Abstract
Tandem duplication gives rise to copy number variation and subsequent functional novelty among genes as well as diversity between individuals in a species. Functional novelty can result from either divergence in coding sequence or divergence in patterns of gene transcriptional regulation. Here, we investigate conservation and divergence of both gene sequence and gene regulation between the copies of the α‐zein gene family in maize inbreds B73 and W22. We used RNA‐seq data generated from developing, self‐pollinated kernels at three developmental stages timed to coincide with early and peak zein expression. The reference genome annotations for B73 and W22 were modified to ensure accurate inclusion of their respective α‐zein gene models to accurately assess copy‐specific expression. Expression analysis indicated that although the total expression of α‐zeins is higher in W22, the pattern of expression in both lines is conserved. Additional analysis of publicly available RNA‐seq data from a diverse population of maize inbreds also demonstrates variation in absolute expression, but conservation of expression patterns across a wide range of maize genotypes and α‐zein haplotypes.
Keywords: copy number variation, gene duplication, maize, opaque2, zein
1. INTRODUCTION
Gene duplications introduce functional novelty to populations. Duplication events may result in the subfunctionalization (the compartmentalizing of a gene function), neofunctionalization (the development of new gene function), or loss of genes (Conant & Wolfe, 2008; Ohno, 2013). The genomes of many plant species, including maize, contain tandem duplications. In B73, the reference genome line of Zea mays, 11.3% of total genes, result from tandem duplication events (Kono et al., 2018). Gene duplication is a potential source of evolutionary novelty. However, successful duplication of a gene and associated regulatory sequence will often result in increased gene product dosage. Different classes of genes appear to encode dose sensitive and dose insensitive products, with dose insensitive genes being more likely to be duplicated in tandem. Duplication of dose sensitive genes will frequently be deleterious with the haplotype containing the duplication being lost from the population (Freeling, 2009). However, selection for increased gene product abundance can produce positive selection for increased copy number at either linked or unlinked positions in the genome, as seen in the case of the evolution of glyphosate resistance in water hemp (Gaines et al., 2010). In other cases, an initial mutation of a neutral gene duplication can confer novel function. An example of neofunctionalization by gene duplication is the independent evolution of venomous compounds in eukaryotes such as venomous members of Arachnida (Wong & Belov, 2012).
One particular family of maize genes, the zeins, have been a focus of long‐term sustained research efforts due to their function as the main storage proteins in maize kernels (Holding & Messing, 2013). The zein proteins can be grouped into four families, α, β, γ, and δ, based on amino acid sequence (Esen, 1987; Thompson & Larkins, 1994). The α‐zein family contains the highest number of genes and is composed of two subfamilies defined by mobility on protein gels: the 19‐kDa (Song & Messing, 2002) and 22‐kDa (Song et al., 2001) α‐zein families. Although the β, γ, and δ families consist of low copy numbers or single genes, the number of α‐zein gene copies ranges from 40 to 48 depending on the specific maize genotype tested.
The products of the α‐zein gene family are the most abundant zein proteins, and the family has been studied extensively. Although α‐zeins make up 70% of endosperm protein content, they lack lysine and tryptophan. The lack of lysine in α‐zein, combined with the high abundance of α‐zein in maize kernels, has a major and detrimental impact on the protein quality of maize grain (Holding, 2014). Maize‐based feed requires lysine supplementation when feeding monogastric livestock, such as swine and poultry. In addition, lysine is an essential amino acid in human diets. As a result, maize is not a complete protein source for humans or livestock. The OPAQUE2 (O2) transcription factor regulates expression of the α‐zein gene family (Schmidt et al., 1990; Vicente‐Carbajosa et al., 1997). o2 mutant kernels have reduced zein accumulation resulting in a higher lysine content, thus improved nutritional quality for animal consumption. However, o2 mutants also have negative agronomic effects such as a chalky kernels and disease susceptibility (Mertz et al., 1964). To circumvent the negative pleiotropy associated with o2 knock‐outs, researchers at CIMMYT developed modified o2 mutants known as quality protein maize (QPM) (Bjarnason & Vasal, 1992). While still an o2 mutant, QPM recovers the pleiotropically affected phenotypes via modifier genes at loci unlinked to o2. The o2 modifiers result in increased expression of the 27‐kDa γ‐zein gene, which is under relatively lower transcriptional control by O2 (Geetha et al., 1991; Holding et al., 2008; Wu et al., 2010). It was previously shown that dosage of 27‐kDa γ‐zein protein is directly proportional to kernel vitreousness (Liu et al., 2016). This variable expression dosage was subsequently shown to result from 27‐kDa γ‐zein duplication and triplication events, the latter of which was superior for modification (Liu et al., 2019). The O2 transcription factor has minimal involvement in 27‐kDa γ‐zein gene expression, rather the ZmbZIP22 transcription factor plays a larger role (Li et al., 2018). Of the two inbred lines used in this study, B73 has the single copy 27‐kDa allele, whereas W22 has the duplicated allele. In addition to O2 and bZIP22, other transcription factors regulate zein expression. Prolamine‐box binding factor 1 (PBF1) binds to a conserved region 20 bp upstream of the O2 binding site and interacts with O2 to facilitate α‐zein expression (Vicente‐Carbajosa et al., 1997). MADS47 also interacts with O2 to facilitate α‐zein expression (Qiao et al., 2016). PBF1 and MADS47 contribute to 27‐kDa γ‐zein expression as well (Li & Song, 2020), interacting with the O2 homologs OHP1 and OHP2 (Xu & Messing, 2008; Zhang et al., 2015).
Previous studies have compared the differential expression of individual α‐zeins between different maize inbreds at a single development stage (Miclaus et al., 2011; Song & Messing, 2003) or observed how zein expression changes over seed development in a single inbred (Feng et al., 2009). Inbred line B73 has been shown to contain 40 α‐zein gene copies (Song & Messing, 2003), whereas inbred W22 contains 43 (Dong et al., 2016) and inbred line BSSS53 contains 48 (Song et al., 2001; Song & Messing, 2002). Here, we sought to track patterns of transcriptional regulation across developmental time resolved at the level of individual α‐zein gene copies. We employed two maize inbreds, B73 and W22, which vary in α‐zein content and for which independent genome assemblies and gene model annotations have been generated (Schnable et al., 2009; Springer et al., 2018). The use of RNA‐seq data made it feasible to track expression of individual gene copies in each inbred over time. The assembly and annotation of tandem α‐zein repeats in published genome assemblies was imperfect; however, it was possible to compensate using previously published manually improved assemblies and/or annotations for the α‐zein regions in B73 and W22 (Dong et al., 2016; Song et al., 2001; Song & Messing, 2002).
2. MATERIALS AND METHODS
2.1. Zein extraction and SDS‐PAGE
Zeins were extracted according to the method of Wallace et al 1989 (Wallace et al., 1990). For developing kernel samples, single whole kernels, previously flash frozen in liquid N2 and stored at −80°C, were weighed and ground with a mortar and pestle in liquid N2 before addition of extraction buffer. Loading was standardized based on kernel fresh weight to account for minor differences in kernel weight. For mature kernel samples, single whole kernels were pulverized to a fine flour using a dental amalgamator, 50 mg (±0.1 mg) of flour was extracted for B73 and W22, and a common volume was loaded. Zeins are labeled as the size in kDa of each respective protein.
2.2. Determining zein gene model sequence and phylogenetic analysis
In order to compare the expression of homologous duplicates, the α‐zein genes from each inbred were matched with their respective homolog. Prior research had identified the α‐zein gene models for W22 (Dong et al., 2016). In addition, sequenced and assembled BAC contigs containing the B73 α‐zein gene models had been previously described (Song et al., 2001; Song & Messing, 2002). In order to determine the α‐zein sequences for B73, the W22 gene models were aligned to the B73 BAC contigs with Bowtie2v2.3 (Langmead & Salzberg, 2012). When conflicts occurred between alignments, the conflict was resolved in favor of the alignment with the highest MAPQ score. In the case of conflicting alignments with equal MAPQ scores, the assignment was determined by reported physical order of zeins (Dong et al., 2016; Song et al., 2001; Song & Messing, 2002, 2003) as well as exclusivity of the respective B73 homolog being considered. Phylogenetic trees produced by MUSCLE (Edgar, 2004) were used to determine the accuracy of homologous pair assignment.
2.3. Simulation study
Simulated gene expression data were generated by generating 10 k pseudofragments each of 50, 200, or 300 base pairs from each annotated α‐zein gene copy in W22. The combined set of psuedofragments of a given length generated for all α‐zein gene copies were aligned to a reference containing the complete set of α‐zein gene copies from W22 using HISAT2 v2.1 (Kim et al., 2019) to a reference containing the full set of W22 zein family members.
2.4. Plant materials and RNA extraction
Maize seed of inbred lines B73 and W22 were planted at the University of Nebraska—Lincoln East Campus research farm in Lincoln, NE during the 2019 growing season. Plants were self‐pollinated, and developing seeds were harvested at 10, 18, and 24 days after pollination (DAP). RNA was extracted from three biological replicates at each of the three timepoints. Each biological replicate consisted of a pool of four randomly selected whole kernels from the same ear.
2.5. RNA‐seq and qPCR
All 18 samples were subjected to Ribozero rRNA reduction and converted to dual‐indexed TruSeq stranded RNA‐seq libraries. An insert size of 300 bp was used. All libraries were combined in a single pool and sequenced in 1 lane of a NovaSeq SP 2 × 150‐bp run.
Expression differences of zeins and zein related genes were validated with real‐time quantitative polymerase chain reaction (qRT‐PCR). qRT‐PCR was conducted on the same whole kernel RNA samples used for RNA‐seq and incorporated three biological replicates and two technical replicates. cDNA synthesis and PCR was conducted similar to a previous study (Holding et al., 2011). Briefly, qRT‐PCR primers are shown in Table S1. The cDNA was synthesized from 1‐μg samples of DNase 1‐treated total RNA using IScript plus (Bio‐Rad, Hercules, CA), according to the manufacturer's instructions. The cDNAs were diluted tenfold in water and amplified using iQ SYBR green super mix (Bio Rad, Hercules, CA) according to the manufacturer's instructions. A My iQ Real‐time PCR thermocycler (Bio Rad, Hercules, CA) was used with the following program: 95°C for 5 min, followed by 45 cycles of 95°C (10 s) and 60°C (10 s) with 20°C per second ramp rates. Melting curves were obtained by heating from 65°C to 95°C with a 0.1°C per second ramp rate. Expression levels of genes were calculated as fold changes in W22 relative to B73 ± standard deviation at each timepoint using the maize gene Zm00001d018145 as an internal control because it was found to have invariant Ct values between all samples. For each gene tested, the average cycle threshold (Ct) value was calculated for the three biological replicate ears of each genotype. The relative expression at each timepoint was calculated using the following equation, where X = gene of interest, C = control gene, W = average Ct of three B73 samples, G = average Ct of W22 samples: 2[(WX‐WC)‐(GX‐GC)].
2.6. Read alignment and transcript quantification
RNA‐seq reads were aligned to reference genomes of B73v5 and W22v2 (Schnable et al., 2009; Springer et al., 2018) using HISAT2 v2.1 (Kim et al., 2019). Reads from B73 and W22 were each aligned to their own reference genome and their counterparts' reference. Both reference genomes associated genomic feature files (GFF) were modified in order to integrate the individual α‐zein gene models without encountering redundancy when quantifying reads. The modification consisted of aligning α‐zein gene models from B73 and W22 to their respective reference, then removing features from the GFF that fall within those alignments. For example, some α‐zein genes are described as multiple exons of a single gene in the reference annotation. Twenty‐seven genomic features were removed from the W22 GFF. Twenty‐four genomic features were removed from the B73 GFF. The modified GFFs and HISAT2 output were used in conjunction with StringTie2 v2.1 (Kovaka et al., 2019) to quantify transcript abundance. Transcript abundance was calculated at the gene level, due to the overall focus of this study being the α‐zein gene family in particular, which lack introns.
2.7. Cross‐referencing of B73 v5 and W22 v2
In order to reduce mapping bias that may occur when comparing two different genotypes to the same reference, equivalent gene models were cross‐referenced between the two genome assemblies. Version 4 of the B73 maize genome (Jiao et al., 2017) had previously been cross‐referenced to W22v2 by MaizeGDB. MaizeGDB has made available chain files between B73v4 and B73v5. These chain files were used in conjunction with CrossMap (Zhao et al., 2014) to create a v5 GFF with v4 genome coordinates. The B73v5 and W22v2 GFF files, cross‐referenced to v4, were compared with one another. Genomic features that contained 50% overlapped coordinates in both references were deemed equivalent to one another.
2.8. Differential expression analysis
In order to determine which genes were differentially expressed between W22 and B73, DESeq2 was used (Love et al., 2014). In order to determine which genes were differentially expressed between the two lines at each timepoint, B73 and W22 were compared by GLM at each of 10, 18, and 24 DAP (y = genotype). Another model, defined as y = genotype + DAP + genotype × DAP, was used to identify genes that are divergent in temporal patterns between each line. This group of genes is referred to below as differentially temporally regulated.
2.9. Deriving α‐zein expression estimates from NAM founder line RNA‐seq data
The two maize inbreds included in this study represent only one example of the diversity that exists in maize. A publicly available RNA expression dataset was utilized to better assess the generality of the data. The nested association mapping (NAM) founder lines are a set of maize inbreds spanning a large amount of the diversity of the species (Yu et al., 2008). Liu et al., 2020 extracted RNA from endosperm tissue at 16 days after self‐pollination (Liu et al., 2020). One hundred and fifty‐base pair cDNA fragments were sequenced via 75‐bp paired end reads. This read size is half the size of the current studies' reads, and therefore, alignment to the α‐zein gene copies may be less accurate than our B73 and W22 data. However, there are also many more samples, thus averaging transcripts per million (TPM) values may alleviate some of this limitation. Reads were aligned with HISAT2 and quantified with StringTie2, in the same fashion as the previously described W22 and B73 data. Zein TPM values for each of the NAM founder samples have been uploaded to https://github.com/jhurst5/W22/_B73/_RNAseq/_paper.
2.10. α ‐Zein nomenclature
α ‐Zein genes are described using a nomenclature consistent with Dong et al.'s annotation of the W22 α‐zein gene family (Dong et al., 2016). The gene name follows a pattern of z1 (Family)(Sub‐Family, if necessary)_(Individual Copy). For example, copy 3 of subfamily 2 of family A would be z1A2_3.
2.11. Code availability
Python scripts used for the simulation study and GFF modification as well as RNAseq library information are available at https://github.com/jhurst5/W22_B73_RNAseq_paper.
3. RESULTS
3.1. Zein extraction and SDS‐PAGE
As a prelude to detailed examination of zein transcript abundance, zein protein abundance in developing kernels at different stages of development was semi‐quantitatively analyzed using SDS‐PAGE in both B73 and W22 (Figure 1). Little zein protein is observed at 10 DAP, although W22 appears to be slightly advanced over B73 with respect to total zein accumulation. This W22 advancement is more pronounced at 18 DAP, although the two zein profiles appear similar at 24 DAP and at seed maturity. The 15‐kDa β‐zein, a transcriptional target of O2, and the 10‐kDa δ‐zein appear more abundant in W22 at 24 DAP.
FIGURE 1.
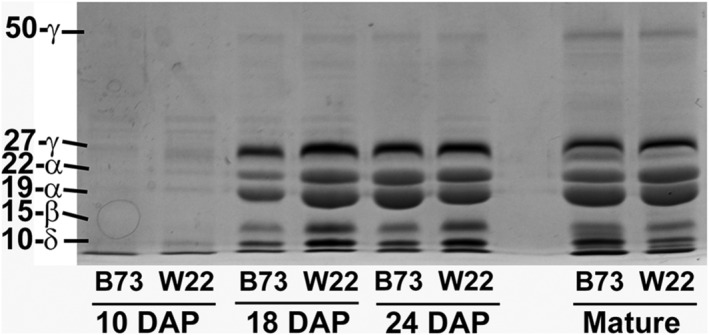
SDS‐PAGE analysis of zein fraction from whole developing kernels [days after pollination (DAP)] and mature kernels of B73 and W22. Each lane for developing kernels represents zeins from 3 mg fresh weight of frozen kernel. Each lane for mature kernels represents zeins from 1 mg of kernel flour
3.2. Matching homologous duplicates and alignment simulation
In order to directly compare homologous gene copies, α‐zein duplicates from W22 were matched to their B73 counterpart based on amino acid alignment. Phylogenetic analysis showed that 35 of the 40 α‐zein genes clustered with their expected counterpart, and zero clustered with duplicate homologs of a non‐expected duplicate (Figure S1 through Figure S5). Of the five that did not cluster as expected, three were on the same node. The two genes that were divergent by phylogenetic analysis were z1B_8 and z1B_9, which paired with each other in their respective genotypes (Figure S3). Because z1B_8 and z1B_9 paired with each other in their respective genotypes, our results indicate that they are more similar to their own genotype than they are to homologs in the other's genotype, potentially as a result of gene conversion.
An in‐silico simulation approach was used to determine the feasibility of uniquely assigning short‐read cDNA sequences to α‐zein gene models. In many cases, simulated reads 50 bp in length could not be confidently and uniquely assigned to a single gene copy of origin. Ninety‐eight percent of 200‐bp fragments could be assigned accurately, with all exceptions resulting from a confusion of z1A1_5, and z1A1_7. For each gene copy, it was possible to correctly identify the origin of 300‐bp simulated reads. It was possible to correctly identify the origin of 99% of 300‐bp simulated reads, with all exceptions resulting from confusion between a conserved region of z1A1_5 and z1A1_7. Based on these results, a sequencing strategy was designed employing 2 × 150 bp sequencing of RNA‐seq libraries constructed using an average fragment length of 300 bp.
3.3. Read alignment and transcript quantification
When reads from B73 and W22 samples were aligned to their respective references, 93.76% of reads aligned (93.42% B73 reads; 94.09% W22 reads). When the reads were aligned to the reference of the other genotype, only 75.23% of reads aligned (75.18% B73 reads; 75.27% W22 reads) (Figure 2). To better account for this bias, a cross‐reference was made between B73v5 and W22v2 reference genomes.
FIGURE 2.
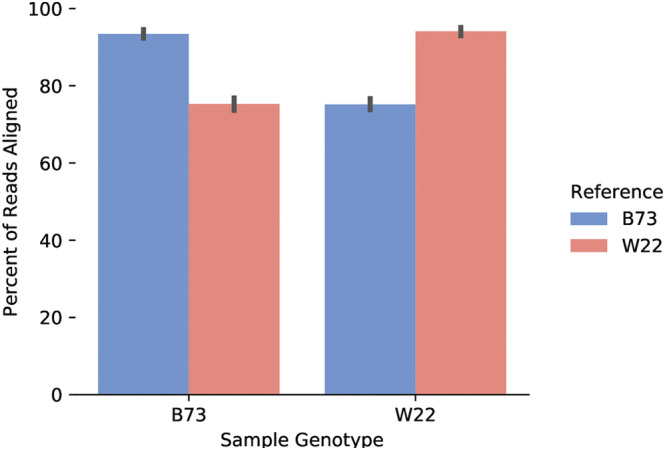
Effect of reference genome on alignment and transcript quantification. The percentage of RNA‐seq reads aligned by HISAT2 differs depending on the reference genome used
Gene‐level transcript abundance was calculated for each sample, across alignments against both references as well as the cross‐referenced gene model set. The cross‐referenced gene model set contained 40% of the B73v5 gene models and 41% of the W22v2 gene models, and consisted of 16,681 total gene models. There are 41,577 gene models in the B73 reference genome, 887 more than the 40,690 in the W22 reference. When B73 samples were aligned to the W22 reference, a 7.56% loss in gene model coverage was observed relative to alignment to the B73 reference. For W22 samples, a 10.56% loss in gene model coverage was observed when aligned to the B73 reference versus the W22 reference. Given the higher alignment rate of the samples to their respective genomes, as well as the loss of gene model coverage when aligned to the non‐origin genome, it is expected that the more accurate calculation of TPM values would occur when using the genome of the sample origin. The drawback of employing alignments to independent reference genomes is that transcriptional regulation patterns of only the subset of 16,681 gene models that could be confidently cross‐referenced between the B73 and W22 genome assembly can be accurately compared. However, as 35 of 40 pairs α‐zein genes could indeed be confidently assigned reciprocal relationships between the B73 and W22 genomes, we chose to proceed with the strategy of quantifying gene expression by aligning RNA‐seq data from a given genetic background to the independent genome assembly generated for that same genetic background.
3.4. Differentially expressed genes at each timepoint
Differential expression was tested at each timepoint sampled to determine expression differences between the two lines. Across 10, 18, and 24 DAP, 7900 gene models were differentially expressed between W22 and B73 at least once. This contains about 47% of the cross referenced gene list. The two largest categories of differentially expressed genes were shared by all three timepoints, 33% of those differentially expressed genes were significant at all three timepoints. A 25.7% of all DE genes were significant at 10 DAP exclusively (Figure 3a). The number of genes differentially expressed exclusively at a single timepoint decreased with each sampling. Whereas the total number of differentially expressed genes decreased with each timepoint, some genes not differentially expressed at one timepoint became significant at the next timepoint (Figure 3b).
FIGURE 3.
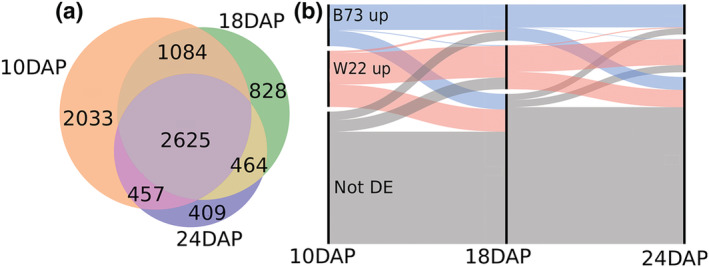
Summary of genes differentially expressed between B73 and W22. (a) Overlap of differentially expressed genes among the three sampling points. (b) Changes in upregulated genes over the course of kernel development. Blue represents genes upregulated in B73. Red indicates genes upregulated in W22. Gray indicates genes not differentially expressed
At 10 DAP, 13 α‐zein genes were differentially expressed between the two lines. Thirty‐two α‐zein genes were differentially expressed at 18 DAP, and 23 α‐zeins at 24 DAP. Eight α‐zein genes were differentially expressed at all three timepoints. The 19‐ and 22‐kDa subfamilies were similiar in that only a subset of each group was expressed (Figures 4 and 5). Despite detecting transcripts for almost all of the α‐zein genes, 50% of the total zein transcripts in B73 and W22 were related to five or six individual zein genes, respectively. W22 consistently showed higher α‐zein expression, which was confirmed by RT‐PCR (Table S1).
FIGURE 4.
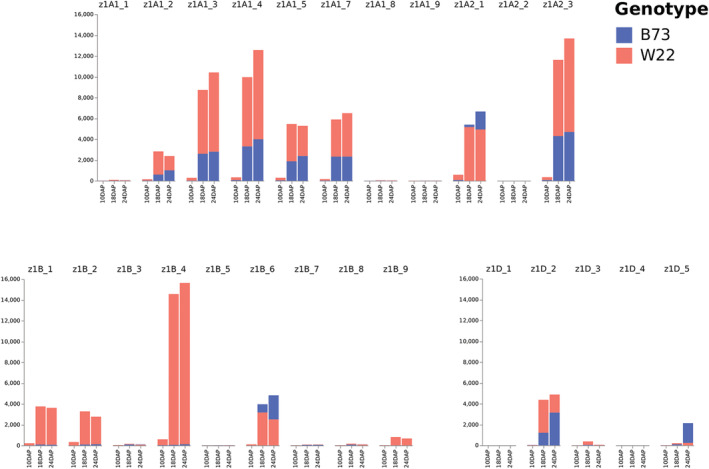
Comparison of transcripts per million (TPM) values of the 19‐kDa α‐zeins between W22 and B73. The bars in this graph are overlapped, not stacked. DAP, days after pollination
FIGURE 5.
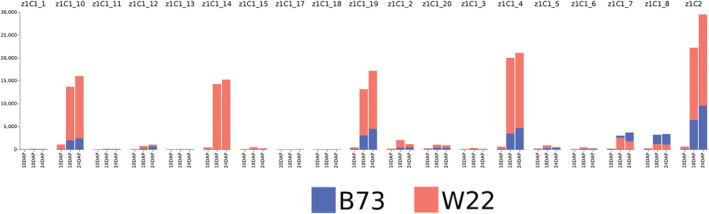
Comparison of transcripts per million (TPM) values of the 22‐kDa α‐zeins between W22 and B73. The bars in this graph are overlapped, not stacked. DAP, days after pollination
3.5. Differences in temporal patterns of zein expression between W22 and B73
Genes that were determined statistically significant in a linear model that included both genotype as well as all three timepoints and the interaction term were defined as differentially temporally regulated. Although several α‐zein genes were differentially expressed at a given timepoint, few were differentially temporally regulated. Five α‐zein genes were determined to be differentially temporally regulated: z1B_3, z1B_5, z1C1_3, z1C1_13, and z1C1_20. All five differentially temporally regulated genes are expressed at low levels relative to the highly expressed α‐zeins. Interestingly, z1C1_14 is one of the most highly expressed α‐zeins in W22, yet is one of the α‐zein duplicates not found in B73.
3.6. β ‐, γ‐, and δ‐zein expression, opaque2, and other prolamin related genes
Consistent with the overlaps in transcriptional control of the entire zein gene family, differential expression of other zein genes (Figure 6) and their related transcription factors was also observed (Figure 7). The 27‐kDa γ‐zein is differentially expressed at 10 and 18 DAP. This is expected, due to the duplication allele present in W22. At 18 DAP, the 50‐kDa γ‐zein was significantly upregulated in W22. The 15‐kDa β‐zein was differentially expressed at all timepoints. The expression differences of these non‐α‐zeins were valiated by qRT‐PCR (Table S1).
FIGURE 6.
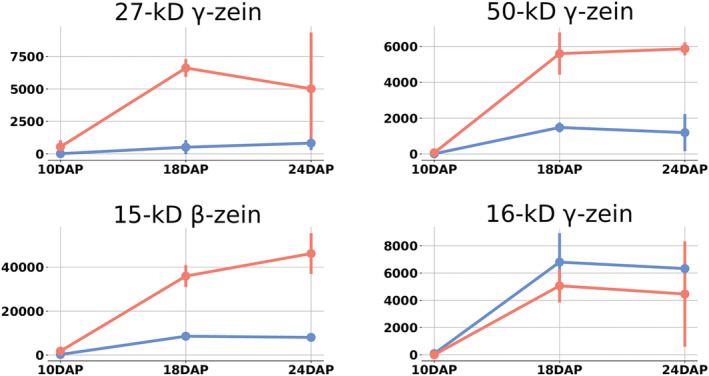
Expression patterns of the non‐α‐zeins in the cross referenced gene set. Values are in transcripts per million (TPM). Error bars are standard deviation. Blue represents B73. Red indicates W22
FIGURE 7.
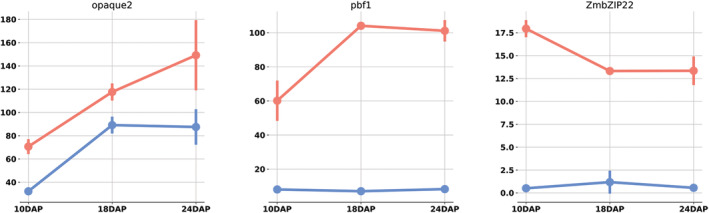
Expression patterns of three key transcription factors that regulate zein gene expression, O2, PBF1, and bZIP22. Values are in TPM. Error bars are standard deviation. Blue represents B73. Red indicates W22
Transcription factor O2, involved in transcription of the zein family, was differentially temporally regulated, as well as differentially expressed at 10 and 24 DAP. O2 was upregulated in W22. PBF1, also essential to α‐zein expression, is upregulated in W22. The differences in O2 and PBF1 expression were validated by RT‐PCR (Table S1).
3.7. Comparison of zein expression in a diverse population
As the two lines compared here, B73 and W22, are only a small representation of maize diversity, publicly available expression data from the NAM founder lines was utilized to get a broader representation of α‐zein expression. To generalize the transcript quantification, the mean TPM across 43 biological reps from 23 NAM genotypes was taken. After aligning to both the B73 and W22 reference genomes, the Spearman correlation of the NAM mean and both W22 and B73 was .9 (p < .0001) (Figure 8). This indicates that the pattern of zein expression is conserved across diverse maize inbreds.
FIGURE 8.
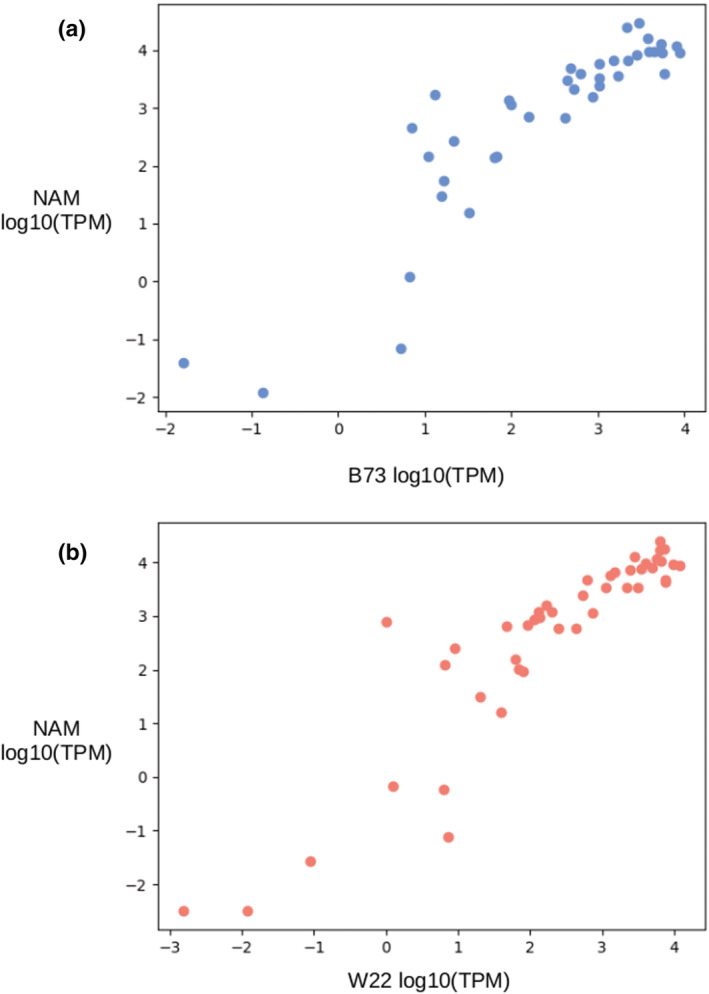
The correlation of α‐zein expression between the nested association mapping (NAM) founder lines and B73 or W22. Each point represents a single α‐zein gene copy. (a) B73, Spearman's R = .91 (p < .001) (b) W22, Spearman's R = .90 (p < .001)
In order to account for differences in insert size between the current RNAseq data and the published NAM data, the B73 and W22 reads were trimmed to 75 bp and aligned to their respective genomes. Comparisons of zein TPM between trimmed and nontrimmed libraries showed a Pearson coeffecient >.98 (p < .0001) indicating the read‐length had minor effect on measured expression values. This is contrast to the previous simulation study, likely due to similar gene models being expressed at low levels. In addition to read length differences between the two datasets, differences in library size were also present. To determine the effect that library size has on zein transcript quantification, the correlation between library size and measured zein expression was taken. Ten gene models were shown to have a statistically significant Pearson correlation. However, when running the analysis without the library size‐affected zein gene models, the resulting Spearman correlation stayed significant and .91 for both B73 and W22.
4. DISCUSSION
Duplicated genes serve as the primary source of new genetic material that can develop new functions in an organism (Moore & Purugganan, 2003). The present study looked into the expression of a tandem duplicate gene family in two separate, recently diverged haplotypes. These results indicate disparity in the overall expression of both homologous α‐zein gene copies between two maize lines, as well as among copies in a single genotype. Also observed was variation in other zeins and in the transcription factors related to prolamin gene expression. Finally, α‐zein expression in a diverse pool of maize inbreds indicate that that although absolute levels of expression vary, the general pattern is largely conserved.
4.1. RNA‐seq confirms a wide variety of α‐zein gene copies are expressed
Early studies had utilized PCR‐based approaches to measure transcript abundance of α‐zein genes, which may have been unsuitable for detecting the gene copies with lower amounts of transcripts. Dong et al. (2016) utilized an endosperm capillary‐sequencing dataset (Lai et al., 2004) to observe which gene copies were expressed. This analysis revealed that a wider range of α‐zein gene copies are expressed than was previously thought.
A challenge to using modern short‐read RNA‐seq for separating transcripts in a highly similar gene family is whether an aligner is able to accurately assign sequencing reads to the correct gene copy. By aligning a set of simulated read sets to the entire gene family, it was determined that reads could be aligned accurately if a larger read length (300 bp) was used. Short‐read RNA‐seq confirms a wide variety of α‐zein gene copies are expressed. However, most of the transcripts come from a subset of gene copies in both haplotypes.
4.2. Disparity between protein profile and transcript abundance
Based solely on transcript abundance, it may be expected that W22 grain contains a different zein protein profile from B73. However, SDS‐PAGE performed on the two inbreds does not suggest drastic differences at the protein level. Although the protein profile was consistent with transcript abundance at 10 and 18 DAP, by 24 DAP, it appeared that B73 and W22 were not much different in protein profile despite the higher zein expression in W22. A similiar zein protein abundance was also observed in fully mature kernels. This indicates substantial transcriptional plasticity exists in zein accumulation and that processes that occur during or after translation, as opposed to during transcription, is the limiting factor of kernel protein content.
Prior work has identified genes that play post‐transcriptional roles in zein accumulation or indirectly affect zein accumulation. For example, mutations in members of the arogenate dehydrogenase gene family, involved in amino acid synthesis, reduce translation of α‐zein protein (Holding et al., 2010). Mutants of an acyl‐CoA synthetase‐like gene display reduced α‐zein accumulation, likely due to posttranslational modification (Miclaus, Wu, et al., 2011). The Floury1 protein regulates zein localization in the protein body. Mutants of the Fl1 gene have altered accumulation of 22‐kDa α‐zeins, whereby they are not deposited in their usual ring on the outer portion of the protein body (Holding et al., 2007). Opaque10 encodes a protein that interacts with the 22‐kDa α‐zeins during formation of the PB. Mutants of O10 ultimately alter the localization of 22‐kDa α‐zeins (Yao et al., 2016).
4.3. W22 expresses α‐zeins at a higher level than does B73 yet the temporal pattern is mostly conserved
Inbred line W22 expressed α‐zeins at a higher TPM than did B73. In each of the four α‐zein subfamilies, there appeared to be a few dominant copies responsible for most of the transcripts. One of these, z1C1_14, only exists in the W22 haplotype, indicating it is a recently duplicated gene copy that is responsible for a substantial amount of α‐zein expression. Interestingly, z1C_14 appears to be a copy of z1C_4, another highly expressed zein.
The pattern of expression is similar in both lines. At 10 DAP, only low levels of expression were seen in both lines. This is a contrast to the overall set of common gene models, where over a third of all differentially expressed genes were differentially expressed at 10 DAP. This may indicate that α‐zein expression is more conserved than other genes; however, as genes with low levels of expression (such as truncated zein copies) produce less transcripts and thus have less statistical power to detect differences, this cannot be inferred from the current results. Major differences in α‐zein transcript abundance were not apparent until after the first sampling date. Five α‐zein genes were identified as differentially temporally regulated. One other, z1C1_14, is a dominantly expressed duplicate that is only present in W22 and not in B73. Of the five differentially temporally regulated α‐zeins found in both haplotypes, none are the top 50% highest expressed α‐zeins.
Based on these observations, it appears that expression of the dominant α‐zeins is conserved. The exception is z1C1_14, one of the most highly expressed zeins in W22 though not found in the B73 haplotype. Additionally, comparing B73 and W22 to patterns of α‐zein expression in diverse maize appears to confirm these results.
4.4. Differences in transcription factor regulation may drive the difference in overall expression
Although the general pattern and dominant gene copies is largely conserved, the overall zein expression was higher in W22. A disparity is also observed in the levels of gene expression among prolamin related transcription factors (Figure 7). Three transcription factors interact to express the α‐zein gene family: O2, MADS47, and PBF1 (Li & Song, 2020). Although no difference was seen in MADS47 expression, O2 and PBF1 were both differentially temporally regulated. Whereas O2 is expressed at roughly 50% higher TPM in W22 compared with B73, PBF1 is expressed at nearly 2000% higher TPM in W22. Given the substantial increase in PBF1 over O2 between each line, it may indicate that PBF1 is a significant limiting factor in zein expression.
A similar observation is made when comparing expression of the 27‐kDa γ‐zein between B73 and W22 (Figure 6). W22 had statistically significant higher expression of the 27‐kDa γ‐zein, which is not surprising given that W22 allele is duplicated. However, ZmbZIP22 is also expressed at much higher levels in W22. ZmbZIP22 is involved, along with O2 and PBF1, in regulation of the 27‐kDa γ‐zein. Although the duplication of the 27‐kDa γ‐zein locus has widely been assumed to be responsible for the higher expression of γ‐zein in the W22 background, the results presented here suggest the potential of role for increased expression of upstream regulators contributing to the higher expression of γ‐zein observed in W22.
5. CONCLUSION
Previous studies have shown that multiple α‐zein haplotypes exist in maize (Dong et al., 2016; Song & Messing, 2003), their expression changes over time (Feng et al., 2009), and that the expression patterns are similar between two inbreds (Miclaus et al., 2011). In this study, an additional inbred, W22, was directly compared with B73 using RNA‐seq reads mapped to their precise haplotype at different developmental stages. The current study found that although temporal regulation of the dominant α‐zeins does not change between the two inbred lines, W22 has greater expression of both the α‐zein gene family and the transcription factors responsible for regulating their expression.
As the number of inbred lines with assembled genomes grows (Liu et al., 2020), future steps will be to index the zein haplotypes in diverse maize genomes. After identifying the α‐zein haplotype diversity, a broader observation of gene copy divergence will become accessible.
CONFLICT OF INTEREST
The Authors did not report any conflict of interest.
Supporting information
Figure S1. Phylogenetic analysis of B73 and W22 z1A1 gene family. Red indicates instances of B73 and W22 gene models not clustering.
Figure S2. Phylogenetic analysis of B73 and W22 z1A2 gene family. Red indicates instances of B73 and W22 gene models not clustering.
Figure S3. Phylogenetic analysis of B73 and W22 z1B gene family. Red indicates instances of B73 and W22 gene models not clustering. Purple indicates gene copies that only exist in W22.
Figure S4. Phylogenetic analysis of B73 and W22 z1C1 gene family. Red indicates instances of B73 and W22 gene models not clustering. Purple indicates gene copies that only exist in W22.
Figure S5. Phylogenetic analysis of B73 and W22 z1D gene family. Red indicates instances of B73 and W22 gene models not clustering.
Table S1. Primer sequence and qRT‐PCR results.
Hurst, P., Schnable, J. C., & Holding, D. R. (2021). Tandem duplicate expression patterns are conserved between maize haplotypes of the α‐zein gene family. Plant Direct, 5(9), e346. 10.1002/pld3.346
REFERENCES
- Bjarnason, M., & Vasal, S. (1992). Breeding of quality protein maize (qpm). Plant Breeding Reviews, 9, 181–216. [Google Scholar]
- Conant, G. C., & Wolfe, K. H. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nature Reviews. Genetics, 9, 938–950. 10.1038/nrg2482 [DOI] [PubMed] [Google Scholar]
- Dong, J., Feng, Y., Kumar, D., Zhang, W., Zhu, T., Luo, M. C., & Messing, J. (2016). Analysis of tandem gene copies in maize chromosomal regions reconstructed from long sequence reads. Proceedings of the National Academy of Sciences, 113, 7949–7956. 10.1073/pnas.1608775113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen, A. (1987). A proposed nomenclature for the alcohol‐soluble proteins (zeins) of maize (Zea mays L.). Journal of Cereal Science, 5, 117–128. [Google Scholar]
- Feng, L., Zhu, J., Wang, G., Tang, Y., Chen, H., Jin, W., Wang, F., Mei, B., Xu, Z., & Song, R. (2009). Expressional profiling study revealed unique expressional patterns and dramatic expressional divergence of maize α‐zein super gene family. Plant Molecular Biology, 69, 649–659. 10.1007/s11103-008-9444-z [DOI] [PubMed] [Google Scholar]
- Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: Tandem, whole‐genome, segmental, or by transposition. Annual Review of Plant Biology, 60, 433–453. 10.1146/annurev.arplant.043008.092122 [DOI] [PubMed] [Google Scholar]
- Gaines, T. A., Zhang, W., Wang, D., Bukun, B., Chisholm, S. T., Shaner, D. L., Nissen, S. J., Patzoldt, W. L., Tranel, P. J., Culpepper, A. S., Grey, T. L., Webster, T. M., Vencill, W. K., Sammons, R. D., Jiang, J., Preston, C., Leach, J. E., & Westra, P. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri . Proceedings of the National Academy of Sciences, 107, 1029–1034. 10.1073/pnas.0906649107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha, K., Lending, C. R., Lopes, M. A., Wallace, J. C., & Larkins, B. A. (1991). Opaque‐2 modifiers increase gamma‐zein synthesis and alter its spatial distribution in maize endosperm. The Plant Cell, 3, 1207–1219. 10.1105/tpc.3.11.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding, D., & Messing, J. (2013). Evolution, structure, and function of prolamin storage proteins. In Seed genomics (pp. 139–158). John Wiley & Sons, Inc. [Google Scholar]
- Holding, D. R. (2014). Recent advances in the study of prolamin storage protein organization and function. Frontiers in Plant Science, 5, 276. 10.3389/fpls.2014.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding, D. R., Hunter, B. G., Chung, T., Gibbon, B. C., Ford, C. F., Bharti, A. K., Messing, J., Hamaker, B. R., & Larkins, B. A. (2008). Genetic analysis of opaque2 modifier loci in quality protein maize. Theoretical and Applied Genetics, 117, 157–170. 10.1007/s00122-008-0762-y [DOI] [PubMed] [Google Scholar]
- Holding, D. R., Hunter, B. G., Klingler, J. P., Wu, S., Guo, X., Gibbon, B. C., Wu, R., Schulze, J. M., Jung, R., & Larkins, B. A. (2011). Characterization of opaque2 modifier qtls and candidate genes in recombinant inbred lines derived from the k0326y quality protein maize inbred. Theoretical and Applied Genetics, 122, 783–794. 10.1007/s00122-010-1486-3 [DOI] [PubMed] [Google Scholar]
- Holding, D. R., Meeley, R. B., Hazebroek, J., Selinger, D., Gruis, F., Jung, R., & Larkins, B. A. (2010). Identification and characterization of the maize arogenate dehydrogenase gene family. Journal of Experimental Botany, 61, 3663–3673. 10.1093/jxb/erq179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holding, D. R., Otegui, M. S., Li, B., Meeley, R. B., Dam, T., Hunter, B. G., Jung, R., & Larkins, B. A. (2007). The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. The Plant Cell, 19, 2569–2582. 10.1105/tpc.107.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y., Peluso, P., Shi, J., Liang, T., Stitzer, M. C., Wang, B., Campbell, M. S., Stein, J. C., Wei, X., Chin, C.‐S., Guill, K., Regulski, M., Kumari, S., Olson, A., Gent, J., Schneider, K. L., Wolfgruber, T. K., May, M. R., Springer, N. M., … Ware, D. (2017). Improved maize reference genome with single‐molecule technologies. Nature, 546, 524–527. 10.1038/nature22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Paggi, J. M., Park, C., Bennett, C., & Salzberg, S. L. (2019). Graph‐based genome alignment and genotyping with hisat2 and hisat‐genotype. Nature Biotechnology, 37, 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono, T. J., Brohammer, A. B., McGaugh, S. E., & Hirsch, C. N. (2018). Tandem duplicate genes in maize are abundant and date to two distinct periods of time. G3: Genes, Genomes, Genetics, 8, 3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka, S., Zimin, A. V., Pertea, G. M., Razaghi, R., Salzberg, S. L., & Pertea, M. (2019). Transcriptome assembly from long‐read rna‐seq alignments with stringtie2. Genome Biology, 20, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J., Dey, N., Kim, C. S., Bharti, A. K., Rudd, S., Mayer, K. F., Larkins, B. A., Becraft, P., & Messing, J. (2004). Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Research, 14, 1932–1937. 10.1101/gr.2780504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., & Salzberg, S. L. (2012). Fast gapped‐read alignment with bowtie 2. Nature Methods, 9(357), 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., & Song, R. (2020). The regulation of zein biosynthesis in maize endosperm. Theoretical and Applied Genetics, 133(5), 1443–1453. 10.1007/s00122-019-03520-z [DOI] [PubMed] [Google Scholar]
- Li, C., Yue, Y., Chen, H., Qi, W., & Song, R. (2018). The zmbzip22 transcription factor regulates 27‐kd γ‐zein gene transcription during maize endosperm development. The Plant Cell, 30, 2402–2424. 10.1105/tpc.18.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Huang, Y., Li, X., Wang, H., Ding, Y., Kang, C., Sun, M., Li, F., Wang, J., Deng, Y., & Yang, X. (2019). High frequency dna rearrangement at qγ27 creates a novel allele for quality protein maize breeding. Communications Biology, 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Shi, J., Sun, C., Gong, H., Fan, X., Qiu, F., Huang, X., Feng, Q., Zheng, X., Yuan, N., Li, C., Zhang, Z., Deng, Y., Wang, J., Pan, G., Han, B., Lai, J., & Wu, Y. (2016). Gene duplication confers enhanced expression of 27‐kda γ‐zein for endosperm modification in quality protein maize. Proceedings of the National Academy of Sciences, 113, 4964–4969. 10.1073/pnas.1601352113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Seetharam, A. S., Chougule, K., Ou, S., Swentowsky, K. W., Gent, J. I., Llaca, V., Woodhouse, M. R., Manchanda, N., Presting, G. G., Kudrna, D. A., Alabady, M., Hirsch, C. N., Fengler, K. A., Ware, D., Michael, T. P., Hufford, M. B., & Dawe, R. K. (2020). Gapless assembly of maize chromosomes using long‐read technologies. Genome Biology, 21(1). 10.1186/s13059-020-02029-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for rna‐seq data with deseq2. Genome Biology, 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz, E. T., Bates, L. S., & Nelson, O. E. (1964). Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science, 145, 279–280. 10.1126/science.145.3629.279 [DOI] [PubMed] [Google Scholar]
- Miclaus, M., Wu, Y., Xu, J.‐H., Dooner, H. K., & Messing, J. (2011). The maize high‐lysine mutant opaque7 is defective in an acyl‐coa synthetase‐like protein. Genetics, 189, 1271–1280. 10.1534/genetics.111.133918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclaus, M., Xu, J.‐H., & Messing, J. (2011). Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes. PLoS Genetics, 7(6), e1002131. 10.1371/journal.pgen.1002131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., & Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proceedings of the National Academy of Sciences, 100, 15682–15687. 10.1073/pnas.2535513100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. (2013). Evolution by gene duplication. New York: Springer Science & Business Media. [Google Scholar]
- Qiao, Z., Qi, W., Wang, Q., Feng, Y.’., Yang, Q., Zhang, N., Wang, S., Tang, Y., & Song, R. (2016). Zmmads47 regulates zein gene transcription through interaction with opaque2. PLoS Genetics, 12, e1005991. 10.1371/journal.pgen.1005991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R. J., Burr, F. A., Aukerman, M. J., & Burr, B. (1990). Maize regulatory gene opaque‐2 encodes a protein with a “leucine‐zipper” motif that binds to zein dna. Proceedings of the National Academy of Sciences, 87, 46–50. 10.1073/pnas.87.1.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., Liang, C., Zhang, J., Fulton, L., Graves, T. A., Minx, P., Reily, A. D., Courtney, L., Kruchowski, S. S., Tomlinson, C., Strong, C., Delehaunty, K., Fronick, C., Courtney, B., … Wilson, R. K. (2009). The b73 maize genome: Complexity, diversity, and dynamics. Science, 326, 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Song, R., Llaca, V., Linton, E., & Messing, J. (2001). Sequence, regulation, and evolution of the maize 22‐kd α zein gene family. Genome Research, 11, 1817–1825. 10.1101/gr.197301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., & Messing, J. (2002). Contiguous genomic dna sequence comprising the 19‐kd zein gene family from maize. Plant Physiology, 130, 1626–1635. 10.1104/pp.012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., & Messing, J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proceedings of the National Academy of Sciences, 100, 9055–9060. 10.1073/pnas.1032999100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, N. M., Anderson, S. N., Andorf, C. M., Ahern, K. R., Bai, F., Barad, O., Barbazuk, B., Bass, H. K., Baruch, K., Ben‐Zvi, G., Buckler, E. S., Bukowski, R., Campbell, M. S., Cannon, E. K. S., Chomet, P., Dawe, R. K., Davenport, R., Dooner, H. K., Du, L. H., … Brutnell, T. P. (2018). The maize w22 genome provides a foundation for functional genomics and transposon biology. Nature Genetics, 50, 1282–1288. 10.1038/s41588-018-0158-0 [DOI] [PubMed] [Google Scholar]
- Thompson, G. A., & Larkins, B. A. (1994). Characterization of zein genes and their regulation in maize endosperm. In The maize handbook (pp. 639–647). Springer. [Google Scholar]
- Vicente‐Carbajosa, J., Moose, S. P., Parsons, R. L., & Schmidt, R. J. (1997). A maize zinc‐finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator opaque2. Proceedings of the National Academy of Sciences, 94, 7685–7690. 10.1073/pnas.94.14.7685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, J. C., Lopes, M. A., Paiva, E., & Larkins, B. A. (1990). New methods for extraction and quantitation of zeins reveal a high content of γ‐zein in modified opaque‐2 maize. Plant Physiology, 92, 191–196. 10.1104/pp.92.1.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E. S., & Belov, K. (2012). Venom evolution through gene duplications. Gene, 496, 1–7. 10.1016/j.gene.2012.01.009 [DOI] [PubMed] [Google Scholar]
- Wu, Y., Holding, D. R., & Messing, J. (2010). γ‐Zeins are essential for endosperm modification in quality protein maize. Proceedings of the National Academy of Sciences, 107, 12810–12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.‐H., & Messing, J. (2008). Diverged copies of the seed regulatory opaque‐2 gene by a segmental duplication in the progenitor genome of rice, sorghum, and maize. Molecular Plant, 1, 760–769. 10.1093/mp/ssn038 [DOI] [PubMed] [Google Scholar]
- Yao, D., Qi, W., Li, X., Yang, Q., Yan, S., Ling, H., Wang, G., Wang, G., & Song, R. (2016). Maize opaque10 encodes a cereal‐specific protein that is essential for the proper distribution of zeins in endosperm protein bodies. PLoS Genetics, 12, e1006270. 10.1371/journal.pgen.1006270 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yu, J., Holland, J. B., McMullen, M. D., & Buckler, E. S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics, 178, 539–551. 10.1534/genetics.107.074245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Yang, J., & Wu, Y. (2015). Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine‐box binding factor, and O2 heterodimerizing proteins. The Plant Cell, 27, 1162–1172. 10.1105/tpc.15.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Sun, Z., Wang, J., Huang, H., Kocher, J. P., & Wang, L. (2014). Crossmap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics, 30, 1006–1007. 10.1093/bioinformatics/btt730 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phylogenetic analysis of B73 and W22 z1A1 gene family. Red indicates instances of B73 and W22 gene models not clustering.
Figure S2. Phylogenetic analysis of B73 and W22 z1A2 gene family. Red indicates instances of B73 and W22 gene models not clustering.
Figure S3. Phylogenetic analysis of B73 and W22 z1B gene family. Red indicates instances of B73 and W22 gene models not clustering. Purple indicates gene copies that only exist in W22.
Figure S4. Phylogenetic analysis of B73 and W22 z1C1 gene family. Red indicates instances of B73 and W22 gene models not clustering. Purple indicates gene copies that only exist in W22.
Figure S5. Phylogenetic analysis of B73 and W22 z1D gene family. Red indicates instances of B73 and W22 gene models not clustering.
Table S1. Primer sequence and qRT‐PCR results.


