad27 function. However, the double mutant rad27-p exo1 was viable (see below), and we observed no significant change in the mutability of these strains compared with the exo1 single mutant (Table 1).
The lack of Rad27 PCNA binding has little effect on MMS sensitivity but amplifies sensitivity in a rad51 mutant.
Unlike a rad27-Δ mutant, rad27-p grew nearly as well as the wild type on 4 mM MMS (Fig. 4A). A slight decrease of MMS resistance of rad27-p compared with wild type was observed when yeast cells were treated with a higher dose of MMS (Fig. 4B). Nevertheless, the resistance of rad27-p to MMS was much greater than that of the rad27-Δ strain (Fig. 4A), which suggests that the Rad27-p protein can make a significant contribution to the repair of alkylation damage.
The rad27-Δ mutation is lethal in combination with an additional mutation in DSB repair genes, such as RAD51 (42, 44). We found that rad27-p rad51-null double mutants were viable (see below). Since null mutations in both rad51 and rad27 genes cause MMS sensitivity, we determined the effect of rad27-p on the MMS sensitivity of a rad51 mutant (Fig. 4B). Even though the effect of rad27-p alone on MMS sensitivity was barely detectable, it amplified dramatically (up to 1,000-fold) the MMS sensitivity of a rad51 strain.
Defect in Rad27 PCNA binding is incompatible with a homozygous or heterozygous pol3-01 mutation in the 3′→5′ exonuclease domain of DNA Pol δ.
We investigated whether rad27-p leads to the same types of incompatibility as rad27-Δ displays with other defects in DNA metabolism. This was done by assessing the requirement for a plasmid expressing wild-type RAD27 to permit cell growth (see Material and Methods). In agreement with earlier studies (20, 42–44), the presence of the mitotically unstable RAD27 plasmid LC80B rescued the growth defect of double mutants that were rad27-Δ and either exo1, rad51, or pol3-01 (Table 2). We note that, in agreement with the findings of Tishkoff et al. (43), we did not observe the loss of the RAD27 plasmid from the rad27-Δ exo1 double mutant. Viable transformants that combine rad27-Δ and exo1-null have been obtained (16) but may have resulted from suppressor mutants.
TABLE 2.
Loss of TRP1 marker of the plasmid containing wild-type RAD27 gene
| Genotype (ploidy) | No. of isolates | Loss of TRP1 marker (%)a | Viability of a double mutant |
|---|---|---|---|
| RAD27 (1n) | 7 | 22–56 | NAb |
| rad27-Δ (1n) | 7 | 11–26 | NA |
| rad27-p (1n) | 9 | 17–56 | NA |
| exo1 rad27-Δ (1n) | 15 | 0 | Nonviable |
| exo1 rad27-p (1n) | 14 | 37–49 | Viable |
| rad51 rad27-Δ (1n) | 4 | 0 | Nonviable |
| rad51 rad27-p (1n) | 6 | 17–22 | Viable |
| pol3-01 rad27-Δ (1n) | 9 | 0–0.06 | Nonviable |
| pol3-01 rad27-p (1n) | 24 | 0–1 | Nonviable |
| pol3-01/POL3 rad27-Δ/RAD27 (2n) | 12 | 29–80 | Viable |
| pol3-01/POL3 rad27-p/RAD27 (2n) | 12 | 33–69 | Viable |
| pol3-01/POL3 rad27-Δ/rad27-Δ (2n) | 15 | 0 | Nonviable |
| pol3-01/POL3 rad27-p/rad27-Δ (2n) | 15 | 0–1 | Nonviable |
Minimal and maximal values of frequencies of the TRP1 loss are shown.
NA, not applicable.
The pol3-01 mutation is weakly dominant in the heterozygote; a sixfold mutator effect was observed in a heterozygous diploid versus a 500-fold mutator activity when homozygous (29). We confirmed this observation with the lys2-A14 reporter system for +1 frameshifts (data not shown), where homozygous pol3-01 caused an approximately 1,000-fold increase in mutation rate, whereas the rate in a heterozygous pol3-01/POL3 diploid was only twofold higher than in homozygous POL3/POL3. The effect of rad27-Δ was profound in that even when pol3-01 was heterozygous in a diploid it required the RAD27 plasmid for viability (Table 2). We note that the wild-type allele POL3 on the ARS-CEN plasmid pBL304 can restore the viability of a rad27-Δ pol3-01 haploid double mutant (reference 20 and unpublished results). The reason that the haploid rad27-Δ pol3-01 double mutant carrying POL3 on a plasmid is viable, whereas the rad27-Δ/rad27-Δ pol3-01/POL3 diploid is nonviable, could be higher expression of Pol δ from the plasmid or an accumulation of several copies of the plasmid containing POL3.
We found that the rad27-p PCNA-binding defect was compatible with either exo1 or rad51, which is in agreement with a subtle change in Rad27 function. However, rad27-p caused inviability of haploid strains that carried a putative defect in the Pol δ 3′→5′ exonuclease (pol3-01). Surprisingly, rad27-p was incompatible even with a heterozygous pol3-01/POL3 defect in a diploid (Table 2). Because the effects of either rad27-p or pol3-01/POL3 alone are very modest, a toxic intermediate might be produced via their interaction.
In most of the experiments addressing RAD27 plasmid loss in incompatible combinations, we were unable to detect any colonies lacking the RAD27 plasmid even after an extended 6- to 7-day incubation. However, in the case of heterozygous pol3-01/POL3+ diploids carrying the rad27-p (but not the rad27-Δ) defect, we observed tiny colonies that appeared after prolonged incubation. When streaked onto fresh medium, these colonies formed rapidly growing progeny which had lost the TRP1 marker of the RAD27 plasmid. In 26 of 32 cases, these rapidly growing isolates also lost the pol3-01 allele, as determined by PCR analyses. Those losses probably occurred either by mitotic recombination or by chromosome loss. The other six might have been due to a secondary mutation that inactivated the pol3-01 allele.
DISCUSSION
PCNA can interact with a variety of proteins, including DNA Pol δ, Pol ɛ, RF-C, Fen1, XPG, DNA ligase I, DNA-(cytosine-5) methyltransferase, p21 (Waf1/Cip1), and p57 (Kip2) (17, 51, 53), indicating that it may have multiple roles in DNA replication, repair, and cell cycle regulation. Several attempts to uncover the in vivo significance of such interactions have used modified PCNA (7, 26), but the likelihood of pleiotropic effects of a PCNA mutation has made it difficult to attribute biological observations unambiguously to individual protein-protein interactions. Many PCNA-binding proteins, including Fen1, use a common motif for interacting with PCNA. Thus, specific mutation of the PCNA binding partner, rather than PCNA itself, may be the best strategy for revealing the biological roles of PCNA interactions with individual proteins.
Hosfield et al. (13) have combined human PCNA and archeal Fen1 structural data and showed that a DNA-PCNA-Fen1 ternary complex is conformationally plausible and could serve to aid flap removal. Because PCNA is a DNA-binding protein that encircles DNA, PCNA binding by Fen1 could serve to bring the nuclease to its substrate. However, Fen1 possesses intrinsic DNA-binding and nuclease activities in vitro in the absence of PCNA. PCNA enhances the nuclease activity of Fen1 in enzymatic assays by using synthetic oligonucleotide substrates. The relevance of these studies to in vivo functions must be considered because the structure of Fen1 substrates in vivo may differ from artificial substrates examined in vitro and because the in vivo reaction environment may have unique features due to a complex of associated proteins. Therefore, we investigated the in vivo effects of the mutation rad27-p, which eliminates PCNA-binding in the yeast Fen1 homolog, Rad27. We tested the effects of this mutation alone and in combination with the nuclease mutation rad27-n. Finally, in order to uncover effects that might be obscured by redundant activities in DNA metabolism, we examined the PCNA binding defect in genetic backgrounds that are known to cause synergistic effects when combined with the rad27-Δ null allele.
An in vivo interaction between Rad27 and PCNA during substrate cleavage.
We observed a dominant-negative effect of the nuclease-defective rad27-n on cell growth and found that this effect required the ability to bind PCNA. Intragenic suppression of the dominant-negative effect of rad27-n by the PCNA binding mutation confirms the functional significance of PCNA binding in cell growth and suggests a coordination of PCNA binding and nuclease activities in Rad27. The distinct phenotypes elicited by the catalytically inactive rad27-n and rad27-n,p show that enhancement of the catalytic rate cannot be the sole function of PCNA binding in Rad27. We suggest that PCNA binding also serves to precisely position Rad27 within a macromolecular assembly. In rad27-n,p cells, both nuclease activity and PCNA-dependent nuclease positioning are absent, so that DNA metabolic processes that utilize Rad27 must rely on secondary enzymes with partially redundant functions. However, in the case of rad27-n, the catalytic defect in conjunction with normal PCNA-dependent nuclease targeting creates an intolerable situation. We propose that the interaction of Rad27-n with PCNA allows the mutant protein to occupy its normal position within multiprotein complexes and thereby hinders access to the incompletely processed DNA. The lack of a dominant-negative effect of the double mutation rad27-n,p suggests that PCNA binding activity is necessary to integrate Rad27 within multiprotein complexes with sufficient stability to exclude redundant enzymes.
It is important to note that excessive titration of PCNA was not the cause of Rad27-n cytotoxicity. A normal level of expression of Rad27-n from the single-copy genomic locus was highly toxic (Fig. 3A), whereas high-level multicopy plasmid expression of wild-type Rad27, which has the same PCNA binding activity as Rad27-n on a molar basis, was well tolerated (Fig. 3B). Only when PCNA binding activity was coupled to the Rad27 nuclease defect was severe cytotoxicity manifested.
Elimination of Rad27-PCNA binding has a minor effect on genomic stability.
In contrast to rad27-Δ, rad27-p did not significantly increase genetic instability (Table 1). However, all rad27-p strains showed a small (up to twofold) increase in mutation or recombination compared with isogenic RAD27 strains. Even if these increases reflect a real tendency, it can be concluded that prevention of these types of genomic instability by Rad27 is nearly normal in the absence of PCNA binding.
We found that rad27-Δ exhibited a strong synergism with pol2-4, a defect in the DNA Pol ɛ proofreading 3′→5′ exonuclease (Table 1). This synergism could be explained either through participation of Rad27 nuclease in postreplication mismatch repair as discussed earlier (15, 43) or through an increased likelihood of frameshift intermediates (20). Regardless of the mechanism of this synergism, rad27-p caused no statistically significant effect or at most a very weak effect (lys2-A12) in this sensitized background. Another genetic background that can be sensitive to subtle rad27 defects is the null mutant in the 5′→3′ exonuclease gene EXO1, which is incompatible with rad27-Δ (reference 43 and Table 2). The exo1-null allele is a mutator at the CAN1 reporter gene and has a strong mutator effect on long homonucleotide runs (references 43 and 45; Table 1), possibly due to participation of Exo1 in mismatch repair. However, rad27-p exo1-null double mutants were viable (Table 2) and exhibited no significant increase in mutation (Table 1).
Role of PCNA binding for Fen1/Rad27 repair function.
Several in vitro studies showed that Fen1/Rad27 is involved in long-patch BER (18, 19). The sensitivity of rad27-Δ yeast to MMS alkylation damage is consistent with a long-patch BER defect, especially because the alternative short-patch BER pathway dependent on Pol β may play only a secondary role in this species (21). MMS sensitivity in yeast cells is also caused by defects in RAD51 and other genes in the RAD52 epistasis group that control recombinational repair of DNA breaks (34). Each of the DSB repair genes is required for the viability of the rad27-Δ mutant (42, 43), which implies that lesions occurring in rad27-Δ are repaired via the recombinational pathway.
Rad27/Fen1-mediated excision activity is facilitated by PCNA during BER in vitro (9), which suggests the importance of this interaction for in vivo repair. A deficiency in Rad27 PCNA binding had little effect on MMS sensitivity, but dramatically amplified the MMS sensitivity of a rad51 null mutant (Fig. 4B). This can be explained by the overlapping roles of BER and DSB proteins in MMS lesion repair. Rad27-p may reduce long-patch BER efficiency, thus placing a greater burden on the alternative DSB repair pathway. Further, because apurinic or apyrimidinic (AP) endonuclease-dependent incision precedes excision by Rad27 in BER, a bottleneck at the excision step can cause an accumulation of breaks that require recombinational repair functions of the DSB pathway (55).
Role of Rad27/Fen1-PCNA binding in DNA replication.
We combined rad27-p with three different mutations which are known to be lethal in combination with rad27-Δ, namely, exo1-null, rad51-null, and pol3-01. rad27-p was lethal only in combination with pol3-01. Strong negative interaction appeared to be specific for pol3-01, because another pol3 allele, the temperature-sensitive pol3-t mutation, was compatible with rad27-Δ and rad27-p defects (reference 20 and unpublished results). Although DNA Pol δ can be involved in both replication and repair, we suggest that the incompatibility of pol3-01 and rad27-p alleles reflects a DNA replication problem. The lack of requirement for the repair and recombination genes EXO1 and RAD51 in the normal growth of rad27-p suggests that any potential DNA repair defect caused by rad27-p was insufficient to produce catastrophic consequences in the absence of exogenous damage, even in repair-compromised cells.
The conserved amino acid changes used to generate the pol3-01 mutation and the genetic properties of this allele indicate that it inactivates a 3′→5′ proofreading exonuclease activity (28–30). The 3′→5′ exonuclease activity associated with DNA Pol δ could not be detected in DNA Pol δ prepared from pol3-01 mutant cells in the same assay that was used to measure the exonuclease activity of wild-type and exonuclease mutant (pol2-4) forms of DNA Pol ɛ (28, 41a). Elevated mutation rates are observed in pol3-01 mutants in agreement with the putative proofreading defect. Was the rad27-p pol3-01 incompatibility due to an intolerably high mutation rate? The mutator mutations pms1, msh2, or exo1, which inactivate mismatch repair, are lethal when combined with pol3-01 mutator in a haploid, but the homozygous diploid strains are viable even though they exhibit extremely high mutation rates (29, 45). This contrasts with the nonviability of pol3-01/pol3-01 rad27-p/rad27-Δ and pol3-01/POL3 rad27-p/rad27-Δ diploids. Low mutation rates in both rad27-p (Table 1) and in pol3-01/POL3 strains (reference 29; see also Results), make it unlikely that the inviability of pol3-01/POL3 rad27-p/rad27-Δ diploids was due to a catastrophic mutation rate. We suggest instead that this nonviability was due to the formation of toxic replication intermediates during lagging-strand synthesis. This hypothesis accounts for the apparent dominance of the pol3-01 allele in pol3-01/POL3 heterozygotes in producing synthetic lethality with rad27-p. Toxic intermediates might be formed at replication forks that use the mutant Pol δ.
A model for interactive effects of DNA Pol δ and Rad27/Fen1 in replication.
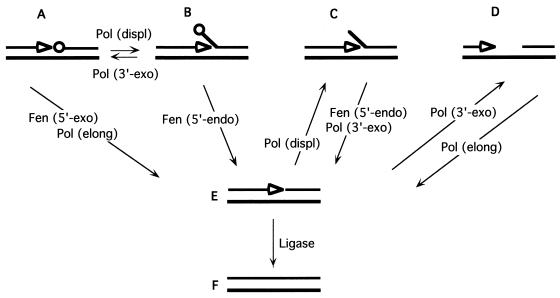
To explain the severe consequences of combining pol3-01 and rad27-p alleles, we propose a model based on the interaction of 5′- and 3′-nuclease defects in lagging-strand synthesis at the replication fork (Fig. 5). 5′-Terminal ribonucleotide removal during Okazaki fragment processing may depend exclusively on Rad27 action (see above); other 5′→3′ exonucleases, such as Exo1, are not known to substitute for Rad27 in this process. DNA Pol δ can fill the gap up to the 5′-terminal ribonucleotide to generate a substrate for Rad27 5′→3′ exonuclease activity (Fig. 5A) or can perform additional strand-displacing DNA synthesis to generate a substrate for Rad27 5′-flap endonuclease activity (Fig. 5B). These replication intermediates must be processed to yield nicked structures (Fig. 5E) that must be ligated (Fig. 5F). The disruption of Rad27-PCNA interaction by the Rad27-p mutation may alter the normal dynamics of processive synthesis at the replication fork. If the 3′→5′ exonuclease activity of Pol δ can degrade the nascent strand under these conditions, then the pol3-01 polymerase mutation can be expected to block the conversion of structure B to structure A. When combined with pol3-01, the rad27-p mutation is lethal. We propose that PCNA binding by Rad27 may be more important for 5′-endonuclease activity during replication (B→E step) than for 5′-exonuclease activity (A→E step), as suggested by in vitro studies with oligonucleotide substrates in which PCNA appears to stimulate Fen1/Rad27 5′-flap endonuclease activity more than 5′→3′ nick exonuclease activity (23, 54). Taking this proposal, the model accounts for the surprising severity of the pol3-01 rad27-p double mutation: conversion of B to A would be blocked as a result of the pol3-01 mutation, and conversion of B to E would be impaired if 5′-endonuclease activity depends on PCNA association. Thus, while either mutation alone might be tolerated, in combination they would produce an unacceptable accumulation of replication intermediate B (unremoved flaps). Furthermore, ligatable intermediate E can undergo an alternative reaction via strand-displacing synthesis to generate structure C, which would also accumulate in the pol3-01 rad27-p double mutant. Unprocessed flap structures such as B and C could result in lethal chromosomal aberrations if allowed to persist until mitosis.
FIG. 5.
Intermediate DNA structures and reactions at the border between two Okazaki fragments. Template strand (bottom) and nascent lagging strand (top) are shown with the 3′ growing end (open arrowheads) and the last unremoved ribonucleotide (open circles). Enzymatic reactions leading to interconversion of various intermediates include the following: Fen1 5′→3′ exonuclease, Fen (5′-exo); Fen1 5′-flap endonuclease, Fen (5′-endo); DNA polymerase strand displacement, Pol (displ); DNA polymerase elongation, Pol (elong); DNA polymerase 3′→5′ exonuclease, Pol (3′-exo); and DNA ligase (Ligase).
Biologically important Rad27/Fen1 polymorphisms and relevance to natural populations.
This study has identified changes in Rad27/Fen1 properties that affect its biological function in DNA replication, repair, and mutation avoidance. Natural polymorphisms that cause subtle alterations of such properties could accumulate in a population, but our results show that combining subtle defects can lead to severe functional consequences. The effects of subtle but potentially deleterious polymorphisms in human Fen1 could be assessed rapidly by characterizing yeast strains with mutations in Rad27 at homologous positions in the sensitized genetic backgrounds identified in the current study. For example, new alleles with slight deficiencies in nuclease activity could be identified by their strong inhibition of yeast growth when overexpressed in rad51 strains. These alleles represent a class of polymorphisms that could be dangerous even when heterozygous. Similarly, assays could be developed that use strains sensitized by rad51 and pol3-01 to identify natural polymorphisms that reduce PCNA binding of Fen1/Rad27. It would be difficult to screen for this genotype without exploiting the effect of combining alleles, but such polymorphisms could be readily identified by their increased MMS sensitivity in a rad51 strain. Although pol3-01 is lethal in combination with rad27-p, we hypothesize that there are rad27 mutants with partial PCNA binding that can survive in a pol3-01 background. Such double mutants might accumulate unprocessed flaps (Fig. 5 and related discussion). As with rad27-Δ, this could lead to genetic instability and could be used for detection of polymorphisms altering PCNA binding. In particular, long unremoved flaps could increase the likelihood of expansion in trinucleotide repeats or other at-risk motifs where flaps might form Fen1-resistant secondary structures (11).
Our results suggest that natural, well-tolerated polymorphisms might have disastrous consequences when combined, which could have implications for genetically based diseases, differential responses to environmental stress, and pharmacogenetics. Understanding the potential effect of allelic variants in combination is a complex problem. Human genetic linkage studies are unlikely to reveal important interactions between alleles that have no effect when present alone. Instead, insight into this problem can come from experimental organisms in which specific mutations can be created that represent entire classes of polymorphisms. Studies in model systems can help to direct epidemiological studies to specific pairs of genes, such as Fen1 and Pol δ, where severe interactions between otherwise neutral polymorphisms are most likely to occur.
ACKNOWLEDGMENTS
We thank M. Budd and J. Campbell for the plasmid YEP24-3a; A. Sugino for providing unpublished data, P. Bradley for help in experiments; and J. Drake, M. Longley, and R. Schaper for discussions and advice on the manuscript.
This work was supported by NIH grant CA71630 (to M.S.P.). K.S.L. is on leave from the Department of Genetics, St. Petersburg State University, St. Petersburg, Russia.
REFERENCES
- 1.Bae S H, Choi E, Lee K H, Park J S, Lee S H, Seo Y S. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J Biol Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 2.Batzer M A, Alegria-Hartman M, Deininger P L. A consensus Alu repeat probe for physical mapping. Genet Anal Tech Appl. 1994;11:34–38. doi: 10.1016/1050-3862(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 3.Budd M E, Campbell J L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang L S, Ian H I, Koh T W, Ng H H, Xu G, Li B F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 5.Clark A B, Cook M E, Tran H T, Gordenin D A, Resnick M A, Kunkel T A. Functional analysis of human MutSalpha and MutSbeta complexes in yeast. Nucleic Acids Res. 1999;27:736–742. doi: 10.1093/nar/27.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMott M S, Shen B, Park M S, Bambara R A, Zigman S. Human RAD2 homolog 5′- to 3′-exo/endonuclease can efficiently excise a displaced DNA fragment containing a 5′-terminal abasic lesion by endonuclease activity. J Biol Chem. 1996;271:30068–30076. doi: 10.1074/jbc.271.47.30068. [DOI] [PubMed] [Google Scholar]
- 7.Eissenberg J C, Ayyagari R, Gomes X V, Burgers P M. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase delta and DNA polymerase epsilon. Mol Cell Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 9.Gary R, Kim K, Cornelius H L, Park M S, Matsumoto Y. PCNA facilitates excision in long-patch base-excision repair. J Biol Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 10.Gary R, Ludwig D L, Cornelius H L, MacInnes M A, Park M S. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 11.Gordenin D A, Kunkel T A, Resnick M A. Repeat expansion—all in a flap? Nat Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 12.Harrington J J, Lieber M R. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 13.Hosfield D J, Mol C D, Shen B, Tainer J A. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 14.Huang L, Rumbaugh J A, Murante R S, Lin R J, Rust L, Bambara R A. Role of calf RTH-1 nuclease in removal of 5′-ribonucleotides during Okazaki fragment processing. Biochemistry. 1996;35:9266–9277. doi: 10.1021/bi9603074. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R E, Kovvali G K, Prakash L, Prakash S. Role of yeast Rth1 nuclease and its homologs in mutation avoidance, DNA repair, and DNA replication. Curr Genet. 1998;34:21–29. doi: 10.1007/s002940050362. [DOI] [PubMed] [Google Scholar]
- 17.Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- 18.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 19.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leem S H, Ropp P A, Sugino A. The yeast Saccharomyces cerevisiae DNA polymerase IV: possible involvement in double strand break DNA repair. Nucleic Acids Res. 1994;22:3011–3017. doi: 10.1093/nar/22.15.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin D S, Bai W, Yao N, O’Donnell M, Tomkinson A E. An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc Natl Acad Sci USA. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Li J, Harrington J, Lieber M R, Burgers P M. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem. 1995;270:22109–22112. doi: 10.1074/jbc.270.38.22109. [DOI] [PubMed] [Google Scholar]
- 24.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 25.Lobachev K S, Shor B M, Tran H T, Taylor W, Keen J D, Resnick M A, Gordenin D A. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAlear M A, Howell E A, Espenshade K K, Holm C. Proliferating cell nuclear antigen (pol30) mutations suppress cdc44 mutations and identify potential regions of interaction between the two encoded proteins. Mol Cell Biol. 1994;14:4390–4397. doi: 10.1128/mcb.14.7.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrill B J, Holm C. The RAD52 recombinational repair pathway is essential in pol30 (PCNA) mutants that accumulate small single-stranded DNA fragments during DNA synthesis. Genetics. 1998;148:611–624. doi: 10.1093/genetics/148.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison A, Bell J B, Kunkel T A, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′→5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison A, Johnston A L, Johnston L H, Sugino A. Pathway correcting DNA replication errors in S. cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison A, Sugino A. The 3′-5′ exonucleases of both DNA polymerase δ and ɛ participate in correcting errors of DNA replication in S. cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 31.Nolan J P, Shen B, Park M S, Sklar L A. Kinetic analysis of human flap endonuclease-1 by flow cytometry. Biochemistry. 1996;35:11668–11676. doi: 10.1021/bi952840+. [DOI] [PubMed] [Google Scholar]
- 32.Price A, Lindahl T. Enzymatic release of 5′-terminal deoxyribose phosphate residues from damaged DNA in human cells. Biochemistry. 1991;30:8631–8637. doi: 10.1021/bi00099a020. [DOI] [PubMed] [Google Scholar]
- 33.Reagan M S, Pittenger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resnick M A, Bennett C, Perkins E, Porter G, Priebe S D. Double-strand breaks and recombinational repair: the role of processing, signalling and DNA homology. In: Wheals A E, Rose A H, Harrison J S, editors. The yeasts. Vol. 6. New York, N.Y: Academic Press, Inc.; 1995. pp. 357–410. [Google Scholar]
- 35.Robins P, Pappin D J, Wood R D, Lindahl T. Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J Biol Chem. 1994;269:28535–28538. [PubMed] [Google Scholar]
- 36.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 37.Scherly D, Nouspikel T, Corlet J, Ucla C, Bairoch A, Clarkson S G. Complementation of the DNA repair defect in xeroderma pigmentosum group G cells by a human cDNA related to yeast RAD2. Nature. 1993;363:182–185. doi: 10.1038/363182a0. [DOI] [PubMed] [Google Scholar]
- 38.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 39.Shen B, Nolan J P, Sklar L A, Park M S. Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J Biol Chem. 1996;271:9173–9176. doi: 10.1074/jbc.271.16.9173. [DOI] [PubMed] [Google Scholar]
- 40.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 41.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 41a.Sugino, A. Personal communication.
- 42.Symington L S. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 1998;26:5589–5595. doi: 10.1093/nar/26.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 45.Tran H T, Gordenin D A, Resnick M A. The 3′→5′ exonucleases of DNA polymerase δ and ɛ and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turchi J J, Huang L, Murante R S, Kim Y, Bambara R A. Enzymatic completion of mammalian lagging-strand DNA replication. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallen E A, Cross F R. Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol Cell Biol. 1995;15:4291–4302. doi: 10.1128/mcb.15.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 50.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 51.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 52.Warbrick E, Lane D P, Glover D M, Cox L S. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe H, Pan Z Q, Schreiber-Agus N, DePinho R A, Hurwitz J, Xiong Y. Suppression of cell transformation by the cyclin-dependent kinase inhibitor p57KIP2 requires binding to proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 1998;95:1392–1397. doi: 10.1073/pnas.95.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X, Li J, Li X, Hsieh C L, Burgers P M, Lieber M R. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–2043. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X, Wang Z. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 1999;27:956–962. doi: 10.1093/nar/27.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]