Abstract
Insect pickpocket (PPK) receptors mediate diverse functions, among them the detection of mechano- and chemo-sensory stimuli. Notwithstanding their relevance, studies on their evolution only focused on Drosophila. We have analyzed the genomes of 26 species of eight orders including holometabolous and hemimetabolous insects (Blattodea, Orthoptera, Hemiptera, Phthiraptera, Hymenoptera, Lepidoptera, Coleoptera, and Diptera), to characterize the evolution of this gene family. PPKs were detected in all genomes analyzed, with 578 genes distributed in seven subfamilies. According to our phylogeny, ppk17 is the most divergent member, composing the new subfamily VII. PPKs evolved under a gene birth-and-death model that generated lineage-specific expansions usually located in clusters, while purifying selection affected several orthogroups. Subfamily V was the largest, including a mosquito-specific expansion that can be considered a new target for pest control. PPKs present a high gene turnover generating considerable variation. On one hand, Musca domestica (59), Aedes albopictus (51), Culex quinquefasciatus (48), and Blattella germanica (41) presented the largest PPK repertoires. On the other hand, Pediculus humanus (only ppk17), bees, and ants (6–9) had the smallest PPK sets. A subset of prevalent PPKs was identified, indicating very conserved functions for these receptors. Finally, at least 20% of the sequences presented calmodulin-binding motifs, suggesting that these PPKs may amplify sensory responses similarly as proposed for Drosophila melanogaster ppk25. Overall, this work characterized the evolutionary history of these receptors revealing relevant unknown gene sequence features and clade-specific expansions.
Keywords: pickpocket receptors, DEG/ENaCs, insects, phylogeny, evolution
Significance
Sensory receptors of the insect pickpocket (PPK) family allow the detection of environmental stimuli like water, salts, or odors. In spite of these fundamental roles for insect biology, their evolution has been poorly studied. We compared the PPK repertoires of 26 insect genomes to study their phylogeny and diversity across insects. Our phylogenetic analysis revealed that ppk17 represents the most divergent member of the PPK family, which has evolved under a gene birth-and-death model that generated gene expansions in mosquitoes, beetles, cockroaches, and bugs. In fact, mosquitoes, the German cockroach, and the house fly presented the largest PPKs repertoires that thus become new targets for pest control.
Introduction
The insect pickpocket (PPK) family belongs to the much larger Degenerin/Epithelial Sodium Channel (DEG/ENaC) gene superfamily, first described when the genetic bases of mechanosensory pathways were studied in the nematode Caenorhabditis elegans (García-Añoveros et al. 1995). This gene superfamily includes seven families, three of which were first described in vertebrates: ENaC, Acid-Sensing Ion Channels (ASICs), and Brain–Liver–Intestine Sodium Channel (BLINaC)/Human Intestine Sodium Channel (hINaC). Other DEG/ENaC families were reported in invertebrates: the Degenerins from C. elegans, the Drosophila PPK channels, the FMRFamide-gated Sodium Channel (FaNaC) from mollusks, and the FLR-1 receptor that was only identified in C. elegans (Kellenberger and Schild 2002). DEG/ENac members encode a diverse array of epithelial Na+ channel proteins related to fundamental functions such as Na+ absorption, neuron membrane potential control, detection of pH variation, and touch (Kellenberger and Schild 2002). DEG/ENaCs form channels through the union of hetero- or homotrimeric subunits (Canessa et al. 1994), whose identities have an important effect on the pharmacological and kinetic properties of the channel (Benson et al. 2002; Xie et al. 2003). A property of many DEG/ENaCs is their sensitivity to amiloride, an antagonist drug that transiently blocks channel activity (Schild et al. 1997).
The sensory abilities of insects have been shaped along their evolution to adapt to a diverse array of ecological conditions. Insects explore their environment through diverse sensory modalities, and chemoreception is probably the best studied to date. Odorant, gustatory, and ionotropic receptors (ORs, GRs, and IRs, respectively) constitute the three main gene families related to insect chemoreception and have received most of the attention in the last two decades (Carey and Carlson 2011; Hansson and Stensmyr 2011; Breer et al. 2019). Insects present a fourth gene family that encodes receptor proteins that can be related with chemosensory processes, the PPK family, which has received less attention. Several PPKs have a fundamental role in mediating the perception of stimuli of diverse modalities, including water, salts, osmotic potential, pheromones, and mechanosensory properties of their environment (Liu et al. 2003; Zhong et al. 2010; Thistle et al. 2012; Pontes et al. 2017; Matthews et al. 2019; Masagué et al. 2020).
The sequences of PPK proteins are characterized by one highly conserved cysteine-enriched domain in their extracellular loop and two transmembrane domains (Liu et al. 2003; Zelle et al. 2013). According to Zelle et al. (2013) the PPKs of Drosophila spp. can be divided into six subfamilies, but our knowledge about the PPKs of other insect species is scarce. The lack of comparative genomic and phylogenetic studies on the PPK gene family hinders our understanding of the extent of their conservation and diversification across different insect orders. This gap of knowledge prevents predicting whether the functional roles described for model organisms like Drosophila can be extrapolated to other insects. As most PPKs described belong to insects, a proper phylogenetic and functional characterization can have practical implications for developing novel tools to control pests causing sanitary or economic damage. Here, we study the PPK sequences found in 26 insect genomes belonging to eight orders to characterize the evolution of the PPK family across insects.
Results
Analysis of PPK Sequences
A total of 578 PPKs were identified in the genomes of the insect species analyzed (supplementary table S1, Supplementary Material online). All PPKs presented the PFAM domain 00858 characteristic of the DEG/ENaC superfamily. The average length of PPKs was 455 amino acids (aa) and Subf-I to VII presented 464, 402, 523, 548, 627, 522, and 436 aa, respectively. Structural analysis revealed the presence of transmembrane domains in most cases, although their numbers were variable: 149 PPKs (26%) showed one transmembrane domain, 323 PPKs (56%) had two, 46 PPKs (8%) presented between three and five, whereas for 60 PPKs (10%), no transmembrane domains were identified. A total of 274 sequences (47.4%) presented between 13 and 14 cysteine residues, 170 (29.4%) between 10 and 12, and 134 (23.2%) nine or less (supplementary table S1, Supplementary Material online), a characteristic feature of the extracellular loop of PPKs.
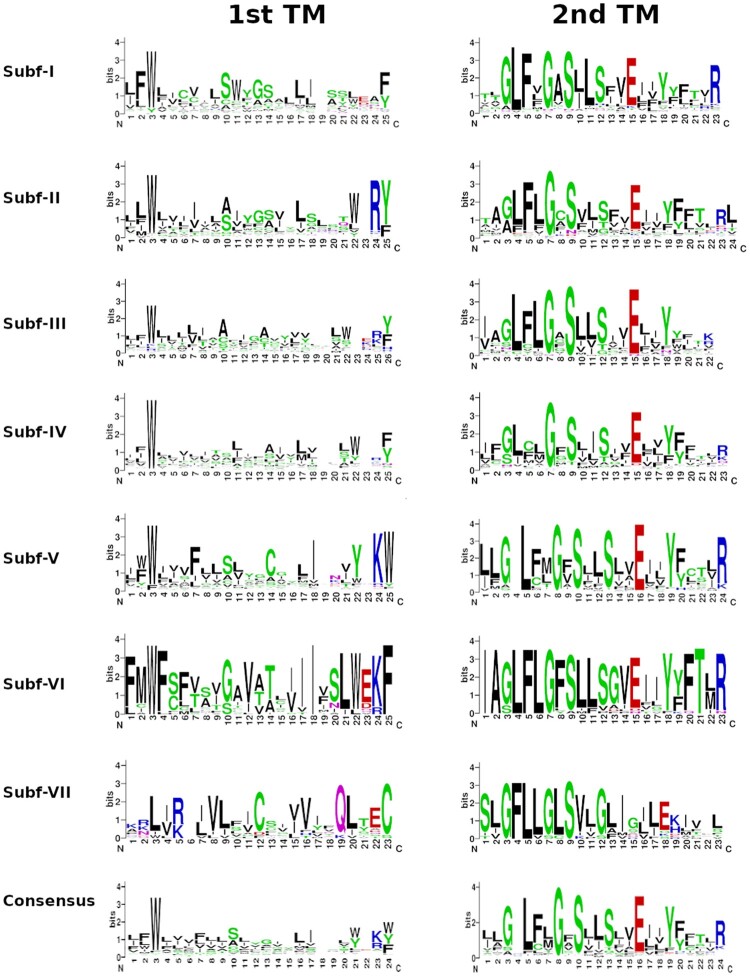
Based on Liu et al. (2003) findings of Drosophila melanogaster PPK sequences, highly conserved amino acid residues were looked for in the PPK sequences found for the species here analyzed (supplementary table S1, Supplementary Material online). Firstly, 43.4% of the sequences presented the “T/S-X-h-H-G” motif (where “h” indicates a hydrophobic residue) anteceding the first transmembrane domain (132 exhibiting an initial threonine and 119 a serine). Secondly, the first transmembrane domain presented a conserved tryptophan residue in 381 (66%) PPK sequences (fig. 1). The second transmembrane domain of 395 (68.3%) PPK sequences presented the “GxS” motif (fig. 1). Interestingly, this region of the receptor was consistently conserved in other positions such as the glycine at position 3 (58%), the leucine at position 5 (66%), the serine at position 13 (59%), the glutamic acid at position 16 (69%), and the tyrosine at position 19 (59%). Additionally, before the first cysteine, a set of 256 sequences (44.2%) presented the “F-P-h-h-T-h-C” motif and 335 sequences (60%) had the “G-X-C-X-X-F-N” motif associated with the fourth cysteine. Alanine was the most common residue (i.e., found in 180 candidates representing 31.1% of the sequences) located in the Degenerin (Deg) site. This residue was followed by valine (137), serine (79), glycine (23), isoleucine (7), threonine (6), cysteine (4), asparagine (3), methionine (3), proline (3), arginine (2), glutamic acid (2), and lysine (2). Leucine and histidine were identified in one sequence each. All PPK protein sequences are listed in supplementary data file S1, Supplementary Material online.
Fig. 1.
Graphical representation of amino acid sequence alignments of the first (left) and second (right) transmembrane domains of the PPK subfamilies and its consensus. These images were generated using WebLogo Version 2.8.2 (https://weblogo.berkeley.edu/logo.cgi; last accessed August 24, 2021).
Finally, the analysis of the Calmodulin-binding motifs (CBMs) revealed that 116 candidates out of 323 sequences with two transmembrane domains had this motif in their sequences. The CBM was located in the N-terminal in 63.7% of the sequences (74 sequences) and in 36.3% of the sequences (42 sequences) it was in the C-terminal region (supplementary table S2, Supplementary Material online). CBMs were identified in PPKs from the seven subfamilies. Some PPK orthogroups, including ppk15, ppk9, ppk6, and in particular Subf-V expansions, such as those of Culex quinquefasciatus (16 sequences) and Tribolium castaneum (eight sequences), had a higher prevalence of CBMs in their sequences. The alignment of CBM sequences had a certain degree of conservation in PPKs belonging to the same subfamily and the presence of positively charged amino acids (red residues in supplementary fig. S1, Supplementary Material online) interspersed among hydrophobic residues (blue residues in supplementary fig. S1, Supplementary Material online), a typical feature of CBMs (Rhoads and Friedberg 1997).
PPK Evolution in Insects
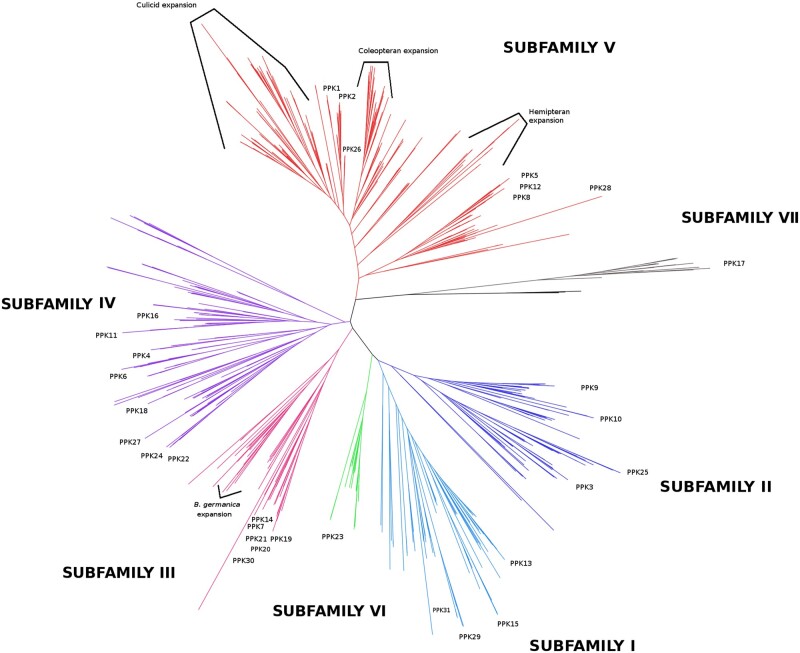
Phylogenetic analyses are summarized in figure 2, which presents a reduced version of the maximum likelihood (ML) tree obtained with IQ-Tree and depicted in supplementary figure S2, Supplementary Material online. The ML tree obtained with RAxML is presented in supplementary figure S3, Supplementary Material online and was almost identical to the IQ-Tree one, with only a few PPKs belonging to Subf-I and II presenting slight changes within some clades. In addition, phylogenetic trees (one “per” subfamily) allowed reinforcing the evolutionary relations observed within Sub-III, Subf-V, Sub-VI, and Sub-VII and resolving ambiguities in the remaining PPK subfamilies (supplementary figs. S4–S10, Supplementary Material online).
Fig. 2.
Phylogeny of the PPK receptor gene family. The phylogenetic tree based on a MAFFT alignment was obtained with IQ-Tree using the WAG+F+R9 best-fit model according to Bayesian Information Criterion. The tree was unrooted and three amiloride-sensitive sodium channels (SCNN) from Rattus norvegicus were used as an outgroup. PPK subfamilies are displayed in different colors. Branch tags and support values were eliminated to facilitate visualization and extended trees are presented in supplementary figures S2 and S3, Supplementary Material online.
Our phylogenetic reconstructions distributed PPKs into seven subfamilies, each being recovered in a well-supported clade (fig. 2), showing that the previous PPK classification into subfamilies (Zelle et al. 2013) is mostly in agreement with the evolutionary history of this family. The PPKs of D. melanogaster were distributed as previously described (Zelle et al. 2013), except for Dmelppk17. Indeed, ppk17 came out as the most divergent PPK and the current analysis suggests that it constitutes the sole member of a novel subfamily named Subf-VII (fig. 2 and supplementary figs. S1 and S4, Supplementary Material online).
The second gene clade to split off from the remaining PPKs was Subf-V (fig. 2). This subfamily presented many duplications and split into two main orthogroups (supplementary fig. S5, Supplementary Material online). One of these orthogroups included ppk28, ppk8, ppk5, and ppk12, whereas the other was composed of ppk1 and ppk2 and a large mosquito expansion.
Two main clades were identified in Subf-II, one including ppk9 and ppk10, and the other one including ppk25 and ppk3 (fig. 2 and supplementary figs. S2 and S6, Supplementary Material online). Interestingly, one-to-one orthologs of ppk9 were found for the different insect orders, the phylogenetic branching pattern resembling the phylogeny of the species studied here.
Three orthogroups were found for Subf-I, the first including the orthologs of ppk15, the second including those of ppk13, and the third group including the orthologs of both ppk29 and ppk31 (fig. 2 and supplementary figs. S2 and S7, Supplementary Material online). As shown by the phylogenetic tree, this subfamily exhibited a consistently dynamic evolutionary history along with duplications in different insect groups (supplementary fig. S7, Supplementary Material online). This contrasts with our findings for Dmelppk23 orthologs forming Subf-VI in which less duplications or losses seemed to have occurred (fig. 2 and supplementary figs. S2 and S8, Supplementary Material online).
Subf-III was basally subdivided into two main clades (fig. 2 and supplementary figs. S2 and S9, Supplementary Material online). The first included hemipteran genes, an expansion of 11 paralogs in Blattella germanica, and one PPK of Camponotus floridanus (the only Subf-III gene found for hymenopterans). The other orthogroup included several D. melanogaster PPKs (Dmelppk7, Dmelppk14, Dmelppk19, Dmelppk20, Dmelppk21, and Dmelppk30), their corresponding orthologs identified in Muscadomestica and Glossina morsitans, and representatives of other insects, mostly hemimetabolous. Notably, no Subf-III PPK was found across lepidopterans, mosquitoes, and beetles (fig. 3a).
Fig. 3.
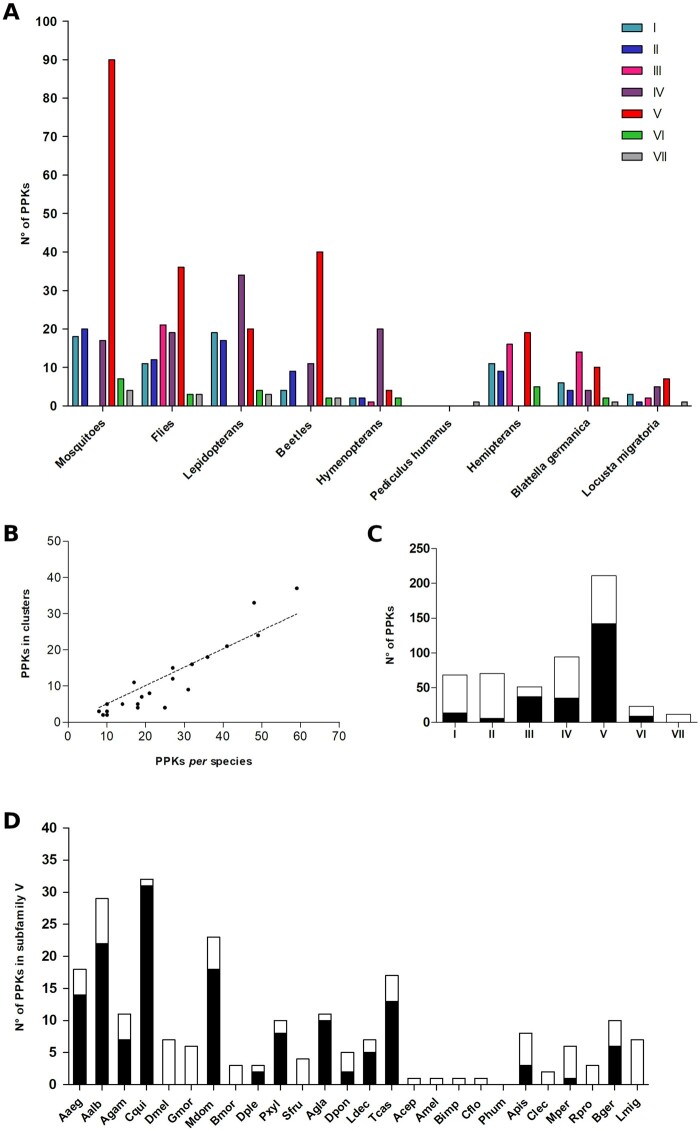
Number of PPKs “per” subfamily in the different taxonomic groups and species analyzed (A); correlation between the number of PPKs in clusters and number of PPK “per” species (B); number of clustered (black) and nonclustered (white) PPKs in each subfamily (C); and number of clustered (black) and nonclustered (white) PPKs in the analyzed species for Subf-V (D). Abbreviations are described in table 2.
Subf-IV encompassed four orthogroups and many lineage losses and duplications were identified (fig. 2 and supplementary figs. S2 and S10, Supplementary Material online). A first clade included orthologs of ppk18, ppk27, ppk24, and ppk22, the second comprised those of ppk4 and ppk6, a third included genes with no known orthologs in D. melanogaster, whereas a fourth clade included orthologs of ppk11 and ppk16.
PPK Distribution along Different Insect Orders
Muscadomestica and B. germanica had the largest PPK repertoires, with 59 and 41 genes respectively, followed by mosquitoes (table 1). Our analysis identified Aaegppk29, an additional gene to be included in the Aedes aegypti PPK repertoire (Matthews et al. 2019). In contrast, at the end of the table are included hymenopteran species that presented six to nine PPKs and Pediculus humanus, for which only ppk17 was found (table 1).
Table 1.
Composition of PPK Subfamilies in the Different Insect Orders Analyzed
| Order | Insect Species | SF I | SF II | SF III | SF IV | SF V | SF VI | SF VII | Total |
|---|---|---|---|---|---|---|---|---|---|
| Diptera | Aedes aegypti | 3 | 4 | 0 | 5 | 18 | 2 | 0 | 32 |
| Aedes albopictus | 8 | 6 | 0 | 2 | 29 | 2 | 2 | 49 | |
| Anopheles gambiae | 4 | 4 | 0 | 5 | 11 | 1 | 1 | 26 | |
| Culex quinquefasciatus | 3 | 5 | 0 | 5 | 32 | 2 | 1 | 48 | |
| Drosophila melanogaster | 4 | 4 | 6 | 8 | 7 | 1 | 1 | 31 | |
| Glossina morsitans | 2 | 2 | 1 | 2 | 6 | 1 | 1 | 15 | |
| Musca domestica | 5 | 6 | 14 | 9 | 23 | 1 | 1 | 59 | |
| Lepidoptera | Bombyx mori | 3 | 4 | 0 | 7 | 3 | 0 | 1 | 18 |
| Danaus plexippus | 4 | 3 | 0 | 7 | 3 | 1 | 0 | 18 | |
| Plutella xylostella | 7 | 5 | 0 | 11 | 10 | 2 | 1 | 36 | |
| Spodoptera frugiperda | 5 | 5 | 0 | 9 | 4 | 1 | 1 | 25 | |
| Coleoptera | Anoplophora glabripennis | 1 | 2 | 0 | 3 | 11 | 0 | 0 | 17 |
| Dendroctonus ponderosae | 1 | 3 | 0 | 3 | 5 | 1 | 1 | 14 | |
| Leptinotarsa decemlineata | 0 | 2 | 0 | 1 | 7 | 0 | 0 | 10 | |
| Tribolium castaneum | 2 | 2 | 0 | 4 | 17 | 1 | 1 | 27 | |
| Hymenoptera | Atta cephalotes | 0 | 0 | 0 | 5 | 1 | 0 | 0 | 6 |
| Apis mellifera | 1 | 1 | 0 | 4 | 1 | 1 | 0 | 8 | |
| Bombus impatiens | 1 | 1 | 0 | 5 | 1 | 1 | 0 | 9 | |
| Camponotus floridanus | 0 | 0 | 1 | 6 | 1 | 0 | 0 | 8 | |
| Phthiraptera | Pediculus humanus | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Hemiptera | Acyrthosiphon pisum | 3 | 2 | 7 | 0 | 8 | 1 | 0 | 21 |
| Cimex lectularius | 2 | 3 | 2 | 0 | 2 | 1 | 0 | 10 | |
| Myzus persicae | 3 | 2 | 6 | 0 | 6 | 2 | 0 | 19 | |
| Rhodnius prolixus | 3 | 2 | 1 | 0 | 3 | 1 | 0 | 10 | |
| Blattodea | Blattella germanica | 6 | 4 | 14 | 4 | 10 | 2 | 1 | 41 |
| Orthoptera | Locusta migratoria | 3 | 1 | 2 | 5 | 7 | 0 | 1 | 19 |
| Total | 74 | 74 | 54 | 110 | 226 | 25 | 15 | 578 |
Note.—SF, subfamily.
Table 2.
Number of PPKs Identified in the Different Insect Genomes and Corresponding Clustering Patterns
| Order | Insect Species | PPKs | PPKs in Clusters (%) | No. Clusters | Average of PPKs “Per” Cluster | No. PPKs Clustered “Per” Subfamily |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SF I | SF II | SF III | SF IV | SF V | SF VI | SF VII | ||||||
| Diptera | Aedes aegypti | 32 | 16 (50%) | 5 | 3.2 | 0 | 0 | — | 0 | 14 (78%) | 2 (100%) | — |
| Aedes albopictus | 49 | 24 (49%) | 7 | 3.4 | 0 | 0 | — | 0 | 22 (76%) | 2 (100%) | 0 | |
| Anopheles gambiae | 26 | 12 (46%) | 4 | 3 | 2 (50%) | 1 (20%) | — | 2 (40%) | 7 (64%) | 0 | 0 | |
| Culex quinquefasciatus | 48 | 33 (69%) | 7 | 3.6 | 0 | 0 | — | 0 | 31 (97%) | 2 (100%) | 0 | |
| Drosophila melanogaster | 31 | 9 (29%) | 3 | 2.9 | 0 | 0 | 6 (100%) | 3 (37%) | 0 | 0 | 0 | |
| Glossina morsitans | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Musca domestica | 59 | 37 (63%) | 11 | 3.4 | 0 | 0 | 12 (86%) | 7 (78%) | 18 (78%) | 0 | 0 | |
| Lepidoptera | Bombyx mori | 18 | 5 (27%) | 2 | 2.5 | 2 (67%) | 0 | — | 5 (43%) | 0 | — | 0 |
| Danaus plexippus | 18 | 4 (22%) | 2 | 2 | 0 | 0 | — | 2 (29%) | 2 (67%) | 0 | — | |
| Plutella xylostella | 36 | 18 (50%) | 9 | 2 | 2 (29%) | 0 | — | 7 (64%) | 8 (70%) | 1 (50%) | 0 | |
| Spodoptera frugiperda | 25 | 4 (16%) | 2 | 2 | 3 (60%) | 0 | — | 1 (11%) | 0 | 0 | 0 | |
| Coleoptera | Anoplophora glabripennis | 17 | 11 (65%) | 3 | 3.3 | 0 | 0 | — | 0 | 10 (90%) | — | — |
| Dendroctonus ponderosae | 14 | 5 (35%) | 2 | 2.5 | 0 | 0 | — | 3 (100%) | 2 (40%) | 0 | 0 | |
| Leptinotarsa decemlineata | 10 | 5 (50%) | 2 | 2.4 | — | 0 | — | 0 | 5 (71%) | — | — | |
| Tribolium castaneum | 27 | 15 (55.5%) | 3 | 5 | 0 | 0 | — | 2 (50%) | 13 (76%) | 0 | 0 | |
| Hymenoptera | Atta cephalotes | 6 | 0 | 0 | 0 | — | — | — | 0 | 0 | — | — |
| Apis mellifera | 8 | 0 | 0 | 0 | 0 | 0 | — | 0 | 0 | 0 | — | |
| Bombus impatiens | 9 | 2 (22%) | 1 | 2 | 0 | 0 | — | 2 (40%) | 0 | 0 | — | |
| Camponotus floridanus | 8 | 3 (38%) | 1 | 3 | — | — | 0 | 3 (50%) | 0 | — | — | |
| Phthiraptera | Pediculus humanus | 1 | 0 | 0 | — | — | — | — | — | — | — | 0 |
| Hemiptera | Acyrthosiphon pisum | 21 | 8 (38%) | 3 | 2.6 | 0 | 0 | 5 (71%) | — | 3 (37%) | 0 | — |
| Cimex lectularius | 10 | 3 (33%) | 1 | 3 | 1 (50%) | 2 (67%) | 0 | — | 0 | 0 | — | |
| Myzus persicae | 19 | 7 (37%) | 3 | 2 | 1 (33%) | 2 (50%) | 3 (67%) | — | 1 (17%) | 0 | — | |
| Rhodnius prolixus | 10 | 2 (20%) | 1 | 2 | 2 (67%) | 0 | — | 0 | 0 | 0 | — | |
| Blattodea | Blattella germanica | 41 | 21 (51%) | 6 | 3.5 | 1 (17%) | 1 (25%) | 11 (79%) | 0 | 6 (60%) | 2 (100%) | 0 |
| Orthoptera | Locusta migratoria | 19 | 0 | 0 | — | 0 | 0 | 0 | 0 | 0 | — | 0 |
Note.— —, families with any PPK; SF, subfamily.
Subf-I and Subf-II genes were found in all insect orders studied, except for Phthiraptera (fig. 3a). Genes belonging to Subf-III were restricted to flies, hemipterans, the German cockroach, the migratory locust, and the ant Cam. floridanus (table 1 and fig. 3a). Interestingly, flies presented several duplications and one M. domestica-specific expansion including six paralogs which are orthologs to ppk19. Few one-to-one orthologs were identified across orders in Subf-IV (fig. 2), which presented no hemipteran members (fig. 3a). Presenting almost 40% of all PPKs analyzed, Subf-V was the largest one, including a total of 226 genes (table 1 and fig. 3a). This family included four lineages representing taxon-specific expansions. The first lineage included 37 beetle members, whereas the second corresponded to 85 mosquito members (Anophelesgambiae: 10, A. aegypti: 17, Aedesalbopictus: 27, and Cul. quinquefasciatus: 31). The third lineage included 13 genes belonging to hemipterans. Finally, the fourth lineage presented a M. domestica expansion related to ppk2 that included 11 paralogs. Subf-VI was composed of a single orthogroup that includes Dmelppk23 and the corresponding orthologs of 19 other insects (table 1). The ppk17 gene was the only member of Subf-VII and was absent in hymenopterans and hemipterans (fig. 3a).
Genomic Organization of PPKs in Insect Genomes
We examined the location of PPK genes along the different insect genomes and found a nonrandom distribution of these genes: 57% of all PPKs (329) shared a scaffold or chromosome with another PPK, most of which (86%) were closely related, that is, members of the same subfamily (supplementary table S3, Supplementary Material online). Next, we studied the distance between PPKs located in the same genomic region and observed that 70% of them were separated by less than 100,000 bp (supplementary table S3, Supplementary Material online). This distance was used as a cut-off reference defining that two or more PPKs belonged to the same genomic cluster (Engsontia et al. 2014). According to this criterion, the PPKs of G. morsitans, Atta cephalotes, Apis mellifera, Locusta migratoria, and P. humanus were not arranged in clusters and thus, were not considered in subsequent analyses. Ten out of 21 remaining insect genomes presented more than 40% of their PPKs into clusters (table 3). Among them, mosquitoes, the house fly, the cockroach, and the moth Plutella xylostella exhibited a higher number of clusters (between 5 and 11 genes “per” cluster) in comparison with the rest of the genomes analyzed. A positive correlation was observed between the number of PPKs and the number of genes located in clusters (Spearman R = 0.91, P < 0.0001, fig. 3b).
Table 3.
Gene Birth and Death Rate Estimates for the Entire PPK Family and for Each Subfamily
| Total Time Rates |
Branch Average Rates |
|||
|---|---|---|---|---|
| Subfamily | Birth | Death | Birth | Death |
| Subf-I | 0.0008 | 0.0016 | 0.0006 | 0.0007 |
| Subf-II | 0.0007 | 0.0011 | 0.0019 | 0.0014 |
| Subf-III | 0.0028 | 0.0026 | 0.0059 | 0.0040 |
| Subf-IV | 0.0027 | 0.0016 | 0.0083 | 0.0028 |
| Subf-V | 0.0069 | 0.0011 | 0.0102 | 0.0018 |
| Subf-VI | 0.0008 | 0.0008 | 0.0021 | 0.0011 |
| Subf-VII | 0.0004 | 0.0016 | 0.0015 | 0.0029 |
| All | 0.0024 | 0.0015 | 0.0047 | 0.0020 |
Most clusters were formed by genes of the same subfamily (72 out of 80), whereas the remaining eight presented genes coming from more than one subfamily (supplementary table S3, Supplementary Material online). Most genes belonging to Subf-III and Subf-V were distributed in clusters (73% and 67%, respectively) (fig. 3c). This high degree of clustering is explained by gene expansions seen in several taxonomic groups for both subfamilies, for example, in M. domestica and B. germanica for Subf-III (supplementary table S3, Supplementary Material online), and in mosquitoes, beetles, and hemipterans for Subf-V (fig. 3d). A second group including Subf-IV and Subf-VI presented an intermediate clustering profile (40%). Finally, most of the PPKs belonging to Subf-I and Subf-II were not included in clusters (21% and 9% of PPKs, respectively).
Family Size Dynamics
The PPK gene family evolved according to a gene birth-and-death evolutionary model (Nei and Rooney 2005). Overall, the PPK family is expanding, as inferred from the comparison of birth and death rates (table 3). Among which Subf-III, Subf-IV, and Subf-V were the most dynamic, presenting birth/death rates compatible with gene expansion (table 3). Although Subf-II and Subf-VI had different rates depending on the estimation methods, their birth and death rates were always similar to each other, suggesting that their sizes are stable across the groups analyzed (table 3). On the other hand, Subf-I and Subf-VII appeared to be contracting, with the latter contracting at a faster rate (table 3).
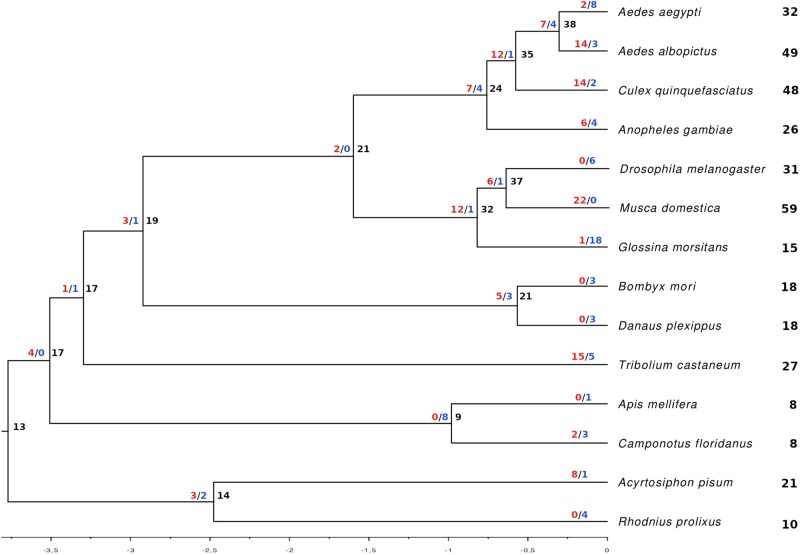
Higher rate estimates obtained with the “branch average” method reflected the multiple duplication and loss events that occurred in short branches of the phylogeny, that is, within a specific order or taxonomic group, as observed for the expansions in M. domestica (fig. 4).
Fig. 4.
Species tree showing the number of gene gains (blue) and losses (red) in each branch and the total number of genes for each terminal.
Selective Pressure Analysis
A total of 20 orthogroups, that is, those composed by orthologs of 16 D. melanogaster PPKs and four taxon-specific expansions, were included in the analysis of selective pressure. The role of natural selection in the evolution of these 20 orthogroups was evaluated by means of a phylogeny based analysis (supplementary table S4, Supplementary Material online). The ω values calculated by means of the M0 were lower than 0.16, except for the hemipteran and B. germanica orthogroups, which exhibited ω values of 0.20 and 0.21, respectively. Interestingly, the ppk17 orthogroup, the most divergent member of the PPK family, presented the lowest ω = 0.01.
Only three genes (ppk6, ppk25, and ppk27) were found to be under positive selection (M7 vs. M8 model comparison, supplementary table S4, Supplementary Material online). The ppk6 orthogroup showed two predicted codons in the C-terminal region with posterior probabilities (PP) >0.95 of being under positive selection. The ppk25 orthogroup presented three codons under positive selection: one codon at the N-terminal region (329) and two codons (565 and 567) at the extracellular loop (i.e., 11 residues downstream of the “G-X-C-X-X-F-N” motif). Finally, the ppk27 orthogroup showed three positions under positive selection that were located at the N-terminal region before the first transmembrane domain.
Discussion
We have performed a large-scale genomic data mining analysis to inquire into the evolution of the PPK gene family across 26 species spanning four hemimetabolous and four holometabolous insect orders. PPKs were detected in all genomes studied and assigned to seven subfamilies, including the new Subf-VII formed by orthologs of the Dmelppk17 gene that seem to be the most divergent PPK family members. Our analysis shows that PPKs evolved under a gene birth-and-death model that generated lineage‐specific expansions by gene duplication events, and the effect of purifying selection. The size of PPK repertoires displayed high variability; whereas M. domestica, B. germanica, and several mosquito species presented the largest gene sets, P. humanus, bees, and ants had very small ones. Whether these important differences are related to specific ecological needs and constraints of these insects, or to clade-specific genetic properties favoring or limiting gene duplication events is not clarified by our analyses.
Ppk17 Formed a New and Divergent Subfamily
According to our phylogenetic analysis, the ppk17 orthogroup is an independent lineage that should not be included in Subf-V as previously suggested (Zelle et al. 2013). Furthermore, the clade formed by proteins of the ppk17 orthogroup represented the most divergent member of the PPK family (fig. 2 and supplementary figs. S2 and S4, Supplementary Material online). Interestingly, ppk17 was identified in all insect orders, except for hymenopterans that apparently lost it (fig. 2 and table 1). This gene presented the strongest level of purifying selection (ω value = 0.01), similar to that reported for the odorant receptor coreceptor, Orco (Soffan et al. 2018), suggesting that its function is likely conserved and fundamental. George et al. (2019) used loss-of-function mutants and RNAi lines to study the role of ion channels in wing development in D. melanogaster and found that Dmelppk17, Dmelppk1, Dmelppk2, Dmelppk25, and Dmelppk30, among others, are likely involved in this process. Functional genetic studies in other experimental models are necessary to confirm this hypothesis and provide further details on its function.
Genetic Mechanisms Driving PPK Evolution
Detailed examination of the relationships of PPKs across several insect orders revealed the expected evolutionary pattern of gene birth-and-death typical of environmentally relevant genes (Nei and Rooney 2005; McBride et al. 2007; Sánchez-Gracia et al. 2011; Vieira and Rozas 2011; Almeida et al. 2014), with lineage‐specific expansions (through duplication) and contractions (through pseudogenization). Consistently with this evolution model, 284 PPKs (49%) shared the same scaffold or chromosome with at least another PPK of the same subfamily, most of them being located in clusters. The fact that these related genes were located in nearby chromosomal regions, suggests that many are products of relatively recent gene duplications. A similar distribution pattern was previously reported for ORs or GRs (Robertson et al. 2003; Engsontia et al. 2008; Wanner and Robertson 2008; Smadja et al. 2009). Gene duplication is fundamental for the diversification of gene family repertoires as it can be followed by neofunctionalization or subfunctionalization processes leading to evolutionary innovation (Sánchez-Gracia et al. 2009). It has been observed that genes within clusters are usually coregulated, which can lead to joint gene expression (Robertson et al. 2003; Guo and Kim 2007; Nozawa and Nei 2007). Therefore, we would expect that PPKs follow a similar pattern: clusters might be subject to common regulatory mechanisms and deal with common or related stimuli, even though their functions cannot be considered exclusively related to sensory processes.
Estimates of gene birth and death rates suggest that the PPK family is generally expanding in the set of species we analyzed, although it could be contracting in particular species or groups. It is important to acknowledge that our gene birth and death rates might be underestimated due to lack of information on pseudogenes and because we used a conservative approach when counting duplication and loss events by gene tree versus species tree reconciliation. The “total time” method was our choice for all comparisons among families discussed herein, because it was the method used to obtain previous estimates for sensory gene families (Sánchez-Gracia et al. 2009), even though it may lead to underestimates. The “branch average” method estimated higher rates, reflecting the availability of genomes of closely related species (i.e., flies, mosquitos) and the large number of duplication and loss events in these groups, as it takes into account each branch length (in units of time). This last approach is more accurate, but also more dependent on the species set used in the analyses. We used estimates obtained for chemosensory gene families in the genus Drosophila for comparisons with the rates obtained herein (see Sánchez-Gracia et al. [2009] and references therein), as no estimates of birth and death rates were available across insect orders. Our rate estimates for the PPK family as a whole were in the lower range of rates obtained for those gene families, but yet it was substantially higher than the genomic average (Hahn et al. 2007).
There was considerable variation in gene birth and death rate estimates among PPK subfamilies (table 3), probably as a consequence of different selection pressures. Although some had very low rates (e.g., Subf-II and Subf-VI), others had rates comparable to those of the most dynamic chemosensory gene families (e.g., Subf-V) suggesting similar evolutionary dynamics that could be linked to functional similarity. Moreover, whereas some families seem to be expanding (e.g., Subf-IV and Subf-V), others are stable (e.g., Subf-II and Subf-VI), and others have a contraction trend (e.g., Subf-I and Subf-VII). As expected, the subfamilies with low turnover rates, represented by ppk23 and ppk17 orthogroups, were also the most conserved across insect orders (tables 1 and 3). The dearth of data on insect PPK function, however, precludes conclusions about these differences among subfamilies.
PPK evolution is mostly driven by purifying selection, as ω values estimated for selected orthogroups were generally low and positively selected sites were detected in only three orthogroups (supplementary table S4, Supplementary Material online). One of these instances was in the orthogroup of ppk25, for which diverse reports have presented evidence supporting its role in sensory activity in D. melanogaster (supplementary table S5, Supplementary Material online). Future functional studies should examine why these specific residues were subject to positive selection across the orthogroup. Despite these few exceptions, ω values were low even when we analyzed genes within an expansion, contrary to the suggestion that purifying selection is relaxed after duplication events. Analyses including more closely related species and specific gene lineages belonging to the expansions described for Subf-V may provide more insights into the evolutionary forces that shaped PPK evolution. It is important to acknowledge that not all orthogroups were included in the selection analysis, and therefore additional PPK genes could escape the general trend observed.
Subf-V Represents the Largest Gene Lineage
Sánchez-Gracia et al. (2009) reported the turnover rates of diverse chemosensory gene families among which GRs were the only showing a turnover rate higher than that seen for Subf-V in our study (table 3). Three large expansions made it the largest subfamily (226 genes), including almost half of the PPKs identified in this study (fig. 2). A similar pattern was described for the 9-exon subfamily of ant ORs that is extremely expanded (Engsontia et al. 2015). The expansions of Subf-V sat into genomic clusters (supplementary table S3, Supplementary Material online) and were taxon-specific: flies, mosquitoes, beetles, and hemipterans (fig. 2). The origin of these expansions may have been a response to rapid adaptation to changing environments, as suggested for sensory receptor families (Smadja et al. 2009; Engsontia et al. 2015).
In the case of mosquitoes, the increase in the number of PPKs is mainly due to the Subf-V expansion of the ppk1–ppk2–ppk26 clade (fig. 2). The functional information available for this set of PPKs is restricted to D. melanogaster larvae (see supplementary table S5, Supplementary Material online), where they are coexpressed mediating mechanosensation, acid sensing (pH between 5 and 9), and locomotion (Adams et al. 1998; Boiko et al. 2012; Tsubouchi et al. 2012; Zelle et al. 2013; Gorczyca et al. 2014). Whether this expansion was driven to mediate mosquito pH sensing in aquatic larval environments deserves experimental assessment. In the end, this large exclusive gene expansion emerges as a potential target to manipulate mosquito behavior and physiology through genetic tools.
Variations in PPK Repertoires
Consistently with their fast evolution, the size and composition of insect PPK repertoires were shown to be very diverse, although phylogenetic proximity is generally associated with similar repertoire sizes (table 1). The impressive species diversity that insects have reached through their evolution attests a high capacity to adapt to diverse environmental conditions. In this sense, the dramatically different PPK repertoires reported herein may reflect specific adaptations to the ecological niches where these animals thrive (Robertson and Wanner 2006; McBride et al. 2007; Sánchez-Gracia et al. 2011; Smadja et al. 2012). The extreme examples of 59 PPK family members found in the genome of M. domestica, and only one in that of P. humanus capture the extent of the plasticity shown by these gene repertoires to adapt to environmental pressures. The reductions observed in the PPK repertoires of hymenopterans and human lice resemble patterns observed in some of their chemosensory receptor families, and could be mainly attributed to increased PPK gene loss in these insects (fig. 4 and supplementary table S6, Supplementary Material online). In the case of the human lice, a general pattern of gene loss has been attributed to its limited foraging range (Kirkness et al. 2010). As for the hymenopterans, it is not clear why the PPK family suffered such a reduction. A denser sampling of species and more data on PPK function could provide more clues on the issue.
On the other extreme, some dipterans and B. germanica presented large PPK expansions that significantly increased their PPK repertoires, a pattern that also resembles the one previously described for their chemosensory receptor repertoires (Scott et al. 2014; Robertson et al. 2018; supplementary table S6, Supplementary Material online). In the case of dipterans, we could observe a trend of PPK gene gains along the evolution of the clade (fig. 4), that culminated in very large numbers of PPKs in most species. Nevertheless, the lack of data for species closely related to the German cockroach precludes us from having a better understanding of the evolution of PPK repertoires in Blattodea. It is not clear what selective pressures have driven the evolution of the diversity of sensory-related genes in these insects and whether it equips them with extreme abilities to sound the informational complexity of the environment. Interestingly, some of the extreme expansions involved synanthropic species, which suggests that a large repertoire of PPKs and chemosensory genes may represent a preadaptation to anthropic environments that offer a broad diversity of nutrient sources, as well as toxins. Highly diversified sensory abilities can be an important asset to tell apart and exploit the complex features of anthropic supplies, providing access to a number of resources and a cosmopolitan distribution.
Expanded species-specific lineages were identified in the Subf-III and Subf-V of B. germanica and M. domestica. In the case of the Subf-III, two expansions were identified, one for B. germanica (11 members) and one for M. domestica (six members). The latter is related to Dmelppk19, which has been shown to be involved in sensing low salt concentrations, acting together with Dmelppk11 (Liu et al. 2003). The expansion of this PPK lineage would probably increase house fly sensitivity or capacity to detect different types of salts or to discriminate concentration ranges. The placement of the B. germanica expansion in the phylogenetic tree depicting Subf-III prevents proposing a putative function for this lineage due to their lack of functional data and direct orthology with D. melanogaster PPKs. A M. domestica expansion including 11 paralogs related to ppk2, a gene that mediates gentle touch in D. melanogaster larvae, was identified in Subf-V (supplementary table S5, Supplementary Material online). Future studies should evaluate whether this expansion provides enhanced mechanosensory abilities to M. domestica larvae (Tsubouchi et al. 2012). In sum, these species-specific expansions represent new specific targets to control these highly synatropic insects.
A Subset of Conserved PPKs
Our data mining analysis evinced a pattern of prevalent versus divergent PPKs. Indeed, we observed that most PPKs are divergent, lacking one-to-one orthology across insect orders. However, several PPK orthogroups were prevalent among the insect orders studied here. Members of the ppk28 orthogroup were identified in 92% of the genomes studied (fig. 2), being absent in only P. humanus, and Leptinotarsa decemlineata. Ppk28 has been reported to mediate water detection in D. melanogaster, whereas it is also reported to mediate responses to water and low salt concentrations in A. aegypti during egg-laying initiation (supplementary table S5, Supplementary Material online). A conserved mechanism for the detection of environmental osmolarity could explain why ppk28 has been conserved through the evolution of insects.
Another prevalent gene, ppk23 is coexpressed with ppk29 and ppk25 in the proboscis and legs of D. melanogaster, where this complex acts as a contact chemoreceptor of cuticular hydrocarbon pheromones controlling the sexual behavior of both sexes (supplementary table S5, Supplementary Material online). Cuticular hydrocarbons mediate sexual, colony recognition, and aggregation behaviors in diverse insect orders (Pavković-Lučić et al. 2012), suggesting that the pheromone receptor role seen for D. melanogaster can be potentially found in other cases. Dmelppk23 is also expressed in GRNs from the labellum where together with Gr64f, Gr66a, Ir94e, Ir76b, and ppk28, mediates salt taste and adjusts responses to salt depending on the fly internal state (Jaeger et al. 2018). It is possible that either of these functions have been maintained through insect evolution.
Notwithstanding the fact that ppk9 was as prevalent as ppk23 (both present in 73% of the genomes analyzed), raising functional hypotheses is not feasible because its role is unknown to date. Other examples of high prevalence are ppk16 (65% of the genomes had orthologs for this gene), ppk13 (58%), and ppk17 (54%). To date, there is no information on the physiological properties and functions of these three conserved PPKs.
Conserved Features of PPK Sequences
The degenerin site (DEG or d position) is a residue located in the segment that precedes the second transmembrane domain and plays a fundamental role in the open state and ion selectivity of the channel (Eastwood and Goodman 2012). This position is most commonly occupied by glycine, alanine, or serine in DEG/ENaC proteins (Eastwood and Goodman 2012). Nevertheless, PPK sequences frequently have a bulkier residue, that is, valine, which could have functional implications, as amino acids with big side chains in the DEG site have been implicated in changes in channel activity in other DEG/ENaC superfamily members (Eastwood and Goodman 2012).
Liu et al. (2003) and Zelle et al. (2013) reported conserved features in PPK sequences of several Drosophila species and Ano.gambiae that according to our analysis are conserved in insects in general. First, the cysteine-rich domain, which seems fundamental in mammals for an efficient transport of assembled channels to the cell membrane (Firsov et al. 1999) is highly conserved (77% of sequences had more than 10 cysteine residues). Second, a tryptophan residue in the first transmembrane domain that is critical to Na+ sensitivity was detected in 66% of the PPK sequences (fig. 1; Pochynyuk et al. 2009). Finally, the “GxS” motif of the second transmembrane domain (fig. 1), which contributes to the channel pore and is fundamental for ion selectivity and surface expression (Eastwood and Goodman 2012), was identified in 68% of the sequences. Our analysis revealed a higher conservation of the second transmembrane domain compared with the first one (fig. 1), which is consistent with its fundamental role in pore formation in DEG/ENaCs (Hong and Driscoll 1994). Besides, additional highly conserved residues, a leucine in position 5 and a glutamic acid in position 16, were identified in the second transmembrane domain (fig. 1), suggesting that they may contribute to pore function.
Recently, Ng et al. (2019) have described the involvement of Dmelppk25 in the amplification of olfactory responses in D. melanogaster. These authors reported that after odor stimulation of Or47b and Ir84a, Ca2+ influx increases and activates Dmelppk25 via an intracellular CBM. CBMs were also identified in other D. melanogaster PPKs, suggesting that they function as Ca2+-activated amplification channels. Our results revealed the presence of CBMs in more than a hundred PPKs from other insect species (supplementary table S2, Supplementary Material online), reinforcing the hypothesis that these receptors may amplify sensory responses through Ca2+ activation.
Final Remarks
In this study, we used data from genome annotations, so it is possible that future TBlastN searches and insect genome reannotations will reveal additional PPKs, as seen with A. aegypti (Matthews et al. 2019). Besides, improving the annotation of the PPKs identified herein by means of RNA-Seq and subsequent functional experiments, as well as extending the analysis to other arthropods, would help further characterizing the PPK family. Recently, Vizueta et al. (2020) have presented evidence of potential PPK sequences in the genomes of Daphnia pulex and Strigamia maritima. Indeed, our preliminary searches using the DEG/ENaC PFAM domain on several arthropod genomes have identified a set of candidate PPK sequences, reinforcing that the PPK family is not exclusive to insects. Further work under development, including new searches on nonarthropod genomes followed by sequencing and phylogenetic analyses will evaluate the origin and provide a more complete understanding of the evolutionary history of PPKs.
Our phylogenetic and evolutionary characterization of PPKs across different insect orders has allowed identifying specificities that turn many of these genes into potential candidates to manipulate insect behavior and physiology. First, PPKs participate in fundamental insect physiological processes, like water, salt, or pheromone detection. Second, they are particularly expanded in several synanthropic insects including M. domestica and B. germanica, suggesting that these receptors may have been a relevant preadaptation to exploit anthropic habitats. Third, the large culicid expansion observed in PPK Subf-V suggests an important role that grants them as very specific targets for developing specific mosquito control tools. Finally, the identification of CBMs in PPK sequences suggests that a large subset of these receptors may be related to the amplification of sensory responses, and tools targeting them could affect a diversity of sensory modalities.
Materials and Methods
Candidate Sequences
Amino acid sequences of PPKs from D. melanogaster (Zelle et al. 2013), A. aegypti, and Ano.gambiae (Matthews et al. 2019) were used as queries in BlastP searches (with e-value threshold = 0.0001 and the low complexity regions filter deactivated) in different databases (details in supplementary table S7, Supplementary Material online) to identify their putative orthologs in: Blattodea (B. germanica), Orthoptera (Locustamigratoria), Hemiptera (Acyrthosiphon pisum, Myzus persicae, and Cimex lectularius), Phthiraptera (P. humanus), Hymenoptera (Attacephalotes, Api. mellifera, Bombus impatiens, and Cam. floridanus), Lepidoptera (Bombyx mori, Danaus plexippus, Plutellaxylostella, and Spodoptera frugiperda), Coleoptera (Anoplophora glabripennis, Dendroctonus ponderosae, Leptinotarsadecemlineata, and T. castaneum), and Diptera (A. albopictus, Cul. quinquefasciatus, G. morsitans, and M. domestica). Iterative searches were conducted with each new PPK protein sequence as a query until no new genes could be identified for each PPK subfamily or lineage. Translated sequences of PPK genes previously described for Rhodnius prolixus (Latorre-Estivalis et al. 2017) were also included in the analysis.
Analysis of PPK Sequences
Protein sequences obtained in the similarity searches were characterized at functional and structural levels. The Pfam (v.27.0) (Finn et al. 2016) database was used to check for the presence of the amiloride-sensitive sodium channel domain (PFAM00858) characteristic of PPKs using the Batch Web CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, last accessed August 24, 2021) from the Conserved Domain Database at NCBI (Lu et al. 2020). The presence and number of transmembrane domains predicted were established using TOPCONS (Bernsel et al. 2009). The Calmodulin Target Database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html, last accessed August 24, 2021) was used to identify CBMs in the N- and C-terminal intracellular regions of PPK sequences. In order to provide the best candidates, this analysis was exclusively performed on PPKs with two transmembrane domains, CBMs longer than seven amino acids and normalized scores ≥8. Finally, protein sequences were aligned using the G-INS-I strategy in MAFFT v.7 (https://mafft.cbrc.jp/alignment/server, last accessed August 24, 2021) and different functional and conserved motifs, previously characterized for D. melanogaster (Liu et al. 2003) were identified in the alignment. In spite of these criteria applied to select PPK sequences for our data set, we consider that many of them might be truncated (as they were extracted from automatic gene prediction databases), and require manual curation for use in future functional studies. Nevertheless, we also consider that this should not significantly affect the quality of our data set considering the phylogenetic scope of our work.
Phylogenetic Analysis
Sequences were aligned with MAFFT using the G-INS-i strategy, and the following settings: unaligned level = 0.1; offset value = 0.12; maxiterate = 1,000; and the option “leave gappy regions.” The alignment was trimmed using trimAl v1.2 (Capella-Gutierrez et al. 2009) with default parameters except for the gap threshold that was fixed at 0.3. Following trimming, two phylogenetic trees were built. The first one was based on the maximum-likelihood approach by using IQ-Tree v 1.6.12 (Nguyen et al. 2015). The branch support was estimated using both the approximate likelihood ratio test (LRT) based on the Shimodaira–Hasegawa (aLRT-SH) and the ultrafast bootstrap or UFBoot (Hordijk and Gascuel 2005; Minh et al. 2013) procedures. The best-fit amino acid substitution model, reached by IQ-Tree, was WAG+F+R9. This model was chosen according to the Bayesian Information Criterion. The second maximum-likelihood phylogenetic tree was obtained using RAxML v8 (Stamatakis 2014) with the PROTCATWAG amino-acid substitution model for tree search and 200 replicates in the bootstrap analysis. Both phylogenetic trees were displayed and edited with FigTree (http://tree.bio.ed.ac.uk/software/figtree, last accessed August 24, 2021). The PPK candidates were annotated based on their relation with those of D. melanogaster. The nomenclature adopted in our study implies that candidates without a clear relationship with D. melanogaster PPK genes retained the original codes from their corresponding databases. Candidate sequences that did not group with any of the PPK families reported were excluded from the analysis (representing less than 1% of all candidate sequences preselected from the 26 genomes).
Selection Analysis
Codeml package from PALM4 (Yang 2007) was used to estimate the nonsynonymous to synonymous substitution rate (dN/dS) ratios (ω) across several PPK lineages following the methodology implemented in Almeida et al. (2014). Lineages bearing genes found in D. melanogaster and having more than four orthologs for each of them were included in the analysis. Exceptionally, the ppk2 and ppk19 lineages of M. domestica and the expansion identified for B. germanica in Subf-III were also included due to their corresponding expansions even though the previous criteria were not met for these lineages. We fitted different codon-based substitution models (M) using a maximum-likelihood approach. Initially, M0 was used to obtain branch lengths and mean ω. Then, we estimated model parameters and the log likelihood (L) of M7 and M8 and compared the L of these two models with the LRT (α = 0.05). To reduce the proportion of false positives of the M7 versus M8 comparison, we also used the M8a, which is considered an alternative null hypothesis for M8 where the highest ω of the distribution across sites is fixed to ω = 1. We assumed 2 and 1 degrees of freedom for the M8 versus M7, and the M8 versus M8a comparisons, respectively. Finally, the Bayes Empirical Bayes (Yang 2005) analysis under M8 was used to identify codons under positive selection. Sites were considered under positive selection when the PP of belonging to the class with ω > 1 was PP > 0.5.
Family Size Evolution
The evolution of PPKs was further analyzed by estimating gene birth and death rates. Each gene set was evaluated for the presence of 1,367 BUSCO genes representative of the Insecta BUSCO database (BUSCO completeness metrics provided in supplementary table S7, Supplementary Material online). Therefore, we obtained phylogenetic trees for each subfamily (using RAxML with 20 independent runs) using a matrix including the sequences of D. melanogaster, M. domestica, G. morsitans, A. aegypti, A. albopictus, Cul. quinquefasciatus, Ano. gambiae, Bombyxmori, Danausplexippus, T. castaneum, Api. mellifera, Cam. floridanus, P. humanus, Acyrthosiphonpisum, and Rhodniusprolixus. Besides using these phylogenetic trees for family size analyses, they were also used to resolve the evolution of PPK subfamilies (complementing the information offered by both trees built for the Phylogenetic Analysis section).
We estimated the number of gene duplications and losses by applying the gene tree versus species tree reconciliation method for each orthogroup as in Almeida et al. (2014). To obtain gene birth and death rates, we applied equations 1 (“total time” approach) and 2 (“branch average” approach) from Almeida et al. (2014), using previously published node divergence times (Vieira et al. 2007; Logue et al. 2013; Misof et al. 2014). Total rates comprise an average across branches, and take into account the number of duplications/losses “per” number of genes in the ancestral node, “per” branch length in time.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors wish to thank the Program for Technological Development in Tools for Health-PDTIS-FIOCRUZ for having facilitated the use of its facilities. Authors are indebted to INCTEM (Project Number: 465678/2014-9), CONFAP-MRC (Project Number: TEC—APQ-00913-16), FIOCRUZ, CNPq (Project Numbers: 308337/2015-8 and 311826/2019-9), Le Studium for a granting research fellowship to M.G.L. (Short Term Contract of Employment No: 2017-2001-179—Y17F16), and Agencia Nacional de Promoción Científica y Tecnológica (Project Number: PICT 2016-3103). J.M.L.E., F.C.A., G.P., H.D., and R.B.B. are CONICET researchers.
Author Contributions
J.M.L.E., F.C.A., and M.G.L. conceived the project and designed the experiments. J.M.L.E. and F.C.A. performed data analyses. All authors wrote the manuscript and provided comments on versions, read, and approved the final manuscript.
Data Availability
The data underlying this article are available in the article and in its online Supplementary Material.
Literature Cited
- Adams CM, et al. 1998. Ripped pocket and pickpocket novel Drosophila DEG/ENaC subunits expressed in early development and mechanosensory neurons. J Cell Biol. 140(1):143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida FC, Sánchez-Gracia A, Campos JL, Rozas J.. 2014. Family size evolution in Drosophila chemosensory gene families: a comparative analysis with a critical appraisal of methods. Genome Biol Evol. 6(7):1669–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson CJ, et al. 2002. Heteromultimers of DEG/ENaC subunits form H+-gated channels in mouse sensory neurons. Proc Natl Acad Sci U S A. 99(4):2338–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BernselA, , ViklundH, , HennerdalA, , Elofsson A.. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37(suppl 2):W465–W468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko N, Kucher V, Stockand JD, Eaton BA.. 2012.Pickpocket1 is an ionotropic molecular sensory transducer. J Biol Chem. 287(47):39878–39886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breer H, Fleischer J, Pregitzer P, Krieger J.. 2019. Molecular mechanism of insect olfaction: olfactory receptors. In: Picimbon J-F, editor. Olfactory concepts of insect control – alternative to insecticides. Switzerland: Springer International Publishing. p. 93–114. [Google Scholar]
- Canessa CM, et al. 1994. Amiloride-Sensitive Epithelial Na+ channel is made of three homologous subunits. Nature 367(6462):463–467. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey AF, Carlson JR.. 2011. Insect olfaction from model systems to disease control. Proc Natl Acad Sci U S A. 108(32):12987–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood AL, Goodman MB.. 2012. Insight into DEG/ENaC channel gating from genetics and structure. Physiology (Bethesda) 27(5):282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engsontia P, et al. 2008. The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol. 38(4):387–397. [DOI] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Chotigeat W, Satasook C.. 2014. Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: implications for their adaptation and speciation. J Mol Evol. 79(1–2):21–39. [DOI] [PubMed] [Google Scholar]
- Engsontia P, Sangket U, Robertson HM, Satasook C.. 2015. Diversification of the ant odorant receptor gene family and positive selection on candidate cuticular hydrocarbon receptors. BMC Res Notes. 8:380–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firsov D, Robert-Nicoud M, Gruender S, Schild L, Rossier BC.. 1999. Mutational analysis of cysteine-rich domains of the Epithelium Sodium Channel (ENaC): identification of cysteines essential for channel expression at the cell surface. J Biol Chem. 274(5):2743–2749. [DOI] [PubMed] [Google Scholar]
- García-Añoveros J, Ma C, Chalfie M.. 1995. Regulation of Caenorhabditis elegans degenerin proteins by a putative extracellular domain. Curr Biol. 5(4):441–448. [DOI] [PubMed] [Google Scholar]
- George LF, et al. 2019. Ion channel contributions to wing development in Drosophila melanogaster. G3 (Bethesda) 9(4):999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorczyca DA, et al. 2014. Identification of Ppk26, a DEG/ENaC channel functioning with Ppk1 in a mutually dependent manner to guide locomotion behavior in Drosophila. Cell Rep. 9(4):1446–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kim J.. 2007. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol. 24(5):1198–1207. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Han MV, Han SG.. 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3(11):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC.. 2011. Evolution of insect olfaction. PLoS Genet. 72(5):698–711. [DOI] [PubMed] [Google Scholar]
- Hong K, Driscoll M.. 1994. A transmembrane domain of the putative channel subunit MEC-4 influences mechanotransduction and neurodegeneration in C. elegans. Nature 367(6462):470–473. [DOI] [PubMed] [Google Scholar]
- Hordijk W, Gascuel O.. 2005. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 21(24):4338–4347. [DOI] [PubMed] [Google Scholar]
- Jaeger AH, et al. 2018. A complex peripheral code for salt taste in Drosophila. Elife 7:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger S, Schild L.. 2002. Epithelial Sodium Channel/Degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 82(3):735–767. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, et al. . 2010. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 107(27):12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Estivalis JM, et al. 2017. The molecular sensory machinery of a Chagas disease vector: expression changes through imaginal moult and sexually dimorphic features. Sci Rep. 7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, et al. 2003. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron 39(1):133–146. [DOI] [PubMed] [Google Scholar]
- Logue K, et al. 2013. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malar J. 12:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, et al. 2020. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48(D1):D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masagué S, Cano A, Asparch Y, Barrozo RB, Minoli S.. 2020. Sensory discrimination between aversive salty and bitter tastes in an haematophagous insect. Eur J Neurosci. 51(9):1867–1880. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Younger MA, Vosshall LB.. 2019. The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. Elife 8:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR, O’Meara BC.. 2007. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177(3):1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346(6210):763–767. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP.. 2005. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 39:121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R, et al. 2019. Amplification of Drosophila olfactory responses by a DEG/ENaC channel. Neuron 104(5):947–959.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Nei M.. 2007. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci U S A. 104(17):7122–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavković-Lučić S, Miličić D, Lučić L.. 2012. Insect hydrocarbons. biology, biochemistry, and chemical ecology. Insect Sci. 19(6):703–704. [Google Scholar]
- Pochynyuk O, et al. 2009. Intrinsic voltage dependence of the epithelial Na+ channel is masked by a conserved transmembrane domain tryptophan. J Biol Chem. 284(38):25512–25521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes G, Pereira MH, Barrozo RB.. 2017. Salt controls feeding decisions in a blood-sucking insect. J Insect Physiol. 98:93–100. [DOI] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F.. 1997. Sequence motifs for calmodulin recognition. FASEB J. 11(5):331–340. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Baits RL, Walden KKO, Wada-Katsumata A, Schal C.. 2018. Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattella germanica. J Exp Zool B Mol Dev Evol. 330(5):265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Wanner KW.. 2006. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 16(11):1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR.. 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100(Suppl 2):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Almeida FC, Rozas J.. 2011. Comparative genomics of the major chemosensory gene families in arthropods. In: Encyclopedia of life sciences. Chichester (United Kingdom: ): John Wiley & Sons, Ltd. [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Rozas J.. 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb). 103(3):208–216. [DOI] [PubMed] [Google Scholar]
- Schild L, Schneeberger E, Gautschi I, Firsov D.. 1997. Identification of amino acid residues in the α, β, and γ subunits of the Epithelial Sodium Channel (ENaC) involved in amiloride block and ion permeation. J Gen Physiol. 109(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JG, et al. 2014. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 15(10):466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C, Shi P, Butlin RK, Robertson HM.. 2009. Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol Biol Evol. 26(9):2073–2086. [DOI] [PubMed] [Google Scholar]
- Smadja CM, et al. 2012. Large-scale candidate gene scan reveals the role of chemoreceptor genes in host plant specialization and speciation in the pea aphid. Evolution 66(9):2723–2738. [DOI] [PubMed] [Google Scholar]
- Soffan A, Subandiyah S, Makino H, Watanabe T, Horiike T.. 2018. Evolutionary analysis of the highly conserved insect odorant coreceptor (ORCO) revealed a positive selection mode, implying functional flexibility. J Insect Sci. 18: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K.. 2012. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149(5):1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi A, Caldwell JC, Tracey WD.. 2012. Dendritic filopodia, ripped pocket, NOMPC, and NMDARs contribute to the sense of touch in Drosophila larvae. Curr Biol. 22(22):2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Rozas J.. 2011. Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 3:476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FG, Sánchez-Gracia A, Rozas J.. 2007. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 8(11):r235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizueta J, et al. 2020. Evolutionary history of major chemosensory gene families across Panarthropoda. Mol Biol Evol. 37(12):3601–3615. [DOI] [PubMed] [Google Scholar]
- Wanner KW, Robertson HM.. 2008. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol Biol. 17(6):621–629. [DOI] [PubMed] [Google Scholar]
- Xie J, Price MP, Wemmie JA, Askwith CC, Welsh MJ.. 2003. ASIC3 and ASIC1 mediate FMRFamide-related peptide enhancement of H+-gated currents in cultured dorsal root ganglion neurons. J Neurophysiol. 89(5):2459–2465. [DOI] [PubMed] [Google Scholar]
- Yang Z.2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22(4):1107–1118. [DOI] [PubMed] [Google Scholar]
- Yang Z.2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zelle KM, Lu B, Pyfrom SC, Ben-Shahar Y.. 2013. The genetic architecture of Degenerin/Epithelial Sodium Channels in Drosophila. G3 (Bethesda) 3(3):441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, Tracey WD.. 2010. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 20(5):429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online Supplementary Material.