Abstract
Numerous oncogenes have been identified to cause leukemia. For example, chromosomal translocation generates various fusion genes of the mixed-lineage leukemia (MLL) gene and a partner gene in leukemia, whose gene products transform primary myeloid progenitors into an immortalized state. To characterize the transforming ability of leukemic oncogenes, researchers in the field have developed an ex vivo murine myeloid transformation assay using retroviral gene transduction and its protocol has been improved over the past 10 years. Here, we provide the detailed procedure for this assay.
Keywords: Mixed lineage leukemia, Transformation, Myeloid, Progenitor, MLL fusion
Background
Chromosomal translocation generates a variety of MLL fusion genes that cause leukemia ( Meyer et al., 2017 ). The wild-type mixed lineage leukemia (MLL) protein functions as a transcriptional regulator that enhances the expression of a set of genes including homeobox (Hox) genes in hematopoietic immature progenitor cells ( Jude et al., 2007 ). During normal hematopoiesis, Hox genes are expressed in the stem/progenitor cell fractions, but are transcriptionally down-regulated throughout differentiation (Somervaille and Cleary, 2006; Yokoyama et al., 2013 ); however, the MLL fusion protein constitutively up-regulates its target genes and blocks differentiation to establish an immortalized state under ex vivo culture conditions. Recently, we reported that the MLL-ENL and MLL-AF10 fusion proteins recruit AF4 to activate transcription and also recruit the DOT1L complex to maintain the transcription of the same target genes to efficiently transform hematopoietic progenitors ( Okuda et al., 2017 ). To investigate the molecular mechanism of leukemogenesis, one can perform a myeloid progenitor transformation assay. By this assay, one can identify essential functional domains of an oncoprotein relatively easily at low cost compared to an in vivo leukemogenesis assay. In this assay, the leukemic oncogene is transduced by a retrovirus into primary murine hematopoietic progenitor cells derived from bone marrow and the transduced cells are cultured in semi-solid medium containing cytokines for the myeloid lineage ( Lavau et al., 1997 ). Cells fully transformed by an oncogene can often be established as a cell line and cultured indefinitely ex vivo. Therefore, one can perform gene knockdown experiments of these immortalized cells by transducing an sh-RNA-carrying lentivirus. Alternatively, one can immortalize progenitors derived from a genetically engineered mouse carrying conditional knockout alleles with loxP sites, and knockout the gene of interest by activating Cre-recombinase. We have shown that the MLL fusion protein associates with various co-factors to form a functional complex. MENIN and lens epithelium-derived growth factor (LEDGF) are essential co-factors for leukemogenic activity ( Yokoyama et al., 2005 ; Yokoyama and Cleary, 2008; Okuda et al., 2014 ). AF4 and ENL family proteins are the most frequent MLL-fusion partners, accounting for two-thirds of MLL-rearranged leukemia incidence ( Huret et al., 2001 ), and form a biochemically stable complex with the SL1 complex and the P-TEFb elongation factor to activate transcription initiation and elongation ( Yokoyama et al., 2010 ; Okuda et al., 2015 ; Okuda et al., 2016 ). MLL fusion proteins appear to activate transcription through the association with these cofactors in immortalized cells. In these previous studies, we used an sh-RNA-mediated knockdown strategy and Cre-mediated knockout strategy with MLL fusion-immortalized cells to demonstrate the importance of cofactors in leukemic transformation. Because this method allows us to generate cell lines immortalized with different oncogenes in a short period of time, the efficacy of drugs can be analyzed on various cell lines that are dependent on different oncogenes ( Grembecka et al., 2012 ). Thus, this assay is a powerful tool for functional analysis of leukemic oncogenes and testing of drug efficacies.
Materials and Reagents
Pipette tips
Tissue culture 10-cm dish (Greiner Bio One International, catalog number: 664160)
Collagen-coated tissue culture 6-cm dishes (Corning, catalog number: 354401)
Terumo syringe® 10 ml (Terumo, catalog number: SS-10ESZ)
Terumo needle 21 G x 1 ½” (Terumo, catalog number: NN-2138S)
Pre-separation filter (Miltenyi Biotec, catalog number: 130-041-407)
MS column (Miltenyi Biotec, catalog number: 130-042-201)
15-ml conical centrifuge tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339650)
Tissue culture 48-well plate (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150687)
Terumo syringe® 5 ml (Terumo, catalog number: SS-05SZ)
Millex-GV 0.45-µm PVDF 33-mm sterile syringe filter (Merck, catalog number: SLHV033RB)
Corning® Cell Lifter (Corning, catalog number: 3008)
Terumo syringe® 1 ml (Terumo, catalog number: SS-01T)
Terumo needle 18 G x 1 ½” (Terumo, catalog number: NN-1838S)
Tissue culture 12-well plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150628)
Bottle-top filter 0.2-µm PVDF (Corning, catalog number: 431098)
Five-week-old female C57BL/6JJcl mice (CLEA Japan, Tokyo, Japan)
Platinum-E packaging (PLAT-E) cell line ( Morita et al., 2000 ) (gifted from Dr. Toshio Kitamura, or Cell Biolabs, catalog number: RV-101)
WEHI-3 (ATCC, catalog number: TIB-68)
pMSCV-neo vector (Takara Bio, ClontechTM, catalog number: 634401)
LipofectamineTM 2000 transfection reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668019)
Opti-MEMTM I medium (Thermo Fisher Scientific, InvitrogenTM, catalog number: 31985070)
CD117 microbeads, mouse (Miltenyi Biotec, catalog number: 130-091-224)
G418 solution (NACALAI TESQUE, catalog number: 16513-84)
Polybrene infection/transfection reagent (10 mg/ml) (Merck, catalog number: TR-1003-G)
Beta-mercaptoethanol (NACALAI TESQUE, catalog number: 21418-84)
RNeasy® mini kit (QIAGEN, catalog number: 74106)
Superscript® III first-strand synthesis system for RT-PCR (Thermo Fisher Scientific, InvitrogenTM, catalog number: 18080051)
Sodium chloride (NaCl) (NACALAI TESQUE, catalog number: 31320-05)
Na2HPO4.12H2O (Wako Pure Chemical Industries, catalog number: 196-02835)
Potassium dihydrogenphosphate (KH2PO4) (Wako Pure Chemical Industries, catalog number: 164-22635)
Potassium chloride (KCl) (Wako Pure Chemical Industries, catalog number: 160-22115)
Fetal bovine serum (FBS) (NICHIREI, Sigma-Aldrich, catalog number: 172012-500ML)
Penicillin-streptomycin-glutamine (P/S) solution (NACALAI TESQUE, catalog number: 06168-34)
Dulbecco’s modified Eagle medium (DMEM) (NACALAI TESQUE, catalog number: 08459-64)
RPMI 1640 (NACALAI TESQUE, catalog number: 30264-56)
Ethylenediaminetetraacetic acid (EDTA) (NACALAI TESQUE, catalog number: 15130-95)
Trypsin-EDTA solution (NACALAI TESQUE, catalog number: 32778-34)
Sodium hydroxide (NaOH) (Wako Pure Chemical Industries, catalog number: 198-13765)
Ammonium chloride (NH4Cl) (Wako Pure Chemical Industries, catalog number: 017-02995)
Potassium hydrogen carbonate (KHCO3) (Wako Pure Chemical Industries, catalog number: 166-03275)
Murine stem cell factor (SCF) (PeproTech, catalog number: 250-03)
Murine interleukin-3 (IL-3) (PeproTech, catalog number: 213-13)
Murine interleukin-6 (IL-6) (PeproTech, catalog number: 216-16)
Murine granulocyte-macrophage colony-stimulating factor (GM-CSF) (PeproTech, catalog number: 315-03)
Bovine serum albumin (BSA) (Wako Pure Chemical Industries, catalog number: 019-23293)
IMDM powder (Thermo Fisher Scientific, GibcoTM, catalog number: 12200036)
Sodium hydrogen carbonate (NaHCO3) (Wako Pure Chemical Industries, catalog number: 195-14515)
Methyl cellulose (viscosity: 4,000 cP) (Sigma-Aldrich, catalog number: M0512)
-
TaqMan® probes against murine Hoxa9 and Gapdh
Hoxa9 (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: Mm00439364_m1)
Gapdh (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: Mm99999915_g1)
TaqMan® fast advanced master mix (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4444557)
25x phosphate-buffered saline Ca2+/Mg2+-free (PBS) (see Recipes)
D10 media (see Recipes)
R10 media (see Recipes)
R10W10 media (see Recipes)
0.5 M EDTA solution (see Recipes)
ACK lysis buffer (see Recipes)
SM buffer (see Recipes)
Cytokine stocks (see Recipes)
AC media (see Recipes)
Equipment
5% CO2 incubator 37 °C
5% CO2 incubator 32 °C
MACS multistand (Miltenyi Biotec, catalog number: 130-042-303)
Pipetman P2, P20, P200, P1000
Portable Pipet-Aid® XP pipette controller (Drummond Scientific, catalog number: 4-000-101)
Microscope
Cell counter
Autoclave
1-L glass bottle
Shaker
Surgical scissors and forceps
Laminar flow cabinet
Centrifuge for 15-ml conical tubes
MiniMACS separator (Miltenyi Biotec, catalog number: 130-042-102)
StepOnePlusTM realtime PCR system (Thermo Fisher Scientific, Applied BiosystemsTM, model: StepOnePlusTM, catalog number: 4376600)
General equipment for Western blotting and SDS-PAGE
Software
GraphPad Prism (GraphPad Software, La Jolla, CA, USA)
Procedure
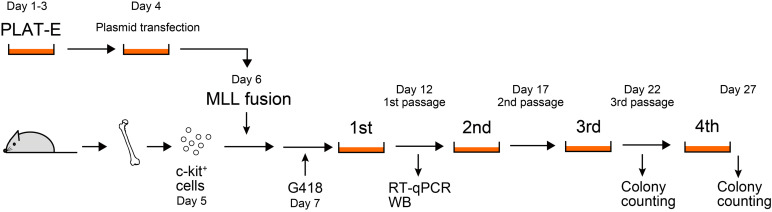
Schedule (see Figure 1):
Figure 1. Schedule of murine myeloid progenitor transformation assay.
Day 1. Start culturing PLAT-E cells from frozen stock
Day 3. Replate PLAT-E cells for transfection
Day 4. Transfect PLAT-E cells
Day 5. Preparation of c-kit-positive cells
Day 6. Transduction of retrovirus into c-kit-positive cells
Day 7. Addition of G418
Day 12. 1st passage
Day 17. 2nd passage
Day 22. Colony counting and 3rd passage
Day 27. Colony counting
-
Virus preparation
On day 1, thaw PLAT-E cells from frozen stock and culture 4 x 106 cells in 10-cm dishes with 10 ml of D10 media (see Recipes) in a 37 °C 5% CO2 incubator.
On day 3, replate 8 x 105 PLAT-E cells in 6-cm collagen-coated dishes with 5 ml of D10 media in a 37 °C 5% CO2 incubator (one positive control [MLL-ENL], one negative control [empty vector; pMSCV-neo], two mock controls for G418 selection, and samples [up to 16 samples]).
-
On day 4, transfect PLAT-E cells with 8 µg of DNA and 20 µl of LipofectamineTM 2000 transfection reagent. At the time of transfection, the cell confluency should be 60-70%. (In detail, dilute 8 µg of DNA in 500 µl of Opti-MEMTM media and dilute 20 µl of LipofectamineTM 2000 transfection reagent in 500 µl of Opti-MEMTM media, separately. After 5 min, combine the DNA solution with the LipofectamineTM 2000 solution. Mix gently and incubate for 20 min at room temperature. Add the DNA-Lipofectamine mixture to each dish and incubate in a 37 °C 5% CO2 incubator.) After 6 h, replace the media with fresh D10 media and incubate in a 32 °C 5% CO2 incubator for 2 days.
Note: Virus supernatant should be freshly prepared in every experiment because retrovirus is unstable for storage.
-
Preparation of c-kit (CD117)-positive progenitor cells
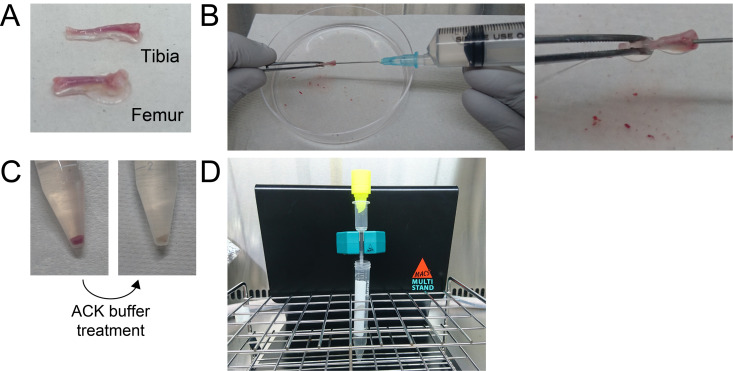
On day 5, harvest the femurs and tibiae and remove the peripheral muscle as thoroughly as possible from a five-week-old C57BL/6J mouse (Figure 2A).
Cut the end of femurs and tibiae and flush bone marrow with 10 ml of PBS (see Recipes) using a 10-ml syringe attached with a 21 G needle (Figure 2B) (Video 1).
Homogenize the bone marrow cells gently by passing through a 21 G needle several times.
Spin down the cells at 400 × g for 5 min at room temperature (Figure 2C left) and then remove as much of the supernatant as possible.
Resuspend the cells in 1 ml of ACK lysis buffer (see Recipes) and incubate for 1 min on ice.
Add 10 ml of R10W10 media (see Recipes) and spin down cells at 400 × g for 5 min at room temperature, and then remove the supernatant.
Resuspend the cells in 10 ml of SM buffer (see Recipes) and spin down cells at 400 × g for 5 min at room temperature (Figure 2C right), and then remove the supernatant.
Resuspend the cells in 0.5 ml of SM buffer, add 20 µl of CD117 microbeads, and incubate for 20 min on ice or in the refrigerator.
Add 10 ml of SM buffer and spin down cells at 400 × g for 5 min at room temperature, and then remove the supernatant to wash the cells.
Wash the cells again as in step B9.
Install an MS column on a magnetic stand and equilibrate with 1 ml of SM buffer.
Place a pre-separation filter on top of the column and load the cells onto the MS column through the pre-separation filter (Figure 2D).
Wash the column twice with 1 ml of SM buffer.
Remove the column from the magnetic stand and place it on a new 15-ml conical tube.
Add 1 ml of SM buffer to the column to elute c-kit-positive cells by gravity flow (optionally using a plunger).
Add 10 ml of R10W10 media and spin down the cells at 400 × g for 5 min at room temperature, and then remove the supernatant.
Resuspend all of the cells in 1 ml of R10W10 and transfer into one well in a 48-well plate.
Add cytokines (10 ng/ml SCF, 10 ng/ml IL-3, 10 ng/ml IL-6 at the final concentration) and incubate the cells in a 37 °C 5% CO2 incubator overnight.
-
Virus transduction and cell culture
On day 6, count the cells. A total of 1-2 x 106 c-kit-positive cells are expected from each mouse.
Add an appropriate amount of R10W10 media to prepare a 1 x 105 cells/ml suspension and add a 1/250 volume of 10 mg/ml polybrene solution.
Aliquot 0.5 ml of cell suspensions into 15-ml conical tubes (additionally, prepare two mock infection controls).
Suck 5 ml of the virus supernatant using a 5-ml syringe, attach a filter (0.45-µm) to the 5-ml syringe, and add the virus supernatant directly to the c-kit-positive cell suspension.
Spin the cell suspension at 1,100 × g for 2.5 h at 32 °C (Spinoculation).
During spinoculation, add 1 ml of PBS onto PLAT-E cells, harvest the cells using a cell lifter, and transfer the cells into a new tube.
Wash the packaging cells with 1 ml of PBS again and prepare the whole cell lysate in 500 µl of WB lysis buffer.
-
After spinoculation, remove the virus supernatant (leaving approximately 200 µl of media), resuspend the cells in residual media, add 1 ml of AC media (see Recipes) using a 1-ml syringe attached with an 18 G needle, and mix by vortexing.
Note: Use a 1-ml syringe and a needle to take 1 ml of AC media because this media is very viscous. A pipet cannot be used.
Incubate the 15-ml conical tubes containing the cells in a 37 °C 5% CO2 incubator overnight.
-
On day 7, add 20 µl of G418 solution, mix by vortexing, and transfer the cells into a 12-well plate using a 1-ml syringe attached with an 18 G needle. For the two mock controls, add G418 in one tube to ensure that G418 selection is working.
Note: Samples should be placed in the central 6 wells in a 12-well plate and PBS should be added to the peripheral 6 wells to prevent the cultures in AC media from drying.
-
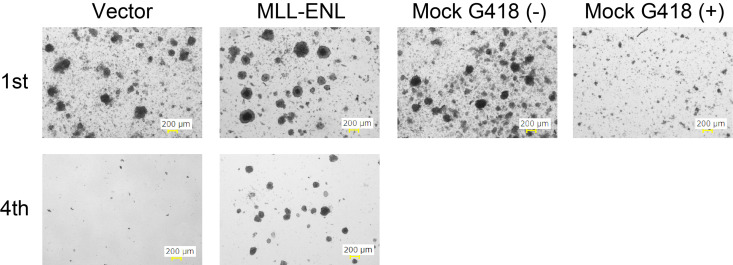
Incubate in a 37 °C 5% CO2 incubator for 5 days (Figure 3). If media turns orange during culture because of excess cells, add an additional 1 ml of AC media with G418.
Note: It is important to maintain good culture conditions. Overgrowth drastically affects colony formation in the next round of culture.
Optionally, one can check the protein expression of transgenes in the packaging cells by WB during the first round of culture.
-
1st passage
On day 12, collect cells with 10 ml of PBS, transfer to a 15-ml conical tube, and spin down at 400 × g for 5 min at room temperature.
-
Wash the cells with 10 ml of PBS.
Note: It is important to suspend thoroughly by vigorous pipetting to remove the debris of AC media.
Resuspend the cells in 1 ml of R10W10 media and count the cells.
Transfer 4 x 104 cells to a new 15-ml conical tube and add an appropriate amount of R10W10 to prepare a 200-µl cell suspension.
-
Add 1 ml of AC media and mix by vortexing.
Transfer the cells into a well in a new 12-well plate using a 1-ml syringe attached with an 18 G needle.
Note: Samples should be placed in the central 6 wells in a 12-well plate and PBS should be added to the peripheral 6 wells to prevent the cultures in AC media from drying.
Incubate the cells in a 37 °C 5% CO2 incubator for 5 days. If the media turns orange during culture, add an additional 1 ml of AC media to the well.
Optionally, collect residual cells into a new tube and lyse the cells in RLT lysis buffer containing 1% beta-mercaptoethanol of an RNeasy® mini kit for RNA isolation as described in Procedure E.
-
Cell collection and analysis
Isolate the RNA using the RNeasy® mini kit and synthesize cDNA using the Superscript® III first-strand synthesis system with oligo-dT primers.
Evaluate gene expression by quantitative PCR. In case of myeloid progenitor transformation by MLL-fusion genes, Hoxa9 gene expression is a good indicator of immortalization.
-
2nd passage
On day 17, harvest and count the cells as in 1st passage.
Culture 2 x 104 cells/well.
-
Colony counting and 3rd passage
-
On day 22, count the colony number under a microscope.
Note: No colonies should be observed in the vector control. It is important to set a standard which defines a colony and use this definition throughout the study. We typically consider a cluster of more than 100 cells as a colony.
Harvest and count the cells as in the 1st passage.
Culture 1 x 104 cells/well.
-
-
Colony counting
On day 27, count the colony number under a microscope (Figure 3).
Figure 2. Representative image of c-kit-positive progenitor cell isolation.
A. Harvested tibia and femur; B. Flushing the bone marrow with PBS; C. Red blood cell removal by ACK buffer treatment; D. c-kit-positive cell isolation by MACS.
Video 1. Bone marrow isolation from tibiae by flushing with PBS.
Figure 3. Image of colonies during the 1st and 4th rounds of culture.
Data analysis
Because of experimental variations, this assay must be performed a minimum of three times.
For statistical analysis, we use GraphPad Prism software. To compare two data sets, we perform Student’s t-test. To compare three or more data sets, we perform one-way analysis of variance and post-hoc Tukey correction.
Recipes
-
25x phosphate-buffered saline Ca2+/Mg2+-free (PBS) (1 L)
Mix 200 g NaCl, 72.4 g Na2HPO4·12H2O, 5 g KH2PO4 and 5 g KCl
Bring to 1 L with distilled H2O
Autoclave for 20 min at 121 °C
Dilute to 1x with distilled H2O for working solution
-
D10 media
Add 55 ml of FBS and 5.5 ml of P/S solution to 500 ml of DMEM
-
R10 media
Add 55 ml of FBS and 5.5 ml of P/S solution to 500 ml of RPMI 1640
-
R10W10 media
Culture WEHI-3 in R10 media to confluence
When media turns orange, collect and filter (0.22-µm) the media
Aliquot and store at -80 °C
Add 55 ml of FBS, 55 ml of WEHI-3 culture media, and 5.5 ml of P/S solution to 500 ml of RPMI 1640
-
0.5 M EDTA solution
Weigh 93.06 g of EDTA
Bring to 1 L with distilled water and adjust pH to 8.0 with NaOH
-
ACK lysis buffer
Mix 8.29 g of NH4Cl and 1 g of KHCO3
Bring to 1 L with distilled water
Add 200 µl of 0.5 M EDTA solution
Filter (0.2-µm) and store at 4 °C
-
SM buffer
Add 15 ml of FBS to 500 ml of 1x PBS
Filter (0.2-µm) and store at 4 °C
-
Cytokine stocks
Dissolve cytokines (SCF, IL-3, IL6, GM-CSF) to 50 µg/ml in PBS + 0.1% BSA
Aliquot and store at -80 °C
-
AC media (alternatively Methocult M3231 from STEMCELL Technologies, Vancouver, Canada)
Dissolve IMDM powder in 500 ml of distilled water, add 3 g of NaHCO3, and filter (0.2-µm)
Weigh 16 g of methyl cellulose in a 1-L glass bottle
Autoclave methyl cellulose powder for 20 min at 121 °C
Dissolve sterile methyl cellulose in 300 ml of sterile water and 500 ml of IMDM in a shaker overnight
Add 200 ml of FBS and 7 µl of beta-mercaptoethanol
Aliquot 100 ml per bottle and store at -20 °C
Before use, add 20 µl SCF, 20 µl IL-3, 20 µl GM-CSF, and 1 ml P/S solution to 100 ml of media
Acknowledgments
This study was supported by JSPS KAKENHI grants to H.O. (number 17H07379) and A.Y. (number 16H05337). This protocol was based on a previous report by Lavau et al. (1997). The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Grembecka J., He S., Shi A., Purohit T., Muntean A. G., Sorenson R. J., Showalter H. D., Murai M. J., Belcher A. M., Hartley T., Hess J. L. and Cierpicki T.(2012). Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 8(3): 277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huret J. L., Dessen P. and Bernheim A.(2001). Leukemia 15(6): 987-989.
- 3.Jude C. D., Climer L., Xu D., Artinger E., Fisher J. K. and Ernst P.(2007). Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1(3): 324-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavau C., Szilvassy S. J., Slany R. and Cleary M. L.(1997). Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J 16(14): 4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer C., Burmeister T., Groger D., Tsaur G., Fechina L., Renneville A., Sutton R., Venn N. C., Emerenciano M., Pombo-de-Oliveira M. S., Barbieri Blunck C., Almeida Lopes B., Zuna J., Trka J., Ballerini P., Lapillonne H., De Braekeleer M., Cazzaniga G., Corral Abascal L., van der Velden V. H. J., Delabesse E., Park T. S., Oh S. H., Silva M. L. M., Lund-Aho T., Juvonen V., Moore A. S., Heidenreich O., Vormoor J., Zerkalenkova E., Olshanskaya Y., Bueno C., Menendez P., Teigler-Schlegel A., Zur Stadt U., Lentes J., Gohring G., Kustanovich A., Aleinikova O., Schafer B. W., Kubetzko S., Madsen H. O., Gruhn B., Duarte X., Gameiro P., Lippert E., Bidet A., Cayuela J. M., Clappier E., Alonso C. N., Zwaan C. M., van den Heuvel-Eibrink M. M., Izraeli S., Trakhtenbrot L., Archer P., Hancock J., Moricke A., Alten J., Schrappe M., Stanulla M., Strehl S., Attarbaschi A., Dworzak M., Haas O. A., Panzer-Grumayer R., Sedek L., Szczepanski T., Caye A., Suarez L., Cave H. and Marschalek R.(2017). The MLL recombinome of acute leukemias in 2017. Leukemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita S., Kojima T. and Kitamura T.(2000). Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7(12): 1063-1066. [DOI] [PubMed] [Google Scholar]
- 7.Okuda H., Kanai A., Ito S., Matsui H. and Yokoyama A.(2015). AF4 uses the SL1 components of RNAP1 machinery to initiate MLL fusion- and AEP-dependent transcription. Nat Commun 6: 8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okuda H., Kawaguchi M., Kanai A., Matsui H., Kawamura T., Inaba T., Kitabayashi I. and Yokoyama A.(2014). MLL fusion proteins link transcriptional coactivators to previously active CpG-rich promoters. Nucleic Acids Res 42(7): 4241-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda H., Stanojevic B., Kanai A., Kawamura T., Takahashi S., Matsui H., Takaori-Kondo A. and Yokoyama A.(2017). Cooperative gene activation by AF4 and DOT1L drives MLL-rearranged leukemia. J Clin Invest 127(5): 1918-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuda H., Takahashi S., Takaori-Kondo A. and Yokoyama A.(2016). TBP loading by AF4 through SL1 is the major rate-limiting step in MLL fusion-dependent transcription. Cell Cycle 15(20): 2712-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somervaille T. C. and Cleary M. L.(2006). Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 10(4): 257-268. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama A. and Cleary M. L.(2008). Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14(1): 36-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama A., Ficara F., Murphy M. J., Meisel C., Hatanaka C., Kitabayashi I. and Cleary M. L.(2013). MLL becomes functional through intra-molecular interaction not by proteolytic processing. PLoS One 8(9): e73649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama A., Lin M., Naresh A., Kitabayashi I. and Cleary M. L.(2010). A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell 17(2): 198-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokoyama A., Somervaille T. C., Smith K. S., Rozenblatt-Rosen O., Meyerson M. and Cleary M. L.(2005). The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123(2): 207-218. [DOI] [PubMed] [Google Scholar]