Abstract
The function of the U3 small nucleolar ribonucleoprotein (snoRNP) is central to the events surrounding pre-rRNA processing, as evidenced by the severe defects in cleavage of pre-18S rRNA precursors observed upon depletion of the U3 RNA and its unique protein components. Although the precise function of each component remains unclear, since U3 snoRNA levels remain unchanged upon genetic depletion of these proteins, it is likely that the proteins themselves have significant roles in the cleavage reactions. Here we report the identification of two previously undescribed protein components of the U3 snoRNP, representing the first snoRNP components identified by using the two-hybrid methodology. By screening for proteins that physically associate with the U3 snoRNP-specific protein, Mpp10p, we have identified Imp3p (22 kDa) and Imp4p (34 kDa) (named for interacting with Mpp10p). The genes encoding both proteins are essential in yeast. Genetic depletion reveals that both proteins are critical for U3 snoRNP function in pre-18S rRNA processing at the A0, A1, and A2 sites in the pre-rRNA. Both Imp proteins associate with Mpp10p in vivo, and both are complexed only with the U3 snoRNA. Conservation of RNA binding domains between Imp3p and the S4 family of ribosomal proteins suggests that it might associate with RNA directly. However, as with other U3 snoRNP-specific proteins, neither Imp3p nor Imp4p is required for maintenance of U3 snoRNA integrity. Imp3p and Imp4p are therefore novel protein components specific to the U3 snoRNP with critical roles in pre-rRNA cleavage events.
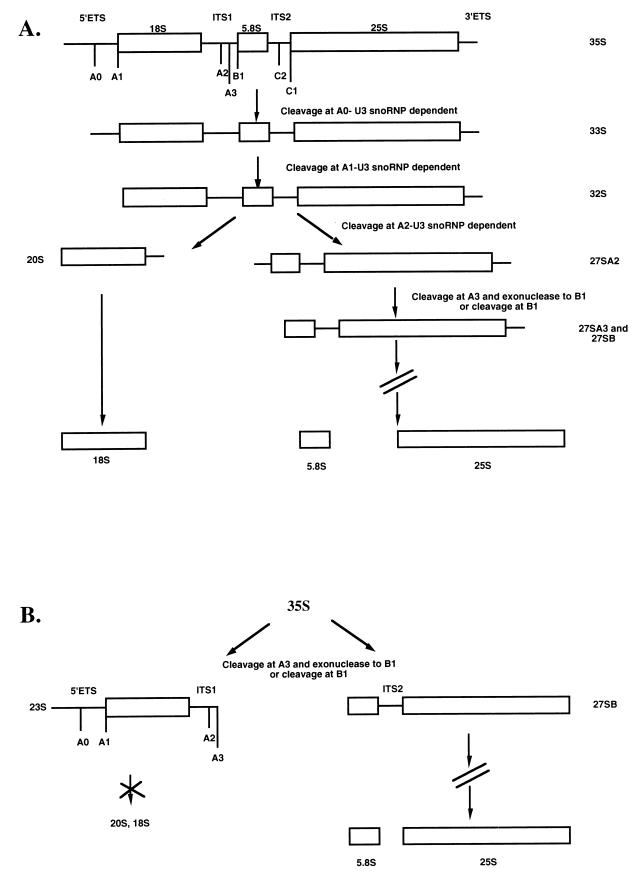
Eukaryotic ribosomes are large ribonucleoproteins (RNPs) composed of four rRNAs and dozens of ribosomal proteins. Three of the four RNA components are processed from a single polycistronic transcript, undergo extensive nucleotide modification, and assemble with the appropriate proteins in the cell nucleolus. In the yeast Saccharomyces cerevisiae, the small-subunit (SSU) rRNA (18S) and the large-subunit (LSU) rRNAs (5.8S and 25S) are processed from the nascent 35S transcript via a series of cleavage steps (Fig. 1A). Successive cleavages at the 5′ end of the precursor rRNA yield the 33S and 32S transcripts, while subsequent cleavage at the A2 site separates the precursors of the SSU rRNA from those of the LSU rRNA. The 20S pre-rRNA is matured to 18S rRNA, while the LSU precursors are terminally processed to the 5.8S and 25S rRNAs.
FIG. 1.
(A) Pre-rRNA processing in the yeast S. cerevisiae. The nascent 35S transcript is cleaved in three U3 snoRNP-dependent reactions at A0, A1, and A2 to separate the precursors to the SSU (18S) rRNA from those to the LSU (5.8S and 25S) rRNAs. (B) Pre-rRNA processing pathway in yeast depleted of the U3 snoRNA. Genetic depletion of the U3 snoRNA causes an apparent increase in the levels of the nascent 35S precursor and accumulation of the 23S precursor resulting from lack of cleavage at A0, A1, and A2. As a result, decreased levels of the 20S and 18S rRNAs occur. Processing of the LSU pre-rRNA occurs via the 27S precursor (18).
Several small nucleolar RNPs (snoRNPs) are required for the cleavage steps that generate the mature 18S rRNA (reviewed in reference 35). Of these, the most widely studied is the U3 snoRNP. In yeast, genetic depletion of the U3 snoRNA results in a deficiency in processing at the A0, A1, and A2 sites, causing a sharp decrease in 20S and 18S rRNA levels and an apparent increase in the presence of the 23S precursor and the 35S nascent transcript (Fig. 1B). A role for the U3 snoRNP in pre-rRNA cleavage events has also been observed in metazoans. Oligonucleotide-mediated depletion of the U3 snoRNA in vivo in Xenopus laevis oocytes and in vitro in mouse cell extracts results in a loss of cleavage events that lead to release of the 18S rRNA (21, 28). The conservation of the requirement for an intact U3 snoRNP in pre-rRNA processing in a wide variety of organisms is a powerful argument that this is an essential function of this RNP.
The U3 snoRNA, whose RNA component was first described for human cells (37), has since been sequenced from a number of different organisms. The size of the U3 snoRNA ranges from 144 nucleotides in the protozoan parasite Trypanosoma brucei to about 330 nucleotides in the yeast S. cerevisiae (14, 19). The U3 snoRNA harbors the consensus box C and box D motifs that classify it with the box C/D family of snoRNAs. Consistent with the other box C/D snoRNAs, the U3 RNA associates with the nucleolar protein fibrillarin (Nop1p in yeast) as well as Nop56p and Nop5p/Nop58p, two new box C/D snoRNP common proteins recently identified in yeast (16, 23, 23a, 29, 39). Although U3 shares protein components with the other box C/D snoRNPs, it is functionally distinct from the overwhelming majority that effect 2′-O-ribose methylation of rRNA, as it is required for cleavage events in the pre-rRNA. Consistent with its unique role, it is noteworthy that so far, the U3 snoRNP is the only box C/D snoRNP for which specific components have been identified. Coimmunoprecipitation has demonstrated that Sof1p, Mpp10p, and Lcp5p associate specifically with the U3 snoRNA and not with any other small RNAs (10, 20, 38).
Thus far, all of the genes encoding the six U3 proteins in yeast have proven to be essential for viability, facilitating the use of genetic depletion to probe, in vivo, their roles in pre-rRNA processing. These studies have demonstrated that the functional consequences of depleting any of the specific components (Sof1p, Mpp10p, and Lcp5p) as well as any one of two common components (Nop1p and Nop5p/Nop58p) are identical to those of U3 snoRNA depletion, that is, loss of pre-18S rRNA processing due to defective cleavage at the A0, A1, and A2 sites (10, 18, 20, 23, 34, 38, 39). Presumably, this common depletion phenotype results from effects on the overall organization of the U3 RNP particle. Depletion of Nop5p/Nop58p leads to underaccumulation of the U3 snoRNA, as well as other box C/D snoRNAs, indicating a function in maintenance of snoRNA stability (23). In contrast, steady-state levels of the U3 snoRNA remain unaffected upon depletion of the U3-specific proteins, indicating a more direct role for these components in pre-rRNA cleavage events (10, 20, 38).
Despite the fact that the effector molecule(s) in processing is unknown, it has been possible to define the placement of the U3 snoRNP on the pre-rRNA for the required cleavage steps in SSU rRNA synthesis. Mutational analysis guided by cross-linking results has suggested that a base-pairing interaction between the U3 snoRNA and the 5′ external transcribed spacer of the pre-rRNA is crucial to generate cleavages at the A0, A1, and A2 sites (4–6). Perhaps this region of complementarity is key in determining proper alignment of the U3 snoRNP on the pre-rRNA. A second base-pairing interaction may occur between box A of the U3 snoRNA and sequences adjacent to the 5′ boundary of the 18S rRNA. Mutational studies have intimated the importance of residues in and proximal to the conserved box A sequence for efficient cleavage only at the A1 and A2 sites (17). These mutations had no effect on cleavage efficiency at the A0 site, however, indicating that the A0 and A1-A2 cleavages are functionally separable.
Interestingly, carboxy-terminal truncation of the U3-specific Mpp10 protein mimics the processing defect caused by the U3 snoRNA with the box A mutations, suggesting that these two mutations may target the same step in processing (24). Furthermore, the Mpp10p truncation work allows preliminary assignment of at least a portion of this protein in mediating the A1 and A2 cleavages, and thus Mpp10p is the only U3 protein component that can be placed in any mechanistic context. Therefore, although we do not yet know which enzyme or components carry out the cleavage reactions, we do know that mutations in a protein subunit can disrupt them. This suggests that some cleavage reactions are, at least in part, mediated by protein components of the U3 snoRNP and reveals the necessity of elucidating and scrutinizing all of the protein components of the U3 snoRNP to better understand how this RNP functions in pre-rRNA processing.
We report here two new U3 snoRNP-specific proteins that were isolated by using the two-hybrid methodology. We searched for proteins that physically interact with Mpp10p, reasoning that we might find new U3 snoRNP components or U3 snoRNP-associated factors or be able to better define Mpp10 interactions with other previously identified U3 snoRNP components. Our screen isolated two previously unidentified proteins that we have named Imp3p and Imp4p (for interacting with Mpp10p). Imp3p has a predicted molecular mass of 22 kDa and has homology to the yeast ribosomal S9 proteins (Escherichia coli S4); Imp4p has a predicted migration of 34 kDa and no similarity to any known protein. We have confirmed that both proteins interact with Mpp10p under physiological conditions and that, like previously described U3 snoRNP-specific components, they are essential for viability and are associated exclusively with the U3 snoRNA. Consistent with their role as U3 snoRNP components, genetic depletion of the Imp proteins results in reduced production of 18S rRNA and accumulation of SSU precursors due to the absence of cleavage at sites A0, A1, and A2. However, as in depletion of the other U3 snoRNP-specific components, U3 snoRNA levels are not affected, suggesting that the Imp proteins are not required for maintenance of RNA integrity but may instead play a more direct role in pre-rRNA cleavage events.
MATERIALS AND METHODS
Yeast strains and media.
All two-hybrid manipulations were carried out with strain MaV103 (MATa) (36). HIS3 and lacZ (encoding beta-galactosidase) serve as reporter genes for this system. Subsequent functional studies were performed with the diploid strain YPH260 (MATa/α ura3-52/ura3-52 lys2-801amber/lys2-801amber ade2-101ochre/ade2-101ochre his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1) or its derivatives YPH258 (MATa ura3-52 lys2-801amber ade2-101ochre his3-Δ200 leu2-Δ1) and YPH259 (MATα ura3-52 lys2-801amber ade2-101ochre his3-Δ200 leu2-Δ1). Yeast was grown in rich medium YPD (1% yeast extract, 2% peptone, 2% dextrose or glucose) or YPG (1% yeast extract, 2% peptone, 2% galactose). When the omission of a certain nutrient(s) was required to maintain plasmid selection, cells were grown in synthetic medium SD (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose) and supplemented with only the appropriate nutrients. SD was prepared according to the specifications outlined by Clontech. Medium in culture plates included 2% Bacto Agar.
Yeast two-hybrid screen.
A two-hybrid screen was carried out with the yeast Mpp10 protein as bait. A fragment, amplified by PCR, containing the intact MPP10 gene was inserted into the SmaI/XmaI and SalI sites of pAS2-1 (TRP1 selectable marker; Clontech), creating an in-frame, N-terminal fusion to the GAL4 DNA binding domain. Because the full-length Mpp10 fusion protein activated the reporter genes (HIS3 and lacZ) in the absence of the prey plasmid (fusion with the GAL4 activation domain), a 95-amino-acid (aa)-carboxy-truncated MPP10 gene (aa 1 to 498; mpp10-5) was cloned into the same sites of pAS2-1. The mpp10-5 fragment was amplified by using ympp10.27 (5′-TCCCCCCGGGCATGTCAGAACTCTTTGGA-3′; SmaI/XmaI site) and ympp10.9a (5′-CCCGCTCGAGTCAGACATTGTATATCTCTTGAGG-3′; stop codon and XhoI site) for the 5′ and 3′ ends, respectively. The fragment was restriction digested with XmaI and XhoI, purified, and then ligated into pAS2-1 that had been sequentially cut with SalI and XmaI. This construct did not autoactivate the reporter genes. The clone was sequenced on an Applied Biosystems 373 Stretch Sequencer at the W.M. Keck sequencing facility through the GAL4 binding domain-mpp10 fusion to confirm the maintenance of the correct reading frame by using oligonucleotide pAS2-1.1 (5′-GACTGTATCGCCGGTATTGC-3′).
mpp10-5 was used as bait to carry out the two-hybrid screen. Initially, pAS2-1-mpp10-5 was introduced into strain MaV103 by lithium acetate transformation (13). A yeast cDNA library (108 transformants), obtained from Stephen Elledge, cloned into pACT2 (LEU2 gene) and fused to a GAL4 activation domain was used as prey to find Mpp10p-interacting proteins. The library was transformed into the MaV103 strain with the mpp10-5 bait plasmid. Transformants were selected on SD containing His, Leu, Trp and 50 mM 3-aminotriazole. Of the estimated 513,000 colonies studied, approximately 43,000 were positive for HIS3 expression in the screen. These transformants were then assayed for lacZ activation as described previously (3), with the exception that the amount of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) used in the detection buffer was doubled. Twenty beta-galactosidase-positive colonies were obtained, each bearing prey proteins that interact with the Mpp10p bait.
To verify that the prey proteins selected by the mpp10-5 bait required interaction with Mpp10p for reporter gene activation, the positive yeast strains obtained in the two-hybrid screen were selected for loss of the mpp10-5 bait plasmid. This was accomplished by repetitive streaking of the positive strains onto SD-Leu plates and then onto YPD plus cycloheximide (2 μg/ml). These yeast strains, which bear only the prey plasmids, were then assayed for beta-galactosidase activity. Of the 20 positive strains, 3 activated lacZ in the absence of the Mpp10p bait. These were false positives.
The identities of the genes encoding the Mpp10p-interacting proteins in the remaining 17 true positives were determined by sequencing PCR-amplified inserts from bait plasmids. Plasmid DNA was extracted from the positive yeast strains by preparing Hirt lysates (2). Insert sequences were amplified with oligonucleotides (provided by X. Sun and D. Stern, Yale University) pACT2.1 (5′-CGCGTTTGGAATCACTACAGG-3′) and pACT2.2 (5′-GAGATGGTGCACGATGCACAGTTG-3′), which recognize the regions that flank the insert in the pACT2 plasmid. The PCR products from some of the positives were sequenced at the W.M. Keck facility, and the obtained sequences were analyzed by using the BLAST search algorithm via the National Center for Biotechnology Information (26a). Sequences representing two different open reading frames (ORFs), YHR148w and YNL075w, were obtained. We have named these IMP3 and IMP4 (for interacting with Mpp10p), respectively. Probes to IMP3 and IMP4 were used for Southern blot analysis of the remaining PCR products to identify and classify all of the 17 positive strains (data not shown).
Other Mpp10p constructs were made for use in a directed two-hybrid screen with the interacting proteins that we identified with Mpp10-5p. Only the plasmid bearing mpp10-6, representing aa 242 to 593, did not activate the reporter genes in the absence of an interacting partner. This construct was made by using PCR and oligonucleotides complementary to the desired 5′ and 3′ ends. The mpp10-6 mutant was cloned with primers ympp10.24 (5′-CCGCAGGCCTCATGAAAGAACCTGTGAAGAAG-3′; StuI site) and ympp10.9a (5′-CCCGCTCGAGTCAGACATTGTATATCTCTTGAGG-3′; stop codon and XhoI site). The amplification product was digested with the indicated enzymes, purified, and ligated into pAS2-1 (sequentially cut with SmaI and SalI). The bait constructs that autoactivated the lacZ reporter gene included Mpp10 proteins representing aa 47 to 498 and 47 to 593.
Disruption of IMP3 and IMP4.
One allele of the IMP3 or IMP4 gene was fully disrupted in YPH260 by homologous recombination with the PCR-based strategy described previously (10). Oligonucleotides used to perform the IMP3 disruption were hr148.1 (5′-ATATAGAACGTTCTAAATAGATAAACCACCAGGCATAATCACAACCAATACTCTTGGCCTCCTCTAG-3′) and hr148.2 (5′-GAAGATGATAAATCGAGTATTGATACAGAAAACGTAAGTTTGTAGTCAATTCGTTCAGAATGACACG-3′). nl075.1 (5′-TTTATCATTGATTCAAGTGTTCCCATAAATAATAAAACAGTTAAATCGAACTCTTGGCCTCCTCTAG-3′) and nl075.2 (5′-TAAGGCCTCATCGGCCTTCTATTTTAACCTTTACAAATCTATAAAAAAATTCGTTCAGAATGACACG-3′) were used to disrupt IMP4.
Expression of Imp3 and Imp4 proteins from a conditional promoter.
IMP3 and IMP4 were put under the control of the GAL1-10 promoter by cloning the genes into the BamHI and NotI sites of pGAD3 (ADE2 HIS3 CEN6 ARSH4) (31). The promoter is active when yeast is grown in galactose medium but repressed when yeast is grown in glucose medium. Fragments of IMP3 and IMP4 were generated by PCR from 25 ng of yeast genomic DNA per reaction. The primers used to amplify IMP3 were hr148.3 (5′-CCGCGGATCCATGGTTAGAAAACTAAAG-3′; BamHI site) and hr148.4 (5′-GCATAGTTTAGCGGCCGCTTATGAAAAATCAAAATCGTC-3′; stop codon and NotI site); the primers used to amplify IMP4 were nl075.3 (5′-CCGGAATTCATGCTAAGAAGACAAGCC-3′; BglII site) and nl075.4 (5′-GCA TAGTTTAGCGGCCGCTGACAAATAGTCTTTTTTATTGGC-3′; stop codon and NotI site). These constructs were transformed into the appropriate diploid strain lacking an allele of either IMP3 or IMP4. These strains were sporulated, and tetrads were dissected. Haploid segregants were selected by streaking onto the appropriate media to identify the disrupted chromosomal IMP gene and the presence of the plasmid carrying a conditionally expressed allele of IMP3 or IMP4. These strains, pGAL1::IMP3 (YPH259 Δimp3::HIS3 pGAL1::IMP3) and pGAL1::IMP4 (YPH258 Δimp4::HIS3 pGAL1::IMP4) were used for experiments requiring depletion of Imp3p or Imp4p.
Depletion of the Imp proteins.
pGAL1::IMP3 and pGAL1::IMP4 were grown in YPG to an optical density at 600 nm (OD600) of 0.7 to 0.8. Cultures were subsequently diluted 1:500 in YPD (for depletion) or YPG. At the indicated time points, the absorbance at 600 nm was recorded.
Northern analysis of pre-rRNA processing.
pGAL1::IMP3 and pGAL1::IMP4 were grown in YPG to an OD600 of 1.0, and total RNA was isolated from 10 OD units of cells by the hot-phenol method (2). This represented the zero time point after the switch to glucose. These cultures were then diluted 1:500 into YPD and grown for 24 h before harvest of total RNA for the 24-h point. RNA was also extracted from YPH259 grown to an OD of 1.0 in YPG. Equal amounts of RNA (4.5 μg) were resolved on a 1% agarose–formaldehyde gel. The blotting and oligonucleotide hybridization protocols were as described previously (10). Oligonucleotides a, b, c, and e are described in reference 6, oligonucleotide z is referred to as oligonucleotide c in reference 22, and oligonucleotide y is 5′-GCCCGTTCCCTTGGCTGTG.
Pulse-chase labelling.
All strains were grown in synthetic medium (SD or SG) supplemented with only adenine (40 μg/ml), histidine (20 μg/ml), leucine (60 μg/ml), lysine (30 μg/ml), and uracil (20 μg/ml). pGAL1::IMP3 and pGAL1::IMP4 were grown in SG to an OD600 of 1.0. Cells were washed and diluted 1:100 in SD. After 48 h of depletion, 20 to 30 OD units of cells was collected for pulse labelling with [methyl-3H]methionine as described previously (10). Thirty OD units of YPH259 (grown directly in SD to an OD600 of 1.0) was similarly handled. Total RNA was prepared by hot-phenol extraction (2). Labelled RNAs (8,500 cpm) were resolved on a 1% agarose–formaldehyde gel and detected as described previously (10).
Epitope tagging of Imp3p and Imp4p.
Imp3p and Imp4p were tagged on their C termini with triple-MYC or triple-HA. PCR fragments of either gene lacking stop codons were inserted directly upstream of epitope sequences in tag cassette vectors derived from p415GPD (26). To create these cassette vectors, a PCR fragment of the triple-MYC or triple-HA epitope was amplified from pMPY-3xMYC and pMPY-3xHA, respectively (obtained from Bruce Futcher) (30). The MYC fragment was generated with C-myc.1 (5′-CCCAAGCTTGTCGACTCTAGAGGTGAACAAAAG-3′; HindIII and SalI sites) and C-myc.2 (5′-CCGCTCGAGTCAAGAAGATCCGTTCAAGTC-3′; stop codon and XhoI site); the hemagglutinin (HA) fragment was produced with oligonucleotides C-HA.1 (5′-CCCAAGCTTGTCGACTACCCATACGATGTTCCT-3′; HindIII and SalI sites) and C-HA.2 (5′-CCGCTCGAGTCAAGCGTAATCTGGAACGTCAT ATGGATAAGATCC-3′; stop codon and XhoI site). Amplified products were digested with HindIII and XhoI, gel purified, and ligated into the HindIII and XhoI sites of p415GPD (Ampr LEU2 CEN/ARS). The inserted tags were verified by automated sequencing with p415GPD.1 (5′-GGTTGAAACCAGTTCCCTG-3′). IMP3 was amplified with hr148.3 and hr148.12 4 (5′-CCCGCTCGAGTGAAAAATCAAAATCGTC-3′; XhoI site); the IMP4 fragment was created by nl075.6 (5′-AGTTCAATGCTAAGAAGACAAGCC-3′; SpeI site) and nl075.12 (5′-CTCGAGCAAATAGTCTTTTTTATTGGC-3′; XhoI site). The fragments were digested with either BamHI or SpeI and XhoI and ligated into the tag cassette vectors cut with BamHI/SpeI and SalI.
Western blotting.
Yeast whole-cell extracts were prepared by glass bead disruption in NET-2 buffer (40 mM Tris-Cl [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40) containing protease inhibitors (aprotinin [2 μg/ml], leupeptin [1 μg/ml], and pepstatin A [1 μg/ml]). After vigorous vortexing (six times for 45 s each), lysates were cleared by centrifugation (10 min at 13,000 rpm in a microcentrifuge). Five microliters of extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. Blots were incubated with one of the primary antibodies anti-Mpp10p (rabbit polyclonal serum, 1:10,000), anti-MYC (mouse monoclonal, ascites, 1:500), and anti-HA (12CA5 mouse monoclonal, culture supernatant, 1:1,000) and the corresponding secondary antibody (Pierce; 1:100,000). The Mpp10p antibody was characterized previously (10). Antibody detection was achieved with a Pierce SuperSignal ULTRA Western Blotting Kit.
Immunoprecipitations.
Three milligrams of protein A–Sepharose CL-4B (Pharmacia) was bound to anti-Mpp10p (50 μl of rabbit serum (10), antifibrillarin (100 μl of culture supernatant 17C12 [Michael Pollard, Scripps]), anti-MYC (100 μl of purified ascites), or anti-HA (100 μl of culture supernatant) by incubation in a total volume of 500 μl with NET-2 (40 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40) at 4°C for 16 h with rotation. In parallel, a mock immunoprecipitation with protein A–Sepharose CL-4B alone (no added antibody) was done. Antibody-bound beads were washed three times in NET-2 and subsequently incubated with whole-cell yeast extract (representative of 5 OD600 units from cells harvested at an OD600 of 0.5) at 4°C for 1 h.
Following incubation, beads were washed five or six times with NET-2. RNA was recovered via phenol-chloroform-isoamyl alcohol (25:24:1) extraction and ethanol precipitation; protein was recovered by boiling the beads at 90°C for 4 min in SDS sample buffer (40 mM Tris [pH 6.8], 7.5% glycerol, 1% SDS, 25 mM dithiothreitol, 1% bromophenol blue). Northern blotting procedures and 3′ end labelling of all immunoprecipitated RNAs were done as described previously (10). Western blotting protocols are detailed above.
RESULTS
Two novel proteins interact with Mpp10p.
A screen for proteins that physically interact with Mpp10p was carried out by using the two-hybrid methodology (11). Because the full-length MPP10 cloned into the pAS2-1 bait plasmid activated transcription of the reporter genes in the MaV103 strain without the presence of an interacting partner, a 95-aa-carboxy-truncated version of the gene (aa 1 to 498; mpp10-5) was used as bait. This plasmid did not autoactivate the reporter genes. Mpp10-5p will complement a null mpp10 allele; however, it confers cold sensitivity and is deficient in cleavage at the A1 and A2 sites, with a phenotype identical to that of a similarly truncated Mpp10p (unpublished results and reference 24). A yeast prey cDNA library, cloned into pACT-2, was transformed into the strain bearing the mpp10-5 bait plasmid. Five hundred thousand transformants were screened for activation of the HIS3 and beta-galactosidase genes. Transformants were first screened for the activation of the HIS3 gene by growth on medium lacking histidine in the presence of 3-aminotriazole. These colonies, in turn, were screened for beta-galactosidase activity (blue color) in the presence of the indicator X-Gal. Of the 20 positive blue colonies, 3 were false positives whose prey plasmids activated the reporter genes in the absence of a bait plasmid. The inserts carried by the prey library plasmids in the remaining 17 strains were enriched by PCR with flanking oligonucleotides, and some were directly sequenced. Computer analysis indicated that two unidentified ORFs were represented: YHR148w and YNL075w. We have named them IMP3 (YHR148w) and IMP4 (YNL075w), for interacting with Mpp10p. Subsequent Southern blot analysis of all of the positive PCR products from the two-hybrid screen indicated that 7 were IMP3 and 10 were IMP4.
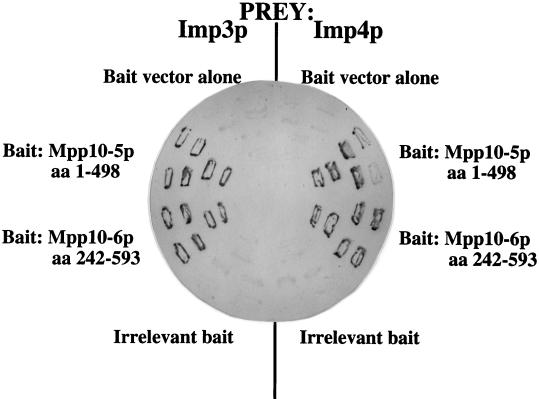
Figure 2 shows the Mpp10p-Imp protein interactions in the two-hybrid screen. The screen was carried out with the Mpp10-5p (aa 1 to 498) fusion protein as bait, and one each (Imp3p and Imp4p) of the original beta-galactosidase-positive blue strains is shown. These strains were cured of the mpp10-5 bait plasmid and then retransformed with the mpp10-6 bait plasmid, which is truncated to aa 242 at the amino terminus (aa 242 to 498). This truncated Mpp10 protein also interacts with both Imp3p and Imp4p in the two-hybrid screen. Taken together, these results demonstrate that aa 242 to 498 of Mpp10p are sufficient for interaction with the Imp proteins. Neither an empty bait vector alone nor an irrelevant bait interacts with the Imp proteins and activates the reporter genes (white strains).
FIG. 2.
Imp3p and Imp4p interact with Mpp10p in a two-hybrid screen. Interactions were assayed by activation of the beta-galactosidase gene, which, if positive, yields a dark blue color. Two different Mpp10p bait constructs, mpp10-5 and mpp10-6, were tested for interaction with the Imp3p and Imp4p prey.
To verify that the products of these ORFs interact with Mpp10p, the full-length sequences for genes encoding Imp3p and Imp4p were cloned into the pAS2-1 bait plasmid and tested for interaction with MPP10 cloned into the pACT-2 prey plasmid. In both cases, the reporter genes were activated (data not shown), demonstrating that Mpp10p interacts with Imp3p or Imp4p when either is bait or prey.
IMP3 and IMP4 are both uncharacterized ORFs in the yeast genome. Imp3p is a protein with a predicted molecular mass of 22 kDa and a calculated isoelectric point of 9.9. It has 20% identity and 33% similarity with the yeast ribosomal S9 proteins (S4 ribosomal protein family) over its full length (Fig. 3) and 26% identity over the amino acid residues that include the predicted S4 RNA binding domain (aa 36 to 164 of Imp3p) (1). It is clearly a member of the S4 family of proteins, as a BLAST search yields significantly low random expectation values when compared with a wide variety of known S4 ribosomal proteins (e.g., 3e-12, 1e-10, and 6e-08 for the Archeoglobus fulgidus, Methanobacterium thermoautotrophicum, and Sulfolobus acidocaldarius S4 ribosomal proteins, respectively). Imp4p is a protein with a predicted molecular mass of 34 kDa and a calculated isoelectric point of 9.2, with no significant homology to any known protein. Other than the S4 RNA binding domain in Imp3p, no conserved motifs in either protein were detected by using Prosite (Swiss-Prot). Importantly, expressed sequence tags (ESTs) representing likely homologs can be identified in mice and humans, suggesting that these proteins and their function are highly conserved.
FIG. 3.
Imp3p has a high similarity with ribosomal protein S9 (Rps9; E. coli S4). Shown is the GAP (Genetics Computer Group) sequence comparison between Imp3p and Rsp9Ap, one of the two ribosomal protein S9s in yeast. The lines indicate identity, and the dots indicate similarity. The predicted S4 RNA binding domain of Rps9 occurs from aa 107 to aa 170 (1).
Imp3p and Imp4p are required for processing of rRNA precursors.
Since both Imp proteins were identified by their interaction with Mpp10p, and because Mpp10p is required for processing of rRNA precursors, we reasoned that the Imp proteins may also be required for pre-rRNA processing. Dissection of tetrads from the heterozygous disruption IMP strains indicated that both genes were essential genes in yeast (data not shown). We cloned each full-length ORF under the control of a galactose-inducible, glucose-repressible promoter (GAL1-10); this plasmid can complement a null allele when strains are grown in galactose (data not shown). Strains designated pGAL1::IMP3 and pGAL1::IMP4 were constructed so that the only Imp3 or Imp4 protein in the cell is that encoded from the glucose-repressible plasmid. To investigate whether either of these proteins is required for pre-rRNA processing, we depleted protein levels by transferring the cells from growth in galactose to growth in glucose. Consistent with previous observations of protein components required for pre-rRNA processing (10, 20), cell growth slows beginning at 12 to 15 h following transfer to glucose (data not shown).
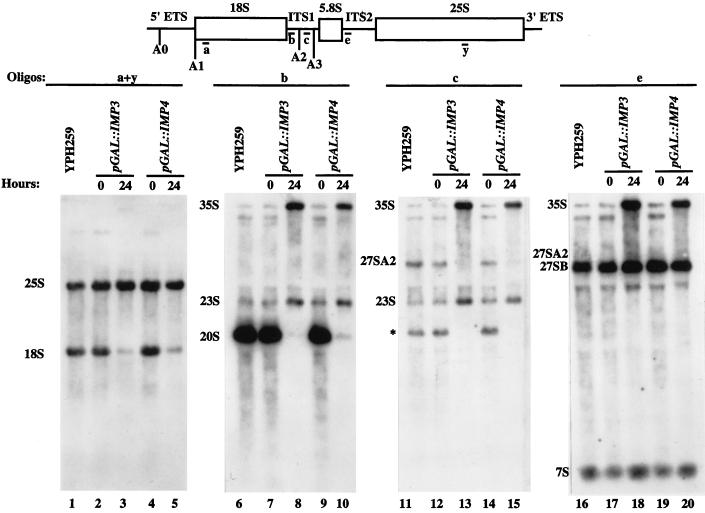
The levels of mature and precursor rRNAs were assessed by Northern blot analysis at 24 h after the shift to glucose. Upon depletion of Imp3p and Imp4p, we observed a large decrease in 18S rRNA levels compared to 25S rRNA levels (Fig. 4, oligonucleotides a and y, lanes 3 and 5), suggesting that both proteins are required for biogenesis of the 18S rRNA. Blotting with oligonucleotides that allowed visualization of the precursor rRNAs indicated that when the Imp proteins were depleted, the levels of the direct precursor to 18S rRNA, the 20S rRNA, were significantly reduced and the levels of the 23S precursor were increased (Fig. 4, oligonucleotide b, lanes 8 and 10). Because the 20S precursor results from cleavage at the A0, A1, and A2 sites and the 23S precursor results from lack of cleavage at the A0, A1, and A2 sites, these observations indicate that the Imp proteins are required for cleavage at these three sites in the pre-rRNA. To verify that cleavage at the A2 site was affected, we hybridized the blot with oligonucleotide c and observed lack of the 27SA2 precursor to the LSU rRNAs in the Imp protein-depleted strains (Fig. 4, lanes 13 and 15). However, the LSU RNAs are produced because cleavage can occur at A3, as shown in the Northern blot probed with oligonucleotide e, where the levels of the 27SB precursor were the same under all conditions (lanes 18 and 20). Similarly, the levels of the 7S precursor to 5.8S rRNA remained the same even in the depleted strains. Hybridization with oligonucleotides b, c, and e also revealed an apparent increase in the nascent 35S precursor when either Imp protein was depleted (Fig. 4, lanes 8, 10, 13, 15, 18, and 20). As seen with other components of the U3 snoRNP, these Northern results argue that Imp3p and Imp4p are required for cleavage of 18S rRNA precursors at the A0, A1, and A2 sites.
FIG. 4.
Imp3p and Imp4p are essential for cleavage at A0, A1, and A2 in the pre-rRNA. Strains bearing Imp3p or Imp4p expressed from a galactose-inducible, glucose-repressible promoter were constructed and designated pGAL1::IMP3 and pGAL1::IMP4. RNA was extracted from these strains either prior to transfer from galactose to glucose (time zero) or 24 h after transfer to glucose. As a control, RNA was also extracted from an otherwise isogenic strain, YPH259. RNA was analyzed by formaldeyde gel electrophoresis followed by Northern blotting with oligonucleotides complementary to the mature 18S and 25S rRNAs (a and y), to ITS1 (b and c), and to ITS2 (e). The asterisk indicates a precursor that we observe in the nondepleted strains and in the control strain but that we have not observed previously (10). Blotting with oligonucleotide z, complementary to sequences between A0 and A1, suggests that it probably extends from site A1 to A3 (data not shown).
To verify that the depletion of the Imp proteins affects pre-rRNA processing, we performed a pulse-chase labelling of the pre-rRNA with [methyl-3H]methionine. The pulse-chase experiments were performed with strains depleted of Imp proteins for 48 h and compared to those with an otherwise isogenic haploid strain, YPH259. The precursor and mature rRNAs were analyzed by formaldehyde gel electrophoresis, transfer of the gel to a Zeta-Probe membrane, and fluorography. Figure 5 indicates that depletion of Imp3p and Imp4p causes defects in pre-rRNA processing. When precursor and mature rRNAs in the nondepleted strain were examined, the 35S, 27S, and 20S precursors visualized at the beginning of the chase were all processed into mature 18S and 25S rRNAs by the end of the chase (Fig. 5; compare lanes 1 and 3). In contrast, for both Imp3p- and Imp4p-depleted strains only the 35S precursor was apparent at the beginning of the chase (Fig. 5, lanes 4 and 7). Midway through the chase, the 27S precursors were detectable (Fig. 5, lanes 5 and 8), but 20S precursors were not. At the end of the chase, while mature 25S rRNA was present, 18S rRNA was not (lanes 6 and 9). A band representing the 23S precursor (resulting from a lack of cleavage at A0, A1, and A2) could also be seen in the depleted strains during the chase (Fig. 5, lanes 5, 6, 8, and 9). These results conclusively implicate the Imp proteins in the cleavage events that process precursor rRNAs to 18S rRNA.
FIG. 5.
Imp3p and Imp4p are required for pre-rRNA processing. pGAL1::IMP3 and pGAL1::IMP4 were grown in glucose for 48 h and then labelled with [methyl-3H]methionine for 2.5 min. A chase was performed with unlabelled methionine, and samples were removed for RNA analysis immediately after addition of the unlabelled methionine (time zero) or 3 or 12 min postaddition. An otherwise isogenic strain, YPH259, was similarly treated. The RNA was analyzed by agarose-formaldehyde gel electrophoresis followed by transfer to a Zeta-Probe membrane which was subsequently treated with En3Hance (NEN).
Furthermore, it is also clear that depletion of the Imp proteins affects the overall kinetics of pre-rRNA processing. At the beginning of the chase (0 min), the nascent 35S precursor is accumulated compared to that in the YPH259 strain (Fig. 5, lanes 4 and 7). Three minutes into the chase, 35S rRNA levels remained unchanged, whereas mature rRNAs are already visualized in the nondepleted strain. Even at the end of the chase (12 min), the 35S nascent transcript is still present (Fig. 5, lanes 6 and 9). This differs from the pulse-chase profile following Mpp10p depletion, where the kinetics of processing the 25S precursors were not so strongly affected (10). Growth curve comparisons reveal that Imp protein depletion affects growth more dramatically than depletion of Mpp10p (data not shown). Whether the results obtained for the kinetics of pre-rRNA processing are due to this difference is not known. However, we have observed that pulse-chase analysis at shorter times after the switch to glucose (18 h) gives identical results.
Epitope-tagged Imp proteins complement the null allele.
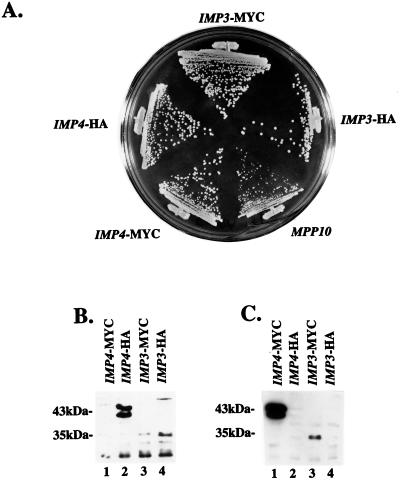
Both Imp proteins were tagged with triple-MYC and triple-HA epitopes at their carboxy termini, cloned into the yeast expression vector p415GPD (26), and shuffled into the appropriate pGAL::IMP strain. In these strains, designated IMP3-MYC, IMP3-HA, IMP4-MYC, and IMP4-HA, the only Imp3 or Imp4 protein is that which is tagged and expressed from the plasmid. These plasmids were tested for their ability to support growth of yeast at 30°C. Figure 6A shows that addition of either epitope tag to either Imp protein did not affect growth under these conditions.
FIG. 6.
(A) Epitope-tagged Imp3 and Imp4 proteins complement their respective null alleles. Either a triple-MYC or a triple-HA tag was placed at the 3′ end of an IMP3 or an IMP4 gene and cloned into the expression plasmid p415GPD. Strains were constructed so that the only expressed Imp3 or Imp4 protein is that with the indicated epitope tag. Growth on YPD of the epitope-tagged Imp protein strains was compared to that of a similarly constructed strain that expresses Mpp10p from p415GPD (24). (B) Western blotting with anti-HA epitope antibodies. Total protein was isolated from strains expressing HA-tagged Imp proteins and analyzed by SDS-PAGE and Western blotting. As a control, protein was isolated from strains expressing MYC-tagged protein. (C) Western blotting with anti-MYC epitope antibodies. Total protein was isolated from strains expressing MYC-tagged Imp proteins and analyzed by SDS-PAGE and Western blotting. As a control, protein was isolated from strains expressing HA-tagged protein.
Expression of the epitope-tagged Imp proteins was analyzed by Western blotting of cell lysates with anti-HA or anti-MYC antibodies. When anti-HA antibodies were reacted with lysates from cells which express HA-tagged Imp4p, two pronounced bands corresponding to the HA-tagged Imp4 protein were visualized at about 43 kDa (Fig. 6B, lane 2). The anti-HA antibodies did not react with these same bands from lysates that contained only the MYC-tagged Imp4p (Fig. 6B, lane 1). When anti-HA antibodies were reacted with lysates from cells which express HA-tagged Imp3p, a band of about 35 kDa, corresponding to the HA-tagged Imp3 protein, was detected (Fig. 6B, lane 4). A band of this intensity was not present in the strains in which Imp3p is tagged with MYC (Fig. 6B, lane 3). Likewise, when anti-MYC epitope antibodies were reacted on Western blots of lysates from Imp4 strains tagged with MYC (IMP4-MYC), two pronounced bands were visualized at about 43 kDa (Fig. 6C, lane 1). These bands were not present in lysates in which Imp4p is tagged with HA (IMP4-HA) (Fig. 6C, lane 2). The anti-MYC antibodies also highlighted a band at 35 kDa in the strain in which Imp3p is tagged with MYC (IMP3-MYC) (Fig. 6C, lane 3) that was not present in the strain in which Imp3 is tagged with HA (IMP3-HA) (Fig. 6C, lane 4). These results indicate that both HA- and MYC-tagged Imp3p and Imp4p are expressed and can be detected by Western blotting. By SDS-PAGE, the tagged Imp4 protein runs as two bands at about 43 kDa (the predicted untagged molecular mass is 34 kDa); we do not know if this results from proteolysis or if there are different forms of Imp4p in this strain. The tagged Imp3 protein runs at 35 kDa on SDS-PAGE (the predicted untagged molecular mass is 22 kDa).
The Imp proteins are complexed with Mpp10p in vivo.
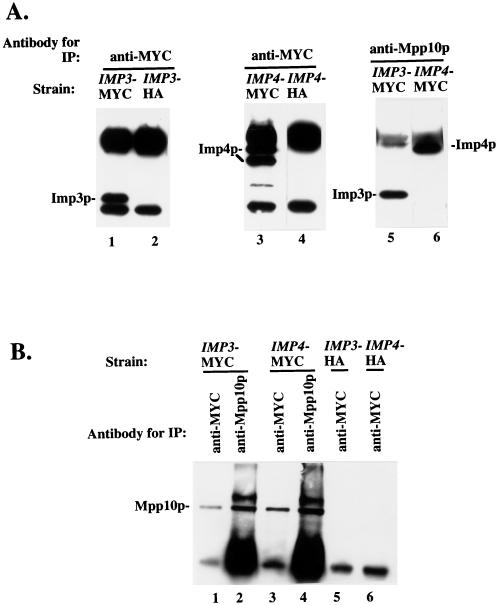
To investigate whether the Mpp10 and Imp proteins were associated in vivo, we performed the following coimmunoprecipitation experiments. Anti-MYC antibodies were used to immunoprecipitate the Imp3 and Imp4 proteins from cell lysates from strains in which the proteins are MYC tagged. Immunoprecipitations were also performed with the anti-MYC antibodies on lysates from strains in which the Imp proteins are HA tagged. The resulting immunoprecipitated proteins were analyzed by Western blotting with anti-MYC antibodies. Both Imp3p and Imp4p were visualized when immunoprecipitations were performed from strains in which they are MYC tagged (Fig. 7A, lanes 1 and 3). As in Fig. 6B and C, Imp4p runs as two bands. The bands visible in the lanes from strains in which the Imp proteins are HA tagged are the immunoglobulin G heavy and light chains (Fig. 7A, lanes 2 and 4). To assess whether Mpp10p and the Imp proteins are found in a stable complex, immunoprecipitations were performed with anti-Mpp10p antibodies and lysates from strains expressing either MYC-tagged Imp3p or Imp4p and analyzed by Western blotting with anti-MYC antibodies. Both Imp3p and Imp4p were detected (Fig. 7A, lanes 5 and 6), indicating that Mpp10p and the Imp proteins are stably associated in yeast cells.
FIG. 7.
The Imp3 and Imp4 proteins are complexed with Mpp10p in vivo. (A) Immunoprecipitation (IP) followed by Western blotting with anti-MYC epitope antibodies. Immunoprecipitations were performed with lysates and the indicated antibody, and the resulting bound proteins were analyzed by SDS-PAGE and Western blotting with anti-MYC epitope antibodies. (B) Immunoprecipitation followed by Western blotting with anti-Mpp10p antibodies. Immunoprecipitations were performed with lysates and the indicated antibody, and the resulting bound proteins were analyzed by SDS-PAGE and Western blotting with anti-Mpp10p antibodies.
Curiously, only the larger of the two Imp4p bands is associated with Mpp10p. If the smaller band results from proteolysis, it is likely to possess a shortened amino terminus, since proteolysis at the carboxy terminus would render this carboxy-tagged protein undetectable. If so, this hints that sequences at the amino terminus of Imp4p may be required for Mpp10p association.
To confirm that Mpp10p and the Imp proteins are associated in vivo, immunoprecipitations were analyzed by Western blotting with anti-Mpp10p antibodies. In strains in which one of the Imp proteins is tagged with MYC, coimmunoprecipitation of Mpp10p was observed (Fig. 7B, lanes 1 and 3). The bands present in the lanes where coimmunoprecipitations were performed on lysates from strains without MYC-tagged Imp proteins represent immunoglobulin G heavy chain (Fig. 7B, lanes 5 and 6). These additional results bolster the argument that Mpp10p is complexed with the Imp proteins in yeast cells.
Both Imp proteins are components of the U3 snoRNP.
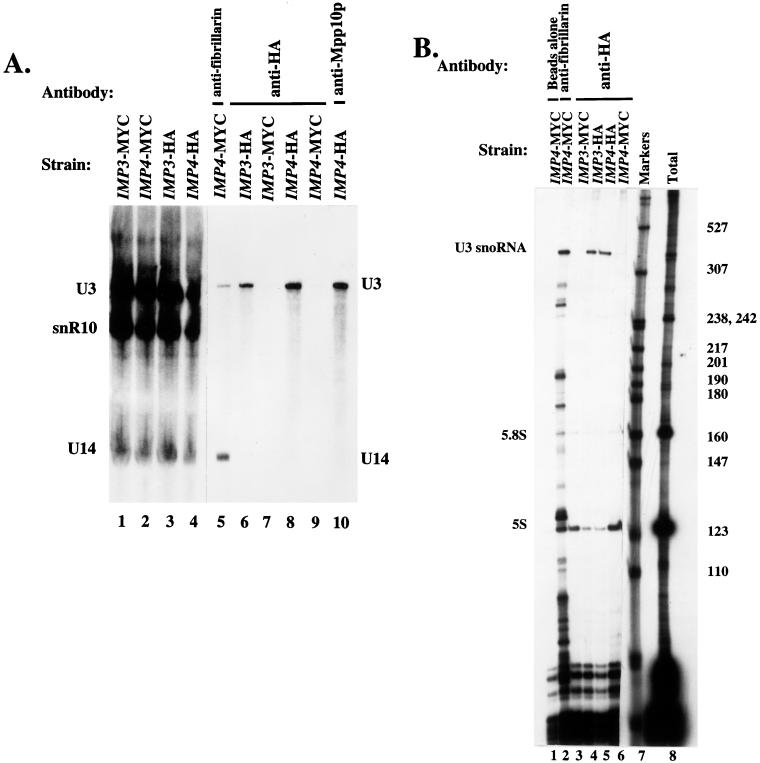
Several experiments were performed to determine whether the Imp3 and Imp4 proteins were components of an RNP particle. Compelling evidence would be coimmunoprecipitation of a snoRNA via the Imp proteins. Since both proteins were identified through their interaction with Mpp10p, a specific component of the U3 snoRNP, we first asked whether they were also components of that snoRNP. Immunoprecipitations were performed with extracts from the IMP3-HA and IMP4-HA strains with anti-HA epitope antibodies. As a control, immunoprecipitations were also performed with the IMP3-MYC and IMP4-MYC strains with the anti-HA antibodies. An antifibrillarin and an anti-Mpp10p immunoprecipitation were included for comparison. RNA was isolated from each pellet, and the presence of the U3 snoRNA, snR10 (a box H/ACA snoRNA), and U14 (a box C/D snoRNA) were assessed by Northern blot analysis. Anti-HA epitope antibodies immunoprecipitated the U3 snoRNA in both the IMP3-HA and IMP4-HA strains (Fig. 8A, lanes 6 and 8), indicating that both Imp proteins are components of the U3 snoRNP. The level of nonspecific background U3 snoRNA binding can be assessed by performing the anti-HA immunoprecipitations with extracts that are not expressing the HA-tagged Imp proteins, that is, by using the MYC-tagged strains. The results of this control immunoprecipitation (Fig. 8A, lanes 7 and 9) strongly support our finding that Imp3p and Imp4p are U3 snoRNP components. As expected, antifibrillarin antibodies immunoprecipitated the U3 and U14 snoRNAs, but not snR10, because both of these snoRNAs are associated with fibrillarin (Fig. 8A, lane 5). Similarly, anti-Mpp10p antibodies immunoprecipitated the U3 snoRNA (Fig. 8A, lane 10). Total RNA for each strain used for the immunoprecipitations was also analyzed for the presence of the U3, snR10, and U14 snoRNAs by Northern blot analysis. All three snoRNAs were present in approximately equivalent amounts in each strain (Fig. 8A, lanes 1 to 4).
FIG. 8.
(A) Imp3p and Imp4p are protein components of the U3 snoRNP. Immunoprecipitations were performed with lysates from the epitope-tagged strains and the indicated antibodies. The RNA from bound RNPs was extracted and analyzed by denaturing gel electrophoresis followed by Northern blotting. Probes were used to detect the U3 and U14 box C/D snoRNAs and snR10, a box H/ACA snoRNA. Lanes 1 to 4 represent total RNA extracted from the same amount of lysate used for immunoprecipitation. (B) Imp3p and Imp4p are specific protein components of the U3 snoRNP. Immunoprecipitations were performed with the indicated antibodies and lysates from the indicated strains. RNAs from the resulting bound RNPs was isolated and analyzed by labelling with [5′-32P]cytidine 3′,5′-bis phosphate and T4 RNA ligase, followed by denaturing gel electrophoresis.
These results demonstrate that the Imp proteins are components of the U3 snoRNP but do not prove that they are associated only with the U3 snoRNA. To do this, we performed an additional set of immunoprecipitations with the IMP3-HA and IMP4-HA strains and the anti-HA antibodies and analyzed the associated small RNAs by labelling them directly. This allowed us to survey for other small RNAs associated with the Imp proteins. When immunoprecipitations were performed with the IMP3-HA and IMP4-HA strains with the anti-HA antibody, the only specific RNA obtained was the U3 snoRNA (Fig. 8B, lanes 4 and 5). The RNAs present in lanes 3 and 6 are the small RNAs bound nonspecifically to the anti-HA antibody and the protein A–Sepharose beads, since they were apparent in immunoprecipitations with extracts from the IMP3-MYC and IMP4-MYC strains with the anti-HA antibody. For comparison, an antifibrillarin immunoprecipitation was included (Fig. 8B, lane 2). This indicated, as expected, that fibrillarin was complexed with the U3 snoRNA and many other box C/D snoRNAs. Taken together, these results forcefully argue that both Imp proteins are specific components of the U3 snoRNP.
Although we are confident that the Imp proteins associate specifically with the U3 snoRNA, we cannot assert that every U3 snoRNA is associated with the Imp proteins. As shown in Fig. 8A, only a small portion of the U3 snoRNA is immunoprecipitable in the Imp-tagged strains with the anti-HA antibodies, even though all immunoprecipitations were performed with an antibody excess. We do not known if this means that only some of the U3 snoRNA is associated with the Imp proteins or if the HA epitope is masked in a fraction of the cellular U3 snoRNP. It is possible that Imp3p and Imp4p may be associated only with the portion of the U3 snoRNA that is actively involved in RNA cleavage if, for example, the Imp proteins play a regulatory role in pre-rRNA processing.
The U3 snoRNA is stable after depletion of the Imp proteins.
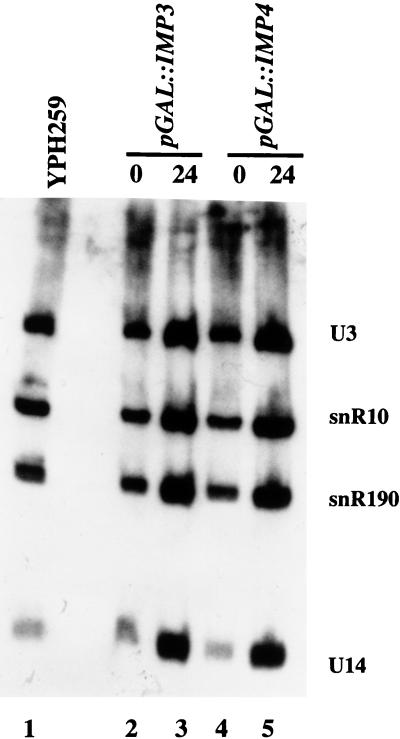
The U3 snoRNA itself is required for cleavage at the A0, A1, and A2 sites in the pre-rRNA. Since both Imp proteins are components of the U3 snoRNP and are also required for cleavage at these three sites, one possible explanation is that the amount of U3 snoRNA is reduced following their depletion. The resulting effects on pre-rRNA cleavage would then be attributable to a lack of the U3 snoRNA. To investigate this possibility, total RNA was isolated from strains depleted of Imp3p and Imp4p for 24 h and analyzed by Northern blotting for the U3, snR10, snR190, and U14 snoRNAs. Levels of these RNAs were compared to those in the nondepleted strains and in an otherwise isogenic strain, YPH259. As shown in Fig. 9 (lanes 3 and 5), the levels of each of the snoRNAs, when compared to each other, remain approximately the same following depletion of either Imp protein. This demonstrates that depletion of the Imp proteins does not affect U3 snoRNA levels.
FIG. 9.
Depletion of the Imp proteins does not affect U3 snoRNA levels. RNA was isolated from pGAL1::IMP3 and pGAL1::IMP4 at the switch from galactose to glucose (time zero) and 24 h after the switch to glucose. As a control, RNA was also isolated from an otherwise isogenic strain, YPH259. Equal amounts of RNA were run on a denaturing polyacrylamide gel, blotted, and hybridized with probes complementary to the U3, snR10, snR190, and U14 snoRNAs.
DISCUSSION
In one approach to identify factors involved in pre-rRNA processing, we used the yeast U3 snoRNP-specific protein, Mpp10p, as bait in a two-hybrid screen to search for interacting proteins. Two proteins representing previously uncharacterized ORFs were recovered multiple times and designated Imp3p and Imp4p, for interacting with Mpp10p. Imp3p (22 kDa) bears some similarity to the yeast ribosomal S9 proteins; in contrast, Imp4p (34 kDa) has no similarity to any characterized protein. The genes encoding both proteins are essential genes in yeast. Both proteins are required for cleavage reactions in the pre-rRNA at sites leading to production of the mature 18S rRNA (A0, A1, and A2). As expected based on the methodology used to identify them, Imp3p and Imp4p are complexed with Mpp10p in vivo. Coimmunoprecipitation analysis suggests that they are both protein components specific to the U3 snoRNP. However, the U3 snoRNA remains stable upon depletion of the Imp proteins, suggesting that they are not required to maintain RNA integrity.
A number of snoRNAs and protein components are required for the A0, A1, and A2 cleavage reactions in the 5′ETS and in ITS1 that lead to the production of the 18S rRNA. Among these are the U3 snoRNA and the proteins tightly associated with it, i.e., Sof1p, Mpp10p, and Lcp5p (10, 20, 38). Other U3 RNP protein components that are required for pre-18S rRNA processing are common to the box C/D snoRNAs and include Nop1p (fibrillarin), Nop56p, and Nop5p (12, 23, 34, 39). The role of the Imp proteins in pre-rRNA processing, then, is similar to that described for the other components of the U3 snoRNP.
The similarity of Imp3p to proteins of known function gives us additional clues regarding its specific function in pre-rRNA processing. Imp3p has significant similarity to the eukaryotic ribosomal S9 proteins (prokaryotic S4) over the portion of the sequence that includes the putative S4 RNA binding domain (aa 107 to 170 in Rps9) (1). The S4 ribosomal proteins of prokaryotes have been extensively studied, and the crystal and solution structures of Bacillus stearothermophilus S4 have recently been solved (9, 25). The S4 ribosomal protein is one of the first to bind to the rRNA as it is being assembled into mature ribosomes and therefore is a primary rRNA binding protein (15). It directly recognizes the base of a multiheaded helix of the SSU rRNA to facilitate correct RNA folding and assembly with the other SSU proteins (27, 33). It also directly associates with a pseudoknot structure in the regulatory sequences of the mRNA of its own operon, thereby modulating expression of itself and other ribosomal proteins (32). The S4 ribosomal protein, therefore, is a protein that binds directly to two distinct RNAs of diverse sequence and structure with remarkable versatility. Because S4 binds directly to two different RNA sequences and structures, and because Imp3p is a member of the S4 family of proteins, we speculate that Imp3p may bind directly to the U3 snoRNA, perhaps facilitating folding of the 5′-most sequences for interaction with the pre-rRNA. Alternatively, Imp3p may bind to both the U3 snoRNA and the pre-rRNA sequences simultaneously to stabilize that interaction for subsequent cleavage of the pre-rRNA. The latter possibility is particularly intriguing, as it has been proposed that the U3 snoRNP may guide correct folding of a pseudoknot in the mature SSU RNA (17).
Imp3p is likely to be another member of a growing class of trans-acting nucleolar factors exapted from ribosomal proteins but required for pre-rRNA processing. A second factor, Rrp5p, which is required for processing of precursors to the mature 18S and 5.8S rRNAs, bears 12 repeats of the S1 ribosomal protein RNA binding motif (8, 34a). Since ribosomal proteins were among the first proteins, it is striking that as these diverse pre-rRNA processing factors evolved they borrowed sequences directly implicated in RNA binding.
Vertebrate ESTs can be identified for both Imp3p (accession no. AA184637) and Imp4p (accession no. AA404192) by BLAST searches, suggesting that they are highly conserved and may also be involved in ribosome biogenesis in more complex organisms. We used the ESTs that we identified to clone the full-length cDNAs for mouse Imp3p and Imp4p (data not shown). Both mouse Imp3p and Imp4p are 54% identical (the Imp3ps are 67% similar; the Imp4ps are 64% similar) to their respective yeast counterparts. Further experimentation is necessary to determine if they are indeed functional homologs of the yeast Imp3 and Imp4 proteins.
BLAST searches indicate that putative homologs for the Imp proteins also exist in Schizosaccharomyces pombe and Caenorhabditis elegans. The likely S. pombe and C. elegans homologs for Imp3p have low random expectation values of 2e-59 and 2e-34, respectively (accession no. Z99531 and U97189). Likewise, the S. pombe and C. elegans homologs for Imp4p have low random expectation values of 2e-85 and 4e-67, respectively (accession no. Z98974 and Z83246). These values are well within range to indicate homologs, as comparison of the well-characterized human and yeast Mpp10 proteins yields a random expectation value of 8e-35 in a BLAST search.
In summary, our search for Mpp10p-interacting proteins has led us to the identification and characterization of two novel components of the U3 snoRNP, Imp3p and Imp4p. Many questions remain about the nature of their interaction with Mpp10p and the U3 snoRNA. For example, it is not known whether either of the Imp proteins associates with Mpp10p or the U3 snoRNA directly. Because Imp3p is similar to the family of S4 ribosomal proteins, which bind to two different RNA sequences and structures in E. coli, it is a likely candidate for direct U3 snoRNA binding. In addition, questions remain about the assembly of Mpp10p, Imp3p, and Imp4p on the U3 snoRNA. Assembly is unlikely to involve interaction between Imp3p and Imp4p, since they do not react by two-hybrid analysis (data not shown), but assembly in the RNP may still be interdependent. These studies, as well as the generation of conditional IMP3 and IMP4 alleles, will allow us to more fully explore the essential role of the U3 snoRNP in ribosome biogenesis.
ACKNOWLEDGMENTS
We thank Stephen Elledge, Bruce Futcher, Michael Pollard, Jeff Settleman, David Stern, and Marc Vidal for plasmids and reagents essential for carrying out this work. We thank Sandra Wolin and her laboratory for use of their tetrad dissection microscope and Barbara Pannone in the Wolin lab for purified anti-MYC antibody. We appreciate the help of Baserga lab members, Steven Wormsley, Karen Wehner, and David Dunbar for comments on the manuscript and Tina Agentis for expert technical assistance. We thank David Tollervey for his thorough review. This work was supported by NIH GM52581 to S.J.B.
REFERENCES
- 1.Aravind L, Koonin E V. Novel predicted RNA-binding domains associated with the translation machinery. J Mol Evol. 1999;48:291–302. doi: 10.1007/pl00006472. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley and Sons, Inc.; 1995. [Google Scholar]
- 3.Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 4.Beltrame M, Henry Y, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. . [Corrected and republished version of Nucleic Acids Res. 22:4057–4065, 1994.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berges T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 9.Davies C, Gerstner R B, Draper D E, Ramakrishnan V, White S W. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998;17:4545–4558. doi: 10.1093/emboj/17.16.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar D A, Wormsley S, Agentis T M, Baserga S J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 12.Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 14.Hartshorne T, Agabian N. RNA B is the major nucleolar trimethylguanosine-capped small nuclear RNA associated with fibrillarin and pre-rRNAs in Trypanosoma brucei. Mol Cell Biol. 1993;13:144–154. doi: 10.1128/mcb.13.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Held W A, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 16.Henriquez R G, Blobel G, Aris J P. Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J Biol Chem. 1990;265:2209–2215. [PubMed] [Google Scholar]
- 17.Hughes J M. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 18.Hughes J M X, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and prevents formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes J M X, Konings D A M, Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of pre-ribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 22.Lafontaine D, Vandenhaute J, Tollervey D. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 1995;9:2470–2481. doi: 10.1101/gad.9.20.2470. [DOI] [PubMed] [Google Scholar]
- 23.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Lafontaine, D. L. J., and D. Tollervey. Personal communication.
- 24.Lee S J, Baserga S J. Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc Natl Acad Sci USA. 1997;94:13536–13541. doi: 10.1073/pnas.94.25.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markus M A, Gerstner R B, Draper D E, Torchia D A. The solution structure of ribosomal protein S4 delta41 reveals two subdomains and a positively charged surface that may interact with RNA. EMBO J. 1998;17:4559–4571. doi: 10.1093/emboj/17.16.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 26a.National Center for Biotechnology Information. 13 April 1999, revision date. BLAST search algorithm. [Online.] http://www.ncbi.nlm.nih.gov/. [10 June 1999, last date accessed.]
- 27.Powers T, Noller H F. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 28.Savino R, Gerbi S A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 31.Sobel S G, Synder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spedding G, Draper D E. Allosteric mechanism for translation repression in the Escherichia coli α operon. Proc Natl Acad Sci USA. 1993;90:4399–4403. doi: 10.1073/pnas.90.10.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern S, Wilson R C, Noller H F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 34.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Venema, J. Personal communication.
- 35.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 36.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J D. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinberg R A, Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968;38:289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- 38.Wiederkehr T, Pretot R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]