Abstract
Long non-coding (lnc) RNAs have been associated with osteoarthritis (OA) progression. The aim of the present study was to investigate the regulatory mechanism of lncRNA LINC01385 in OA in vitro. The mRNA expression level of LINC01385, microRNA(miR)-140-3p, and Toll-like receptor 4 (TLR4) was detected using reverse transcription-quantitative PCR, while ELISA was used to determine the concentration of different inflammatory factors [tumor necrosis factor-α (TNF-α), IL-6, and prostaglandin E2 (PGE2)]. The viability of human articular chondrocytes (HC-a) was measured using a MTT assay and western blot analysis was performed to quantify the protein expression level of TLR4. The associations between miR-140-3p and LINC01385/TLR4 were confirmed using a dual-luciferase reporter assay. LINC01385 mRNA expression level was increased in OA tissues and IL-1β-induced HC-a. LINC01385 knockdown and miR-140-3p mimics reduced the concentration of inflammatory factors in IL-1β-induced HC-a and promoted cell survival. In addition, it was confirmed that LINC01385 targeted miR-140-3p, while TLR4 was a target gene of miR-140-3p. Negative correlations between LINC01385 and miR-140-3p, and between miR-140-3p and TLR4 were observed in OA tissues. Low mRNA expression level of miR-140-3p and high protein expression level of TLR4 reversed the inhibitory effect of LINC01385 knockdown on the inflammatory responses of IL-1β-induced HC-a and exhibited a stimulating effect on cell viability. LINC01385 knockdown reduced the progression of OA by modulating the miR-140-3p/TLR4 axis in vitro; thus, LINC01385 may be a therapeutic target for OA.
Keywords: osteoarthritis, long non-coding RNA LINC01385, microRNA-140-3p, Toll-like receptor 4, in vitro
Introduction
Osteoarthritis (OA) is a type of degenerative joint disease that is most common in the elderly (aged ≥60 years), typically originates from chondrocytes (1,2). Patients with OA typically present with evidence of chondrocytes apoptosis, joint inflammation and cartilage sclerosis (3). The main treatment methods, such as acupuncture, drugs and electromagnetic therapies, are used to relieve pain or control symptoms; however, they cannot cure OA (4-8). Therefore, novel treatment strategies are required to cure OA.
An increasing number of studies have investigated long non-coding (lnc) RNAs and OA progression, and have revealed that lncRNAs play important roles in some cellular processes of OA (9-11). Tang et al (9) reported that silencing of lncRNA-p21 significantly increased cell viability and inhibited apoptosis in human chondrocytes. Both Hu et al (10) and Luo et al (11) discovered that in the human C28/I2 cartilage cell line, cell viability was promoted, whereas apoptosis and inflammatory cytokines levels were suppressed following transfection with short inhibiting RNA targeting H19 or MFI2-AS1. In addition, Li and Zhang (12) reported that LINC01385 may have a enhancing effect on the proliferation of nasopharyngeal carcinoma. Xiao et al (13) found that LINC01385 mRNA expression level was increased in human OA tissues using microarray and bioinformatics analyses. However, the detailed regulatory mechanism of LINC01385 in OA progression remains unclear.
Emerging evidence has confirmed that microRNAs (miRNAs/miR), such as miR-451(9), miR-130a-3p (10,11,14), miR-142-5p (15), miR-137(16), and miR-20b (17), play protective roles against the development of OA. In addition, an increasing amount of research has been directed towards understanding the anti-osteoarthritic role of miR-140 (18-20). Tardif et al (18) showed that miR-140 was regulated by NFAT3 and transforming growth factor-β/SMAD3 to inhibit the progression of OA. Furthermore, Tardif et al (19) demonstrated that the suppressive role of miR-140 in human OA was associated with the protein level of insulin-like growth factor-binding protein 5 and matrix metalloproteinase 13. Notably, Wang et al (20) showed that miR-140 exhibited its anti-osteoarthritic effects by promoting cell survival and suppressing inflammation; however, the ability of miR-140 to interact with LINC01385 to modulate OA progression requires further investigation.
In the present study, the effects of the LINC01385/miR-140-3p/TLR4 axis on the development and progression of OA was investigated and the results could identify a potential therapeutic target for OA.
Materials and methods
Tissues samples
A total of 25 patients with OA (males, 7; females, 18; mean age, 51.34±16.27 years) and 25 age-matched patients (males, 9; females, 16; mean age, 50.12±14.38 years) with a femoral fracture without OA or rheumatoid arthritis were selected from Liaocheng People's Hospital (Shandong, China) between December 2016 and July 2018. The inclusion criteria were: i) Patients who were diagnosed as OA (21); ii) patients who were willing to participate; iii) patients who fully understood the experimental protocol. The exclusion criteria were as follows: i) Patients with other disease complications, such as chronic inflammatory diseases; ii) patients who were treated within 3 months before admission. OA cartilage was collected from the patients with OA, who had undergone total knee replacement surgery, whereas the normal cartilage tissues were obtained from the knee joints of patients without OA or rheumatoid arthritis. All the cartilage samples were obtained in accordance with the diagnostic criteria of osteoarthritis from the Orthopaedic Society of the Chinese Medical Association (21). All participants provided written informed consent and the protocols of the present study were approved by the ethical committee of Liaocheng People's Hospital (approval no. 2020037).
Cell culture, transfection, and induction
The human articular chondrocytes (HC-a) were purchased from Tongpai Biotech, Co., Ltd., then cultured in DMEM containing 10% FBS at 37˚C in a humidified incubator with 5% CO2. To establish OA in vitro model, HC-a cells were treated with 10 ng/ml of IL-1β (Sigma-Aldrich; Merck KGaA) for 24 h at 37˚C and HC-a cells without IL-1β treatment were served as the controls. Afterwards, the short hairpin (sh)RNA targeting LINC01385 (sh-LINC01385), toll-like receptor 4 (sh-TLR4), and the non-targeting negative control (sh-NC) were purchased from VectorBuilder Inc. The linear pSIH1-H1-copGFP shRNA Vector was obtained from System Biosciences, LLC. Overexpression TLR4 (pcDNA-TLR4), overexpression LINC01385 (pcDNA-LINC01385), and their respective NCs (pcDNA3.1); miR-140-3p mimics (5'-UACCACAGGGUAGAACCACGG-3') and its NC (NC mimics, 5'-GCAAGAGACAAGCGCUUAGCC-3'); and miR-140-3p inhibitor (5'-CCGUGGUUCUACCCUGUGGUA-3') and its NC (NC inhibitor, 5'-GGUCCUGAUUCGUGCUACUCG-3') were obtained from Hunan Fenghui Biotechnology Co., Ltd. The aforementioned agents (20 nM) were transfected into the IL-1β-induced HC-a (2x106) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C. Subsequently, 48 h after transfection, the transfected HC-a cells were harvested to perform the following experiments.
Reverse transcription (RT)-quantitative PCR (RT-qPCR)
Total RNA was extracted from the OA and normal cartilage tissues, and the transfected HC-a using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). RNA was then quantified using a NanoDrop® 1000 (Thermo Fisher Scientific, Inc.) and RT was performed using a FastQuant cDNA First Chain Synthesis kit (Tiangen Biotech, Co., Ltd.) from 2 µg total RNA, according to the manufacturer's instructions. The conditions of RT were as follows: 10 min at 25˚C, 35 min at 50˚C and 15 min at 80˚C. qPCR was performed using Fast SYBR™ Green Master Mix (Thermo Fisher Scientific, Inc.) and an ABI 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used: Initial denaturation at 94˚C for 10 min, followed by 40 cycles at 94˚C for 10 sec, 60˚C for 20 sec and 72˚C for 1 min. GADPH and U6 were used as the internal references. Gene expression was quantified using the 2-ΔΔCq method (22). The following primer sequences were used: LINC01385 forward, 5'-TGTTTCTCGAGTGTGGGCAG-3' and reverse, 5'-GGCACTCGCGTTTTCTTCTG-3'; miR-140-3p forward, 5'-CACTCCAGCTGGGAGGCGGGGCGCCGCGGGA-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3'; TLR4 forward, 5'-AGTTGATCTACCAAGCCTTGAGT-3' and reverse, 5'-GCTGGTTGTCCCAAAATCACTTT-3'; GAPDH forward, 5'-CCAGGTGGTCTCCTCTGA-3' and reverse, 5'-GCTGTAGCCAAATCGTTGT-3' and U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'.
MTT assay
The viability of the HC-a was detected using a MTT assay. Briefly, the transfected and IL-1β-induced HC-a were seeded into a 96-well plate (2x105 cells per well) and incubated for 48 h at 37˚C. Subsequently, 20 µl MTT (Nanjing KeyGen Biotech Co., Ltd.) was added to each well and the cells were then incubated for 2 h at 37˚C. Following which, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to dissolve the formazan crystal. The viability (measured at 450 nm) was analyzed using a Multiskan Spectrum microplate reader (Thermo Fisher Scientific, Inc.).
ELISA
The concentrations of the inflammatory cytokines [IL-6 (cat. no. 70-EK106/2-96), tumor necrosis factor- α (TNF-α, cat. no. 70-EK182HS-96), and prostaglandin E2 (PGE2, cat. no. 70-EK8103/2-48; http://www.liankebio.com/gallery.html?scontent=n,70-EK8103%2F2-48)] in IL-1β-induced HC-a were measured using the corresponding ELISA kits [Hangzhou Multi Sciences (Lianke) Biotech Co., Ltd.] according to the manufacturer's protocols. The absorbance at 450 nm was measured using a Multiskan Spectrum microplate reader (Thermo Fisher Scientific, Inc.).
Target prediction
The miRNA targets of LINC01385 were predicted using the StarBase database version 2.0 (http://starbase.sysu.edu.cn/) and LncBase Predicted v.2 database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index-predicted). In addition, the mRNA targets of miR-140-3p were predicted using the StarBase database. TLR4 was selected for further analysis due to its important role in OA (23-25) and unknown relationship with miR-140-3p in OA.
Dual luciferase reporter (DLR) assay
LINC01385 with wild-type (WT) or mutant (mut) miR-140-3p-binding sites were ligated into the pGL3 vector (Promega Corporation). In addition, the 3'-untranslated region from TLR4 containing the predicted miR-140-3p recognition sequence was amplified by PCR using Platinum™ Direct PCR Universal Master Mix (cat. no. A44647100; Thermo Fisher Scientific, Inc.). The DNA template was isolated from the normal cartilage tissues of the patients without OA or rheumatoid arthritis using DNAzol™ Reagent (Thermo Fisher Scientific, Inc., cat. no. 10503027). The primer sequences were: forward, 5'-TGTATAGCAGAGTTCGTAT-3' and reverse 5'-ACTGAATTTTGTTGTCT-3'. The following thermocycling conditions were used: Initial denaturation at 94˚C for 5 min, followed by 30 cycles at 94˚C for 5 sec, 60˚C for 10 sec and 72˚C for 30 sec. The amplified product was ligated into the pGL3 vector. HC-a (2x105) were then co-transfected with LINC01385-mut/TLR4-mut or LINC01385-WT/TLR4-WT (80 ng) and miR-140-3p mimics or NC mimics (80 ng) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C. Following transfection for 48 h, Renilla and firefly luciferase activities were measured using the dual-luciferase reporter assay system (Promega Corporation). The activity of firefly luciferase was normalized to the activity of Renilla luciferase.
Western blot analysis
Following protein extraction from the HC-a using RIPA buffer containing protease inhibitor, the BCA protein assay kit (Abcam) was used to detect the protein concentrations. Proteins (~50 µg) were separated using 10% SDS-PAGE, then transferred onto PVDF membranes and blocking with 5% bovine serum albumin (Thermo Fisher Scientific, Inc.) at room temperature for 2 h. Following which, the membranes were incubated with primary antibodies against TLR4 (1:1,000; Abcam, cat. no. ab22048), and α-tubulin (1:1,000; Abcam, cat. no. ab7291) overnight at 4˚C. The membranes were then washed a minimum of three times using TBS-Tween-20 (TBST; Tween-20, 0.05%) and incubated with the HRP-conjugated anti-mouse IgG secondary antibody (1:5,000; Abcam, cat. no. ab6728) for 1 h at room temperature. Tubulin was used as the internal reference. The membranes were visualized using a ECL kit (Invitrogen; Thermo Fisher Scientific, Inc.) and quantified using ImageLab software version 1.46 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analysis was performed using SPSS v23.0 (IBM Corp). The differences between two groups or among multiple groups was analyzed using either a Student's t-test (unpaired) or one-way ANOVA followed by Tukey's multiple comparison test, respectively. The data are presented as the mean ± SD. The correlation was determined using Pearson's correlation analysis. P<0.05 was considered to indicate a statistically significant difference. All the experiments were conducted in triplicate, from at least three independent experiments.
Results
LINC01385 is highly expressed in OA tissues and in IL-1β-induced HC-a
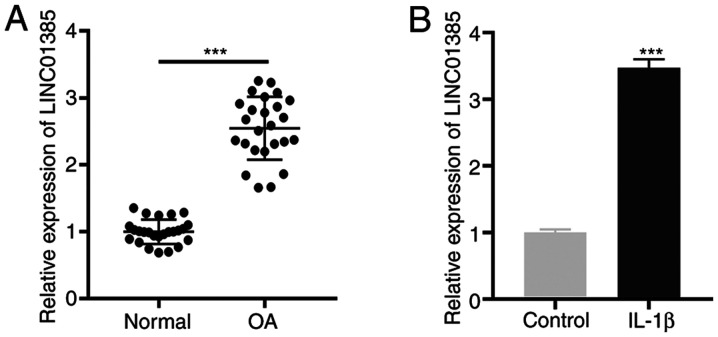
First, the mRNA expression level of LINC01385 in OA tissues was determined and the results showed that LINC01385 was significantly expressed in OA tissues compared with that in normal tissues (P<0.001; Fig. 1A). Similarly, an increased mRNA expression level of LINC01385 was observed in IL-1β-induced HC-a compared with that in the controls (P<0.001; Fig. 1B).
Figure 1.
LINC01385 is highly expressed in OA tissues and IL-1β-induced HC-a. (A) The mRNA expression level of LINC01385 in OA (n=25) and normal tissues (n=25) was detected using RT-qPCR. ***P<0.001 vs. normal. (B) The mRNA expression level of LINC01385 in IL-1β-induced HC-a was detected using RT-qPCR. ***P<0.001 vs. control. RT-qPCR, reverse transcription-quantitative PCR; OA, osteoarthritis.
LINC01385 knockdown suppresses inflammatory protein concentrations and promotes cell viability in the IL-1β-induced HC-a
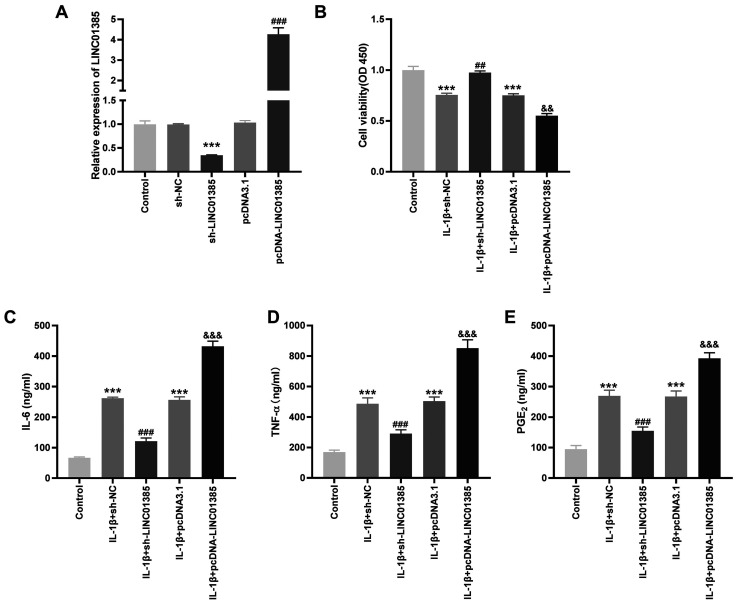
To investigate the potential role of LINC01385 on OA progression in vitro, the transfection efficiency of sh-LINC01385 and pcDNA-LINC01385 was evaluated. The mRNA expression level of LINC01385 was notably decreased following transfection with sh-LINC01385, whereas it was increased following transfection with pcDNA-LINC01385 (P<0.001; Fig. 2A). Following which, the viability of the IL-1β-induced HC-a was measured. Cell viability was inhibited in the IL-1β + sh-NC and IL-1β + pcDNA3.1 groups compared with that in the control (P<0.001; Fig. 2B). However, the viability of the HC-a was increased in the IL-1β + sh-LINC01385 group compared with that in the IL-1β + sh-NC group, whereas it was suppressed in the IL-1β + pcDNA-LINC01385 group compared with that in the IL-1β + pcDNA3.1 group (P<0.01; Fig. 2B). The concentrations of the inflammatory cytokines, which were measured using ELISA, revealed contrasting results. As shown in Fig. 2C-E, transfection of sh-NC or pcDNA3.1 increased the secretion of the inflammatory cytokines compared with that in the control. Compared with that in the sh-NC group, transfection with sh-LINC01385 significantly reduced the concentration of the inflammatory cytokines in the IL-1β-induced HC-a, while transfection with pcDNA-LINC01385 significantly increased the concentration of inflammatory cytokines compared with that in the pcDNA3.1 group (P<0.001).
Figure 2.
LINC01385 knockdown decreases the concentration of inflammatory factors and increases cell viability in the IL-1β-induced HC-a. (A) The mRNA expression level of LINC01385 following transfection with sh-LINC01385 or pcDNA-LINC01385 and their respective NCs into the HC-a was detected using reverse transcription-quantitative PCR. ***P<0.001 vs. the sh-NC group. ###P<0.001 vs. the pcDNA3.1 group. (B) The viability of the IL-1β-induced HC-a was measured using a MTT assay. ***P<0.001 vs. the Control group. ##P<0.01 vs. the IL-1β + sh-NC group; &&P<0.01 vs. the IL-1β + pcDNA3.1 group. The concentration of (C) IL-6, (D) TNF-α and (E) PGE2 in IL-1β-induced HC-a was measured using ELISA. ***P<0.001 vs. the Control group. ###P<0.001 vs. the IL-1β + sh-NC group. &&&P<0.001 vs. the IL-1β + pcDNA3.1 group. NC, negative control; sh, short hairpin; OD, optical density; TNF, tumor necrosis factor; PGE2, prostaglandin E2.
Identification miR-140-3p as a target of LINC01385
To investigate the downstream target of LINC01385, the StarBase database was used to predict potential binding sites between LINC01385 and miRNAs and miR-140-3p was found to be a target (Fig. 3A). DLR verified that the luciferase activity was significantly reduced in the presence of WT LINC01385 and miR-140-3p mimics; but not with miR-NC, suggesting that miR-140-3p was a target of LINC01385 (P<0.001; Fig. 3B). As illustrated in Fig. 3C, decreased mRNA expression level of miR-140-3p was detected in OA tissues compared with that in the normal tissues (P<0.001). In addition, there a negative correlation between the mRNA expression levels of LINC01385 and miR-140-3p in OA tissues (R=-0.805; P<0.0001; Fig. 3D). To further confirm the target relationship between LINC01385 and miR-140-3p, miR-140-3p mRNA expression level was detected following transfection with pcDNA-LINC01385 or sh-LINC01385 in the HC-a. The results from RT-qPCR revealed that miR-140-3p mRNA expression level was significantly decreased following transfection with pcDNA-LINC01385, while it was increased following transfection with sh-LINC01385 (P<0.001; Fig. 3E), which demonstrated that miR-140-3p was negatively regulated by LINC01385.
Figure 3.
Identification miR-140-3p as a target of LINC01385. (A) The predicted complementary binding site between LINC01385 and miR-140-3p. (B) The luciferase activity in the HC-a cells, co-transfected with pGL3-LINC01385 WT or pGL3-LINC01385 MUT with miR-140-3p mimics or NC mimics was determined using a dual luciferase reporter assay. ***P<0.001 vs. the NC mimics group. (C) The mRNA expression level of miR-140-3p in OA (n=25) and normal (n=25) tissues was detected using RT-qPCR. ***P<0.001 vs. normal. (D) The correlation between LINC01385 and miR-140-3p mRNA expression level. (E) The mRNA expression level of miR-140-3p following transfection with sh-LINC01385 or pcDNA-LINC01385-SNHG1 and their respective controls into the HC-a was detected using qRT-PCR. ***P<0.001 vs. the pcDNA3.1 group. ###P<0.001 vs. the sh-NC group. RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; NC, negative control; miR, microRNA; WT, wild-type; MUT, mutant; OA, osteoarthritis.
Overexpression of miR-140-3p reduces the concentration of the inflammatory cytokines in IL-1β-induced HC-a
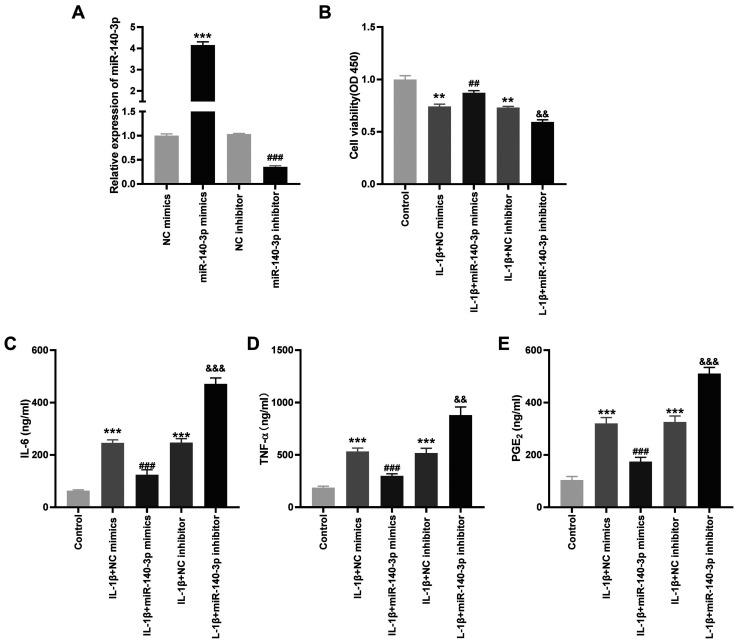
To investigate the role of miR-140-3p on the biological functions of OA in vitro, the transfection efficiency of miR-140-3p mimics and inhibitors was detected. miR-140-3p mRNA expression level was significantly increased by miR-140-3p mimics, whereas it was decreased by the miR-140-3p inhibitor (P<0.001; Fig. 4A). As shown in Fig. 4B-E, it was found that transfection with miR-140-3p mimics significantly increased cell viability and reduced the concentration of the inflammatory cytokines compared with that in the control group, whereas miR-140-3p inhibitor significantly reduced cell viability and increased the concentration of the inflammatory cytokines (P<0.01).
Figure 4.
Overexpression of miR-140-3p reduces the concentration of the inflammatory cytokines in the IL-1β-induced HC-a. (A) The mRNA expression level of miR-140-3p following transfection with miR-140-3p mimics or miR-140-3p inhibitor or their respective NCs into the HC-a was detected using reverse transcription-quantitative PCR. ***P<0.001 vs. the NC mimics group. ###P<0.001 vs. the NC inhibitor group. (B) The viability of the IL-1β-induced HC-a was measured using a MTT assay. **P<0.01 vs. the Control group. ##P<0.01 vs. the IL-1β + NC mimics group. &&P<0.01 vs. the IL-1β + NC inhibitor group. The concentration of (C) IL-6, (D) TNF-α and (E) PGE2 in IL-1β-induced HC-a was measured using ELISA. ***P<0.001 vs. the Control group. ###P<0.001 vs. the IL-1β + NC mimics group. &&&P<0.001 vs. the IL-1β + NC inhibitor group. NC, negative control; miR, microRNA; TNF, tumor necrosis factor; PGE2, prostaglandin E2.
miR-140-3p targets TLR4
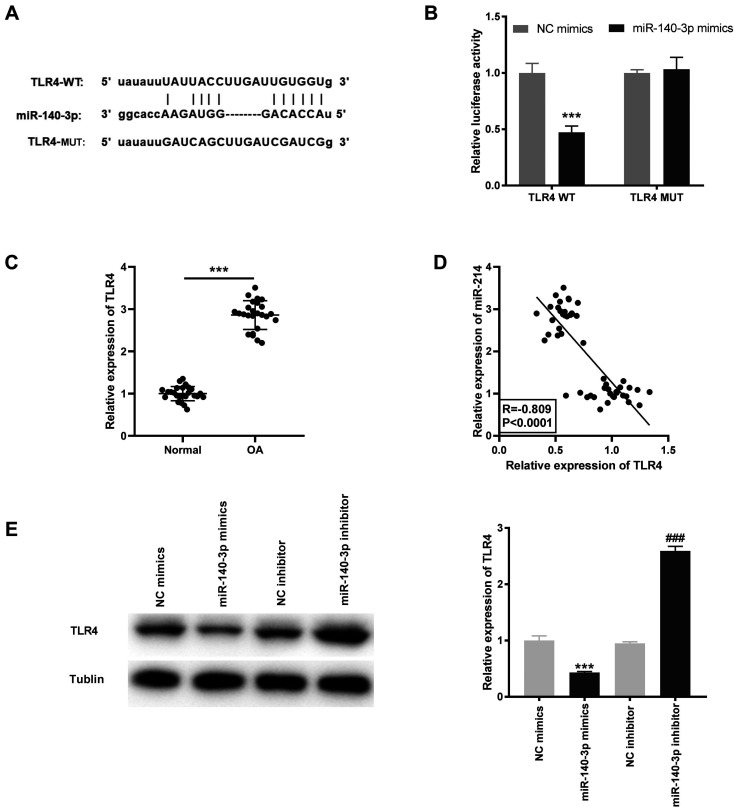
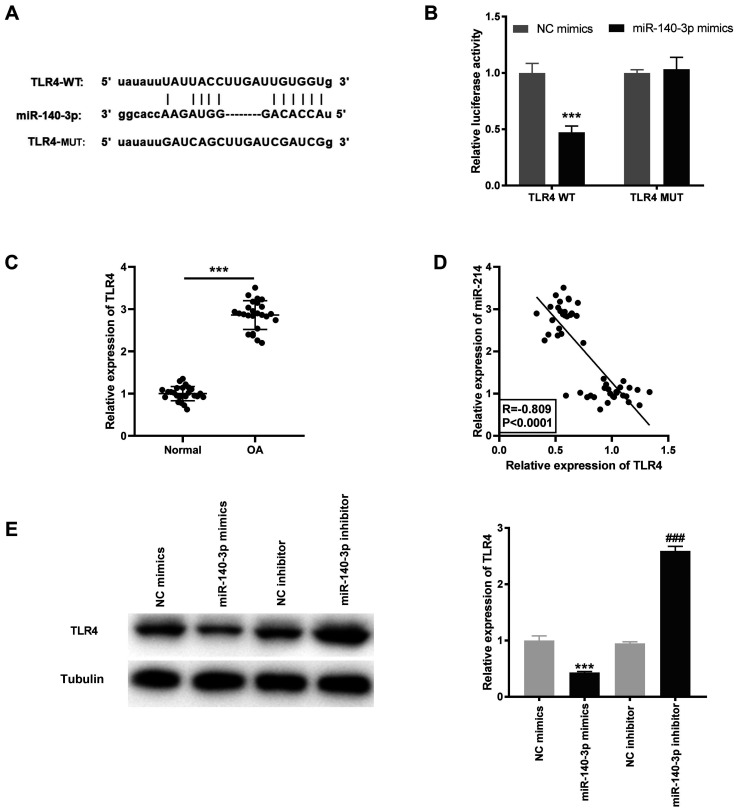
Based on the results of the StarBase database, the potential binding site between miR-140-3p and TLR4 was identified (Fig. 5A). Using the DLR assay, it was found that the luciferase activity was significantly decreased in the TLR4 WT/miR-140-3p mimics group compared with that in the TLR4 WT/miR-NC group (P<0.001; Fig. 5B). TLR4 mRNA expression level was significantly higher in OA tissues compared with that in the normal tissues (P<0.001; Fig. 5C) and was negatively correlated with miR-140-3p mRNA expression levels (R=-0.809; P<0.0001; Fig. 5D). Furthermore, the TLR4 protein expression level was determined following transfection with miR-140-3p mimics or inhibitor into the HC-a to further confirm the interaction between miR-140-3p and TLR4. The results showed that the TLR4 protein expression level was significantly inhibited by miR-140-3p mimics and increased by miR-140-3p inhibitor (P<0.001; Fig. 5E).
Figure 5.
TLR4 is a target gene of miR-140-3p. (A) The predicted complementary binding site between miR-140-3p and TLR4. (B) The luciferase activity in the HC-a, co-transfected with pGL3-TLR4 WT or pGL3-TLR4 MUT and with either miR-140-3p mimics or NC mimics was determined using a dual luciferase reporter assay. ***P<0.001 vs. the NC mimics group. (C) The mRNA expression level of TLR4 in OA (n=25) and normal (n=25) tissues was detected using reverse transcription-quantitative PCR. ***P<0.001 vs. normal. (D) The correlation between TLR4 and miR-140-3p expression level. (E) The protein expression level of TLR4 following transfection with miR-140-3p mimics or miR-140-3p inhibitor and their respective controls into the HC-a was measured using western blot analysis. ***P<0.001 vs. the NC mimics group, ###P<0.001 vs. the NC inhibitor group. NC, negative control; miR, microRNA; WT, wild-type; MUT, mutant; OA, osteoarthritis; TLR4, toll-like receptor 4.
Decreased TLR4 mRNA expression level suppresses the concentration of the inflammatory cytokines in OA in vitro
To investigate the effect of TLR4 on OA inflammation in vitro, the transfection efficiency of sh-TLR4 and pcDNA-TLR4 was initially determined. TLR4 mRNA expression level in the HC-a transfected with sh-TLR4 was decreased, while it was increased following transfection with pcDNA-TLR4 (P<0.001; Fig. 6A). Similarly, the protein expression level of TLR4 was significantly reduced following transfection with sh-TLR4 and was increased by pcDNA-TLR4 (P<0.001; Fig. 6B). As illustrated in Fig. 6C-E, the concentrations of IL-6, TNF-α, and PGE2 were significantly reduced following TLR knockdown and increased following overexpression of TLR (P<0.01).
Figure 6.
Decreased TLR4 is identified as a suppressor of OA inflammatory factors in vitro. (A) The mRNA and (B) protein expression level of TLR4 following transfection with sh-TLR4 or pcDNA-TLR4 and their respective negative controls into the HC-a was detected using reverse transcription-quantitative PCR and western blot analysis respectively. ***P<0.001 vs. the sh-NC group. ###P<0.001 vs. the pcDNA3.1 group. The concentration of (C) IL-6, (D) TNF-α and (E) PGE2 in IL-1β-induced HC-a was measured using ELISA. ***P<0.001 vs. the Control group. ###P<0.01 vs. the IL-1β + sh-NC group. &&P<0.01 vs. the IL-1β + pcDNA3.1 group. &&&P<0.001 vs. the IL-1β + pcDNA3.1 group. NC, negative control; miR, microRNA; TNF, tumor necrosis factor; PGE2, prostaglandin E2; TLR4, toll-like receptor 4.
Knockdown of LINC01385 suppresses the development of OA by regulating the miR-140-3p/TLR4 axis in vitro
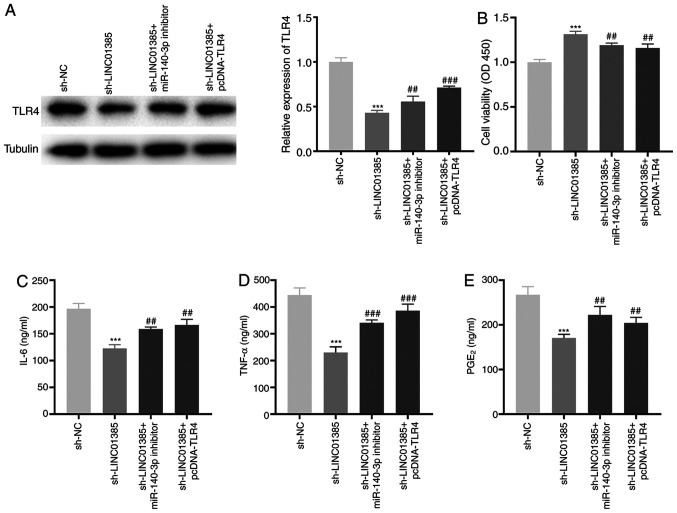
To investigate the regulatory mechanisms of LINC01385, miR-140-3p and TLR4 on OA in vitro, sh-LINC01385, sh-LINC01385 + miR-140-3p inhibitor or sh-LINC01385 + pcDNA-TLR4 was transfected into the HC-a. The protein expression level of TLR4 was decreased following transfection with sh-LINC01385, whereas it was increased following transfection with sh-LINC01385 + miR-140-3p inhibitor or sh-LINC01385 + pcDNA-TLR4 (P<0.01; Fig. 7A). Following which, rescue experiments revealed that transfection with miR-140-3p inhibitor and TLR4 overexpression vector reversed the enhancing effect of LINC01385 knockdown on cell viability, and the inhibitory effects on secretion of the inflammatory cytokines in IL-1β-treated HC-a (P<0.01; Fig. 7B-E).
Figure 7.
Knockdown of LINC01385 suppresses the development of OA by regulating the miR-140-3p/TLR4 axis in vitro. (A) The protein expression level of TLR4 following transfection with sh-LINC01385, sh-LINC01385 + miR-140-3p inhibitor or sh-LINC01385 + pcDNA-TLR4 into the HC-a was measured using western blot analysis. ***P<0.001 vs. the sh-NC group. ##P<0.01, ###P<0.001 vs. the sh-LINC01385 group. (B) The viability of the IL-1β-induced HC-a was measured using a MTT assay. ***P<0.001 vs. the sh-NC group. ##P<0.01 vs. the sh-LINC01385 group. The concentration of (C) IL-6, (D) TNF-α and (E) PGE2 in IL-1β-induced HC-a was measured using ELISA. ***P<0.001 vs. the sh-NC group. ##P<0.01, ###P<0.001 vs. the sh-LINC01385 group. NC, negative control; miR, microRNA; sh, short hairpin; TNF, tumor necrosis factor; PGE2, prostaglandin E2; TLR4, toll-like receptor 4.
Discussion
OA is a common joint disease, which causes suffering and inconvenience to the patients, due to joint pain on a daily basis (26). Furthermore, it is a leading cause of disability and shortening of an adult working life globally (26). Numerous lncRNAs have been associated with the development of OA (27-29). For example, lncRNA anti-differentiation non-coding RNA expression was increased in plasma specimens from patients with OA and CHON-001 human chondrocytes (27). The mRNA expression level of lncRNA MALAT1 was increased in OA tissues and in a time-dependent manner in IL-1β-induced chondrocytes (28). Furthermore, increased mRNA expression of FOXD2-AS1 was detected in OA cartilage tissue (29). Similar results were obtained in the present study, as LINC01385 mRNA expression was found to be higher in OA tissues and in IL-1β-induced HC-a, suggesting that LINC01385 may play a pathogenic role in OA.
Over the past decade, lncRNAs have been shown to play crucial roles in regulating proliferation, apoptosis and inflammatory reactions in OA progression (10,11,15). For example, MFI2-AS1 silencing reversed the inhibitory effect of lipopolysaccharide (LPS)-induced OA on cell viability and the promoting effect on apoptosis and inflammation in chondrocytes (11). In addition, knockdown of XIST played a stimulatory role in the proliferation of human SW1353 chondrocytes (15). The cell viability of LPS-treated human C28/I2 cartilage cells was facilitated by H19 knockdown, whereas apoptosis and the concentrations of inflammatory cytokines (IL-6, IL-1β, and TNF-α) were suppressed (10). Consistent with this previous study, in the present study it was found that transfection with sh-LINC01385 significantly reduced cell viability in IL-1β-induced HC-a, whilst it decreased the concentration of IL-6, TNF-α and PGE2. However, contrasting results were obtained following transfection with pcDNA-LINC01385. Therefore, we hypothesized that silencing of LINC01385 protected against OA.
Numerous studies have indicated that miRs serve as suppressors in the progression of OA (11,15,16). Luo et al (11) demonstrated that miR-130a-3p was minimally expressed in OA tissues, whereas overexpression of miR-130a-3p notably promoted cell viability and inhibited inflammation in LPS-induced C28/I2 cells. Sun et al (15) showed that the mRNA expression level of miR-142-5p was decreased in IL-1β-stimulated SW1353 chondrocytes, while the proliferation of these cells was increased following transfection with miR-142-5p mimics. A recent study conducted by Wang et al (16) detected low mRNA expression level of miR-137 in OA tissues and increase of miR-137 expression in LPS-treated human chondrocytes significantly elevated cell viability, suppressed apoptosis and reduced the concentration of inflammatory cytokines (16). In the present study, decreased mRNA expression level of miR-140-3p was found in OA tissues. Overexpression of miR-140-3p significantly increased cell viability, whereas it reduced the concentration of inflammatory factors in IL-1β-induced HC-a, suggesting that miR-140-3p could be an anti-inflammatory miRNA in OA. In accordance with the results from the present study, Wang et al revealed that miR-140 expression level was reduced in OA tissues, and transfection of miR-140 mimics into IL-1β-induced chondrocytes increased cell viability and reduced the concentration of IL-6 and TNF-α (20). By contrast, it was shown in the present study that transfection with miR-140-3p inhibitor reduced cell viability and increased the concentration of inflammatory factors. Simultaneously, miR-140-3p was verified as a target of LINC01385 and was found to be negatively regulated by it. We hypothesized that silencing of LINC01385 attenuated OA progression by negatively regulating miR-140-3p. The results from the present study verified that knockdown of miR-140-3p reversed the promoting effect of LINC01385 knockdown on cell viability and the inhibitory effect on inflammation in IL-1β-induced HC-a cells. Therefore, miR-140-3p was negatively regulated by LINC1385 and found to be associated with the progression of OA.
TLR4, a member of the TLR family, has been associated with inflammation, including in OA (30,31). Liu et al (32) showed that TLR4 mRNA expression was increased in human OA chondrocytes. In addition, other studies have demonstrated that TLR4 was overexpressed in OA tissues (23-25). In accordance with these studies, it was found that mRNA expression level of TLR4 was increased in OA tissues compared with that in normal tissues. These results suggested that TLR4 might be a pro-inflammatory gene in OA. The ELISA results showed that the concentration of IL-6, TNF-α and PGE2 in IL-1β-induced HC-a was inhibited following transfection with sh-TLR4; however, it was increased following transfection with pcDNA-TLR4, which supports the aforementioned hypothesis. Furthermore, TLR4 was found to be a target gene of miR-140-3p. As aforementioned, the results from the present study showed that LINC01385 decreased OA progression by regulating miR-140-3p. Therefore, we hypothesized that silencing of LINC01385 ameliorated OA by regulating the miR-140-3p/TLR4 axis. The results of rescue experiments showed that TLR4 overexpression reversed the effect of LINC01385 knockdown on increasing cell viability and the inhibitory effect on the increase in the concentration of inflammatory factors in IL-1β-induced HC-a, which confirmed our hypothesis. In conclusion, the results of the present study suggested that silencing of LINC01385 inhibited OA progression by modulating the miR-140-3p/TLR4 axis.
However, there are some limitations to the present study. First, OA progression was investigated at a cellular level and in vivo experiments should be performed. Second, OA pathogenesis is complex and the LINC01385/miR-140-3p/TLR4 may not be the sole regulatory axis involved. Therefore, further elucidation of alternate pathways and conduction of in vivo experiments are warranted.
In summary, the present study elucidated that LINC01385 acted as an endogenous sponge of miR-140-3p to promote cell viability and suppress inflammation in IL-1β-induced HC-a. In addition, TLR4 was found to be associated with OA progression, as a target gene of miR-140-3p. The present study demonstrated that the LINC01385/miR-140-3p/TLR4 axis could be important in the development of OA, providing a potential therapeutic target for OA.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author's contributions
ZW and CH were involved in the conception and design of the study, analyzed the data, and drafted the manuscript. CZ, HZ and ZZ made substantial contributions to analysis and interpretation of data and revised the article critically for important intellectual content. DX made substantial contributions to conception and design and revised the article. ZW and CH confirm the authenticity of all the raw data. All the authors performed the experiments and approved the final version of the manuscript.
Ethics approval and consent to participate
This study was conducted after obtaining ethics approval from the Liaocheng People's Hospital's Ethics Committee (no. 2020037).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Neogi T, Zhang Y. Epidemiology of osteoarthritis. Rheum Dis Clin North Am. 2013;39:1–19. doi: 10.1016/j.rdc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markstedt K, Mantas A, Tournier I, Martinez Avila H, Hagg D, Gatenholm P. 3D bioprinting human chondrocytes with nanocellulose-alginate bioink for cartilage tissue engineering applications. Biomacromolecules. 2015;16:1489–1496. doi: 10.1021/acs.biomac.5b00188. [DOI] [PubMed] [Google Scholar]
- 3.Cross M, Smith E, Hoy D, Nolte S, Ackerman I, Fransen M, Bridgett L, Williams S, Guillemin F, Hill CL, et al. The global burden of hip and knee osteoarthritis: Estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73:1323–1330. doi: 10.1136/annrheumdis-2013-204763. [DOI] [PubMed] [Google Scholar]
- 4.Zhang W, Nuki G, Moskowitz RW, Abramson S, Altman RD, Arden NK, Bierma-Zeinstra S, Brandt KD, Croft P, Doherty M, et al. OARSI recommendations for the management of hip and knee osteoarthritis: Part III: Changes in evidence following systematic cumulative update of research published through January 2009. Osteoarthritis Cartilage. 2010;18:476–499. doi: 10.1016/j.joca.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Majeed MH, Sherazi SA, Bacon D, Bajwa ZH. Pharmacological treatment of pain in osteoarthritis: A descriptive review. Curr Rheumatol Rep. 2018;20(88) doi: 10.1007/s11926-018-0794-5. [DOI] [PubMed] [Google Scholar]
- 6.Wollheim FA. Current pharmacological treatment of osteoarthritis. Drugs. 1996;52 (Suppl 3):27–38. doi: 10.2165/00003495-199600523-00006. [DOI] [PubMed] [Google Scholar]
- 7.Chun-Lei Y. Clinical observation on treatment of knee osteoarthritis with internal and external therapies in Chinese medicine. Liaoning J Tradit Chin Med. 2010;9:1744–1745. (In Chinese) [Google Scholar]
- 8.Baker CL Jr, Ferguson CM. Future treatment of osteoarthritis. Orthopedics. 2005;28 (Suppl 2):s227–s234. doi: 10.3928/0147-7447-20050202-08. [DOI] [PubMed] [Google Scholar]
- 9.Tang L, Ding J, Zhou G, Liu Z. LncRNAp21 promotes chondrocyte apoptosis in osteoarthritis by acting as a sponge for miR451. Mol Med Rep. 2018;18:5295–5301. doi: 10.3892/mmr.2018.9506. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, Li S, Zou Y. Knockdown of lncRNA H19 relieves LPS-induced damage by modulating miR-130a in osteoarthritis. Yonsei Med J. 2019;60:381–388. doi: 10.3349/ymj.2019.60.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo X, Wang J, Wei X, Wang S, Wang A. Knockdown of lncRNA MFI2-AS1 inhibits lipopolysaccharide-induced osteoarthritis progression by miR-130a-3p/TCF4. Life Sci. 2020;240(117019) doi: 10.1016/j.lfs.2019.117019. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Zhang F. Novel long noncoding RNA LINC01385 promotes nasopharyngeal carcinoma proliferation via the miR-140-3p/Twist1 signaling pathway. Cell Cycle. 2020;19:1352–1362. doi: 10.1080/15384101.2020.1750133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao K, Yang Y, Bian Y, Feng B, Li Z, Wu Z, Qiu G, Weng X. Identification of differentially expressed long noncoding RNAs in human knee osteoarthritis. J Cell Biochem. 2019;120:4620–4633. doi: 10.1002/jcb.27750. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Li J, Shao W, Shen N. LncRNA CTBP1-AS2 is upregulated in osteoarthritis and increases the methylation of miR-130a gene to inhibit chondrocyte proliferation. Clin Rheumatol. 2020;39:3473–3478. doi: 10.1007/s10067-020-05113-4. [DOI] [PubMed] [Google Scholar]
- 15.Sun P, Wu Y, Li X, Jia Y. miR-142-5p protects against osteoarthritis through competing with lncRNA XIST. J Gene Med. 2020;22(e3158) doi: 10.1002/jgm.3158. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Fang L, Ye L, Ma S, Huang H, Lan X, Ma J. miR-137 targets the inhibition of TCF4 to reverse the progression of osteoarthritis through the AMPK/NF-κB signaling pathway. Biosci Rep. 2020;40(BSR20200466) doi: 10.1042/BSR20200466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Zhang L, Li E, Zhang G, Hou Y, Yuan W, Qu W, Ding L. Long-chain non-coding RNA HOTAIR promotes the progression of osteoarthritis via sponging miR-20b/PTEN axis. Life Sci. 2020;253(117685) doi: 10.1016/j.lfs.2020.117685. [DOI] [PubMed] [Google Scholar]
- 18.Tardif G, Pelletier JP, Fahmi H, Fahmi H, Hum D, Zhang Y, Kapoor M, Martel-Pelletier J. NFAT3 and TGF-β/SMAD3 regulate the expression of miR-140 in osteoarthritis. Arthritis Res Ther. 2013;15(R197) doi: 10.1186/ar4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tardif G, Hum D, Pelletier JP, Duval N, Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC Musculoskelet Disord. 2009;10(148) doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Shen S, Li Z, Li W, Weng X. miR-140-5p affects chondrocyte proliferation, apoptosis, and inflammation by targeting HMGB1 in osteoarthritis. Inflamm Res. 2020;69:63–73. doi: 10.1007/s00011-019-01294-0. [DOI] [PubMed] [Google Scholar]
- 21.Wang MY. Review and prospect: 2009's annual report of Orthopaedic Trauma Society of Chinese Medical Association. Chinese Journal of Orthopaedic Trauma. 2010;1(3) (In Chinese) [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.De Nardo D. Toll-like receptors: Activation, signalling and transcriptional modulation. Cytokine. 2015;74:181–189. doi: 10.1016/j.cyto.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Wang P, Zhu F, Tong Z, Konstantopoulos K. Response of chondrocytes to shear stress: Antagonistic effects of the binding partners Toll-like receptor 4 and caveolin-1. FASEB J. 2011;25:3401–3415. doi: 10.1096/fj.11-184861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HA, Cho ML, Choi HY, Yoon CS, Jhun JY, Oh HJ, Kim HY. The catabolic pathway mediated by toll-like receptors in human osteoarthritic chondrocytes. Arthritis Rheum. 2006;54:2152–2163. doi: 10.1002/art.21951. [DOI] [PubMed] [Google Scholar]
- 26.Schiphof D, van den Driest JJ, Runhaar J. Osteoarthritis year in review 2017: Rehabilitation and outcomes. Osteoarthritis Cartilage. 2018;26:326–340. doi: 10.1016/j.joca.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Li Q, Zhang Z, Guo S, Tang G, Lu W, Qi X. LncRNA ANCR is positively correlated with transforming growth factor-β1 in patients with osteoarthritis. J Cell Biochem. 2019;120:14226–14232. doi: 10.1002/jcb.28881. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Wang F, Chen G, He R, Yang L. LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis. Cell Biosci. 2019;9(54) doi: 10.1186/s13578-019-0302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Cao L, Wang Q, Huang J, Xu S. LncRNA FOXD2-AS1 induces chondrocyte proliferation through sponging miR-27a-3p in osteoarthritis. Artif Cells Nanomed Biotechnol. 2019;47:1241–1247. doi: 10.1080/21691401.2019.1596940. [DOI] [PubMed] [Google Scholar]
- 30.Herrero-Beaumont G, Perez-Baos S, Sanchez-Pernaute O, Roman-Blas JA, Lamuedra A, Largo R. Targeting chronic innate inflammatory pathways, the main road to prevention of osteoarthritis progression. Biochem Pharmacol. 2019;165:24–32. doi: 10.1016/j.bcp.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 31.Gómez R, Villalvilla A, Largo R, Gualillo O, Herrero-Beaumont G. TLR4 signalling in osteoarthritis-finding targets for candidate DMOADs. Nat Rev Rheumatol. 2015;11:159–170. doi: 10.1038/nrrheum.2014.209. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, Gu H, Liu H, Jiao Y, Li K, Zhao Y, An L, Yang J. Protective effect of resveratrol against IL-1β-induced inflammatory response on human osteoarthritic chondrocytes partly via the TLR4/MyD88/NF-κB signaling pathway: an ‘in vitro study’. Int J Mol Sci. 2014;15:6925–6940. doi: 10.3390/ijms15046925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.