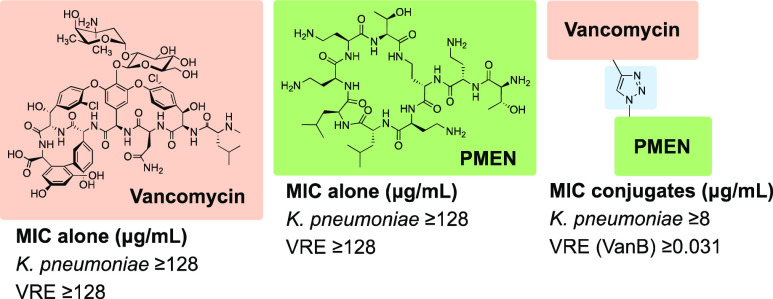
Abstract
Vancomycin functions by binding to lipid II, the penultimate bacterial cell wall building block used by both Gram-positive and Gram-negative species. However, vancomycin is generally only able to exert its antimicrobial effect against Gram-positive strains as it cannot pass the outer membrane (OM) of Gram-negative bacteria. To address this challenge, we here describe efforts to conjugate vancomycin to the OM disrupting polymyxin E nonapeptide (PMEN) to yield the hybrid “vancomyxins”. In designing these hybrid antibiotics, different spacers and conjugation sites were explored for connecting vancomycin and PMEN. The vancomyxins show improved activity against Gram-negative strains compared with the activity of vancomycin or vancomycin supplemented with PMEN separately. In addition, the vancomyxins maintain the antimicrobial effect of vancomycin against Gram-positive strains and, in some cases, show enhanced activity against vancomycin-resistant strains. The hybrid antibiotics described here have reduced nephrotoxicity when compared with clinically used polymyxin antibiotics. This study demonstrates that covalent conjugation to an OM disruptor contributes to sensitizing Gram-negative strains to vancomycin while retaining anti-Gram-positive activity.
Keywords: Gram-negative, outer membrane, vancomycin, polymyxin, Gram-positive, click ligation
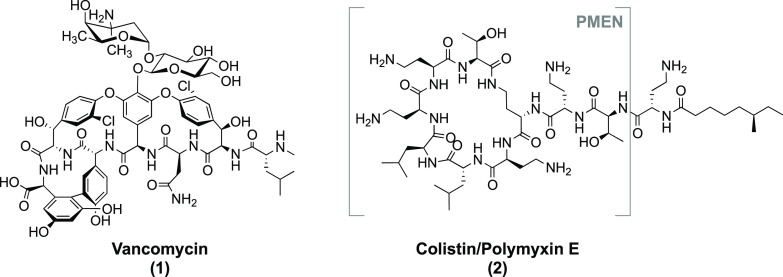
Vancomycin (Figure 1) is the most prominent clinically used glycopeptide antibiotic and exhibits potent activity against Gram-positive bacteria. It functions by inhibiting cell-wall biosynthesis by targeting the peptidoglycan precursor lipid II and specifically by binding the d-Ala-d-Ala terminus of the lipid II pentapeptide via a network of five hydrogen bonds. In binding to this peptidoglycan precursor, vancomycin prevents cell-wall polymerization by bacterial transpeptidases and transglycosylases which leads to decreased bacterial cell-wall integrity, eventually resulting in lysis of the bacterial cell.1,2 Furthermore, binding of vancomycin to lipid II is enhanced by cooperative dimerization which increases the binding affinity of vancomycin to lipid II and enhances its antimicrobial activity.3−5 While lipid II is also present in Gram-negative bacteria, vancomycin is unable to access it due to the presence of the additional outer membrane (OM) found in Gram-negatives.6 The OM is characterized by an inner leaflet of phospholipids and an outer leaflet decorated by lipopolysaccharide (LPS).7 Notably, the ability of vancomycin to also bind to Gram-negative lipid II from E. coli was confirmed by Shlaes and co-workers. Furthermore, this study suggested that defects in the LPS core can revert resistance of Gram-negative strains to large hydrophilic molecules such as vancomycin.6 Additionally, in a recent investigation, Bardoel and co-workers showed that serum can sensitize multidrug resistant (MDR) K. pneumoniae to vancomycin, a process facilitated by the membrane attack complex (MAC) of the complement system found in human serum. The MAC forms pores in the OM causing disruption, allowing otherwise Gram-positive specific antimicrobials to also exert their action against Gram-negative strains.8 These studies, and others carried out in the same area, highlight the potential of vancomycin to be effective against Gram-negative bacteria when the integrity of the OM is compromised.
Figure 1.
Structures of clinically used antibiotics vancomycin and colistin (polymyxin E), with the colistin-derived polymyxin E nonapeptide (PMEN) also indicated.
Different strategies to sensitize Gram-negative bacteria to antibiotics which typically only work against Gram-positive pathogens have been explored in the literature.9−11 The two main approaches used most often rely on (a) covalent attachment of OM-disrupting or OM-bypassing moieties and (b) coadministration with “adjuvants”, which can either affect the OM integrity or impair the bacteria’s efflux system.7,12,13 Previously described covalent conjugates include those reported by Miller and co-workers wherein vancomycin was linked to an iron sequestering siderophore mimetic to yield hybrids with slightly reduced activity against Gram-positive strains but with enhanced activity toward a hypersensitive strain of Pseudomonas aeruginosa under iron depleted conditions.14 More recently, the group of Haldar reported a lipophilic cationic vancomycin analogue, VanQAmC10, which was shown to be bactericidal against MDR A. baumannii.15 Another recent vancomycin derivative, developed by the groups of Wender and Cegelski, involves the introduction of an arginine-amide moiety at the vancomycin C-terminus, significantly enhancing activity against E. coli (MIC 8–16 μM) including resistant strains.16,17 Notably, this arginine-vancomycin conjugate was demonstrated to successfully reduce bacterial burden >6-log fold compared to vehicle and vancomycin in a murine thigh E. coli infection model.16 In addition to such covalent approaches to enhance anti-Gram-negative activity, agents capable of potentiating or synergizing with Gram-positive-specific antibiotics also present an attractive option. In this regard, many OM-disrupting cyclic or linear cationic peptides have been reported to sensitize Gram-negative pathogens to anti-Gram-positive antibiotics including vancomycin.10,18,19
Among the most notable OM disrupting agents are the so-called polymyxin nonapeptides which are derived from the clinically used antibiotics polymyxin B and polymyxin E (colistin). Readily obtained by enzymatic degradation of the full-length antibiotic, the cyclic nonapeptides lack the fatty acyl tail and N-terminal Dab residue present in the parent polymyxins (Figure 1). Due to its associated (nephro)toxicity when administered systemically, colistin has traditionally only been used as an antibiotic of last resort.20 However, given increasing rates of resistance, the use of colistin is now on the rise.20 By comparison, the polymyxin nonapeptides are significantly less toxic than the parent compounds21,22 but still maintain the capacity to bind to Gram-negative bacteria by recognition of the Lipid A unit of LPS.23,24 Given their high positive charge, polymyxin nonapeptides displace the divalent cations responsible for stabilizing membrane packing in the Gram-negative OM25 resulting in disruption of the OM.20 Notably, while polymyxin nonapeptides retain little-to-none of the activity of the parent antibiotic,26 they function effectively as synergistic agents and can improve the activity of otherwise Gram-positive specific antibiotics including vancomycin.18,19
To date, a small number of studies have explored the effect of conjugating polymyxins to antibiotic agents with the aim of using the covalently attached OM disruptor as an adjuvant for the antimicrobial agent. Generally speaking, these studies have focused on conjugation with Gram-negative active antibiotics. In a recent example, Schweizer and co-workers described the ligation of full-length polymyxin B to the aminoglycoside tobramycin.27 The resulting hybrid did not outperform either polymyxin or tobramycin in direct activity but interestingly did potentiate other antibiotics toward several P. aeruginosa strains, including MDR isolates.27 In another even newer development, researchers at Polyphor described bicyclic hybrids comprising a monocyclic β-hairpin peptidomimetic of protegrin I and PMEN.28 While neither monocyclic peptide exhibits significant activity on its own, the bicyclic hybrids demonstrated extremely potent activity both in vitro and in mouse models of infection with a range of Gram-negative pathogens. Notably, these bicyclic constructs are proposed to target the extracellular part of the OM protein BamA, thereby avoiding the need to pass the OM.28
Given previous reports showing that covalent attachment of siderophores, LPS binding moieties, or positively charged moieties to vancomycin can lead to improved antimicrobial activity against Gram-negative strains,15−17,29,30 we hypothesized that conjugation of vancomycin to the highly positively charged OM disruptor PMEN could sensitize Gram-negative strains. Our interest in exploring vancomycin-PMEN conjugates was further spurred given that OM disruption has also previously been demonstrated to enable anti-Gram-negative activity for vancomycin.6,8,10,18,19 Also of note are recent reports showing that the covalent attachment of cationic moieties to vancomycin is an effective means to resensitize clinically relevant vancomycin-resistant Gram-positive strains.15,31−35 For these reasons, we anticipated that vancomycin-PMEN conjugates might exhibit enhanced activity toward drug-resistant Gram-positive strains as well. Here, we report the synthesis and evaluation of the vancomyxins: a new class of vancomycin-PMEN hybrid antibiotics. The antimicrobial activities of the vancomyxins against Gram-positive and Gram-negative bacteria (including drug-resistant clinical isolates) as well as an assessment of their toxicity toward eukaryotic cells is here reported.
Results and Discussion
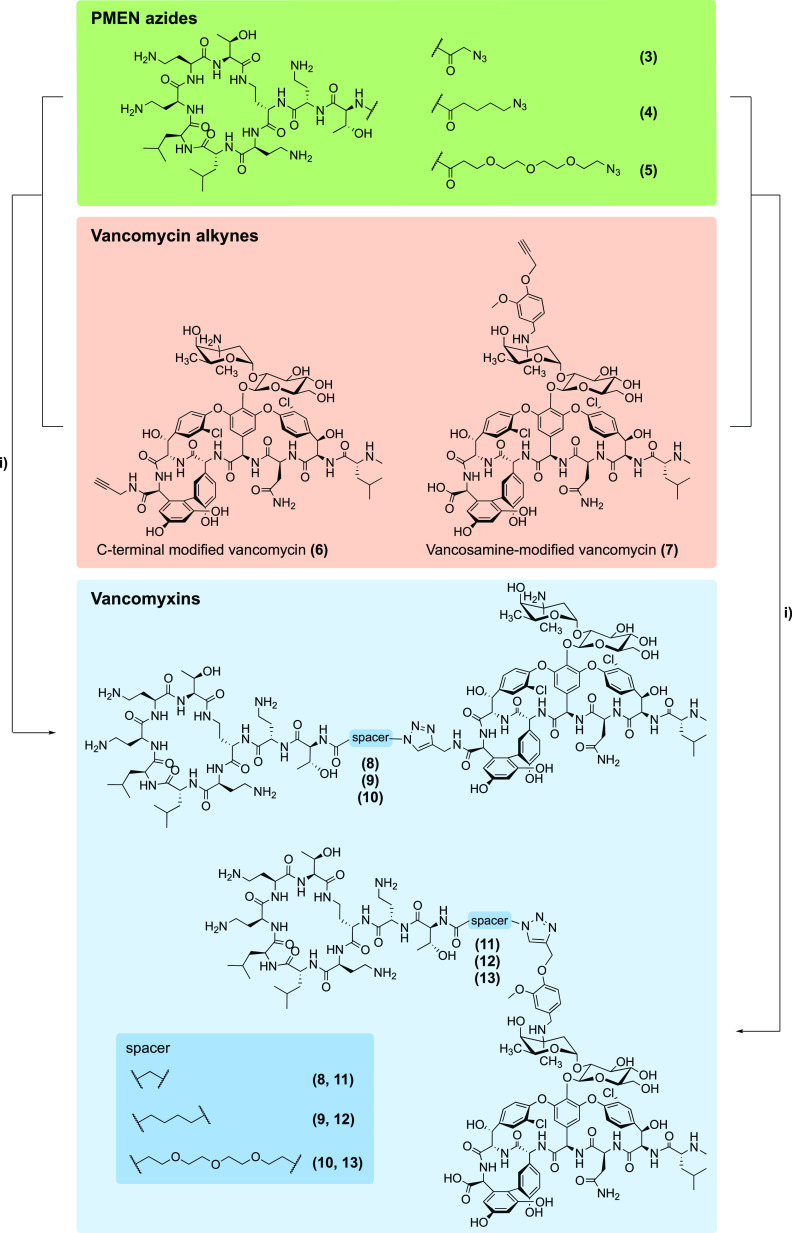
As a strategy for preparing the vancomycin/PMEN conjugates we envisioned the use of so-called “click chemistry”, wherein complementary azide and alkyne containing precursors are covalently linked by means of the well-established copper-catalyzed azide–alkyne cycloaddition reaction.36−38 In pursuing this approach, we opted to add the required azide handle to the N-terminus of the PMEN moiety while the alkyne functionality was incorporated into the vancomycin structure at two different locations (Scheme 1).
Scheme 1. Vancomyxins Prepared in Present Study.
Reagents and conditions used: (i) CuSO4 pentahydrate, sodium ascorbate, H2O.
The azido-modified PMEN building blocks were obtained via a convenient semisynthetic approach starting from colistin. In short, degradation of colistin using the readily available enzyme ficin yielded PMEN which was subsequently converted to PMEN-Boc4 wherein the four Dab side chains are selectively protected and the N-terminus remains free.39 Subsequently, azido-carboxylic acids of varying lengths were coupled to the N-terminus of PMEN-Boc4 using BOP/DIPEA. Following Boc-deprotection and HPLC purification, the azide-modified PMEN building blocks 3–5 where obtained (Scheme 1). In the case of the alkyne-modified vancomycin partners, we followed a previously described protocol reported by Sharpless and co-workers who used click chemistry approaches in preparing various dimers of vancomycin.38 To this end, an alkyne handle was incorporated at either the vancomycin C-terminus or at the vancosamine moiety. Given that neither of these modifications impacts the lipid II binding core of vancomycin, it was expected that structural alterations at these sites would not impair the ability of vancomycin to recognize its target. To install the alkyne at the C-terminus, vancomycin was treated with propargyl amine and HBTU/DIPEA resulting in building block 6. For the preparation of building block 7, installation of the alkyne handle at the vancosamine moiety was achieved via reductive amination using a known alkyne-containing aromatic aldehyde (see Scheme 1 and Supporting Information Schemes S1–S2).31,38 With the required PMEN-azides and vancomycin-alkynes in hand, the conjugation step involving triazole formation was achieved by means of copper catalysis.36−38 In all cases, the ligation reactions proceeded cleanly and rapidly to yield the expected vancomyxins (8–13). As indicated in Scheme 1, compounds 8 and 11 feature the shortest spacer deriving from the azido glycine modified PMEN, compounds 9 and 12 contain a 5-carbon moiety, while compounds 10 and 13 include a longer and more hydrophilic PEG3 spacer.
PMEN azides 3-5, the alkyne modified vancomycins 6 and 7, and the resulting hybrid vancomyxins 8-13 were all assessed for antibacterial activity. As expected, compounds 3-5 were significantly less active than colistin (see Supporting Information Table S1) while compounds 6 and 7 showed activity comparable to vancomycin (see Supporting Information Table S2). The vancomyxins 8-13 were initially assessed against two Gram-negative and two Gram-positive strains (Table 1). Notably, against the Gram-negative E. coli ATCC25922 strain used, the vancomyxins displayed improved antimicrobial activity, with a >8-fold reduction in MIC compared to vancomycin observed for compounds 8, 9, 11, 12 (MIC going from >128 to 16 μg/mL). The decrease in MIC values was less pronounced for the compounds with PEG3-based spacers 10 and 13. Against the K. pneumoniae ATCC27736 strain used, vancomyxin 11 was found to have an MIC of 8 μg/mL, a > 16-fold improvement with respect to vancomycin, while the other vancomyxins were also found to have increased potencies in the range of >4- to >8-fold (Table 1). In the case of the Gram-positive B. subtilis and S. aureus ATCC29213 strains used in this preliminary screen, it was found that conjugation of PMEN to vancomycin did not significantly impair anti-Gram-positive activity relative to the parent vancomycin.
Table 1. Preliminary Antibacterial Activity Assessment of Vancomyxins 8-13.
|
MIC (μg/mL)a |
|||||||
|---|---|---|---|---|---|---|---|
| Strain ID | Vancomycin | 8 | 9 | 10 | 11 | 12 | 13 |
| Gram-negative bacteria | |||||||
| E. coli ATCC25922 | >128 | 16 | 16 | 32 | 16 | 16 | 32 |
| K. pneumoniae ATCC27736 | >128 | 32 | 16 | 64 | 8 | 16 | 32 |
| Gram-positive bacteria | |||||||
| B. subtilis 168 | 0.25 | 0.5 | 0.25 | 0.5 | 0.25 | 0.25 | 1 |
| S. aureus ATCC29213 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.5 |
MIC = Minimum inhibitory concentration.
In assessing these preliminary results, it was notable that compounds 10 and 13 consistently exhibited higher MIC values compared to the other vancomyxins, indicating that shorter spacers are preferable. In addition, the location used for attachment of the PMEN moiety to vancomycin (C-terminus or vancosamine) was found to have minimal impact on antibacterial activity, with MIC values differing by no more than 2-fold for the same spacers. Compound 11, wherein the PMEN motif is connected to the vancosamine functionality, performed particularly well. On the basis of these results against Gram-negative and Gram-positive strains, conjugates 8, 9, 11, and 12 were selected for further assessment and vancomyxin 11 for further modification.
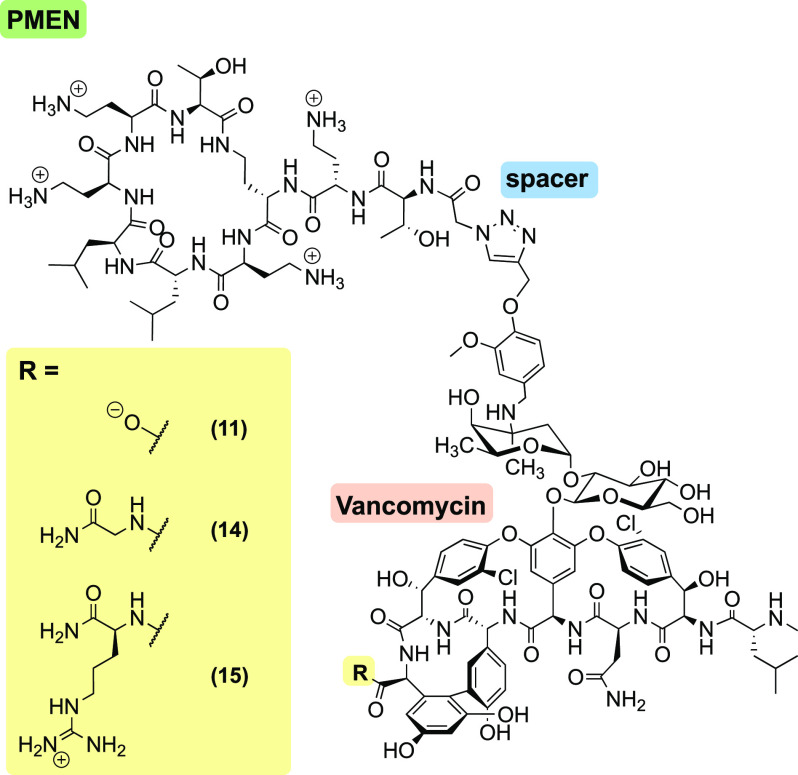
As noted above, recent studies by the groups of Wender and Cegelski have shown that C-terminal modification of vancomycin with a positively charged amino acid (arginine-amide) leads to a significant improvement of anti-Gram-negative activity. These reports prompted us to synthesize two additional compounds building upon vancomyxin 11 wherein either a glycine amide or arginine amide was coupled to the C-terminus of 11. In doing so, it is possible to probe the influence of charge at the C-terminus: while the parent vancomyxin 11 contains a negative charge, analogues 14 and 15 are neutral and positively charged, respectively (Figure 2). Vancomyxins 14 and 15 were prepared by coupling either glycine amide or arginine amide to the C-terminus of vancomycin alkyne building block 7 (see Supporting Information Scheme S2). Subsequent copper-catalyzed click ligation to azido-PMEN building block 3 yielded analogues 14 and 15. An expanded antibacterial activity assessment using a variety of Gram-negative and Gram-positive strains was then performed for vancomyxins 14 and 15 along with compounds 8, 9, 11, and 12, which were identified as most promising in the initial screen (Table 2).
Figure 2.
Structures of the vancomyxins 11, 14, and 15 with ionization state at neutral pH.
Table 2. Expanded Assessment of 8, 9, 11, 12, 14, and 15 against Gram-Negative and Gram-Positive Bacteriaa.
| |
MIC (μg/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain ID | Vancomycin | PMEN | Vancomycin +8 μg/mL PMEN | 8 | 9 | 11 | 12 | 14 | 15 | |
| Gram-negative bacteria | ||||||||||
| E. coli | ATCC25922 | >128 | >128 | 32 | 16 | 16 | 16 | 16 | 16 | 16 |
| ATCC35218 | 128 | >128 | 32 | 16 | 16 | 16 | 16 | 8 | 8 | |
| K. pneumoniae | ATCC13883 | >128 | >128 | 128 | 32 | 32 | 8 | 8 | 16 | 32 |
| ATCC27736 | >128 | >128 | 128 | 32 | 16 | 8 | 16 | 16 | 16 | |
| A. baumannii | ATCC17978 | >128 | >128 | 128 | 128 | 64 | 32 | 32 | 128 | 128 |
| BAA-747 | >128 | >128 | 128 | 32 | 64 | 32 | 32 | 16 | 32 | |
| P. aeruginosa | ATCC10145 | >128 | >128 | 16 | >128 | >128 | 64 | 64 | 32 | 32 |
| ATCC27853 | >128 | >128 | 4 | 16 | 32 | 16 | 16 | 16 | 16 | |
| Gram-positive bacteria | ||||||||||
| B. subtilis | 168 | 0.25 | >128 | ND | 0.5 | 0.25 | 0.25 | 0.25 | ≤0.008 | ≤0.008 |
| S. simulans | 22 | 0.125 | >128 | ND | ≤0.008 | 0.031 | 0.031 | 0.016 | ≤0.008 | ≤0.008 |
| S. aureus | MSSA ATCC29213 | 0.125 | >128 | ND | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 1 |
| MRSA USA300 | 0.25 | >128 | ND | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 | 1 | |
| VISA LIM-2 | 4 | >128 | 4 | 2 | 2 | 2 | 4 | 2 | 8 | |
| VISA NRS402 | 8 | >128 | 8 | 16 | 8 | 8 | 2 | 8 | 4 | |
| VRSA 2 (vanA) | 128 | >128 | 128 | 32 | 64 | 64 | 128 | >128 | >128 | |
| VRSA 3b (vanA) | >128 | >128 | >128 | 32 | >128 | 32 | >128 | >128 | >128 | |
| E. faecalis | VRE E1246 (vanA) | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| VRE E7406(vanB) | 32 | >128 | 32 | 8 | 64 | 64 | 128 | 128 | 128 | |
| E. faecium | VSE E980 | 0.5 | >128 | ND | 0.25 | 0.25 | 0.25 | 0.25 | 0.125 | 0.125 |
| VRE E155 (vanA) | >128 | >128 | >128 | 64 | 64 | 64 | 64 | 64 | 8 | |
| VRE E7314 (vanB) | 128 | >128 | 128 | 2 | 8 | 2 | 8 | 0.5 | 0.031 | |
MIC = Minimum inhibitory concentration, ND = not determined, PMEN = Polymyxin E nonapeptide, MSSA = Methicillin-sensitive S. aureus, MRSA = Methicillin-resistant S. aureus, VISA = Vancomycin-intermediate S. aureus, VRSA = Vancomycin-resistant S. aureus, VSE = Vancomycin-sensitive Enterococci, VRE = Vancomycin-resistant Enterococci.
Activity against Gram-Negative Bacteria
As anticipated, neither vancomycin nor PMEN showed any activity against the Gram-negative strains tested when administered on their own. However, the addition of 8 μg/mL PMEN to vancomycin resulted in increased activity against E. coli (≥4-fold), K. pneumoniae and A. baumannii (both ≥2-fold), as well as P. aeruginosa (≥8/16-fold). This synergistic effect of PMEN with vancomycin is in accordance with previous studies.18,19 In the case of vancomyxins, 8, 9, 11, 12, 14, and 15, however, the covalent linking of the vancomycin and PMEN units was found to enhance the activity against most of the strains tested. Against the two E. coli strains used, all six vancomyxins showed improved activity relative to the vancomycin/PMEN combination. Notably, the C-terminally modified 14 and 15 exhibited the greatest enhancement with a 4-fold improvement in activity against E. coli ATCC35218, reaching MICs as low as 8 μg/mL. When tested against K. pneumoniae, the vancomyxins typically displayed a 4- to 8-fold improved activity compared with the vancomycin/PMEN combination, which itself showed little effect. In this case, 11 was found to be the most active compound with an MIC of 8 μg/mL against both Klebsiella strains tested and 12 with an MIC of 8 μg/mL against K. pneumoniae ATCC13883 (a 16-fold enhancement relative vancomycin + 8 μg/mL PMEN). Similarly, in assays with A. baumannii, the vancomycin/PMEN combination was only moderately active (MIC 128 μg/mL) while the vancomyxins showed 2- to 8-fold improvements in antibacterial activity. Interestingly, while the covalently linked vancomyxins show enhanced activity compared to vancomycin supplemented with PMEN against E. coli, K. pneumoniae, and A. baumannii, the opposite pattern was observed in the case of P. aeruginosa. Against P. aeruginosa ATCC10145 vancomycin supplemented with 8 μg/mL PMEN exhibited an MIC of 16 μg/mL (an >8-fold enhancement) while the vancomyxyins showed little to no improvement relative to vancomycin. A similar trend was observed with P. aeruginosa ATCC27853. While the vancomyxins in this case generally displayed an >8-fold reduction in MIC compared to vancomycin alone (16 μg/mL vs >128 μg/mL), PMEN supplementation strongly synergized with vancomycin reducing its MIC to 4 μg/mL. The greater sensitivity of the P. aeruginosa strains to the vancomycin/PMEN combination versus the larger covalently linked vancomyxins may be attributable to the known low-permeability of the P. aeruginosa outer membrane.40 Furthermore, previous investigations have established that Pseudomonas strains are particularly sensitive to the polymyxin antibiotics as well as the synergistic effects of the corresponding nonapeptides when coadministered with other Gram-positive specific antibiotics.41,42
Activity against Gram-Positive Bacteria
In assessing the activity of the vancomyxins against Gram-positive bacteria, a number of vancomycin-sensitive (MIC ≤ 2 μg/mL), vancomycin-intermediate (MIC 4–8 μg/mL), and vancomycin-resistant (≥16 μg/mL) strains were selected (Table 2). For the vancomycin-intermediate and vancomycin-resistant strains, the effect of PMEN addition at 8 μg/mL was also investigated which, not surprisingly, had no impact on the reduced potency of vancomycin. This lack of synergy was expected given that PMEN serves as an OM disruptor, a barrier only present in Gram-negative strains. Among the vancomycin-sensitive strains tested (MSSA, MRSA, and VSE) the vancomyxins were found to exhibit a similar or slightly enhanced activity relative to vancomycin. Notably, when assessed against B. subtilis, compounds 8, 9, 11, and 12 were found to be as active as vancomycin, while 14 and 15 were found to be extremely potent with activities below 0.008 μg/mL, a >32-fold improvement. Similarly, all the vancomyxins showed significantly increased potency compared to vancomycin against the strain of S. simulans tested. When tested against VISA, the vancomyxins demonstrated potencies similar or slightly enhanced relative to vancomycin. Interestingly, however, against VRSA, compounds 9, 12, 14, and 15 were found to show no enhancement of activity while compounds 8 and 11 displayed improved antibacterial potencies with a ≥4-fold reduction of MIC values compared to vancomycin. Among the VRE strains tested, the activities of the vancomyxins relative to vancomycin were found to be highly variable. Against vancomycin-resistant E. faecalis, the vancomyxins showed little-to-no enhancement whereas against vancomycin-resistant E. faecium, particularly the VanB type, the vancomyxins exhibited potent antibacterial activity. Specifically, for the VanB type E. faecium strain tested, vancomyxins 9 and 12 have an MIC of 8 μg/mL and vancomyxins 8 and 11 have MIC values of 2 μg/mL, while 14 and 15 demonstrated even more impressive potencies with MICs of 0.5 and 0.031 μg/mL, respectively.
The enhanced antimicrobial activities observed for the vancomyxins against vancomycin-resistant strains suggest that our novel compounds are able to (partially) compensate for the reduced binding of the vancomycin core to the d-Ala-d-Lac lipid II motif, common to VREs containing either vanA or vanB resistance genes. It is well established that mutation of the d-Ala-d-Ala unit found in wild type lipid II to the d-Ala-d-Lac unit found in VanA or VanB positive strains results in a >1000-fold reduction in binding affinity for vancomycin and loss of antimicrobial activity.1,2,4,43 The finding that the vancomyxins overcome this resistance, especially in the case of VanB positive E. faecium, indicates that other structural features are contributing to their enhanced activity. Specifically, the large positive net charge introduced by conjugating the PMEN motif to vancomycin may facilitate electrostatic interactions with the negatively charged bacterial membrane. This in turn may lead to increased membrane anchoring and lipid II engagement thereby interfering with cell wall synthesis and lowering the MIC of the vancomyxins toward otherwise vancomycin-resistant strains. In this regard, it is noteworthy that previous reports have also described the addition of positively charged moieties to vancomycin as means of overcoming vancomycin resistance.4,15,31−35 Furthermore, conjugation of vancomycin to bacteriocin nisin (1–12) has previously shown to reduce the MIC against VRE.44
As described above, vancomyxins 14 and 15 were synthesized to assess if a difference in charge at the C-terminus (neutral and positive) compared to 11 (negative) would enhance activity. Against Gram-negative strains, this did not appear to have a large effect on activity as the MICs of vancomyxins 11, 14, and 15 are generally similar, differing by no more than 2-fold. Against Gram-positive strains, however, there are notable species-dependent differences; against VRSA, the negatively charged C-terminus of 11 results in activity superior to that of the neutral 14 and positive-charged 15, while against vancomycin-resistant E. faecium, the positively charged 15 exhibits enhanced activity.
From a mechanistic perspective, we hypothesized that the enhanced anti-Gram-negative activity of the vancomyxins compared to vancomycin might be due to the ability of PMEN to bind LPS and disrupt the outer membrane.23,24 To investigate this, we performed an LPS antagonization assay which indeed points to an interaction of compound 11 with LPS, as its MIC against E. coli ATCC25922 increased significantly from 16 μg/mL to >128 μg/mL when incubated with exogenous LPS (see Supporting Information Table S4).
Cell-based toxicity studies
In parallel to the antibacterial activity assays, the hemolytic properties of the vancomyxins were also assessed, revealing them to be nonhemolytic up to the highest concentration tested (512 μg/mL). These findings are in line with our expectations given the nonhemolytic nature of both vancomycin and colistin (see Supporting Information Figure S1). We next turned our attention to evaluating the nephrotoxicity of the vancomyxins. Reports in the literature indicate that both polymyxins and vancomycin can impair kidney function.45,46 In particular, proximal tubule epithelial cells are known to be sensitive to the polymyxins as a result of extensive reabsorption and intracellular accumulation.46 In addition, proximal tubule cells have been previously used to characterize the cytotoxic effects of vancomycin.47 To compare the hybrid vancomyxins with the corresponding parent compounds, their nephrotoxicity was assessed by means of a viability assay using conditionally immortalized proximal tubule epithelial cells (ciPTECs), with relative mitochondrial activity after 24 h as the end point measurement. In these assays, polymyxin B was found to exhibit relatively high nephrotoxicity (TC50 = 0.07 mM) while vancomycin and PMEN were significantly less toxic (>50% viability at a concentration of 1 mM for both). By comparison, the vancomyxins were found to exhibit intermediate toxicity toward ciPTECs with TC50 values ranging from 0.11 mM for compound 15 to 0.37 mM for compound 8 (see Supporting Information Figure S2 for full details), concentrations generally multiple orders of magnitude higher than the corresponding MIC values.
Conclusion
While vancomycin is an important antibiotic for the treatment of hospitalized patients with Gram-positive infections, it has little activity against Gram-negative bacteria due to the inaccessibility of its target. Given that vancomycin can bind to the Gram-negative form of lipid II,6 OM disruptors present a possible means for enhancing the activity of vancomycin against Gram-negative pathogens.8,10,18,19 To this end, combination strategies involving polymyxin nonapeptide or other OM disruptors have been explored.10,18,19 However, the covalent conjugation of vancomycin to the OM disrupting PMEN motif has not been previously described. In this study, we report a new class of vancomycin-PMEN hybrids, the vancomyxins. The vancomyxins maintain the activity of vancomycin against vancomycin-sensitive strains and, in some cases, also overcome vancomycin-resistance in Gram-positive organisms. Against Gram-negative organisms, the vancomyxins also show enhanced activity that was generally superior to that observed with a simple combination of vancomycin with PMEN. It is worth noting that when comparing the activities of the hybrid vancomyxins to vancomycin or the combination of vancomycin/PMEN, the conventional concentration units of μg/mL was used. However, given that the molecular weights of the vancomyxins are approximately double that of vancomycin, the differences in MIC are even more pronounced when comparing the appropriate molar concentrations (see Supporting Information Tables S1–S3). In addition, LPS binding of the vancomyxins is confirmed. Furthermore, the vancomyxins are not hemolytic and exhibit lower toxicity against kidney cells compared to the clinically used polymyxin B. In summary, these findings indicate that the covalent attachment of an OM disrupting PMEN motif to vancomycin is a viable strategy for enhancing anti-Gram-negative activity.
Methods
Synthetic Procedures
The synthesis of all intermediates and of the vancomyxins is described in detail in the Supporting Information. Given here is the general procedure for the copper-catalyzed azide–alkyne cycloaddition used to prepare the vancomyxins: The ligation protocol used generally followed that previously described by Silverman et al.38 In short, to a solution of the vancomycin alkyne (0.03 mmol, 1 equiv) in water (1.5 mL), the PMEN azide (0.03 mmol, 1 equiv) in water (1.5 mL) was added. Subsequently, sodium ascorbate (0.008 mmol, 0.25 equiv) and CuSO4·5H2O (0.003 mmol, 0.1 equiv) were added and the mixture was allowed to stir at RT for 16 h. The reaction mixture was directly purified using preparative high performance liquid chromatography (HPLC) using a C18 column (25 × 250 mm, 10 μm) with UV detection at 214 nm. The following method was used: flow rate = 12 mL/min; solvent A, 0.1% TFA in H2O/acetonitrile 95:5, and solvent B, 0.1% TFA in H2O/acetonitrile 5:95. The gradient elution was as follows: 95:5 (A/B) for 5 min, 95:5 to 40:60 (A/B) over 50 min, 40:60 to 0:100 (A/B) for 1 min, 0:100 (A/B) for 2 min, then reversion back to 95:5 (A/B) over 1 min, 95:5 (A/B) for 2 min. Fractions were immediately freeze-dried and subsequently analyzed by LCMS. Pure product containing fractions were redissolved, pooled, and lyophilized to yield the vancomyxins as white powders.
Broth Microdilution Assays
From glycerol stocks, bacteria were plated out on blood agar plates overnight at 37 °C. One colony was transferred to growth media and grown at 37 °C at 200 rpm to exponential growth phase as determined by OD600. The growth media used for most strains was cation adjusted Mueller Hinton Broth (0.5 mM Mg2+ and Ca2+) + 0.002% of polysorbate 80 (p80), except for all Enterococci and VISA/VRSA strains, for which the positive control was unable to grow well in this media; therefore, TSB + 0.002% p80 was used in these cases. For VRSA, the media was supplemented with 6 μg/mL vancomycin at this stage. At OD600 = 0.5, the bacteria were diluted 100-fold and 50 μL was added to a 2-fold serial dilution series of test compound (50 μL) to reach a total volume of 100 μL per well. The 96-well polypropylene plates were incubated at 37 °C at 600 rpm overnight (18–20 h for Gram-negative strains, 20–24 h for Gram-positive strains), and plates were inspected for visual bacterial growth. Synergy experiments were performed in a similar manner as the MIC assay, except 8 μg/mL PMEN final concentration was added to the wells. LPS antagonization assays were performed in a similar manner except that a final concentration of 1 mg/mL LPS was added to the wells. In this case, colistin was used as a control. MICs are reported as the median of triplicates.
Hemolysis Assays
Defibrinated whole sheep blood was centrifuged for 15 min at 4 °C (400 g). The top layer was discarded, and the bottom layer was washed with phosphate buffered saline (PBS) and centrifuged for 15 min at 4 °C (400 g). Washing cycles were repeated at least three times. In polypropylene 96-well microtiter plates, 10-fold serial dilutions of antibiotics in PBS with 0.002% p80 in biological triplicates were added (75 μL) and an equal volume of packed blood cells diluted 25x in PBS with 0.002% p80 (75 μL) was added to all wells. Plates were incubated for 20 h at 37 °C with continuous shaking (500 rpm). After incubation, plates were centrifuged for 5 min (800 g) and 25 μL of supernatant was transferred to a clear UV-star flat-bottom polystyrene 96-well plate already containing 100 μL H2O per well. Absorption was measured at 415 nm. Data were corrected by subtraction of the background response of 1% DMSO in the presence of cells with no antibiotic and normalized using the absorbance of 0.1% Triton X-100 with blood cells as 100% hemolysis control.
PTECs Assay
Cell culture
ciPTECs overexpressing organic anion transporter 1 (OAT-1)48,49 were cultured in DMEM/F12 medium, supplemented with fetal calf serum (10%), insulin (5 μg/mL), transferrin (5 μg/mL), selenium (5 μg/mL), hydrocortisone (35 ng/mL), Epidermal Growth Factor (10 ng/mL), and tri-iodothyronine (40 pg/mL). Cells were cultured at 33 °C for sustained proliferation. For the experiment, cells were washed with HBSS and detached by incubating them with Accutase solution for 5 min at 37 °C. Density was adjusted to 2.0 × 105 cells/mL of which 100 μL was added to each well of a 96-well plate. Seeded cells were incubated for 24 h at 33 °C, followed by 6 days incubation at 37 °C to allow them to fully differentiate. Medium was refreshed every second or third day. All cells were grown in a humidified atmosphere containing 5% (v/v) CO2.
Cell Viability Assay
Cytotoxicity was assessed using PrestoBlue cell viability reagent. Compounds were dissolved and diluted in serum free medium. Differentiated ciPTECs were washed once with HBSS and exposed to the compounds for 24 h at 37 °C. Afterward, cells were washed with HBSS and incubated with 10% PrestoBlue reagent in HBSS at 37 °C for 1 h in the dark. Fluorescence was recorded using excitation wavelength of 530 nm and emission wavelength of 590 nm. Raw data were corrected for PrestoBlue background fluorescence and reported relative to the no-treatment control (cells with medium only). Data were fitted with Graphpad Prism software by nonlinear regression with 0 as constraint to obtain TC50 values. Presented data are based on triplicates and presented as mean ± SEM.
Acknowledgments
Financial support provided by the European Research Council (ERC consolidator grant to NIM, grant agreement no. 725523).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.1c00318.
Synthetic schemes and synthesis of all building blocks; analytical RP-HPLC traces, HRMS analysis and yields for all new compounds synthesized; additional MIC analysis of building blocks and control compounds; MIC analysis in μM; hemolytic and PTEC toxicity data (PDF)
Author Contributions
# These authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Pootoolal J.; Neu J.; Wright G. D. (2002) Glycopeptide Antibiotic Resistance. Annu. Rev. Pharmacol. Toxicol. 42 (1), 381–408. 10.1146/annurev.pharmtox.42.091601.142813. [DOI] [PubMed] [Google Scholar]
- Walsh C. T.; Fisher S. L.; Park I. S.; Prahalad M.; Wu Z. (1996) Bacterial Resistance to Vancomycin: Five Genes and One Missing Hydrogen Bond Tell the Story. Chem. Biol. 3 (1), 21–28. 10.1016/S1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- Beauregard D. A.; Williams D. H.; Gwynn M. N.; Knowles D. J. (1995) Dimerization and Membrane Anchors in Extracellular Targeting of Vancomycin Group Antibiotics. Antimicrob. Agents Chemother. 39 (3), 781–785. 10.1128/AAC.39.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovich M. A. T.; Hansford K. A.; Butler M. S.; Jia Z.; Mark A. E.; Cooper M. A. (2018) Developments in Glycopeptide Antibiotics. ACS Infect. Dis. 4, 715–735. 10.1021/acsinfecdis.7b00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J. P.; Gerhard U.; Beauregard D. A.; Williams D. H.; Westwell M. S.; Searle M. S. (1994) Glycopeptide Antibiotic Activity and the Possible Role of Dimerization: A Model for Biological Signaling. J. Am. Chem. Soc. 116 (11), 4581–4590. 10.1021/ja00090a006. [DOI] [Google Scholar]
- Shlaes D. M.; Shlaes J. H.; Davies J.; Williamson R. (1989) Escherichia Coli Susceptible to Glycopeptide Antibiotics. Antimicrob. Agents Chemother. 33 (2), 192–197. 10.1128/AAC.33.2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J. C.; Zimmerman S. M.; Crofts A. A.; Boll J. M.; Kuhns L. G.; Herrera C. M.; Trent M. S. (2016) The Power of Asymmetry: Architecture and Assembly of the Gram-Negative Outer Membrane Lipid Bilayer. Annu. Rev. Microbiol. 70 (1), 255–278. 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- Heesterbeek D. A. C.; Martin N. I.; Velthuizen A.; Duijst M.; Ruyken M.; Wubbolts R.; Rooijakkers S. H. M.; Bardoel B. W. (2019) Complement-Dependent Outer Membrane Perturbation Sensitizes Gram-Negative Bacteria to Gram-Positive Specific Antibiotics. Sci. Rep. 9 (1), 3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J. M.; MacNair C. R.; Ilyas B.; French S.; Côté J.-P.; Bouwman C.; Farha M. A.; Sieron A. O.; Whitfield C.; Coombes B. K.; Brown E. D. (2017) Pentamidine Sensitizes Gram-Negative Pathogens to Antibiotics and Overcomes Acquired Colistin Resistance. Nat. Microbiol. 2, 17028. 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Cebrián R.; Montalbán-López M.; Ren H.; Wu W.; Kuipers O. P. (2021) Outer-Membrane-Acting Peptides and Lipid II-Targeting Antibiotics Cooperatively Kill Gram-Negative Pathogens. Commun. Biol. 4 (1), 31. 10.1038/s42003-020-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M.; Lin Y.-M.; Miller P. A.; Möllmann U.; Boggess W. C.; Miller M. J. (2018) Siderophore Conjugates of Daptomycin Are Potent Inhibitors of Carbapenem Resistant Strains of Acinetobacter Baumannii. ACS Infect. Dis. 4 (10), 1529–1535. 10.1021/acsinfecdis.8b00150. [DOI] [PubMed] [Google Scholar]
- Li X.-Z.; Plésiat P.; Nikaido H. (2015) The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 28 (2), 337–418. 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabawa T. P.; Pucci M. J.; Parr T. R.; Lister T. (2016) Treatment of Gram-Negative Bacterial Infections by Potentiation of Antibiotics. Curr. Opin. Microbiol. 33, 7–12. 10.1016/j.mib.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Ghosh M.; Miller M. J. (1996) Synthesis and in Vitro Antibacterial Activity of Spermidine-Based Mixed Catechol- and Hydroxamate-Containing Siderophore--Vancomycin Conjugates. Bioorg. Med. Chem. 4 (1), 43–48. 10.1016/0968-0896(95)00161-1. [DOI] [PubMed] [Google Scholar]
- Sarkar P.; Samaddar S.; Ammanathan V.; Yarlagadda V.; Ghosh C.; Shukla M.; Kaul G.; Manjithaya R.; Chopra S.; Haldar J. (2020) Vancomycin Derivative Inactivates Carbapenem-Resistant Acinetobacter Baumannii and Induces Autophagy. ACS Chem. Biol. 15 (4), 884–889. 10.1021/acschembio.0c00091. [DOI] [PubMed] [Google Scholar]
- Neville L. F.; Shalit I.; Warn P. A.; Scheetz M. H.; Sun J.; Chosy M. B.; Wender P. A.; Cegelski L.; Rendell J. T. (2021) In Vivo Targeting of Escherichia Coli with Vancomycin-Arginine. Antimicrob. Agents Chemother. 65 (4), e02416–20. 10.1128/AAC.02416-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonoplis A.; Zang X.; Wegner T.; Wender P. A.; Cegelski L. (2019) Vancomycin-Arginine Conjugate Inhibits Growth of Carbapenem-Resistant E. Coli and Targets Cell-Wall Synthesis. ACS Chem. Biol. 14 (9), 2065–2070. 10.1021/acschembio.9b00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M. (1992) Agents That Increase the Permeability of the Outer Membrane. Microbiol. Rev. 56 (3), 395–411. 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek I.; Cohen S.; Rahmani R.; Kabha K.; Tamarkin D.; Herzig Y.; Rubinstein E. (1994) Antibacterial Synergism of Polymyxin B Nonapeptide and Hydrophobic Antibiotics in Experimental Gram-Negative Infections in Mice. Antimicrob. Agents Chemother. 38 (2), 374–377. 10.1128/AAC.38.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T.; Thompson P. E.; Nation R. L.; Li J. (2010) Structure--Activity Relationships of Polymyxin Antibiotics. J. Med. Chem. 53 (5), 1898–1916. 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiharha S.; Ito A.; Yahata M.; Tobita T.; Koyama Y. (1974) Chemical Synthesis, Isolation and Characterization of α-N-Fattyacyl Colistin Nonapeptide with Special Reference to the Correlation between Antimicrobial Activity and Carbon Number of Fattyacyl Moiety. Agric. Biol. Chem. 38 (3), 521–529. 10.1080/00021369.1974.10861184. [DOI] [Google Scholar]
- Keirstead N. D.; Wagoner M. P.; Bentley P.; Blais M.; Brown C.; Cheatham L.; Ciaccio P.; Dragan Y.; Ferguson D.; Fikes J.; Galvin M.; Gupta A.; Hale M.; Johnson N.; Luo W.; McGrath F.; Pietras M.; Price S.; Sathe A. G.; Sasaki J. C.; Snow D.; Walsky R. L.; Kern G. (2014) Early Prediction of Polymyxin-Induced Nephrotoxicity with next-Generation Urinary Kidney Injury Biomarkers. Toxicol. Sci. 137 (2), 278–291. 10.1093/toxsci/kft247. [DOI] [PubMed] [Google Scholar]
- Vaara M. (1983) Polymyxin B Nonapeptide Complexes with Lipopolysaccharide. FEMS Microbiol. Lett. 18 (1–2), 117–121. 10.1111/j.1574-6968.1983.tb00461.x. [DOI] [Google Scholar]
- Brandenburg K.; Moriyon I.; Arraiza M. D.; Lewark-Yvetot G.; Koch M. H. J.; Seydel U. (2002) Biophysical Investigations into the Interaction of Lipopolysaccharide with Polymyxins. Thermochim. Acta 382 (1), 189–198. 10.1016/S0040-6031(01)00731-6. [DOI] [Google Scholar]
- Domalaon R.; Idowu T.; Zhanel G. G.; Schweizer F. (2018) Antibiotic Hybrids: The Next Generation of Agents and Adjuvants against Gram-Negative Pathogens?. Clin. Microbiol. Rev. 31 (2), e00077–17. 10.1128/CMR.00077-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwe A. K.; Rupar C. A.; Horsman G. B.; Vas S. I. (1986) In Vitro Cytotoxicity and Antibiotic Activity of Polymyxin B Nonapeptide. Antimicrob. Agents Chemother. 30 (2), 340–341. 10.1128/AAC.30.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domalaon R.; Yang X.; Lyu Y.; Zhanel G. G.; Schweizer F. (2017) Polymyxin B(3)-Tobramycin Hybrids with Pseudomonas Aeruginosa-Selective Antibacterial Activity and Strong Potentiation of Rifampicin, Minocycline, and Vancomycin. ACS Infect. Dis. 3 (12), 941–954. 10.1021/acsinfecdis.7b00145. [DOI] [PubMed] [Google Scholar]
- Luther A.; Urfer M.; Zahn M.; Müller M.; Wang S.-Y.; Mondal M.; Vitale A.; Hartmann J.-B.; Sharpe T.; Monte F. Lo; Kocherla H.; Cline E.; Pessi G.; Rath P.; Modaresi S. M.; Chiquet P.; Stiegeler S.; Verbree C.; Remus T.; Schmitt M.; Kolopp C.; Westwood M.-A.; Desjonquères N.; Brabet E.; Hell S.; LePoupon K.; Vermeulen A.; Jaisson R.; Rithié V.; Upert G.; Lederer A.; Zbinden P.; Wach A.; Moehle K.; Zerbe K.; Locher H. H.; Bernardini F.; Dale G. E.; Eberl L.; Wollscheid B.; Hiller S.; Robinson J. A.; Obrecht D. (2019) Chimeric Peptidomimetic Antibiotics against Gram-Negative Bacteria. Nature 576 (7787), 452–458. 10.1038/s41586-019-1665-6. [DOI] [PubMed] [Google Scholar]
- Guan D.; Chen F.; Qiu Y.; Jiang B.; Gong L.; Lan L.; Huang W. (2019) Sulfonium, an Underestimated Moiety for Structural Modification, Alters the Antibacterial Profile of Vancomycin Against Multidrug-Resistant Bacteria. Angew. Chem., Int. Ed. 58 (20), 6678–6682. 10.1002/anie.201902210. [DOI] [PubMed] [Google Scholar]
- Shi W.; Chen F.; Zou X.; Jiao S.; Wang S.; Hu Y.; Lan L.; Tang F.; Huang W. (2021) Design, Synthesis, and Antibacterial Evaluation of Vancomycin-LPS Binding Peptide Conjugates. Bioorg. Med. Chem. Lett. 45, 128122. 10.1016/j.bmcl.2021.128122. [DOI] [PubMed] [Google Scholar]
- Yarlagadda V.; Akkapeddi P.; Manjunath G. B.; Haldar J. (2014) Membrane Active Vancomycin Analogues: A Strategy to Combat Bacterial Resistance. J. Med. Chem. 57 (11), 4558–4568. 10.1021/jm500270w. [DOI] [PubMed] [Google Scholar]
- Okano A.; Isley N. A.; Boger D. L. (2017) Peripheral Modifications of [Ψ[CH2NH]Tpg4]Vancomycin with Added Synergistic Mechanisms of Action Provide Durable and Potent Antibiotics. Proc. Natl. Acad. Sci. U. S. A. 114, 5052–5061. 10.1073/pnas.1704125114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaskovich M. A. T.; Hansford K. A.; Gong Y.; Butler M. S.; Muldoon C.; Huang J. X.; Ramu S.; Silva A. B.; Cheng M.; Kavanagh A. M.; Ziora Z.; Premraj R.; Lindahl F.; Bradford T. A.; Lee J. C.; Karoli T.; Pelingon R.; Edwards D. J.; Amado M.; Elliott A. G.; Phetsang W.; Daud N. H.; Deecke J. E.; Sidjabat H. E.; Ramaologa S.; Zuegg J.; Betley J. R.; Beevers A. P. G.; Smith R. A. G.; Roberts J. A.; Paterson D. L.; Cooper M. A. (2018) Protein-Inspired Antibiotics Active against Vancomycin- and Daptomycin-Resistant Bacteria. Nat. Commun. 9 (1), 1–17. 10.1038/s41467-017-02123-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.-C.; Cameron M. D.; Boger D. L. (2020) Vancomycin C-Terminus Guanidine Modifications and Further Insights into an Added Mechanism of Action Imparted by a Peripheral Structural Modification. ACS Infect. Dis. 6 (8), 2169–2180. 10.1021/acsinfecdis.0c00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar P., Basak D., Mukherjee R., Bandow J. E., and Haldar J.. Alkyl-Aryl-Vancomycins: Multimodal Glycopeptides with Weak Dependence on the Bacterial Metabolic State. J. Med. Chem. 202164 ( (14), ), 10185−10202 10.1021/acs.jmedchem.1c00449 [DOI] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. (2002) A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem., Int. Ed. 41 (14), 2596–2599. . [DOI] [PubMed] [Google Scholar]
- Tornøe C. W.; Christensen C.; Meldal M. (2002) Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J. Org. Chem. 67 (9), 3057–3064. 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Silverman S. M.; Moses J. E.; Sharpless K. B. (2017) Reengineering Antibiotics to Combat Bacterial Resistance: Click Chemistry [1,2,3]-Triazole Vancomycin Dimers with Potent Activity against MRSA and VRE. Chem. - Eur. J. 23 (1), 79–83. 10.1002/chem.201604765. [DOI] [PubMed] [Google Scholar]
- O’Dowd H.; Kim B.; Margolis P.; Wang W.; Wu C.; Lopez S. L.; Blais J. (2007) Preparation of Tetra-Boc-Protected Polymyxin B Nonapeptide. Tetrahedron Lett. 48 (11), 2003–2005. 10.1016/j.tetlet.2007.01.071. [DOI] [Google Scholar]
- Chevalier S.; Bouffartigues E.; Bodilis J.; Maillot O.; Lesouhaitier O.; Feuilloley M. G. J.; Orange N.; Dufour A.; Cornelis P. (2017) Structure, Function and Regulation of Pseudomonas Aeruginosa Porins. FEMS Microbiol. Rev. 41 (5), 698–722. 10.1093/femsre/fux020. [DOI] [PubMed] [Google Scholar]
- Moore R. A.; Bates N. C.; Hancock R. E. (1986) Interaction of Polycationic Antibiotics with Pseudomonas Aeruginosa Lipopolysaccharide and Lipid A Studied by Using Dansyl-Polymyxin. Antimicrob. Agents Chemother. 29 (3), 496–500. 10.1128/AAC.29.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viljanen P.; Vaara M. (1984) Susceptibility of Gram-Negative Bacteria to Polymyxin B Nonapeptide. Antimicrob. Agents Chemother. 25 (6), 701–705. 10.1128/AAC.25.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy V. L.; Lessard I. A.; Roper D. I.; Knox J. R.; Walsh C. T. (2000) Vancomycin Resistance in Enterococci: Reprogramming of the D-Ala-D-Ala Ligases in Bacterial Peptidoglycan Biosynthesis. Chem. Biol. 7, 109–119. 10.1016/S1074-5521(00)00116-2. [DOI] [PubMed] [Google Scholar]
- Arnusch C. J.; Bonvin A. M. J. J.; Verel A. M.; Jansen W. T. M.; Liskamp R. M. J.; de Kruijff B.; Pieters R. J.; Breukink E. (2008) The Vancomycin–Nisin(1–12) Hybrid Restores Activity against Vancomycin Resistant Enterococci. Biochemistry 47 (48), 12661–12663. 10.1021/bi801597b. [DOI] [PubMed] [Google Scholar]
- van Hal S. J.; Paterson D. L.; Lodise T. P. (2013) Systematic Review and Meta-Analysis of Vancomycin-Induced Nephrotoxicity Associated with Dosing Schedules That Maintain Troughs between 15 and 20 mg per Liter. Antimicrob. Agents Chemother. 57 (2), 734–744. 10.1128/AAC.01568-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad M. A. K.; Roberts K. D.; Yu H. H.; Liu B.; Schofield A. V.; James S. A.; Howard D. L.; Nation R. L.; Rogers K.; de Jonge M. D.; Thompson P. E.; Fu J.; Velkov T.; Li J. (2015) Significant Accumulation of Polymyxin in Single Renal Tubular Cells: A Medicinal Chemistry and Triple Correlative Microscopy Approach. Anal. Chem. 87 (3), 1590–1595. 10.1021/ac504516k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y.; Yano T.; Hanada Y.; Takeshita A.; Inagaki F.; Masuda S.; Matsunaga N.; Koyanagi S.; Ohdo S. (2017) Vancomycin Induces Reactive Oxygen Species-Dependent Apoptosis via Mitochondrial Cardiolipin Peroxidation in Renal Tubular Epithelial Cells. Eur. J. Pharmacol. 800, 48–56. 10.1016/j.ejphar.2017.02.025. [DOI] [PubMed] [Google Scholar]
- Jansen J.; Schophuizen C. M. S.; Wilmer M. J.; Lahham S. H. M.; Mutsaers H. A. M.; Wetzels J. F. M.; Bank R. A.; van den Heuvel L. P.; Hoenderop J. G.; Masereeuw R. (2014) A Morphological and Functional Comparison of Proximal Tubule Cell Lines Established from Human Urine and Kidney Tissue. Exp. Cell Res. 323 (1), 87–99. 10.1016/j.yexcr.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Nieskens T. T. G.; Peters J. G. P.; Schreurs M. J.; Smits N.; Woestenenk R.; Jansen K.; van der Made T. K.; Röring M.; Hilgendorf C.; Wilmer M. J.; Masereeuw R. (2016) A Human Renal Proximal Tubule Cell Line with Stable Organic Anion Transporter 1 and 3 Expression Predictive for Antiviral-Induced Toxicity. AAPS J. 18 (2), 465–475. 10.1208/s12248-016-9871-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.