Abstract
hsHec1p, a Homo sapiens coiled-coil-enriched protein, plays an important role in M-phase progression in mammalian cells. A Saccharomyces cerevisiae protein, identical to Tid3p/Ndc80p and here designated scHec1p, has similarities in structure and biological function to hsHec1p. Budding yeast cells deleted in the scHEC1/NDC80 allele are not viable, but this lethal phenotype can be rescued by hsHEC1 under control of the endogenous scHEC1 promoter. At the nonpermissive temperature, significant mitotic delay, chromosomal missegregation, and decreased viability were observed in yeast cells with temperature-sensitive (ts) alleles of hsHEC1. In the hshec1-113 ts mutant, we found a single-point mutation changing Trp395 to a stop codon, which resulted in the expression of a C-terminally truncated 45-kDa protein. The binding of this mutated protein, hshec1-113p, to five identified hsHec1p-associated proteins was unchanged, while its binding to human SMC1 protein and yeast Smc1p was ts. Hec1p also interacts with Smc2p, and the binding of the mutated hshec1-113p to Smc2p was not ts. Overexpression of either hsHEC1 or scHEC1 suppressed the lethal phenotype of smc1-2 and smc2-6 at nonpermissive temperatures, suggesting that the interactions between Hec1p and Smc1p and -2p are biologically significant. These results suggest that Hec1 proteins play a critical role in modulating chromosomal segregation, in part, through their interactions with SMC proteins.
Chromosome segregation during the cell division cycle results from the cooperation of many complex mechanisms. The physical segregation of pairs of sister chromatids into two daughter cells is modulated precisely during M phase. The ultimate goal of this process is to ensure high-fidelity transmission of replicated DNA to offspring. Each step during M-phase progression is coordinated by a group of structural and regulatory proteins. Many of these proteins are highly conserved in all eukaryotes from Saccharomyces cerevisiae to Homo sapiens.
The structural proteins required for the reorganization of chromosomes after replication have been identified and found to be essential for chromosome segregation. During this chromosomal reorganization process, linkage of duplicated DNA molecules, termed sister chromatid cohesion, is established, and paired sister chromatids undergo condensation. Sister chromatid cohesion and chromosome condensation are required for proper spindle attachments and chromosome movements. Moreover, the separation of paired chromatids is triggered by the dissolution of sister chromatid cohesion at the metaphase-anaphase transition. All of these steps must be precisely regulated to ensure the faithful transmission of chromosomes (19, 21, 24, 40, 41, 45, 53, 54, 56–59). A better understanding of the structural components involved in these chromatin reassembly processes has been achieved by the identification and characterization of the SMC (structural maintenance of chromosomes) protein family. SMC proteins, with members highly conserved from yeast to human, may be classified into four major subfamilies, SMC1- to SMC4-type proteins (24, 40, 56). SMC1- and SMC3-type proteins have been suggested to be essential for sister chromatid cohesion (21, 24, 41, 45, 56). On the other hand, chromosome condensation depends on SMC2- and SMC4-type proteins (24, 25, 27, 40, 53, 56, 57).
Proteins associated with kinetochore assembly are also essential for faithful chromosome segregation (33). Kinetochore proteins are responsible for attachments and movements of chromosomes along the microtubules of the mitotic spindle. Besides centromere-binding proteins, the components of microtubules, spindle pole bodies, microtubule-based motor proteins, and other microtubule-binding proteins are also required for the proper chromosome movement (2, 31, 42).
The structural dynamics of chromosomes must be coordinated with the cell cycle regulatory machinery to execute the precise order of mitotic events. Ubquitin-dependent proteolysis appears to be a major regulatory mechanism modulating M-phase progression as well as other mitotic events. Yeast genes including CDC16, CDC23, and CDC27, which encode components of the anaphase-promoting complex (34), and SUG1/CIM3 and CIM5, which encode the 26S proteasome subunits (4), play pivotal roles in the completion of mitosis (16, 34). The anaphase-promoting complex has been demonstrated to be essential for the degradation of mitotic cyclins (34, 38). Moreover, it controls the onset of sister chromatid separation and the metaphase-anaphase transition by degradation of specific regulators of chromosome transmission such as Pds1p, Cut2p, and Ase1p (10, 15, 30, 36, 39). cim3 and cim5 mutants have been shown to arrest yeast cells at G2/M phase, and a defect in the degradation of mitotic cyclins has also been proposed for these mutants (16).
Through our investigation of the molecular events of mitosis and the origin of the chromosomal abnormalities observed in malignant cells, a novel human nuclear protein, hsHec1p, was previously identified. hsHec1p was originally isolated as a retinoblastoma protein (Rb)-associated protein (13), and it apparently plays an important role in chromosome segregation in mammalian cells (8). Our studies showed that upon inactivation of hsHec1p by microinjection of hsHec1p-specific antibodies, chromosome congression is severely disturbed. Moreover, hsHec1p has been found to interact with a group of proteins important for G2/M progression (9), including sb1.8, the human SMC1 protein (49); p45/Trip1, the S8 subunit of the human 26S proteasome (4) and the human homolog of S. cerevisiae Sug1p/Cim3p; MSS1, the S7 subunit of the human 26S proteasome (4) and the human homolog of S. cerevisiae Cim5p; and NEK2, a human homolog of NIMA kinase (14). Collectively, these results have provided circumstantial evidence suggestive of a role for hsHec1p as a regulator of M-phase progression.
To elucidate how hsHec1p is involved in M-phase progression, a functional homolog of hsHec1p was identified in the budding yeast S. cerevisiae. This protein, previously isolated as Tid3p/Ndc80p (12, 61) and designated scHec1p here, can be complemented by hsHec1p. By expressing temperature-sensitive (ts) mutants of human hsHEC1 in yeast strains in which scHEC1 has been deleted, we demonstrate that Hec1p functions in chromosomal segregation, at least in part, through interactions with SMC proteins.
MATERIALS AND METHODS
Strains, reagents, and media.
Yeast strains are described in Table 1. Chemicals and medium components were purchased from Sigma and Difco Laboratories. Standard media were made as described elsewhere (47). S. cerevisiae strains used in this study were grown in complete medium (YPD; 1% yeast extract, 2% peptone, 2% dextrose) or in supplemented minimal medium (SMM) lacking appropriate amino acids. To induce the GAL1 promoter, yeast cells were grown at 25°C to log phase in complete medium or SMM containing 2% glucose and then shifted to the same medium containing 2% galactose. For all the other purposes, the yeasts were cultured in YPD or SMM containing 2% glucose. Yeast transformation, plasmid, and genomic DNA isolation have been described elsewhere (51).
TABLE 1.
Yeast strains used
| Strain | Genotype | Source |
|---|---|---|
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | P. Hieter |
| YPH501 | MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ | P. Hieter |
| WHL101 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 schec1Δ::URA3 hsHEC1 (YCpPA-HSHEC1::TRP1) | This study |
| WHL102 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 schec1Δ::hsHEC1::URA3 | This study |
| WHL103 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 schec1Δ::URA3 scHEC1 (YCpPA-SCHEC1::TRP1) | This study |
| WHL113 | MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 schec1Δ::URA3 hshec1-113 (YCpPA-hshec1-113::TRP1) | This study |
| WHL1713 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 schec1Δ::hshec1-113::URA3 (CFIII HIS3 SUP11) | This study |
| YPH1017 | MATα ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 (CFIII HIS3 SUP11) | P. Hieter |
| WHL9702 | MATa/α lys2-801/lys2-801 ura3-52/ura3-52 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 schec1Δ::URA3/schec1Δ::URA3 hsHEC1 (YcpPA-HSHEC1::TRP1) (CFIII HIS3 SUP11) | This study |
| WHL9713 | MATa/α lys2-801/lys2-801 ura3-52/ura3-52 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 schec1Δ::URA3/schec1Δ::URA3 hshec1-113 (YcpPA-hshec1-113::TRP1) (CFIII HIS3 SUP11) | This study |
| CH2080 | MATα lys2-801 ade2-101 trp1-Δ63 leu2-Δ1 | C. Holm |
| CH2082 | MATa lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 (CFIII TRP1 SUP11) | C. Holm |
| WHL2003 | MATa/α lys2-801/lys2-801 ade2-101/ade2-101 leu2-Δ1/leu2-Δ1 scHEC1/scHEC1 (CFIII TRP1 SUP11) | This study |
| 1-1bAS172 | MAT × lys2 his3 leu2 smc1-Δ2::HIS3 ura3::smc1-2 | D. Koshland |
| 2aAS283 | MAT × ura3 lys2 ade2 his3 leu2-Δ1 smc2-6 | D. Koshland |
Cloning and disruption of scHEC1.
The open reading frame (ORF) of scHEC1 and its upstream promoter region were amplified by PCR using yeast genomic DNA isolated from YPH499 as the template. The 2.0-kb DNA fragment of the coding region was subcloned into pBluescript SK (Stratagene), generating pBSK-SCHEC1. The 500-bp fragment of the promoter region with HindIII-EcoRI sites, together with a EcoRI/blunt DNA fragment containing the URA3 gene, were inserted into pBSK-SCHEC1 to replace the 1.5-kb scHEC1 coding region between HindIII and EcoRV, creating plasmid pBSK-SCHKO. The XhoI-BamHI fragment from pBSK-SCHKO containing the target allele, in which amino acids (aa) 1 to 549 of the scHEC1 ORF were replaced by the URA3 gene, was transformed into diploid strain YPH501 to disrupt the scHEC1 allele. Ura+ prototrophs were selected, sporulated, and dissected. To confirm the gene disruption, genotyping by PCR methods with primers outside the targeted region and primers inside of the URA3 genes were performed.
Replacement of the scHEC1 gene with hsHEC cDNA.
The same strategy was used to disrupt the scHEC1 allele in the haploid strain YPH499. To complement this scHEC1 disruption, YCpLac22 (17) was inserted by using the 500-bp fragment of the scHEC1 promoter region flanked with EcoRI-BamHI sites at ends; the 2.0-kb DNA fragment containing the complete ORF of scHEC1 was inserted in the BamHI site. Meanwhile, YCpLac22 was inserted by using the same 500-bp fragment of the scHEC1 promoter but flanked with EcoRI-HindIII sites, and the 2.0-kb hsHEC1 cDNA (8) was inserted in the HindIII site. The resultant constructs (YCpPA-SCHEC1 and YCpPA-HSHEC1, respectively) with the selection marker, TRP1, were transformed into YPH499 together with the scHEC1 disruption construct. Trp+ prototrophs were selected and genotyped by PCR to confirm the gene disruption and replaced into the given haploid strains. To replace the scHEC1 ORF with hsHEC1 cDNA directly on the chromosome, pBSK-FB was created by insertion of the HindIII fragment of hsHEC1 full-length cDNA driven by the scHEC1 promoter on plasmid pBSK-SCHKO. The KpnI-NotI fragment from pBSK-FB was transformed into YPH499, and Ura+ prototrophs were selected.
2μm or CEN plasmids (pGAL1-SCHEC1 or pGAL1-HSHEC1) with LEU2 selection, containing the GAL1-inducible promoter and the same expressing genes as in YCpPA-SCHEC1 and YCpPA-HSHEC1, or the vector (pGAL1) containing the GAL1 promoter alone, were constructed from a modified form of pGAD10 (Clontech, Palo Alto Calif.) or pOC29 (10). They were transformed into 1-1bAS172, 2aAS283, or YPH499 in the assay for suppressing the smc1-2 or smc2-6 mutants.
Generation of conditional mutant alleles of human hsHEC1 in yeast.
The ts mutant alleles were isolated as described elsewhere (1). The 1.8-kb BamHI-SalI fragment of hsHEC1 cDNA on plasmid YCpPA-HSHEC1 was treated with 1 M hydroxylamine at 75°C for 90 min to mutate the hsHEC1 cDNA randomly and then ligated to the YCpPA-HSHEC1 vector linearized by BamHI-SalI. Approximately 106 ampicillin-resistant Escherichia coli DH5α transformants were obtained from this ligation reaction. Mutagenized plasmid DNA was extracted from them and used to transform YPH499 together with the scHEC1 disruption fragment. About 5,000 transformants of Trp+ Ura+ prototrophs were selected on SMM plates. Each plate was duplicated; one was cultured at 25°C, and the other was cultured at 37°C. Eleven clones grew at 25°C but not at 37°C. The plasmids from these mutant clones were recovered and retransformed to verify the ts phenotype. One of the ts alleles of hshec1, hshec1-113, was sequenced with an Applied Biosystems model 377A sequencer. Yeast strains with an integrated hshec1-113 allele on the scHEC1 locus were generated by transforming the same KpnI/XhoI DNA fragment carrying the hshec1-113 mutation.
Cell growth and synchronization.
For the growth property studies shown in Fig. 1 and 2, a 24-h culture grown in YPD or SMM at 25°C was diluted with fresh YPD or SMM containing 2% glucose at a starting density of 4 × 106 cells/ml. Then cells were grown at 25°C with vigorous shaking. Change to the nonpermissive temperature occurred during log phase or immediately after release from synchronization. Aliquots of the culture were removed at the indicated times for determining growth densities, colony formation on solid plates, and individual cell morphology. Growth density was measured by spectrophotometric methods and converted to cell number according to standards measured on a hemacytometer.
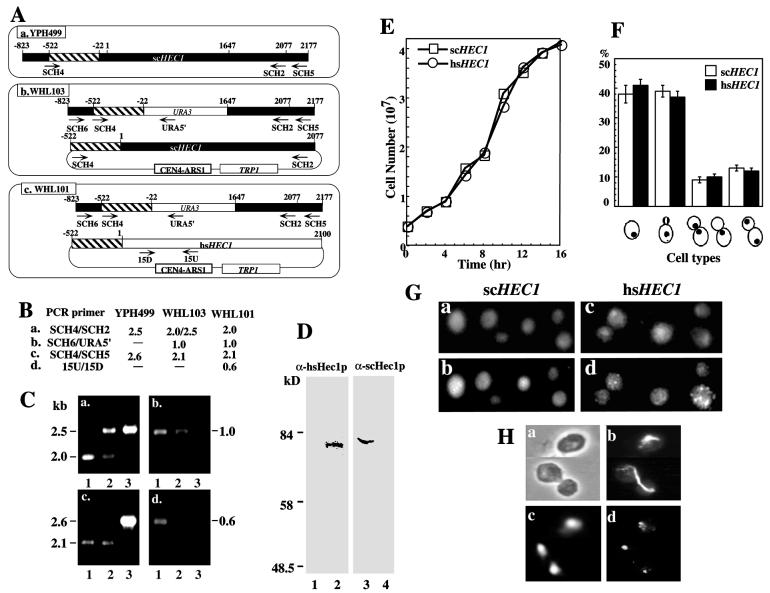
FIG. 1.
Yeast cells with the schec1 null mutation can be rescued by human hsHEC1 (A) Schematic diagram of wild-type cell (a. YPH499) and yeast cells rescued by scHEC1 (b. WHL103) or hsHEC1 (c. WHL101). The yeast scHEC1 promoter region is marked by hatched boxes; arrows indicate the primers used in PCR genotyping. (B) Predicted DNA fragment sizes determined by PCR genotyping using the primers shown in panel A. (C) DNA fragments of PCR genotyping analyzed by agarose gel electrophoresis. Lane 1, WHL101; lane 2, WHL 103; lane 3, YPH499. (D) Immunoblot analysis of total cell lysates from YPH499 (lanes 1 and 3) and WHL101 (lanes 2 and 4), using anti-hsHec1p MAb 9G3 and mouse anti-scHec1p sera, respectively. (E) Growth-density curve of the yeast strain carrying the scHEC1 allele (YPH499) and the isogenic yeast strain in which the scHEC1 allele is replaced by hsHEC1 (WHL102) at 25°C. (F) The cell type distributions of yeast grown at 25°C for 4 h in (E) were observed with fluorescence microscopy. The percentages of cells at different phases of the cell cycle were determined according to budding morphologies, nuclear patterns, and spindle formation, and presented as histograms. (G) Immunostaining of Hec1p in yeast cells carrying the scHEC1 (a and b) or hsHEC1 (c and d) allele. scHec1p was detected by mouse polyclonal anti-scHec1p antibodies (b); hsHec1p was detected by purified rabbit polyclonal anti-hsHec1p antibodies (d). Corresponding FITC-conjugated secondary antibodies were used. DNA was stained with DAPI (a and c). (H) Immunostaining of hsHec1p in yeast cells. a, phase-contrast; b, spindle stained by rat antitubulin antibodies and FITC-conjugated secondary antibodies; c, nuclear DNA strained by DAPI; d, hsHec1p stained by rabbit anti-hsHec1p antibodies and Texas red-conjugated secondary antibodies.
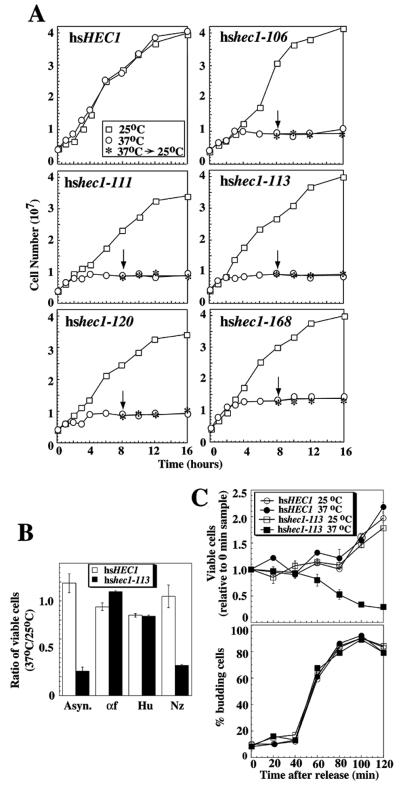
FIG. 2.
Temperature sensitivity of mutated hshec1 alleles. (A) Growth-density curves of yeast strains carrying different mutant alleles or the wild-type allele of hsHEC1 were measured by spectrophotometric methods. Log-phase cells originally growing at 25°C were inoculated and cultured at 25 or 37°C, respectively. After 8 h, half of the cultures grown at 37°C were shifted back to 25°C, as indicated by arrows. (B) Log-phase cells from hsHEC1 or hshec1-113 strains were arrested with α-factor at 5 μg/ml, hydroxyurea at 0.1 M, or nocodazole at 20 μg/ml or were left untreated for 3 h at 25°C. The percentages of arrested cells, determined by DAPI staining and observation by fluorescence microscopy, were between 85 to 95%. While arrested, half of each culture was shifted to 37°C and the remaining was kept at 25°C. After another 3-h incubation, cells were released and plated at 25°C. The ratios between the numbers of colonies formed by the cells exposed to 37°C and those by the cells exposed to 25°C are shown by histograms. Asyn., asynchronous; αf, α-factor; Hu, hydroxyurea; Nz, nocodazole. (C) Log-phase cells from hsHEC1 or hshec1-113 strains were arrested with α-factor at 5 μg/ml for 3 h at 25°C. Immediately after release, half of each culture was shifted to 37°C and the remaining half was kept at 25°C. The numbers of colonies formed by equal aliquots taken from these cultures every 20 min were counted from duplicated plates. Numbers of colonies formed for each sample relative to the 0-min sample were calculated to generate the curves in the upper panel. The lower panel shows the percentages of budding cells in each sample.
Synchronization of yeast cells at G1, at early S phase, or at metaphase was performed as follows. Cells were inoculated into YPD or SMM at 4 × 106 cells/ml, grown at 25°C for 6 h, and then treated either with α-factor at 5 μg/ml, hydroxyurea at 0.1 M, or nocodazole at 20 μg/ml for the periods of time indicated. Synchronized cultures were released from cell cycle arrest by quick spin followed by washing.
Immunostaining and fluorescence microscopy.
Cell preparation for immunostaining was modified from the standard procedures already described (48). Aliquots of cultured yeast containing approximately 107 cells were fixed by the addition of 16% EM-grade formaldehyde to a final concentration of 4% for 20 min, followed by incubation with yeast fixation buffer (40 mM potassium phosphate [pH 6.5] containing 0.5 M MgCl2 and 4% formaldehyde) for 24 h. Fixed cells were washed twice with solution A (40 mM potassium phosphate [pH 6.5], 0.5 M MgCl2, 1.2 M sorbitol). Cells were spheroplasted in 1 ml of solution A containing 10 μl of 2-mercaptoethanol and 55 μl of Glusulase (Du Pont NEN) for 2 h at 36°C with gentle agitation. Cells were washed twice in solution A and resuspended in 0.2 ml of solution A. Twenty microliters of this suspension was spotted into a gelatin-coated cover slide. The immunostaining procedures were as described elsewhere (8). Microtubules were detected by monoclonal antitubulin antibody YOL134 (Accurate Scientific) diluted 1:50, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-rat immunoglobulin G (Fisher) diluted 1:100. hsHec1p or hshec1-113p was detected by anti-hsHec1p monoclonal antibody (MAb) 11A5 (1:250) or affinity-purified rabbit polyclonal anti-hsHec1p antibodies (1:250 to 1:500), and scHec1p was detected by mouse polyclonal anti-scHec1p antibodies (1:250), followed by FITC- or Texas red-conjugated secondary antibodies as indicated. The cells were further stained with 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) per ml in phosphate-buffered saline (PBS) containing 1 mM p-phenylenediamine and mounted in Permafluor (Lipshaw-Immunon Inc.). Cells were observed using a standard fluorescence microscope (Axiophot photomicroscope; Zeiss).
Flow cytometry.
Cells were prepared for flow cytometry as described elsewhere (23). Cells were resuspended in PBS and fixed in 70% ethanol. After being washed twice in 50 mM Tris (pH 8.0), cells were treated with RNase (1 mg/ml) for 2 h at 37°C and then incubated in proteinase K (40 μg/ml) for another 1 h at 50°C. Cells were pelleted and resuspended in propidum iodide (0.2 mg/ml in PBS). Before analysis, cells were sonicated for 5 s at the lowest setting. Fifty thousand cells from each sample were analyzed by FACScalibur (Becton Dickinson).
Generation of polyclonal antibodies against scHec1p or Smc1p.
scHec1p was detected by mouse anti-Hec1p polyclonal antisera raised against a glutathione S-transferase–scHec1p fusion protein expressed in E. coli.
Two kinds of mouse anti-Smc1p polyclonal antisera, anti-Smc1B and anti-Smc1C, were raised against a glutathione S-transferase protein fused with aa 634 to 847 and aa 1021 to 1225, respectively, of Smc1p. Specificities of the antibodies compared with that of rabbit anti-Smc1p antisera (kindly provided by A. Strunnikov) (58), were checked. The antibodies recognized a 165-kDa cellular protein of the same size as described for Smc1p. The assays represented in Fig. 6 were repeated using rabbit anti-Smc1p antisera (data not shown).
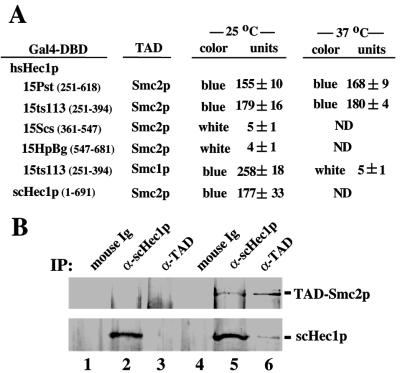
FIG. 6.
Hec1p interacts with Smc2p. (A) Domain mapping and temperature sensitivity of the interaction between Hec1p and Smc2p. The same constructs of Hec1p as used for Fig. 5 were used in yeast two-hybrid assays to test their binding ability to yeast Smc2p expressed as a Gal4 TAD fusion protein. Binding ability was tested at 25 or 37°C. Gal4-DBD, Gal4 DNA-binding domain; ND, not determined. (B) In vivo interaction between Hec1p and Smc2p. Equal amounts of cell lysates from the log-phase culture of strain Y153 alone (lanes 1 to 3) or Y153 expressing TAD-Smc2p (lanes 4 to 6) were immunoprecipitated (IP) by the antibodies indicated. The resultant immunocomplexes were separated by SDS-PAGE followed by immunoblotting. The upper panel was blotted by polyclonal anti-TAD antibodies, and the lower panel was stained by anti-scHec1p. Ig, immunoglobulin.
Immunoprecipitation and Western blotting.
Yeast cell lysates were prepared as described elsewhere (51), with the following modifications. Cell pellets were washed once and resuspended in 200 μl of yeast lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) plus protease inhibitors. Two volumes of glass beads (0.5-mm diameter) were added, and the suspensions were vortexed at the highest speed for six periods totaling 3 min. The clarified lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting analysis as described previously (8). hsHec1p was detected by anti-hsHec1p MAb 9G3. For immunoprecipitation, the lysates were incubated with anti-scHec1p (diluted 1:500), 9G3 (1:2,000), anti-Smc1C (1:300), or anti-Gal4 transactivation domain (TAD; 1:100; Santa Cruz) at 4°C for 3 h; then protein A-Sepharose beads were added. After another 2 h of incubation, beads were washed with lysis 250 buffer (50 mM Tris [pH 7.5], 250 mM NaCl, 50 mM sodium fluoride, 0.5% Nonidet P-40) for immunoprecipitation or lysate 175 (175 mM NaCl) for coimmunoprecipitation. Finally, beads were boiled in 2× SDS sample buffer and subjected to SDS-PAGE followed by immunoblotting analysis with the antibodies indicated.
Colony sectoring assay.
hsHEC1 and hshec1-113 diploid strains for colony sectoring assay were mated from two haploid strains, YPH499 (Mata) and YPH1017 (Matα) (Table 1), in which scHEC1 was deleted and rescued by hsHEC1 or hshec1-113 in the YCp-PA vector as described above. The resultant diploid strains contain a homozygous schec1 null mutation marked by URA3, a homozygous ade2-101 ochre color mutation, a chromosome fragment carrying a copy of SUP11, and a wild-type allele of hsHEC1 or a ts allele hshec1-113 in a CEN-ARS1 vector marked by TRP1. The scHEC1 diploid strain used in this assay was mated from two wild-type haploid strains, CH2082 and CH2080 (29). Five single pink colonies of each diploid strain were picked and cultured in histidine-free SMM at 25°C for 3 days. Equal numbers of cells from each strain were inoculated at 37 or 25°C into fresh histidine-containing SMM and then cultured further for 3 h before plating on medium containing 6 mg of adenine per liter. The plates were incubated at 25°C for 6 days and 4°C overnight before observation.
RESULTS
hsHEC1 can functionally substitute for yeast scHEC1.
Using the hsHec1p sequence, we identified in a search of the current GenBank protein database a potential homologous protein, scHec1p, encoded by an ORF in the S. cerevisiae genome. Interestingly, this gene has been independently isolated twice, once as TID3 in a screen for proteins interacting with S. cerevisiae Dmc1p (13) and again as NDC80 in an attempt to identify spindle pole components. Ndc80p was previously proposed to be a potential homolog of hsHec1p, based on structural similarity (61). Consistently, we found that human and yeast Hec1p are 36% identical in their N-terminal regions, and their C-terminal regions constitute similar coiled-coil enriched structures. Such homologous sequences were also identified in fission yeast (61), Caenorhebditis elegans, and mouse genomes (data not shown).
Inactivation of NDC80 has been shown to be lethal in budding yeast (61), which is consistent with our previous finding that hsHEC1 is essential for cell viability since inactivation of hsHec1p in mammalian cells by specific antibodies leads to cell death. The structural and functional similarities between hsHec1p and scHec1p/Ndc80p indicated that scHec1p may be the hsHec1p homolog. Because of a marked overall structural divergence between the two proteins, however, it was necessary to test whether hsHEC1 could functionally substitute for scHEC1 in yeast cells. To this end, the endogenous scHEC1 was inactivated by one-step gene disruption (53). The URA3 marker gene, flanked by a 0.5 kb of 5′ sequence from the putative promoter region of scHEC1 and 1.6 kb of 3′ sequence of scHEC1 (Fig. 1A), was first used to replace one wild-type allele of scHEC1 in a diploid strain, YPH501. The mutated diploid strain exhibited segregation of two viable (Ura−) and two lethal spores by tetrad analysis. Similar scHEC1 disruption was then performed with a haploid strain, YPH499. No yeast colonies survived after disruption of the scHEC1 allele; however, cotransformation of the scHEC1 disruption fragment with scHEC1 DNA in a TRP1-marked CEN-ARS1 vector rescued the lethal phenotype and generated strain WHL103 (Table 1; Fig. 1A). We therefore introduced the hsHEC1 cDNA, in a TRP1-marked CEN-ARS1 vector, into selected haploid yeast cells in which the scHEC1 allele was disrupted (Fig. 1A). Expression of hsHEC1 cDNA, under transcriptional control of the yeast scHEC1 promoter, was able to rescue the lethal phenotype resulting from scHEC1 disruption, thereby generating the strain WHL101 (Table 1; Fig. 1A). Similar results were obtained when hsHEC1 was integrated into the scHEC1 locus to generate strain WHL102 (Table 1). Genotyping by PCR methods confirmed the replacements (Fig. 1B and C). Yeast cells in which scHEC1 was disrupted and reconstituted by hsHEC1 expressed only hsHec1p; endogenous scHec1p was no longer detected (Fig. 1D).
To determine whether the hsHec1p can functionally substitute for scHec1p during yeast cell cycle progression, growth properties as well as cell morphology, spindle shape, and DNA content of the parental and rescued yeast strains were examined. Colony formation in solid plates and growth kinetics in liquid culture appeared to be indistinguishable in the strains with scHEC1 or with hsHEC1 at 25°C (Fig. 1E). Fluorescence microscopy of cells stained with antitubulin antibodies and DAPI demonstrated similarities in morphology, spindle shape, and DNA content in the two strains; the distributions of cells in different phases of the cell cycle were also indistinguishable in these two strains (Fig. 1F). Neither the growth properties nor the morphologies of cells expressing hsHec1p were changed upon cultured at 37°C (data not shown). These results demonstrated that hsHec1p is indeed the functional homolog of scHec1p. Moreover, two yeast strains in which scHEC1 is replaced by the hsHEC1 gene in either an episomal form (WHL101) or an integrated form (WHL102) were established for further studies.
Hec1p localizes to the nucleus.
Our previous study showed that hsHec1p localized to the nucleus and that a portion of it moved to the centromere regions during mitosis (8). To determine whether hsHec1p localizes similarly in yeast cells, we performed immunostaining of Hec1p in yeast cells carrying hsHEC1 or scHEC1. As shown in Fig. 1G, a nuclear staining pattern was observed in yeast cells carrying hsHEC1, and brighter speckles can be seen in nuclei. Costaining of spindles suggested that these brighter speckles were concentrated in the vicinity of the spindle pole body and confined to the periphery of the chromosomal DNA mass in unbudded or large budded anaphase cells (Fig. 1H). This pattern reflected the position of the centromeric DNA in these cells, as previously revealed by fluorescence in situ hybridization (20, 55), and was similar to the staining of centromere proteins, such as Mif2p (43), Cse4p (44), and Ctf19p (32). Consistently, the nuclear staining pattern and brighter speckles were also seen in the cells carrying scHEC1 (Fig. 1G).
Isolation of ts mutants of hsHEC1.
The yeast strain in which the essential function is sustained by the hsHEC1 gene has allowed us to explore the precise role of this human protein in mitotic division through isolating conditional alleles of hsHEC1 in yeast. We performed random mutagenesis on an hsHEC1 cDNA fragment, subcloned the resultant fragments in the CEN-ARS1 vector, and transformed the scHEC1-disrupted strain for isolation of conditional lethal mutants. Eleven ts mutants which grew at 25°C but not 37°C were isolated, and their growth in liquid culture was examined. At 25°C, the growth pattern of the mutant yeast appears no different from that of the wild-type strain (WHL101). When the cultures were shifted to 37°C, however, the ts mutants stopped growing after the first or second division (Fig. 2A). Like mammalian cells in which hsHec1p is inactivated by specific antibodies, yeast cells without functional hsHec1p fail to arrest completely during mitosis and instead apparently proceed to cell division and cytokinesis. These mutant cells did not regain growth potential when shifted from the nonpermissive to the permissive temperature. Samples from these mutants cultured at 37°C for 3 h (approximately 1.5 generation times) were plated and incubated at 25°C. Only 30 to 40% of cells were able to form colonies on solid plates, whereas the wild-type strain showed no loss of viability at 37°C. These results suggested that mutations of hsHEC1 lead to cell death at the nonpermissive temperature.
To determine whether the lethality at the nonpermissive temperature is specifically due to the failure of a cell cycle event, one of the ts mutants, hshec1-113, was further examined. Treatment of exponentially growing cultures of hshec1-113 cells or cogenic wild-type hsHEC1 cells with α-factor, hydroxyurea, or nocodazole at the permissive temperature resulted in cell cycle arrest at G1, early S, or G2/M phase, respectively. While arrested, these cells were shifted to 37°C for 3 h to inactivate hsHec1 proteins and then released by plating at 25°C. As controls, equivalent numbers of the arrested cells were cultured at 25°C for the same period of time. The ratio of viable cells at the nonpermissive temperature to those at the permissive temperature was used to determine viability. When the hsHec1p activity was depleted in G2/M phase, this ratio dropped to 0.3, or approximately the same as the ratio in unsynchronized cells (Fig. 2B). In contrast, when the hsHec1p activity was depleted in either G1 or S phase, the cell viability did not decrease. These results suggested that the function of hsHec1p is essential for cell viability during M phase. Increased lethality during M phase has also been observed in cells defective in proteins involved in sister chromatid cohesion or chromosome condensation, including Top2p (28), Smc1p (58), and Mcd1p (22). This resemblance is consistent with a relationship between Hec1p and these proteins suggested below.
The function of hsHec1p in M phase was further determined by following the viability of cells as they synchronously traversed a dynamic cell cycle (28). The hshec1-113 mutant and wild-type hsHEC1 strains were synchronized in G1 phase by α-factor at 25°C for 3 h and then released to fresh medium at 37 and 25°C as controls. At 20-min intervals, equal portions of the 37°C culture were shifted to 25°C and plated immediately to assess cell viability (Fig. 2C; upper panel). Cell cycle progression was monitored according to cell-budding morphologies (Fig. 2C; lower panel) and DNA content analysis (data not shown). During the initial 60 min, the viability of both strains at both temperatures remained relatively constant. After 60 min, however, the number of viable cells from the hshec1-113 strain grown at 37°C began to decrease. At 100 to 120 min after release from G1 phase, the viability of the hshec-113 culture at 37°C continued to decrease; in contrast, the numbers of viable cells from all the other cultures doubled because their buds were released from the mother cells. Thus, death of the mutant cells could be prevented by a shift back to the permissive temperature during the initial 60 min, suggesting that the essential role of hsHec1p was not required during that period of time immediately following release from G1. However, at 80 to 100 min after release (i.e., during M phase), cell death could no longer be prevented in cells containing inactivated hsHec1p. These results suggested that the essential function of hsHec1p is required when cells enter M phase.
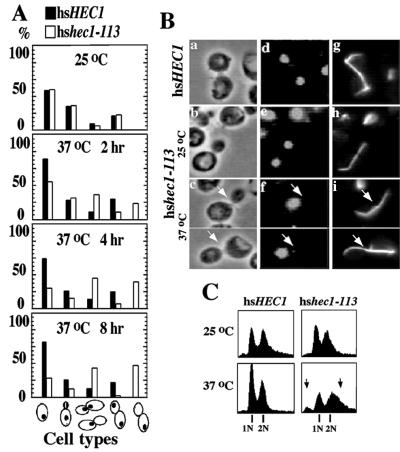
hshec1 mutants demonstrate mitotic delay and unequal chromosome segregation at the nonpermissive temperature.
Cytological analysis of the hshec1 mutant revealed accumulation of large-budded cells at the nonpermissive temperatures (Fig. 3A). The subpopulation of cells at anaphase consisted predominately of the cells in which DNA failed to separate into two discrete masses. Some cells with large buds contained DNA evident only at one pole (Fig. 3B). DNA content analysis showed that hshec1-113 mutant cells accumulated with a G2 DNA content after shifting to 37°C; the shoulder of the G2 peak was extended to reflect a DNA content greater than 2n, and a small peak representing DNA content less than 1n appeared. This profile suggested the cells with the hshec1-113 allele become aneuploid at the nonpermissive temperature (Fig. 3C). Therefore, the phenotypes of hshec1 mutants are very similar to those observed in mutants of its yeast homolog (61), suggesting that human hsHec1p also plays a role in chromosome segregation.
FIG. 3.
Phenotypes of ts hshec1-113 mutants. (A) Characterization of cell cycle progression of hshec1-113 and wild-type alleles. Unsynchronized log-phase cells originally growing at 25°C were shifted to 37°C. At 2, 4, and 8 h, 300 to 500 cells were categorized and placed in different phases of cell cycle according to their budding morphologies, nuclear patterns, and spindle formation under microscopic observation. The percentages of different types of cells are shown by histograms. (B) Morphologies of hshec1-113 cells growing at 25°C (b, e, and h) and 37°C (c, f, and i) for 4 h and of wild-type hsHEC1 cells growing at 37°C (a, d, and g). (C) DNA content analysis of unsynchronized log-phase cells carrying a wild-type hsHEC1 or mutant hshec1-113 allele. Cells were cultured at 25 or 37°C for 6 h. Arrows indicate shoulders representing DNA content of less than 1n or more than 2n. (D) Morphologies of synchronized hshec1-113 (g to l) and wild-type (a to f) cells after release from S phase for 1 and 2 h. The cells were synchronized with 0.1 M hydroxyurea for 5 h at 25°C and shifted to 37°C when released. (E) DNA content analysis of the cells shown in panel D, before release (0 h) and 1, 2, and 3 h after release. Note that 2 h after release, wild-type cells exited from M phase and exhibited a 1n peak, but hshec1-113 cells did not exhibit such a peak (arrowheads). In panels B (a to c) and D (a, d, g, and j), cell morphology is shown by phase-contrast. DNA was stained with DAPI (B, d to f; D, b, e, h, and k). The spindle was stained by antitubulin antibodies and FITC-conjugated secondary antibodies (B, g to i; D, c, f, i, and l). The large-budded cells with unevenly divided nuclei are marked by arrows.
Cell cycle progression of hshec1-113 mutant cells was monitored following synchronization with 0.1 M hydroxyurea at 25°C and release to fresh cultures at 37°C (Fig. 3D). Both wild-type and mutant cells entered M phase 1 h after release from hydroxyurea-induced S-phase arrest. After 2 h, however, only the wild-type cells exited from M phase. The hshec1-113 cells were still dominated by large-budded morphologies. Cells with unequal nuclear division and disperse DNA morphology were also observed. Results from DNA content analysis of the same cell samples were consistent with the cytological studies (Fig. 3E). A mitotic delay in hshec1-113 cells was thus suggested.
hshec1 mutations increase the rate of chromosome missegregation.
Both cytological observations and DNA content analysis suggested that mutations of HEC1/NDC80 cause a defect in chromosome segregation. To examine the rates at which this defect occurred in an hshec1 mutant, we used a colony sectoring assay in which diploid strains carried a homozygous ade2 (ochre) color mutation and a single copy of ochre suppressor (SUP11) on a dispensable chromosome fragment (CF) (26). Exponentially growing cells from hsHEC1 and hshec1-113 diploid strains were cultured at 25 or 37°C for 3 h and then plated at the permissive temperature. The total numbers of pink colonies representing 1:1 segregation, half-pink/half-red sectored colonies representing 1:0 segregation of CF, and half-white/half-red sectored colonies representing 2:0 segregation of CF were determined. The rates of chromosome loss and nondisjunction in the first division were determined by the frequencies of half-pink/half-red colonies and half-white/half-red colonies, respectively. Results suggested that the diploid strain carrying the wild-type hsHEC1 allele exhibited very low rates of both chromosome loss and nondisjunction, which were indistinguishable from those for the strain carrying the wild-type scHEC1 allele (Table 2). The diploid strain carrying mutant hshec1-113 allele at 25°C, however, exhibited a 65-fold increase in the rate of chromosome loss and an 18-fold increase in nondisjunction compared to that carrying the wild-type alleles. After transient exposure to 37°C, the rate of chromosome missegregation was increased further. However, this increment was not dramatic, presumably because of excessive cell death at the nonpermissive temperature. These results showed that hshec1-113 mutant strain has missegregated chromosomes; the more severe phenotype represents chromosome loss and the moderate phenotype nondisjunction events.
TABLE 2.
Chromosome loss and nondisjunction in hshec1-113 diploids
| Genotype | Temp (°C) | No. of coloniesa | Missegregation rate
|
|
|---|---|---|---|---|
| % 1:0 eventsb | % 2:0 eventsc | |||
| scHEC1 | 25 | 10,021 | 0.02 | 0.01 |
| hsHEC1 | 25 | 6,096 | 0.02 | 0.01 |
| 37 | 6,827 | 0.03 | 0.01 | |
| hshec1-113 | 25 | 4,299 | 1.35 | 0.37 |
| 37 | 5,138 | 1.85 | 0.41 | |
Total number of pink colonies that carry one chromosome fragment.
Number of half-red/half-pink colonies divided by the total number of pink colonies.
Number of half-red/half-white colonies divided by the total number of pink colonies.
Although defects in DNA metabolism may lead to large increases only in the rate of chromosome loss (47), the hshec1-113 mutant predominantly has a 2n DNA content at the nonpermissive temperature, making it unlikely that a failure of bulk DNA synthesis is responsible for the increasing rate of chromosome loss. Interestingly, the observed phenotype is consistent with roles of several key proteins in the process of chromosome segregation. For example, one member of a family of ctf (chromosome transmission fidelity) mutants, spt4, exhibits a similar increase in chromosome loss events, apparently due to a defect in kinetochore function (3). Another member, ctf13-30, has abnormal kinetochores and displays a comparable increase in both chromosome loss and nondisjunction events (11).
hshec-113 contains a nonsense mutation and encodes a 45-kDa truncated protein.
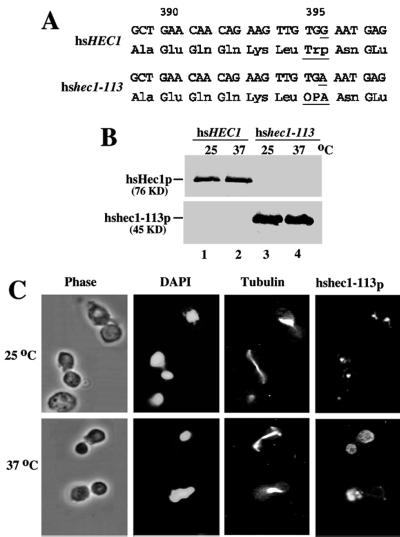
To determine the mutations of hsHEC1 leading to the ts phenotype, the plasmid DNA containing the hshec1-113 mutant allele was recovered from the yeast mutant strain WHL113 for sequence analysis. The hshec1-113 mutant allele was found to have a nonsense mutation, changing Trp395 to a stop codon in its translational sequence and resulting in the expression of a C-terminally truncated, 45-kDa protein (Fig. 4A). To further substantiate this finding, the hshec1-113 yeast lysates were analyzed by immunoprecipitation followed by Western blot analysis using an anti-hsHec1p MAb as probe. A 45-kDa protein, hshec1-113p, consistent with the predicted size, was detected in the mutant cells (Fig. 4B). The relative abundance of hshec1-113p did not appear to be significantly different at either 37 or 25°C. The ts phenotypes are, therefore, probably not the result of changes in protein expression at the nonpermissive temperature.
FIG. 4.
Characterization of the mutant hshec1-113p. (A) Partial nucleotide and protein sequences of wild-type hsHEC1 and mutant hshec1-113 alleles. The nucleotide substitution in hshec1-113p (G to A) and the corresponding amino acid change at position 395 (Trp to a stop codon OPA) are underlined. (B) Identification of hshec1-113p in mutant cells. Cell lysate prepared from wild-type (lanes 1 and 2) and hshec1-113 (lanes 3 and 4) cells growing at 25°C (lanes 1 and 3) or 37°C (lanes 2 and 4) for 4 h were immunoprecipitated by mouse anti-hsHec1p MAbs, followed by SDS-PAGE and Western blotting with the same antibodies. (C) Subcellular localization of the mutant hshec-113p. Log-phase cells of hshec1-113 were grown at 25°C or shifted to 37°C for 2 h. hshec-113p was stained by purified rabbit anti-hsHec1p antibodies and Texas red-conjugated secondary antibodies. Nuclear DNA of the same cells was stained by DAPI; spindle was stained by rat antitubulin antibodies and FITC-conjugated secondary antibodies.
The staining pattern of hshec1-113p at 25°C was not distinguishable from that of wild-type Hec1p (Fig. 4C). The brighter speckles of hshec1-113p appeared to be restricted to the region proposed for the centromere localization. However, at 37°C, these brighter speckles were no longer confined to this region but randomly distributed. It remains to be explored whether this abnormal pattern is reminiscent of the mislocalization of mutant hec1p or a rearrangement of centromeres in the mutant cells.
hshec1-113p interacts with hsHec1p-associated proteins but not with SMC1 proteins at the nonpermissive temperature.
hsHec1p has been shown to interact with several proteins important for M-phase progression by yeast two-hybrid screening (9). To test whether scHec1p can bind to the hsHec1p-associated proteins, a yeast two-hybrid assay was used to score for β-galactosidase activity. The results showed that scHec1p can indeed interact with most of the hsHec1p-associated proteins, including sb1.8, p45, NEK2, and other novel proteins (Table 3). Interestingly, scHec1p does not interact with human Rb and MSS1; therefore, the interaction of Hec1 proteins with these proteins may be restricted to higher organisms.
TABLE 3.
Interaction between scHec1p and hsHec1p-associated proteins
| hsHec1p-associated protein | Binding with hsHec1pa (Ub) | Binding with scHec1p
|
Homolog | |
|---|---|---|---|---|
| (Ub) | Color | |||
| MSS1 | 371.5 ± 19.3 | White | Cim5p | |
| 15A2 | 468.9 ± 65.8 | 1,066 ± 40 | Blue | |
| sb1.8 | 273.3 ± 10.0 | 348 ± 20 | Blue | Smc1p |
| NEK2 | 239.4 ± 32.6 | 126 ± 2 | Blue | NIMA |
| 15A20 | 319.1 ± 16.6 | 309 ± 1 | Blue | |
| Subunit p45 of 26S proteasome | 1,105.2 ± 159.3 | 122 ± 2 | Blue | Sug1p/Cim3p |
| Rb | 8,345 ± 3,086 | White | ||
Some data adapted from reference 9.
β-Galactosidase activity units.
Since most hsHec1p-associated proteins appear to have homologs in budding yeast, it is reasonable to postulate that hsHec1p may interact with the yeast homologs of these HEC-associated proteins. Mutant hshec1-113p contains the first complete coiled-coil domain but is missing most of the second domain. The first domain is sufficient for binding to many associated proteins (NEK2, sb1.8, and p45) but is insufficient for binding to MSS1 (9). The interaction between the truncated hshec1-113p and hsHec1p-associated proteins was tested at either 25 or 37°C. Mutant derivatives containing the first coiled-coil domain and a small part of the second domain (aa 251 to 394), which resembled the truncated C-terminal coiled-coil region of hshec1-113p, were able to interact with all of the associated proteins assayed at 25°C. However, the interaction with sb1.8 was specifically abolished at 37°C, while binding to other proteins was not changed (Fig. 5A). This result suggested that mutant hec1-113p may interact with yeast Smc1p in a ts manner similar to its binding to the human SMC1 protein in the yeast two-hybrid assay.
FIG. 5.
The interaction between the mutated hshec1-113p and Smc1p is ts. (A) Binding between hsHec1-113p and hsHec1p-associated proteins (hsHec1p-Aps) at permissive and nonpermissive temperatures. 15ts113, the cDNA encoding the deletion mutant (aa 251 to 394) of hsHec1p that is truncated at the same position as the hshec1-113p mutant, was generated by in-frame fusion to the Gal4 DNA-binding domain. The same constructs in Table 3 were used to express hsHec1p-associated proteins that were fused with the Gal4 TAD. The interactions at 25 or 37°C were tested in yeast two-hybrid assays. Units represent β-galactosidase activity. (B) Domain mapping of the interaction between Hec1p and SMC1 proteins. The schematic structures of the regions in Hec1p that were fused with the Gal4 DNA-binding domain are shown. Shaded boxes are the three leucine heptad repeat-enriched regions. sb1.8/human SMC1 and Smc1p/yeast SMC1 were expressed as Gal4 TAD fusion proteins and used to test for interaction with Hec1 fusion proteins in yeast two-hybrid assays. (C) Log-phase yeast cells carrying an hshec1-113 or hsHEC1 allele were transferred to fresh medium and grown at either 25 or 37°C. After 3 h, cells were collected and lysed, and equal amounts of protein extracts (about 15 mg) were immunoprecipitated (IP) with mouse preimmune serum (lane 1 to 4) or anti-hsHec1p MAb 9G3 (lane 5 to 8). The immunoprecipitates were then resolved by SDS-PAGE followed by immunoblotting with mouse anti-Smc1p antiserum for Smc1p (a), 9G3 for hsHec1p (b), or hshec1-113p (c). Equal amounts of the same lysates (about 2 mg) were separated by SDS-PAGE followed by immunoblotting with anti-Smc1p antiserum for detecting the expression of Smc1p (d, lanes 9 to 12). Lanes 1, 5, and 9, hsHEC1 at 25°C; lanes 2, 6, and 10, hsHEC1 at 37°C; lanes 3, 7, and 11, hshec1-113 at 25°C; lanes 4, 8, and 12, hshec1-113 at 37°C.
To determine which region(s) of Hec1p is necessary for interaction with the SMC1 protein, four hsHec1p deletion derivatives containing different coiled-coil regions fused to the Gal4 DNA-binding domain as previously described (9) were used in the yeast two-hybrid assay. The fragment of the human SMC1, sb1.8, originally cloned from yeast two-hybrid screening contains a 0.6-kb region (aa 621 to 836) encompassing the second coiled-coil domain. The putative homologous region of this 0.6-kb fragment (aa 634 to 847) of the yeast Smc1p, corresponding to the second coiled-coil domain, was also cloned. These two fragments, fused in frame to the Gal4 TAD, were used in the yeast two-hybrid assay. Both the first (aa 251 to 431) and the second (aa 361 to 547) coiled-coil regions of Hec1p are sufficient for interaction with human or yeast SMC1. The remaining region (aa 547 to 618), in which the coiled coil domain is poorly formed, although enriched in leucine heptad repeats, is not able to bind SMC1 (Fig. 5B). Similarly, the scHec1p appears to interact with both yeast and human SMC1.
The specific interaction between Hec1p and yeast Smc1p allows us to test whether the binding of the mutated hshec1-113p to Smc1p is temperature dependent in yeast cells. In assays using anti-hsHec1p MAbs, Smc1p was coimmunoprecipitated from the hshec1-113 mutant cells cultured at the permissive temperature. Little Smc1p, however, was detectable in the immunocomplex precipitated by anti-hsHec1p antibodies from the cells cultured at the nonpermissive temperature (Fig. 5C). This result further confirmed that hshec113p fails to interact with Smc1p at nonpermissive temperatures, although the abundance of Smc1p is not changed.
Hec1p also interacts with Smc2p.
The study of Xenopus condensin and cohesin complexes suggests that the SMC proteins of the SMC1/SMC3 subgroup and the SMC2/SMC4 subgroup have distinct functions in sister chromatid cohesion and chromosome condensation, respectively (41). Studies of S. cerevisiae, however, indicated that there are certain links between cohesion and condensation in certain organisms (21, 24, 56). Since Hec1p is associated with SMC1, it is possible that Hec1p is associated with other SMC proteins; in yeast, this association may also extend to condensation proteins such as Smc2p. To test this hypothesis, the same hsHec1p deletion derivatives were used to test their ability to bind to Smc2p. The full-length coding region of SMC2 was cloned by PCR amplification from S. cerevisiae genomic DNA and fused to the Gal4 TAD (aa 768 to 881). As shown in Fig. 6A, the first coiled-coil-enriched region (aa 251 to 361) of hsHec1p was sufficient for binding to Smc2p; however, unlike the binding to Smc1p, the second coiled-coil enriched region (aa 361 to 547) was unable to bind Smc2p. These results suggested that Hec1p also interacts with Smc2p, albeit in a manner distinct from its interaction with Smc1p. Consistently, the binding ability of the first coiled-coil enriched region (aa 251 to 394) within the hshec1-113p mutant showed no differences at various temperatures (Fig. 6A). Therefore, the role of Hec1p in the modulation of SMC2 function may not be affected in hec1 mutants at nonpermissive temperatures by the dissociation of these two proteins.
The interaction between scHec1p and Smc2p was further verified in vivo in yeast strain Y153 (SMC2 gal4Δ) (13). Smc2p was expressed in episomal form, tagged by TAD, and driven by the ADH1 promoter as described above. Smc2p can be coimmunoprecipitated by anti-scHec1p antibodies and subsequently detected by immunoblot analysis using anti-TAD, an antibody that specifically recognize the TAD from cells expressing tagged Smc2p (Fig. 6B, lanes 4 to 6), but not from Y153 cells. On the other hand, anti-TAD can precipitate the TAD-Smc2p and coprecipitate scHec1p. These results suggested that Hec1p interacts with Smc2p in vivo.
Genetic suppression of the smc mutants by overexpression of hsHEC1 or scHEC1.
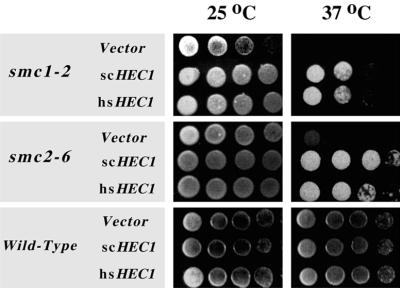
The in vivo interaction between Hec1p and SMC1 or SMC2 may have biological significance. Both smc1 and smc2 mutations cause failure of chromosome segregation, leading to cell death at nonpermissive temperatures (57, 58). Such phenotypes are also observed with hshec1 mutations. If the interaction between Hec1p and Smc1p or Smc2p is critical for the Smc1p or Smc2p function in chromosomal segregation, the overexpression of HEC1 may overcome the ts smc1 or smc2 phenotype. To test this possibility, we introduced an episomal plasmid containing hsHEC1 or scHEC1 under control of the GAL1 promoter into smc1-2 and smc2-6 ts mutants. After induction by galactose, the overexpression of hsHEC1 or scHEC1 appeared to be able to suppress the lethal phenotype of both the smc1-2 and smc2-6 mutants at nonpermissive temperatures, while no suppression was observed with vector alone (Fig. 7). This result indicates that Hec1p proteins may modulate both sister chromatid cohesion and chromosome condensation, and that its interaction with Smc1p or Smc2p is crucial for chromosomal segregation.
FIG. 7.
Suppression of ts phenotype of smc1 or smc2 by overexpressing hsHEC1 or scHEC1. smc1-2, smc2-6, or wild-type cells were transformed by hsHEC1 or scHEC1 in a GAL1-inducible vector or by the vector alone. Different dilutions of log-phase cells grown in 2% glucose at 25°C were inoculated on the plates with 2% galactose and incubated at 25 or 37°C.
DISCUSSION
hsHec1p plays an essential role in mammalian cell mitosis. Cells injected with antibodies specific for hsHec1p exhibit several features of abnormal mitotic phenotypes, including the formation of multiple spindle poles and disordered metaphase chromosome alignment. These cells complete all aspects of division, including anaphase, cytokinesis, and re-formation of the nuclear envelope, but nonetheless missegregate chromosomes to daughter cells (8). To elucidate the molecular bases of hsHec1p in M-phase progression, an S. cerevisiae homolog of the hsHEC1 gene was identified and characterized. Consistent with previous observations of Wigge et al. (61), we found that scHEC1/NDC80 encodes an 80-kDa cellular protein that is 36% identical to the human hsHec1p protein in its N-terminal one-third. Although the overall homology between the two proteins is not high, both proteins contain a long stretch of leucine heptad repeats that constitute two coiled-coil enriched domains in their otherwise divergent C-terminal regions. Genes with similar structure were also identified in fission yeast, C. elegans, and mouse genomes (data not shown). Moreover, the hsHEC1 gene can complement the function of its yeast counterpart in cells deleted for scHEC1. The result suggests that the essential functions of Hec1p proteins have been conserved throughout eukaryotic evolution.
The conservation of Hec1p function provided a useful tool to explore in a simple unicellular organism the molecular mechanisms by which hsHec1p functions. Our results suggest that hsHec1p function is essential for cell viability and M-phase progression. Its function is required during M phase, since conditional hshec1 mutations led to the mitotic delay, accumulation of large-budded cells, and increased lethality during M-phase arrest. Given the structural divergence that exists between hsHec1p and scHec1p, it was unexpected that the phenotypes of the mutants of these two proteins would be almost identical. This observation, however, strongly suggests that Hec1 proteins are functionally very conserved from yeast to human, and it is therefore feasible to study the in vivo properties of a human protein in an organism as simple as the budding yeast.
The chromosome missegregation resulting from mutated hshec1 may stem from a defect in the interaction between hsHec1p and Smc1p. The interaction between hsHec1p and the second coiled-coil domain of the human SMC1 protein was originally indicated by yeast two-hybrid screening. Later, the interaction of hsHec1p with yeast Smc1p was also found to be mediated by the corresponding region in this yeast homolog (Fig. 5). The coiled-coil domains of SMC proteins have been implicated in oligomerization or interaction with other proteins (40). The SMC1-type proteins, yeast Smc1p and Xenopus XSMC1, have been implicated in sister chromatid cohesion (21, 24, 41, 45, 56). The association of hsHec1p or scHec1p with Smc1p might therefore link one of the functions of Hec1p to the regulatory machinery controlling the process of chromatin assembly. At nonpermissive temperatures, the mutant hshec1-113p fails to bind to Smc1p, and this may cause a defect in sister chromatid separation, leading to chromosome missegregation. Consistently, a defect in sister chromatid cohesion has been proposed as the cause leading to chromosome missegregation. Our observation that overexpression of both yeast and human Hec1p can suppress the temperature sensitivity of the smc1 mutant suggests that increasing amounts of Hec1 proteins may augment their affinity for the mutant smc1p and help maintain the proper function of sister chromatid cohesion. Thus, the interaction between Hec1p and SMC1 protein is essential for the physiological function of Hec1p in chromosome segregation and cell viability.
Hec1p is also associated with Smc2p in yeast. Several lines of evidence have revealed the connection between sister chromatid cohesion and chromosome condensation (21, 24, 56). A subunit (Scc1p/Mcd1p) of the Smc1-containing cohesion complex is proposed to function as a linker molecule that connects these two different chromatin-structuring activities on yeast mitotic chromosomes (22). S. cerevisiae Trf4p, a protein required for rDNA condensation, interacts with both Smc1p and Smc2p (5), and a trf4 mutant also exhibits a cohesion defect (24). Like these proteins, Hec1p may modulate both chromatin-structuring activities through association with Smc1p or Smc2p.
The biochemical basis of how Hec1p binds to Smc1p or Smc2p and participates in the activities of SMC proteins remains to be elucidated. It has been suggested by the in vitro activity of Xenopus condensins that SMC proteins may act as motors facilitating formation of chromosome loops in an energy-dependent fashion (22, 24, 37). The ATP-binding ability and ATPase activities of SMC protein complexes are proposed to provide them with motor energy and modulate their functions. Interestingly, other hsHec1p-associated proteins such as MSS1 and p45 of the 26S proteasome were also suggested to have ATPase activities (4), and hsHec1p is able to down-regulate the in vitro ATPase activity of MSS1, the human Cim5p homolog (9). Whether Hec1p can modulate ATPase activities of SMC proteins remains to be tested.
Besides a potential role in the modulation of sister chromatid cohesion or chromosome condensation, a kinetochore function can not be excluded for Hec1p in chromosome segregation since hsHec1p has been shown to localize to centromere regions in mammalian cells during M phase (8). The phenotypes of the mutant forms of both human Hec1p (Fig. 3) and the yeast Hec1p homolog (61) are similar to those of ndc10-1, a mutant form of a centromere-binding protein (18). The staining pattern observed for Hec1p/Ndc80p in this study and that previously reported by Wigge et al. (61) is reminiscent of the staining of the spindle pole body and is consistent with centromere localization. Moreover, the staining of Ndc80p on microtubules adjacent to the spindle pole body and along the short spindle, an important characteristic observed with centromere proteins including Ndc10p (18) and Cse4p (44), was also demonstrated in a previous report (50). A recent study suggested that Hec1p/Ndc80p interacts genetically with Ctf19p, which is a centromere protein (32). The yeast Hec1p/Ndc80p, however, was initially purified from the spindle pole (61), suggesting that Hec1p/Ndc80p may also localize to the spindle pole region. In addition, Hec1p may be distributed among other chromatin regions since cohesion and condensation occur at multiple places along the entire chromosome other than the centromere.
The interaction between Hec1p and SMC proteins may reflect a novel function for the SMC proteins. It is evident that SMC proteins are involved not only in sister chromatid cohesion or chromosome condensation but also in other activities such as DNA replication, recombination, and repair (24, 35, 54, 56, 59). The chromatin assembly activities involved in the cohesion and condensation processes that occur at the centromere have the potential to function in the structural remodeling of centromeric chromatin during mitosis. In higher eukaryotes, the assembly of highly ordered centromeric chromatin structure is suggested to be essential for kinetochore functions. In S. cerevisiae, specialized chromatin structures of the centromeres also appear to be important for kinetochore assembly (60). Therefore, it is, possible that the interaction between Hec1p and SMC proteins is involved in the process of centromeric chromatin assembly and modulation of kinetochore function.
Hec1p may play multiple roles in M-phase progression since it has been shown to associate with other proteins such as the human homologs of Sug1p/Cim3p, Cim5p, and NIMA. It has been shown that Sug1p/Cim3p, Cim5p, and NIMA are important M-phase players because mutants of these proteins lead to G2/M arrest (16, 46). Interestingly, the interactions between Hec1 proteins and their associated proteins appear to have been conserved (Table 3). These interactions are likely to be common modes for regulation of M-phase progression in all eukaryotes.
Although hsHec1p may serve as a regulator of multiple mitotic pathways, it is itself regulated by higher-level modulators. The retinoblastoma protein Rb, as an hsHec1p-associated protein, is likely to be one of these modulators. hsHec1p is not the first protein linking Rb to M-phase progression. The association of Rb with mitosin/CENP-F (62), H-nuc/CDC27 (7), and protein phosphatase 1α (13) has provided circumstantial evidence that Rb has an important role in M-phase progression. The higher-level regulatory function that we propose for Rb is less conserved in lower eukaryotes. First, no gene with sequence similarity to Rb exists in the entire S. cerevisiae genome. Second, the specific I-C-E motifs found in hsHec1p sequences could well serve as Rb-binding domains (6). The protein sequence of either the budding or fission yeast homolog, however, contains no I-C-E motif, consistent with the lack of interaction between budding yeast Hec1p and human Rb in the yeast two-hybrid system (Table 3). The lack of Rb in yeast will allow us to address Rb function by using yeast machinery as a powerful assay tool, without interference from endogenous Rb. The strain in which the scHEC1 gene has been replaced by the hsHEC1 gene will be available for future studies of the in vivo interaction between Rb and hsHec1p and the biological consequences of such an interaction.
ACKNOWLEDGMENTS
The first two authors contributed equally to this report.
We thank O. Cohen-Fix, R. D. Gietz, D. Koshland, P. Hieter, C. Holm, A. M. Hoyt, C. Mann, A. Murray, and A. Strunnikov for yeast strains, antibodies, plasmid vectors, and assistance with yeast genetic analysis, Z. D. Sharp, D. J. Riley, T. Boyer, and H. Chew for critical reading of the manuscript, D. Jones, P. Garza, and S.-Y. Lee for assistance in the preparation of yeast scHec1p antibodies, and C.-F. Chen for cloning of scHEC1 by PCR.
This work was supported by NIH grants EY05758 and CA58318.
REFERENCES
- 1.Araki H, Ropp P A, Johnson A J, Johnston L H, Morrison A, Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase epsilon, replicates chromosomal DNA in the yeast S. cerevisiae. EMBO J. 1992;11:733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton N R, Goldstein L S B. Going mobile: microtubule motors and chromosome segregation. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basrai M A, Kinsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosomal transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 5.Castano I B, Brzoska P M, Sadoff B U, Chen H, Christman M F. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- 6.Chen P-L, Riley D J, Lee W-H. The retinoblastoma protein as a fundamental mediator of growth and differentiation signals. Crit Rev Eukaryotic Gene Expr. 1995;5:79–95. [PubMed] [Google Scholar]
- 7.Chen P-L, Ueng Y-C, Durfee T, Chen K-C, Yang-Feng T, Lee W-H. Identification of a human homologue of yeast nuc2 which interacts with the retinoblastoma protein in a specific manner. Cell Growth Differ. 1995;6:199–210. [PubMed] [Google Scholar]
- 8.Chen Y, Riley D J, Chen P-L, Lee W-H. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Sharp Z D, Lee W-H. HEC binds to the seventh regulatory subunit of the 26 S proteasome and modulates the proteolysis of mitotic cyclins. J Biol Chem. 1997;272:24081–24087. doi: 10.1074/jbc.272.38.24081. [DOI] [PubMed] [Google Scholar]
- 10.Cohen-Fix O, Peters J, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 11.Doheny K F, Sorger P K, Hyman A A, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dresser M E, Ewing D J, Corrad M N, Dominguez A M, Bastead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Lee W-H, Elledge S J. The retinoblastoma gene product associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.Fry A M, Schultz S J, Bartek J, Nigg E A. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J Biol Chem. 1995;270:12899–12905. doi: 10.1074/jbc.270.21.12899. [DOI] [PubMed] [Google Scholar]
- 15.Funanbiki H, Yamano H, Kumada K, Nagao T, Hunt T, Yanagida M. Cut2 proteolysis required for sister chromotid separation in fission yeast. Nature. 1996;349:132–138. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 16.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 17.Gietz R D, Sugino A. New yeast-E. coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 18.Goh P Y, Kilmartin J. NDC10, a gene involved in chromosome segregation in S. cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1996;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guacci V, Yamamoto A, Strunnikov A, Kingsbury J, Hogan E, Meluh P, Koshland D. Structure and function of chromosomes in mitosis of budding yeast. Cold Spring Harbor Symp Quant Biol. 1993;58:677–685. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- 23.Hanna D E, Rethinaswamy A, Glover C V C. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J Biol Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- 24.Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- 25.Hirano T, Mitchison T J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 26.Hieter P, Mann C, Synder M, Davis R. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 27.Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 28.Holm C, Guto T, Wang J C, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 29.Houman F, Holm C. DBF8, an essential gene required for efficient chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6350–6360. doi: 10.1128/mcb.14.9.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyt M A. Eliminating all obstacles: regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 31.Hoyt M A, Geiser J R. Genetic analysis of the mitotic spindle. Annu Rev Genet. 1996;30:7–33. doi: 10.1146/annurev.genet.30.1.7. [DOI] [PubMed] [Google Scholar]
- 32.Hyland K M, Kingsbury J, Koshland D, Hieter P. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyman A A, Sorger P K. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;111:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- 34.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;61:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 35.Jessberger R, Riwar B, Baechtold H, Akhmedov A T. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- 36.Juang Y-L, Huang J, Peters J-M, McLaughlin M E, Tai C-Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 37.Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condesin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 38.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 39.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 40.Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- 41.Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda H. The formation and functioning of yeast mitotic spindles. Bioessays. 1994;17:45–51. doi: 10.1002/bies.950170110. [DOI] [PubMed] [Google Scholar]
- 43.Meluh P B, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meluh P B, Yang P, Glowczewski L, Koshland D, Smith M M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 45.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 46.Oakley B R, Morris N R. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J Cell Biol. 1983;96:1155–1158. doi: 10.1083/jcb.96.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer R E, Hogan E, Koshland D. Mitotic transmission of artificial clones in cdc mutants of the yeast, Saccharomyces cerevisiae. Genetics. 1990;125:763–774. doi: 10.1093/genetics/125.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 49.Rocques P J, Clark J, Ball S, Crew J, Gill S, Christodoulou Z, Borts R H, Louis E J, Davies K E, Cooper C S. The human SB1.8 gene (DXS423E) encodes a putative chromosome segregation protein conserved in lower eukaryotes and prokaryotes. Hum Mol Genet. 1995;4:243–249. doi: 10.1093/hmg/4.2.243. [DOI] [PubMed] [Google Scholar]
- 50.Rout M P, Kilmartin J V. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 52.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–300. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 53.Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skibbens R V, Corson L B, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Straight A F, Marshall W F, Sedat J W, Murray A W. Mitosis in living budding yeast. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- 56.Strunnikov A V. SMC proteins and chromosome structure. Trends Cell Biol. 1998;8:454–459. doi: 10.1016/s0962-8924(98)01370-1. [DOI] [PubMed] [Google Scholar]
- 57.Strunnikov A V, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 58.Strunnikov A V, Larionov V L, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesion complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiens G R, Sorger P K. Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell. 1998;93:313–316. doi: 10.1016/s0092-8674(00)81157-5. [DOI] [PubMed] [Google Scholar]
- 61.Wigge P A, Jensen O N, Holmes S, Soues S, Mann M, Kilmartin J V. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/Ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu X, Mancini M A, Chang K-H, Liu K-Y, Chen C-F, Shan B, Jones D, Yang-Feng T L, Lee W-H. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol Cell Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]