Abstract
This protocol describes how to measure interaction between Notch receptors and their ligands by cell-based assay using Dynabeads. We have used the protocol to determine binding capacity between Notch1-transfected HEK293T cells and ligand-coated Dynabeads. Expression of Eogt in Notch1-expressing cells promoted binding toward DLL4-coated beads, but not JAG1-coated beads. The Notch-ligand assay using Dynabeads suggested that Eogt facilitates DLL4-Notch1 interaction ( Sawaguchi et al., 2017 ).
Keywords: Notch, Ligand binding assay, Dynabeads, Dll4-Fc
Background
The Notch signal pathway regulates many types of cellular events such as proliferation, cell fate determination, and cellular differentiation in all metazoans (Mumm and Kopan, 2000). To initiate Notch signaling, extracellular domains of Notch receptors engage their ligands, Delta like (DLL) ligands or Jagged (JAG) ligands, presented on opposing cells.
Epidermal growth factor (EGF)-like domains of Notch receptors are critical for the ligand binding and modified by specific glycans including O-fucose, O-glucose, and O-GlcNAc glycans (Stanley and Okajima, 2010). Some of these glycans serve as regulators of Notch signaling pathway by modulating physical interaction between Notch receptors and ligands ( Moloney et al., 2000 ). To investigate whether O-GlcNAc regulates Notch-ligand interaction, we developed a novel Dynabeads-based Notch-ligand binding assay. In this assay, Notch receptors expressed on HEK293T cells are incubated with Dynabeads Protein A coated with DLL4-Fc or JAG1-Fc. Unlike soluble ligands used for other binding assay, direction of Notch ligands is fixed on the beads so that they behave like ligand-expressing cells. Thus, the detected binding represents trans-binding rather than cis-binding, which occurs when Notch receptors and their ligands are expressed in the same cells. This assay demonstrated that O-GlcNAc modification of Notch1 by Eogt potentiates Notch1 binding to DLL4 without affecting JAG1 binding ( Sawaguchi et al., 2017 ).
Materials and Reagents
Microtube (INA•OPTIKA, Bio-Bik, catalog number: ST-0150F)
Multiwell culture plates 6 wells (Greiner Bio One International, catalog number: 657160)
15 ml conical centrifuge tubes (Greiner Bio One International, catalog number: 188271)
Syringe filter with a 0.22 µm pore size membrane (Pall, catalog number: 4192)
HEK293T cell
-
Notch1 expressing vector (pTracer-CMV/Notch1) ( Sawaguchi et al., 2017 )
Note: NOT commercially available.
-
Eogt expressing vector (pSecTag2/Hygro/Eogt) ( Sawaguchi et al., 2017 )
Note: NOT commercially available.
GFP expressing vector (pMAX-GFP) (Addgene, catalog number: VDF-1012)
Dynabeads Protein A (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10002D)
Opti-MEM (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070)
4% paraformaldehyde (Wako Pure Chemical Industries, catalog number: 163-20145)
Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: 172012-500ML)
Sodium phosphate dibasic (Na2HPO4) (Wako Pure Chemical Industries, catalog number: 196-02835)
Potassium phosphate monobasic (KH2PO4) (Wako Pure Chemical Industries, catalog number: 164-04295)
Sodium chloride (NaCl) (Wako Pure Chemical Industries, catalog number: 197-01667)
Potassium chloride (KCl) (Wako Pure Chemical Industries, catalog number: 163-03545)
DLL4-Fc (Thermo Fisher Scientific, catalog number: 10171H02H)
JAG1-Fc (Thermo Fisher Scientific, catalog number: 11648H02H)
Dulbecco’s modified Eagle medium (NISSUI PHARMACEUTICAL, catalog number: 05915)
Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122)
Polyethylenimine, linear, MW 25,000 (PEI 25000) (Polysciences, catalog number: 23966)
Fetal bovine serum (FBS) (see Recipes)
Phosphate-buffered saline (PBS) (see Recipes)
100 ng/µl DLL4-Fc/PBS and 100 ng/µl JAG1-Fc/PBS (see Recipes)
Complete culture media (see Recipes)
PEI solution (see Recipes)
Equipment
Pipettes (various sizes) (GILSON)
6-Tube magnetic separation rack (New England Biolabs, catalog number: S1506S)
Tube rotator (Taiyo, model: RT-50) (Figure 1A)
CO2 incubator (SANYO, catalog number: MCO-175)
Orbital rotator (Oriental Instruments, catalog number: KS-6300) (Figure 1B)
Self-made aspirator (Figures 1C and 1D)
Box-type fluorescence imaging device (Olympus, catalog number: FSX100)
Figure 1. Equipment.
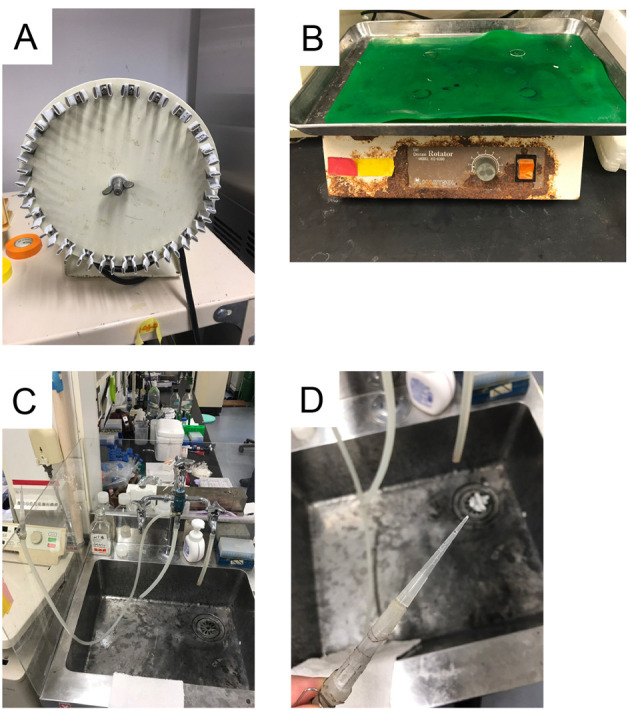
A. Tube rotator RT50; B. Orbital rotator; C and D. Self-made aspirators.
Procedure
-
Preparation of ligand-coated Dynabeads
-
20 µl of Dynabeads Protein A is washed with 1 ml of PBS (see Recipes) in a microtube.
Note: The standard procedure can be scaled up for multiple samples.
Beads are collected using Magnetic Separation Rack.
The solution is removed from the beads.
The beads are resuspended in 100 µl of PBS.
2 µl of 100 ng/µl DLL4-Fc/PBS (see Recipes) or JAG1-Fc/PBS (see Recipes) is added to the tube
The beads are incubated for 20 min at room temperature or overnight at 4 °C using a tube rotator (Video 1).
Beads are collected using a Magnetic Separation Rack.
The solution is removed from the beads.
Beads are washed 3 times with 1 ml of PBS.
The solution is removed from the beads.
100 µl of PBS is added to the tube.
Just before use, 900 µl of ice-cold complete culture media is added to the tube.
-
-
Preparation of Notch-expressing cells
HEK293T cells are seeded in a 6-well plate at 6 x 105 cells per well in complete culture media (see Recipes).
Next day, the culture media are replaced with 800 µl of Opti-MEM.
Cells are incubated for 30 min at 37 °C in a CO2 incubator.
-
Plasmid DNA (2 µg) is diluted into 200 µl of Opti-MEM in a microtube.
Note: To identify transfected cells, GFP expression vector (e.g., pMAX-GFP) is included at 1/8 amount of total plasmids.
The tube is vortexed gently.
6 µl of 1 mg/ml PEI solution (see Recipes) is added to the DNA solution.
The mixture is incubated for 30 min at room temperature.
The DNA/PEI mixtures are gently dropped onto each well of the plate.
Cells are incubated for 4 h at 37 °C in a CO2 incubator.
The culture media are replaced with 2 ml of complete culture media.
Cells are incubated for 48 h at 37 °C in a CO2 incubator.
-
Binding assay (Figure 2)
The culture media are replaced with 1 ml of ice-cold complete culture media containing DLL4-Fc or JAG1-Fc beads (Figure 2C).
The cells are incubated in cold room for 30 min.
The culture media are removed from the culture dish using aspirator.
Cells are fixed with 4% paraformaldehyde for 20 min at room temperature (Figure 2D).
The fixed cells are gently washed with 5 ml of PBS for 5 min using an orbital rotator (Video 2).
-
PBS is removed from the culture plate using aspirator (Video 3).
Note: The beads accumulated in the center of each well can be removed using aspirator.
Cells are washed 5 times in total by repeating steps C5 and C6.
1 ml of PBS is added to each well of the dish (Figure 2I).
Phase contrast and fluorescence images are captured using FSX100 (Figure 3).
-
The number of bound beads on GFP-positive cell is counted (Figure 4).
Note: 50 GFP-positive cells were counted for quantification. Floating beads were excluded for counting.
Video 1. Dynabeads Protein A was incubated at room temperature using a tube rotator.

Note that Dynabeads have brown color in the video.
Figure 2. Binding assay.
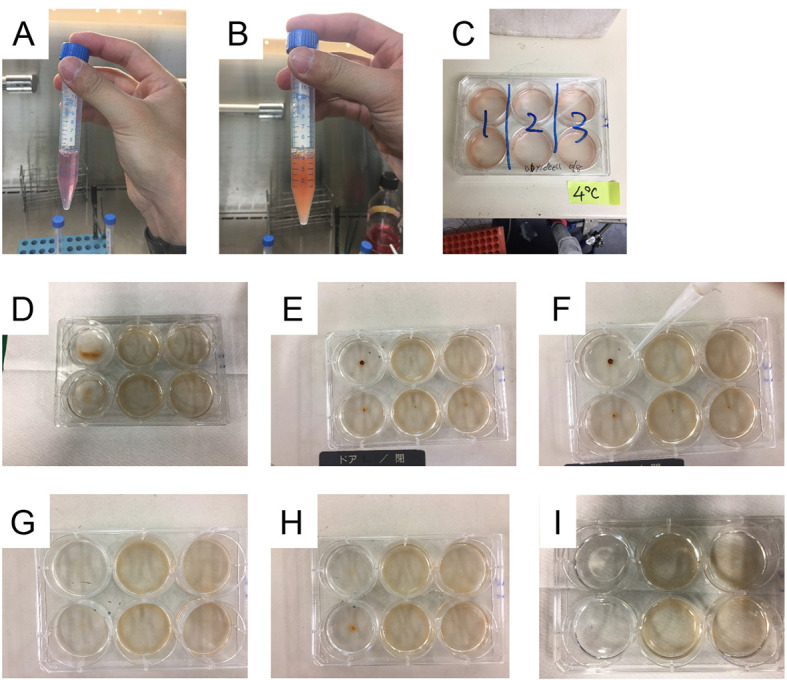
A. Complete culture media in a 15 ml tube; B. Dynabeads resuspended in complete culture media; C. HEK293T cells in a 6-well plate were incubated with ligand-coupled beads in a cold room. D. The culture plate during fixation with paraformaldehyde; E. The culture plate before removing PBS; F. PBS was removed from the culture plate using self-made aspirator. G. The culture plate after removing PBS; H. The culture plate after washing three times with PBS; I. The culture plate after washing five times with PBS.
Video 2. The fixed cells were washed with PBS for using an orbital rotator.

Video 3. PBS was removed from the culture plate using aspirator.
Figure 3. Counting of Dynabeads-bound cells.
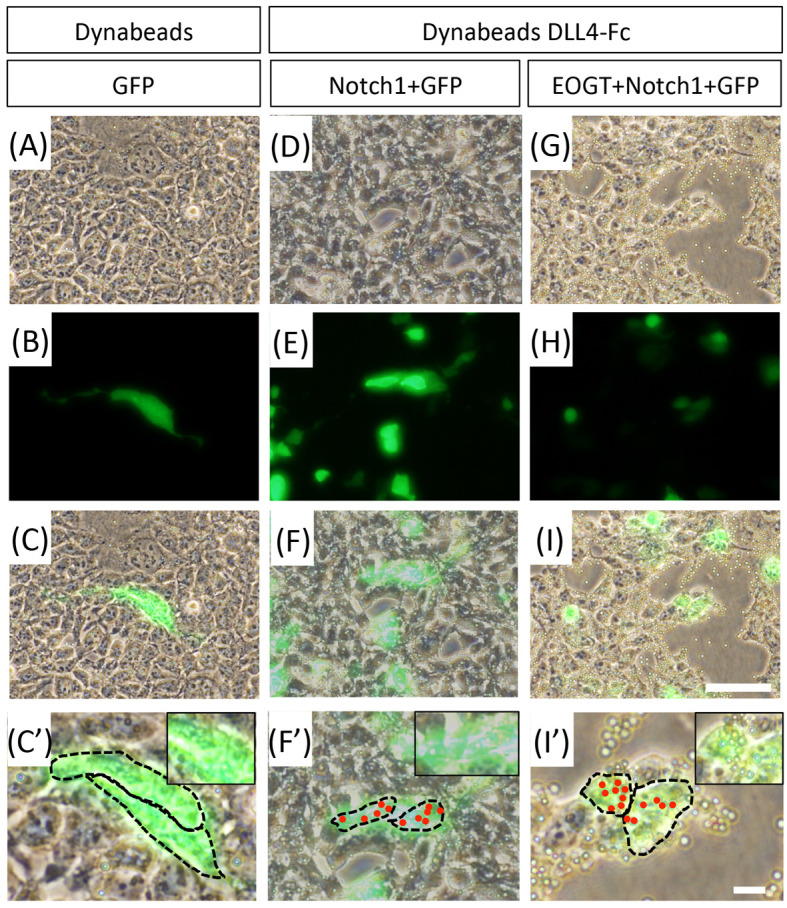
A-C. HEK293T cells transfected to express GFP were cultured with control Dynabeads. D-F. HEK293T cells transfected to express GFP, Notch1 were incubated with DLL4-Fc-bound beads. G-I. HEK293T cells transfected to express GFP, Eogt, and Notch1 were incubated with DLL4-Fc-bound beads. A, D and G. Phase contrast images; B, E and H. Fluorescent Images; C, F and I. Merged images. Scale bars = 60 µm. C’, F’ and I’. Higher magnification of boxed area in images (C, F, I). Scale bars = 60 µm. C”, F” and I”. Same as C’, F’, and I’. Red dots represent bound beads on GFP-positive cells. The dotted line shows outline of the cells. Scale bars = 60 µm.
Figure 4. Analysis of counting of Dynabeads-bound cells.
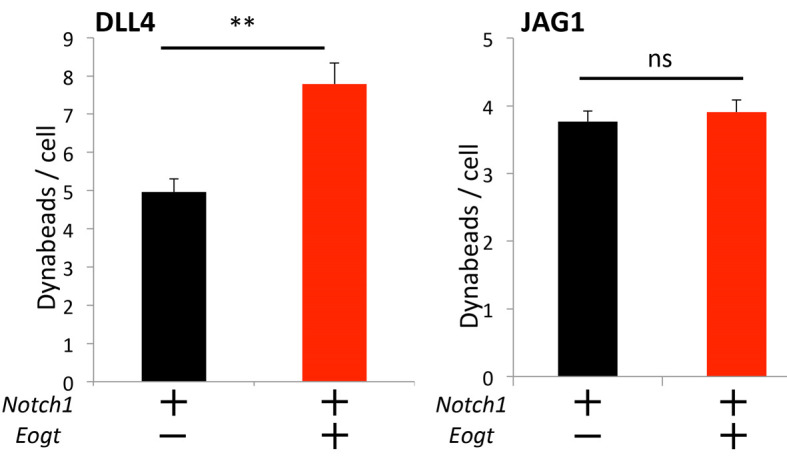
HEK293T cells or cells transiently transfected with Notch1 with or without Eogt, were incubated with DLL4 or JAG1 beads. The number of Dynabeads bound per transfected cell marked by GFP expression was determined (n = 50). Data are mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; bar by Welch’s t-test.
Data analysis
In the previously published experiments ( Sawaguchi et al., 2017 ), data were shown as mean ± SD from three independent experiments. In each experiment, 50 GFP-positive cells are analyzed. Welch’s t-test was used.
Notes
This protocol provides high reproducibility. Counting 50 GFP-positive cells gives low variability in most experiments.
Recipes
-
Fetal bovine serum (FBS)
The bottle containing FBS is incubated at 56 °C for 30 min before use
-
Phosphate-buffered saline (PBS)
10 mM Na2HPO4
1.8 mM KH2PO4
137 mM NaCl
2.7 mM KCl
-
100 ng/µl DLL4-Fc/PBS and 100 ng/µl JAG1-Fc/PBS
DLL4-Fc or JAG1-Fc (5 µg) is dissolved in 50 µl of PBS
Note: 100 ng/µl DLL4-Fc/PBS and 100 ng/µl JAG1-Fc/PBS can be stored at 4 °C for 3 months.
-
Complete culture media
DMEM containing 10% FBS and penicillin-streptomycin
-
PEI solution
100 mg of PEI is dissolved in 100 ml of Milli-Q water
The solution is sterilized by passing through a 0.22 µm membrane
Aliquots are stored at -30 °C
Acknowledgments
This protocol was modified from the previously published article ( Sawaguchi et al., 2017 ). This work was supported by Japan Society for the Promotion of Science grants # JP15K15064 to TO and MO, #JP26110709 to TO, #JP26291020 to TO, #JP15K18502 to MO, #JP16J00004 to MO; Takeda Science Foundation to TO; Japan Foundation for Applied Enzymology to TO; YOKOYAMA Foundation for Clinical Pharmacology #YRY-1612 to MO.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Moloney D. J., Panin V. M., Johnston S. H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K. D., Haltiwanger R. S. and Vogt T. F.(2000). Fringe is a glycosyltransferase that modifies Notch. Nature 406(6794): 369-375. [DOI] [PubMed] [Google Scholar]
- 2.Mumm J. S. and Kopan R.(2000). Notch signaling: from the outside in. Dev Biol 228(2): 151-165. [DOI] [PubMed] [Google Scholar]
- 3.Sawaguchi S., Varshney S., Ogawa M., Sakaidani Y., Yagi H., Takeshita K., Murohara T., Kato K., Sundaram S., Stanley P. and Okajima T.(2017). O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. Elife 6: e24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley P. and Okajima T.(2010). Roles of glycosylation in Notch signaling. Curr Top Dev Biol 92: 131-164. [DOI] [PubMed] [Google Scholar]


