Abstract
Since hyperthermophilic Archaea (HA) thrive in high-temperature environments, which accelerate the rates of deamination of base in DNA, their genomic stability is facing a severe challenge. Hypoxanthine (Hx) is one of the common deaminated bases in DNA. Generally, replication of Hx in DNA before repaired causes AT → GC mutation. Biochemical data have demonstrated that 3-methyladenine DNA glycosylase II (AlkA) and Family V uracil DNA glycosylase (UDG) from HA could excise Hx from DNA, thus triggering a base excision repair (BER) process for Hx repair. Besides, three endonucleases have been reported from HA: Endonuclease V (EndoV), Endonuclease Q (EndoQ), and Endonuclease NucS (EndoNucS), capable of cleaving Hx-containing DNA, thereby providing alternative pathways for Hx repair. Both EndoV and EndoQ could cleave one DNA strand with Hx, thus forming a nick and further initiating an alternative excision repair (AER) process for the follow-up repair. By comparison, EndoNucS cleaves both strands of Hx-containing DNA in a restriction endonuclease manner, thus producing a double-stranded break (DSB). This created DSB might be repaired by homologous recombination (HR) or by a combination activity of DNA polymerase (DNA pol), flap endonuclease 1 (FEN1), and DNA ligase (DNA lig). Herein, we reviewed the most recent advances in repair of Hx in DNA triggered by DNA glycosylases and endonucleases from HA, and proposed future research directions.
Keywords: hyperthermophilic archaea, DNA glycosylase, endonuclease, hypoxanthine, base deamination
Introduction
DNA is constantly damaged by endogenous and environmental factors. Base deamination is a typical pathway for producing DNA damage. Generally, adenine, cytosine, and guanine are deaminated to hypoxanthine (Hx), uracil (U), and xanthine (X), respectively. Base deamination can occur spontaneously under physiological conditions, and are also accelerated by ionizing radiation, high temperature, aerobic respiration, and nitrosative stress (Chatterjee and Walker, 2017). Since they are mutagenic, deaminated bases in DNA need to be repaired to maintain cellular genomic DNA stability.
Hyperthermophilic Archaea (HA) can grow optimally above 80°C (Stetter, 2013), which are predominantly isolated from high temperature environments, such as deep-sea hydrothermal vents, volcanic craters, and terrestrial hot springs (van der Oost et al., 1998). HA have become a research hotspot because they possess a large number of unknown biological processes and a variety of highly thermostable enzymes that can be potentially applied in biotechnology (Cabrera and Blamey, 2018), and are a simplified model organism for uncovering eukaryotic DNA replication and repair mechanisms (Zatopek et al., 2018). Since high temperature accelerates the rates of base deamination (Lindahl and Nyberg, 1974), HA might have higher levels of U and Hx in their genomic DNA than mesophiles, since they thrive in high temperature environments. Surprisingly, HA display spontaneous mutational frequencies similar to mesophiles (Grogan et al., 2001), thereby suggesting that they possess more efficient repair capability than mesophiles. However, our understanding on DNA damage repair of HA remains incomplete.
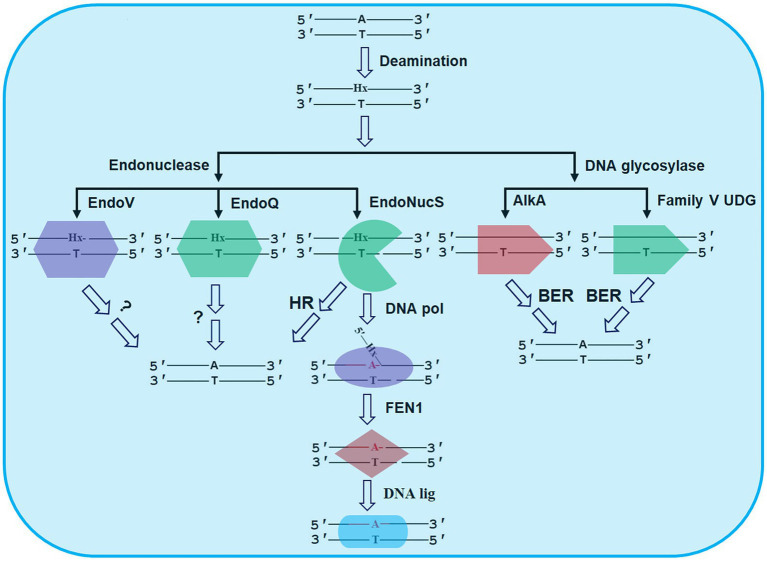
Hypoxanthine is a common DNA damage base, which can be formed by two pathways: one is deamination of adenine in DNA, and the other is dHxTP (a product of dATP deamination) incorporation by DNA polymerase (DNA pol). In the first pathway, an Hx/C pairing would be formed after a round of DNA replication, and then a G/C pairing would be produced after the next round of replication, thereby leading to AT→GC mutation (Kuraoka, 2015). In the second pathway, dHxTP can be incorporated opposite the DNA template C since Hx can pair with cytosine, thus forming Hx in DNA. Although the Hx/C pairing in DNA is no-mutagenic (Budke and Kuzminov, 2006), a large amount of Hx accumulation in DNA might trigger the activity of repair-related enzymes, such as EndoV, further causing single-strand breaks (Kuraoka, 2015). Therefore, Hx in DNA is harmful to cells, which needs to be repaired.
Fortunately, cells have evolved several DNA glycosylases, including 3-methyladenine DNA glycosylase II (AlkA), Family V uracil DNA glycosylase (UDG), and Family VI UDG, capable of excising Hx from DNA, thus initiating a typical base excision repair (BER) process to repair Hx in DNA.
Genomic analyses have shown that AlkA and Family V UDG are encoded in HA. Additionally, three endonucleases have been reported from HA: Endonuclease V (EndoV), Endonuclease Q (EndoQ), and Endonuclease NucS (EndoNucS), which are able to cleave Hx-containing DNA, thereby providing alternative pathways for repair of Hx in DNA. Herein, we focused on the most advances of repair of Hx in DNA triggered by two DNA glycosylases (AlkA and Family V UDG) and three endonucleases (EndoV, EndoQ, and EndoNucS) from HA. Furthermore, we proposed a few prospects for future research directions.
Excision of Hx From DNA by Archaeal AlkA
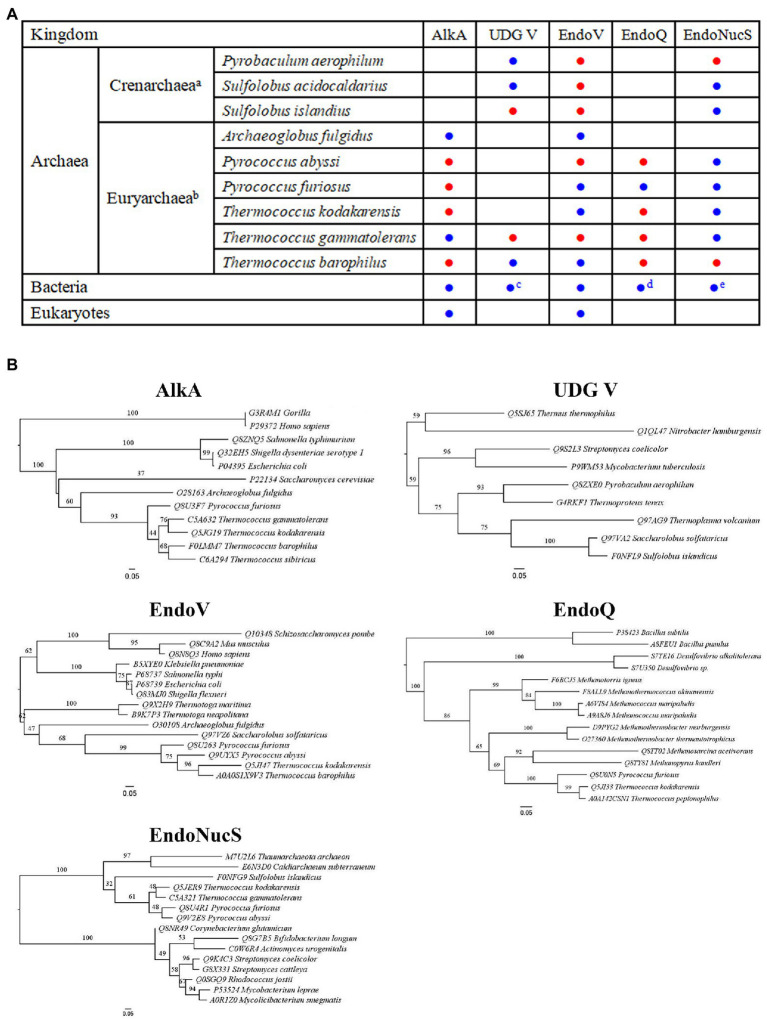
Base excision repair, which is triggered by DNA glycosylase, is thought to be an important pathway for repair of Hx in DNA (Wallace, 2014). AlkA is a DNA glycosylase that is able to remove methylated bases from DNA. Analysis of genomic sequences has shown that AlkA is ubiquitous in most of bacteria and eukaryotes, and a few Euryarchaea rather than Crenarchaea (Figure 1A). In addition to removal of methylated bases from DNA, AlkA can also possess the activity toward excising Hx from DNA (Figure 2; Evensen and Seeberg, 1982). A number of structural and biochemical studies have revealed damaged base recognition and removal mechanisms of Escherichia coli AlkA (Nakabeppu et al., 1984; Bjelland and Seeberg, 1996; Labahn et al., 1996; Yamagata et al., 1996; O’Brien and Ellenberger, 2004), thus enabling the enzyme to be a model alkylated DNA glycosylase. Currently, only two archaeal AlkA homologs have been reported from the hyperthermophilic euryarchaeon Archaeoglobus fulgidus and Thermococcus gammatolerans (Figure 1; Birkeland et al., 2002; Mansfield et al., 2003; Leiros et al., 2007; Jiang et al., 2021). Thus, our understanding on biochemical function and catalytic mechanism of archaeal AlkA remains incomplete.
Figure 1.
Distribution and phylogenetic analyses of DNA repair proteins involved in Hypoxanthine (Hx) repair. (A) Distribution of 3-methyladenine DNA glycosylase II (AlkA), uracil DNA glycosylase (UDG) V, Endonuclease V (EndoV), Endonuclease Q (EndoQ), and Endonuclease NucS (EndoNucS). a and b: only includes archaea which at least one enzyme among five target enzymes has been characterized; c: includes a few hyperthermophilic bacteria; d: includes partial bacteria, such as Bacillus and Desulfovibrio; and e: includes partial bacteria that lack mismatch repair pathway, such as Actinobacteria. The characterized and uncharacterized enzymes are shown with blue and red circle, respectively. (B) Phylogenetic analyses of AlkA, UDG V, EndoV, EndoQ, and EndoNucS from hyperthermophilic Archaea. Note that only partial Archaea, bacteria, and eukaryotes are included in these five phylogenetic trees due to the space limit.
Figure 2.
Possible repair pathways of Hx in DNA revealed by DNA glycosylases and endonucleases from HA. The details are seen the text. AlkA, 3-methyladenine DNA glycosylase II; BER, base excision repair; DNA lig, DNA ligase; DNA pol, DNA polymerase; EndoNucS, endonuclease NucS; EndoQ, endonuclease Q; EndoV, endonuclease V; FEN1, flap endonuclease 1; HR, homologous recombination; and UDG, uracil DNA glycosylase.
Biochemical data have demonstrated that A. fulgidus AlkA and T. gammatolerans AlkA are able to remove Hx from dsDNA (Mansfield et al., 2003; Jiang et al., 2021). Although, the two protein hosts are hyperthermophiles, T. gammatolerans AlkA displays biochemical characteristics distinct from A. fulgidus AlkA. Firstly, T. gammatolerans AlkA is able to excise Hx from dsDNA rather than from ssDNA (Jiang et al., 2021), suggesting that the enzyme might be critical for repair of Hx in genomic DNA. By comparison, A. fulgidus AlkA can remove Hx from ssDNA (Mansfield et al., 2003). Additionally, T. gammatolerans AlkA is a bi-functional DNA glycosylase, harboring the activity for not only excising Hx from dsDNA but further cleaving the generated apurinic/apyrimidinic (AP) site (Jiang et al., 2021), while A. fulgidus AlkA is monofunctional DNA glycosylase, only removing Hx from DNA (Mansfield et al., 2003). Overall, effective excision of Hx from DNA by the two archaeal AlkA proteins might provide an alternative pathway for repair of Hx in DNA.
Analysis of amino acid sequences of AlkA homologs suggests that these AlkA proteins harbor a conserved helix-hairpin-helix (HhH)-GPD motif that is composed of an HhH motif for binding DNA and a glycine/proline-rich loop followed by an invariable Asp (GPD; Kuznetsov and Fedorova, 2020). Mutational analyses have demonstrated that the conserved Asp residue in the HhH-GPD motif serves as a catalytic residue in T. gammatolerans AlkA and A. fulgidus AlkA (Mansfield et al., 2003; Jiang et al., 2021), which is also observed in Helicobacter pylori AlkA (Eichman et al., 2003). Additionally, residue W204 in T. gammatolerans AlkA is another essential residue for catalysis since the replacement of residue W204 with alanine abolishes the enzyme activity (Jiang et al., 2021). Note that residue W204 in T. gammatolerans AlkA corresponds to residue W218 in E. coli AlkA. The mutant E. coli cells harboring the AlkA W218A mutant display the sensitivity to methylmethane sulfonate (MMS), whereas the wild-type cells are resistant to MMS, suggesting that residue W218 is part of the active site residues of the enzyme (Yamagata et al., 1996). The crystal structure of E. coli AlkA demonstrates that residue W218 is positioned behind the ribose of the flipped out the damaged base for the attack on the back of the glycosylic bond (Yamagata et al., 1996; Hollis et al., 2004).
Genetic studies suggest that Pseudomonas putida AlkA plays an essential role in repairing induced lesions by MMS or N-methyl-N'-nitro-N-nitrosoguanidine in vivo (Mielecki et al., 2013). Additionally, the exposure to alkylating agents leads to about 100-fold increase in the E. coli alkA gene transcription (Samson and Cairns, 1977; Evensen and Seeberg, 1982; Nakabeppu et al., 1984), suggesting that induced AlkA is involved in repair of alkylation lesion in vivo. Similar effect is observed in Saccharomyces cerevisiae AlkA (Chen et al., 1990). However, no genetic analysis on archaeal AlkA has been currently reported.
Excision of Hx From DNA by Archaeal Family V UDG
Uracil DNA glycosylase is thought to be responsible for repair of uracil in DNA. Based on amino acid sequence similarity, UDG is classified into six families, and each family possesses specific characteristics (Schormann et al., 2014). Hx DNA glycosylase, which belongs to Family VI UDG, was first reported from the archaeon Methanosarcina barkeri, capable of excising Hx from DNA (Lee et al., 2011). However, no Hx DNA glycosylase homolog has been found in HA to date.
Family V UDG is present in hyperthermophilic bacteria, such as Thermus and Mycobacterium, and hyperthermophilic Crenarchaea and Euryarchaea (Figure 1B; Kosaka et al., 2007; Xia et al., 2014), capable of removing Hx from DNA in addition to uracil excision (Figure 2). At least three different UDGs are encoded in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum: Pa-MIG (mismatch DNA glycosylase), Pa-UDGa, and Pa-UDGb. Pa-UDGb was the first reported as an archaeal Family V UDG member (Sartori et al., 2002), displaying the activity toward removal of Hx from DNA in addition to uracil, hydroxymethyluracil, and fluorouracil opposite from guanine as well. Thus, removal of Hx from DNA by P. aerophilum UDGb might potentially function in avoiding A to G transition mutations in the archaeal cells, which needs to be confirmed in vivo.
The Family V UDG from Thermus thermophilus HB8 is capable of removing uracil, hypoxanthine and xanthine from DNA (Xia et al., 2014). Genetic studies suggest that T. thermophilus UDG plays a crucial role removing uracil from DNA in vivo (Sakai et al., 2008). Additionally, the udgB knockout strain from Mycobacterium smegmatis shows approximately 2-fold higher mutation rates than the wild-type strain, suggesting that M. smegmatis UDGb is involved in repair of deaminated bases. Further genetic studies suggest a synergistic effect of M. smegmatis UDGb and UNG (uracil N-glycosylase, Family I UDG) in DNA repair (Malshetty et al., 2010). Currently, no archaeal family V UDG function has been described in vivo.
Cleavage of Hx-Containing DNA by Archaeal EndoV
In addition to BER, alternative excision repair (AER) is also thought to be another important pathway for repair of Hx in DNA. AER is generally initiated by an endonuclease that can nick DNA (Yasui, 2013). However, the complete AER pathway has not been clarified to date. EndoV is the first reported endonuclease that triggers the AER process for repair of Hx in DNA (Cao, 2013).
Endonuclease V is a DNA repair enzyme encoded by the nfi gene, which was first reported in E. coli (Eco-EndoV), capable of cleaving the second phosphodiester bond downstream of the deaminated damaged base (Figure 2; Gates and Linn, 1977; Demple and Linn, 1982). EndoV is ubiquitously present in three domains: bacteria, Archaea, and eukaryotes (Figure 1B). Alignment of amino acid sequences of EndoV homologs from bacteria, Archaea, and eukaryotes has shown that EndoV has seven conserved motifs (Wang et al., 2018), suggesting similar catalytic mechanism.
Since the hyperthermophilic euryarchaeon A. fulgidus EndoV (Afu-EndoV) was first reported (Liu et al., 2000), two other archaeal EndoV homologs have been characterized biochemically, including the hyperthermophilic euryarchaeon Pyrococcus furiosus EndoV (Pfu-EndoV; Kiyonari et al., 2014), and Thermococcus barophilus Ch5 EndoV (Tba-EndoV; Wang et al., 2018). No relevant structure of archaeal EndoV has been solved to date. Afu-EndoV displays 39% amino acid sequence identity and 55% similarity with Eco-EndoV (Liu et al., 2000). Whereas Eco-EndoV cleaves DNA substrates with other damaged bases in addition to Hx (Demple and Linn, 1982), Afu-EndoV only cleaves DNA containing Hx (Liu et al., 2000). Overall, cleaving Hx-containing DNA is the most primitive activity of EndoV homologs from bacteria and Archaea.
Biochemical data demonstrate that three archaeal EndoV homologs are able to cleave Hx-containing DNA at high temperature, and their optimal reaction temperature are 70~90°C. However, these three archaeal EndoV homologs display distinct biochemical characteristics. Firstly, Pfu-EndoV can not only cleave Hx-containing DNA, but also cleave Hx-containing RNA (Kiyonari et al., 2014), suggesting that this enzyme may be involved in RNA metabolism. By comparison, Afu-EndoV and Tba-EndoV act on only Hx-containing DNA (Liu et al., 2000; Wang et al., 2018). Secondly, Tba-EndoV can bind normal DNA and Hx-containing DNA, and display higher binding Hx-containing DNA (Wang et al., 2018), while Pfu-EndoV can only bind Hx-containing DNA (Kiyonari et al., 2014). Furthermore, Pfu-EndoV is inactive to cleave DNA at pH 6.0 (Kiyonari et al., 2014), while Tba-EndoV still possesses strong cleavage activity at pH 6.0 (Wang et al., 2018). Additionally, Tba-EndoV is more resistant to NaCl than Pfu-EndoV (Kiyonari et al., 2014; Wang et al., 2018).
Genetic studies suggest that Eco-EndoV plays a significant role in repair of Hx and abasic site in DNA (Guo and Weiss, 1998). Compared with the wild-type, the nfi mutant of E. coli displays about 8-fold lower Hx repair levels in vivo, and the exonuclease activity of E. coli DNA polymerase I is required for the Eco-EndoV repair pathway (Su et al., 2018). Additionally, the analysis of effect of EndoV deletion in Bacillus subtilis on spontaneous mutation rates suggests the presence of this pathway in vivo, which counteracts the genotoxic effects of base deamination (Patlán et al., 2019). To data, no genetic study on archaeal EndoV function has been currently reported.
Cleavage of Hx-Containing DNA by EndoQ From HA
In addition to EndoV, a second endonuclease was recently identified in P. furiosus as Pfu-EndoQ, capable of cleaving damaged DNA, including U, Hx, and AP site (Shiraishi et al., 2015). Pfu-EndoQ is able to recognize and cleave the first phosphodiester bond on the 5'-side of damaged bases in DNA to form a nick (Figure 2; Shiraishi et al., 2015). Recently, molecular mechanism of recognition of damaged bases in DNA by Pfu-EndoQ was revealed, demonstrating that the damaged bases are flipped out from the DNA duplex by the enzyme, thus leading to the base pair instability of the damaged bases and further enhancing the cleavage activity of the enzyme (Shiraishi and Iwai, 2020).
Genomic analyses show that EndoQ are only found in Euryarchaea and a few bacteria including Bacillus and Desulfovibrio (Figure 1B), whereas EndoV is widespread in all organisms. The limited distribution of EndoQ suggests that the enzyme is involved in the specific repair pathway of damaged bases. Although EndoQ appears to be conserved in a limited number of species, the substrate specificities of the EndoQ homologs seem to be highly conserved. The archaeal EndoQ homologs from Thermococcus kodakarensis and the methanogen Methanosarcina acetivorans and the bacterial EndoQ homolog from Bacillus pumilus have been shown to create a nick on DNA strands containing uracil, Hx, and AP site (Shiraishi et al., 2015, 2017, 2018).
Further biochemical data have shown that Pfu-EndoQ interacts with the sliding clamp proliferating cell nuclear antigen (PCNA) protein (Shiraishi et al., 2016), a central protein, which coordinates DNA replication and DNA repair, and other biological processes (Moldovan et al., 2007), which may help to recruit the enzymes or proteins to the replication fork. The nick created by the enzyme is repaired by a combination activity of DNA helicase and endonuclease flap endonuclease 1 (FEN1), DNA pol, and DNA ligase (DNA lig). Additionally, the Pfu-EndoQ activity is stimulated by PCNA, and the physical interaction has been confirmed between the two proteins through a PIP-motif of EndoQ and the toroidal structure of PCNA, thus providing a clue to elucidate a unique DNA repair system in Archaea (Shiraishi et al., 2016).
EndoV cleaves the second phosphodiester bond downstream of Hx in DNA, while EndoQ cleaves the first phosphodiester bond on the 5'-side of Hx in DNA (Demple and Linn, 1982; Shiraishi et al., 2015), thereby suggesting that Hx in DNA might be repaired by EndoV and EndoQ together. However, no direct interaction is confirmed between EndoQ and EndoV in P. furiosus no matter whether Hx-containing DNA is present or not (Ishino et al., 2015). Compared with Pfu-EndoV, Pfu-EndoQ has a higher affinity for DNA containing Hx. In addition, the expression levels of Pfu-EndoQ in P. furiosus cells are higher than those of Pfu-EndoV, suggesting that Pfu-EndoQ might participate in repair of Hx in DNA in vivo.
Recently, the crystal structures of P. furiosus EndoQ bound to DNA substrates containing U, Hx, or an AP lesion have been solved (Shi et al., 2021), demonstrating the mechanisms of recognition and cleavage of deaminated bases in DNA by this endonuclease. The structures of the P. furiosus EndoQ-DNA complexes show that a deep active-site pocket in the enzyme would be shaped by a concerted swing motion of its zinc-binding and C-terminal helical domains (Shi et al., 2021), which allows the extruded deaminated bases to be accommodated. Furthermore, U and Hx bases associate with amino acid residues in this pocket, which is coordinated with an essential magnesium ion (Shi et al., 2021). Thus, the EndoQ-DNA complex structures provide mechanistic insights into damaged DNA recognition and cleavage by this endonuclease, which is helpful for understanding how EndoQ recognizes and cleaves these structurally diverse damaged DNA substrates rather than undamaged DNA substrate.
Cleavage of Hx-Containing dsDNA by EndoNucS From HA
Endonuclease NucS was identified from HA as the third endonuclease, capable of cleaving Hx-containing DNA. EndoNucS was the first reported from the hyperthermophilic euryarchaeon Pyrococcus abyssi (Ren et al., 2009), displaying the activity on branched and splayed DNA. Later, the EndoNucS homologs from the hyperthermophilic euryarchaeon T. kodakarensis and T. gammatolerans, and the hyperthermophilic crenarchaeon Sulfolobus islandius REY15A have been reported, possessing the activity on mismatched and Hx-containing DNA (Ishino et al., 2016; Ahmad et al., 2020; Zhang et al., 2020a). Intriguingly, these three EndoNucS homologs from HA harbor distinct cleavage sites, as reviewed in our recent publication (Zhang et al., 2020b).
Biochemical data suggest that these three reported EndoNucS homologs from HA cleave both strands of Hx-containing dsDNA in a restriction endonuclease manner, thereby forming a double-stranded break (DSB), which is sharply distinct from EndoQ and EndoV as discussed above. Since they might pose a severe damage for archaeal cells, the DSBs potentially generated by archaeal EndoNucS need to be repaired by homologous recombination (HR) or a combinational activity of DNA pol, FEN1, and DNA lig (Figure 2). These hypotheses need to be verified by genetic analysis in vivo. Overall, DNA cleavage by archaeal EndoNucS provides a possible alternative pathway for repair of Hx in DNA.
Endonuclease NucS is distributed in Euryarchaea, Crenarchaea, and a few bacteria, especially the bacteria that lack a typical mismatch repair pathway, such as Actinobacteria (Figure 1B). The analysis of amino acid sequences of EndoNucS homologs suggests that this endonuclease possesses several conserved motifs that are composed of mostly negatively and positively charged amino acid residues (Zhang et al., 2020a). Structural and mutational studies have provided insight into catalytic mechanism of archaeal EndoNucS. The complex structure of T. kodakarensis EndoNucS dimer with mismatched dsDNA was solved (Nakae et al., 2016), demonstrating that the mismatched bases are flipped out into binding sites, which resembles the overall architecture of most restriction endonucleases.
Mutational data have demonstrated that residues Y41, N76, and W77 are essential for DNA binding, and residues D165, E179, and K181 are critical for catalysis (Ishino et al., 2016). Additionally, we revealed that the replacement of residue D163 in T. gammatolerans EndoNucS, which is analogous to residue D165 in T. kodakarensis EndoNucS, with alanine leads to the partial loss of the enzyme activity on U- and Hx-containing dsDNA, suggesting that residue D163 is essential for catalysis (Zhang et al., 2020a).
Genetic data have demonstrated that the nucS deletion strain from the actinobacterium Corynebacterium glutamicum leads to a drastic increase of spontaneous transition mutations, thereby suggesting that C. glutamicum EndoNucS protein plays an essential role in repair mismatch (Ishino et al., 2018). Similar effects are observed in M. smegmatis, Streptomyces coelicolor, and S. islandicus REY15A (Castaneda-Garcia et al., 2017; Takemoto et al., 2018; Ahmad et al., 2020). By comparison, the nucS deletion strain from S. acidocaldarius does not cause the increased mutation rates (Suzuki and Kurosawa, 2019). Further genetic data suggest that this endonuclease associates with XPF endonuclease via the HR-mediated stalled-fork to remove helix-distorting DNA lesions, such as intrastrand crosslinks (Suzuki and Kurosawa, 2019). Thus, more work needs to be done to investigate the archaeal EndoNucS function in vivo.
Conclusion and Future Directions
Adenine deamination produces Hx, which is one of common deamination types. Since it is mutagenic, Hx in DNA needs to be repaired. HA encodes AlkA and Family V UDG, triggering the BER process for repairing Hx in DNA. Besides, some HA encode three endonucleases: EndoV, EndoQ, and EndoNucS, which has been confirmed that they are capable of cleaving Hx-containing DNA. Interestingly, these archaeal endonucleases cleave Hx-containing DNA in distinct manners. Overall, a combination of these potential repair pathways initiated with DNA glycosylases and endonucleases might enable HA to counteract the potentially increased mutations caused by high temperature, thus maintaining spontaneous mutation frequencies similar to other mesophiles.
AlkA and Family V UDG from HA can excise Hx from DNA in vitro. However, their physiological function in vivo still remains unclear. Whether AlkA or Family V UDG is a major DNA glycosylase for repair of Hx in DNA in archaeal cell is unknown. A combination of structural, biochemical, and genetic analyses for these two DNA glycosylases would provide new insights into Hx repair in HA.
Archaeal EndoV and EndoQ cleave Hx-containing DNA, thus forming a nick. Generally, the 3’-OH of the nick generated by archaeal EndoV and EndoQ can be utilized by other nucleic acid enzymes to complete subsequent repairs. Recently, EndoV, ExoA, and PolA are jointly involved in repair of DNA containing deaminated bases in B. subtilis (Patlán et al., 2019). However, how the nicks created by archaeal EndoV and EndoQ are repaired remains elusive. It would be interesting to investigate molecular mechanism of repair of Hx in DNA triggered by EndoV and EndoQ in vivo.
As discussed above, the EndoNucS homologs from the hyperthermophilic T. kodakarensis, S. islandius, and T. gammatolerans are able to cleave Hx-containing dsDNA in a restriction endonuclease manner in vitro, leading to the formation of DSB. The evidence for the generated DSBs by archaeal EndoNucS needs to be provided to confirm whether the DSB is in fact generated in vivo and the nuclease activity is directed in a strand-specific manner similar to MutL proteins in Bacteria.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding. This work was supported by Earmarked Fund for Modern Agro-Industry Technology Research System in Hebei Province, China (HBCT2018080205) to ZY; the Natural Science Foundation of Jiangsu Province (No. BK20191219), High Level Talent Support Program of Yangzhou University, and the Academic Leader of Middle and Young People of Yangzhou University Grant to LZ.
References
- Ahmad S., Huang Q., Ni J., Xiao Y., Yang Y., Shen Y. (2020). Functional analysis of the NucS/EndoMS of the hyperthermophilic archaeon Sulfolobus islandicus REY15A. Front. Microbiol. 11:607431. 10.3389/fmicb.2020.607431, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland N. K., Anensen H., Knaevelsrud I., Kristoffersen W., Bjorås M., Robb F. T., et al. (2002). Methylpurine DNA glycosylase of the hyperthermophilic archaeon Archaeoglobus fulgidus. Biochemistry 41, 12697–12705. 10.1021/bi020334w, PMID: [DOI] [PubMed] [Google Scholar]
- Bjelland S., Seeberg E. (1996). Different efficiencies of the tag and AlkA DNA glycosylases from Escherichia coli in the removal of 3-methyladenine from single-stranded DNA. FEBS Lett. 397, 127–129. 10.1016/S0014-5793(96)01166-0, PMID: [DOI] [PubMed] [Google Scholar]
- Budke B., Kuzminov A. (2006). Hypoxanthine incorporation is nonmutagenic in Escherichia coli. J. Bacteriol. 188, 6553–6560. 10.1128/JB.00447-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M. Á., Blamey J. M. (2018). Biotechnological applications of archaeal enzymes from extreme environments. Biol. Res. 51:37. 10.1186/s40659-018-0186-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. (2013). Endonuclease V: an unusual enzyme for repair of DNA deamination. Cell. Mol. Life Sci. 70, 3145–3156. 10.1007/s00018-012-1222-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda-Garcia A., Prieto A. I., Rodriguez-Beltran J., Alonso N., Cantillon D., Costas C., et al. (2017). A non-canonical mismatch repair pathway in prokaryotes. Nat. Commun. 8:14246. 10.1038/ncomms14246, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N., Walker G. C. (2017). Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263. 10.1002/em.22087, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. (1990). Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 9, 4569–4575. 10.1002/j.1460-2075.1990.tb07910.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Linn S. (1982). On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J. Biol. Chem. 257, 2848–2855. 10.1016/S0021-9258(19)81041-4, PMID: [DOI] [PubMed] [Google Scholar]
- Eichman B. F., O’Rourke E. J., Radicella J. P., Ellenberger T. (2003). Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J. 22, 4898–4909. 10.1093/emboj/cdg505, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. (1982). Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature 296, 773–775. 10.1038/296773a0, PMID: [DOI] [PubMed] [Google Scholar]
- Gates F. T., Linn S. (1977). Endonuclease V of Escherichia coli. J. Biol. Chem. 252, 1647–1653. 10.1016/S0021-9258(17)40598-9, PMID: [DOI] [PubMed] [Google Scholar]
- Grogan D. W., Carver G. T., Drake J. W. (2001). Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. U. S. A. 98, 7928–7933. 10.1073/pnas.141113098, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Weiss B. (1998). Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol. 180, 46–51. 10.1128/JB.180.1.46-51.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis T., Ichikawa Y., Ellenberger T. (2004). DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 19, 758–766. 10.1093/emboj/19.4.758, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino S., Makita N., Shiraishi M. (2015). EndoQ and EndoV work individually for damaged DNA base repair in Pyrococcus furiosus. Biochimie 118, 264–269. 10.1016/j.biochi.2015.06.015, PMID: [DOI] [PubMed] [Google Scholar]
- Ishino S., Nishi Y., Oda S., Uemori T., Sagara T., Takatsu N., et al. (2016). Identification of a mismatch-specific endonuclease in hyperthermophilic archaea. Nucleic Acids Res. 44, 2977–2986. 10.1093/nar/gkw153, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino S., Skouloubris S., Kudo H., Hermitte-Stead C., Es-Sadik A., Lambry J. C., et al. (2018). Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res. 46, 6206–6217. 10.1093/nar/gky460, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Zhang L., Dong K., Gong Y., Oger P. (2021). Biochemical characterization and mutational studies of a novel 3-methlyadenine DNA glycosylase II from the hyperthermophilic Thermococcus gammatolerans. DNA Repair 97:103030. 10.1016/j.dnarep.2020.103030, PMID: [DOI] [PubMed] [Google Scholar]
- Kiyonari S., Egashira Y., Ishino S. (2014). Biochemical characterization of endonuclease V from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biochem. 155, 325–333. 10.1093/jb/mvu010, PMID: [DOI] [PubMed] [Google Scholar]
- Kosaka H., Hoseki J., Nakagawa N., Kuramitsu S., Masui R. (2007). Crystal structure of family 5 uracil-DNA glycosylase bound to DNA. J. Mol. Biol. 373, 839–850. 10.1016/j.jmb.2007.08.022, PMID: [DOI] [PubMed] [Google Scholar]
- Kuraoka I. (2015). Diversity of endonuclease V: from DNA repair to RNA editing. Biomolecules 5, 2194–2206. 10.3390/biom5042194, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov N. A., Fedorova O. S. (2020). Kinetic milestones of damage recognition by DNA glycosylases of the helix-hairpin-helix structural superfamily. Adv. Exp. Med. Biol. 1241, 1–18. 10.1007/978-3-030-41283-8_1, PMID: [DOI] [PubMed] [Google Scholar]
- Labahn J., Scharer O. D., Long A., Ezaz-Nikpay K., Verdine G. L., Ellenberger T. E. (1996). Structural basis for the excision repair of alkylation-damaged DNA. Cell 86, 321–329. 10.1016/S0092-8674(00)80103-8, PMID: [DOI] [PubMed] [Google Scholar]
- Lee H. W., Dominy B. N., Cao W. G. (2011). New family of deamination repair enzymes in uracil-DNA glycosylase superfamily. J. Biol. Chem. 286, 31282–31287. 10.1074/jbc.M111.249524, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiros I., Nabong M. P., Grosvik K., Ringvoll J., Haugland G. T., Uldal L., et al. (2007). Structural basis for enzymatic excision of N1-methyladenine and N3-methylcytosine from DNA. EMBO J. 26, 2206–2217. 10.1038/sj.emboj.7601662, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. (1974). Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13, 3405–3410. 10.1021/bi00713a035, PMID: [DOI] [PubMed] [Google Scholar]
- Liu J., He B., Qing H. (2000). A deoxyinosine specific endonuclease from hyperthermophile, Archaeoglobus fulgidus: a homolog of Escherichia coli endonuclease V. Mutat. Res. 461, 169–177. 10.1016/s0921-8777(00)00054-9, PMID: [DOI] [PubMed] [Google Scholar]
- Malshetty V. S., Jain R., Srinath T., Kurthkoti K., Varshney U. (2010). Synergistic effects of UdgB and Ung in mutation prevention and protection against commonly encountered DNA damaging agents in Mycobacterium smegmatis. Microbiology 156, 940–949. 10.1099/mic.0.034363-0, PMID: [DOI] [PubMed] [Google Scholar]
- Mansfield C., Kerins S. M., McCarthy T. V. (2003). Characterisation of Archaeglobus fulgidus AlkA hypoxanthine DNA glycosylase activity. FEBS Lett. 540, 171–175. 10.1016/S0014-5793(03)00257-6, PMID: [DOI] [PubMed] [Google Scholar]
- Mielecki D., Saumaa S., Wrzesiński M., Maciejewska A. M., Żuchniewicz K., Sikora A., et al. (2013). Pseudomonas putida AlkA and AlkB proteins comprise different defense systems for the repair of alkylation damage to DNA - in vivo, in vitro, and in silico studies. PLoS One 8:e76198. 10.1371/journal.pone.0076198, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G. L., Pfander B., Jentsch S. (2007). PCNA, the maestro of the replication fork. Cell 129, 665–679. 10.1016/j.cell.2007.05.003, PMID: [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Miyata T., Kondo H., Iwanaga S., Sekiguchi M. (1984). Structure and expression of the alkA gene of Escherichia coli involved in adaptive response to alkylating agents. J. Biol. Chem. 259, 13730–13736. 10.1016/S0021-9258(18)89806-4, PMID: [DOI] [PubMed] [Google Scholar]
- Nakae S., Hijikata A., Tsuji T., Yonezawa K., Kouyama K., Mayanagi K., et al. (2016). Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure 24, 1960–1971. 10.1016/j.str.2016.09.005, PMID: [DOI] [PubMed] [Google Scholar]
- O’Brien P. J., Ellenberger T. (2004). The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J. Biol. Chem. 279, 26876–26884. 10.1074/jbc.M403860200, PMID: [DOI] [PubMed] [Google Scholar]
- Patlán A. G., Ayala-García V. M., Valenzuela-García L. I., Meneses-Plascencia J., Vargas-Arias P. L., Barraza-Salas M., et al. (2019). YwqL (EndoV), ExoA and PolA act in a novel alternative excision pathway to repair deaminated DNA bases in Bacillus subtilis. PLoS One 14:e0211653. 10.1371/journal.pone.0211653, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Kuhn J., Meslet-Cladiere L., Briffotaux J., Norais C., Lavigne R., et al. (2009). Structure and function of a novel endonuclease acting on branched DNA substrates. EMBO J. 28, 2479–2489. 10.1038/emboj.2009.192, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Tokishita S., Mochizuki K., Motomiya A., Yamagata H., Ohta T. (2008). Mutagenesis of uracil-DNA glycosylase deficient mutants of the extremely thermophilic eubacterium Thermus thermophilus. DNA Repair 7, 663–669. 10.1016/j.dnarep.2008.01.006, PMID: [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. (1977). A new pathway for DNA repair in Escherichia coli. Nature 267, 281–283. 10.1038/267281a0, PMID: [DOI] [PubMed] [Google Scholar]
- Sartori A. A., Fitz-Gibbon S., Yang H. J., Miller J. H., Jiricny J. (2002). A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 21, 3182–3191. 10.1093/emboj/cdf309, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schormann N., Ricciardi R., Chattopadhyay D. (2014). Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 23, 1667–1685. 10.1002/pro.2554, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K., Moeller N. H., Banerjee S., McCann J. L., Carpenter M. A., Yin L., et al. (2021). Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q. Proc. Natl. Acad. Sci. U. S. A. 118:e2021120118. 10.1073/pnas.2021120118, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M., Ishino S., Cann I., Ishino Y. (2017). A functional endonuclease Q exists in the bacterial domain: identification and characterization of endonuclease Q from Bacillus pumilus. Biosci. Biotechnol. Biochem. 81, 931–937. 10.1080/09168451.2016.1277946, PMID: [DOI] [PubMed] [Google Scholar]
- Shiraishi M., Ishino S., Heffernan M., Cann I., Ishino Y. (2018). The mesophilic archaeon Methanosarcina acetivorans counteracts uracil in DNA with multiple enzymes: EndoQ, ExoIII, and UDG. Sci. Rep. 8:15791. 10.1038/s41598-018-34000-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M., Ishino S., Yamagami T., Egashira Y., Kiyonari S., Ishino Y. (2015). A novel endonuclease that may be responsible for damaged DNA base repair in Pyrococcus furiosus. Nucleic Acids Res. 43, 2853–2863. 10.1093/nar/gkv121, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M., Ishino S., Yoshida K., Yamagami T., Cann I., Ishino Y. (2016). PCNA is involved in the EndoQ-mediated DNA repair process in thermococcales. Sci. Rep. 6:25532. 10.1038/srep25532, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi M., Iwai S. (2020). Molecular basis of substrate recognition of endonuclease Q from the euryarchaeon Pyrococcus furiosus. J. Bacteriol. 202, e00542–e00519. 10.1128/JB.00542-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter K. O. (2013). A brief history of the discovery of hyperthermophilic life. Biochem. Soc. Trans. 41, 416–420. 10.1042/BST20120284, PMID: [DOI] [PubMed] [Google Scholar]
- Su K. Y., Lin L. I., Goodman S. D., Yen R. S., Wu C. Y., Chang W. C., et al. (2018). DNA polymerase I proofreading exonuclease activity is required for endonuclease V repair pathway both in vitro and in vivo. DNA Repair 64, 59–67. 10.1016/j.dnarep.2018.02.005, PMID: [DOI] [PubMed] [Google Scholar]
- Suzuki S., Kurosawa N. (2019). Endonucleases responsible for DNA repair of helix-distorting DNA lesions in the thermophilic crenarchaeon Sulfolobus acidocaldarius in vivo. Extremophiles 23, 613–624. 10.1007/s00792-019-01120-9, PMID: [DOI] [PubMed] [Google Scholar]
- Takemoto N., Numata I., Su’etsugu M., Miyoshi-Akiyama T. (2018). Bacterial EndoMS/NucS acts as a clamp-mediated mismatch endonuclease to prevent asymmetric accumulation of replication errors. Nucleic Acids Res. 46, 6152–6165. 10.1093/nar/gky481, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J., Ciaramella M., Moracci M., Pisani F. M., Rossi M., de Vos W. M. (1998). Molecular biology of hyperthermophilic archaea. Adv. Biochem. Eng. Biotechnol. 61, 87–115. 10.1007/BFb0102290, PMID: [DOI] [PubMed] [Google Scholar]
- Wallace S. S. (2014). Base excision repair: a critical player in many games. DNA Repair 19, 14–26. 10.1016/j.dnarep.2014.03.030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Zhu X., Li Y., Shi H., Oger P., et al. (2018). Biochemical characterization of a thermostable endonuclease V from the hyperthermophilic euryarchaeon Thermococcus barophilus Ch5. Int. J. Biol. Macromol. 117, 17–24. 10.1016/j.ijbiomac.2018.05.155, PMID: [DOI] [PubMed] [Google Scholar]
- Xia B., Liu Y. L., Li W., Brice A. R., Dominy B. N., Cao W. G. (2014). Specificity and catalytic mechanism in family 5 uracil DNA glycosylase. J. Biol. Chem. 289, 18413–18426. 10.1074/jbc.M114.567354, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y., Kato M., Odawara K., Tokuno Y., Nakashima Y., Matsushima N., et al. (1996). Three-dimensional structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II from Escherichia coli. Cell 86, 311–319. 10.1016/S0092-8674(00)80102-6, PMID: [DOI] [PubMed] [Google Scholar]
- Yasui A. (2013). Alternative excision repair pathways. Cold Spring Harb. Perspect. Biol. 5, 239–249. 10.1101/cshperspect.a012617, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatopek K. M., Gardner A. F., Kelman Z. (2018). Archaeal DNA replication and repair: new genetic, biophysical and molecular tools for discovering and characterizing enzymes, pathways and mechanisms. FEMS Microbiol. Rev. 42, 477–488. 10.1093/femsre/fuy017, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang L., Jiang D., Wu M., Yang Z., Oger P. M. (2020b). New insights into DNA repair revealed by NucS endonucleases from hyperthermophilic archaea. Front. Microbiol. 11:1263. 10.3389/fmicb.2020.01263, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shi H., Gan Q., Wang Y., Wu M., Yang Z., et al. (2020a). An alternative pathway for repair of deaminated bases in DNA triggered by archaeal NucS endonuclease. DNA Repair 85:102734. 10.1016/j.dnarep.2019.102734, PMID: [DOI] [PubMed] [Google Scholar]