Abstract
Studies have indicated that collagen α-1 (IV) chain (COL4A1) has an indispensable regulatory role in the complex pathological mechanisms of numerous types of malignant tumor. However, its role in the development of glioma has remained elusive. Therefore, the present study sought to determine the association between the expression levels of COL4A1 and the clinical characteristics of gliomas by analyzing large samples. First, analysis of thousands of glioma tissue samples collected from the Gene expression profiling interactive analysis, Gene Expression Omnibus database, the Ivy glioblastoma atlas, The Human Protein Atlas, Chinese Glioma Genome Atlas and The Cancer Genome Atlas. In addition, glioma tissues and normal brain tissues from patients with glioma and epilepsy undergoing surgical resection were collected. These samples, which were subjected to a variety of different detection techniques (including sequencing data, chip data, reverse transcription-quantitative PCR, cell lines and tissue samples, in situ hybridization and immunology) revealed that COL4A1 expression was not only increased at the mRNA level but also at the protein level as compared with that in normal brain tissue. Furthermore, Kaplan-Meier analysis revealed that COL4A1 expression was associated with reduced overall survival of patients, particularly those with World Health Organization grade III glioma. Receiver operating characteristic analysis suggested that COL4A1 had a moderate diagnostic value for glioma. In addition, the Mann-Whitney U-test or Kruskal-Wallis test indicated that the expression levels of COL4A1 were positively associated with the histological type and historical grade of the tumor, patient age, ‘Primary, Recurrent, Secondary’ type and the chemotherapy status, and negatively associated with isocitrate dehydrogenase mutation and 1p19q co-deletion (P<0.001). Gene-set enrichment analysis indicated that overexpression of COL4A1 promoted cancer-associated pathways, such as the JAK/STAT signaling pathway and cell cycle regulation. Finally, an MTT assay, immunohistochemical analysis of the cell cycle regulator KI67 and a wound-healing assay further confirmed that knockdown of COL4A1 inhibited the proliferation and migration ability of glioma cells. In conclusion, COL4A1, as a novel oncogene, is a marker for poor prognosis in patients with glioma. The present study expanded the understanding of the pathogenesis of glioma and identified COL4A1 as a potential target for the diagnosis and treatment of gliomas.
Keywords: collagen α-1 (IV) chain, glioma, intracranial malignancies, Chinese Glioma Genome Atlas, prognosis
Introduction
Gliomas are among the most common types of intracranial malignancies (1). Although the global incidence of gliomas is only 3-5.5 per 100,000 individuals, gliomas have high mortality rates (2,3). Although the prognosis of gliomas has improved with advancements in surgical technologies and chemotherapies, the 5-year survival rate for advanced glioma is still <36% (4,5). The poor prognosis of glioma is mainly due to its highly invasive capabilities, rendering it difficult to completely resect, with high rates of recurrence (6). In addition, the prognosis of glioma is closely associated with pathological grade, histological type and abnormal expression of a number of oncogenes, including chitinase 3 like 1, MET and PTEN (7).
Studies have indicated that abnormally expressed genes are closely associated with the prognosis of gliomas. For instance, several studies have reported that the malignant progression of gliomas involved an EGFR variant, whose abnormal expression is involved in the infiltration and proliferation of tumor cells (4). Furthermore, TGF-β influences the proliferation and migration of tumor cells by affecting T-cell function (8). In addition, certain studies have reported that in glioma patients undergoing chemotherapy, CD133 expressed by the HOX gene is a marker for poor prognosis (9). Hale et al (10) revealed that CD36 promotes the growth of cancer stem cells, thereby improving the growth and immune resistance of gliomas, and is associated with poor prognosis. However, these biomarkers are still controversial and hence, novel biomarkers urgently require to be identified.
Collagen α-1 (IV) chain (COL4A1) is an important component of the basement membrane of numerous tissues and cell types in the human body (11). The basement membrane is a vascular and extracellular scaffold that supports and partially regulates cell behavior (12). A previous study reported that mutations in COL4A1 may affect glycine residues in the protein, which is not conducive for its structural and functional stability (13). COL4A1 mutations may cause several diseases, including those pertaining to the cardiovascular, cerebrovascular, renal, gastrointestinal and circulatory systems (14-19). Studies have indicated that abnormal expression of COL4A1, as a novel oncogene, is closely associated with the occurrence, development and outcome of a variety of cancer types (14,16,20,21). However, the role of COL4A1 in the diagnosis and prognosis of glioma has remained to be determined.
In the present study, thousands of glioma tissue samples were collected through a variety of different detection techniques [such as sequencing data, chip data, reverse transcription-quantitative (RT-q)PCR, in situ hybridization and immunology] to reveal that COL4A1 expression was not only increased at the mRNA level, but also markedly higher at the protein level in comparison with normal brain tissue. It was attempted to determine the relationship between COL4A1 expression levels and the clinical characteristics of glioma, and also the possible role of COL4A1 in the pathological process of gliomas. The present study was the first to report that high expression of COL4A1, as a novel oncogene, is significantly associated with the prognosis of gliomas. Furthermore, by performing gene-set enrichment analysis (GSEA) analysis, the possible carcinogenic pathways of COL4A1 were identified. Therefore, it may be indicated that COL4A1 is a valuable potential biomarker for the diagnosis and treatment of gliomas.
Materials and methods
Data collection
The glioma RNA sequencing (RNA-seq) dataset and matched clinical information were downloaded from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) and the Chinese Glioma Genome Atlas (CGGA; http://www.cgga.org.cn/) databases. In the CGGA, the information of 1,018 glioma patients was collected. Clinical information included gender, age, grade, Primary, Recurrent, Secondary (PRS) type, radiotherapy status, chemotherapy status and histological grade of glioma, while gene expression information included 1p19 co-deletion status and isocitrate dehydrogenase (IDH) mutation status. Samples with incomplete clinical information were excluded. The sequencing data of 749 glioma samples from CGGA were further analyzed. Furthermore, sequencing data of 655 glioma samples from TCGA and microarray data of 268 glioma samples from CGGA, which were separate from the 749 samples aforementioned, were used for verification of the association between the expression of COL4A1 and the prognosis of glioma.
In addition, the datasets of the four glioma gene chips were obtained from Gene Expression Omnibus database (GEO; https://www.ncbi.nlm.nih.gov/geo/), namely GSE2223, GSE4290, GSE50161 and GSE116520. The GSE2223 dataset included 50 glioma tissue samples and 4 normal brain tissue samples; the GSE4290 dataset comprised 77 glioma tissue samples and 23 normal brain tissue samples; the GSE50161 dataset included 34 glioma tissue samples and 13 normal brain tissue samples; and the GSE116520 dataset comprised 34 glioma tissue samples and 8 normal brain tissue samples. The four glioma datasets included a total of 195 glioma tissue samples and 48 normal brain tissue samples. These samples were used to detect changes in the expression levels of COL4A1 in tumor tissues compared with normal samples.
Certain online data analysis platforms were also used in the present study. Gene expression profiling interactive analysis (GEPIA; http://gepia.cancer-pku.cn/) is an online public data analysis platform that was used to detect changes in the expression levels of COL4A1 in various human malignant tumor tissues. The Human Protein Atlas (https://www.proteinatlas.org/) is a proteomics database that was used to detect changes in protein expression levels of COL4A1 in gliomas vs. normal brain tissues. The database was used to evaluate immunohistochemical results for the protein expression of COL4A1 in 2 normal brain tissues and 11 glioma samples. Images of immunohistochemical sections of 2 normal brain tissues and 2 glioma tissues were obtained for display. The Ivy Glioblastoma Atlas (http://glioblastoma.alleninstitute.org/) is a database focused on glioma research, which contains multiple data types such as in situ hybridization (ISH) and H&E staining. The Ivy Glioblastoma Atlas database was used by the present study to detect the expression levels of COL4A1 in gliomas. From this database, 8 glioma samples with ISH staining for COL4A1 containing tumor boundaries were obtained. H&E staining and annotated images of anatomical boundaries were also obtained for the corresponding samples.
GSEA
Gene set enrichment analysis (GSEA) is a bioinformatics analysis tool that is widely used to annotate and predict gene functions. To reveal the potential impact of COL4A1 on the prognosis of gliomas, an enrichment analysis was performed using GSEA version 3.0 software (https://www.gsea-msigdb.org/gsea/index.jsp). The important pathways that may be involved in the pathological mechanisms of glioma in the group with high and low COL4A1 expression were elucidated. In this analysis, 1,000 was set as the number of gene set permutations. The gene sets with a normal P<0.05 and false discovery rate (FDR) <0.25 were considered to be significantly enriched. FDR indicates false positive discovery rate that may be included and the nominal P-value describes the statistical significance of the enrichment score obtained for a subset of functional genes (22).
RT-qPCR analysis
To confirm COL4A1 expression in glioma patients and healthy controls, RT-qPCR analysis was performed. Tissues of glioma patients who underwent surgical resection in Henan Provincial People's Hospital (Zhengzhou, China) from March 2017 to June 2020 were collected. A total of 30 glioma tissues and 15 normal brain tissues were collected. Normal brain tissue was obtained from patients who underwent surgical resection for primary epilepsy. All patients signed an informed consent to use their organization for relevant research.
Three glioma cell lines (U87, U251 and LN229) and human-derived astrocytes (HA) were used to detect changes in the expression levels of COL4A1 by RT-qPCR. Total RNA was isolated from the tissues using Tri®Reagent (Sigma-Aldrich; Merck KGaA). RNA quality and quantity were determined using the NanoDrop One spectrophotometer (Thermo Fisher Scientific, Inc.). Complementary DNA (cDNA) was prepared using the Transcriptor First Stand cDNA Synthesis Kit (Roche Diagnostics) under the protocol of 55˚C for 30 min and 85˚C for 5 min. FastStart™ SYBR® Green (Roche Diagnostics) was used for qPCR experiment according to the manufacturer's protocols in the QuantStudio™ 3 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation at 95˚C for 10 min, followed by 40 cycles of denaturation at 95˚C for 10 sec, annealing and extension at 60˚C for 30 sec. GAPDH was used as the internal reference and the primer sequence was 5'-CAAGGTCATCCATGACAACTTTG-3' (forward) and 5'-GTCCACCACCCTGTTGCTGTAG-3' (reverse). The primer sequence for COL4A1 was 5'-CTGCCTGGAGGAGTTTAGAAG-3' (forward) and 5'-GAACATCTCGCTCCTCTCTATG-3' (reverse). The relative COL4A1 expression levels were determined using the 2-∆ΔCq method (23). An unpaired t-test was performed for analyzing the data of the two groups and a P-value <0.05 was considered to indicate statistical significance.
Cell culture and transfection
The human glioma cell line lines (U87-MG, U251 and LN229) and human-derived astrocytes (HA, cat. no. 3111C0001CCC000525; http://www.cellresource.cn/fdetail.aspx?id=2446) were purchased from the Chinese Academy of Sciences. Of these, U87-MG is a glioblastoma cell line of unknown origin (cat. no. TCHu138). All cells were authenticated by STR analysis and were cultured in DMEM plus 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured at 37˚C in a humidified atmosphere with 5% carbon dioxide in an incubator. Subsequent cell passages were performed every 2-3 days. The short interfering (si)RNA to COL4A1 was purchased from Shanghai GenePharma Co., Ltd. The siRNA sequence was as follows: sense, 5'-CCCACCUGGAAUUGUUAUATT-3' and antisense, 5'-UAUAACAAUUCCAGGUGGGTT-3'. The negative control (NC) siRNA sequence was sense, 5'-UUCUCCGAACGUGUCACGUTT-3' and antisense, 5'-ACGUGACACGUUCGGAGAATT-3'. Cells were transfected using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. After 24 h of transfection, the knockdown efficiency was detected by RT-qPCR technology.
Cell viability assay
Cell proliferation was quantified by an MTT assay. The transfected cells were plated onto 96-well plates (2,000 cells/well). Cell proliferation assays were performed at 12, 24, 48 and 72 h according to the manufacturer's protocol. First, 20 µl of MTT (5 mg/ml) was added to each well and after 4 h of incubation, the cell supernatant was discarded. Subsequently, 150 µl of DMSO was added to each well and the plate was agitated evenly for 15 min at room temperature. Finally, the absorbance of the solution in each well was measured at 490 nm on a spectrophotometer.
Cell immunofluorescence staining
The treated cells were fixed with 4% paraformaldehyde for 30 min, and subsequently, 0.1% Triton X-100 was used for cell permeation for 10 min at room temperature. After blocking with 5% BSA (Beijing Solarbio Science & Technology Co., Ltd.) for 30 min, cells were incubated with a primary antibody against Ki-67 (cat. no. ab15580; 1:1,000 dilution; Abcam) at 37˚C for 2 h. The cells were washed three times with PBS for 5 min each time and then incubated with DyLight 594-conjugated AffiniPure Goat Anti-rabbit IgG (cat. no. BA1142; 1:500 dilution; Boster Biological Technology) in a humidified atmosphere at room temperature in the dark for 1 h. Subsequently, DAPI was used to stain the nuclei. Finally, images were acquired under a fluorescence microscope.
Wound-healing assay
The LN229 cells were seeded in a six-well plate at 5x105 cells/well. Wounding was achieved by performing a linear scratch using a 200-µl sterile pipette tip. Cells that did not reattached were thoroughly washed away with PBS. After adding 2 ml of serum-free medium, the cells were incubated in a humid environment at 37˚C and 5% CO2 to allow the cells to migrate into the scratched area for 48 h. The periphery of the wound was observed under a general light microscope and images are acquired at 0, 24 and 48 h. The wound areas were analyzed using Image J software (v.1.52r; National Institutes of Health). Calculation formula of healing rate: Healed area/total wound area.
Statistical analysis
Data were analyzed by using SPSS version 22.0 (IBM Corp.) and R software (v.3.6.1). Student's t-test was used to compare the two groups. A Mann-Whitney U-test was used to determine the COL4A1 expression levels in the glioma and healthy brain tissue samples. Uni- and multivariate logistic Cox regression and the Kaplan-Meier method were used to determine the influence on COL4A1 expression levels on the overall survival rate. ROC method was used by R software (v.3.6.1) to detect whether COL4A1 may be used as an independent prognostic factor for glioma. Mann-Whitney U-test or Kruskal-Wallis test was used to examine the association between COL4A1 expression levels and the clinical data of glioma patients obtained from the databases. P<0.05 was considered to indicate a statistically significant difference.
Results
COL4A1 is abnormally highly expressed in gliomas
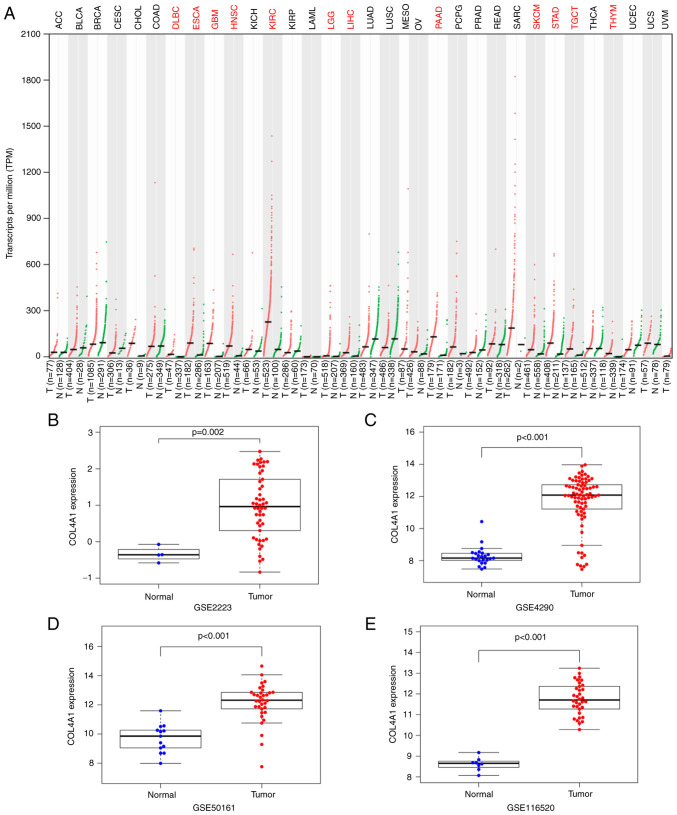
In order to detect changes in the expression levels of COL4A1 in tumor tissues, data on COL4A1 expression were first retrieved from the GEPIA database. The database contained 163 glioblastoma multiforme (GBM) tissue samples, 518 low-grade glioma (LGG) tissue samples and 207 normal brain tissues. It was revealed that the expression levels of COL4A1 were abnormally increased in various human tumor tissue types, including GBM and LGG (Fig. 1A).
Figure 1.
Expression levels of COL4A1 in glioma compared to normal brain tissue. (A) COL4A1 expression in various tumor types based on the GEPIA database. Red indicates that the expression level of COL4A1 in tumor tissues is significantly higher than that in normal tissues. Black indicates no difference in expression between tumor and normal tissues. Expression of COL4A1 in the Gene Expression Omnibus datasets (B) GSE2223, (C) GSE4290, (D) GSE50161, (E) GSE116520. COL4A1, collagen α-1 (IV) chain; T, tumor samples; N, normal samples; DLBC, lymphoid neoplasm diffuse large b-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THYM, thymoma.
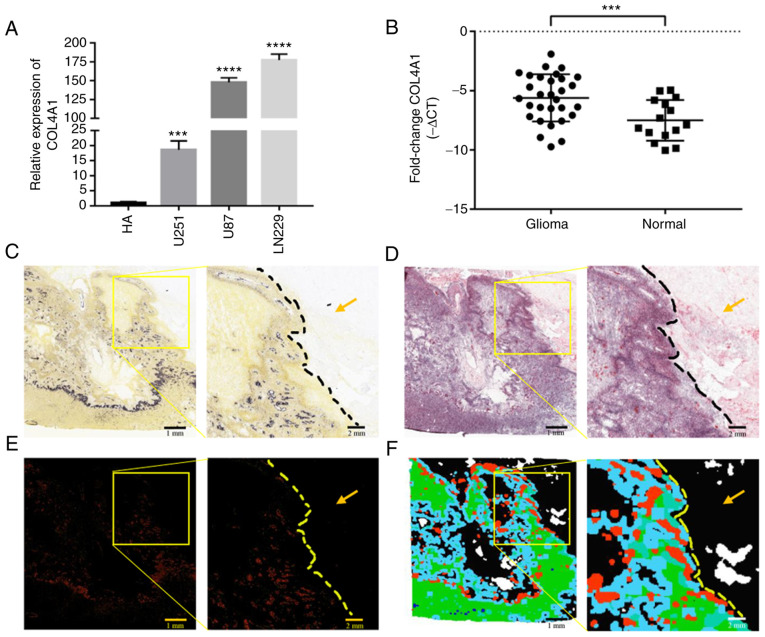
In order to improve the reliability of the results, four glioma gene chip datasets (GSE2223, GSE4290, GSE50161 and GSE116520) were further obtained from the GEO database to detect changes in the expression levels of COL4A1. The results indicated that the expression levels of COL4A1 were indeed abnormally increased in gliomas, as presented in Fig. 1B-E. The expression levels of COL4A1 were also detected in the glioma cell lines LN229, U87 and U251 and compared with those in HA cells by using RT-qPCR, and it was revealed that the expression levels of COL4A1 in the three glioma cell lines was also significantly increased compared with that in HA cells (Fig. 2A). Furthermore, the expression levels of COL4A1 were detected in 30 glioma samples and 15 healthy brain samples by RT-qPCR and it was indicated that COL4A1 was significantly increased in the glioma samples (Fig. 2B). The glioma group consisted of 16 males and 14 females, aged 23-72 years, with an average age of 48.51±13.77 years. Normal brain tissues from epileptic patients were obtained from 8 males and 7 females, aged 7-66 years, with an average age of 32.2±17.37 years.
Figure 2.
Expression levels of COL4A1 in glioma are abnormally increased. (A) The expression levels of COL4A1 in glioma cell lines were significantly higher than those in astrocytes based on RT-qPCR detection. (B) COL4A1 expression levels in glioma tissue samples were significantly higher than those in normal brain tissue according to RT-qPCR detection. ***P<0.001; ****P<0.0001. (C-F) COL4A1 detection in samples from the Ivy Glioblastoma Atlas database. Scale bars, 1 mm. (C) Histological section of glioblastoma under the light microscope. (D) Histological section of glioblastoma and adjacent tissue stained with H&E. (E) Detection of COL4A1 expression levels in tumor and paratumoral tissue by in situ hybridization. (F) Tumor feature annotation of histological section of glioblastoma and adjacent tissue; Green, Cellular Tumor; Black, non-tumor cell infiltration area; Red, Area of vascular proliferation; Blue, Perinecrotic zone; Gray-green and purple, Pseudopalisading cells. The yellow arrows represent non-neoplastic infiltrates. COL4A1, collagen α-1 (IV) chain; RT-qPCR, reverse transcription-quantitative PCR; HA, human-derived astrocytes.
Data aforementioned confirmed that the expression levels of COL4A1 were indeed increased in glioma. In order to make the data of this study more objective and comprehensive, microscopy images on the changes in the expression levels of COL4A1 in GBM were further obtained from the Ivy Glioblastoma Atlas. As presented in Fig. 2C-F, Fig. 2C and E shows the in situ hybridization experiment and related features. The expression of COL4A1 in the tumor area is higher than that in the non-tumor infiltrated area. Fig. 2D and F shows the image and anatomical tumor characteristics of H&E-stained tissue sections. In Fig. 2D, the tissue structure in the non-tumor infiltrated area is clear without obvious abnormality, whilst in the tumor area, the cell density is high and the morphology is fusiform. According to the anatomical region annotations in Fig. 2F, it can be determined that the upper right corner of the sample is the region of non-tumor cell infiltration and the left side is the tumor region. As shown in the figure, the expression level of COL4A1 is higher in tumor areas than in non-tumor cell infiltration areas.
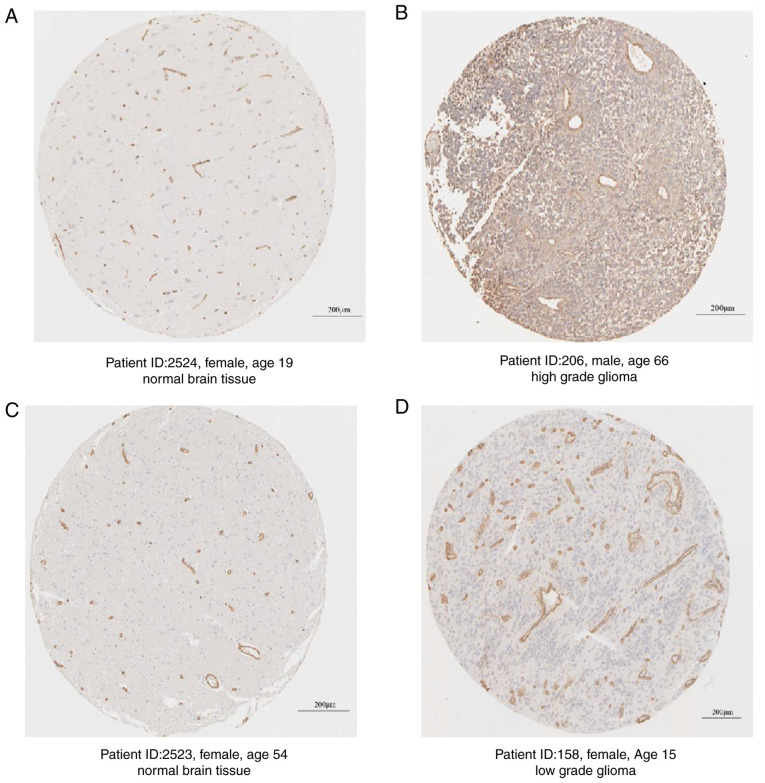
Furthermore, the protein expression levels of COL4A1 in glioma tissues and normal brain tissues were obtained from The Human Protein Atlas and the results also suggested that the expression of COL4A1 at the protein level was markedly higher than that in normal brain tissues from non-glioma patients (Fig. 3).
Figure 3.
Protein expression levels of COL4A1 compared between glioma tissues and normal brain tissues by immunohistochemical detection in samples from the human protein atlas. (A) Normal brain tissue; (B) high-grade glioma tissue; (C) Normal brain tissue; (D) low-grade glioma tissue. Yellow-brown areas represent positive staining results. Scale bars, 200 µm. COL4A1, collagen α-1 (IV) chain.
Association between COL4A1 expression and clinicopathological variables
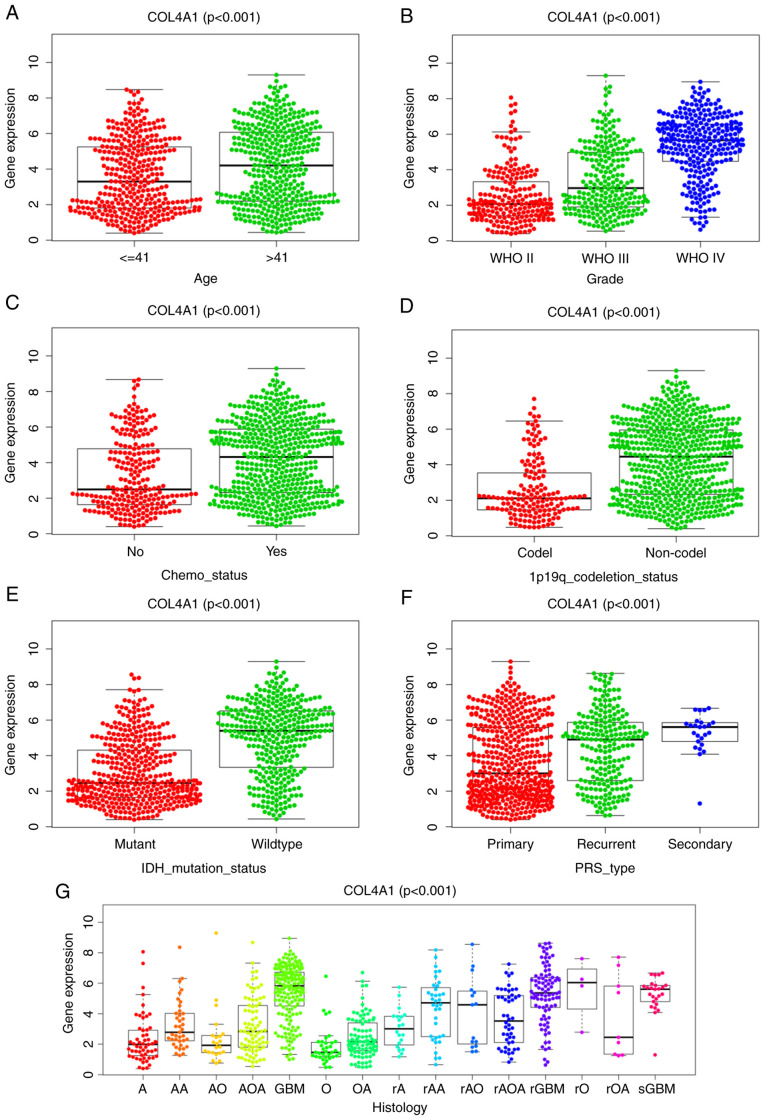
As presented in Fig. 4A-G, COL4A1 expression was significantly correlated with the clinical data obtained from the CGGA database. Increased expression levels of COL4A1 were positively associated with the histological type (P<0.001) and histological grade (P<0.001) of the tumor, patient age (P<0.001), PRS type (P<0.001) and chemotherapy status (P<0.001), while it was negatively associated with IDH mutation (P<0.001) and 1p19q co-deletion (P<0.001). The specific clinical information of the patients is presented in Table SI.
Figure 4.
In the CGGA database, COL4A1 expression in glioma according to different clinicopathological characteristics. Association of COL4A1 expression with (A) patient age (years), (B) tumor grade, (C) chemotherapy status, (D) 1p19q codel status, (E) IDH mutation status, (F) PRS type and (G) Histology. COL4A1, collagen α-1 (IV) chain; IDH, isocitrate dehydrogenase; codel, codeletion; WHO, World Health Organization; PRS, Primary, Recurrent, Secondary; A, strocytoma; AA, anaplastic astrocytoma; AO, anaplastic oligodendroglioma; AOA, anaplastic oligoastrocytoma; GBM, glioblastoma multiforme; O, oligodendroglioma; OA, oligoastrocytoma; rA, recurrence of strocytoma; rAA, recurrence of anaplastic astrocytoma; rAO, recurrence of anaplastic oligodendroglioma; rAOA, recurrence of anaplastic oligoastrocytoma; rGBM, recurrence of glioblastoma multiforme; rO, recurrence of oligodendroglioma; rOA, recurrence of oligoastrocytoma; sGBM, secondary glioblastoma.
Survival analysis and diagnostic value
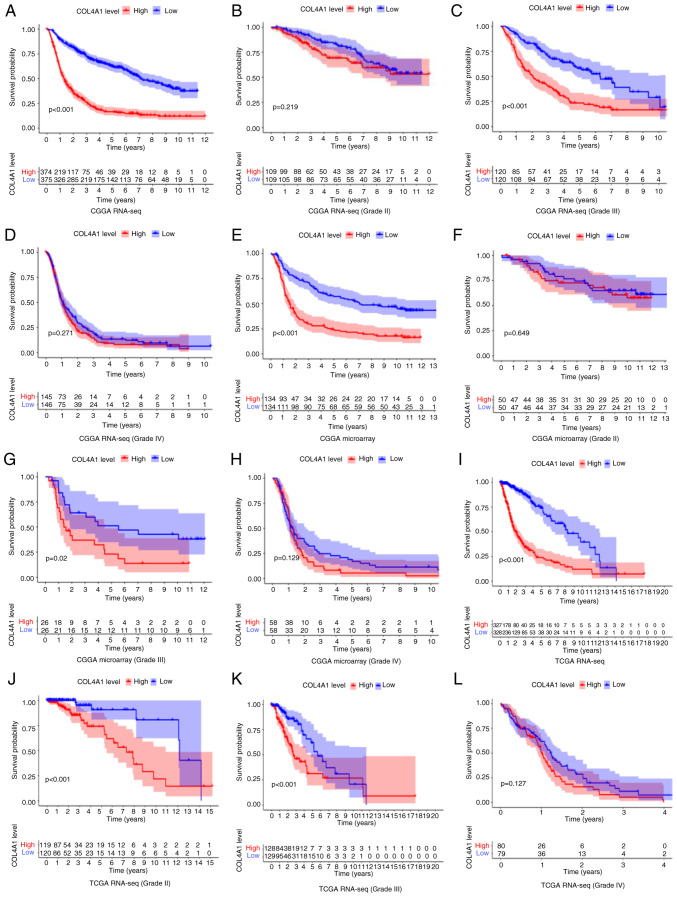
Although it has been clarified that the expression of COL4A1 was significantly increased in gliomas and has a close association with various clinical features of patients, the prognostic impact of COL4A1 on glioma patients remained elusive. Therefore, in order to investigate the effect of COL4A1 on the overall survival of glioma patients, three sets of data were analyzed to determine this (Fig. 5). The overall survival analysis of glioma patients of all grades in the CGGA RNA-seq, CGGA microarray and TCGA RNA-seq datasets is provided in Fig. 5A, E and I, respectively. The results obtained with the three datasets all suggested that the survival time of patients was significantly shortened in the COL4A1 high expression group (P<0.001). In the World Health Organization (WHO) grade III category, the three sets of data had consistent results in terms of high expression of COL4A1 being associated with a reduction in patient survival (Fig. 5C, G and K). Among the remaining data, only the TCGA RNA-seq (Grade II) results indicated that high expression of COL4A1 is associated with poor prognosis of patients (Fig. 5J), while the results obtained with the other datasets for Grade II and all datasets for Grade IV were not statistically significant (Fig. 5B, D, F, H and L). To further evaluate the diagnostic value of COL4A1 for patients with glioma, receiver operating characteristic curves were plotted based on the CGGA RNA-seq data (Fig. 6), using survival time and survival status as cutoff levels. The area under the curve values for COL4A1 to predict one-, three- and five-year survival were 0.725, 0.792 and 0.803, respectively, further reiterating the observation that the prognosis of patients with glioma and high COL4A1 expression levels was poor.
Figure 5.
Effect of COL4A1 expression levels on overall survival of glioma patients at all levels based on three different databases [(A-D) CGGA RNA-seq; (E-H) CGGA RNA-microarray; (I-L) TCGA RNA-seq]. COL4A1, collagen α-1 (IV) chain; RNA-seq, RNA sequencing; TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas.
Figure 6.
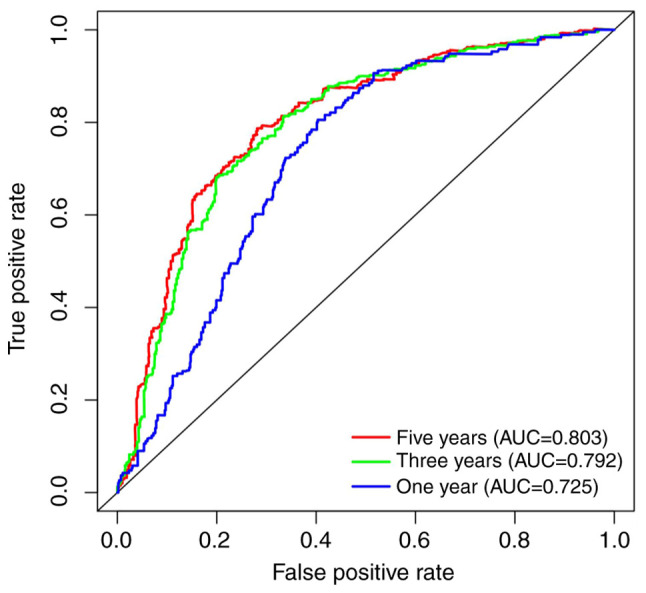
Diagnostic value of COL4A1 expression in glioma. The AUC of the receiver operating characteristic diagnostic accuracy curve for COL4A1 was 0.725 (1 year), 0.792 (3 years) and 0.803 (5 years). COL4A1, collagen α-1 (IV) chain; AUC, area under the curve.
Univariate and multivariate analyses
As presented in Fig. 7A, univariate regression analysis confirmed the result that prognosis was poor in patients with high COL4A1 expression levels in their glioma tissues [hazard ratio (HR): 1.343, 95% CI: 1.289-1.400, P<0.001]. Furthermore, PRS-type (HR: 2.123, 95% CI: 1.818-2.478, P<0.001), Histology (HR: 4.487, 95% CI: 3.695-5.449, P<0.001), high grade (HR: 2.883, 95% CI: 2.526-3.291, P<0.001), age (HR=1.624, 95% CI: 1.345-1.960, P<0.001), chemotherapy (HR: 1.647, 95% CI: 1.328-2.044, P<0.005), IDH mutation status (HR: 0.317, 95% CI: 0.262-0.384, P<0.001) and patients with 1p19q codeletion status (HR: 0.231, 95% CI: 0.169-0.315, P<0.001) significantly influenced the survival prognosis. Multivariate regression analysis was also performed (Fig. 7B), and the results indicated that COL4A1 expression levels (HR: 1.100, 95% CI: 1.046-1.157, P<0.001), PRS-type (HR: 1.891, 95% CI: 1.606-2.226, P<0.001), high grade (HR: 2.584, 95% CI: 1.886-3.540, P<0.001), age (HR: 1.241, 95% CI: 1.017-1.514, P=0.033), chemotherapy (HR: 0.688, 95% CI: 0.541-0.874, P=0.002), IDH mutation status (HR: 0.663, 95% CI: 0.522-0.841, P<0.001) and 1p19q codeletion status (HR: 0.408, 95% CI: 0.292-0.569, P<0.001) were independently associated with survival. These results indicated that COL4A1 is a potential prognostic factor for glioma and increased expression levels of COL4A1 may lead to poor prognosis.
Figure 7.
Univariate and multivariate regression analyses of prognosis in glioma patients. (A) Univariate regression analysis, used to analyze the independent impact of each factor on the prognosis of glioma patients. (B) Multivariate regression analysis. Considering the impact of all factors on the prognosis of glioma patients, the direction and degree of the impact of each factor change on the prognosis of patients were analyzed. COL4A1, collagen α-1 (IV) chain; IDH, isocitrate dehydrogenase; PRS, Primary, Recurrent, Secondary; chemo, chemotherapy; radio, radiotherapy.
GSEA identifies a COL4A1-associated signaling pathway
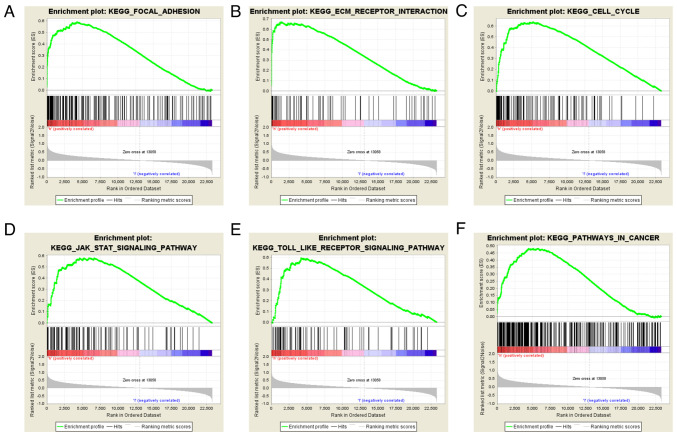
GSEA is a commonly used gene enrichment analysis tool, and in the present study, GSEA was performed to identify signaling pathways associated with glioma development and compare datasets with low and high COL4A1 expression. An FDR value of <0.05 and a NOM P-value of <0.05 were considered to indicate statistical significance. Among the factors analyzed, extracellular matrix (ECM) receptor interaction, JAK/STAT signaling pathway, cell cycle, focal adhesion and Toll-like receptor signaling pathway exhibited significant enrichment in the COL4A1 high expression group (Fig. 8; Table I). These results suggested that COL4A1 has an important role in the pathogenesis of glioma through these biological pathways.
Figure 8.
Enrichment plots from the gene set enrichment analysis. (A) Focal adhesion, (B) ECM receptor interaction, (C) cell cycle, (D) JAK/STAT signaling pathway, (E) Toll-like receptor signaling pathway and (F) pathways in cancer. ECM, extracellular matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Table I.
Gene set enrichment in the high collagen α-1 (IV) chain expression phenotype.
| Gene set name | NES | NOM Q-value | FDR Q-value |
|---|---|---|---|
| KEGG_FOCAL_ADHESION | 2.09 | <0.01 | <0.01 |
| KEGG_ECM_RECEPTOR_INTERACTION | 2.02 | <0.01 | <0.01 |
| KEGG_CELL_CYCLE | 1.93 | <0.01 | 0.02 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 1.83 | <0.01 | 0.05 |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 1.79 | <0.01 | 0.05 |
| KEGG_PATHWAYS_IN_CANCER | 1.81 | <0.01 | 0.06 |
NES, normalized enrichment score; NOM, nominal; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix.
Knockdown of COL4A1 inhibits the malignant behavior of LN229 cells
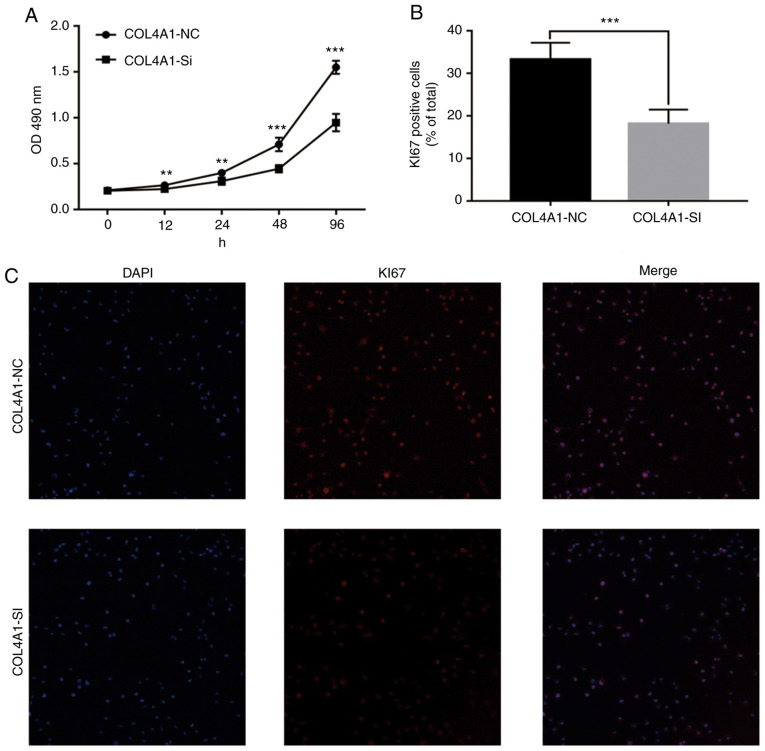
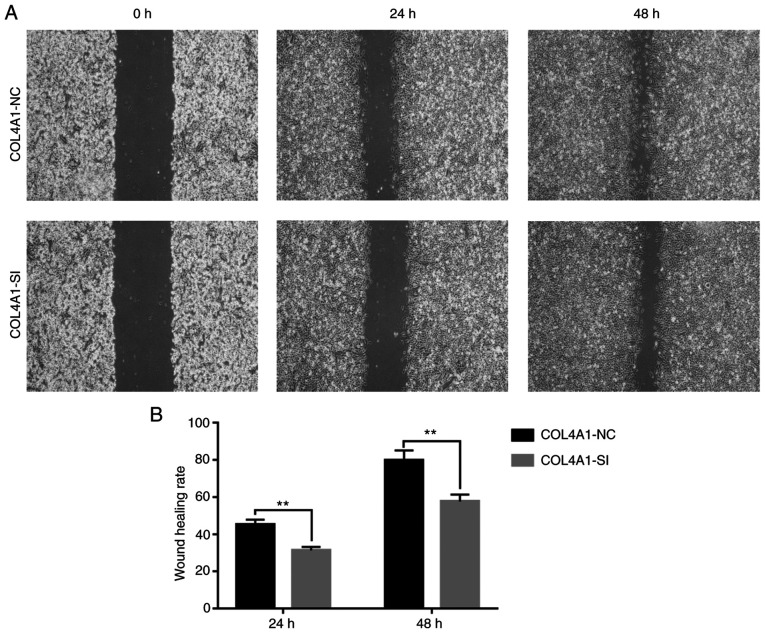
To assess the effect of COL4A1 on the malignant behavior of glioma cell lines, siRNA was used to interfere with the expression of COL4A1. The knockdown efficacy is provided in Fig. S1. An MTT assay then indicated that the proliferation ability of cells in the experimental group was significantly reduced (Fig. 9A). Furthermore, immunofluorescence staining suggested that the expression levels of the cell proliferation marker KI67 were significantly lower than those in the control group (Fig. 9B and C). Finally, a wound-healing assay indicated that compared with the control group, the scar healing ability in the experimental group was significantly reduced (Fig. 10).
Figure 9.
Effect of knockdown of COL4A1 on the expression levels of the proliferation marker KI67 in LN229 cells based on immunofluorescence detection. (A) Effect of knockdown of COL4A1 on the cell proliferation ability of LN229 cells determined by MTT assay. (B) Quantified results of Ki-67 staining are provided. (C) Representative fluorescence microscopy images for Ki-67 staining and magnification, x10. **P<0.01; ***P<0.001. COL4A1, collagen α-1 (IV) chain; NC, negative control; SI, short interfering RNA.
Figure 10.
Effect of knockdown of COL4A1 expression on the migration ability of LN229 cells determined by a wound-healing assay. (A) Representative microscopy images and magnification, x4. (B) Quantified results are provided. **P<0.01. COL4A1, collagen α-1 (IV) chain; NC, negative control; SI, short interfering RNA.
Discussion
Various studies have indicated that COL4A1 is involved in the pathological process of a variety of cancer types and has a key role in their occurrence, development and metastasis (24-26). In addition, certain studies suggested that COL4A1 is a novel oncogenic transcription regulator that is closely associated with the prognosis of glioma (27). The present study aimed to determine the role of COL4A1 in glioma using a variety of different data types and experimental techniques, including sequencing data, chip data, RT-PCR, in situ hybridization and immunohistochemistry. The results indicated that the expression levels of COL4A1 in glioma tissues were significantly higher than those in normal brain tissues. This is supported by previous reports that COL4A1 expression is increased in malignant tumors (16,24,25,28). For instance, COL4A1 expression is also increased in numerous types of malignant tumor tissue (such as esophageal squamous cell carcinoma, renal cell carcinoma, urothelial carcinoma of the bladder and breast cancer) and has an important impact on the occurrence and development of cancer (16,20,24,28).
Of note, COL4A1 has also been reported to be increased in LGG. Jiang et al (27) indicated that COL4A1 has synergistic effects with COL1A2, COL3A1, COL1A1, COL4A2 and COL5A2 and is associated with poor prognosis of patients. However, that study only relied on a single data source and only revealed the expression changes of COL4A1 in LGG and the prognosis of patients, but did not fully explain the role of COL4A1 in different grades of gliomas. In addition, since Jiang et al (27) did not explore the clinical diagnostic value of COL4A1, whether COL4A1 was associated with clinical characteristics of glioma patients remains unknown. Finally, the six genes reported by the authors were suggested to have regulatory effects in LGG, but in the mechanistic analysis, certain results indicated no association with the occurrence of cancer, and further investigation is required to empirically determine their effect. Although the present study and that performed by Jiang et al (27) are based on the analysis of public data, the present study is the first to explore the role of COL4A1 in glioma. In addition, the present study also performed experiments using glioma cells and clinical samples.
The present study objectively confirmed the increased expression level of COL4A1 in glioma tissues through public database data from GEPIA, GEO, Ivy and HPA in addition to tissue and cell lines. The relationship between COL4A1 and the prognostic diagnostic value and clinical characteristics of glioma patients also warrants further exploration. Therefore, the CGGA database was used to obtain clinically relevant traits of glioma patients and the association of the expression of COL4A1 with clinicopathological features was assessed through a Mann-Whitney U-test or Kruskal-Wallis test. Of note, the expression level of COL4A1 increased with the WHO grade and was significantly increased in recurrent and secondary glioma. Previous studies have also indicated that abnormally high expression of COL4A1 in gastric carcinoma may lead to decreased overall survival of patients and has a close relationship with tumor recurrence (16).
Furthermore, the present study revealed for the first time that COL4A1 expression, particularly in WHO grade III tumors, is negatively associated with patient prognosis, which confirms that COL4A1 is associated with poor prognosis in glioma patients. In addition, COL4A1 was revealed to be an independent risk factor leading to poor prognosis in the multivariate analysis. The above results suggest that COL4A1 may be involved in the malignant progression of glioma. Therefore, an MTT assay, KI67 staining and wound-healing assay further confirmed that knockdown of COL4A1 inhibited the proliferation and migration ability of glioma cells. Taken together, the present results suggest that COL4A1 is indeed an oncogene in gliomas, and a similar role has been reported in breast cancer (14).
To further understand the role of COL4A1 in the development and progression of gliomas and the cellular pathways involved, a functional annotation by GSEA was performed. The results suggested that in the high-expression COL4A1 group, focal adhesion, ECM receptor interaction, JAK/STAT signaling pathway, Toll-like receptor signaling pathway and cell cycle were significantly enriched. Focal adhesion is overexpressed in numerous tumor types and has an important role in cell cycle regulation, adhesion, migration and formation through various signaling pathways (29-31). Compared with normal brain tissue, FAK expression was higher in glioma cells, and it was negatively correlated with survival time (32). Some studies have shown that focal adhesion proteins and proline-containing proteins were linked together by a tyrosine phosphatase to form a complex, which in turn controls the invasion of glioblastoma cells (33). Another study has also indicated that focal adhesion kinase promotes the formation of small blood vessels in high-grade malignant gliomas (34). The occurrence, development, invasion and metastasis of malignant tumors are frequently accompanied by changes in the ECM and its cell surface receptor expression. Previous studies have indicated that ECM components are connected to glioma cells through hyaluronic acid receptor molecules, and their interaction is mainly controlled by glycosidase and protease. These enzymes regulate the adhesion of the ECM to the intracellular matrix and also activate growth molecules and chemokines in glioma cells (35). The JAK/STAT signaling pathway is a common signal transduction pathway for numerous cytokines. It is widely involved in cell proliferation, differentiation, apoptosis and inflammation, and promotes the occurrence and development of various diseases, including inflammatory diseases, lymphoma, leukemia and formation of solid tumors. Henrik Heiland et al (36) indicated that JAK/STAT pathway activation was significantly increased in glioma samples compared to healthy tissue samples, which is consistent with the present results. Furthermore, several studies suggested that inhibition of the JAK/STAT signaling pathway may inhibit the proliferation, migration and invasion of glioma cells (37-39). Toll-like receptor signaling is a well-known cancer pathway. Numerous human tumors and tumor cell lines express Toll-like receptors, which have an important role in the progression from precancerous lesions to tumors. Hu et al (40) indicated that versican released by glioma cells promotes tumor proliferation through Toll-like receptor 2 signal transduction and expression of membrane type-1 matrix metalloproteinase. It has been suggested that deregulation of miRNAs in glioma cells may promote tumor proliferation by directly acting on key cell-cycle regulators. For instance, lncRNA breast cancer anti-estrogen resistance 4, the earliest anti-estrogen resistance found in breast cancer, was reported to accelerate the progression of glioma cells by affecting the cell cycle and inhibiting apoptosis (41). Therefore, COL4A1 promoting the activation of oncogenic pathways may be a major cause of poor prognosis in glioma patients.
Although a large amount of data was analyzed in the present study to demonstrate that COL4A1, as a novel oncogene, is involved in the pathological processes of glioma, there are various limitations. First, there are certain shortcomings in the public database platform, including the lack of clinical characteristics for various patients, and it was not possible to include the details of clinical treatment for all patients. However, the database has unique advantages, such as multi-center cohorts, a large sample size and ethnic diversity, which may provide information that may improve the prediction, diagnosis, treatment and monitoring of tumors. Furthermore, COL4A1 as a novel oncogene in glioma may be involved in its molecular mechanisms. By using GSEA, it was indirectly revealed that multiple signaling pathways of COL4A1 are involved in the pathological processes of glioma. However, it is well known that a gene may have a variety of roles in organisms. The present study provides an index for further research to study the role of COL4A1 as an oncogene in the pathological processes of glioma.
In conclusion, through a series of analytical methods, the present study confirmed that abnormally high expression of COL4A1 is an independent predictor of poor prognosis in glioma patients, and possible molecular pathways underlying its oncogenic effect were elucidated. The present study revealed part of the complex pathological process of glioma from the perspective of molecular biology and provided COL4A1 as an effective potential diagnostic and therapeutic target.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by the 2018 Henan Provincial Medical Science and Technology Tackling Program Provincial-ministerial Co-construction Project (grant no. SBGJ2018076) and the Central Plains Thousand Talents Plan of Henan Province (grant no. ZYQR201912122).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HBW, YZG and ZDL conceived and designed the study. AL is responsible for collecting clinical samples and collating raw data from public databases. JLW, JTL, BFL and XYL performed the experiments. BZ, BP and LYL analyzed the data and prepared figures and/or tables. YZG contributed reagents/materials/analysis tools, wrote and reviewed drafts of the manuscript and approved the final manuscript. HBW and ZDL can authenticate the raw data in the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in the study were in accordance with the ethical standards of the Ethical Committee of the Henan Provincial People's Hospital (Zhengzhou, China). The experimental scheme was also approved by the Ethics Committee of Henan Provincial People's Hospital (Zhengzhou, China). All patients signed an informed consent and agreed to use their glioma tissues or normal brain tissues after surgical resection for relevant studies.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
References
- 1.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. NeuroOncol. 2013;15 (Suppl 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 3.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, et al. The epidemiology of glioma in adults: A ‘state of the science’ review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claus EB, Black PM. Survival rates and patterns of care for patients diagnosed with supratentorial low-grade gliomas: Data from the SEER program, 1973-2001. Cancer. 2006;106:1358–1363. doi: 10.1002/cncr.21733. [DOI] [PubMed] [Google Scholar]
- 5.Yang P, Wang Y, Peng X, You G, Zhang W, Yan W, Bao Z, Wang Y, Qiu X, Jiang T. Management and survival rates in patients with glioma in China (2004-2010): A retrospective study from a single-institution. J Neurooncol. 2013;113:259–266. doi: 10.1007/s11060-013-1103-9. [DOI] [PubMed] [Google Scholar]
- 6.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: A review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 7.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 8.Lohr J, Ratliff T, Huppertz A, Ge Y, Dictus C, Ahmadi R, Grau S, Hiraoka N, Eckstein V, Ecker RC, et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-β. Clin Cancer Res. 2011;17:4296–4308. doi: 10.1158/1078-0432.CCR-10-2557. [DOI] [PubMed] [Google Scholar]
- 9.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, et al. Stem cell-related ‘self-renewal’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 10.Hale JS, Otvos B, Sinyuk M, Alvarado AG, Hitomi M, Stoltz K, Wu Q, Flavahan W, Levison B, Johansen ML, et al. Cancer stem cell-specific scavenger receptor CD36 drives glioblastoma progression. Stem Cells. 2014;32:1746–1758. doi: 10.1002/stem.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollner R, Schmidt C, Fischer G, Kühn K, Pöschl E. Cooperative and competitive interactions of regulatory elements are involved in the control of divergent transcription of human Col4A1 and Col4A2 genes. FEBS Lett. 1997;405:31–36. doi: 10.1016/s0014-5793(97)00152-x. [DOI] [PubMed] [Google Scholar]
- 12.Yurchenco PD. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011;3(a004911) doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labelle-Dumais C, Schuitema V, Hayashi G, Hoff K, Gong W, Dao DQ, Ullian EM, Oishi P, Margeta M, Gould DB. COL4A1 mutations cause neuromuscular disease with tissue-specific mechanistic heterogeneity. Am J Hum Genet. 2019;104:847–860. doi: 10.1016/j.ajhg.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin R, Shen J, Zhang T, Liu Q, Liao C, Ma H, Li S, Yu Z. The highly expressed COL4A1 genes contributes to the proliferation and migration of the invasive ductal carcinomas. Oncotarget. 2017;8:58172–58183. doi: 10.18632/oncotarget.17345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitzler TM, Schneider R, Kohl S, Kolvenbach CM, Connaughton DM, Dai R, Mann N, Nakayama M, Majmundar AJ, Wu CW, et al. COL4A1 mutations as a potential novel cause of autosomal dominant CAKUT in humans. Hum Genet. 2019;138:1105–1115. doi: 10.1007/s00439-019-02042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, Wang NN, Chang X, Wang SL, Wang LS, Yao J, Li ZS, Bai Y. Bioinformatics analysis suggests that COL4A1 may play an important role in gastric carcinoma recurrence. J Dig Dis. 2019;20:391–400. doi: 10.1111/1751-2980.12758. [DOI] [PubMed] [Google Scholar]
- 17.Lin S, Xia C, He S, Yang J, Li H, Zheng J, Liu M, You C. Genetic variations of the COL4A1 gene and intracerebral hemorrhage risk: A case-control study in a Chinese han population. World Neurosurg. 2018;112:e527–e533. doi: 10.1016/j.wneu.2018.01.072. [DOI] [PubMed] [Google Scholar]
- 18.Raza ST, Abbas S, Eba A, Karim F, Wani IA, Rizvi S, Zaidi A, Mahdi F. Association of COL4A1 (rs605143, rs565470) and CD14 (rs2569190) genes polymorphism with coronary artery disease. Mol Cell Biochem. 2018;445:117–122. doi: 10.1007/s11010-017-3257-9. [DOI] [PubMed] [Google Scholar]
- 19.Saskin A, Sillon G, Palfreeman N, Buhas D. COL4A1/2 CNVs and cerebral small vessel disease: Narrowing in on the critical chromosomal region. Neurology. 2018;90:1026–1028. doi: 10.1212/WNL.0000000000005601. [DOI] [PubMed] [Google Scholar]
- 20.Chen FF, Zhang SR, Peng H, Chen YZ, Cui XB. Integrative genomics analysis of hub genes and their relationship with prognosis and signaling pathways in esophageal squamous cell carcinoma. Mol Med Rep. 2019;20:3649–3660. doi: 10.3892/mmr.2019.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Z, Li L, Fang Q, Zhang Y, Hu X, Qian Y, Huang P. Analysis of dynamic molecular networks for pancreatic ductal adenocarcinoma progression. Cancer Cell Int. 2018;18(214) doi: 10.1186/s12935-018-0718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) methods. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Wang SM, Chen PM, Sung YW, Huang WC, Huang HS, Chu PY. Effect of COL4A1 expression on the survival of neoadjuvant chemotherapy breast cancer patients. J Oncol. 2020;2020(5209695) doi: 10.1155/2020/5209695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Jin H, Hu J, Li X, Ruan H, Xu H, Wei L, Dong W, Teng F, Gu J, et al. COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK-Src signaling. J Exp Clin Cancer Res. 2020;39(148) doi: 10.1186/s13046-020-01650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu F, Li F, Lin X, Xu F, Cui RR, Zhong JY, Zhu T, Shan SK, Liao XB, Yuan LQ, Mo ZH. Exosomes increased angiogenesis in papillary thyroid cancer microenvironment. Endocr Relat Cancer. 2019;26:525–538. doi: 10.1530/ERC-19-0008. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y, He J, Guo Y, Tao H, Pu F, Li Y. Identification of genes related to low-grade glioma progression and prognosis based on integrated transcriptome analysis. J Cell Biochem. 2020;121:3099–3111. doi: 10.1002/jcb.29577. [DOI] [PubMed] [Google Scholar]
- 28.Miyake M, Morizawa Y, Hori S, Tatsumi Y, Onishi S, Owari T, Iida K, Onishi K, Gotoh D, Nakai Y, et al. Diagnostic and prognostic role of urinary collagens in primary human bladder cancer. Cancer Sci. 2017;108:2221–2228. doi: 10.1111/cas.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aboubakar Nana F, Vanderputten M, Ocak S. Role of focal adhesion kinase in small-cell lung cancer and its potential as a therapeutic target. Cancers (Basel) 2019;11(1683) doi: 10.3390/cancers11111683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giaginis CT, Vgenopoulou S, Tsourouflis GS, Politi EN, Kouraklis GP, Theocharis SE. Expression and clinical significance of focal adhesion kinase in the two distinct histological types, intestinal and diffuse, of human gastric adenocarcinoma. Pathol Oncol Res. 2009;15:173–181. doi: 10.1007/s12253-008-9120-2. [DOI] [PubMed] [Google Scholar]
- 31.Ozkal S, Paterson JC, Tedoldi S, Hansmann ML, Kargi A, Manek S, Mason DY, Marafioti T. Focal adhesion kinase (FAK) expression in normal and neoplastic lymphoid tissues. Pathol Res Pract. 2009;205:781–788. doi: 10.1016/j.prp.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Ding L, Sun X, You Y, Liu N, Fu Z. Expression of focal adhesion kinase and phosphorylated focal adhesion kinase in human gliomas is associated with unfavorable overall survival. Transl Res. 2010;156:45–52. doi: 10.1016/j.trsl.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Morales JE, Guerrero PA, Sun H, McCarty JH. PTPN12/PTP-PEST regulates phosphorylation-dependent ubiquitination and stability of focal adhesion substrates in invasive glioblastoma cells. Cancer Res. 2018;78:3809–3822. doi: 10.1158/0008-5472.CAN-18-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haskell H, Natarajan M, Hecker TP, Ding Q, Stewart J Jr, Grammer JR, Gladson CL. Focal adhesion kinase is expressed in the angiogenic blood vessels of malignant astrocytic tumors in vivo and promotes capillary tube formation of brain microvascular endothelial cells. Clin Cancer Res. 2003;9:2157–2165. [PubMed] [Google Scholar]
- 35.Ferrer VP, Moura Neto V, Mentlein R. Glioma infiltration and extracellular matrix: Key players and modulators. Glia. 2018;66:1542–1565. doi: 10.1002/glia.23309. [DOI] [PubMed] [Google Scholar]
- 36.Henrik Heiland D, Ravi VM, Behringer SP, Frenking JH, Wurm J, Joseph K, Garrelfs NWC, Strähle J, Heynckes S, Grauvogel J, et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun. 2019;10(2541) doi: 10.1038/s41467-019-10493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Peng X, Zhang J, Wang Z, Meng J, Cen B, Ji A, He S. LncRNA-135528 inhibits tumor progression by up-regulating CXCL10 through the JAK/STAT pathway. Apoptosis. 2018;23:651–666. doi: 10.1007/s10495-018-1482-7. [DOI] [PubMed] [Google Scholar]
- 38.Xu CH, Liu Y, Xiao LM, Chen LK, Zheng SY, Zeng EM, Li DH, Li YP. Silencing microRNA-221/222 cluster suppresses glioblastoma angiogenesis by suppressor of cytokine signaling-3-dependent JAK/STAT pathway. J Cell Physiol. 2019;234:22272–22284. doi: 10.1002/jcp.28794. [DOI] [PubMed] [Google Scholar]
- 39.Zhang P, Chen FZ, Jia QB, Hu DF. Upregulation of microRNA-133a and downregulation of connective tissue growth factor suppress cell proliferation, migration, and invasion in human glioma through the JAK/STAT signaling pathway. IUBMB Life. 2019;71:1857–1875. doi: 10.1002/iub.2126. [DOI] [PubMed] [Google Scholar]
- 40.Hu F, Dzaye O, Hahn A, Yu Y, Scavetta RJ, Dittmar G, Kaczmarek AK, Dunning KR, Ricciardelli C, Rinnenthal JL, et al. Glioma-derived versican promotes tumor expansion via glioma-associated microglial/macrophages Toll-like receptor 2 signaling. Neuro Oncol. 2015;17:200–210. doi: 10.1093/neuonc/nou324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei L, Yi Z, Guo K, Long X. Long noncoding RNA BCAR4 promotes glioma cell proliferation via EGFR/PI3K/AKT signaling pathway. J Cell Physiol. 2019;234:23608–23617. doi: 10.1002/jcp.28929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.