Abstract
Organoid cultures represent a unique tool to investigate the developmental complexity of tissues like the human retina. NRL is a transcription factor required for the specification and homeostasis of mammalian rod photoreceptors. In Nrl‐deficient mice, photoreceptor precursor cells do not differentiate into rods, and instead follow a default photoreceptor specification pathway to generate S‐cone‐like cells. To investigate whether this genetic switch mechanism is conserved in humans, we used CRISPR/Cas9 gene editing to engineer an NRL‐deficient embryonic stem cell (ESC) line (NRL −/−), and differentiated it into retinal organoids. Retinal organoids self‐organize and resemble embryonic optic vesicles (OVs) that recapitulate the natural histogenesis of rods and cone photoreceptors. NRL −/− OVs develop comparably to controls, and exhibit a laminated, organized retinal structure with markers of photoreceptor synaptogenesis. Using immunohistochemistry and quantitative polymerase chain reaction (qPCR), we observed that NRL −/− OVs do not express NRL, or other rod photoreceptor markers directly or indirectly regulated by NRL. On the contrary, they show an abnormal number of photoreceptors positive for S‐OPSIN, which define a primordial subtype of cone, and overexpress other cone genes indicating a conserved molecular switch in mammals. This study represents the first evidence in a human in vitro ESC‐derived organoid system that NRL is required to define rod identity, and that in its absence S‐cone‐like cells develop as the default photoreceptor cell type. It shows how gene edited retinal organoids provide a useful system to investigate human photoreceptor specification, relevant for efforts to generate cells for transplantation in retinal degenerative diseases.
Keywords: cone photoreceptor, NRL, optic vesicles, retinal organoids, stem cells
Precursor cells differentiate as rod photoreceptor cells upon expression of the NRL gene; in its absence, the default photoreceptor cell type is a blue opsin‐expressing cone. Parental embryonic stem cells differentiated into retinal organoids generate mostly rods and a small proportion of blue and red/green cones, whereas the engineered NRL‐deficient cells generate rod‐less organoids that contain red/green cones and increased numbers of blue cones.
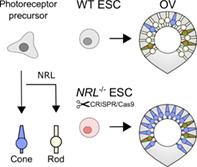
Significance statement.
Photoreceptor cells located in the retina are essential for vision and their degeneration leads to a large proportion of global blindness. Rod photoreceptors needed for night vision are prevalent, whereas cone photoreceptors needed for high acuity daylight vision are rare. This study has engineered a human pluripotent embryonic stem cell line that lacks NRL gene function. When differentiated in vitro into 3D retinal organoids, the engineered organoids contain cone cells, but no rods. This study showed that NRL is required to differentiate rod photoreceptors and represents a powerful tool to generate enriched populations of cone photoreceptor cells in the laboratory.
1. INTRODUCTION
During retinal histogenesis, the different cell types of the neural retina, including the photoreceptor cells, are each born within defined developmental time periods from multipotential retinal progenitor cells.1 Rod photoreceptors are generated later and in greater numbers, than the cone photoreceptors needed for daylight vision. The NRL gene encodes the neural retina leucine zipper protein,2 a conserved bZIP transcription factor that in mouse is initially expressed in nascent rod photoreceptor precursor cells after the terminal progenitor division, and persists in rod photoreceptors thereafter.3 Genetic studies have shown that Nrl is required for the acquisition of rod identity, differentiation and homeostasis, acting via upregulation of downstream rod target genes, such as the Rhodopsin gene, Rho, or Nr2e3, which plays a complementary role in the repression of cone specification genes.4, 5, 6, 7, 8 Nrl acts synergistically with the cone rod homeobox transcription factor, Crx to regulate Rho transcription7 whereas ectopic expression of Nrl under the control of the Crx promoter leads to a rod‐only retina.9 From an evolutionary perspective, the emergence of the Nrl gene is thought to have facilitated the evolution of mammalian rod photoreceptors from short wavelength‐sensitive, S‐cone photoreceptors (also known as blue cones), which are considered to represent an ancient photoreceptor fate.10 Conversely, in the retina of Nrl −/− mice, rods are absent and photoreceptors appear cone‐like; Nrl −/− animals exhibit super‐normal cone function mediated by S‐cones, with atypically elevated patterns of blue light detection, concomitant with deficient scotopic vision caused by rod absence.7, 11, 12, 13, 14 Expression of several key molecular markers are altered in Nrl −/− retinae, which supports the hypothesized model that progenitors generate S‐cones by a default pathway, and that Nrl acts like a master regulator required to induce the rod differentiation pathway and suppress the cone fate.6, 10
In humans, heterozygous missense mutations in the NRL gene are associated with dominant retinitis pigmentosa phenotypes15, 16, 17, 18, 19; gain‐of‐function mutations (at codons 49, 50, 51, 56 in the NRL transactivation domain) lead to reduced NRL phosphorylation and enhanced activation of the rhodopsin promoter.20 Such patients present with scotopic vision deficits at a young ages, progressing to loss of photopic response with age; clinical signs that point towards a pattern of early loss of rod photoreceptors, followed by subsequent cone cell death later in life.19 By contrast, homozygous and compound heterozygous loss‐of‐function NRL mutations cause recessive enhanced S‐cone syndrome (ESCS). ESCS is characterized by an abnormally increased perception of blue light stimuli, coincident with night blindness and abnormal pigmentation patterns.21, 22 The recessive NRL mutations reported in ESCS patients are often nonsense or frameshift mutations.23, 24, 25, 26 Unaffected relatives carrying such mutations indicate that heterozygous NRL loss‐of‐function is not pathogenic.24, 25, 26 ESCS has also been frequently associated with NR2E3 mutation. Histological analysis performed on a postmortem retina from a 77‐year‐old individual with ESCS, caused by recessive NR2E3 mutation (previously diagnosed as retinitis pigmentosa) showed degeneration, lack of rods, and increased numbers of S‐opsin positive cells and reduced L/M‐opsin positive cells.27 Similar analysis of NRL‐deficient human retina has not been performed.
Because of its apparent importance in rod generation, models to study NRL function in human development are needed. Retinal organoids generated from human pluripotent stem cells represent an invaluable tool to model human retinal development and pathologies, as they are able to recapitulate typical features of retinogenesis such as an organized multilayered tissue structure, and long human developmental differentiation times.28, 29, 30, 31, 32 In the present study, we generated a homozygous human embryonic stem cell (ESC) line deficient for NRL, taking advantage of CRISPR/Cas9 gene editing to design and introduce a biallelic nonsense mutation into the NRL gene. To address whether NRL has a conserved role in the establishment of the human rod photoreceptor fate, and if the S‐cone pathway represents a default photoreceptor differentiation pathway as it is in mouse, NRL −/− ESCs were differentiated to form retinal organoids. We analyzed cell identity at different time points, using immunohistochemistry and gene expression via quantitative PCR. We find that NRL −/− optic vesicles (OVs) exhibit a drastic reduction in expression at the RNA and protein level, not only of NRL, but also of known NRL‐target genes and characteristic rod markers, particularly at late differentiation time points. Additionally, cone markers are highly upregulated, particularly S‐OPSIN, defining an increased number of S‐cone‐like cells. Our results suggest that the rod photoreceptor identity‐defining function of NRL is conserved in this human in vitro model system, and that in the absence of NRL, human photoreceptor precursors are directed toward a default S‐cone‐like cell fate.
2. RESULTS
2.1. Generation of NRL −/− ESCs and retinal organoid in vitro differentiation
To generate a pluripotent ESC line deficient in NRL, we took advantage of CRISPR/Cas9 technology to target exon number 2, the first coding exon of the NRL gene. We designed a small 127 bp single‐stranded donor oligonucleotide (ssODN)33 to introduce a stop codon at a.a. position 74, as well as an EcoRI restriction site to facilitate screening (Figure 1A). This mutation, c.221_222insAATTC p.(Trp74*) is predicted to yield a truncated version of the NRL N‐terminus, containing only 73 a.a. of the minimal 96 a.a. transactivation domain, while the functional basic Leucine‐zipper DNA binding domain is eliminated. Human ESCs (MShef10; University of Sheffield34) were electroporated with a plasmid containing spCas9, a puromycin resistance gene and a single‐guide RNA (sgRNA) scaffold sequence35, together with the ssODN; cells were selected and recovered colonies screened using PCR amplification of the targeted region followed by EcoRI digestion, to identify the de novo restriction site (Figure 1B). We identified one clone homozygous for the vector insertion, and confirmed its genotype by subcloning and Sanger sequencing analysis (Figure 1C). Off target effects were not detected at three predicted off‐target sites for the sgRNA and the edited line retained a normal karyotype (Figure S1). Targeted cells were expanded, and displayed a normal growth rate and morphology and maintained pluripotency marker expression as determined by immunocytochemistry analysis (Figure 1D).
FIGURE 1.
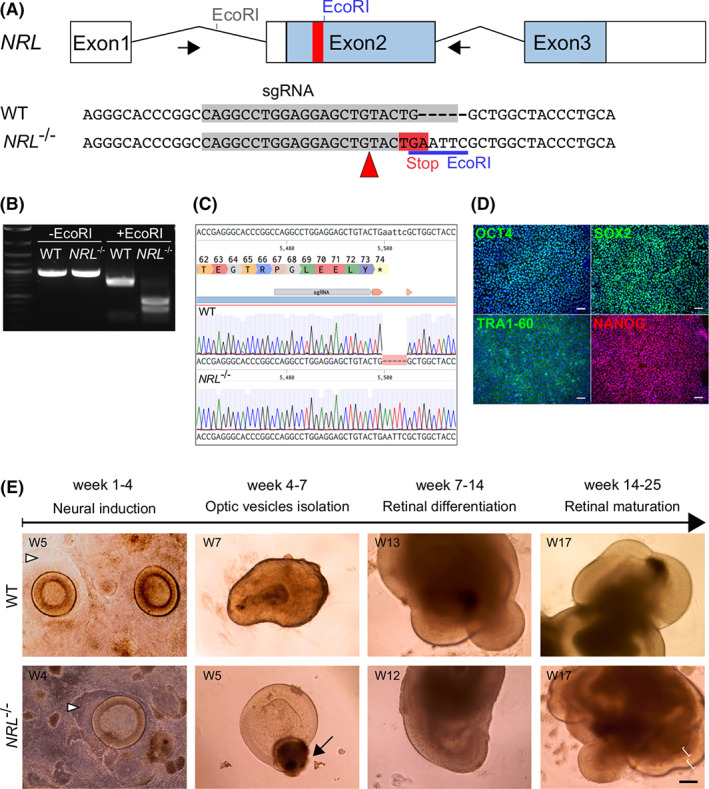
A, The human NRL gene comprises three exons, with the translation initiation codon located in the second exon (blue bars). Schematic shows the gene editing strategy using a 127 nucleotide single strand oligonucleotide donor to introduce a nonsense mutation in exon 2 at amino acid position 74 and a new EcoRI restriction site (in blue), 221 bp downstream of the translational start site; a specific guide RNA (gray bar) directed a simultaneous, Cas9‐double strand break. B, Targeted ESCs clones were analyzed by gDNA extraction and PCR‐amplification of the targeted area (indicated by arrows in A), followed by EcoRI digestion to identify donor integration. C, Sanger sequencing confirmed the correct NRL gene sequence editing. D, CRISPR‐edited NRL −/− ESCs maintained a healthy morphology and growth rate, and expressed pluripotency markers OCT4, SOX2, TRA‐1‐60 and NANOG, similar to the parental embryonic stem cell line (not shown). DAPI, blue. Scale bar = 100 μm. E, Parental isogenic (wild type [WT]) and NRL −/− ESCs were directed toward retinal differentiation in vitro. Week 1 to 4: Optic vesicles (OVs) displayed a round 3D neuroepithelial structure over a patch of immature retinal pigmented epithelium (arrowheads). Week 4 to 7 OVs were excised and transferred to 96 well plates, to allow maturation as floating organoids. Week 7 to 14: OVs grew in size over time. Week 14 to 25: Lamination was evident from week 15 onward. Scale bar = 200 μm. ESC, embryonic stem cell. DAPI, 4',6‐diamidino‐2‐phenylindole; gDNA, genomic DNA; PCR, polymerase chain reaction
The NRL −/− ESCs and the parental ESC line (referred to as wild type [WT]) were differentiated toward three‐dimensional retinal organoids using a previously published protocol.28 Briefly, ESCs were expanded until confluent, then deprived of pluripotency‐supporting factors fibroblast growth factor (FGF) and transforming growth factor beta (TGFB), followed by a period of neural induction to promote forebrain identity. After 3 weeks, round structures of organized neuroepithelium started to be visible, and were termed OVs. OVs were harvested between week 4 and week 7 and cultured individually in nonadherent 96‐well plates, in retinal differentiation medium (RDM) supplemented with taurine and retinoic acid (see Supporting Information Methods). Using this protocol, we previously showed NRL localized in rod precursors by OV culture week 10, which closely mimics human fetal retinal development.28, 38, 39
No obvious differences were detected between the WT and the NRL −/− lines in terms of ability to generate OVs, vesicle number, or size of vesicles (Figure S2). OVs displayed neuroepithelial morphology as early as differentiation week 3, often growing over a patch of presumptive retinal pigmented epithelium (RPE) (Figure 1E, arrowheads). After isolation, the RPE tissue typically formed a small bundle of pigmented cells at the proximal end of the OVs (arrow). The vesicles continued to grow over several weeks, and in some cases developed additional lobes of neuroepithelium; sometimes more than one organized layer was visible under the light microscope (Figure 1E, brackets).
2.2. Characterization of early and mid‐stage NRL −/− OVs
We harvested OVs at week 7 and 14 of differentiation, and performed immunohistological analysis on OV sections to compare the course of differentiation. Both WT and NRL −/− OVs displayed regions of laminated neuroepithelium (WT: n = 96 individual OVs obtained from one independent differentiation passage; NRL −/−: n > 400 OVs, from five independent differentiation passages), with an apical outer nuclear layer (ONL)‐like layer. At week 7, the outer layer of cells showed colocalization of the retinal progenitor marker VSX2 and the proliferation maker Ki67 (Figure 2A,A′). The early marker for committed photoreceptor cells CRX was also detectable at this stage, colabeling cells positive for Recoverin (RCVRN), a protein involved in the visual phototransduction cascade (Figure 2B,B′). Some RXRγ‐positive cells were identified, indicating the presence of cone precursors at a comparable stage to that of human fetal development38 (Figure 2C,C′). We used ZO‐1 staining to label adherens junctions that revealed apical polarization of the OVs at the presumptive photoreceptor layer (Figure 2D,D′). As expected, no NRL protein was detected at week 7 of differentiation (Figure S2), consistent with OV formation occurring similarly in both WT and NRL−/−.
FIGURE 2.
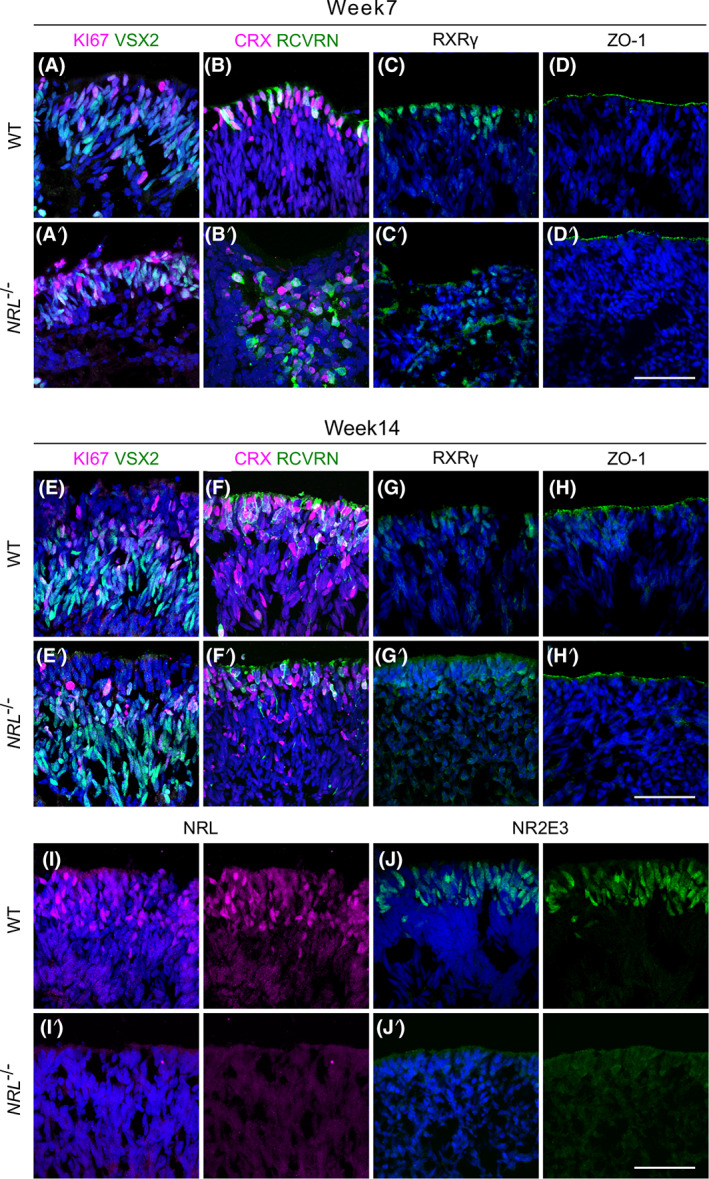
Immunostaining of OV sections using retinal markers showed recapitulation of human retinal development in both WT and NRL −/− OVs. Markers used at week 7 include the retinal progenitor cell marker VSX2, proliferation marker Ki67 (A, A′), early photoreceptor markers CRX and Recoverin (RCVRN) (B, B′), and cone photoreceptor precursor cell marker RXRγ (C, C′). ZO‐1 staining revealed the apical surface and polarization of the OV neuroepithelium (D, D′). At week 14 of differentiation, OVs showed more complex organization with Ki67+ cells, and the localization of VSX2+ cells in a deeper layer, consistent with a transition from retinal progenitors to bipolar cells (E, E′). Pan‐photoreceptor markers CRX and RCVRN stained cells on the outermost layer of the organoids, the presumptive outer nuclear layer (F, F′). RXRγ was used to stain cone‐precursor cells (G, G′) and ZO‐1 to reveal the apical neuroepithelium (H, H′). At week 14 of differentiation, specific markers for rod photoreceptor were used. NRL (I, I′) and NR2E3 (J, J′) are located in the outermost layer corresponding to the photoreceptors in WT control samples, and undetectable in the NRL −/− OVs (N = 2; n = 4). Scale bar = 50 μm. DAPI, blue. DAPI, 4',6‐diamidino‐2‐phenylindole; OV, optic vesicles; WT, wild type
VSX2 is also a marker for bipolar cells that connect with rod photoreceptors and arise later during retinogenesis. Notably, at 14 weeks of differentiation, the Ki67 and VSX2 labeling became more prominent in the layer of cells located immediately underneath the presumptive ONL (Figure 2E,E′). This pattern of expression was consistent with the development of an inner nuclear layer containing bipolar cells. A more organized ONL was apparent by week 14, with CRX and RCVRN staining present in the defined outer layer (Figure 2F,F′). Similarly, RXRγ cone precursor cells continued to organize on the outermost layer of cells, apically labeled with ZO‐1 (Figure 2G,G′,H,H′). Taken together, these immunohistological data support the ability of our gene edited and parental cell lines to form OVs, that display lamination and retinal markers consistent with the human retinogenesis time frame.36, 37, 38
At week 14, NRL was evident in rod precursor cells in the presumptive ONL of the control vesicles, as well as its direct target NR2E3, also expressed in rod photoreceptors and their immature precursors (Figure 2I,J). Strikingly, the NRL −/− retinal organoids showed a downregulation of NRL and NR2E3 immunoreactivity, with a complete absence in the ONL or elsewhere of NRL‐positive cells. Similarly, NR2E3, whose expression is directly regulated by NRL, was not detected in the NRL −/− OVs (Figures 2I′,J′). This indicates the efficiency of the gene editing knockout approach to eliminate functional NRL from retinal three‐dimensional organoids.
2.3. Late stage NRL −/− OVs lack rod photoreceptors and generate S‐like cones instead
To characterize the long‐term differentiation of retinal organoids, some of the markers used in early samples were tested in OV cultures at week 25 of differentiation. Very few, Ki67‐positive dividing cells were observed, and VSX2, which labels both retinal progenitor cells and bipolar cells, was at this stage restricted to cells under the ONL, which are presumed to be the latter cell type (Figure 3A,A′). Costaining with ON bipolar cell marker PKCα, confirmed the presence of VSX2+ PKCα+ double positive cells in both WT and NRL −/− OVs (Figure 3B,B′). Virtually all cells within the presumptive ONL expressed the pan‐photoreceptor markers CRX and Recoverin (Figure 3C,C′). OVs showed a distinct organization, with a defined ONL‐like structure of more tightly packed cells at the outermost layer, typically 4 to 6 cells thick. The tight junction marker ZO‐1 remained labeling the apical side of the OVs, indicating the exterior of the laminated structure (Figure 3D,D′).
FIGURE 3.
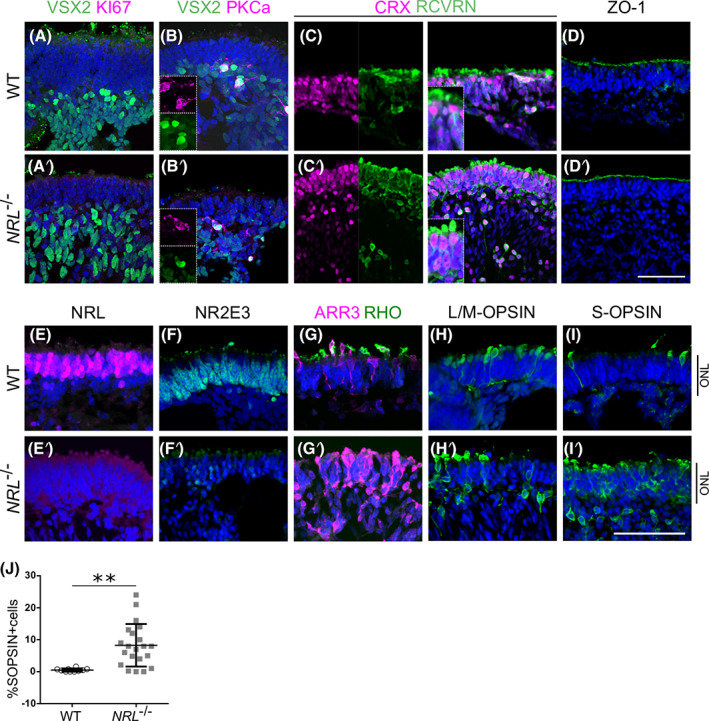
Immunostaining of OV sections at retinal differentiation week 25. OVs present defined lamination and organization of cellular layers, and a differential marker expression between WT and NRL −/− OVs. A,A′, VSX2 was restricted to the cells underlying the ONL, suggesting a presence of bipolar cells that connect with photoreceptors, whereas the proliferation marker Ki67 was undetectable, for both the control and NRL −/− samples. B, B’, VSX2+ population contains a subset of double positive cells for the rod bipolar cell marker PKCa. C, C′, CRX and Recoverin stained the photoreceptor cells populating the ONL of both types of OVs. D, D′, ZO‐1 labeled the apical surface indicating polarization of the OV neuroepithelium. E, NRL localized in the nuclei of the presumptive ONL only in WT OVs. E′, Lack of NRL detected in the NRL −/− line. F, The NRL downstream target NR2E3 localized across the WT OV external cell layer (presumptive ONL) but is absent in the NRL −/− OV (F′). G, Cone photoreceptor marker Cone Arrestin (ARR3) is present in both WT and NRL −/− OVs, which show typical morphology of developing cones cells; but rod‐specific marker Rhodopsin (RHO) was present only in developing photoreceptor cell outer segments in WT OVs and could not be detected in NRL −/− OVs (G′). H, H′, L/M‐OPSIN labels the red and green cone subtypes, and distribution of positive cells is comparable in WT and NRL −/− OVs. I, WT OVs develop a few S‐cones, labeled with S‐OPSIN, in line with human retinal development. I′, Strikingly, NRL −/− OVs present a high proportion of S‐opsin‐positive cells, suggesting a shift in the genetic specification of newly generated photoreceptor cells. Scale bar = 50 μm. DAPI, blue. NRL −/− differentiations, N = 3, n = 4 to 10 samples; WT, N = 2, n = 4. J, Quantification of S‐OPSIN‐ positive cells (mean ± SD, 8.24% ± 1.45 in NRL −/−, n = 21 fields from nine samples, three experiments, compared with 0.49% ± 0.15 in WT OVs, n = 10 from three samples, one experiment). ONL, outer nuclear layer; OV, optic vesicles; WT, wild type. DAPI, 4',6‐diamidino‐2‐phenylindole
We analyzed the proportion of VSX2‐positive and Ki67 VSX2 double positive cells within organoids at weeks 14 and 25 (Figure S3A,B). There was no significant difference between the percentage of VSX2‐positive cells between WT and NRL‐deficient OVs at either time point (Figure S3A). The proportion of proliferating Ki67 VSX2‐positive progenitors at 14 weeks (18.50% ± 14.66; 16.44% ± 14.68, mean% ± SE, for WT and NRL−/−, respectively) decreased to negligible levels in both lines by week 25 (Figure S3B), suggesting the OVs comprised postmitotic cells by this stage. Variance between samples likely reflected lamination heterogeneity within, and between, organoids.
Because NRL is proposed as a crucial player in the establishment and homeostasis of rod photoreceptor identity7 we explored how lack of functional protein affected rod and cone photoreceptor differentiation (Figure 3E‐J). As expected, control OVs expressed retinal markers characteristic of rod and cone photoreceptor cell types; cells throughout the ONL showed positive nuclear staining for the rod factors NRL and NR2E3 (Figure 3E,F), indicating the presence of large numbers of rods. Rod‐specific opsin, Rhodopsin (RHO), and cone arrestin (ARR3), expressed in cone photoreceptors, also labeled the cells of the ONL, following a mutually exclusive pattern (Figure 3G). Mature cone markers for cone cell subtypes, L/M‐OPSIN or S‐OPSIN, labeled a proportion of cells in the ONL (Figure 3H,I). The OVs differentiated from NRL −/− ESCs, despite displaying organization with an external ONL formed by closely packed cell bodies, as seen in the controls, showed a drastic change in their protein distribution pattern. Most notable was the disappearance of NRL‐positive cells from the ONL and across the entire OV structure, demonstrating the effective gene knockout strategy (Figure 3E′). Accordingly, NR2E3 labeling disappeared, suggesting loss due to the lack of functional NRL, which is required to activate its expression7, 12, 39 (Figure 3F′). Consistent with the deficiency of rod photoreceptor transcription factors (NRL, NR2E3), the expression of Rhodopsin was undetectable in the NRL‐deficient samples, whereas ARR3‐positive cells were present in both WT and NRL −/− samples (Figure 3G,G′).
L/M‐OPSIN labels a subtype of cone cells regulated by pathway components different to that of S‐cones.40, 41 The immunohistochemical analysis of NRL −/− OVs showed comparable expression of L/M‐OPSIN to the controls (Figure 3H,H′), suggesting that cone genesis and specification of L/M‐opsin cones is not affected by the loss of NRL. Strikingly, the number of S‐OPSIN‐ positive cells in the NRL −/− OVs increased dramatically compared to the control ones, with abundant positive cells found in most areas that present a retinal neuroepithelial ONL structure (Figure 3I′). The quantification of S‐OPSIN‐positive cells at week 25 is shown in Figure 3J. While some OV areas did not develop an ONL and therefore did not have any positive cells, those that generated a laminated retinal structure contained higher numbers of S‐OPSIN‐positive cells in NRL −/− samples compared to those from the WT controls.
2.4. Gene expression confirms a tendency to cone gene overexpression at expense of rod markers in NRL −/− OVs
To quantify the transcriptional changes caused by the lack of functional NRL, we analyzed the gene expression of a number of retinal genes by quantitative quantitative real time PCR (qRT‐PCR) at weeks 7, 14 and 25 of differentiation (Figure 4). VSX2 is expressed in retinal progenitor cells at early developmental stages. In our OVs, no differences in VSX2 expression were observed at week 7. VSX2 then becomes restricted over time to the bipolar and Müller cell populations in the mature retina.42, 43, 44 Coincident with the temporal acquisition of bipolar cell identity,45 VSX2 expression was significantly reduced in the NRL −/− samples at weeks 14 and 25. The early pan‐photoreceptor marker CRX was markedly downregulated at week 7 in the NRL‐deficient samples, but not at later time points.
FIGURE 4.
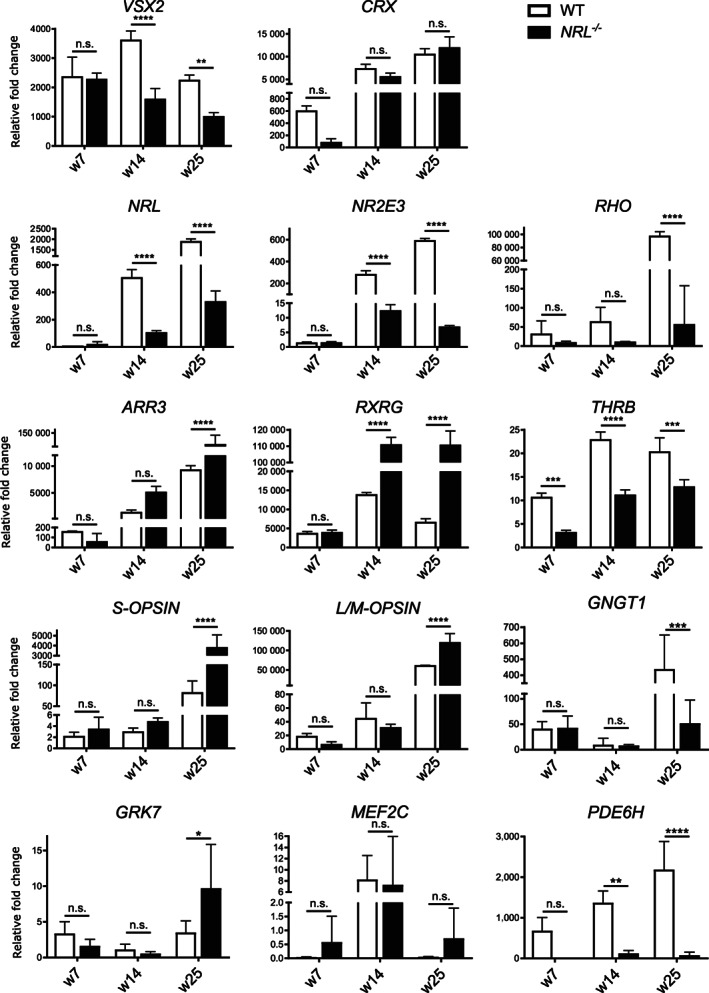
qRT‐PCR analysis of expression of a panel of photoreceptor genes and the neural retinal marker VSX2 in WT and NRL −/− OVs at three time points, week 7, 14, and 25. Markers used were the pan‐photoreceptor marker cone/rod‐homeobox CRX, the rod markers NRL and NR2E3 and cone markers THRB, RXRγ, OPN1MW, OPN1SW, and ARR3 (NRL −/− N = 4 differentiations w7, n = 6; w14, n = 4; w25 n = 6. WT, N = 3 differentiations; n = 4 for each time point). Other genes analyzed were GNGT1, expressed in rod outer segments, GRK7, involved in the cone photoresponse, cone‐specific phosphodiesterase PDE6H and MEF2C, putative regulator of rod to cone identity (NRL −/− N = 2 differentiations w7, n = 4; w14, n = 4; w25 n = 4. WT, N = 2 differentiations; n = 4 for each time point). Samples were normalized to GAPDH and d0 (undifferentiated stem cells), plot mean ± SD. **P < .01, ***P < .001, ****P < .0001. GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; OV, optic vesicles; qRT‐PCR, quantitative real time PCR; WT, wild type
The reduction in VSX2 gene expression in NRL −/− OVs at later stages was not expected as VSX2 expression in progenitors precedes NRL expression in post mitotic rods, crosstalk between these genes has not been reported, and we showed that the number of VSX2‐positive cells was similar between WT and NRL −/− OVs at 14 and 25 weeks (Figure S3). As VSX2‐positive cells are postmitotic (Ki67‐negative) at later stages (Figure S3B) together these data suggest that VSX2 gene expression levels are reduced in differentiating cells (bipolar or Müller glia) in the post mitotic NRL‐deficient retina, while the number of VSX2‐positive cells appears unchanged.
We then studied the expression of a panel of genes involved in rod development and function. NRL, as expected, showed a clear downregulation at week 14 in NRL −/− OVs, with expression showing a 10‐fold reduction relative to control by week 25, most likely due to nonsense mediated decay caused by the introduced mutation. The NRL target and rod‐specific transcription factor gene NR2E3 followed a similar trend, with low, nondifferential expression at week 7, and a drastic reduction in expression in NRL −/− OVs at week 14, falling to a 100‐fold difference in samples by week 25. Finally, this same trend in differential expression between samples was also observed with the rod‐specific photopigment gene RHO, whose expression is regulated synergistically by NRL and CRX5, 46; expression in NRL −/− OVs was reduced relative to controls at week 14, and this difference was highly significant by week 25.
A panel of cone markers was used to investigate whether the increase in the number of cells positive for S‐OPSIN in NRL −/− OVs observed by immunohistochemistry, was coincident with an increase in cone marker expression at the RNA level. By weeks 14 and 25, ARR3 expression in the edited NRL −/− line was significantly higher than in controls. Similarly, expression of RXRγ, which is expressed by cone precursor cells and cooperates with Thyroid Hormone Receptor Beta (THRB) to suppress S‐cone fate and promote M‐cone generation in mouse,40 is comparable between mutants and controls at week 7, but significantly upregulated in NRL −/− OVs at later time points. THRB, on the contrary, displays an inverted, but weaker pattern, with a reduction in NRL −/− OVs relative to controls at every time point analyzed.
Expression of the cone photopigment genes for S‐OPSIN (OPN1SW) and L/M‐OPSIN (OPN1MW), which define the cone subtype populations, were analyzed. In line with the protein observations, S‐OPSIN displayed a progressive and marked trend of upregulation upon the loss of NRL, significant from week 14 and reaching 100‐fold difference at week 25. L/M‐OPSIN showed a significant upregulation at week 25, pointing to a possible influence of NRL loss in the generation of these cone subtypes.
Finally, we examined expression of four genes that were recently reported to be altered in OVs derived from an induced pluripotent stem cell (iPSC) line from a patient with an NRL mutation47: GNGT1 (G Protein Subunit Gamma Transducin 1), the gamma subunit of transducin, found in rod outer segments,48 was downregulated in the edited NRL −/− line at differentiation week 25; GRK7, a retina‐specific G protein‐coupled receptor kinase involved in the cone photoresponse,49 was slightly upregulated, at week 25 in the NRL −/− line; MEF2C, member of the MADS (MCM1‐agamous‐deficiens serum response factor) family of transcription factors, did not show significant differential expression at any stage; PDE6H, a cone‐specific phosphodiesterase, was downregulated in the NRL −/− samples at week 14 and 25.
2.5. NRL−/−OVs establish comparable synaptic connections and show similar glial marker expression at late stages of differentiation compared to WT
Previous studies in the Nrl −/− mouse showed that in the absence of rod photoreceptors, the rod bipolar cells form synaptic connections with cones.14 As OVs represent a complex three‐dimensional in vitro retinal culture, capable of developing stratified layers of different cell types50 we investigated the ability of the NRL −/− cells to establish interlayer connectivity. We explored the potential for synaptic generation using immunostaining for various synaptic markers on 25‐week‐old OV samples. We detected colocalization of the synaptic vesicle protein Synaptophysin (SYN) and the postsynaptic marker PSD95 basally to the photoreceptor layer, between the ONL and the inner layer of cells, defining a presumptive outer plexiform layer. Both control and NRL‐deficient sections displayed positive staining of these markers in a comparable fashion (Figure 5A,A′). We also used CACNA1F, a calcium channel subunit expressed in photoreceptor cells, and detected positive signal colocalization with Synaptophysin. The pattern of expression for these synaptic markers was indistinguishable between control and NRL −/− sections (Figure 5B,B′). In addition, we used cone arrestin together with RIBEYE, the main component of the synaptic ribbon,51 to visualize the cone photoreceptor cell synapses organized in the presumptive outer plexiform layer underlying the polarized photoreceptor cell bodies (Figure 5C,C′).
FIGURE 5.
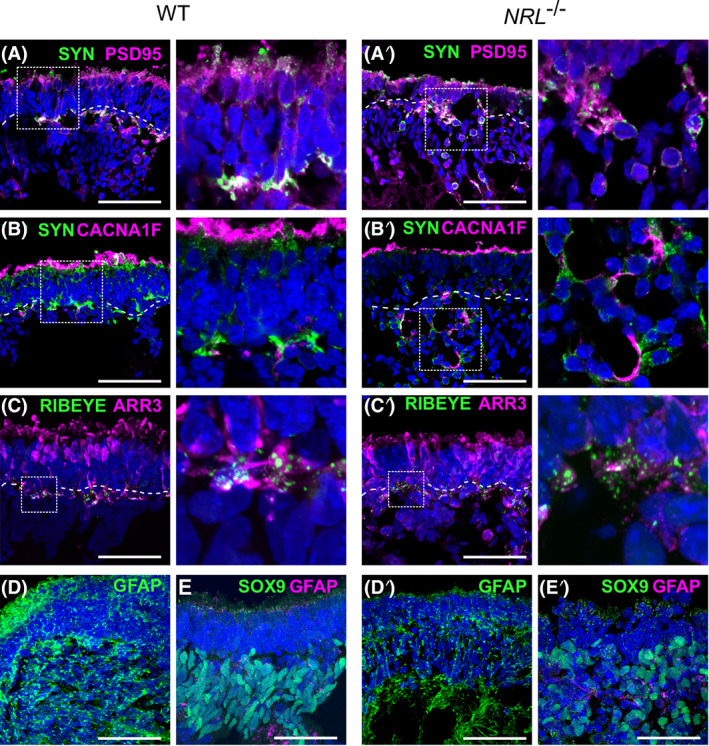
Immunostained OVs at retinal differentiation week 25 from WT and NRL −/− ESCs. A,A′, Synaptic markers Synaptophysin (SYN, green) and PSD95 (magenta) define the putative plexiform layer between presumptive ONL and inner nuclear layer cells. B,B′, Synaptic markers Synaptophysin (green) and calcium channel subunit CACNA1F (magenta) appear adjacent in the processes of the photoreceptor cells. C,C′, Cone marker cone arrestin (ARR3, magenta) and the synaptic protein RIBEYE (green) label photoreceptor layer and underlying synaptic ribbon. Panels to the right are high magnification insets outlined by dotted line. D,D′, Glial marker GFAP (green) with nuclei in blue was used as a marker for stress resulting in gliosis across the OV structure (N = 1, n = 2). E,E′, Müller glia marker SOX9 (green) together with GFAP (magenta) labeling cells of the inner layer. Scale bars = 50 μm. ESC, embryonic stem cell; qRT‐PCR, quantitative real time PCR; ONL, outer nuclear layer; OV, optic vesicles; WT, wild type
To investigate whether the lack of NRL might have a detrimental effect on OV integrity, resulting in reactive gliosis, a common feature of retinal dystrophies,52 we stained mature retinal organoids sections for activated retinal Müller glia cell marker, glial fibrillary acidic protein (GFAP).53 The staining pattern was similar across the two genotypes, being mostly evident beneath the ONL and throughout the core of the organoids. No evidence of enhanced gliosis was observed in the presumptive ONL of NRL‐deficient organoids compared to WT (Figure 5D,D′). To further characterize the inner cell layer of the OVs, we used SOX9, a marker of Müller glia cells, in addition to GFAP.53, 54 In both WT and NRL −/− samples, the SOX9 positive cells were located internal to the presumptive ONL, consistent with the native retinal organization of Müller glial cells (Figure 5E,E′).
3. CONCLUSION
Mammalian retinogenesis is a highly conserved biological process,55 although striking differences remain between humans and the mouse models available. The recent expansion of CRISPR/Cas9‐gene editing, human iPSCs and organoid technologies has opened up an extensive platform for researchers to model a myriad of developmental and pathological processes.56, 57, 58, 59 Here we report the first NRL‐deficient human ESC line, which has been used to investigate the role of NRL in human photoreceptor development and show that human rods share a highly conserved developmental pathway, where NRL is required to form rod photoreceptors, and in its absence increased numbers of photoreceptor precursors acquire an S‐cone‐like phenotype instead. This work allows the future exploration of preclinical potential therapeutic strategies using these supernumerary human cones‐like cells.60
4. DISCUSSION
By generating a human ESC line with an NRL homozygous null mutation, we were able to directly study its effect on retinogenesis in vitro. We introduced a mutation creating a nonsense mutation p.(Trp74*), close to the predicted null NRL allele, p.(L75fs), previously reported in ESCS cases with signs of retinal degeneration, clumped pigmentary retinopathy, and preservation of blue cone function.24, 25 We observed in the organoid model that the loss of functional NRL increases the number of S‐OPSIN positive cells and abolishes NR2E3 expression, which mimics the molecular mechanism behind ESCS; generation of OVs was not affected, consistent with no published reports of disruption of embryonic eye development in ESCS cases. The lack of NRL did not appear to affect directly the viability of retinal organoids, since we did not observe an increase in degeneration at early or later culture stages, and NRL −/− OVs have been maintained up to 30 weeks in culture regardless of the cell line genotype (Figure 1 and data not shown). No increase of gliosis was observed in GFAP‐stained OVs, suggesting that the lack of functional NRL in human photoreceptors drives a change of fate in these cells, without compromising the integrity of the retinal organoids (Figure 5D,D′). The OVs mimic neural retinal tissue and include Müller glia cells labeled with SOX954, 61 (Figure 5E,E′). ESCS patients often present with clumped pigmentary retinal degeneration, characterized by large clumped pigmented deposits in the RPE of the fundus. Such cases have been described in individuals with recessive mutations in NR2E3 or NRL, suggesting a common genetic basis.22, 24, 26, 62, 63 The protocol used in our study recapitulates the formation of the neural retina, which lacks a juxtaposed RPE layer, making it not very suitable to address RPE phenotypes like clumped pigmentary retinal degeneration, nevertheless we did not observe any abnormal pigmentation arising within the NRL −/− OV structures.
In mouse, the rodless Nrl −/− retina transiently develops whorls or “rosettes” in the ONL and the photoreceptor outer segments were shorter than those of the WT.7, 11 These structural changes may result from a conformational alteration due to the loss of rods, leaving supernumerary cones lacking organization and RPE contact, causing degeneration, although in a time‐restricted manner, before stabilization.11, 64, 65 Similar structural abnormalities were described in retinal tissue from a patient affected by NR2E3 mutations, and attributed to the excessive number of cones produced instead of rods, which pushes the tissue and produces convex folds.11, 27 In the organoid model, the loss of NRL did not cause an abnormal histological organization of the ESC‐derived OVs, nor did it have an effect on epithelial polarity compared to the controls, as seen in the histological analysis using apical marker ZO‐1 (Figure 3C,C′). By week 25, both NRL‐deficient and control organoids lacked mature outer segment formation, but show clear evidence of nascent segment structures extending beyond the ONL (Figure 3G‐I′). Occasionally, OVs presented “rosette‐like” structures, with round doughnut‐like organization of neuroepithelium inside of the organoid; however, we observed this phenomenon in several lines (control, edited, and other PSCs, data not shown) and therefore attributed it to the variability of the OV organization, not the NRL mutation.
We report the consequences of edited NRL loss‐of‐function mutation in organoids on the expression profile of several retinal genes and markers of cell fate. VSX2, expressed by bipolar cells at later time points, is reduced relative to controls. This is noteworthy for several reasons: development of bipolar cells and rod photoreceptors are closely related in mammals66; a direct relationship between VSX2 and NRL has not been described, although it has been previously proposed that VSX2 represses photoreceptor differentiation in mouse.66, 67 Our human data suggest a feedback loop, whereby absence of NRL at later stages leads to reduced VSX2 gene expression. The cone rod homeobox gene, CRX, that interacts with NRL,46 shows comparable levels of expression by week 14 and 25. The dramatically reduced expression of the NRL targets NR2E3 and RHO in the NRL‐deficient organoids relative to controls indicating loss of differentiated rod cells is consistent with NRL acting as a key regulator for rod specification. Consequently, cone photoreceptor genes showed an elevated expression in NRL −/− OVs compared to controls. ARR3, RXRƔ, and THRB are expressed earlier in development, and the latter two are known to be involved in cone specification in mouse.68 Expression of ARR3 and RXRƔ is higher in NRL‐deficient OVs than controls at the RNA level, which is consistent with an enhanced cone identity of cells in the NRL‐deficient samples. Notably, in contrast to the direct regulation of the Mef2c promoter activity by Nrl in mice suggesting its role in rod development,69 we found no evidence supporting NRL regulation of MEF2C. MEF2C expression was unchanged in the human NRL−/− OVs organoids and Kallman et al47 showed MEF2C was enriched in cone photoreceptors, suggesting MEF2C may play a different role, and is differently regulated, in humans and mice.
Nrl is known to interact with cone‐specific genes to repress their activity, and has been shown to bind the promoter region of ThrB, responsible for M‐cone specification9; this de‐repression effect might explain the results of reduced THRB gene expression in NRL −/− organoid samples. Previous studies in mouse have reported variable impact of Nrl loss on expression of M‐cone specific marker Opn1mw; earlier studies indicate no changes in Opn1mw protein expression,7, 64 whereas others report a slight increase in gene expression levels27, 70 and the presence of both S and M‐OPSIN pigment in the physiological responses of Nrl −/− photoreceptors.11, 13 Here, we observed a large increase in OPN1SW expression and a modest increase in OPN1MW gene expression in late time point NRL −/− OVs (Figure 4), although L/M‐OPSIN protein distribution appeared equivalent (Figure 3H,H′). These data suggest a role for NRL, whether direct or indirect, in influencing L/M‐cone specification in mammals, in addition to the established repression of S‐cone identity.
Assessment of the ultrastructural, molecular, and electrophysiological features of photoreceptors cells from Nrl −/− mice showed remarkable similarity to bona‐fide cones, while other studies suggested the generation of intermediates between rods and cones.11, 13, 70 A study on retinal organoids derived from a patient iPSC line with an NRL mutation similar to that engineered here in ESC was recently published.47 This study characterized cone and rod photoreceptors using immunostaining and single cell RNA sequencing techniques. Consistent with our work, an increase in S‐OPSIN+ cells was observed in the iPSC line, which lacks expression of rod proteins. Interestingly, the single cell analysis revealed two S‐OPSIN+ populations, one similar to WT S‐cones, and a second one that displays some rod photoreceptor features. Consistent with mouse studies, these results point to the existence of a hybrid population termed “cods” and indicate aspects of human rod photoreceptor specification are independent of NRL.
Most features measured in Nrl −/− cells in the mouse model suggest they are healthy and functional,11 and they connect to rod bipolar cells, implying that the type of photoreceptor connections is not pre‐established based on cell type.14 We observed that NRL −/− OVs form organized layers that express synaptic markers, similar to control organoids, and therefore it appears that the main consequence of losing functional NRL is the change in photoreceptor identity, without compromising cellular homeostasis. Our system therefore represents an in vitro human model that generates supernumerary S‐cone‐like cells and L/M‐cones, but not rods, rather than a human retinitis pigmentosa model, associated with harmful gain of function mutations and characterized by degenerative features.
Several groups have recently explored tampering with Nrl as a therapeutic approach to stimulate the endogenous generation of cones capable of rescuing degeneration and cone‐specific visual function. Using various methods to repress, eliminate or generate a loss‐of‐function Nrl mutation in mouse models of retinal degeneration, retinal degeneration was prevented by reprogramming rods to cones, accompanied by a recovery of ONL architecture and an increase in cone marker expression.71, 72, 73, 74 Nrl‐deficient cone‐like mouse photoreceptors have also been transplanted subretinally in a model of retinal dystrophy and shown to restore some photopic vision.75
The culture of human cells as organoids presents an opportunity to generate virtually unlimited specific cells for transplantation therapy, provided that these are physiologically functional. Photoreceptors must be capable of establishing synapses with second order retinal cells in order to be a relevant product for cell therapy. Previously, a study using Nrl −/− mice reported that expression of the synaptic calcium channel component CACNA1F is altered in the mutant adult, but not newborn retinas, suggesting a role for Nrl in its maturation.67 By contrast, Nrl overexpression in mouse results in cone‐deficient retinas that maintain synaptic organization with expression of the postsynaptic protein PSD95.9 In the present work, we showed that OVs at 25 weeks, equivalent to a mid‐gestational stage develop distinct layers of cells that organize in a comparable manner to the human retina. The photoreceptor cells in the NRL −/− OVs showed evidence of synaptic connections by organized distribution of several key human synaptic markers (PSD98,29, 32 synaptophysin,50 and RIBEYE51 in a presumptive plexiform layer, suggesting the generation of cone cells capable of establishing connectivity across cell layers; Figure 5A,A′,B,B′,C,C′). We did not detect differences in synaptic marker distribution in control and NRL‐deficient OVs suggesting capacity for photoreceptor connectivity was not diminished at this stage in the cone‐rich human organoids, although specific markers for human cone synapses are not available.
In summary, our study indicates the value of using genetically modified cell lines to characterize the retinal developmental events in humans and to generate new disease models for in vitro study. Modified OVs also represent a potential source of abundant cone photoreceptor cells for use in generating a cell product suitable for cone cell replacement transplantation therapy. However, clinical application would be contingent upon regulatory approval of such genetically modified human ESCs.
5. METHODS
Additional details are provided in Supporting Information Methods.
5.1. ESC culture
Human ESCs were routinely maintained on Laminin‐coated 6 well plates on NutriStem medium. When at 80% to 90% confluence, cells were lifted using EDTA 0.5 mM and replated at 10 000 cells/cm2.
5.2. CRISPR/Cas9 gene ablation of NRL
To generate an NRL‐deficient ESC line, we designed a small asymmetric ssODN33 to insert a STOP codon at the a.a. position 74 of NRL. We electroporated the ssODN donor together with a plasmid containing the NRL sgRNA and a Cas9 sequence in ESCs; selected colonies were screened for the insertion of the donor sequence, and one clone homozygous for the designed mutation (NRL −/−) was identified.
5.3. Retinal differentiation
Both parental MShef10 and NRL −/− ESCs were subjected to retinal differentiation using a previously published protocol.28 Briefly, cells were cultured to confluence and cultured in Neural Inductive Media until beginning to form three‐dimensional OVs. These are picked before week 7 of differentiation and further cultured in RDM.
5.4. Immunohistochemistry
OVs were fixed in a 4% paraformaldehyde (PFA) solution, and cryopreserved to generate 12 μm sections. Slides were incubated with the primary antibody O/N at 4°C, followed by secondary antibody incubation 2 hours at R/T and 4',6‐diamidino‐2‐phenylindole (DAPI) counterstaining. Antibodies used are listed in Table S1.
5.5. Quantitative PCR
Triplicate samples of independently cultured OVs were isolated at week 7, 14, and 25; undifferentiated ESCs were used as control. RNA was extracted using the RNeasy Micro Kit and cDNA was synthesized using the RevertAid H Minus First Strand cDNA Synthesis Kit. qRT‐PCR was performed using Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as reference gene and Ct values were analyzed as previously described.76 Primers sequences are in Table S2.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
E.C.: conception and design, provision of study material, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; J.T.: collection and/or assembly of data; D.H.: conception and design, provision of study material, collection and/or assembly of data, manuscript writing; A.A.: collection and/or assembly of data, data analysis and interpretation; J.L.: conception and design, provision of study material; J.C.S.: conception and design, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript
Supporting information
Supplementary Figure 1 CRISPR off‐target effects
Supplementary Figure 2. Representative examples of OVs at differentiation week 14 and NRL immunostaining in week 7 OVs
Supplementary Figure 3. Proportion of VSX2+ progenitor cells at differentiation week 14 and 25 in WT and NRL−/− OVs
Table 1. Antibodies list
Table 2. qRT‐PCR primers list
Table 3. CRISPR/Cas9 Off‐target effects
Table 4. Primers to amplify off‐target regions
ACKNOWLEDGMENTS
This study was supported by the Medical Research Council MR/M015688/1, Great Ormond Street Hospital Children's Charity and the National Institute for Health Research (NIHR) Great Ormond Street Hospital Biomedical Research Centre, Fight for Sight, the Saudi Arabian Cultural Bureau (SACB), and the Wellcome Trust (109053/Z/15/Z). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. We thank Harry Moore at the Centre for Stem Cell Biology, The University of Sheffield, Sheffield, UK for ESC lines, Hassan Rashidi and UCL Genomics for karyotype analysis and Dale Moulding at the UCL Great Ormond Street Institute of Child Health Imaging Facility for technical support.
Cuevas E, Holder DL, Alshehri AH, Tréguier J, Lakowski J, Sowden JC. NRL−/− gene edited human embryonic stem cells generate rod‐deficient retinal organoids enriched in S‐cone‐like photoreceptors. Stem Cells. 2021;39:414–428. 10.1002/stem.3325
Daniel Holder and Ashwak Alshehri contributed equally to this study.
Funding information Fight for Sight UK; Great Ormond Street Hospital Charity; Medical Research Council, Grant/Award Number: MR/M015688/1; National Institute for Health Research; NIHR Great Ormond Street Hospital Biomedical Research Centre; Saudi Arabia Cultural Bureau in London; Wellcome Trust, Grant/Award Number: 109053/Z/15/Z
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199‐205. [DOI] [PubMed] [Google Scholar]
- 2.Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina‐specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89:266‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akimoto M, Cheng H, Zhu D, et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow‐sorted photoreceptors. Proc Natl Acad Sci USA. 2006;103:3890‐3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farjo Q, Jackson AU, Xu J, et al. Molecular characterization of the murine neural retina leucine zipper gene, Nrl. Genomics. 1993;18:216‐222. [DOI] [PubMed] [Google Scholar]
- 5.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif‐leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swain PK, Hicks D, Mears AJ, et al. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem. 2001;276:36824‐36830. [DOI] [PubMed] [Google Scholar]
- 7.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447‐452. [DOI] [PubMed] [Google Scholar]
- 8.Kautzmann MAI, Kim DS, Felder‐Schmittbuhl M‐PP, et al. Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene Nrl, revealed by in vivo promoter analysis. J Biol Chem. 2011;286:28247‐28255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh ECT, Khan N, Novelli E, et al. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci USA. 2007;104:1679‐1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JW, Yang HJ, Oel AP, et al. Recruitment of rod photoreceptors from short‐wavelength‐sensitive cones during the evolution of nocturnal vision in mammals. Dev Cell. 2016;37:520‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniele LL, Lillo C, Lyubarsky AL, et al. Cone‐like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005;46:2156‐2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida S, Mears AJ, Friedman JS, et al. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004;13:1487‐1503. [DOI] [PubMed] [Google Scholar]
- 13.Nikonov SS, Daniele LL, Zhu X, Craft CM, Swaroop A, Pugh EN Jr. Photoreceptors of Nrl−/− mice coexpress functional S‐and M‐cone opsins having distinct inactivation mechanisms. J Gen Physiol. 2005;125:287‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strettoi E, Mears AJ, Swaroop A. Recruitment of the rod pathway by cones in the absence of rods. J Neurosci. 2004;24:7576‐7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bessant DAR, Payne AM, Mitton KP, et al. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet. 1999;21:355‐356. [DOI] [PubMed] [Google Scholar]
- 16.Bessant DAR, Payne AM, Plant C, Bird AC, Swaroop A, Bhattacharya SS. NRL S50T mutation and the importance of “founder effects” in inherited retinal dystrophies. Eur J Hum Genet. 2000;8:783‐787. [DOI] [PubMed] [Google Scholar]
- 17.DeAngelis MM, Grimsby JL, Sandberg MA, Berson EL, Dryja TP. Novel mutations in the NRL gene and associated clinical findings in patients with dominant retinitis pigmentosa. Arch Ophthalmol. 2002;120:369‐375. [DOI] [PubMed] [Google Scholar]
- 18.Martinez‐Gimeno M, Maseras M, Baiget M, et al. Mutations P51U and G122E in retinal transcription factor NRL associated with autosomal dominant and sporadic retinitis pigmentosa. Hum Mutat. 2001;17:520. [DOI] [PubMed] [Google Scholar]
- 19.Bessant DAR, Holder GE, Fitzke FW, Payne AM, Bhattacharya SS, Bird AC. Phenotype of retinitis pigmentosa associated with the Ser50Thr mutation in the NRL gene. Arch Ophthalmol. 2003;121:793‐802. [DOI] [PubMed] [Google Scholar]
- 20.Kanda A, Friedman JS, Nishiguchi KM, Swaroop A. Retinopathy mutations in the bZIP protein NRL alter phosphorylation and transcriptional activity. Hum Mutat. 2007;28:589‐598. [DOI] [PubMed] [Google Scholar]
- 21.Khan AO, Aldahmesh M, Meyer B. The enhanced S‐cone syndrome in children. BMJ Case Rep. 2009;91:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Zhang X, Chen LJ, et al. Association of NR2E3 but not NRL mutations with retinitis pigmentosa in the chinese population. Invest Ophthalmol Vis Sci. 2010;51:2229‐2235. [DOI] [PubMed] [Google Scholar]
- 23.Newman H, Blumen SC, Braverman I, et al. Homozygosity for a recessive loss‐of‐function mutation of the NRL gene is associated with a variant of enhanced S‐cone syndrome. Invest Ophthalmol Vis Sci. 2016;57:5361‐5371. [DOI] [PubMed] [Google Scholar]
- 24.Nishiguchi KM, Friedman JS, Sandberg MA, et al. Recessive NRL mutations in patients with clumped pigmentary retinal degeneration and relative preservation of blue cone function. Proc Natl Acad Sci USA. 2004;101(51):17819‐17824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright AF, Reddick AC, Schwartz SB, et al. Mutation analysis of NR2E3 and NRL genes in enhanced S cone syndrome. Hum Mutat. 2004;24:439. [DOI] [PubMed] [Google Scholar]
- 26.Littink KW, Stappers PTY, Riemslag FCC, et al. Autosomal recessive NRL mutations in patients with enhanced s‐cone syndrome. Genes. 2018;9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milam AH, Rose L, Cideciyan AV, et al. The nuclear receptor NR2e3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc Natl Acad Sci USA. 2002;99:473‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez‐Cordero A, Kruczek K, Naeem A, et al. Recapitulation of human retinal development from human pluripotent stem cells generates transplantable populations of cone photoreceptors. Stem Cell Reports. 2017;9:820‐837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellough CB, Collin J, Khazim M, et al. IGF‐1 signaling plays an important role in the formation of three‐dimensional laminated neural retina and other ocular structures from human embryonic stem cells. Stem Cells. 2015;33:2416‐2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer JS, Howden SE, Wallace KA, et al. Optic vesicle‐like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206‐1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong X, Gutierrez C, Xue T, et al. Generation of three‐dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano T, Ando S, Takata N, et al. Self‐formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:1‐18. [DOI] [PubMed] [Google Scholar]
- 33.Richardson CD, Ray GJ, DeWitt MA, et al. Enhancing homology‐directed genome editing by catalytically active and inactive CRISPR‐Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339‐344. [DOI] [PubMed] [Google Scholar]
- 34.Canham MA, Van Deusen A, Brison DR, et al. The molecular karyotype of 25 clinical‐grade human embryonic stem cell lines. Sci Rep. 2015;5:17258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodgkins A, Farne A, Perera S, et al. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015;31:3078‐3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendrickson A, Bumsted‐O'Brien K, Natoli R, et al. Rod photoreceptor differentiation in fetal and infant human retina. Exp Eye Res. 2008;87:415‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Brien KMB, Schulte D, Hendrickson AE. Expression of photoreceptor‐associated molecules during human fetal eye development. Mol Vis. 2003;9:401‐409. [PubMed] [Google Scholar]
- 38.Welby E, Lakowski J, Di Foggia V, et al. Isolation and comparative transcriptome analysis of human fetal and iPSC‐derived cone photoreceptor cells. Stem Cell Reports. 2017;9:1898‐1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H, Khanna H, Oh ECT, Hicks D, Mitton KP, Swaroop A. Photoreceptor‐specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563‐1575. [DOI] [PubMed] [Google Scholar]
- 40.Roberts MR, Srinivas M, Forrest D, Morreale de Escobar G, Reh TA. Making the gradient: thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc Natl Acad Sci USA. 2006;103:6218‐6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eldred KC, Hadyniak SE, Hussey KA, et al. Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science. 2018;362:eaau6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferda Percin E, Ploder LA, Yu JJ, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397‐401. [DOI] [PubMed] [Google Scholar]
- 43.Eastlake K, Wang W, Jayaram H, et al. Phenotypic and functional characterization of Müller glia isolated from induced pluripotent stem cell‐derived retinal organoids: improvement of retinal ganglion cell function upon transplantation. Stem Cells Translational Medicine. 2019;8:775‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271(2):388‐402. [DOI] [PubMed] [Google Scholar]
- 45.Kim DS, Matsuda T, Cepko CL. A core paired‐type and POU homeodomain‐containing transcription factor program drives retinal bipolar cell gene expression. J Neurosci. 2008;28:7748‐7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. J Biol Chem. 2000;275:29794‐29799. [DOI] [PubMed] [Google Scholar]
- 47.Kallman A, Capowski EE, Wang J, et al. Investigating cone photoreceptor development using patient‐derived NRL null retinal organoids. Commun Biol. 2020;3:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolesnikov AV, Rikimaru L, Hennig AK, et al. G‐protein βγ‐complex is crucial for efficient signal amplification in vision. J Neurosci. 2011;31:8067‐8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cideciyan AV, Jacobson SG, Gupta N, et al. Cone deactivation kinetics and GRK1/GRK7 expression in enhanced S cone syndrome caused by mutations in NR2E3. Invest Ophthalmol Vis Sci. 2003;44:1268‐1274. [DOI] [PubMed] [Google Scholar]
- 50.Singh RK, Mallela RK, Cornuet PK, et al. Characterization of three‐dimensional retinal tissue derived from human embryonic stem cells in adherent monolayer cultures. Stem Cells Dev. 2015;24:2778‐2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitz F, Königstorfer A, Südhof TC. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857‐872. [DOI] [PubMed] [Google Scholar]
- 52.Hippert C, Graca AB, Barber AC, et al. Müller glia activation in response to inherited retinal degeneration is highly varied and disease‐specific. PLoS One. 2015;10:e0120415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lupien C, Brenner M, Guérin SL, Salesse C. Expression of glial fibrillary acidic protein in primary cultures of human Müller cells. Exp Eye Res. 2004;79:423‐429. [DOI] [PubMed] [Google Scholar]
- 54.Sun W, Cornwell A, Li J, et al. SOX9 is an astrocyte‐specific nuclear marker in the adult brain outside the neurogenic regions. J Neurosci. 2017;37:4493‐4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heavner W, Pevny L. Eye development and retinogenesis. Cold Spring Harb Perspect Biol. 2012;4:a008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA‐guided Cas9 nucleases. Nat Biotechnol. 2013;31:827‐832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861‐872. [DOI] [PubMed] [Google Scholar]
- 59.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671‐687. [DOI] [PubMed] [Google Scholar]
- 60.Collin J, Zerti D, Queen R, et al. CRX expression in pluripotent stem cell derived photoreceptors marks a transplantable subpopulation of early cones. Stem Cells. 2019;37:609‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kautzman AG, Keeley PW, Nahmou MM, Luna G, Fisher SK, Reese BE. Sox2 regulates astrocytic and vascular development in the retina. Glia. 2018;66:623‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja TP. Shared mutations in NR2E3 in enhanced S‐cone syndrome, Goldmann‐Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol. 2003;121:1316‐1323. [DOI] [PubMed] [Google Scholar]
- 63.Schorderet DF, Escher P. NR2E3 mutations in enhanced S‐cone sensitivity syndrome (ESCS), Goldmann‐Favre syndrome (GFS), clumped pigmentary retinal degeneration (CPRD), and retinitis pigmentosa (RP). Hum Mutat. 2009;30:1475‐1485. [DOI] [PubMed] [Google Scholar]
- 64.Roger JE, Campos MM, Hiriyanna A, et al. Preservation of cone photoreceptors after a rapid yet transient degeneration and remodeling in cone‐only Nrl−/− mouse retina. J Neurosci. 2012;32:528‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stuck MW, Conley SM, Naash MI. Defects in the outer limiting membrane are associated with rosette development in the Nrl −/− retina. PLoS One. 2012;7:e32484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livne‐Bar I, Pacal M, Cheung MC, et al. Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc Natl Acad Sci USA. 2006;103:4988‐4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J‐W, Yang H‐J, Brooks MJ, et al. NRL‐regulated transcriptome dynamics of developing rod photoreceptors. Cell Rep. 2016;17:2460‐2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aavani T, Tachibana N, Wallace V, Biernaskie J, Schuurmans C. Temporal profiling of photoreceptor lineage gene expression during murine retinal development. Gene Expr Patterns. 2017;23‐24:32‐44. [DOI] [PubMed] [Google Scholar]
- 69.Hao H, Tummala P, Guzman E, et al. The transcription factor neural retina leucine zipper (NRL) controls photoreceptor‐specific expression of myocyte enhancer factor Mef2c from an alternative promoter. J Biol Chem. 2011;286:34893‐34902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smiley S, Nickerson PE, Comanita L, et al. Establishment of a cone photoreceptor transplantation platform based on a novel cone‐GFP reporter mouse line. Sci Rep. 2016;6:22867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu W, Mookherjee S, Chaitankar V, et al. Nrl knockdown by AAV‐delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Montana CL, Kolesnikov AV, Shen SQ, Myers CA, Kefalov VJ, Corbo JC. Reprogramming of adult rod photoreceptors prevents retinal degeneration. Proc Natl Acad Sci USA. 2013;110:1732‐1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreno AM, Fu X, Zhu J, et al. In situ gene therapy via AAV‐CRISPR‐Cas9 mediated targeted gene regulation. Mol Ther. 2018;26:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu J, Ming C, Fu X, et al. Gene and mutation independent therapy via CRISPR‐Cas9 mediated cellular reprogramming in rod photoreceptors. Cell Res. 2017;27:830‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santos‐Ferreira T, Llonch S, Borsch O, Postel K, Haas J, Ader M. Retinal transplantation of photoreceptors results in donor–host cytoplasmic exchange. Nat Commun. 2016;7:13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pfaffl MW. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 2001;29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 CRISPR off‐target effects
Supplementary Figure 2. Representative examples of OVs at differentiation week 14 and NRL immunostaining in week 7 OVs
Supplementary Figure 3. Proportion of VSX2+ progenitor cells at differentiation week 14 and 25 in WT and NRL−/− OVs
Table 1. Antibodies list
Table 2. qRT‐PCR primers list
Table 3. CRISPR/Cas9 Off‐target effects
Table 4. Primers to amplify off‐target regions
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


