Abstract
Glioma is a common malignant tumor of the central nervous system, accounting for ~50% of intracranial tumors. The current standard therapy for glioma is surgical resection followed by postoperative adjuvant radiotherapy and temozolomide (TMZ) chemotherapy. However, resistance to TMZ is one of the factors affecting prognosis. It has been reported that TNF receptor-associated protein 1 (TRAP1) is overexpressed in numerous types of tumor and that interfering with its function may abrogate chemotherapy resistance. TRAP1 inhibitor Gamitrinib triphenylphosphonium (G-TPP) and shRNA were used in the present study to suppress the function of this molecule in glioblastoma multiforme (GBM) cell lines. MTT assay was performed to evaluate the combined effect of G-TPP and TMZ treatment. To investigate the underlying mechanism responsible for this combined effect, the mitochondrial unfolded protein response (mtUPR), mitophagy, mitochondrial fusion and reactive oxygen species (ROS) were quantified using western blotting and immunofluorescence techniques. TMZ treatment induced apoptosis in GBM cells by activating the p53 pathway, whilst simultaneously downregulating mitophagy and enhancing mitochondrial fusion. The latter may occur in order to compensate for the defect caused by downregulated mitophagy. Suppressing the function of TRAP1 disturbed this compensatory mechanism by inducing mtUPR, which resulted in a burst of ROS formation and sensitized the GBM cells to the effects of TMZ treatment. Thus, suppressing the function of TRAP1 sensitized GBM cells to TMZ lysis by inducing mtUPR and the subsequent ROS burst. TRAP1 is therefore considered to be a promising target for GBM therapy.
Keywords: glioblastoma multiforme, temozolomide, TNF receptor-associated protein 1, mitochondrial unfolded protein response, reactive oxygen species
Introduction
Glioblastoma multiforme (GBM) accounts for 40-50% of all gliomas (1,2). Patients with GBM have a progression-free survival of only 6.2-7.5 months after diagnosis and a median survival of 14.6-16.7 months (3-5), while the 5-year survival rate is only 10% (6). As a highly malignant tumor, recurrence of GBM following surgery is inevitable and adjuvant chemotherapy following GBM resection is essential. Temozolomide (TMZ), a second-generation alkylating agent, methylates the O6 guanine site, which causes a G-T mismatch during DNA replication (7). DNA repair mechanisms, such as mismatch repair systems are then activated, thereby triggering cell cycle checkpoint protein production and stress response-related proteins, finally resulting in cell cycle arrest or apoptosis (8). Large-scale clinical studies have demonstrated that adherence to a TMZ chemotherapy regime following surgery prolongs the survival of patients with GBM by 2-4 months (3,9). However, due to the instability of the tumor cell genome, the expression of O6-methylguanine-DNA methyltransferase and the involvement of various different mechanisms facilitating apoptosis escape, chemotherapy resistance is the main factor affecting the efficacy of TMZ (10). Of the multiple studies aimed at enhancing TMZ efficacy, the present study has focused on heat shock protein 90 (Hsp90)-related research.
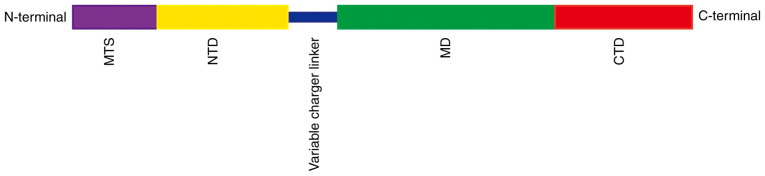
According to the latest classification, the Hsp90 family includes five members, of which Hsp90AA1, Hsp90AA2 and Hsp90AB1 are mainly located in the cytoplasm, Hsp90B1 in the endoplasmic reticulum and TNF receptor-associated protein 1 (TRAP1) in the mitochondrial matrix with a few molecules present in the intermembrane space of mitochondria (11-13). Although the members of the Hsp90 family have a similar structure, TRAP1 has its own specific characteristics. It consists of four domains, mitochondria targeting domain (MTS), N-terminal domain (NTD), middle domain (MD) and C-terminal domain (CTD; Fig. 1) (11) of which it is MTS that is the mitochondrial targeting sequence. Thus, TRAP1 precursors are targeted to mitochondria and, after passing through the inner membrane, the MTS sequence is cleaved by mitochondrial processing peptidase to form mature TRAP1. The other domains have different functions as follows: NTD binds and hydrolyzes ATP (11,14), MD acts as a substrate protein or chaperone-binding domain (11,15) and CTD contains a homodimerization binding domain that regulates N-terminal ATPase activity (11,16). This is necessary for the ATP binding and ADP release cycle. Changes to the dimer conformation of Hsp90 members help to shape the structure of the substrate proteins (11).
Figure 1.
Domain structure of TNF receptor-associated protein 1. MTS, mitochondria targeting domain; NTD, N-terminal domain; MD, middle domain; CTD, C-terminal domain.
A previous study demonstrated that TRAP1 has multiple functions in mitochondria, for example, playing a role in the intra-mitochondria protein control system (17), regulating the opening of the permeability transition pore by interacting with cyclophilin D (13,18) and influencing the mitochondrial respiratory process by interacting with mitochondrial respiratory chain complexes (19). In the present study, suppressing the function of TRAP1 was shown to sensitize GBM cells to TMZ therapy. Furthermore, treatment with the mitochondrial-targeting Hsp90 inhibitor Gamitrinib triphenylphosphonium (G-TPP) together with TMZ exerted an additive effect in reducing the viability of glioblastoma cells.
Materials and methods
Cell culture
SHG44, U251-MG and U87-MG (glioblastoma of unknown origin) glioblastoma cell lines were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and were grown in DMEM culture medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.).
Reagents and antibodies
TMZ and G-TPP were purchased from MedChemExpress company and the MTT assay was obtained from MilliporeSigma. The MitoTracker™ (cat. no. 7512), Hoechst 33342 (cat. no. H3570) and LysoTracker™ (cat. no. L7526) were all obtained from Thermo Fisher Scientific, Inc. RIPA buffer (cat. no. P0013B) and horse serum (cat. no. C0262) were purchased from Beyotime Institute of Biotechnology. The antibodies used in the present study were as follows: anti-Hsp10 (cat. no. ab210681; Abcam), anti-phosphorylated-ubiquitin [serine 65 (Ser65) cat. no. ABS1513-I] from Merck KGaA, anti-ubiquitin (cat. no. 13-1600) from Thermo Fisher Scientific, Inc., anti-β actin (cat. no. 60008-1-Ig), anti-Bax (cat. no. 50599-2-Ig), anti-PTEN-induced kinase 1 (PINK1; cat. no. 23274-1-AP), anti-caspase-3 (cat. no. 19677-1-AP), anti-caspase-9 (cat. no. 10380-1-AP), anti-Hsp60 (cat. no.15282-1-AP) and anti-p53 (cat. no. 60283-2-Ig) from ProteinTech Group, Inc., goat anti-mouse secondary fluorescent antibody (cat. no. F2761) and goat anti-rabbit secondary fluorescent antibody (cat. no. A-11012) from Thermo Fisher Scientific, Inc. and goat anti-mouse, HRP-conjugated secondary antibody (cat. no. SA00001-1) and goat anti-rabbit, HRP-conjugated secondary antibody (cat. no. SA00001-2) from ProteinTech Group, Inc.
MTT assays
Cells were seeded into 96-well plates at a density of 1x104 cells/well and cultured with TMZ or G-TPP, after which 20 µl MTT solution (5 mg/ml) was introduced into each well. After 4 h incubation in the CO2 incubator at 37˚C, 150 µl DMSO was added to each well. The plates were assessed using a CLARIO star microplate reader (BMG Labtech GmbH) at an absorbance of 570 nm.
Immunofluorescence (IF) assay
Cells grown on coverslips in 24-well plates were treated with the different agents. After staining with MitoTracker for 30 min in CO2 incubator at 37˚C, the coverslips were rinsed with PBS and fixed with 4% (w/v) paraformaldehyde for 25 min followed by a PBS rinse. Permeabilization with 0.1% (v/v) Triton X-100 for 7 min was followed by blockade with 10% horse serum for another 30 min at room temperature. Following incubation with anti-p53 or anti-Hsp60 antibodies (1:100) at 4˚C for 12 h, the coverslips were incubated with fluorescent secondary antibodies (1:200) for 30 min at room temperature, then rinsed with cold PBS and stained with Hoechst 33342 according to the manufacturer's instructions for 7 min before rinsing with PBS again. A Revolve Hybrid Microscope (Discover ECHO) was used for image capture. Images were further processed by ImageJ Software version 1.52s (National Institutes of Health) to assess the intensity of IF, as well as the mitochondrial parameters. For the evaluation of mitochondrial fusion, three images in each treatment group from three independent experiments were evaluated using the ImageJ Software and three parameters were used to assess the level of mitochondrial fusion as follows: The mean size of mitochondrial network (MS), the mean length of mitochondria (ML) and the number of mitochondrial nets/individual mitochondria (N/I).
Western blot (WB) analysis
Cells exposed to the different agents were harvested, washed at room temperature with cold PBS and lysed with 80 µl RIPA buffer. Cell lysates were centrifuged at 4,500 x g for 10 min at 4˚C and the supernatant proteins were harvested. A total of 10 µg proteins were separated on 12 or 15% (w/v) SDS-polyacrylamide gels and transferred to Immobilon™-P transfer membranes (EMD Millipore). Next, 10% (w/v) skimmed milk was used to block the membranes for 30 min at room temperature, before they were incubated with primary antibodies (anti-Hsp10, anti-phosphorylated-ubiquitin, anti-ubiquitin, anti-β-actin, anti-Bax, anti-PINK1, anti-caspase-3, anti-caspase-9, anti-Hsp60 or anti-p53; 1:1,000) overnight at 4˚C. The next day, after rinsing with PBST, the membranes were incubated with goat anti-mouse or goat anti-rabbit HRP-conjugated secondary antibodies (1:1,000) for 2 h at room temperature. After washing with PBST for three times, 10 min each, protein bands were visualized using the Syngene Bio Imaging System (Synoptics). β-actin was used as loading control. Quantification of protein bands was performed using the ImageJ Software version 1.52s (National Institutes of Health).
Knocking down TRAP1
shRNA sequences targeting human TRAP1 and the empty vector were purchased from Shanghai GenePharma Co., Ltd. The TRAP1 shRNA sequence was as follows: 5'-CCGGCAGAGCACTCACCCTACTATGC-TCGAGCATAGTAGGGTGAGTGCTCTGTTTTTG-3'. Plasmids were transduced into cells using transfection reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Briefly, SHG44 cells grown in 6-well plates were transfected with 4 µg TRAP1-shRNA or empty vector control and 6 µl transfection reagent was added to each well. Cells were harvested 24 h later and samples were further analyzed by WB analysis.
Apoptosis analysis
In order to evaluate the efficacy of TMZ and G-TPP treatment on apoptosis, SHG44 cells grown in 6-well plates were exposed to 400 µM TMZ (20), 15 µM G-TPP or 400 µM TMZ + 15 µM G-TPP for 24 h, then washed with cold PBS for three times and trypsinized at room temperature for 5 min by 0.25% trypsin solution without EDTA. Apoptosis assays were performed using the Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's instructions. Cells were then counted using a Guava® easyCyte flow cytometer (Merck KGaA).
Effects of combination treatment
A combination index (CI) is used to evaluate the synergistic effects of drugs according to the Chou-Talalay method (21) as follows: Synergism, CI<1; additive effect, CI=1; antagonism, CI >1. In the present study, growth inhibition effects were evaluated by MTT assays on SHG44 cells. To evaluate the effect of a combination treatment of TMZ and G-TPP, independent assays were conducted with different concentrations of TMZ (200, 400, 800, 1,000 and 1,600 µM) and G-TPP (5, 10, 15, 20, 30 and 40 µM) for 24 h and the growth inhibition effect of each treatment group was evaluated by MTT assays. Moreover, 15 µM G-TPP was combined with difference concentrations of TMZ (200, 400 and 800 µM) and in each independent assay, the two agents were added together simultaneously. The growth inhibition effect was evaluated by MTT following 24 h of treatment. The CI was calculated using CalcuSyn version 1.0.1 (Biosoft).
Statistical analysis
Data from three independent experiments were collected and analyzed using SPSS 22.0 (IBM Corp.) and Student's t-test. One-way ANOVA followed by Dunnett's post hoc test was also used to conduct multiple comparisons using Graphpad Prism 7.00 (GraphPad Software, Inc.). Data are presented as the means ± standard deviation and P<0.05 was considered to indicate a statistically significant difference.
Results
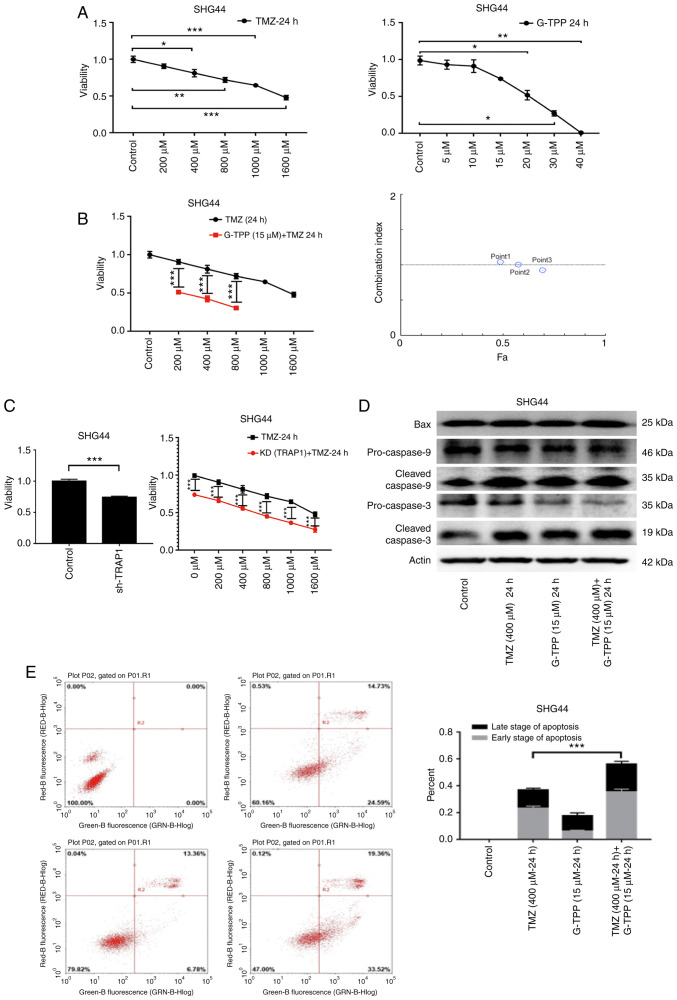
Inhibition of TRAP1 sensitizes GBM cells to TMZ
G-TPP combines the ATPase inhibition module of the Hsp90 inhibitor, 17-allylamino-17-demethoxy-geldanamycin and the mitochondrial targeting portion of TPP. This enables the drug to target and concentrate in the mitochondria and also exert the Hsp90 inhibitory function (22). In the MTT assay, G-TPP and TMZ mediated a significant concentration-dependent inhibitory effect on the viability of the GBM cell line, SHG44 (Fig. 2A). When the two agents were combined, cell viability was significantly more inhibited, and the CI analysis indicated an additive effect (Fig. 2B). When TRAP1 was knocked down by shRNA, the GBM cells were sensitized to TMZ (Fig. 2C) and WB revealed that the levels of expression of apoptosis-related proteins were increased (Fig. 2D). In the flow cytometric apoptosis assay, the degree of apoptosis in the combination treatment group was significantly increased (Fig. 2E). These results demonstrate that inhibition of TRAP1 sensitizes GBM cells to TMZ and indicate the existence of an underlying complementary pathophysiological process that is responsible.
Figure 2.
(A) Following treatment of GBM cell line SHG44 with TMZ and G-TPP the inhibition of cell viability was shown to be concentration-dependent. (B) Combination treatment with TMZ and G-TPP significantly inhibited the viability of GBM cells. Combination index analysis indicated that there was a synergistic effect of treatment with the two agents. Point 1: G-TPP (15 µM) + TMZ (200 µM) for 24 h; Point 2: G-TPP (15 µM) + TMZ (400 µM) for 24 h; Point 3: G-TPP (15 µM) + TMZ (800 µM) for 24 h. (C) Knocking down TRAP1 with shRNA may enhance the chemotherapeutic efficacy of TMZ to GBM cells. (D) Western blot confirmed that the combined treatment led to increased expression of the apoptosis-related proteins Bax, cleaved caspase-9 and cleaved caspase-3. (E) Flow cytometry demonstrated that the proportion of apoptosis in the group treated with both drugs increased significantly when compared with that of the TMZ group. *P<0.05; **P<0.01; ***P<0.001. GBM, glioblastoma multiforme; G-TPP, Gamitrinib triphenylphosphonium; TMZ, temozolomide; Fa, fraction affected; KD, knockdown; TRAP1, TNF receptor-associated protein 1.
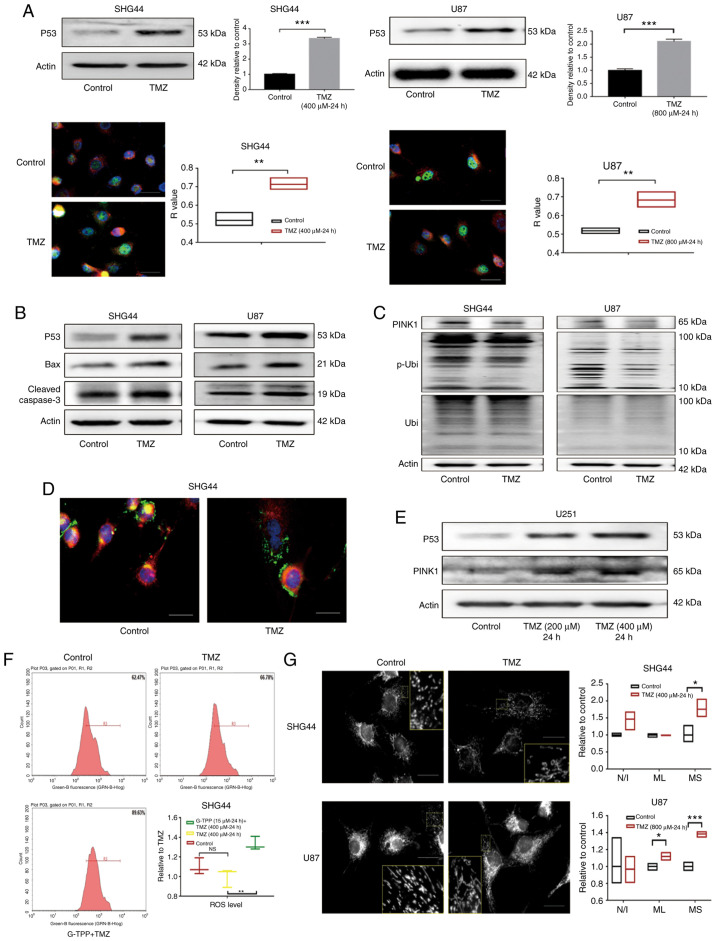
TMZ induces apoptosis of GBM cells by activating the p53 pathway and concurrently downregulates PINK1
The mechanism of TMZ-induced apoptosis in GBM cells was evaluated. After exposing GBM cell lines, SHG44 and U87 to titrated concentrations of TMZ, a significant increase in the expression of stress protein p53 was observed, which was confirmed by WB and IF assays. The co-localization analysis indicated that the p53 protein predominantly accumulated in the nucleus (Fig. 3A), which is consistent with the known role of p53 as an important nuclear transcription factor.
Figure 3.
(A) The expression of p53 in glioblastoma cell lines SHG44 and U87 was significantly upregulated following TMZ treatment. In the cell immunofluorescence assay, p53, nucleus and mitochondria were stained green, blue and red, respectively, which indicated that the upregulated p53 predominantly accumulated in the nucleus following TMZ treatment (scale bar, 80 µm). (B) Following treatment with TMZ, the upregulation of p53 and the activation of its downstream apoptotic pathways mediated by the Bcl-2 family was observed. (C) In GBM cells stimulated by TMZ, downregulation of PINK1 was observed, as well as a decreased level of Ser-65 site phosphorylated ubiquitin that indicated the impaired function of PINK1. (D) Following treatment with TMZ, the co-localization of lysosome (green) and mitochondria (red) decreased, which indicated the downregulation of mitophagy. The nucleus was stained blue (scale bar, 80 µm). (E) In the p53 mutant U251 cells, TMZ treatment increased the level of p53 and PINK1. (F) A total of 62.47% cells of the control group were included in the R3 gate, while 66.78% of the TMZ treatment group and 89.63% of the TMZ + G-TPP group were in the same R3 gate. The level of ROS in GBM cells in the TMZ treatment group was not statistically different from that in the control group (P=0.2892), while the level of ROS in the G-TPP and TMZ combined treatment group was significantly higher than the TMZ group (P=0.0041). (G) Following TMZ treatment, the level of mitochondrial fusion of glioblastoma cells was increased. The mean size of the MS was elevated in SHG44 and U87 cells, and the mean ML in U87 cells was also increased, although the difference in the number of N/I was not significantly different between the two groups (scale bar, 80 µm). *P<0.05; **P<0.01; ***P<0.001. GBM, glioblastoma multiforme; G-TPP, Gamitrinib triphenylphosphonium; TMZ, temozolomide; CI, combination index; TRAP1, TNF receptor-associated protein 1; PINK1, PTEN-induced kinase 1; p, phosphorylated; Ubi, ubiquitin; ROS, reactive oxygen species. MS, mitochondrial network; ML, length of mitochondria; N/I, mitochondrial nets/individual mitochondria.
Downstream, upregulation of the pro-apoptotic Bcl-2 family member Bax was observed. Previous studies have reported that p53 induces apoptosis by promoting the transcription of Bax (23,24). Downstream of Bax the activation of the apoptosis-executing protein caspase-3 was observed (Fig. 3B). In addition, PINK1 expression was markedly downregulated by TMZ in GBM cells. The function of PINK1 was evaluated with an antibody specific for phosphorylated-ubiquitin (Ser65). The phosphorylating activity of PINK1 was identified to be downregulated by TMZ (Fig. 3C). Furthermore, staining of lysosomes and mitochondria demonstrated a decreased co-localization of the two organelles in the TMZ treatment group, which indicated the downregulation of mitophagy (Fig. 3D). Notably, when the expression levels of p53 and PINK1 were evaluated in the p53 mutant GBM cell line, U251, the upregulation of p53 and PINK1 was observed under the TMZ treatment, which indicated that p53-PINK1 regulation may only exist in cell line with wild-type p53 (Fig. 3E).
Mitophagy is an important aspect of the mitochondrial quality control system. To evaluate mitochondrial function under the condition of downregulated mitophagy, intracellular ROS levels following TMZ treatment were assessed, but no difference was identified when compared with the control (Fig. 3F). Furthermore, mitochondrial morphology was assessed and the level of mitochondrial fusion in SHG44 and U87 cells was observed to be elevated following TMZ treatment (Fig. 3G). Currently, it is considered that mitochondrial fusion is a compensatory mechanism for impaired mitochondrial function, which can achieve functional complementation between dysfunctional mitochondria. This restores mitochondrial function to a certain extent (25) and reduces ROS generated by mitochondrial dysfunction and the subsequent damaging reactions.
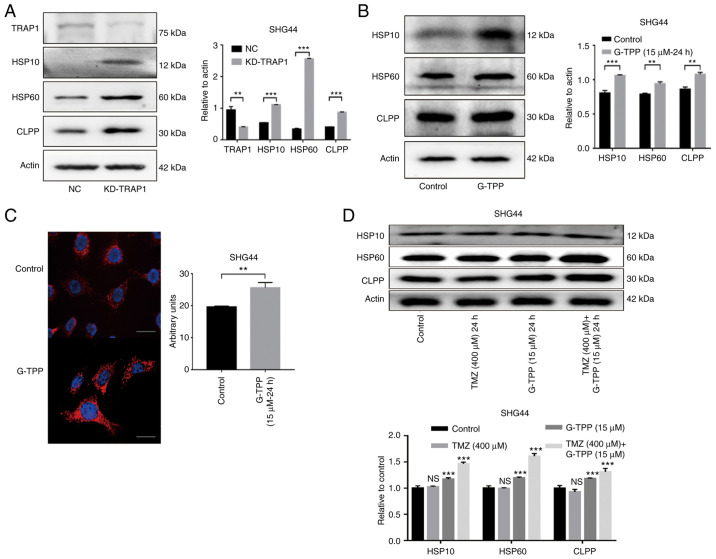
TRAP1 inhibition and TMZ treatment induce an exacerbated mitochondrial unfolded protein response (mtUPR) in GBM cells
The mtUPR was evaluated following TRAP1 knockdown using shRNA (Fig. 4A). The results indicated that TRAP1 plays an important role in the protein quality control system in mitochondria. SHG44 cells were then treated with G-TPP and a typical mtUPR was observed (Fig. 4B). IF assays to evaluate the level of Hsp60 expression also confirmed this effect (Fig. 4C). These findings are consistent with previous studies (26,27). Treatment with a combination of G-TPP and TMZ resulted in a higher mtUPR when compared to treatment with G-TPP alone, although no definite mtUPR occurred in cells that were treated with TMZ alone (Fig. 4D). This indicates that G-TPP and TMZ act together in aggravating the mtUPR in GBM cells.
Figure 4.
(A) The increased expression of Hsp10, Hsp60 and CLPP indicated that knocking down TRAP1 induced mtUPR. The efficiency of TRAP1 knockdown was ~60%. (B) G-TPP treatment of glioblastoma cells induced mtUPR. (C) Cell immunofluorescence assay confirmed that the expression level of Hsp60 (red) in glioblastoma cells was significantly increased following G-TPP treatment. Nuclei were stained blue (scale bar, 80 µm). (D) Increased expression of Hsp10, Hsp60 and CLPP was observed in the combined TMZ and G-TPP treatment group. **P<0.01; ***P<0.001 vs. the indicated groups or control. TRAP1, TNF receptor-associated protein 1; mtUPR, mitochondrial unfolded protein response; Hsp, heat shock protein; CLPP, caseinolytic mitochondrial matrix peptidase proteolytic subunit; G-TPP, Gamitrinib triphenylphosphonium; TMZ, temozolomide; NC, negative control; NS, not significant.
Combined treatment with G-TPP and TMZ significantly increases the level of ROS in glioblastoma cells
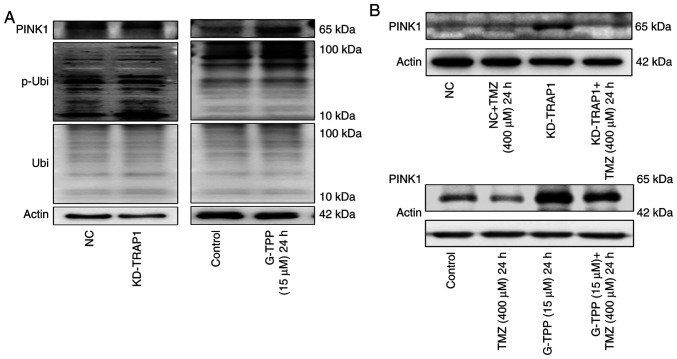
It is generally hypothesized that upon exposure to mtUPR-inducing factors, cells will increase mitophagy to remove damaged mitochondria, reduce the production of harmful metabolites and recycle material. This is exactly what was observed when TRAP1 was knocked down or G-TPP was administered in the present study, particularly, elevated PINK1 expression and phosphorylated ubiquitin at the Ser65 site (Fig. 5A). However, due to the inhibitory effect of TMZ on the expression of PINK1, mitophagy (an essential aspect of the mitochondrial quality control system) could not take place in the presence of a combination treatment with G-TPP and TMZ (Fig. 5B). When the extent of mtUPR exceeds the capacity of the compensatory mechanisms to maintain homeostasis in mitochondria, dysfunction of the mitochondrial system, such as fusion, occurs. This may be the underlying mechanism responsible for the increased level of mtUPR in cells treated with the combination of agents. Numerous damaged mitochondria with mtUPR that cannot be normally removed will affect mitochondrial metabolism and result in the production of increased quantities of deleterious factors, such as ROS (28). Therefore, the ROS level was evaluated and TMZ treatment alone did not affect the level of intracellular ROS, although this increased significantly when TMZ was combined with G-TPP (Fig. 3F). This is consistent with the above-mentioned inference. It is believed that a large quantity of ROS damages mitochondria and nuclear DNA and induces apoptosis (29) and that there may also be a cyclic amplification effect consisting of: ROS, DNA damage, ROS (29). Aggravated mtUPR and the following ROS burst may be the mechanism by which TRAP1 inhibition sensitizes GBM cells to TMZ treatment.
Figure 5.
(A) Knocking down TRAP1 or treatment with G-TPP may upregulate the expression of PINK1 and p-Ubi (Ser65). (B) When G-TPP was combined with TMZ, the expression level of PINK1 was lower than the G-TPP treatment group. TRAP1, TNF receptor-associated protein 1; p-Ubi, phosphorylated ubiquitin; G-TPP, Gamitrinib triphenylphosphonium; TMZ, temozolomide; KD, knockdown; NC, negative control.
Discussion
As the most malignant type of primary tumor of the central nervous system, glioblastoma is associated with a particularly poor prognosis (30). In the current regime for comprehensive therapy, chemotherapy plays an indispensable role and TMZ, as an effective chemotherapeutic agent verified by large-scale clinical trials, remains the dominant drug for GBM chemotherapy (30,31). However, prolonged TMZ chemotherapy results in the development of GBM resistance, which is an important factor affecting the efficacy of the drug (8,10). Therefore, there is an urgent need to develop appropriate adjuvant drugs for combination with TMZ chemotherapy to increase sensitivity and decrease resistance. TRAP1 became the focus of the present study as it is a molecule with an important role in the physiological and pathological processes of tumor cell biology. Studies have shown that TRAP1 is highly expressed in numerous types of cancer, indicating that this molecule may influence various shared behaviors of tumor cells (32,33).
Previous studies have shown that TRAP1 is involved in the protein quality control system in mitochondria. Knocking down TRAP1 causes a typical mtUPR (17), which is consistent with the findings of the present study. Furthermore, studies have reported that TRAP1 reduces the accumulation of ROS in cells (34,35). These characteristics indicate that TRAP1 may represent an alternative target for sensitizing GBM to TMZ chemotherapy. In the present study, it was found that TMZ induced apoptosis by activating p53 and its downstream pathways. Moreover, two other effects induced by TMZ are of note.
Firstly, on treatment with TMZ, tumor cells downregulate PINK1 expression and activity, which may in turn downregulate mitophagy. As one of the main pathways for initiating mitophagy, PINK1 anchors to the surface of mitochondria, phosphorylates the ubiquitin at Ser65 and recruits Parkin to conduct mitophagy (36). The observed downregulation of PINK1 expression and phosphorylating ability implies that the effect of TMZ downregulates mitophagy. Studies have shown that p53 binds to the PINK1 promoter region and blocks its transcription, which results in the downregulation of mitophagy (37). This is consistent with the TMZ-induced upregulation of p53 expression and aggregation in the nucleus in present study.
Secondly, the ROS level of tumor cells did not increase significantly while the level of mitochondrial fusion did increase in present study. Typically, decreased mitophagy will be accompanied by the reciprocal accumulation of damaged mitochondria, thus increasing the level of harmful metabolites, such as ROS in cells (38). However, in the case of TMZ treatment, it is possible that the elevated level of mitochondrial fusion compensates for the downregulation of mitophagy in sustaining a healthy mitochondrial system. Suppressing the function of TRAP1 in the present study, either by G-TPP treatment or by shRNA, may have overloaded this compensatory effect of mitochondrial fusion by inducing the mtUPR, which further exacerbated mitochondrial dysfunction and damage. The accumulation of dysfunctional or damaged mitochondria is always accompanied by the production of deleterious metabolites, such as ROS (39), which damage cells and induce apoptosis, as reported by a large number of earlier studies (40,41). In addition to inducing mitochondrial damage and nuclear DNA damage (42), ROS also activates the downstream JNK pathway and other pathways, thereby inducing apoptosis (43). It is hypothesized that this is the underlying mechanism for sensitizing GBM cells with wild-type p53 to TMZ, that is to say, by TRAP1 inhibition and the additive effect of TMZ and G-TPP treatment (Fig. 6). However, as p53 is frequently mutated in GBM, the present study aimed to establish whether this mechanism continues to work in p53 mutant GBM. U251, a cell line with mutant p53, was treated with TMZ and concentration-dependent, increased expression levels of p53 and PINK1 were observed. The point mutation in the DNA binding domain of p53 in U251 cells impairs the affinity of p53 to DNA, which weakens the downregulation of PINK1 by p53. This result strengthens the hypothesis that p53 downregulates the transcription of PINK1 in the form of nucleus transcriptional factor and also indicates that the above-mentioned mechanism may be ineffective in p53 mutant cell lines.
Figure 6.
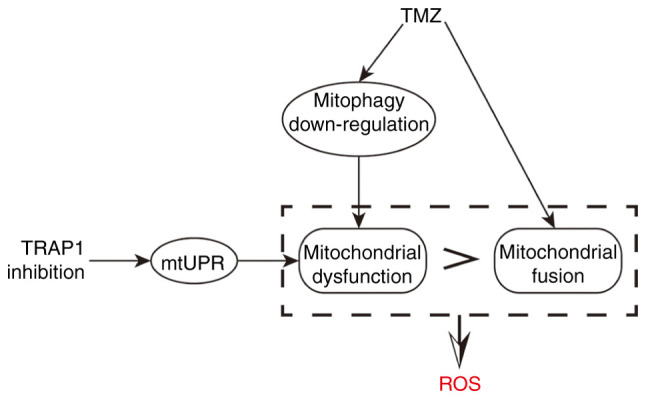
Research line of the present study. TMZ downregulated the mitophagy flux and promoted mitochondrial fusion. The downregulation of mitophagy leaded to the accumulation of damaged mitochondria, while mitochondrial fusion was a compensatory mechanism to counterbalance the damaged mitochondria. Suppressing TRAP1 on the basis of TMZ treatment induced mtUPR and aggravated the damage of the mitochondria. When the damage of the mitochondrial system surpassed the compensatory ability of mitochondrial fusion, the dysfunction of the mitochondrial system was observed with the increase of ROS levels. TMZ, temozolomide; TRAP1, TNF receptor-associated protein 1; mtUPR, mitochondrial unfolded protein response; ROS, reactive oxygen species.
The connections between the various pathways in tumor cells are complex. The present study cannot reveal all of the changes caused by TMZ treatment or interference of TRAP1, however, as a key molecule linking the mitochondrial protein quality control system and mitochondrial metabolism, TRAP1 plays important roles in many physiological and pathophysiological cellular processes. This in turn indicates that the present study on TRAP1 has great potential and practical value in the diagnosis and treatment of tumors.
Inhibiting the function of TRAP1 by G-TPP treatment or shRNA induces the mtUPR, while TMZ treatment downregulates mitophagy in addition to inducing apoptosis of GBM cells via the p53 pathway. The combination of these two effects aggravates the damage to the mitochondrial system and increases the accumulation of harmful metabolites and ROS in GBM cells, thus inducing adverse events, even apoptosis. Therefore, TRAP1, as a key molecule that affects the mitochondrial protein quality control system and mitochondrial metabolism, represents a promising target in the treatment of GBM. However, certain limitations exist to the present study. The mechanism revealed in the current study requires confirmation in additional cell lines and in vivo studies, and the mechanism of TMZ-induced mitochondrial fusion will be investigated in future studies.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was financially supported by the Department of Science and Technology of Jilin Province (grant no. 20180101136JC), the Department of Finance of Jilin Province (grant no. 2018SCZ030), the Education Department of Jilin Province (grant no. JJKH20190005KJ), Development and Reform Commission Engineering Laboratory Project of Jilin Province (grant no. 2019C031), the Norman Bethune Program of Jilin University (grant no. 2015218), the Lateral Research Funds of Jilin University (grant no. 2015377), the Excellent Talents Training Plan of China-Japan Union Hospital (grant no. YXZN-201803), the National Natural Science Foundation of China (grant nos. 81772794 and 81472419) and the Jilin Provincial Industrial Innovation Project (grant no. 2018C052-7).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NW conducted experiments and wrote the original draft of the manuscript. PZ and RH assisted with the experiments and figures. LS and DD reviewed and edited the manuscript and participated in the design of the study. YG conceptualized the project, performed project administration and acquired the funding. YG and DD confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol. 2014;16 (Suppl 4):iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wirsching HG, Galanis E, Weller M. Glioblastoma. Handb Clin Neurol. 2016;134:381–397. doi: 10.1016/B978-0-12-802997-8.00023-2. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert MR, Wang M, Aldape KD, Stupp R, Hegi ME, Jaeckle KA, Armstrong TS, Wefel JS, Won M, Blumenthal DT, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: A randomized phase III clinical trial. J Clin Oncol. 2013;31:4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu P, Li P, Lei T, Qu L, Huang H, Mu Q. Acute lymphoblastic leukemia following temozolomide treatment in a patient with glioblastoma: A case report and review of the literature. Oncol Lett. 2018;15:8663–8668. doi: 10.3892/ol.2018.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strobel H, Baisch T, Fitzel R, Schilberg K, Siegelin MD, Karpel-Massler G, Debatin KM, Westhoff MA. Temozolomide and other alkylating agents in glioblastoma therapy. Biomedicines. 2019;7(69) doi: 10.3390/biomedicines7030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oike T, Suzuki Y, Sugawara K, Shirai K, Noda SE, Tamaki T, Nagaishi M, Yokoo H, Nakazato Y, Nakano T. Radiotherapy plus concomitant adjuvant temozolomide for glioblastoma: Japanese mono-institutional results. PLoS One. 2013;8(e78943) doi: 10.1371/journal.pone.0078943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3:198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoter A, El-Sabban ME. The HSP90 family: Structure, regulation, function, and implications in health and disease. Int J Mol Sci. 2018;19(2560) doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altieri DC. Hsp90 regulation of mitochondrial protein folding: From organelle integrity to cellular homeostasis. Cell Mol Life Sci. 2013;70:2463–2472. doi: 10.1007/s00018-012-1177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altieri DC, Stein GS, Lian JB. TRAP-1, the mitochondrial Hsp90. Biochim Biophys Acta. 2012;1823:767–773. doi: 10.1016/j.bbamcr.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 15.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 16.Soti C, Vermes Á, Haystead TA, Csermely P. Comparative analysis of the ATP-binding sites of Hsp90 by nucleotide affinity cleavage: A distinct nucleotide specificity of the C-terminal ATP-binding site. Eur J Biochem. 2003;270:2421–2428. doi: 10.1046/j.1432-1033.2003.03610.x. [DOI] [PubMed] [Google Scholar]
- 17.Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, Ross AH, Plescia J, Altieri DC. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J Clin Invest. 2011;121:1349–1360. doi: 10.1172/JCI44855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang BH, Altieri DC. Compartmentalized cancer drug discovery targeting mitochondrial Hsp90 chaperones. Oncogene. 2009;28:3681–3688. doi: 10.1038/onc.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masgras I, Sanchez-Martin C, Colombo G. The chaperone TRAP1 as a modulator of the mitochondrial adaptations in cancer cells. Front Oncol. 2017;7(58) doi: 10.3389/fonc.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, Dai D, Zhou M, Li Z, Wang C, Lu Y, Li Y, Wang J. Inhibition of Cyclin D1 expression in human glioblastoma cells is associated with increased temozolomide chemosensitivity. Cell Physiol Biochem. 2018;51:2496–2508. doi: 10.1159/000495920. [DOI] [PubMed] [Google Scholar]
- 21.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 22.Im CN. Past, present, and emerging roles of mitochondrial heat shock protein TRAP1 in the metabolism and regulation of cancer stem cells. Cell Stress Chaperones. 2016;21:553–562. doi: 10.1007/s12192-016-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchenko ND. Mitochondrial death functions of p53. Mol Cell Oncol. 2014;1(e955995) doi: 10.1080/23723548.2014.955995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 25.Kulek AR, Anzell A, Wider JM, Sanderson TH, Przyklenk K. Mitochondrial quality control: Role in cardiac models of lethal ischemia-reperfusion injury. Cells. 2020;9(214) doi: 10.3390/cells9010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baqri RM, Pietron AV, Gokhale RH, Turner BA, Kaguni LS, Shingleton AW, Kunes S, Miller KE. Mitochondrial chaperone TRAP1 activates the mitochondrial UPR and extends healthspan in Drosophila. Mech Ageing Dev. 2014;141-142:35–45. doi: 10.1016/j.mad.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takemoto K, Miyata S, Takamura H, Katayama T, Tohyama M. Mitochondrial TRAP1 regulates the unfolded protein response in the endoplasmic reticulum. Neurochem Int. 2011;58:880–887. doi: 10.1016/j.neuint.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Wang X, Cueto R, Effi C, Zhang Y, Tan H, Qin X, Ji Y, Yang X, Wang H. Biochemical basis and metabolic interplay of redox regulation. Redox Biol. 2019;26(101284) doi: 10.1016/j.redox.2019.101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perillo B, Di Donato M, Pezone A, Di Zazzo E, Giovannelli P, Galasso G, Castoria G, Migliaccio A. ROS in cancer therapy: The bright side of the moon. Exp Mol Med. 2020;52:192–203. doi: 10.1038/s12276-020-0384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marenco-Hillembrand L, Wijesekera O, Suarez-Meade P, Mampre D, Jackson C, Peterson J, Trifiletti D, Hammack J, Ortiz K, Lesser E, et al. Trends in glioblastoma: Outcomes over time and type of intervention: A systematic evidence based analysis. J Neurooncol. 2020;147:297–307. doi: 10.1007/s11060-020-03451-6. [DOI] [PubMed] [Google Scholar]
- 31.Weller M, Le Rhun E, Preusser M, Tonn JC, Roth P. How we treat glioblastoma. ESMO Open. 2019;4 (Suppl 2)(e000520) doi: 10.1136/esmoopen-2019-000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietrafesa M, Maddalena F, Possidente L, Condelli V, Zoppoli P, Li Bergolis V, Rodriquenz MG, Aieta M, Vita G, Esposito F, Landriscina M. Gene copy number and Post-transductional mechanisms regulate TRAP1 expression in human colorectal carcinomas. Int J Mol Sci. 2019;21(145) doi: 10.3390/ijms21010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lettini G, Maddalena F, Sisinni L, Condelli V, Matassa DS, Costi MP, Simoni D, Esposito F, Landriscina M. TRAP1: A viable therapeutic target for future cancer treatments? Expert Opin Ther Targets. 2017;21:805–815. doi: 10.1080/14728222.2017.1349755. [DOI] [PubMed] [Google Scholar]
- 34.Hua G, Zhang Q. Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J Biol Chem. 2007;282:20553–20560. doi: 10.1074/jbc.M703196200. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Lu Y, Yu D, Zhang D, Hu W. TRAP1 provides protection against myocardial ischemia-reperfusion injury by ameliorating mitochondrial dysfunction. Cell Physiol Biochem. 2015;36:2072–2082. doi: 10.1159/000430174. [DOI] [PubMed] [Google Scholar]
- 36.Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goiran T, Duplan E, Rouland L, El Manaa W, Lauritzen I, Dunys J, You H, Checler F, Alves da Costa C. Nuclear p53-mediated repression of autophagy involves PINK1 transcriptional down-regulation. Cell Death Differ. 2018;25:873–884. doi: 10.1038/s41418-017-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Q, Li S, Jiang N, Shao X, Zhang M, Jin H, Zhang Z, Shen J, Zhou Y, Zhou W, et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol. 2019;26(101254) doi: 10.1016/j.redox.2019.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A, Kukreti R, Saso L, Kukreti S. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules. 2019;24(1583) doi: 10.3390/molecules24081583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozben T. Oxidative stress and apoptosis: Impact on cancer therapy. J Pharm Sci. 2007;96:2181–2196. doi: 10.1002/jps.20874. [DOI] [PubMed] [Google Scholar]
- 41.Simon HU, Haj-Yehia A, Levi-Schaffer F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis. 2000;5:415–418. doi: 10.1023/a:1009616228304. [DOI] [PubMed] [Google Scholar]
- 42.Poetsch AR. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput Struct Biotechnol J. 2020;18:207–219. doi: 10.1016/j.csbj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan F, Shao Z, Jiang S. Erlotinib induces the human non-small-cell lung cancer cells apoptosis via activating ROS-dependent JNK pathways. Cancer Med. 2016;5:3166–3175. doi: 10.1002/cam4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.