Abstract
In previous reports we showed that the long 5′ untranslated region (5′ UTR) of c-sis, the gene encoding the B chain of platelet-derived growth factor, has translational modulating activity due to its differentiation-activated internal ribosomal entry site (D-IRES). Here we show that the 5′ UTR contains three regions with a computer-predicted Y-shaped structure upstream of an AUG codon, each of which can confer some degree of internal translation by itself. In nondifferentiated cells, the entire 5′ UTR is required for maximal basal IRES activity. The elements required for the differentiation-sensing ability (i.e., D-IRES) were mapped to a 630-nucleotide fragment within the central portion of the 5′ UTR. Even though the region responsible for IRES activation is smaller, the full-length 5′ UTR is capable of mediating the maximal translation efficiency in differentiated cells, since only the entire 5′ UTR is able to confer the maximal basal IRES activity. Interestingly, a 43-kDa protein, identified as hnRNP C, binds in a differentiation-induced manner to the differentiation-sensing region. Using UV cross-linking experiments, we show that while hnRNP C is mainly a nuclear protein, its binding activity to the D-IRES is mostly nuclear in nondifferentiated cells, whereas in differentiated cells such binding activity is associated with the ribosomal fraction. Since the c-sis 5′ UTR is a translational modulator in response to cellular changes, it seems that the large number of cross-talking structural entities and the interactions with regulated trans-acting factors are important for the strength of modulation in response to cellular changes. These characteristics may constitute the major difference between strong IRESs, such as those seen in some viruses, and IRESs that serve as translational modulators in response to developmental signals, such as that of c-sis.
Eukaryotic gene expression is regulated at the transcriptional and posttranscriptional levels. Regulation at the level of translation initiation is becoming increasingly evident. Most mRNAs encoding oncoproteins and factors related to cell proliferation possess a long, GC-rich, structured mRNA leader sequence with one or more AUG triplets upstream of the translation initiation codon. Such extraordinary features suggest that they are involved in translational control (16). In order to examine the role of these cis elements, about which little is known, we undertook to study the gene encoding the B chain of platelet-derived growth factor (PDGF). PDGF is a potent mitogen of all cells of mesenchymal origin and has a major role in wound healing as well as in embryogenesis and development. PDGF consists of homo- or heterodimers of two protein chains, PDGF-A and -B, whose gene expression is tightly controlled at multiple levels (9). The B chain is the product of the c-sis proto-oncogene, which leads to neoplastic transformation when it is overexpressed (24).
The c-sis mRNA leader contains a 1,022-nucleotide (nt) 5′ untranslated region (5′ UTR) with 3 upstream silent AUG codons and highly stable secondary structures. This configuration poses a major barrier to 5′ cap-dependent translation initiation and ribosomal scanning (47). We demonstrated previously that the 5′ UTR-mediated translational inhibition was relieved during megakaryocytic differentiation by an induced mechanism of translation initiation from an internal site within the structured 5′ UTR. This differentiation-induced internal ribosomal entry site of the c-sis 5′ UTR was termed D-IRES (3, 4). c-sis was the first proto-oncogene demonstrated to use the internal translation mechanism and the first cellular gene whose IRES was shown to be activated by a physiological signal. Thus, the long and complex 5′ UTR of c-sis provided a model for studying the cis elements and trans-acting factors involved in mediating an IRES mechanism which is sensitive to the cellular milieu.
The internal ribosome binding mechanism, independent of the 5′-cap structure, was first observed in poliovirus and encephalomyocarditis virus (EMCV) and later in other viruses including aphthovirus, rhinovirus, hepatitis A virus, hepatitis C virus, and Moloney murine leukemia virus (11, 20, 54). Obviously, the IRES-dependent mechanism offers a clear advantage to viruses that compete with the host conventional ribosomal scanning mechanism for translation initiation. Yet IRES elements have also been found in some cellular mRNAs. These include the mammalian BiP, insulin-like growth factor 2, c-myc (16), eukaryotic translation initiation factor 4G (eIF4G) (14), fibroblast growth factor 2 (55), vascular endothelial growth factor (1, 52), Kv1.4 (38), and MYT2 (22) genes and the Drosophila melanogaster genes Antennapedia (39) and Ultrabithorax (59). IRES elements of cellular genes are thought to take part in translational modulation, especially in response to changes in physiological conditions. The developmentally regulated IRESs of Drosophila (Antennapedia and Ultrabithorax) and the megakaryocytic differentiation-induced IRES of the human PDGF2 gene, c-sis, were demonstrated to confer translational modulation in vivo.
Molecular mechanisms by which cellular IRESs mediate internal translation have not been clarified. However, studies of viral 5′ UTRs strongly suggest that IRES elements are composed of highly ordered RNA structures that are recognized directly, by the 40S initiating ribosomes, or indirectly, via specific trans-acting factors. It is believed that presentation of certain sequences in appropriate conformation is required for protein recognition. A few secondary and tertiary RNA structures, as well as primary sequences, are thought to serve as necessary cis-acting components for viral IRES activity: an oligopyrimidine tract located at the 3′ border of the IRES at a fixed distance (∼20 nt) upstream of an AUG codon (2); sequences complementary to 18S rRNA within the polypyrimidine tract and surrounding the AUG codon (43); and a Y-shaped structure, located upstream of an AUG codon, that is occasionally involved in a pseudoknot interaction (25, 26, 29). These features also exist in the D-IRES element of the c-sis and are therefore believed to play a role in its activity (4).
Intense efforts are invested in identifying proteins that mediate internal initiation of translation. Interestingly, cellular proteins have been shown to mediate the function of viral IRES elements (2). In addition to factors known for their other roles in RNA metabolism, a set of canonical translation initiation factors, including eIF4A, eIF4G, eIF4B, eIF2, and eIF3 (40–42), were shown to be essential to the internal translation of certain picornaviruses. Among the noncanonical factors are the La autoantigen, known as the transcription termination factor of RNA polymerase III; polypyrimidine tract binding protein (pPTB), known as a negative regulator of pre-RNA splicing; and poly(rC) binding protein (PCBP), also known as hnRNP E (2, 6, 13). The exact roles of these proteins as mediators of viral IRES activity remain an enigma. Compared to viral IRESs, little is known about IRES elements of cellular mRNAs. Information about the boundaries of cellular IRES elements is sparse, and there are no data on the requirements for RNA-protein interactions.
The present study was conducted to find the cis elements within the c-sis 5′ UTR required for IRES activity (the basal level of internal translation in nondifferentiated cells) and for D-IRES activity (differentiation-induced enhancement of IRES performance) and to learn about the trans-acting factors that mediate D-IRES activity. We found the following: the maximal basal IRES activity in nondifferentiated cells is conferred by the full-length 5′ UTR; the sensitivity to differentiation (D-IRES) is conferred by a 630-nt fragment spanning nt 215 to 846 of the 5′ UTR; and a ribosome-associated 43-kDa protein, identified as hnRNP C, binds in a differentiation-induced manner to the D-IRES region.
MATERIALS AND METHODS
Plasmid construction.
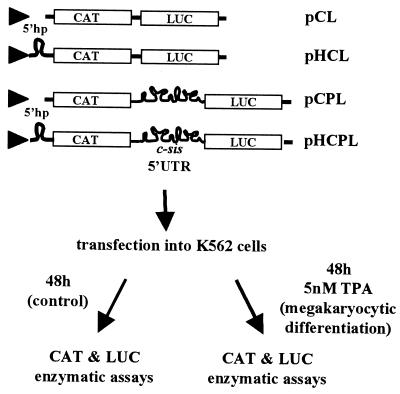
Intact or truncated c-sis 5′ UTRs were inserted upstream of the luciferase (LUC) reporter gene in the pBS-LUC plasmid (4) by ligating the pBS-LUC SpeI-NcoI fragment with PCR fragments generated by using psis4.0 (47) as a template and oligonucleotide primers homologous to specific c-sis 5′ UTR sequences. The following oligonucleotides which bear synthetic SpeI or NcoI sites were used: JB7 (5′-CCCCACTAGTGGCAACTTCTCCTCC-3′) and OS70 (5′-GCGAGCCATGGCTGCTCCGG-3′) to amplify the region spanning nt 1 to 227 (region 1-227) (truncation 2); OS64 (5′-CCCCACTAGTAACCGGAGCAGCCGCAGC-3′) and OS65 (5′-CCAACCATGGCTTTGCAACGGCAGC-3′) to amplify region 215-497 (truncation 3); OS63 (5′-CCCCACTAGTCCGGCGGGCAGAGGA-3′) and OS35 (5′-CCCCCCATGGCGACTCCGGGCCCGGCCC-3′) to amplify region 601-1022 (truncation 4); OS66 (5′-GGGTACTAGTGCTGCCGTTGC-3′) and OS61 (5′-CAGTCCATGGTTCGTCTTCACTCGC-3′) to amplify region 475-649 (truncation 5); OS66 and OS35 to amplify region 475-1022 (truncation 6); N1-1 (5′-CCCCCCGGGACTAGTCTCGAGCTGCCGTTG-3′) and N1-2 (5′-CCCCGCATGCCCATGGCTCCAACCTCCAAGAG-3′) to amplify region 475-685 (truncation 7); JB7 and N1-2 to amplify region 1-685 (truncation 8); OS63 and OS67 (5′-CTCACCCCCATGGCCCCGGC-3′) to amplify region 601-846 (truncation 9); OS66 and OS62 (5′-CAGGCCATGGGTCCGAGGCCGGCTA-3′) to amplify region 475-797 (truncation 10); OS64 and OS67 to amplify region 215-846 (truncation 11); JB7 and OS67 to amplify region 1-846 (truncation 12); OS64 and OS35 to amplify region 215-1022 (truncation 13); and OS68 (5′-GCGGACTAGTCGGGGGCATGCG-3′) and OS35 to amplify region 862-1022 (truncation 14). The ATG of the NcoI site was designed to match exactly with the ATG of LUC. Each SpeI-SacI fragment containing the truncated 5′ UTR fused to LUC was then ligated with the 4.1-kb SpeI-SacI fragment of pBI-FC1 or pHP-FC1 (55) to create the plasmids without or with the 5′ hairpin (5′hp), respectively. The plasmids containing the full-length c-sis 5′ UTR between the chloramphenicol acetyltransferase (CAT) reporter gene and LUC correspond to the previously described pCPL and pHCPL plasmids. Similarly, pCL and pHCL correspond to plasmids that contain the CAT-LUC transcription unit under the control of the cytomegalovirus (CMV) promoter, without any c-sis 5′ UTR sequences between the cistrons (4).
Computer prediction of RNA folding.
The evolutionarily conserved structure of the updated human c-sis 5′ UTR sequence was predicted by a combination of phylogenetic, thermodynamic, and statistical methods as described previously (4). The prediction of specific structural elements within the truncated fragments was reconfirmed by the MFOLD and EFFOLD software programs. MFOLD, based on a dynamic algorithm, generates a set of suboptimal structures close to the lowest free energy (21), whereas EFFOLD computes a set of the lowest-free-energy structures ranked by their frequency of recurrence in a simulation based on fluctuating thermodynamic parameters (27). Both programs were used because items within a predicted lowest-free-energy structure which have a high ratio of recurrence are considered more robust. Thus, evolutionarily conserved structures originally predicted in the full-length 5′ UTR that were robust in the context of the truncated background were considered valid in the truncated fragments.
Cells and megakaryocytic differentiation.
The human chronic myelogenous leukemia cell line K562 (31) was grown in RPMI 1640 medium (Gibco) supplemented with 50 U of penicillin/ml, 50 mg of streptomycin/ml, and 10% fetal calf serum (FCS). Megakaryocytic differentiation was induced by dilution of cells at a density of 1.2 × 106/ml, to a final concentration of 5 × 105/ml, with medium containing 12-O-tetradecanoylphorbol-13-acetate (TPA) (Calbiochem) at a concentration of 5 nM for 48 h.
Plasmid transfections.
Fifteen to 60 μg of supercoiled plasmid DNA per a total of 107 K562 cells resuspended in 0.8 ml of RPMI 1640 without serum was used for each electroporation pool, with an electric pulse of 240 V and 1,500 μF (Easy Ject+ electroporator; Equibio). Immediately following the electric pulse, the cells were transferred to RPMI 1640 medium supplemented with 20% FCS. Twenty-four hours after electroporation, the cells were diluted to a final concentration of 5 × 105 living cells (as determined by using trypan blue for counting) per ml of RPMI 1640 supplemented with 10% FCS, with or without 5 nM TPA, for 48 h.
CAT and LUC assays.
The TPA-treated and control transfected cells were harvested simultaneously for CAT and LUC assays. For the enzymatic activity assays, the cell pellet was lysed by four freeze-thaw cycles in 100 μl of 0.1 M Tris (pH 8.0). CAT activity was determined by a phase extraction assay that quantifies butyrylated 3H-labeled chloramphenicol products by liquid scintillation counting following xylene extraction (pCAT Reporter Gene System; Promega). LUC activity was determined by using a TD-20e-Luminometer (Turner) following a 15-s incubation of 5 to 10 μl of lysate with 470 μM luciferin (Sigma) and 270 μM coenzyme A (Sigma) in 20 mM K-HEPES (pH 7.8)–1 mM EDTA–4 mM magnesium acetate–1 mg of bovine serum albumin/ml–530 μM ATP.
Cell fractionation and preparation of extracts.
All steps were carried out at 4°C. K562 cells were collected by a 2-min spin at 250 × g followed by two washes with phosphate-buffered saline (PBS). For cytoplasm-nucleus separation, cells were resuspended in 4 packed cell volumes of hypotonic buffer containing 10 mM HEPES (pH 7.9), 1.5 mM magnesium chloride, 10 mM potassium chloride, 0.5 mM dithiothreitol, 1 mM sodium orthovanadate, 50 mM NaF, and Complete protease inhibitor (Boehringer Mannheim), followed by a 15-min incubation to allow swelling. Cells were then Dounce homogenized (20 strokes). Nuclei were collected by a 5-min spin at 500 × g and saved on ice for further use. The supernatant was centrifuged at 100,000 × g for 1 h and then saved as the cytoplasmic S100 fraction. The ribosomal pellet was resuspended with 0.2 supernatant volume of ribosomal salt wash (RSW) buffer containing 0.25 M sucrose, 0.5 M potassium chloride, 1 mM dithiothreitol, 0.1 mM EDTA, 1 mM sodium orthovanadate, 50 mM NaF, and Complete. The mixture was then Dounce homogenized by 10 strokes followed by 2 h of incubation and a further 2 h of centrifugation at 120,000 × g. The supernatant was used as the RSW fraction. The previously collected nuclear pellet was resuspended in 0.5 packed nuclear volume of low-salt buffer, followed by dropwise addition of 0.5 packed nuclear volume of high-salt buffer with gentle stirring. The low- and high-salt buffers contained 20 mM HEPES (pH 7.9), 1.5 mM magnesium chloride, 0.2 mM EDTA, 0.5 mM dithiothreitol, 25% glycerol, 1 mM sodium orthovanadate, 50 mM NaF, Complete, and either 10 mM (low salt) or 0.6 M (high salt) potassium chloride. The nuclei were gently mixed and allowed to extract for 30 min, followed by 30 min of centrifugation at 20,000 × g. The supernatant was used as the nuclear fraction. All extracts were dialyzed against a buffer containing 20 mM HEPES (pH 7.9), 20 mM potassium chloride, 0.2 mM EDTA, 0.5 mM dithiothreitol, and 50 mM NaF. Immediately after dialysis, sodium orthovanadate (to a final concentration of 1 mM) and Complete were added. The extracts were further cleared by 5 min of centrifugation at 5,000 × g. All extracts were immediately aliquoted and stored at −70°C.
In vitro transcription reactions.
pBS-LUC plasmids containing truncated c-sis 5′ UTRs downstream of the T7 promoter and upstream of LUC were used as templates for in vitro transcription following their linearization at the 3′ end of the 5′ UTR by NcoI digestion. For [32P]UTP labeling, the transcription reaction mixture in a final volume of 10 μl contained 0.25 mM (each) ATP, GTP, and CTP, 4 μM UTP, 1 μM Br-UTP (Sigma), 6.25 μM [32P]UTP (800 ci/mmol; Amersham), 2.5 mM dithiothreitol, 28 U of RNasin (Promega), 1 μg of linear DNA template, and 20 U of T7 RNA polymerase (Epicentre) with its commercial buffer. For [32P]CTP or [32P]ATP labeling, 6.25 μM [32P]CTP (800 ci/mmol; Amersham) or [32P]ATP (400 ci/mmol; Amersham) was used instead of [32P]UTP, and the concentrations of unlabeled UTP and CTP or ATP were changed to 0.25 mM and 5 μM, respectively. The transcription reaction mixtures were incubated on ice for 2 h, followed by addition of 1 U of RNase-free DNase (Ambion) for a 15-min incubation at 37°C. Unlabeled RNAs were transcribed by using the T7 high-yield transcription kit (AmpliScribe; Epicentre). All RNA transcripts were extracted with phenol-chloroform and filtered through Sephadex G-50 (Pharmacia). The total amount of 32P-labeled RNA probes was calculated based on the incorporated radioactivity quantified following trichloroacetic acid precipitation. The integrity of 32P-labeled RNA, or cold RNA, was verified by 4% polyacrylamide–7 M urea or 1.5% agarose–formaldehyde gel electrophoresis, respectively. Cold RNA concentrations were quantified by using 1D image analysis software (Kodak Digital Science).
UV cross-linking assay.
32P-labeled RNAs (5 fmol) were incubated for 10 min at 37°C with 20 μg of protein extract in protein binding buffer containing 10 mM HEPES (pH 7.6), 3 mM magnesium chloride, 30 mM potassium chloride, 1 mM dithiothreitol, 5% glycerol, 1.3 mM ATP, 1 μg of yeast tRNA (Boehringer Mannheim), and 14 U of RNasin (Promega) in a final volume of 20 μl. For competition experiments, 5 μg of protein extract was used, and the unlabeled RNA competitor was added 10 min prior to the RNA probe addition. RNA was cross-linked to the bound proteins by 15 min of 312-nm radiation (4.5 J/cm2) using a UV cross-linker (UltraLum). RNase A (Boehringer Mannheim) was then added to a final concentration of 2 mg/ml, for 60 min of incubation at 37°C. The cross-linked proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide), followed by autoradiography. A Rainbow 14C-labeled protein molecular weight marker at the range of 14.3 to 220 kDa (Amersham) was used. The electrophoretic mobilities and relative intensities of the cross-linked signals were determined by using 1D image analysis software.
Immunoprecipitation of UV-cross-linked proteins.
Following RNase A treatment, the samples were precleared by centrifugation in a microcentrifuge for 5 min at 20,000 × g. A total of 1.5 μl of the anti-hnRNP C monoclonal antibody 4F4 (from G. Dreyfuss) and 450 μl of PBS with 0.01% CaCl2, 0.01% MgCl2, and 1% Nonidet P-40 were added to the supernatant and incubated for 1 h on ice. Next, 1.5 μl of rabbit anti-mouse polyclonal antibody was added for a further incubation of 1 h on ice. Then a 30-μl packed volume of protein A-Sepharose was added, and the samples were incubated overnight on a rotary shaker. After two washes of the Sepharose beads, the eluted proteins were separated by SDS–10% PAGE.
Determination of hnRNP C phosphorylation level.
At 46 h following TPA treatment, 1.2 × 106 control or differentiated K562 cells were washed twice with saline and resuspended in 2 ml of Dulbecco’s modified Eagle medium lacking sodium phosphate (Sigma) and supplemented with 10% dialyzed FCS. The cells were labeled for 2 h with 0.2 mCi of 32Pi (Amersham catalog no. PBS13)/ml, followed by two washes with cold PBS. Whole-cell extract was prepared by using 60 μl of radioimmunoprecipitation assay buffer containing 50 mM NaF, 1 mM sodium orthovanadate, and Complete (Boehringer). Immunoprecipitation of hnRNP C was performed by using the monoclonal antibody 4F4, in a final volume of 0.4 ml, as described above, in the presence of 1 mM sodium orthovanadate and Complete. Following separation by SDS–10% PAGE, the proteins were blotted to a nitrocellulose membrane and quantified by using a phosphorimager. Then the same membrane was analyzed by Western analysis according to standard procedures, with the 4F4 antibody.
RESULTS
The entire 5′ UTR is necessary for maximal IRES activity.
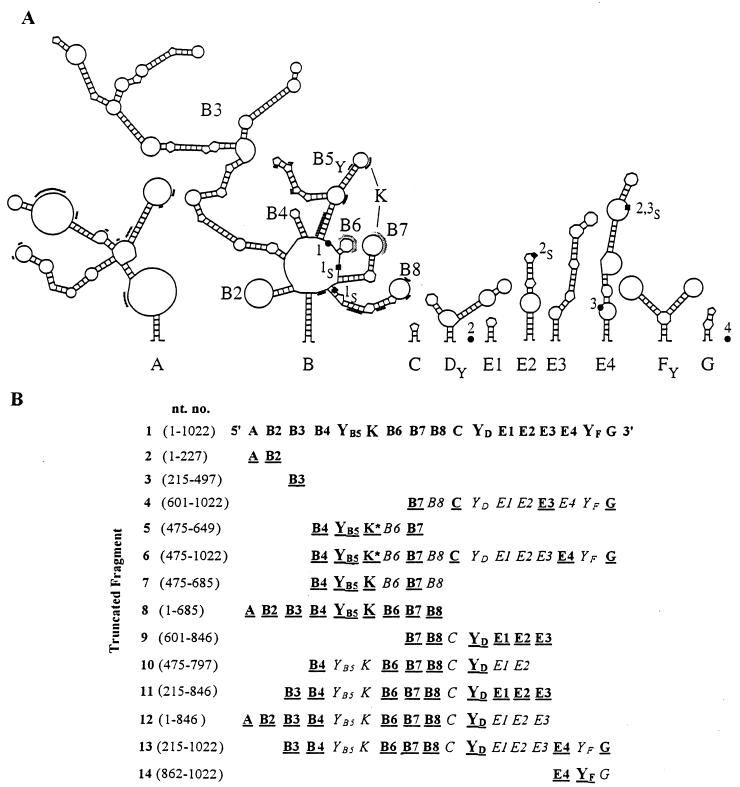
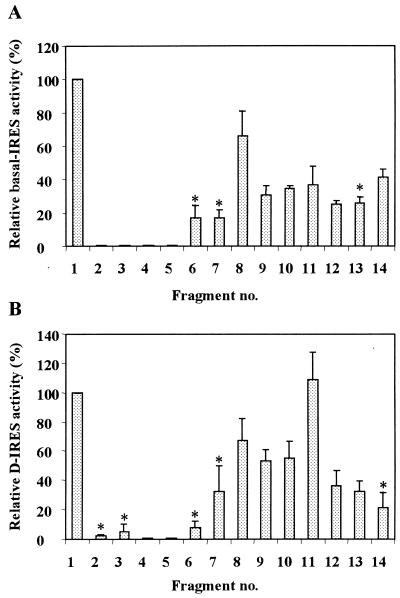
Using deletion mapping of the evolutionarily conserved structural domains, we focused at the boundaries of the IRES—the element required for basal internal initiation in nondifferentiated cells—and the D-IRES—the element required for the enhanced IRES activity upon differentiation. Preparation of the truncated constructs revealed discrepancies compared to the GenBank sequence. Referring to the RNA start site as nt 1 (which corresponds to nt 398 of human c-sis, accession no. M19719), the updated sequence includes the following seven changes: nt 706 and 707 are GG (not CC), nt 921 is C (not U), nt 756 (G) and nt 786 (C) are absent, and immediately following nt 863 and nt 868 there is an insertion of C. The nucleotide numbering throughout the present study includes the above corrections. Interestingly, reexamination of the aligned sequences of the human, mouse, and feline c-sis 5′ UTRs (see our previous study [4]) revealed that the modifications in the human sequence described above exist in the mouse and feline sequences. This greater similarity among the human, mouse, and feline sequences further enhances the credibility of the computer-aided model of the evolutionarily conserved structure presented in Fig. 1A. Specific structural regions were synthesized by PCR amplification and placed in the intercistronic spacer (ICS) region of a bicistronic plasmid vector. The computer prediction of specific structural elements within the context of the truncated fragments was reconfirmed. Evolutionarily conserved structures originally predicted in the full-length 5′ UTR that were robust in the context of the truncated background were considered valid, as shown in Fig. 1B. We used the bicistronic transcriptional unit expressing the Escherichia coli CAT and firefly LUC reporter genes as the first and second cistrons, respectively (Fig. 2). Two versions of each truncated plasmid were made, with and without a 5′hp upstream of the first cistron. The plasmid pCL, lacking a c-sis sequence in the intercistronic space, is referred to as the “empty” plasmid (Fig. 2). In a previous study we demonstrated the integrity of a bicistronic RNA containing the full-length c-sis 5′ UTR between CAT and LUC (4). The integrity of the RNA indicated the absence of cryptic promoters or specific cleavage sites in the 5′ UTR. Similarly, in the present study, both CAT and LUC enzymatic activities reflect the relative translation efficiencies of the first and second cistrons from an intact bicistronic mRNA, regardless of the absolute mRNA level in the sample. The basal IRES value of each fragment in nondifferentiated cells was determined by comparing the inhibitory effect of the 5′hp on the translation of the first cistron (CAT) relative to its effect on the translation of the second cistron (LUC). This was achieved by transfection of both versions of each plasmid (with and without the 5′hp) into nondifferentiated K562 cells. CAT and LUC activities expressed from the empty plasmid, pCL, were inhibited to the same extent by the 5′hp. In contrast, when the full-length 5′ UTR was located in the ICS, the translation of CAT was inhibited while that of LUC was not. The ratio between the 5′hp effect on LUC and the 5′hp effect on CAT, normalized to the ratio obtained from the empty vector, was termed the basal IRES value. In each transfection experiment, the basal IRES value of each truncated fragment was compared to that of the full-length 5′ UTR. Figure 3A shows the basal IRES value of each of the truncated fragments presented in Fig. 1B. Although some truncated 5′ UTR fragments functioned as IRES elements, only the full-length 5′ UTR was able to confer the maximal activity.
FIG. 1.
(A) Updated structural model of the 5′ UTR of human c-sis. Based on the updated sequence, a conserved structure of the c-sis 5′ UTR was predicted by a combination of phylogenetic, thermodynamic, and statistical methods as described previously (4). RNA pseudoknot interaction is indicated by the letter K. The three upstream AUG codons are marked as 1●, 2●, and 3●, and their in-frame stop codons are marked as 1S■, 2S■, and 3S■. AUG4, the translation initiator codon, is positioned at nt 1023 to 1025 at the very 3′ end and is marked as 4●. U- and A-rich sequences are marked by additional light or heavy lines, respectively. (B) Schematic presentation of the truncated fragments. Nucleotide numbering refers to the 5′ and 3′ borders of each of the truncated fragments and corresponds to the human c-sis 5′ UTR. The folding of each fragment was analyzed as described in Materials and Methods. Evolutionarily conserved structures originally predicted in the full-length 5′ UTR that were robust in the context of the truncated background are boldfaced and underlined. The pseudoknot interaction is indicated by the letter K; an asterisk indicates that the pseudoknot is not the authentic one.
FIG. 2.
Schematic presentation of the experimental system. The bicistronic transcriptional unit expressing the E. coli CAT and firefly LUC reporter genes as the first and second cistrons, respectively, under the control of the CMV promoter (triangle) was used. The full-length or truncated c-sis 5′ UTR was placed in the intercistronic space of the bicistronic unit. Two versions of each plasmid were made, with and without a 5′hp upstream of the first cistron, as detailed previously (4). Plasmids lacking c-sis sequences, without or with the 5′hp, were termed pCL or pHCL, respectively, whereas plasmids containing the full-length c-sis 5′ UTR were termed pCPL or pHCPL. K562 cells were transfected with each of the recombinant plasmids, followed by a 48-h incubation in medium with or without TPA, as explained in Materials and Methods. CAT and LUC enzymatic activities were then determined.
FIG. 3.
(A) Each of the truncated 5′ UTR segments described in Fig. 1B was placed in the bicistronic expression unit between CAT and LUC. Both versions of each plasmid, with and without the 5′hp, were transfected into nondifferentiated K562 cells, followed by analysis of CAT and LUC activities. The basal IRES value represents the ratio between the 5′hp effect on LUC and the 5′hp effect on CAT, normalized to the ratio obtained from the empty vector. In each transfection experiment, the IRES value of each truncated fragment was compared to that of the full-length 5′ UTR (fragment 1), which was taken as 100%. Each IRES value is the average ± standard error (SE) from two to four independent experiments. The analysis of variance (ANOVA) procedure was used to verify the statistical significance of the results. Insignificant IRES values (P > 0.05) are marked by asterisks. (B) Each of the truncated bicistronic plasmids described in Fig. 1B without the 5′hp was transfected into K562 cells. After 48 h of incubation under control or differentiation conditions, CAT and LUC activities were determined. The D-IRES value represents the LUC/CAT ratio obtained in the differentiated cells relative to that in the control cells, normalized to the same ratio obtained from the empty vector (pCL). The D-IRES value of the full-length 5′ UTR (fragment 1) was taken as 100%. Each value is the average ± SE from two to four independent experiments. The ANOVA procedure was used to verify the statistical significance of the results. Insignificant D-IRES values (P > 0.05) are marked by asterisks.
IRES-conferring entities within the 5′ UTR.
As shown in Fig. 1A, the c-sis 5′ UTR is predicted to contain three Y-shaped structures, termed B5, D, and F, which are located upstream of AUG1, AUG3 and AUG4, respectively. YB5 is predicted to be involved in a pseudoknot interaction with the B7 stem-loop structure. The statistical simulation, computed by the Turner energy rule, revealed significance scores of −1.75, −1.87, and −2.39 standard deviations for the folding regions of B5, D, and F, respectively. Such lower score values indicate that the predicted RNA structures are more stable than the random sequences of the same base composition and may imply a structural role for the sequence information. Interestingly, the common structural motif shared by many viral and cellular IRES elements is a conserved Y-shaped stem-loop structure upstream of an AUG codon which is occasionally involved in a pseudoknot interaction (1, 25, 26, 28, 29). To check the potential relevance of such a motif to c-sis basal IRES activity, we analyzed the ability of the truncated fragments (Fig. 1) containing a single predicted Y motif (YB5, YD, or YF) to confer internal translation. Interestingly, fragment 2 or 3, which contains domains A and B2 or domain B3, respectively, does not harbor any Y-shaped structure upstream of an AUG codon, and both were totally inactive as IRESs (Fig. 3A). Moreover, fragment 4, for which folding analysis failed to predict any stable Y-shaped structures, also had no basal IRES activity. However, the short fragment 14, containing the predicted structure YF as a single Y-shaped motif, had a basal IRES value of 41%. Fragments containing the predicted YD as the single Y motif (fragments 9 through 13) were also active as IRESs (31, 34, 37, 26, and 26%, respectively). The low IRES values of these fragments, which are spread throughout the complete 5′ UTR, may reflect the stability of the YD structure within each fragment, the importance of the YD context among other structures, or the structural integrity of other elements. Fragments 5 through 7, which harbor the predicted YB5 element, were hardly active as IRESs (0, 17, and 17%, respectively). However, fragment 8, which harbors the YB5 element in the context of predicted structures A through B8, had an IRES value of 66%. Note that fragment 8 was predicted to fold similarly to the corresponding region of the full-length 5′ UTR.
In summary, the data suggest that the 5′ UTR of c-sis contains three operative modules. Each module confers some degree of internal translation and may contain a Y motif among other structures. The Y-shaped structures, which need to be confirmed experimentally, appear to be necessary but not sufficient to confer basal IRES activity. Understanding of the importance of the predicted Y-shaped structures to IRES function awaits extensive mutational analysis. However, the data clearly show that only the full-length 5′ UTR can confer the maximal basal IRES activity in nondifferentiated cells.
D-IRES-conferring entities within the 5′ UTR.
We previously showed that the translation initiation driven by the c-sis 5′ UTR is enhanced upon megakaryocytic differentiation of K562 cells (3, 4). This differentiation-induced IRES was termed D-IRES. To measure the D-IRES capacities of the truncated 5′ UTR fragments, each of the bicistronic plasmids lacking the 5′hp was transfected into K562 cells, which were then induced to differentiate as illustrated in Fig. 2. CAT activity was used as an internal control to normalize LUC activity in the transfected cells. The D-IRES value of each transfected bicistronic plasmid represents the ratio of the above LUC activity obtained from differentiated cells relative to that in control cells, normalized to the same ratio obtained from the empty vector (pCL). Thus, the D-IRES value specifically reflects the c-sis 5′ UTR-dependent enhancement of second-cistron translation due to differentiation. The full-length c-sis 5′ UTR conferred 3.7-fold enhancement of the second-cistron translation, and this D-IRES value was taken as 100%. The D-IRES value obtained from each truncated bicistronic plasmid was compared to the D-IRES value of the full-length 5′ UTR in each transfection experiment. As presented in Fig. 3B, all the truncated fragments except fragment 11 were less active as D-IRES elements than the full-length 5′ UTR. Fragment 14, which harbors the predicted YF as a single Y motif, was able to confer some basal internal translation in nondifferentiated cells but had a very low and insignificant D-IRES activity. Thus, the region spanning 160 nt at the 3′ end of the 5′ UTR seems to harbor a weak IRES element that is not affected by cellular changes. Fragments 2 through 5, which did not confer any IRES activity in nondifferentiated K562 cells (Fig. 3A), also were not able to serve as D-IRES elements (Fig. 3B). Similarly, fragments 6 and 7 conferred low and insignificant D-IRES activity, in accordance with their inability to support efficient basal IRES activity. Fragment 8, which contains the 5′ portion of the 5′ UTR (structural elements A through B8), had a D-IRES value of 68%, similar to its basal IRES value (66%). Fragments 9 and 10, which share structural elements B7 through YD, were also able to confer significant D-IRES activity (53 to 55%). Thus, the IRESs of these fragments were sensitive to cellular differentiation. The fusion of the B3 structure to the regions represented by fragments 9 and 10 to yield fragment 11 resulted in a very efficient D-IRES activity (108%). Note that although the IRES activity of fragment 11 was very modest (37% [Fig. 3A]), it was sensitive to differentiation, like that of the full-length 5′ UTR. The folding analysis of fragment 11 revealed that its predicted B7-through-YD structural elements are significantly robust. Thus, the predicted structure B3 may be important for D-IRES activity by stabilizing other motifs. Structure B3 may play an additional role, such as attracting specific RNA binding proteins to the IRES elements. The addition of either region 1-215 or region 846-1022 to fragment 11 (region 215-846) to yield fragment 12 or 13, respectively, had a negative effect on D-IRES activity, which was prevented by the addition of both regions together. This might indicate a relationship between the structural elements, or it might simply reflect the stability of the structures important for the D-IRES activity.
In summary, the data presented in Fig. 3B further demonstrate that specific secondary structures, rather than primary sequence, are important for D-IRES activity. Since the D-IRES activity of fragment 11 was similar to that of the full-length 5′ UTR, we reasoned that entities important for efficient D-IRES activity within the c-sis 5′ UTR are located in the region spanning domains B3 through E3.
Interactions of proteins with the mRNA leader of c-sis.
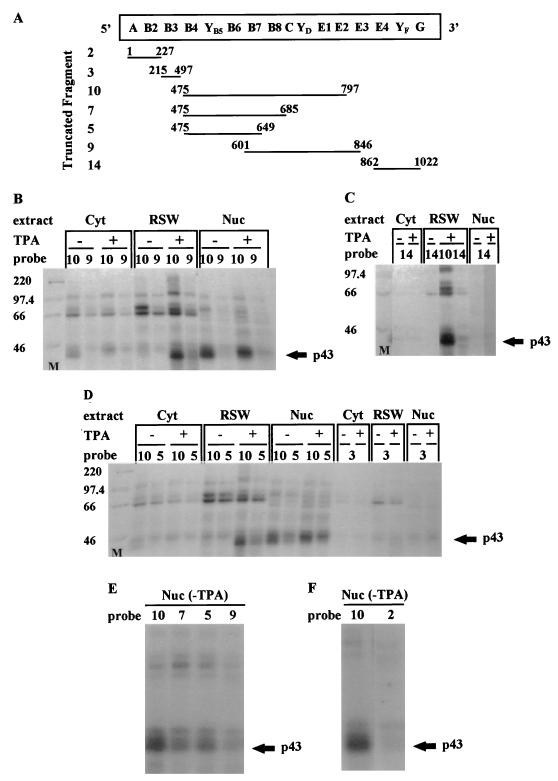
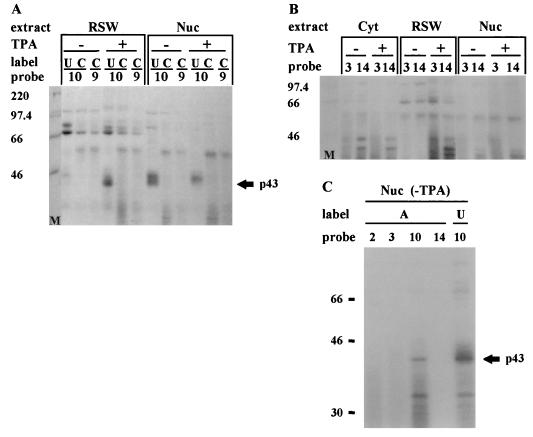
In order to find trans-acting factors required for D-IRES activity, we first analyzed the binding of the c-sis 5′ UTR with proteins from control and megakaryocytic differentiated cells. Proteins extracted from the nuclei, cytoplasm (S100), or RSW of control or TPA-treated K562 cells were UV cross-linked to 32P-labeled RNA probes presenting different structural regions of the 5′ UTR (Fig. 4A). Only fragments of 300 nt or less were used for the UV cross-linking experiments. A set of such experiments (Fig. 4B through D) yielded information about the molecular weights of the binding proteins, the regions to which they bind within the 5′ UTR, and the effect of megakaryocytic differentiation on the binding profile. A broad range of proteins was cross-linked to the 5′ UTR-derived RNA probes. Under the condition used, the majority of binding was observed when fragment 10 was utilized as the probe. Probes 9 and 5, which are truncated versions of probe 10, resulted in less-prominent bands, whereas other probes barely labeled any protein (Fig. 4B through F). As mentioned above, fragment 10 conferred 35% basal IRES activity and 53% D-IRES activity (Fig. 3). The specific binding of RSW-derived proteins to this region supports the notion of their involvement in mediating the internal translation of c-sis.
FIG. 4.
UV cross-linking. (A) Schematic presentation of the predicted structural domains within the c-sis 5′ UTR. Truncated fragments as detailed in Fig. 1B, which were used as 32P-labeled probes for the UV-cross-linking experiments, are indicated. Nucleotide numbering refers to the 5′ and 3′ borders of each truncated fragment. (B through F) Cytoplasmic S100 (Cyt), RSW, and nuclear (Nuc) extracts were prepared from control (−TPA) or differentiated (+TPA) K562 cells. Twenty micrograms of protein was cross-linked to 5 fmol of a [32P]UTP-labeled RNA probe representing a truncated 5′ UTR fragment as listed in Fig. 4A. The proteins were separated on an SDS–10% PAGE gel. Molecular weight markers (M) are indicated at the left.
Effect of differentiation on protein binding.
The increased or decreased intensity of some protein bands upon differentiation reflects a change in their abundance and/or their RNA binding capacity. Interestingly, most of these differentiation-linked changes were exhibited by proteins associated with the ribosomal fraction. The most extreme differentiation-driven change in protein band intensity was exhibited by a 43-kDa protein that was both nucleus and ribosome associated. As shown in Fig. 4B and D, the nuclear signal of p43 was reduced, whereas the RSW-associated signal was dramatically intensified, upon differentiation. Thus, the p43 signal was mostly nuclear prior to differentiation, whereas after TPA treatment a significant portion of the p43 binding activity was associated with the ribosomal fraction. Assuming that the nuclear and RSW p43 bands represent the same protein, most of its RNA binding activity appears to have been relocated from the nucleus to the cytoplasm upon differentiation.
Mapping of the p43 binding site.
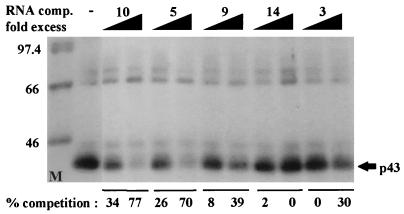
The binding site of p43 was analyzed by using different regions of the 5′ UTR. The most efficient labeling of p43 was to the region spanning nt 475 to 797 (probe 10 [Fig. 4]). A significant decrease in p43 labeling efficiency was observed following deletion of either region 686-797 (probe 7 [Fig. 4E]) or region 650-797 (probe 5 [Fig. 4D and E]) from the 3′ end of this segment. Deletion from the 5′ end of this segment impaired p43 labeling even more severely. Probe 9, which lacks region 475-600, barely labeled p43 (Fig. 4B and E). Therefore, it seems that the integrity of the region spanning nt 475 to 797 (domains B4 through E2) is important for efficient p43 binding. The p43 binding site was further confirmed by RNA probes labeled with [32P]UTP, [32P]CTP, or [32P]ATP for the UV-cross-linking experiments (Fig. 5). p43 was visualized by probe 10 only when it was labeled with [32P]UTP or [32P]ATP, not when it was labeled with [32P]CTP (Fig. 5A and C). Probes 2, 3, 9, and 14 were not able to detect p43, regardless of the labeled nucleotide (Fig. 4 and 5), suggesting that p43 interacts with U- and A-rich sequences located within fragment 10. Indeed, U and A stretches are located within the region spanning nt 500 to 680 of the 5′ UTR, which includes the predicted structures B5 through B8 (Fig. 1A). However, since probe 2, which also contains U and A stretches, does not bind p43 (Fig. 4F), it seems that the high-affinity binding of p43 to the B5-through-B8 domain is due to specific sequence and/or structure within this region and not to random clustering of uridines, which are known to be highly photoreactive. The binding site of p43 was also confirmed by competition experiments using the corresponding fragments as cold RNA competitors (Fig. 6). However, although fragments 3 and 9 barely labeled p43 by UV cross-linking, when they were present in 50-fold molar excess they were able to compete to some extent with probe 10 for p43 binding. The results of these UV-cross-linking and competition experiments point to a high-affinity binding site within region B4 through B8 and to lower-affinity binding sites within regions B8 through E2 and B3. As discussed above, important elements for D-IRES activity within the c-sis 5′ UTR reside within domains B3 through E2 (fragment 11), raising the possibility of p43 importance for D-IRES function. Yet the data presented are not completely consistent with the notion of p43 involvement, since it does not cross-link to fragment 9, which has D-IRES activity, whereas fragment 5 can compete for p43 binding although it does not function as a D-IRES. This discrepancy may be explained by a possible requirement for p43 to stabilize a specific RNA conformation in a specific sequence context, and it may be required but not sufficient for D-IRES function.
FIG. 5.
Twenty micrograms of protein of the cytoplasmic S100 (Cyt), RSW, and nuclear (Nuc) extracts from control (−TPA) or differentiated (+TPA) K562 cells were UV cross-linked to 5 fmol of RNA probes representing the truncated fragments listed in Fig. 4A. The probes were labeled with [32P]UTP or [32P]CTP (A), [32P]CTP (B), or [32P]UTP or [32P]ATP (C). The RNA-protein complexes were separated on an SDS–10% polyacrylamide gel. The molecular weights of the markers (in kilodaltons) are indicated at the left.
FIG. 6.
Five femtomoles of [32P]UTP-labeled RNA probe 10 and 5 μg of protein of RSW from differentiated cells were used for each UV-cross-linking reaction. A 10- or 50-fold molar excess of unlabeled RNA fragment 3, 5, 9, 10, or 14, as listed in Fig. 4A, was included as a cold competitor. Following SDS–10% PAGE and autoradiography, quantitative analysis of the relative p43 band intensity was performed with 1D image analysis software. The percent competition was defined by 100[1 − (x/y)], where x is the p43 signal obtained in each competition reaction and y is the maximum p43 signal obtained without competitors. The molecular weights of the markers (in kilodaltons) are indicated at the left.
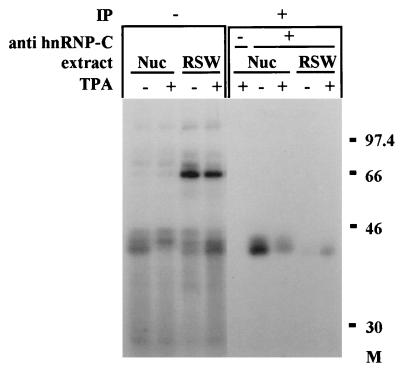
Identification of hnRNP C determinants on p43.
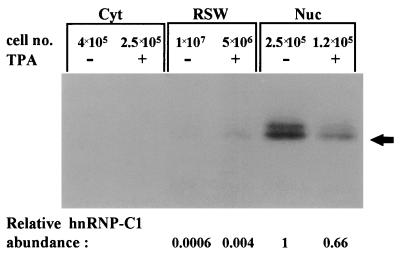
The molecular weight of p43, its RNA binding specificity, and its nuclear localization in undifferentiated cells, have raised the possibility that p43 is a member of the C-type heterogenous nuclear ribonucleoproteins (hnRNP C). The C proteins, C1 and C2, are major components of hnRNPs and appear to play a role in RNA splicing. C1 and C2 are antigenically related phosphoproteins that are generated by alternative splicing and differ in primary structure by the presence of a 13-amino-acid insert sequence in C2 (10, 58). To determine whether p43 is a member of the C proteins, a specific monoclonal antibody against hnRNP-C was used for immunoprecipitation of nuclear and RSW proteins following their UV cross-linking to probe 10. The results, presented in Fig. 7, confirmed the presence of hnRNP C antigenic determinants on p43. An additional nuclear protein, p44, was also immunoprecipitated by the antibodies against hnRNP C. We assume that p43 and p44 correspond to hnRNP C1 and C2, respectively. However, the label of p44 was nuclear both before and after the differentiation of K562 cells, whereas the label of p43 was partly translocated from the nucleus to the ribosomal fraction upon differentiation (Fig. 7). Since hnRNP C proteins are mostly known as nuclear proteins (10, 36), we wished to verify their cellular localization within K562 cells before and after differentiation. Polyclonal antibodies against hnRNP C were used for Western analysis of the cytoplasmic, RSW, and nuclear extracts of control and differentiated K562 cells. Figure 8 shows clearly that the majority of hnRNP C is nuclear both before and after differentiation. When 40-fold-more cells were used, faint bands were also detected in the RSW fractions. As indicated in Fig. 8, there is an overall reduction in hnRNP C biosynthesis in differentiated cells, which is mostly reflected in the nucleus. In contrast, in the ribosomal fraction of these cells, an elevated level of hnRNP C compared to the level in nondifferentiated cells was observed. However, even under differentiation conditions, the relative abundance of the nuclear protein was about 160-fold higher than that of the RSW-associated protein, yet the intensities of the nucleus- and ribosome-derived UV-cross-linking signals were similar (probe 10 [Fig. 4]). This further emphasizes that the intense UV-cross-linking signal of the RSW-associated hnRNP C is conferred by a very small amount of hnRNP C protein, which upon megakaryocytic differentiation may gain binding affinity towards the c-sis mRNA leader.
FIG. 7.
Identification of hnRNP C determinants on p43. Eighty micrograms of protein of nuclear (Nuc) or RSW extracts from control (−TPA) or differentiated (+TPA) K562 cells were UV cross-linked to 9 fmol of [32P]UTP-labeled RNA probe 10, as detailed in Fig. 4. Following UV cross-linking, the samples were immunoprecipitated (IP) by using a monoclonal antibody against hnRNP C (4F4). In the control samples monoclonal antibody 4F4 was omitted (−). Samples before immunoprecipitation are also shown (−IP). The RNA-protein complexes were separated on an SDS–10% polyacrylamide gel.
FIG. 8.
Cellular distribution of hnRNP C proteins in control and differentiated K562 cells. Polyclonal antibodies against hnRNP C were used for the Western analysis of 30 μg of cytoplasmic (Cyt), 30 μg of RSW, and 10 μg of nuclear (Nuc) extracts of control (−TPA) and differentiated (+TPA) K562 cells. The number of cells represented in each lane is indicated. Quantitative analysis of the relative band intensities was performed with 1D image analysis software. The calculated relative hnRNP C1 abundance (per cell) is indicated in arbitrary units.
Differentiation-induced phosphorylation of hnRNP C.
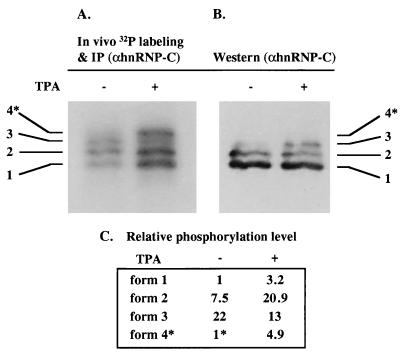
Next we wished to examine the function of hnRNP C as a mediator of D-IRES activity. For that purpose, purified recombinant hnRNP C protein made in E. coli was added to rabbit reticulocyte lysate to monitor its effect on c-sis IRES-mediated translation. The addition of recombinant hnRNP C did not enable the cell-free translation system to support c-sis IRES activity (data not shown). These negative results are not surprising, since the activity of the c-sis IRES is restricted to a specific cellular milieu, although hnRNP C is highly abundant in all cell types, including nondifferentiated K562 cells. Therefore, it seems likely that upon megakaryocytic differentiation, hnRNP C undergoes specific modifications that render it active as a mediator of IRES function. To find such modifications, we looked for differentiation-induced changes in its phosphorylation status. Control and differentiated K562 cells were metabolically labeled with inorganic 32Pi and immunoprecipitated with a monoclonal antibody specific for hnRNP C for detection of the phosphorylated forms. The total hnRNP C protein level was analyzed by Western analysis. As seen in Fig. 9, the overall phosphorylation status of hnRNP C was significantly increased upon differentiation. This is reflected both by the elevated level of 32P labeling per protein and by the appearance of hyperphosphorylated, slower-migrating forms. We speculate that a specific phosphorylated form is functional in IRES activation. Therefore, direct evidence of hnRNP C as a mediator of IRES function awaits identification of the involved kinase.
FIG. 9.
Differentiation-induced hyperphosphorylation of hnRNP C. Control and differentiated K562 cells (− and + TPA, respectively) were metabolically labeled with 32Pi, followed by immunoprecipitation using an antibody specific for hnRNP C, separation by SDS-PAGE, and blotting onto a nitrocellulose membrane. (A) The membrane was exposed to a phosphorimager for quantification of the phosphoproteins. (B) The same membrane was analyzed for hnRNP C total protein level by Western analysis. (C) The intensity of each band in panel A, as quantified by TINA software (Raytest), was compared to that of the corresponding band in panel B, as quantified by 1D image analysis software. The phosphorylation status per protein level of form 1 within the control cells was set at 1. Since the hyperphosphorylated form 4, marked by an asterisk, was not visualized by Western analysis, its 32P band intensity in panel A was compared to the total level of all hnRNP C forms in the corresponding lane in panel B. Thus, the phosphorylation status of form 4 in the control cells was represented as 1∗.
DISCUSSION
Regulation of translation initiation plays a pivotal role in the control of cell proliferation and differentiation. We have shown previously that translational control of the proto-oncogene c-sis is mediated by its cumbersome 5′ UTR, which contains an IRES that functions in a differentiation-induced manner (D-IRES) (3, 4). The observation that other known cellular and viral IRES elements vary in sequence, structure, and efficiency suggests that the details of their internal translation mechanisms also vary. In addition, the differing sensitivities of IRESs to the environmental conditions in various cell types or cell-free systems (7, 8) point to the yet-unresolved structure-function relationships between cis elements and trans-acting factors. The IRES of EMCV, the prototype of picornaviral type II IRESs, bears the initiator AUG codon at the 3′ boundary of the element and is well known for its high efficiency and low sensitivity to cellular conditions. Indeed, it was found to be dependent only upon canonical translation initiation factors (40, 41). In contrast, the inefficient type I and III IRESs are more sensitive to cellular conditions and are located upstream of the actual translation initiation codon, although an AUG codon marks their 3′ boundary (11, 20). The activity of these IRESs is expected to require cellular factors that are differentially expressed in various cell types. Although much less is known about cellular IRESs, distinct subgroups based on IRES structure and performance will emerge as data accumulates. To date there appear to be two major functional groups: (i) translational enhancers under conditions that inhibit cap-dependent translation and (ii) translational modulators for fine tuning of gene expression in response to cellular changes. The c-sis 5′ UTR belongs to the second group, which explains its high complexity and low basal activity.
Since the c-sis IRES is responsive to differentiation, we decided to study the structure-function relationships responsible for its basal activity and for its differentiation-induced activity. Deletion mapping revealed that the c-sis 5′ UTR contains three operative modules. Each module by itself can confer some level of basal internal translation, but only the full-length 5′ UTR confers maximal basal IRES activity. This is similar to the long and complex 5′ UTR of the Antennapedia gene of D. melanogaster (39), whose IRES was also shown to be responsive to development-induced cellular changes (59). While the entire c-sis 5′ UTR was required for maximal basal IRES activity, the elements responsible for the differentiation-sensing ability were mapped to a smaller fragment in the central portion of the 5′ UTR. Since the D-IRES activity conferred by fragment 11 was similar to that of the full 5′ UTR (Fig. 1B and 3B), we assume that the “differentiation sensor” entities are located within the region spanning domains B3 through E3. Notably, the addition of either the upstream or the downstream domain to the central minimal D-IRES region was inhibitory. Full recovery of responsiveness to differentiation required the coexistence of both flanking domains, creating the full-length 5′ UTR. These variations might reflect merely the stability of the structural elements important for D-IRES activity, or they may indicate some long-range interactions between nonadjacent sequences in the context of the entire 5′ UTR. It is noteworthy that although the region responsible for IRES activation (D-IRES) was mapped to the central portion of the 5′ UTR, only the full-length 5′ UTR is capable of mediating the maximal translation efficiency in differentiated cells. This was evidenced by the fact that only the entire 5′ UTR was able to confer the maximal IRES activity in nondifferentiated cells.
Although there is no sequence homology between different IRESs, common motifs shared by many viral and cellular IRESs have been suggested to be functionally important for internal initiation of translation. Among these motifs is a Y-type structural element upstream of an AUG (1, 25, 26, 28, 29). Interestingly, each of the operative modules that confers internal translation harbors a predicted Y-shaped stem-loop structure (designated YB5, YD, or YF) followed by an AUG triplet. In addition to the Y-shaped motif, the central domain of the c-sis 5′ UTR contains additional meaningful elements (Fig. 1A). A Y-shaped structure followed by a downstream oligo(A) loop, found in the YB5-through-B6 region, was shown to contain the binding site for eIF4G in the EMCV IRES (23). A polypyrimidine tract (PPT), required for the activity of picornaviral IRESs (2), as well as for the cellular IRES of eIF4G (15), is present as clusters of evolutionarily conserved U stretches in the B5-through-B8 region. Within the same domain, a pseudoknot interaction between the B5 and B7 loops was predicted. A pseudoknot was shown to play an important part in the IRESs of pestiviruses and hepatitis C virus (42, 48, 56). By affecting tertiary interactions in RNA molecules, a pseudoknot can contribute to protein recognition (45). Another common element in picornaviral IRESs is an unpaired GNRA motif, which was suggested to be essential for the IRES function of aphthoviruses (30). Such a motif (GAAA) is also located in the B7 loop of the c-sis IRES. The collection of so many unique elements within a small region (510 to 680 nt) suggests the importance of this domain to the internal initiation conferred by the c-sis 5′ UTR. Consistent with this notion is the differentiation-induced binding to this region of the 43-kDa ribosome-associated protein, identified as an hnRNP C.
trans-acting factors might facilitate IRES activity by stabilizing important secondary or tertiary structures and/or by attracting the ribosomes via direct or indirect interactions. As an initial step to identify the trans-acting factors involved in the activation of the c-sis IRES, we looked specifically for RSW-associated proteins that interact with the IRES upon differentiation. The most profound differentiation-linked enhancement of the UV-cross-linking signal was exhibited by a 43-kDa protein, whereas the label of a nuclear 43-kDa protein was reduced upon differentiation (Fig. 4). Using a monoclonal antibody, we were able to identify hnRNP C antigenic determinants on both the nucleus- and the ribosome-associated p43, as well as on nuclear p44 (Fig. 7). It seems that p43, which partly exits the nucleus upon differentiation (Fig. 4B and D, 5A, and 7), represents hnRNP C1, whereas p44, which remains nuclear both before and after differentiation, represents hnRNP C2. The binding site of hnRNP C was mapped to U-rich sequences located in the region spanning nt 500 to 680 of the 5′ UTR, which includes structural elements B4 through B8. However, since region 1-227 (probe 2) also contains U stretches, it seems that the high-affinity binding of p43 to the B4-through-B8 domain is due to a specific sequence and/or structure within this region and not to random clustering of uridines, although it has been reported that C proteins show a preference for binding to poly(U) (53). Recently, it was demonstrated that hnRNP C possesses no enhanced affinity for U-rich oligonucleotides but does have an enhanced affinity for a unique structure or context of uridines and guanosines (51). Interestingly, such a SELEX-identified sequence, rAGUAUUUUUGUGGA (51), is similar to a sequence present within the high-affinity binding region of the c-sis D-IRES, i.e., AGUUUUUUGGGGGAGA and GAUUUUGGGGG of the B4 and B5 elements. This notion awaits mutational analysis. The integrity of region 500-790, including elements B4 through E2, seems to be important for more-efficient binding. The specific binding of hnRNP C to the D-IRES region (Fig. 3B), the differentiation-induced hyperphosphorylation (Fig. 9), and the nuclear export of some of its RNA binding capacity (Fig. 4 and 5) suggest a possible involvement of hnRNP C as a trans-acting factor. We speculate that a specific phosphorylated form binds to the D-IRES region, stabilizes certain RNA structures, and mediates the binding of additional proteins. Note that although fragment 9 (nt 601 to 846) did not efficiently bind p43, it was able to confer a D-IRES activity of 53%. Perhaps when this region is separated from the context of the full-length 5′ UTR, it acquires the appropriate structures without the help of p43. Moreover, since a few fragments that exhibited p43 binding conferred neither IRES nor D-IRES activity (fragments 5 and 7 [Fig. 4 and 5]), p43 binding may be necessary but not sufficient for differentiation-induced internal translation.
Nuclear proteins that shuttle to the cytoplasm have been shown before to be involved in mediating the IRES activities of some picornaviruses. This refers to the predominantly nuclear proteins pPTB and La autoantigen, whose cytoplasmic distribution was enhanced by viral infection (19, 34), and PCBP, also called hnRNP E (6, 13). The C proteins, known as ultimate nuclear proteins, were shown to contain a nuclear localization signal as well as a nuclear retention sequence (NRS) (10, 36). Nevertheless, a growing number of studies show the cytoplasmic interaction of hnRNP C proteins with AU-rich sequences of certain labile mRNAs (17, 35, 37, 57, 60). Interestingly, cytoplasmic localization of hnRNP C RNA binding activity was correlated with TPA treatment of peripheral blood mononuclear cells (46) and of LLC-PK1 cells (37). We show here that following TPA-induced differentiation of K562 cells, most of the hnRNP C protein remained nuclear (Fig. 8). The total hnRNP C level decreased due to differentiation (Fig. 8), as was previously reported for HL-60 cells (5). The UV-cross-linking experiments, on the other hand, demonstrate that a small portion of its binding activity to the c-sis 5′ UTR was transported outside the nucleus upon differentiation. We conclude that the intense UV-cross-linking signal of the RSW-associated hnRNP C is conferred by a very small amount of protein. The intensity of the UV-cross-linking signal reflects several factors in addition to the amount of the cross-linked protein: the RNA binding affinity, the number of cross-linking sites, the particular amino acid that contacts the RNA, and the number and nature of protected labeled nucleotides. Thus, it is possible that the binding of the cytoplasmic and nuclear forms of hnRNP C to the c-sis 5′ UTR exhibits different characteristics. This is consistent with a report of variation in the RNA binding capacity of hnRNP A1 as a function of its subcellular location (18). The RSW-associated form of hnRNP C might have a higher affinity to the c-sis 5′ UTR than to the nuclear form. It is well documented that hnRNP C can undergo different levels of phosphorylation (12, 32, 44). Moreover, serine/threonine phosphorylation was found to regulate hnRNP C RNA binding capacity (33). hnRNP C also contains a potential glycosylation site (36). Posttranslational modifications could also induce the nuclear export of hnRNP C, perhaps by masking the NRS. Furthermore, three proteins were recently reported to interact with hnRNP C via a proline-rich motif located in the NRS. Interestingly, the interactions with two of these proteins, Vav and Grb3-3, were enhanced by the presence of poly(U) RNA (49, 50). It is possible that the small population of hnRNP C molecules exported from the nucleus upon differentiation binds the c-sis transcript in the cytoplasm and mediates D-IRES activity. Alternatively, hnRNP C may bind c-sis mRNA immediately following its transcription in the nucleus and mediate its export to the cytoplasm and hence its translation only during differentiation. Current experiments are designed to verify the potential role of hyperphosphorylated forms of hnRNP C in the regulation of c-sis expression during differentiation.
In view of the activity of the c-sis 5′ UTR as a translational modulator in response to cellular changes, it seems that the large number of cross-talking structural entities and the interactions with regulated trans-acting factors are important for its modulation strength and responsiveness to cellular changes. These characteristics may be the major difference between constitutively strong IRES elements, such as those of some picornaviruses, and IRESs that serve as translational modulators in response to developmental signals, such as that of c-sis. Since most mRNAs encoding growth factors, cytokines, and transcription factors and many mRNAs encoding oncoproteins possess extraordinarily long and structured 5′ UTRs, it is conceivable that such mRNA leaders harbor an inducible, cell-type-specific IRES element that is conceptually similar to that of c-sis. Following this line of thought, deregulation of the D-IRES function may lead to cellular transformation.
ACKNOWLEDGMENTS
We thank G. Dreyfuss for monoclonal antibody (4F4) against hnRNP C and Dana Gelbaum for technical help and useful comments.
This work was supported by the Israel Science Foundation, administered by the Israel Academy of Science and Humanities—the Charles H. Revson Foundation. O. Sella acknowledges support from the Charles Clore Foundation doctoral fellowship program.
REFERENCES
- 1.Akiri G, Nahari D, Finkelstein Y, Le S-Y, Elroy-Stein O, Levi B-Z. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–237. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 2.Belsham G J, Sonenberg N. RNA-protein interactions in the regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein J, Shefler I, Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J Biol Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein J, Sella O, Le S-Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 5.Biamonti G, Bassi M T, Cartegni L, Mechta F, Uuvoli M, Cobianchi F, Riva S. Human hnRNP protein A1 gene expression—structural and functional characterization of the promoter. J Mol Biol. 1993;230:77–89. doi: 10.1006/jmbi.1993.1127. [DOI] [PubMed] [Google Scholar]
- 6.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman A M, Bailly J-L, Girard M, Kean K M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borman A M, Mercier P L, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dirks R P H, Bloemers H P J. Signals controlling the expression of PDGF. Mol Biol Rep. 1996;22:1–24. doi: 10.1007/BF00996300. [DOI] [PubMed] [Google Scholar]
- 10.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J E B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–574. [Google Scholar]
- 12.Fung P A, Labrecque R, Pederson T. RNA-dependent phosphorylation of a nuclear RNA binding protein. Proc Natl Acad Sci USA. 1997;94:1064–1068. doi: 10.1073/pnas.94.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–898. [PMC free article] [PubMed] [Google Scholar]
- 14.Gan W, Rhoads R E. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 15.Gan W, La Celle M, Roads R E. Functional characterization of the ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 16.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton B J, Nagy E, Malter J S, Arrick B A, Rigby W F C. Association of heterogenous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- 18.Hamilton B J, Burns C M, Nichols R C, Rigby W F C. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes—the roles of cytoplasmic location, transcription, and phosphorylation. J Biol Chem. 1997;272:28732–28741. doi: 10.1074/jbc.272.45.28732. [DOI] [PubMed] [Google Scholar]
- 19.Hellen C U T, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 21.Jeager J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J G, Amstrong R C, Berndt J A, Kim N W, Hudson L D. A secreted DNA-binding protein that is translated through an internal ribosome entry site (IRES) and distributed in a discrete pattern in the central nervous system. Mol Cell Neurosci. 1998;12:119–140. doi: 10.1006/mcne.1998.0701. [DOI] [PubMed] [Google Scholar]
- 23.Kolupaeva V G, Pestova T V, Hellen C U T, Shatsky I N. Translation eukaryotic initiation factor 4G recognizes specific structural element within the internal ribosome entry site of encephalomycarditis virus RNA. J Biol Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 24.LaRochelle W J, Fleming T P, Aaronson S A. PDGF in cell transformation. In: Westermark B, Sorg C, editors. Biology of platelet-derived growth factor. Basel, Switzerland: Karger Press; 1993. pp. 129–145. [Google Scholar]
- 25.Le S-Y, Chen J-H, Sonenberg N, Maizel J V. Conserved tertiary structure elements in the 5′ untranslated region of human enteroviruses and rhinoviruses. Virology. 1992;191:858–866. doi: 10.1016/0042-6822(92)90261-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le S-Y, Chen J-H, Sonenberg N, Maizel J V. Conserved tertiary structural elements in the 5′ nontranslated region of cardiovirus, aphthovirus and hepatitis A virus RNAs. Nucleic Acids Res. 1993;21:2445–2451. doi: 10.1093/nar/21.10.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le S-Y, Chen J-H, Maizel J V. Prediction of alternative secondary structures based on fluctuating thermodynamic parameters. Nucleic Acids Res. 1993;21:2173–2178. doi: 10.1093/nar/21.9.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le S-Y, Maizel J V. A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le S-Y, Maizel J V. Evolution of a common structural core in the internal ribosome entry sites of picornavirus. Virus Genes. 1998;16:25–38. doi: 10.1023/a:1007941524143. [DOI] [PubMed] [Google Scholar]
- 30.Lopez de Quinto S, Martínez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozzio C B, Lozzio B B. Human chronic myelogenous cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 32.Mayrand S H, Fung P A, Pederson T. A discrete 3′ region of U6 small nuclear RNA modulates the phosphorylation cycle of the C1 heterogeneous nuclear ribonucleoprotein particle protein. Mol Cell Biol. 1996;16:1241–1246. doi: 10.1128/mcb.16.3.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayrand S H, Dwen P, Pederson T. Serine/threonine phosphorylation regulates binding of C hnRNP proteins to pre-mRNA. Proc Natl Acad Sci USA. 1993;90:7764–7768. doi: 10.1073/pnas.90.16.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamaki T, Imamura J, Brewer G, Tsuruoka N, Koeffler H P. Characterization of adenosine-uridine-rich RNA binding factors. J Cell Physiol. 1995;165:484–492. doi: 10.1002/jcp.1041650306. [DOI] [PubMed] [Google Scholar]
- 36.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nanbu R, Montero L, D’Orazio D, Nagamine Y. Enhanced stability of urokinase-type plasminogen activator mRNA in metastatic breast cancer MDA-MB-231 cells and LLC-PK1 cells down regulated for protein kinase C. Eur J Biochem. 1997;247:169–174. doi: 10.1111/j.1432-1033.1997.00169.x. [DOI] [PubMed] [Google Scholar]
- 38.Negulescu D, Leong L E, Chandy K G, Semler B L, Gutman G A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 39.Oh S K, Scott M P, Sarnow P. Homeotic gene Antennapedia mRNA contains 5′-noncoding sequences that confer translational initiation by internal ribosome binding. Genes Dev. 1992;6:1643–1653. doi: 10.1101/gad.6.9.1643. [DOI] [PubMed] [Google Scholar]
- 40.Pestova T, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova T, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova T, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Prokaryotic-like cis-elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 44.Piñol-Roma S, Dreyfuss G. Cell cycle-regulated phosphorylation of the pre-mRNA-binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol Cell Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puglisi J D, Wyatt J R, Tinoco I. A pseudoknotted RNA oligonucleotide. Nature. 1998;331:283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- 46.Rajagopalan L E, Westmark C J, Jarzembowski J A, Malter J S. hnRNP C increases amyeloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 1998;26:3418–3423. doi: 10.1093/nar/26.14.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao C D, Pech M, Robbins K C, Aaronson S A. The 5′ untranslated sequence of the c-sis/platelet derived growth factor 2 transcript is a potent translational inhibitor. Mol Cell Biol. 1988;8:284–292. doi: 10.1128/mcb.8.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rijnbrand R, van der Straaten T, van Rijn P, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Romero F, Germani A, Puvion E, Camonis J, Varin-Blank N, Gisselbrecht S, Fischer S. Vav binding to heterogeneous nuclear ribonucleoprotein (hnRNP) C—evidence for Vav-hnRNP interactions in an RNA-dependent manner. J Biol Chem. 1998;273:5923–5931. doi: 10.1074/jbc.273.10.5923. [DOI] [PubMed] [Google Scholar]
- 50.Romero F, Ramos-Morales F, Domingues A, Rios R M, Schweighoffer F, Tocque B, Pintor-Toro J A, Fischer S, Tortolero M. Grb2 and its apoptotic isoform Grb3-3 associate with heterogeneous nuclear ribonucleoprotein C, and these interactions are modulated by poly(U) RNA. J Biol Chem. 1998;273:7776–7781. doi: 10.1074/jbc.273.13.7776. [DOI] [PubMed] [Google Scholar]
- 51.Soltaninnassab S R, McAfee J G, Shahied-Milam L, LeStourgeon W M. Oligonucleotide binding specificities of the hnRNP C protein tetramer. Nucleic Acids Res. 1998;26:3410–3417. doi: 10.1093/nar/26.14.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vagner S, Waysbort A, Marenda M, Gensac M-C, Amalric F, Prats A-C. Alternative translation initiation of the Molony murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 55.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang C, Le S-Y, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X-Y, Hoyle P E, McCubrey J A. Characterization of proteins binding the 3′ regulatory region of the IL-3 gene in IL-3-dependent autocrine-transformed hematopoietic cells. Leukemia. 1998;12:520–531. doi: 10.1038/sj.leu.2400975. [DOI] [PubMed] [Google Scholar]
- 58.Weighardt F, Biamonti G, Riva S. The roles of heterogenous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 59.Ye X, Fong P, Iizuka N, Choate D, Cavener D. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaidi S H E, Malter J S. Nucleolin and heterogenous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyeloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]