Verlhac previews work from Mori et al. that describes mechanisms by which the paternal chromosomes are segregated until the completion of egg meiosis.
Abstract
Fertilization often triggers the final step of haploidization of the female gamete genome. In this issue, Mori et al. (2021. J. Cell Biol. https://doi.org/10.1083/jcb.202012001) identify two successive actin-dependent mechanisms that delay fusion of maternal and paternal chromosomes, preventing inadvertent elimination of paternal chromosomes together with maternal ones.
Species propagating via sexual reproduction produce haploid gametes through meiosis, a mode of division that culminates with two consecutive rounds of divisions with no intervening DNA replication. Fusion of the two haploid gametes, the sperm and the egg, restores the ploidy of the species. In most mammals, fertilization engages the second round of sister chromatid division of the egg, the unwanted chromosomes being extruded into a small degenerating polar body. In this issue, Mori et al. (1) identify mechanisms that prevent the inadvertent encounter of male and female chromosomes until the mouse egg finishes its second meiotic division. These processes are essential to avoid inadvertent elimination of the male genome together with the haploid maternal set.
Mouse eggs are ovulated while arrested in metaphase of the second meiotic division (metaphase II). Fertilization triggers meiosis resumption and anaphase II. It has been observed that the binding of the sperm is favored where the metaphase II–arrested egg is covered with microvilli (2) and disfavored in the region above the second meiotic spindle, named the actin cap, which is enriched in F-actin and devoid of microvilli (Fig. 1 A; 3). However, the mechanisms behind these observations have been poorly investigated. By following the early events of sperm binding to the egg, Mori et al. discover potential new roles for two membrane proteins previously implicated in sperm/egg binding, namely Juno (4) and CD9 (5, 6, 7).
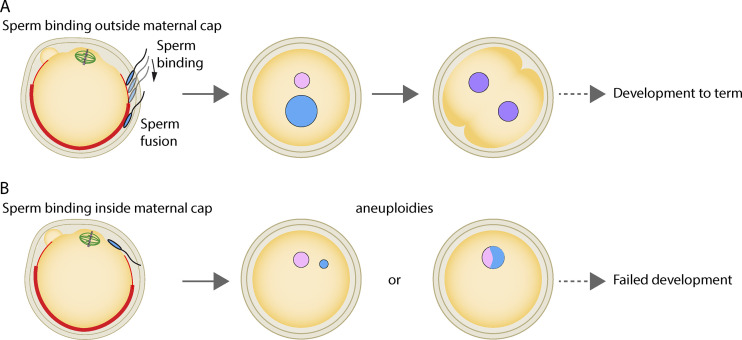
Figure 1.
Sperm binding is favored outside the maternal actin cap for successful embryo development. The density of microvilli (bold red area) increases away from the metaphase II egg spindle, in correlation with more important Juno and CD9 staining patterns. (A) Sperm binding outside the maternal actin cap. A sperm bound in the transition zone (lighter red area), with fewer microvilli and more lamellipodia-like structures, will move toward the denser microvilli area where it will fuse to the egg, preventing its elimination during second polar body extrusion. This mechanism favors correct ploidy of the zygote and its development to term. (B) Sperm binding inside the maternal actin cap. Binding and fusion in the microvilli-depleted region will produce aneuploid zygotes with reduced chances of successful development. Maternal chromosomes appear in pink, paternal ones in blue, parental genome mixing in purple, and spindle microtubules in green.
In a precise follow-up of sperm entry sites, the authors confirm previous observations that the fusion of the sperm occurs in a region >24 µm away from the maternal chromosomes. Simulations acknowledge a clear deviation of the in vivo sperm entry sites from a random configuration. Live imaging uncovers a rapid drift in the sperm’s peripheral position after its entry into the egg. These observations argue that not only does the sperm bind apart from the maternal chromosomes, but that it is also rapidly moved away to prevent further mixing while the second meiotic spindle rotates and the egg becomes truly haploid. This raises two questions that the authors address here: (a) how is the favored binding site—far away from the maternal spindle—determined, and (b) how is the sperm shifted from its original position once fused to the egg?
Mori et al. (1) first show that two key proteins in sperm fusion to the egg, Juno (4) and CD9 (5, 6, 7), display a punctate distribution on the egg membrane with a density correlating with that of the microvilli; greater away from maternal chromosomes and lower above the maternal set. Intriguingly, Juno and CD9 are also abundant in the transition zone, rich in lamellipodia-like structures, between the microvilli-poor and -dense regions. In this transition zone, Juno and CD9’s labeling is dynamic, moving toward the denser microvilli area. When a fluorescently labeled sperm binds in this ring-like zone, it was observed to move away from maternal chromosomes before fusion to the egg. Conversely, if it binds in the dense microvilli portion of the egg, the sperm remains still (Fig. 1 A). Considering the major roles of Juno and CD9 in regulating sperm hooking, it is tempting to speculate that these two proteins promote sperm motion in the ring-like zone before its fusion, trafficking it away from the maternal genome.
Next, the authors investigate the mechanisms behind Juno and CD9 local accumulation on the egg membrane and its potential role in regulating sperm binding. The region overhanging the maternal chromosomes is deprived of microvilli but enriched in F-actin (3). Owing in part to the fact that chromosomes accumulate the Ran GTPase in its GTP-bound active form, a Ran GTP gradient culminating at chromosomes is created around them (8). This Ran GTP gradient plays a major role in local F-actin accumulation and egg polarization (9). Not surprisingly, using an F-actin destabilizing drug as well as overexpression of a dominant negative form of the Ran GTPase, the authors observe that both F-actin and Ran control the accumulation of Juno and CD9 outside the F-actin cap overhanging maternal chromosomes. When the Ran GTP gradient is abolished, this bias in sperm fusion sites on the egg is no longer observed. If F-actin is disrupted using Latrunculin B, fusion sites occur closer to the chromosomes than in controls but do not appear to be random, arguing that F-actin may have other functions that require deeper investigation. The use of other tools to disrupt F-actin (e.g., cytochalasin D or Arp2/3 complex inhibitors) could help identify those F-actin–specific roles that are independent of Ran GTPase.
Finally, the authors use intracytoplasmic sperm injection (ICSI) to introduce and position chromosomes at different locations inside the egg, allowing them to test the importance of this proposed sequestration away from maternal chromosomes until egg anaphase II is achieved. In about two thirds of the cases, having paternal chromosomes in a 20-µm proximity to maternal ones is deleterious for the one-cell zygote, since they are either eliminated into the second polar body or they fuse with the egg haploid set into a single pronucleus, preventing correct independent paternal genome preparation (Fig. 1 B). However, this is not observed when ICSI is performed 20 µm away from the maternal genome, thus demonstrating clearly the importance of sperm binding outside the maternal F-actin cortical cap. These ICSI experiments also show that in 20% of the cases, even if paternal chromosomes fuse inside the cortical cap area, they can be moved away by F-actin–dependent mechanisms.
Altogether, Mori and colleagues (1) illustrate here the importance of delaying the fusion of mouse maternal and paternal genomes to ensure proper zygotic development. While the two pronuclei meet in center of the zygote via F-actin and microtubule-dependent mechanisms (10), it is only in interphase of the two-cell embryo that both parental genomes will potentially mix (11), attesting to the idea that this occurs quite late in the process. The authors show here that two mechanisms, sperm binding and sperm secondary motion inside the egg—both dependent on F-actin—promote the delay in genome fusion. Interestingly, the importance of F-actin in delaying the parental chromosome encounter is also observed in the Caenorhabditis elegans zygote (12), arguing that the process is evolutionary conserved. It will be interesting to determine whether Juno and CD9 also actively participate in egg polarization in a positive feedback loop or if they are only following egg polarity. This might be tested by forcing their localization at various points across the egg membrane, even in the cortical cap area. Ultimately, it might prove important to identify which regulators control their dynamic distribution in the ring-like transition zone on the egg membrane. It might be worth testing whether hydrodynamic forces triggered by the rotation of the second meiotic spindle participate in their oriented mobility toward the microvilli-dense region and maybe potentially in the sperm secondary motility (13), which remains to be further investigated.
References
- 1.Mori, M., et al. 2021. J. Cell Biol. 10.1083/jcb.202012001 [DOI] [Google Scholar]
- 2.Yanagimachi, R., and Noda Y.D.. 1970. J. Ultrastruct. Res. 10.1016/S0022-5320(70)90163-2 [DOI] [Google Scholar]
- 3.Longo, F.J., and Chen D.Y.. 1985. Dev. Biol. 10.1016/0012-1606(85)90320-3 [DOI] [Google Scholar]
- 4.Bianchi, E., et al. 2014. Nature. 10.1038/nature13203 [DOI] [Google Scholar]
- 5.Kaji, K., et al. 2000. Nat. Genet. 10.1038/73502 [DOI] [PubMed] [Google Scholar]
- 6.Le Naour, F., et al. 2000. Science. 10.1126/science.287.5451.319 [DOI] [Google Scholar]
- 7.Miyado, K., et al. 2000. Science. 10.1126/science.287.5451.321 [DOI] [Google Scholar]
- 8.Dumont, J., et al. 2007. J. Cell Biol. 10.1083/jcb.200605199 [DOI] [Google Scholar]
- 9.Deng, M., et al. 2007. Dev. Cell. 10.1016/j.devcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 10.Chaigne, A., et al. 2016. Nat. Commun. 10.1038/ncomms10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichmann, J., et al. 2018. Science. 10.1126/science.aar7462 [DOI] [Google Scholar]
- 12.Panzica, M.T., et al. 2017. J. Cell Biol. 10.1083/jcb.201702020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, H., et al. 2020. Sci. Adv. 10.1126/sciadv.aaz5004 [DOI] [Google Scholar]