Abstract
Various complexes that contain the core subunits of RNA polymerase II associated with different transcription factors have been isolated from eukaryotes; their precise molecular constitution depends on the purification procedure. We estimated the numbers of various components of such complexes in an HeLa cell by quantitative immunoblotting. The cells were lysed with saponin in a physiological buffer; ∼140,000 unengaged polymerases (mainly of form IIA) were released. Only ∼4,000 of these soluble molecules sedimented in glycerol gradients as holoenzyme-sized complexes. About 180,000 molecules of polymerases (∼110,000 molecules of form IIO) and 10,000 to 30,000 molecules of each of TFIIB, TFIIEα, TFIIEβ, TFIIF-RAP74, TFIIF-RAP30, and TFIIH-MAT1 remained tightly associated with the nuclear substructure. Most proteins and run-on activity were retained when ∼50% of the chromatin was detached with a nuclease, but ∼45,000 molecules of bound TATA binding protein (TBP) were detached. Similar results were obtained after cross-linking living cells with formaldehyde. The results provide little support for the existence of a large pool of soluble holoenzyme; they are consistent with TBP-promoter complexes in nuclease-sensitive chromatin being assembled into preinitiation complexes attached to the underlying structure.
Much of our thinking about eukaryotic RNA polymerases (Pols) stems from seminal work on bacteria, where purification led to the isolation of a core enzyme. This core initiates poorly at promoters, and association of an additional factor gives a holoenzyme that initiates more efficiently (6, 7). An analogous approach led to the isolation of a multisubunit eukaryotic Pol, Pol II, that was responsible for the transcription of most genes (69). Again, additional factors known as the general transcription factors (GTFs), which include TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, promoted specific initiation by the core (reviewed in references 47 and 53). Subsequent work showed that a fully functional preinitiation complex could be assembled in vitro at promoters by the successive addition of TFIID (or TATA binding protein [TBP]), TFIIB, Pol II-TFIIF, and TFIIE-TFIIH. Conversion of the preinitiation complex to an elongating one is probably accompanied by the phosphorylation of the C-terminal domain of the largest (catalytic) subunit in the core (13). (Hypo- and hyperphosphorylated forms of the Pol are known as forms IIA and IIO, respectively [13]).
Recently, various types of preformed holoenzyme have been isolated in eukaryotes, which suggests that the preinitiation complex might be formed in one or a few steps (8, 36). For example, a large yeast complex contains a core and mediator, which contains Srbs and promotes activator-dependent transcription (17, 33, 35, 39, 60). These Srbs play an essential role in vivo (16, 25, 61). Many other holoenzymes with molecular constitutions that depend on the purification procedure have now been isolated; some contain TFIIB, TFIIF, and TFIIH; others contain only TFIIF; and still others lack the GTFs or Srbs (34, 35, 38, 63). Therefore, it is not yet clear whether different isolates reflect the purification of distinct complexes or the fragmentation of one larger complex during isolation.
The situation is equally complex in mammalian cells, where different holoenzyme preparations contain some or all of the GTFs, Srbs, DNA-remodelling complexes, DNA repair proteins, and splicing and polyadenylation factors (9–11, 40, 42, 45, 48, 49, 55). Two interrelated factors further complicate the analysis. First, Pol II activity is found in both soluble and insoluble cellular fractions (3, 66), but engaged Pols are found in the latter fraction (29). Moreover, inactive Pols are widely dispersed but engaged Pols are concentrated in several thousand discrete sites (diameters of 40 to 80 nm) that are tightly associated with the underlying structure (20, 27, 31). Second, Pols are often extracted with hypo- and/or hypertonic buffers. Hypotonic conditions are used to isolate nuclei as a first step in the procedure; such conditions are used because breaking even a few nuclei during isolation releases long chromatin strands that tend to aggregate into an unworkable gel in more physiological buffers (12). Hypertonic conditions are used because they extract more protein. Therefore, it is possible that much of the variation seen results from differential extraction and/or aggregation during purification.
A further complicating factor concerns terminology. The bacterial holoenzyme, but not the core, can initiate specifically at promoters. However, many complexes described above cannot initiate specifically unless supplemented with additional factors; therefore, it has been argued that the term “holoenzyme” should be reserved for complexes for which specific initiation in vitro has been demonstrated (10). Since the looser definition has such a wide currency, we use the term “holoenzyme” to describe any complex containing the largest subunit in the core associated with Srbs and/or some of the GTFs.
Given the above, we wished to determine the numbers of molecules of the Pol and its associated factors in isolates prepared under conditions as close to physiological conditions as possible. (Estimates of numbers in the yeast complex isolated by various different procedures have been collated by Lee and Young [37]). Therefore, we developed a simple and rapid procedure for cell fractionation and determined the numbers of protein molecules in the different fractions by quantitative immunoblotting. Cells are lysed with the detergent saponin in a physiological buffer (30); the detergent is sufficiently gentle that little chromatin is released, and the buffer contains roughly the concentrations of sodium, potassium, and magnesium ions found in the cell. Importantly, essentially all magnesium ions are bound to the natural chelating agent, ATP, so that endogenous nucleases remain relatively inactive. Lysis releases some Pol II without affecting run-on activity. Engaged Pols remain tightly associated with the nuclear substructure; these are still able to run on along endogenous templates. About 140,000 Pol molecules per cell were soluble, and about 180,000 were tightly bound. Only ∼4,000 Pol molecules in the soluble fraction (i.e., 3%) sedimented in glycerol gradients as Pol II complexes with the size of the holoenzyme. Therefore, our results provide little evidence for the existence of a large soluble pool of holoenzyme. If such a complex exists, it is probably found with engaged Pols associated with the nuclear substructure.
MATERIALS AND METHODS
Cell fractionation.
Suspension cultures of HeLa cells were grown in minimal essential medium (S-MEM) (Gibco) supplemented with 5% fetal calf serum (Gibco). They were usually grown (for ∼20 h) in [methyl-3H]thymidine (9.25 kBq/ml; 2.92 TBq/mmol [Amersham]) to label DNA uniformly; this enabled (i) corrections to be made if cell loadings varied, and (ii) determination of the amount of chromatin detached by nucleases (both calculated from the 3H counts per minute remaining [29]). All buffers used during and after lysis were ice-cold, unless stated otherwise. The cells were washed twice with physiological buffer (PB) (100 mM potassium acetate, 30 mM KCl, 10 mM Na2HPO4, 1 mM MgCl2, 1 mM disodium ATP, 1 mM dithiothreitol), resuspended (5 × 106/ml) in PBI (PB supplemented with a cocktail of protease inhibitors [complete, EDTA free] from Boehringer), and lysed for 5 min in 1% saponin (Sigma S-4521). Permeabilized cells were centrifuged (500 to 1,000 × g for 3 min at 4°C), and the supernatant was collected (saponin supernatant). The pellet was washed in PBI and resuspended in PBI at 5 × 106 cells/ml to give the saponin pellet. For nuclease digestion (see Fig. 3), 106 cells/ml were incubated (30 to 34°C for 20 min) with or without HaeIII (New England Biolabs). (Less chromatin is detached when higher cell concentrations are used [see Fig. 1]). After treatment, the cells were chilled on ice and centrifuged (1,000 × g for 3 min), and the supernatant was collected (HaeIII supernatant). After being washed in PB, the pellet was resuspended in the original volume of PB (see Fig. 1), or 1/10 volume of PB (see Fig. 3) to give the HaeIII pellet. Permeabilized cells were also sonicated (Sanyo Soniprep 150 with microprobe at level 10 for 10 to 20 s) until >99% nuclei were disrupted as judged by staining with trypan blue (Sigma); they were then centrifuged (10,000 × g for 20 min at 4°C) to yield sonicated supernatant and sonicated pellet. Whole-cell extract was prepared by a modification (49) of the method of Manley et al. (41), and nuclear extract was prepared by the method of Dignam et al. (14). For the experiments in Fig. 5E and F, the saponin supernatant was dialyzed against 10% glycerol in 10 mM HEPES-NaOH (pH 7.9)–50 mM NaCl–0.1 mM EDTA–0.1 mM dithiothreitol and clarified by centrifugation (10,000 × g for 20 min at 4°C).
FIG. 3.
Retention of proteins after detaching chromatin. (A) Immunoblots. Cells were grown in [3H]thymidine for 24 h to label DNA, lysed, incubated with or without HaeIII, and centrifuged; the proteins in the pellet were resolved in gels and blotted; and the filters were probed with antibodies directed against the proteins indicated. The percentage of [3H]DNA remaining in each fraction is shown at the top. Lanes 1 to 4: 1/8×, 1/4×, 1/2×, and 1× loading of lysed cells; lanes 5 to 8: 1× loading of lysed cells treated with 0, 0.1, 0.5, or 2.5 U of HaeIII per ml. (B) DNA and proteins remaining in pellets (average of three experiments). The intensities of bands like those in panel A were measured and are expressed as percentages of those seen in cells treated without HaeIII (lanes 1 to 4 in panel A). Histone levels were determined from gels stained with Coomassie blue. Levels of nascent RNA were determined as follows: cells were allowed to make RNA in [32P]UTP, treated with HaeIII or left untreated, and pelleted, and the amount of [32P]RNA remaining was expressed relative to that in untreated controls. The amount of [3H]DNA remaining is also shown.
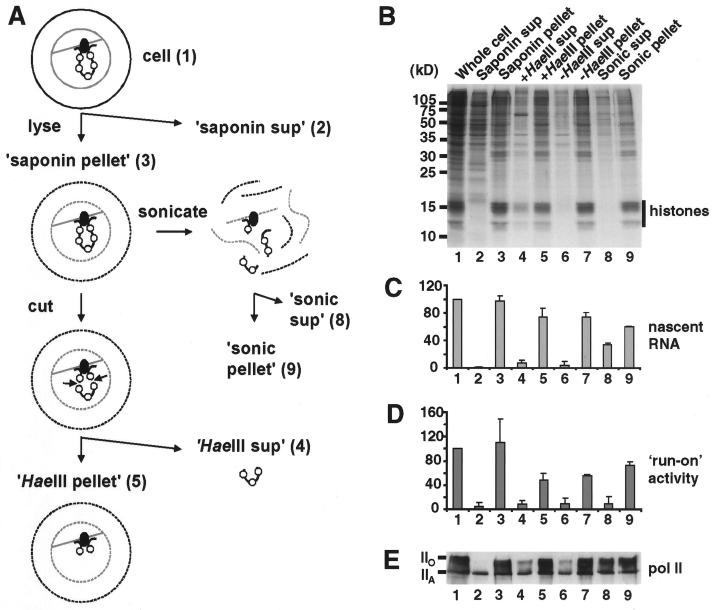
FIG. 1.
Distribution of RNA Pol II and nascent RNA in cell fractions. (A) Schematic representation of fraction preparation. HeLa cells (structure 1) were permeabilized with saponin in a physiological buffer and centrifuged to yield the saponin supernatant (structure 2) and pellet (structure 3). After treatment with HaeIII to cut and detach chromatin from the underlying structure, recentrifugation yielded a HaeIII supernatant (structure 4) and pellet (structure 5). Sonication and recentrifugation also gave a sonic supernatant (structure 8) and pellet (structure 9). In all cases, pellets were resuspended in the original volume of physiological buffer. The numbers refer to samples applied to the corresponding lanes in panels B to E. (B) Protein content (photograph of a Coomassie blue-stained gel). Proteins in the various fractions were resolved on an SDS–15% polyacrylamide gel. (C) Nascent RNA. Cells were grown in [3H]uridine for 2.5 min to label nascent RNA before fractionation, and the amount (average of three experiments with standard deviation) of [3H]RNA in each fraction is expressed relative to that in whole cells. (D) Polymerizing activity. After the addition of Sarkosyl, engaged Pols were allowed to incorporate [32P]UTP, and the amount (average of three experiments with standard deviation) of [32P]RNA made is expressed relative to that made by whole cells. (E) Content of RNA Pol II (photograph of an immunoblot). Proteins were separated on a 6% polyacrylamide gel, blotted, and the blot was probed with an antibody that recognized both forms of the largest subunit of Pol II.
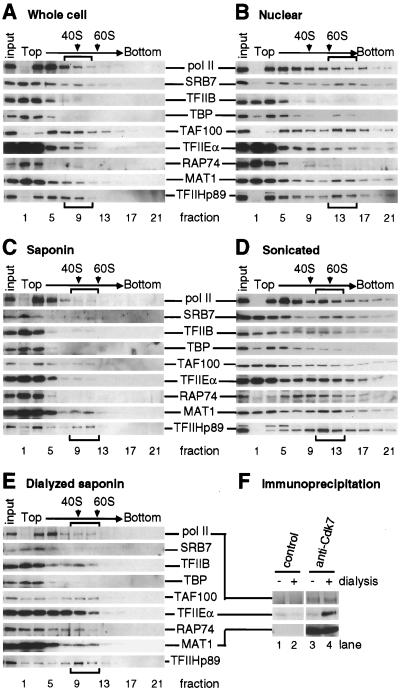
FIG. 5.
Protein complexes detected in glycerol gradients (A to E) and by immunoprecipitation (F). Extracts were prepared in different ways; in panels E and F, they were also dialyzed against a hypotonic buffer and clarified by centrifugation. For analysis in sucrose gradients (A to E), extracts were centrifuged, fractions were collected, and the protein contents of equal volumes of different fractions were determined by immunoblotting with antibodies directed against the various proteins indicated in the center. The content of 1/10 the input applied to the gradient is shown on the left of each blot. Arrows and brackets show positions of 40S and 60S ribosomal subunits and large complexes containing SRB7, respectively. For immunoprecipitation (F), undialyzed or dialyzed extracts were incubated with different antibodies bound to beads, the beads were pelleted, and the protein content in the pellet was determined by immunoblotting as above. (A) Whole-cell (Manley) extract. (B) Nuclear (Dignam) extract. (C) Saponin supernatant (Fig. 1, structure 2). A total of 3, 7, 2, <1, 20, 3, <1, 3, and 32% of Pol II, SRB7, TFIIB, TBP, TAF100, TFIIEα, RAP74, MAT1, and p89, respectively, were found in fractions 9 and 11. (D) Sonicated supernatant (Fig. 1, structure 8). (E) As in panel C but dialyzed against a hypotonic buffer and clarified by centrifugation. A total of 16, 7, 14, 3, 44, 20, 19, 3, and 55% of Pol II, SRB7, TFIIB, TBP, TAF100, TFIIEα, RAP74, MAT1, and p89, respectively, were found in fractions 9 and 11. (F) Proteins in undialyzed (−) and dialyzed (+) saponin extracts (as in panels C and E) were immunoprecipitated with a control mouse IgG (lanes 1 and 2) or an anti-Cdk7 antibody (lanes 3 and 4), and proteins in the pellet were detected by immunoblotting.
Retention of nascent transcripts and transcriptional activity.
Nascent transcripts were labelled in vivo by growing cells (∼5 × 107/ml) for 2.5 min in [3H]uridine (3.7 MBq/ml; 1.55 TBq/mmol [Amersham]), incorporation was stopped by addition of 100 volumes of ice-cold PB, and the cells were washed, resuspended in PB, and lysed. After cell fractionation, the amount of [3H]RNA was measured by scintillation counting (29). Nascent transcripts were also extended in vitro (see Fig. 2C); permeabilized cells were incubated (5 min at 33°C) in PB supplemented with 100 μM ATP, 100 μM CTP, 100 μM GTP, 5 μM UTP (all nucleotides from Pharmacia), [32P]UTP (0.37 MBq/ml; 111 TBq/mmol [Amersham]), 305 μM MgCl2, and 10 U of RNasin (Boehringer) per ml. After addition of 5 volumes of ice-cold PB, the cells were washed twice, resuspended in PB, and treated with HaeIII, and the amount of [32P]RNA remaining was measured by scintillation counting (29).
FIG. 2.
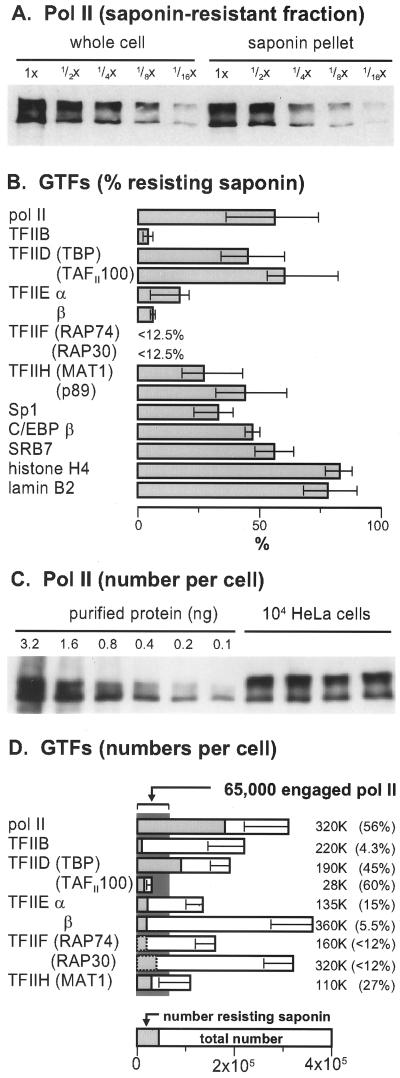
Number of molecules of the largest subunit of Pol II in a cell. (A) Fraction of Pol II in the saponin pellet. Various loadings of proteins from whole cells and the saponin pellet were resolved on a gel and blotted, and the blot was probed with the Pol II antibody. The two bands seen in the saponin pellet have roughly half the intensity of those seen in whole cells. (B) Percentage of various GTFs resisting extraction with saponin (determined from at least three experiments like that in panel A; error bars show the range). (C) Absolute numbers of Pol II molecules obtained by comparing band intensities given by four different preparations of 104 whole cells with those given by dilutions of known amounts of pure Pol II. (D) Absolute numbers of molecules of various proteins (obtained from at least three experiments like that in panel C; error bars show standard deviation). The total numbers and fraction resistant to saponin are shown.
The amount of run-on transcription remaining during cell fractionation (see Fig. 1E) was determined by adding an equal volume of 0.5% Sarkosyl (N-lauroylsarcosine, sodium salt [Sigma]) to a cell fraction and then determining the incorporation of [32P]UTP into acid-insoluble RNA as described above.
Immunoblotting.
All immunoblotting procedures were conducted at room temperature unless stated otherwise. Samples were mixed with 2× sodium dodecyl sulfate (SDS)-gel-loading buffer (54), and the proteins were denatured (95°C for 10 min), resolved on SDS-polyacrylamide gels, and blotted onto nitrocellulose filters (Schleicher & Schuell). The filters were washed in TBST (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20), blocked (30 min) with TBSTM (TBST containing 5% skim milk), incubated (1 to 3 h) with primary antibody diluted in TBSTM, washed four times in TBST over 1 h, incubated (1 h) with a 1/1,000 to 1/4,000 dilution of secondary antibody (goat anti-mouse immunoglobulin G [IgG] or anti-rabbit IgG conjugated with horseradish peroxidase [Amersham]) in TBSTM, and washed four times with TBST over 1 h. Signals were developed with the ECL detection kit and Hyperfilm ECL (Amersham). Digital images were collected by scanning the film, and band intensities were measured with Photoshop (Adobe) or ImageQuant (Molecular Dynamics).
The following primary antibodies were used: mouse monoclonal antibodies directed against the C-terminal domain of the largest subunit of RNA Pol II (clone 7C2 used at 1/10,000 for the experiments in Fig. 1 to 4 [4]; clone 8WG16 used at 1/1,000 for the experiments in Fig. 5 [BabCo] [see reference 50 for an analysis of the epitopes detected]), lamin B2 (1/50 [Serotec]), TFIIB (1/250 [Transduction Lab]), TBP (1/50 to 1/100 [Oncogene Science]), Sp1 (1/25 [Santa Cruz Biotechnology]); rabbit polyclonal antibodies against Srb7 (1/100 to 500 [46]), TAFII100 (1/500 [59]), TAFII135 (1/50 [59a]), TFIIEα (1/200 [Santa Cruz Biotechnology]), TFIIEβ (1/100 [Santa Cruz Biotechnology]), TFIIF-RAP30 (1/100 [32]), TFIIF-RAP74 (1/100 [17a]), TFIIH-p89 (1/50 to 1/100 [Santa Cruz Biotechnology]), TFIIH-MAT1 (1/100 [Santa Cruz Biotechnology]), and C/EBPβ (1/100 [Santa Cruz Biotechnology]).
FIG. 4.
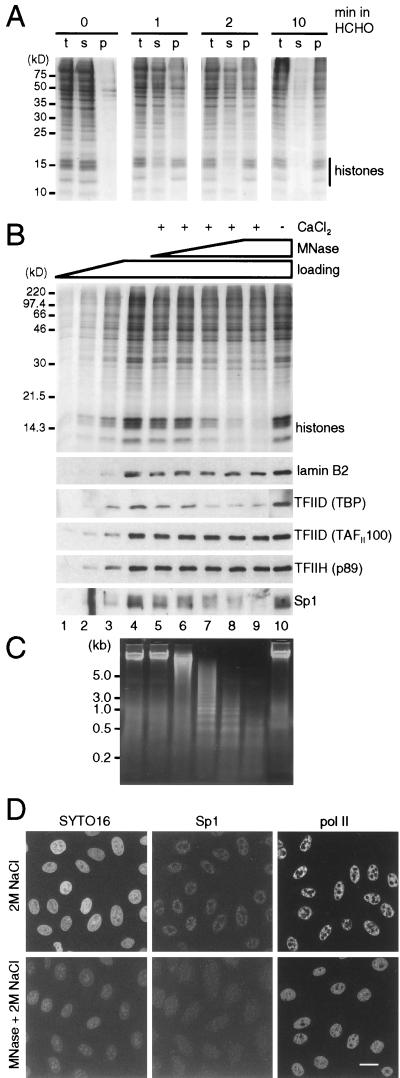
Protein complexes detected after cross-linking with formaldehyde. (A) Establishment of conditions for cross-linking. Cells were either untreated or treated with 1% formaldehyde for 1, 2, or 10 min and then with 0.1% formaldehyde for 3 min, extracted with 2 M NaCl, and centrifuged. Total (t) proteins in extracted cells and those in supernatant (s) and pellet (p) were resolved in a 15% gel and stained with Coomassie blue. After 0, 1, 2, and 10 min in 1% formaldehyde, 0, 65, 80, and 95%, respectively, of the histones were recovered in the pellet. (B) Proteins remaining after treatment of fixed cells with micrococcal nuclease (MNase). Cells were fixed with 1% formaldehyde for 2 min and then with 0.1% formaldehyde for 3 min, lysed with saponin, incubated with or without micrococcal nuclease ± CaCl2 at 37°C, chilled on ice, extracted with 2 M NaCl, and pelleted; then the proteins in the pellet were resolved on a gel, stained with Coomassie blue (top), or blotted and probed with antibodies directed against the proteins indicated. Selected regions of the blots are shown below. Lanes 1 to 4: 1/8×, 1/4×, 1/2×, and 1× loading of cells treated similarly but incubated on ice; lanes 5 to 9: samples incubated with 0, 0.0016, 0.008, 0.04, or 0.2 U of MNase per ml and 1 mM CaCl2; lane 10, sample incubated with 0.2 U of MNase per ml without CaCl2. (C) Photograph of DNA fragments from fixed cells treated with micrococcal nuclease. DNA fragments in samples 4 to 10 in panel B were run on an agarose gel and stained with ethidium. (D) Micrographs illustrating the distribution of Sp1 and Pol II. Cells on coverslips were fixed, treated with micrococcal nuclease or left untreated (as in panel B, lanes 4 and 9), extracted with 2 M NaCl, and refixed with 4% formaldehyde. Then Sp1 and the largest subunit of Pol II were indirectly immunolabelled with Cy3, nucleic acids were counterstained with SYTO16, and equatorial sections through cells were obtained under a confocal microscope. Both Sp1 and Pol II survive extraction with 2 M NaCl and are found in many small foci throughout the nucleoplasm but not in nucleoli (top row). After nuclease treatment, considerable amounts of nucleic acids and Sp1 are extracted, but most of the Pol II remains. Bar, 20 μm.
Number of protein molecules per cell.
The amount of a protein in a fraction relative to that in whole cells was obtained as illustrated in Fig. 2A. HeLa cells were washed twice in PB, resuspended in PB (2 × 107/ml), and mixed with an equal volume of 2× SDS gel-loading buffer; after heat denaturation (95°C for 10 min), proteins in 10-μl samples of whole cells (104 to 105 cells) were resolved on gels and blotted onto filters and the filters were probed with a specific antibody. After digital images were collected, a standard curve relating loading to signal intensity was constructed. The relative amounts of protein in a fraction were determined by linear interpolation with at least two intensity values that (i) were determined from the same blot and (ii) lay on the linear part of the reference curve.
Absolute numbers of protein molecules per cell were obtained in two steps. The first step involved determining the numbers of molecules in purified preparations by reference to bovine serum albumin (Sigma A4503) as a standard. About 100 ng of test protein and 50, 100, 200, 500, and 1,000 ng of albumin were resolved on a gel and stained with Coomassie brilliant blue. After collecting a digital image, band intensities were measured as above and the amount of test protein in the sample was determined by linear interpolation with reference to albumin intensities. In the second step, protein numbers in cell fractions were determined (by immunoblotting) by reference to these purified proteins, again as described above. The proteins analyzed were the largest subunit of Pol II (62), TFIIB, glutathione S-transferase (GST)-TBP, TFIIEα, TFIIEβ, and His-MAT1 (Santa Cruz Biotechnology), TFIIF-RAP74 and TFIIF-RAP30 (18), and GST-TAFII100 (59), which were assumed to have molecular masses of 217, 33, 65, 57, 34, 38, 74, 30, and 60 kDa, respectively.
Formaldehyde cross-linking and digestion with micrococcal nuclease.
For the experiment in Fig. 4A, cells (∼5 × 106/ml) were incubated (25°C) in Dulbecco’s MEM (DMEM) plus 1% formaldehyde (freshly made from paraformaldehyde), 9 volumes of DMEM was added, and the mixture was centrifuged (400 × g for 3 min at 25°C). The 0.1% formaldehyde present during the centrifugation contributes to some cross-linking. Then the cells were washed three times in 50 ml of DMEM containing 25 mM glycine and incubated (10 min at 0°C) in PBI containing 2 M NaCl, and a sample was collected for analysis of total protein. After centrifugation (5,000 × g for 1 min), the supernatant was collected and the pellet was suspended in the original volume of PBI plus 2 M NaCl. After addition of an equal volume of 2× SDS gel-loading buffer (54) and incubation (95°C for 1 h) to reverse the cross-links, the proteins were resolved on an SDS–15% polyacrylamide gel.
For the experiment in Fig. 4B, cells were fixed with formaldehyde and washed with glycine as above. Then they were washed once in PBI, resuspended (∼5 × 106/ml) in PBI without dithiothreitol, lysed in 1% saponin (5 min at 25°C), and washed twice and resuspended (∼2 × 106/ml) in PBI without dithiothreitol, and 0.6-ml samples were incubated (30 min at 37°C) with micrococcal nuclease (Sigma) with or without 1 mM CaCl2. After the mixture was chilled on ice for 5 min, 0.4 ml of 5 M NaCl was added to give a final concentration of 2 M, the cells were incubated (10 min at 0°C) and centrifuged (10,000 × g for 1 min), and the pellet was resuspended in 100 μl of PBI without dithiothreitol. Then 80 μl (for protein analysis) or 15 μl (for DNA analysis) was mixed with an equal volume of the loading buffer (as above) and incubated (95°C for 1 h or 65°C for 16 h, respectively) to reverse the cross-links. Proteins and DNA were resolved on SDS-polyacrylamide or 1.5% agarose gels, respectively.
For immunofluorescence (see Fig. 4D), cells were grown on coverslips overnight, fixed with 1% formaldehyde for 2 min and then with 0.1% formaldehyde for 3 min, washed, lysed, treated with 0.2 U of micrococcal nuclease per ml plus 1 mM CaCl2 or left untreated, and extracted with 2 M NaCl as above. Then the cells were refixed with 4% formaldehyde in 250 mM HEPES (pH 7.4) for 15 min, indirectly immunolabelled with primary antibodies directed against Sp1 (1/25) or Pol II (clone 7C2; 1/5,000) and a secondary goat anti-mouse Ig conjugated with Cy3 (1/500 [Jackson Laboratories]), and counterstained with 0.5 μM SYTO16 (Molecular Probes), and equatorial optical sections were obtained under a confocal microscope (Bio-Rad MRC1000); all procedures for immunolabelling have been described previously (51).
Glycerol gradient centrifugation.
For the experiment in Fig. 5, cell extracts (0.2 ml) prepared as described above were centrifuged (50,000 rpm for 90 min at 4°C in a Beckman SW55 Ti rotor) on 15 to 45% glycerol gradients (5 ml in 10 mM HEPES-NaOH [pH 7.9]–50 mM NaCl–0.1 mM EDTA–0.1 mM dithiothreitol [49]), and 21 fractions (0.25 ml) collected. The proteins in every other fraction were analyzed by electrophoresis and immunoblotting (see above); rRNA in the fractions was also detected by ethidium bromide staining after electrophoresis in agarose gels.
Immunoprecipitation.
For the experiment in Fig. 5F, proteins in the saponin supernatant and dialyzed saponin supernatant were immunoprecipitated (23) in PB containing 20% glycerol by using a normal (control) mouse IgG (Pierce) or an anti-Cdk7 (Sigma) (48) bound to protein G-Sepharose (Pharmacia), and proteins in the pellet were detected by immunoblotting as described above.
RESULTS
Nascent transcripts and some Pol II molecules are tightly bound.
Figure 1A illustrates our fractionation procedure. HeLa cells were lysed with saponin in a physiological buffer to release soluble proteins and centrifuged to yield a saponin supernatant and pellet. The pellet was treated with HaeIII and recentrifuged; the supernatant then contained any chromatin fragments detached from the underlying nuclear structure and able to diffuse out of the cell. In some cases, HeLa cells were grown in [3H]thymidine for ∼20 h before lysis, so that the amount of detached [3H]DNA (and so chromatin) could be determined. This approach is a simplified version of that used previously, in which detached fragments were removed by electrophoresis from cells encapsulated in agarose (29). In some cases, the saponin pellet was also disrupted by sonication and recentrifuged to give another supernatant and pellet. The distribution of proteins in these different fractions is illustrated in Fig. 1B; in this experiment, HaeIII treatment detached ∼30% of the DNA (Fig. 1B) and histones (Fig. 1B, lane 4). Note that it proved difficult to remove more than 50% of the chromatin when high cell densities (required to enable the detection of individual proteins by immunoblotting) and short incubations (required to maintain Pol activity) were used.
We first localized nascent RNA within the different fractions (Fig. 1C). Cells were grown in [3H]uridine for 2.5 min and lysed. Essentially all the [3H]RNA was found in the pellet (lane 3), with little in the supernatant (lane 2); saponin did not detach growing transcripts from engaged Pols. Treatment with sufficient HaeIII to detach 30 to 70% of the chromatin also detached little [3H]RNA (lanes 5 and 6). (The incubation required for nuclease digestion led to an equivalent release of [3H]RNA [lanes 6 and 7]). However, sonication could release some [3H]RNA into the supernatant (lanes 8 and 9). These results show that nascent transcripts cannot be detached from the underlying structure by weak detergents or nucleases.
We next localized Pol activity by using Sarkosyl (Fig. 1D). This strong detergent disassembles nuclei and strips histones from the template; however, it leaves >95% of the engaged Pol II, which is still able to run on along the now naked template (21, 24, 31). Pol activity was retained through the fractionation procedure like nascent transcripts; for example, little activity was found in the HaeIII supernatant (Fig. 1D, lane 4). (Again, incubation led to some loss of activity [lanes 6 and 7].) Sonication released little activity (lane 8) but released some [3H]RNA (Fig. 1C, lane 8).
We also localized Pol II in the different fractions by immunoblotting (Fig. 1E). The largest subunit of this enzyme contains the polymerizing site and a C-terminal domain that becomes hyperphosphorylated when transcription begins (13). The hypo- and hyperphosphorylated forms have apparent molecular masses on gels of ∼220 and ∼240 kDa, respectively, and both were detected (Fig. 1E). Saponin extracted some form IIA but little form IIO (Fig. 1E, lane 2). HaeIII then detached some IIA and IIO (lane 4), but similar amounts were released during the control incubation (lane 6). Considerable amounts of IIO were so tightly bound that they were still found in the pellet after sonication (lane 9). The extraction profile of form IIO, but not IIA, generally mirrors the profiles of nascent RNA and Pol activity.
These results obtained with three markers, i.e., nascent [3H]RNA, activity, and form IIO, show that the polymerizing complex is tightly associated with the nuclear substructure. They are similar to results obtained previously (29) and provide no support for the view that active Pols can be easily detached with chromatin by nucleases.
Numbers of protein molecules in HeLa cells.
The relative amount of Pol II remaining in the pellet after lysis with saponin was estimated by quantitative densitometry (Fig. 2A), and the results for other proteins are summarized in Fig. 2B. (We generally used 1% saponin here; lower concentrations extract less protein but higher concentrations do not extract more, run-on transcription remains constant after extraction with 0.01 to 1% saponin, 1% saponin extracts roughly the same amount of protein as 0.1% Triton X100, and little extra protein is extracted by further treatments with 1% saponin [data not shown].) When purified proteins were available, absolute numbers were calculated by using known amounts of those proteins as standards (Fig. 2C), and average values obtained from three or more different experiments are illustrated in Fig. 2D. The relative proportions of the GTFs seen here are about the same as those seen in yeast (37).
Recent estimates show that an HeLa cell contains ∼65,000 engaged Pol II molecules (31, 51). Our results are consistent with this. Thus, each cell also contains ∼320,000 copies of the largest subunit of Pol II, of which ∼180,000 resist extraction with saponin (Fig. 2D). About 60% of this saponin-resistant fraction, equivalent to ∼110,000 molecules, is pol IIO (Fig. 2A and D). While pol IIO is probably the elongating form (13), it is unlikely that all of it is active; some is found in inactive mitotic cells (5), and some can be solubilized by sonication or Sarkosyl treatment without removing much run-on activity (Fig. 1E) (31). Therefore, ∼60% of the saponin-resistant form IIO may be engaged.
Cells also contain 100,000 to 400,000 molecules of each of the GTFs (i.e., TFIIB, TFIIEα, TFIIEβ, TFIIF-RAP74, TFIIF-RAP30, and TFIIH-MAT1 [Fig. 2D]). If active Pols resist extraction, it is attractive to suppose that transcription factors in a preinitiation complex also resist extraction. Indeed, a very different proportion, but a roughly constant number (i.e., 10,000 to 30,000), of molecules of each of the different factors is found in the saponin-resistant fraction. Moreover, roughly similar numbers of molecules of the two TFIIE subunits, which function as α2β2 heterotetramers, also resist extraction. However, two components of TFIID (i.e., TBP and TAFII100) are present in very different molar ratios (Fig. 2D). (TBP is also present in transcription complexes containing Pols I and III.)
Association of different factors with nuclease-sensitive chromatin.
Since run-on activity is found in the saponin pellet (Fig. 1C and D) and transcriptionally active chromatin is sensitive to nuclease digestion (28, 67), we next investigated whether HaeIII could release Pols and their transcription factors with detached chromatin. After nuclease digestion, the proteins remaining in the pellet were resolved on gels and stained with Coomassie blue. Incubation with progressively more HaeIII detached progressively more histones (Fig. 3A, histones; compare lanes 5 to 8 with the different loadings of unextracted cells in lanes 1 to 4). The relative amounts of other proteins were also obtained after immunoblotting (Fig. 3A). Average values from three independent experiments are plotted in Fig. 3B, which also illustrates the way [3H]DNA was detached by HaeIII.
Individual proteins were detached by HaeIII in a characteristic manner (Fig. 3B). As expected, histone H4 and [3H]DNA were detached at about the same rate, while essentially no lamin B2 could be detached. Pol II, Srb7 (a component of the mediator), and most general transcription factors (i.e., TFIIEα, TFIIEβ, TAFII100, TFIIB, TFIIH-p89, and TFIIH-MAT1) were all relatively resistant to detachment. In contrast, TBP and Sp1 proved very sensitive, with >40% being detached by the lowest concentration of HaeIII used; they appear to be detached much like nuclease-sensitive chromatin. However, another transcription factor, C/EBP (a CAAT box binding protein), was released at the same rate as [3H]DNA. Similar findings were obtained with Sau3AI and AluI (not shown).
Nascent RNA made in vitro resists detachment by HaeIII.
The above approach was also used to evaluate the effect of HaeIII treatment on run-on transcription; nascent RNA made in vitro resisted detachment (Fig. 3B), much like nascent RNA made in vivo (Fig. 1D).
Cross-linking in vivo confirms results obtained in vitro.
The above results suggest that Pol II and all GTFs except TBP are associated with some underlying nuclear structure. Since these associations could arise by nonspecific aggregation after lysis (15), we investigated whether they were also seen in vivo by using the cross-linking agent formaldehyde (19, 56). This approach has several advantages: formaldehyde is so short that initially only molecules in close proximity become cross-linked, and cross-links can be broken subsequently to allow analysis.
We first established the minimum cross-linking required to attach most histones to DNA. Cells were treated with formaldehyde for different times, extracted with 2 M NaCl, and pelleted. All histones in untreated cells were extracted from the pellet into the supernatant (Fig. 4A, 0 min, s); conversely, after 10 min in formaldehyde, 95% of histones were retained in the pellet (10 min, p). After 2 min in formaldehyde, 80% of histones remained in the pellet (2 min, p), and this treatment was used subsequently. It is sufficient to preserve the general shape of nuclei from the destructive effects of 2 M NaCl (Fig. 4D, top left panel). However, it is insufficient to generate many internucleosomal cross-links. This was demonstrated by treating fixed cells with micrococcal nuclease before extraction with 2 M NaCl. This nuclease is inactive in the absence of calcium ions, and so most histones remain in the pellet; they are cross-linked to DNA, which is, in turn, attached to the underlying structure (Fig. 4B, top, lane 10). However, in calcium the nuclease fragments DNA into a nucleosomal ladder (Fig. 4C, lanes 5 to 9), so that histones can be extracted from the pellet by 2 M NaCl (Fig. 4B, lanes 5 to 9). These results show that the level of cross-linking is sufficient to attach most histones to DNA but insufficient to generate so many cross-links that strings of about five nucleosomes cannot escape from the nuclei.
We next examined whether the associations seen in lysed cells in the physiological buffer were also found in vivo. TBP and Sp1 were easily detached by HaeIII from nuclei after lysis in the physiological buffer (Fig. 3). They were also detached by micrococcal nuclease plus 2 M NaCl from cells fixed in formaldehyde (Fig. 4B, bottom, lanes 5 to 9). Other markers that were resistant to HaeIII (i.e., lamin B2, TFIID-TAFII100, TFIIH-p89, TFIIB, TFIIEα, and Pol II) were also resistant to micrococcal nuclease plus 2 M NaCl (Fig. 4B bottom, lanes 5 to 9, and data not shown) under conditions where ∼90% of histones were removed (Fig. 4B, top, lanes 5 to 9). The sensitivity of Sp1 and the resistance of Pol II, to micrococcal nuclease plus 2 M NaCl were confirmed by immunofluorescence (Fig. 4D). Therefore, the associations seen in lysed cells in the physiological buffer were similar to those seen in vivo, making it less likely that they arose artifactually.
Few holoenzyme complexes are found in any soluble fraction.
Holoenzymes sediment at 60S to 80S in glycerol gradients; therefore, we determined whether saponin released any such complexes (Fig. 5). Before doing so, we confirmed that we could detect large complexes in Manley extracts of whole cells (41). Using such extracts, Pan et al. (49) purified a complex containing CDK8(Srb10), TFIIB, TFIIE, TFIIF, TFIIH, TBP, and TAFIIs; this complex contained 45% TAFIIs, 40% TFIIF(RAP74), and 5 to 12% Pol II and the other GTFs. We centrifuged such a whole-cell extract on a glycerol gradient and determined the protein content in different parts of the gradient by immunoblotting (Fig. 5A). Small monomeric proteins were found in fraction 1 at the top. Most Pol II sedimented in fractions 3 to 5, probably as a core complex; ∼12% also sedimented at ∼40S in fractions 7 to 11 (the position of the possible holoenzyme in fractions 7 to 11 is indicated by brackets). Srb7, a protein found in the holoenzyme, sedimented in two broad peaks, one at the top and the other at ∼40S. Between 1 and 10% of TFIIB, TBP, TFIIEα, TFIIF(RAP74), and TFIIH-MAT1, ∼30% of TAFII100, and ∼20% of TFIIH(p89) also sedimented at ∼40S. Therefore, the ∼40S complex probably represents the holoenzyme isolated by Pan et al. (49). A holoenzyme associated with additional repair proteins (e.g., DNA-PK catalytic subunit, HRAD51, RPA, and DNA polymerase ɛ) has also been purified from Dignam’s nuclear extract (10, 14, 40); we could also detect polydisperse complexes of ∼80S in this extract (Fig. 5B, fractions 13 to 15).
Having confirmed that we could detect a large Pol II complex in extracts used by others, we examined whether any could be seen after lysis in our physiological buffer. However, only 3% of soluble Pol II sedimented at ≥40S (Fig. 5C, fractions 9 to 11). Calculations based on data in Fig. 2D show that the holoenzyme, if it exists under these conditions, can contain a maximum of ∼4,000 molecules of Pol II per cell, ∼2,000 of TAFII100, and ∼4,000 of TFIIEα. Moreover, the majority of other transcription factors tested sedimented like monomers in fractions 1 to 3 (Fig. 5C).
We also tried to release holoenzymes by sonicating the saponin pellet; this treatment released some Pol II (Fig. 1B to E, lanes 8 and 9) and most Srb7 (data not shown). Unfortunately, the released enzyme and factors sedimented as a broad peak (Fig. 5D). The sedimentation profiles were unaffected by pretreatment with DNase I or addition of ethidium bromide to the gradient (data not shown), and so it is unlikely that these proteins remained attached to DNA. However, we cannot exclude the possibility that they remained attached to some residual nuclear structure.
The different holoenzymes that were isolated could arise by breakdown of a larger complex or aggregation of smaller complexes, perhaps during dialysis in hypotonic buffers. Therefore, we tested whether dialysis could generate larger complexes and found that it could (compare Fig. 5C and E; also see the legend to Fig. 5). For example, the proportion of Pol II in fractions 9 and 11 increased from 3 to 16% after dialysis and that of TFIIEα increased from 3 to 20%. Only TFIIH-MAT1 and Srb7 showed no change in this region of the gradient.
The high sedimentation rates seen in glycerol gradients could result from self-aggregation of individual proteins and not from the presence of multiprotein complexes containing different protein species. Therefore, we confirmed that at least some complexes contained more than one protein by immunoprecipitation with an antibody directed against a component of TFIIH (i.e., Cdk7); this antibody is known to pull down the holoenzyme (48). As expected, it pulled down its partner in TFIIH (i.e., MAT1) and some Pol II from the saponin extract (Fig. 5F, lane 3). This is consistent with the small amount of Pol II sedimenting at ∼40S (Fig. 5C) being complexed with Cdk7. After dialysis, the antibody pulled down similar amounts of its partner, MAT1; it also pulled down some more Pol II and significantly more TFIIEα (Fig. 5F, lane 4). This shows that dialysis creates some Pol II-Cdk7 and TFIIEα-Cdk7 aggregates.
Taken together, the results in Fig. 5 provide little evidence for the existence of a substantial soluble pool of Pol II complexes with the size expected of a holoenzyme; they also show that dialysis generates larger complexes, which might be confused with the holoenzyme. If substantial amounts of holoenzyme exist, they are likely to be found in the insoluble fraction.
DISCUSSION
Analyzing nucleoprotein complexes in a physiological buffer.
Polymerases are usually isolated in hypo- and hypertonic buffers; hypotonic conditions are used initially because accidental nuclear breakage releases long chromatin strands that tend to aggregate into an unworkable gel, while hypertonic conditions are used subsequently to extract more protein (12). Previously we developed an alternative approach that allowed the use of more physiological conditions (29). Cells were encapsulated in a protective coat of agarose before being lysed in Triton X-100 in a physiological buffer; the resulting encapsulated nuclei were accessible to molecular probes like nucleases and antibodies and retained essentially all the replicative and transcriptional activity of the living cell (30). We now extend this approach, dispensing with the need for encapsulation. Cells are permeabilized with a gentler detergent, saponin, to minimize chromatin release and aggregation. Then, essentially all run-on transcription is retained (Fig. 1D, compare lanes 1 and 3), and this activity is closely associated with the underlying structure (see below). Since this association could result from artifactual aggregation (15), we also analyzed living cells treated with the reversible cross-linking agent, formaldehyde (19, 56). Using conditions that cross-linked most histones to DNA without generating internucleosomal networks, we confirmed that many associations seen in the physiological buffer were also found in vivo (Fig. 4). (Associations tested involved Pol II, TFIIB, TFIID-TBP, TFIID-TAFII100, TFIIEα, TFIIH-p89, Sp1, and lamin B2 [Fig. 4 and data not shown].) Although biochemists can always be criticized for generating artifacts when they break open a cell, the retention of function and all associations tested makes it likely that the complexes seen here in lysed cells have counterparts in vivo.
Active Pols are tightly associated with the nuclear substructure.
A subtetraploid HeLa cell contains ∼80,000 different genes (43) and ∼15,000, ∼65,000, and ∼10,000 nascent transcripts made by Pols I, II, and III, respectively (31, 51). We estimate that a cell also contains ∼320,000 molecules of Pol II, of which ∼140,000 (mainly form IIA) can be released by saponin (Fig. 2D). Of the remainder, ∼70,000 and ∼110,000 molecules are forms IIA and IIO, respectively. Although there are slightly more saponin-resistant IIO molecules than nascent Pol II transcripts, it seems likely that not all will be engaged; sonication releases some IIO without a corresponding loss of run-on activity (Fig. 1D), and Sarkosyl extracts two-thirds of all IIO but leaves >95% of all run-on activity (31). Most engaged Pols, nascent transcripts made in vivo and in vitro, and Pol IIO remain associated with the underlying nuclear structure even when 30 to 70% of the chromatin is detached by nucleases (Fig. 1C and D and 3C). These results confirm earlier results (29) and provide no support for the view that active Pols track around chromatin loops that can be detached with nucleases.
Numbers of GTFs.
Most GTFs in the preinitiation complex are thought to dissociate when elongation begins (70); therefore we might expect to find fewer GTFs than elongating Pols in the saponin pellet. Indeed, our results are consistent with this. Thus, whole cells contained roughly the same numbers of molecules of most GTFs and Pols (i.e., 100,000 to 360,000 and 320,000, respectively); however, 60% of Pols but only 4 to 25% of GTFs resisted extraction with saponin (Fig. 2B), and both could not then be detached by HaeIII (Fig. 3 and 4). As a result, the ∼65,000 molecules of engaged Pols outnumber the 10,000 to 30,000 molecules of each of the GTFs in the saponin pellet. In contrast, TFIIF functions during both initiation and elongation in vitro (52, 70); therefore, we might expect its numbers in the saponin pellet to equal those of the engaged Pols. However, there were fewer molecules of TFIIF in the pellet (Fig. 2B and D).
A significant amount of TBP is associated with nuclease-sensitive chromatin.
Transcriptionally active chromatin is especially sensitive to nuclease digestion (28, 67). Significantly, about half the ∼90,000 molecules of TBP in the saponin pellet were readily detached by a low concentration of HaeIII (Fig. 2D and 3B). This is consistent with in vitro data showing that TBP binds stably to its DNA target and is the only factor that remains bound at the promoter after initiation by Pol II (26, 70). Perhaps this bound fraction is maintained by the presence of a large pool of unbound TBP. Two other DNA binding factors, Sp1 and C/EBPβ, were also sensitive to detachment by HaeIII (Fig. 3B). Therefore, these three proteins may mark potentially active promoters in the absence of the other GTFs. Note that the association of TBP and Sp1 (but not TAFII100 or TFIIH) with nuclease-sensitive chromatin was also found in whole cells cross-linked with formaldehyde (Fig. 4B, bottom, lanes 5 to 9). Note also that a significant fraction of TBP is involved in initiation by Pols I and III (37) and might also mark their promoters.
One of the partners of TBP in TFIID, TAFII100, has quite different properties from TBP; it is less abundant, both in the cell and in the saponin pellet (with ∼28,000 and ∼17,000 molecules, respectively [Fig. 2D]), and it also resists detachment with HaeIII (Fig. 3B). Moreover, this resistance to nucleolytic detachment is also found in whole cells cross-linked with formaldehyde (Fig. 4B, bottom, lanes 5 to 9). Clearly, most TBP is unassociated with TAFII100 in TFIID complexes, just as in yeast (37). Moreover, if nuclease-sensitive TBP is bound at promoters, little TFIID can be associated with it there. It is generally assumed that a role of the TAFII is to recruit TBP to the promoter (47, 53), and so the few TAFII100 molecules would have to assist the loading of many TBP molecules. Alternatively, the few TAFII100 molecules might load only a subset of promoters, consistent with recent data (1, 22, 44, 46, 58, 64, 65). There is sufficient saponin-resistant TBP both to mark potentially active promoters and to form complexes with the relevant TAFs (i.e., TAFI, TAFII100, and TAFIII) (38).
Existence of the holoenzyme in vivo.
Various holoenzymes have been isolated from mammalian cells; the exact composition depends on the isolation procedure (9, 40, 45, 48, 49, 55). This variation could result from the use of different starting materials and purification methods or from the existence of different complexes in vivo. Here we analyzed complexes in the saponin supernatant within 1 h of lysis and sized them on glycerol gradients. However, only 2,000 to 4,000 molecules of Pol II and GTFs per cell had the size expected of a holoenzyme; the rest sedimented like individual components (Fig. 5C). Most SRB7 was also found at the top of the gradient (Fig. 5C). These results should be compared with those obtained in yeast, where 65% of SRBPs are associated with Pol II (35). Significantly, the core Pol, SRB7, and many of the GTFs aggregated on dialysis in a hypotonic buffer to give large complexes (Fig. 5E). This raises the possibility that some holoenzymes seen in vitro arose by aggregation in the hypotonic buffers often used during isolation.
Taken together, our results provide little evidence for a substantial pool of soluble holoenzyme in vivo; only ∼3% of Pol II was seen as a ∼40S complex (Fig. 5C). One possible interpretation of our results is that all soluble Pol II in vivo is in the form of core enzyme and that this spontaneously aggregates to give ∼3% of the larger aggregates in our physiological buffer and ∼16% after dialysis (Fig. 5C and E). If this is so, it is difficult to establish which interactions between components in the larger complex have counterparts in vivo, in the absence of additional data on those interactions from genetic or cross-linking studies. Although we were unable to analyze the ∼60% of Pol that is insoluble, this fraction may well contain a holoenzyme since so many large complexes were seen after sonication (Fig. 5D). Moreover, the higher proportion of large complexes seen in Dignam extracts could also reflect the extraction of holoenzyme from this insoluble fraction. (The preparation of a Dignam extract involves an initial isolation of nuclei, which releases soluble proteins, followed by treatment with 0.25 M NaCl, which could detach a holoenzyme from the underlying structure.)
Models for transcriptional initiation.
As described above, a significant fraction of TBP is bound to nuclease-sensitive chromatin, presumably at promoters. We can incorporate this finding into various models for the formation of the preinitiation complex. In all the models, the existence of a large fraction of TBP bound to its target implies that the next step in the pathway is rate limiting (2, 16, 57, 68). In one, individual components are added progressively (Fig. 6A); in another, a holoenzyme binds as a preformed complex (Fig. 6B). Then we would expect transcription complexes to be released into the supernatant by HaeIII treatment; however, the nuclease detaches essentially no run-on activity, nascent RNA, or GTFs (Fig. 1C and D and 3) (29). Importantly, no large preinitiation complexes containing GTFs and Pol II were detected on nuclease-sensitive chromatin after in vivo cross-linking (Fig. 4B). Alternatively, the preinitiation complex might assemble on the underlying nuclear structure, either progressively or in one or a few steps (Fig. 6C and D); then it would resist nucleolytic detachment. Once transcription had begun, the elongating Pol IIO and the nascent transcript would remain tightly associated with the underlying structure (Fig. 1C and D and 3). Therefore, our results are consistent with either of the models in Fig. 6C and D. We might then expect different preinitiation complexes or holoenzymes to be extracted from the underlying structure by different purification procedures.
FIG. 6.
Models for the formation of the preinitiation complex. In all models, TBP (oval) is shown bound to the promoter, and transcription begins only once the complete complex has formed. (A) Individual components (squares) are added in a stepwise manner. (B) Individual components bind as a preformed holoenzyme. (C) A chromatin loop is shown attached to the underlying nuclear structure (thick grey line); the preinitiation complex is assembled progressively on the underlying structure by an initial attachment of the TBP-promoter complex and subsequent addition of individual components. (D) The TBP-promoter complex attaches to a preformed holoenzyme on the substructure.
ACKNOWLEDGMENTS
We thank M. Vigneron for kindly supplying the Pol II antibody.
This work was supported by the Human Frontier Science Program and the Wellcome Trust.
REFERENCES
- 1.Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 2.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 3.Beebee T J C. A comparison of methods for extracting ribonucleic acid polymerases from rat liver nuclei. Biochem J. 1979;183:43–54. doi: 10.1042/bj1830043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besse S, Vigneron M, Pichard E, Puvion-Dutilleul F. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: the role of interchromatin granules. Gene Expression. 1995;4:143–161. [PMC free article] [PubMed] [Google Scholar]
- 5.Bregman D B, Du L, van der Zee S, Warren S L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess R R. Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem. 1969;244:6168–6176. [PubMed] [Google Scholar]
- 7.Chamberlin M J. The selectivity of transcription. Annu Rev Biochem. 1974;43:721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- 8.Chang M, Jaehning J A. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao D M, Gadbols E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 10.Cho H, Maldonado E, Reinberg D. Affinity purification of a human RNA polymerase II complex using monoclonal antibodies against transcription factor IIF. J Biol Chem. 1997;272:11495–11502. doi: 10.1074/jbc.272.17.11495. [DOI] [PubMed] [Google Scholar]
- 11.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook P R. The nucleoskeleton: artefact, passive framework or active site? J Cell Sci. 1988;90:1–6. doi: 10.1242/jcs.90.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 15.Evan G I, Hancock D C. Studies on the interaction of the human c-myc protein with cell nuclei: p62c-myc as a member of a discrete subset of nuclear proteins. Cell. 1985;43:253–261. doi: 10.1016/0092-8674(85)90030-3. [DOI] [PubMed] [Google Scholar]
- 16.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 17.Flanagan P M, Kelleher R J, Sayre M H, Tschochner H, Kornberg R D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 17a.Fondell, J., and R. G. Roeder. Unpublished data.
- 18.Ge H, Martinez E, Chiang C M, Roeder R G. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 1996;274:57–71. doi: 10.1016/s0076-6879(96)74008-9. [DOI] [PubMed] [Google Scholar]
- 19.Göhring F, Fackelmayer F O. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry. 1997;36:8276–8283. doi: 10.1021/bi970480f. [DOI] [PubMed] [Google Scholar]
- 20.Grande M A, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- 21.Green M H, Buss J, Gariglio P. Activation of nuclear RNA polymerase by sarkosyl. Eur J Biochem. 1975;53:217–225. [Google Scholar]
- 22.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 24.Hawley D K, Roeder R G. Functional steps in transcriptional initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 25.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 26.Hoopes B, LeBlanc J, Hawley D. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J Biol Chem. 1992;267:11539–11546. [PubMed] [Google Scholar]
- 27.Iborra F J, Pombo A, Jackson D A, Cook P R. Active RNA polymerases are localized within discrete transcription ’factories’ in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 28.Igo-Kemenes T, Horz W, Zachau H G. Chromatin. Annu Rev Biochem. 1982;51:89–121. doi: 10.1146/annurev.bi.51.070182.000513. [DOI] [PubMed] [Google Scholar]
- 29.Jackson D A, Cook P R. Transcription occurs at a nucleoskeleton. EMBO J. 1985;4:919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson D A, Yuan J, Cook P R. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J Cell Sci. 1988;90:365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- 31.Jackson D A, Iborra F J, Manders E M M, Cook P R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato H, Sumimoto H, Pognonec P, Chen C H, Rosen C A, Roeder R G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 33.Kelleher R J, Flanagan P M, Kornberg R D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 35.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 36.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 37.Lee T I, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Bjorklund S, Kim Y J, Kornberg R D. Yeast RNA polymerase II holoenzyme. Methods Enzymol. 1996;273:172–175. doi: 10.1016/s0076-6879(96)73017-3. [DOI] [PubMed] [Google Scholar]
- 39.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van-Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 40.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 41.Manley J L, Fire A, Samuels M, Sharp P A. In vitro transcription: whole cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 42.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 43.Miklos G L G, Rubin G M. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 44.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 45.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 47.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 48.Ossipow V, Tassan J-P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 49.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 50.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 51.Pombo A, Jackson D A, Hollinshead M, Wang Z, Roeder R G, Cook P R. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J, 1999;18:2241–2253. doi: 10.1093/emboj/18.8.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solomon M J, Larsen P L, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1998;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 57.Stargell L A, Struhl K. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tao Y, Guermah M, Martinez E, Oelgeschlager T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder R G. Specific interactions and potential functions of human TAFII100. J Biol Chem. 1997;272:6714–6721. doi: 10.1074/jbc.272.10.6714. [DOI] [PubMed] [Google Scholar]
- 59a.Tao, Y., and R. G. Roeder. Unpublished data.
- 60.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 61.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson N E, Aronson D B, Burgess R R. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography: elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J Biol Chem. 1990;265:7069–7077. [PubMed] [Google Scholar]
- 63.Wade P A, Werel W, Fentzke R C, Thompson N E, Leykam J F, Burgess R R, Jaehning J A, Burton Z F. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expression Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- 64.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 65.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weil P A, Luse D S, Segall J, Roeder R G. Selective and accurate initiation of transcription at the Ad2 major late promoter in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979;18:469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- 67.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 68.Xiao H, Lis J T, Jeang K T. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol Cell Biol. 1997;17:6898–6905. doi: 10.1128/mcb.17.12.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 70.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]