Abstract
After a sudden shift to high osmolarity, Saccharomyces cerevisiae cells respond by transiently inducing the expression of stress-protective genes. Msn2p and Msn4p have been described as two transcription factors that determine the extent of this response. In msn2 msn4 mutants, however, many promoters still show a distinct rise in transcriptional activity upon osmotic stress. Here we describe two structurally related nuclear factors, Msn1p and a newly identified protein, Hot1p (for high-osmolarity-induced transcription), which are also involved in osmotic stress-induced transcription. hot1 single mutants are specifically compromised in the transient induction of GPD1 and GPP2, which encode enzymes involved in glycerol biosynthesis, and exhibit delayed glycerol accumulation after stress exposure. Similar to a gpd1 mutation, a hot1 defect can rescue cells from inappropriately high HOG pathway activity. In contrast, Hot1p has little influence on the osmotic stress induction of CTT1, where Msn1p appears to play a more prominent role. Cells lacking Msn1p, Msn2p, Msn4p, and Hot1p are almost devoid of the short-term transcriptional response of the genes GPD1, GPP2, CTT1, and HSP12 to osmotic stress. Such cells also show a distinct reduction in the nuclear residence of the mitogen-activated protein kinase Hog1p upon osmotic stress. Thus, Hot1p and Msn1p may define an additional tier of transcriptional regulators that control responses to high-osmolarity stress.
The HOG (high-osmolarity glycerol response) mitogen-activated protein (MAP) kinase pathway plays a pivotal role in the adaptation of Saccharomyces cerevisiae to conditions of high external osmolarity. This is best illustrated by the fact that cells deficient in this pathway cannot proliferate on media containing high levels of osmotically active molecules (6). On the other hand, ectopic activation of the pathway is also detrimental for cell growth (19, 31). This situation has allowed an extensive genetic analysis, leading to the identification of many positive and negative elements of the HOG signaling pathway (19, 21, 24, 30, 31, 37–41, 60). The system relies on the input of two independent and structurally unrelated sensors whose signals converge at the level of the MAP kinase kinase Pbs2p, the direct activator of the MAP kinase Hog1p (reviewed in references 19 and 39). In addition to countering the effects of persistent high-osmolarity conditions, the HOG pathway also mediates short-term responses to fluctuations in the osmolarity of the environment. Sudden shifts to high osmolarity result in the accumulation of intracellular glycerol due to a combination of HOG pathway-regulated stimulation of glycerol synthesis (1, 6) and decreased glycerol efflux that seems to occur independently of the HOG MAP kinase (29, 53). One of the early HOG pathway-triggered events is the nuclear accumulation of Hog1p kinase (16, 43) followed by a distinct transcriptional response that over a time period of about 90 min to several hours (depending on the extent of osmotic challenge) transiently induces expression of genes encoding enzymes involved in glycerol synthesis, such as GPD1 and GPP2 (HOR2) (1, 2, 20, 36, 44). In addition, the HOG-mediated signal affects expression of genes whose products have a more general role in the protection from stress-induced damage, such as HSP104, which encodes a chaperonin (28); CTT1, which encodes cytoplasmic catalase (49); and HSP12, whose precise molecular function is unknown (51, 57). One of the major unresolved problems concerning this transcriptional response is the identity of the molecular targets recognized by the signaling pathway (19). This problem further extends to the question of whether and how short-term responses and long-term adaptation, both requiring the HOG pathway (44), are mechanistically connected.
Many stress-responsive genes in yeast are regulated via a common promoter element called the stress response element (STRE). This element appears to be sufficient for mediating the transcriptional response to many different environmental challenges (33). Two redundantly acting transcription factors, Msn2p and Msn4p, that recognize this sequence have been identified (35, 48). In response to a large variety of stress situations, including osmotic stress, these factors are translocated to the nucleus, where they bind and activate target promoters such as those of the CTT1 or HSP12 gene (18). Although Msn2p and Msn4p enhance the expression of these genes, suggesting some connection to the HOG pathway, HOG pathway-mediated transcriptional activation can still be found in the absence of Msn2p and Msn4p function. The extent of Msn2p- and Msn4p-independent activation varies from promoter to promoter (this work and reference 35). Consequently, this pair of transcription factors cannot be the only ones responsible for high-osmolarity-induced gene expression. In the case of some yeast promoters, induction appears to be mediated by HOG pathway-dependent inactivation of Sko1p (34, 42, 45). Sko1p is a transcriptional repressor, which binds to CRE-like DNA elements and appears to tether the general repressors Tup1p and Ssn6p to promoters (34, 42). This system does not control expression of GPD1 (44, 45), and hence it is still unclear by which molecular mechanisms the Hog1p MAP kinase triggers induction of transcription of this type of target genes.
This study shows that the hitherto-undescribed nuclear protein Hot1p is important for HOG pathway-dependent osmotic induction of genes encoding enzymes involved in yeast glycerol biosynthesis. A protein with a similar putative DNA-binding domain, Msn1p, contributes to both osmotic and heat stress induction of GPD1, CTT1, and HSP12. MSN1 has previously been isolated as a high-copy-number suppressor of snf1 mutants (14) and has been implicated in the regulation of iron uptake (12) and of pseudohyphal growth (26, 27). It has been shown to be a nuclear protein and to function as a transcriptional activator when fused to the DNA-binding domain of LexA (14). Our data suggest that together Hot1p and Msn1p mediate the bulk of the Msn2p- and Msn4p-independent osmotic stress activation of the genes GPD1, GPP2, CTT1, and HSP12.
MATERIALS AND METHODS
Yeast strains and general methods.
The yeast strains used in this study are summarized in Table 1. Except for L40, they all have the W303-1A genetic background (54). The complete coding region of the HOT1 gene (YMR172w) was deleted by the microhomology PCR method (32) with the kanMX4 cassette (59). Deletion was verified by PCR analysis of chromosomal DNA with specific primers. Strain YMR56 was obtained by crossing and sporulating YSH818 and UG62, and YMR119 and YMR120 were constructed by crossing and sporulating UG44 and UG62. General cloning methods are described by Sambrook et al. (47). Yeast media, growth conditions, and procedures were used as presented by Rose et al. (46). To monitor gene expression after exposure to stress, cells were pregrown at 30°C in rich glucose-containing medium to mid-exponential phase (optical density at 600 nm [OD600] of between 0.7 and 1.5). Osmotic stress was applied by adding NaCl to the cultures to the concentrations indicated in the descriptions of the experiments. Heat stress was applied by mixing the culture with 1/3 volume of medium at 50°C, resulting in a temperature of about 37°C. The culture was then incubated in a water bath at 37°C during the rest of the experiment.
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype | Reference |
|---|---|---|
| W303-1A | MATa leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 GAL SUC2 mal0 | 54 |
| YM24 | MATa msn2-3::HIS3 msn4-1::TRP1 | 15 |
| UG43 | MATα hot1::kanMX4 | This work |
| UG44 | MATa msn1::URA3 | 14 |
| K4327 | MATa hog1::TRP1 | 9 |
| YSH 818 | MATa hog1::LEU2 | 44 |
| YMR 56 | MATa hog1::LEU2 hot1::kanMX4 | This work |
| UG62 | MATα msn2-3::HIS3 msn4-1::TRP1 hot1::kanMX4 | This work |
| UG48 | MATa msn1::URA3 hot1::kanMX4 | This work |
| YMR119 | MATa msn1::URA3 msn2-3::HIS3 msn4-1::TRP1 | This work |
| YMR120 | MATa hot1::kanMX4 msn1::URA3 msn2-3::HIS3 msn4-1::TRP1 | This work |
| VRY140 | MATa hot1::HOT1-GFP (pRS306) | This work |
| YSH392 | MATa gpd1::TRP1 | 1 |
| L40 | MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ GAL4 gal80 | 58 |
All strains except L40 are isogenic to and possess the markers of W303-1A.
Northern blotting techniques.
Total RNA was isolated at the time points indicated in the figures and separated by electrophoresis as described previously (10). Blots were hybridized in buffer containing 7% sodium dodecyl sulfate (SDS), 0.5 M sodium phosphate buffer (pH 7.0), and 1 mM EDTA. The signal was quantified by using a phosphorimager (Fuji BAS-1000). The lacZ probe was a PvuII fragment internal to the lacZ open reading frame. All other probes were generated by PCR from chromosomal DNA of W303-1A. The GPD1 probe includes sequences from position −527 to +95 relative to the start codon (this excludes sequences homologous to GPD2). The probes for CTT1 (−14 to 1007) and HSP12 (−139 to +493) were kindly provided by J. H. de Winde (Leuven, Belgium). As a loading control we used an IPP1 probe including sequences from position +16 to +846 relative to the start codon (44). Probe fragments were labelled with High Prime (Boehringer Mannheim).
Glycerol measurements.
To determine glycerol production rates during an osmotic upshift, samples of a yeast culture were taken at different time points after the shift. The samples were heated for 10 min at 95°C, cells were sedimented, and total glycerol in the supernatant was measured. To obtain the glycerol production rate, differences in glycerol concentrations between subsequent time points were divided by the time interval and the mean of the OD600s at the two time points.
Internal glycerol was determined by harvesting cells by filtration and measuring the glycerol released by boiling the filters for 10 min. Dry weight was determined by drying filters with cells at 80°C overnight. Glycerol concentrations were measured with a glycerol determination kit (Boehringer Mannheim).
Plasmid construction.
The bait vector for the hybrid screen was constructed in the following way. The HOG1 coding sequence was amplified by PCR with oligonucleotides designed according to the sequences corresponding to the N terminus, TTATAAAGGATCCATATGACCACTAAC (the BamHI site and the start are codon underlined), and the C-terminal part, AAACCTGGATCCGTAGTCCGTAATGGTG (the BamHI site is underlined). The PCR product was digested with BamHI and cloned into the BamHI site of pBTM116 (kindly provided by Paul Bartel and Stan Fields), resulting in pLexA-HOG1Δ. This plasmid contains the full-length lexA gene linked in frame to a truncated HOG1 gene coding for the first 343 amino acids of the protein (creating a deletion of the last 93 amino acids from its C terminus).
A GPD1-lacZ fusion construct, pGPD1-lacZ, was made in the following way. Part of the GPD1 promoter, from position −693 to −177 relative to the ATG, was amplified by PCR by using primers with BglII and XhoI linkers (AAAAAGATCTCAAAGGCCTCTCCTGG [the BglII site is underlined] and AAAACTCGAGAGCCAGTCTACGTGCG [the XhoI site is underlined]). This fragment was cloned into the CYC1 promoter lacking the upstream activation sequence (UAS) in front of lacZ in pJS205BXX, an episomal plasmid with URA3 as selectable marker (a kind gift from H.-J. Schüller, Greifswald, Germany) (50).
Creation of a Hot1-GFP fusion.
Hot1p was tagged with green fluorescence protein (GFP) by using the chromosomal gene replacement technique (61). First, a plasmid construct with a NotI site in front of the stop codon of the HOT1 gene was prepared. Oligonucleotides 1 AATATCTCGAGTAAAATCAGGATAGCACG (partly homologous to the sequence around 700 bp upstream of the HOT1 stop codon; the XhoI site is underlined, and the double-underlined sequence indicates the stop codon that serves to prevent the expression of the truncated part of the HOT1 gene, which results from the gene replacement event) and 2 TATTATTTTATTGCGGCCGCCTATTCCAGCAAGGCTCTC (partly homologous to the sequence upstream of the stop codon of the HOT1 coding sequence; the NotI site is underlined) were used to prepare PCR product A (700 bp). Another pair of oligonucleotides, 3 ATAATTATATATGCGGCCGCTAGGCACGTACGATAG (partly homologous to the sequence downstream of the stop codon of the HOT1 coding sequence; the NotI site is underlined) and 4 AATATGAGCTCTTTTTTCACCTTCCC (partly homologous to the region around 250 bp downstream of HOT1 stop codon; the SacI site is underlined) were used to prepare PCR product B (250 bp). PCR product A was digested with XhoI and NotI, combined with an equimolar amount of PCR product B digested with NotI and SacI, and cloned in one step into pRS306 (52) vector treated with XhoI and SacI. The resulting plasmid, p603NotI, was digested with NotI, and a GFP cassette (18) flanked by NotI sites was cloned into p603NotI, resulting in p603GFP. This plasmid was linearized with HindIII (cuts 523 bp upstream of the HOT1 stop codon) and used to transform yeast. The correct integration was checked by analysis of chromosomal DNA by PCR with specific primers.
Two-hybrid screen.
Three genomic yeast libraries (YL2H-C1, YL2H-C2, and YLH2H-C3, constructed in modified versions of pGAD424 [25]), were used to transform yeast strain L40 (58) carrying the LexA-Hog1 (residues 1 to 343) fusion in pBTM116 (pLexA-HOG1Δ). Around 2 million transformants obtained after transformation with each of the three libraries were replica plated from plates selective for the plasmids and containing 25 mM 3-aminotriazole onto nitrocellulose filters and incubated with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) chromogenic substrate (5). Around 200 clones reproducibly produced positive signals in this assay after retransformation with plasmids recovered from candidates isolated in the first round of selection. The second round of selection with 45 mM 3-aminotriazole resulted in the isolation of 14 groups of candidates with different fragments of DNA fused to the GAL4 activation domain. A representative of the group with the highest β-galactosidase activity was selected for further characterization. The plasmid recovered from this strain contained an in-frame fusion of the GAL4 activation domain to codons 140 to 645 of S. cerevisiae reading frame YMR172w.
Fluorescence microscopy.
Cells were grown to logarithmic phase (OD600 ∼ 0.8 to 1.0) in synthetic complete medium with components used as selective markers omitted. 4,6-Diamidino-2-phenylindole (DAPI) (final concentration, 2 g/ml) was added as a DNA dye 15 min prior to microscopy. The number of cells with a signal was expressed relative to the sum of cells showing fluorescence (total number of cells examined, >300) after growth under normal conditions or after exposure to 0.4 M NaCl for 5 min. For time course experiments, cells were scored in a similar manner, using the camera pictures of samples taken at the time points indicated. Living cells were viewed with a Zeiss Axioplan 2 fluorescence microscope, and images were scanned with a Quantix charge-coupled digital camera and IP LAB software.
Protein extract preparation and Western blot analysis.
Cells were grown in appropriate synthetic medium to an OD600 of ∼0.8 to 1.0. For osmotic stress, NaCl was added at room temperature to a final concentration of 0.4 M for the time periods indicated. To prepare protein extracts, cultures were rapidly cooled in ice water and harvested by centrifugation. The cell pellet was resuspended in ice-cold lysis buffer (0.1 M morpholineethanesulfonic acid [MES], 10 mM EGTA, 1% dimethyl sulfoxide, 1 mM dithiothreitol, pH 6.5) containing a complete protease inhibitor mix (Boehringer) and phosphatase inhibitors (0.1 mM Na vanadate, 10 mM NaF) and vortexed with glass beads three times for 5 min each. Samples were centrifuged twice at 14,000 rpm in a microcentrifuge for 10 min each. All operations were performed at 4°C. Protein concentrations in the supernatants were determined by using a Bio-Rad protein assay kit before the supernatants were frozen in liquid nitrogen. Extracts (100 μl) were boiled with 50 μl of 4× sample buffer (0.12 M Tris, 20% [vol/vol] glycerol, 0.286 M 2-mercaptoethanol, 0.086 mM bromophenol blue, 5% SDS, pH 6.8) for 3 min. Samples were resolved by SDS–7.5% polyacrylamide gel electrophoresis (Bio-Rad Mini Protean) and transferred onto nitrocellulose membranes (Schleicher & Schuell) in a semidry blotting apparatus (Bio-Rad). Phosphorylated Hog1-GFP was immunodetected by phospho-specific p38 MAP kinase (T180/Y182) antibody (New England Biolabs), and GFP was visualized with GFP polyclonal antibody (Clontech). Immunoblots were developed by using a horseradish peroxidase-conjugated protein ECL detection kit (Amersham). Probes for immunodetection were removed by incubation in stripping buffer according to recommendations of the suppliers before application of another primary antibody to the same membrane.
RESULTS
Identification of a putative Hog1p-interacting factor with similarity to the transcription factors Msn1p and Gcr1p.
In an attempt to find target proteins of the Hog1p MAP kinase, we carried out a two-hybrid screen with HOG1 coding sequences as bait. To minimize the background signal, we chose a lexA-HOG1 fusion construct encoding only the first 343 residues of Hog1p. Expression of this fusion from a truncated ADH1 promoter indeed exhibited low intrinsic activation. Using libraries expressing yeast genomic fragments fused to a transcription activation domain, we screened around 6 million transformants for increased LexA-dependent expression. Of 200 candidates that reproducibly yielded a positive signal, we selected the clone with the highest β-galactosidase activity (see Materials and Methods for details). Further characterization of the clone showed that it contained codons 140 to 645 of the uncharacterized open reading frame YMR172w fused in frame to the GAL4 activation domain. We called this gene HOT1 for high-osmolarity-induced transcription (see below). The interaction between Hot1p and Hog1p seemed to be specific, as DNA-binding domain fusions with other MAP kinases did not raise the transcriptional readout (not shown). Various attempts to obtain biochemical evidence for Hog1p-Hot1p interaction by coprecipitation assays or by immunokinase assays failed (data not shown).
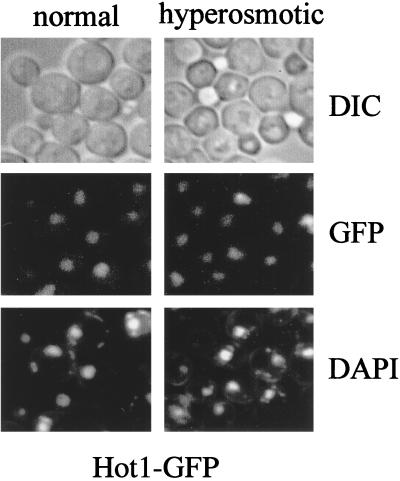
A homology search of the predicted amino acid sequence of Hot1p (718 amino acid residues) did not uncover any close overall match in the databases. However, Msn1p (382 amino acids) and Gcr1p (785 amino acids), two previously identified S. cerevisiae proteins, showed similarity to Hot1p in their C-terminal regions (Fig. 1). The similarity is most pronounced within a stretch of 64 residues in which Hot1p and Msn1p exhibit 42% identity and 64% similarity and Hot1p and Gcr1p exhibit 27% identity and 59% similarity (Fig. 1). The similarity between Hot1p and Msn1p can even be extended beyond this region to encompass 95 residues in total (41% identity and 61% similarity). The C-terminal region has been identified as a DNA-binding domain in the case of Gcr1p (22, 23), a factor required for high expression levels of glycolytic genes (3, 4, 7, 8). Although specific DNA binding has not been proven for Msn1p, the protein can function as a transcriptional activator when fused to the LexA DNA-binding domain (14). Consistent with such a function, Msn1p has been reported to be a resident nuclear protein (14). To localize Hot1p in the cell, we made use of a carboxy-terminal GFP tag. The construct was shown to be functional by complementation of the sensitivity of the hot1 mutant to severe stress (see below). A strong fluorescence signal colocalized with nuclear DNA under normal and hyperosmotic (Fig. 2) and hypo-osmotic (data not shown) conditions.
FIG. 1.
Hot1p is structurally related to Msn1p and Gcr1p. Pairwise alignments of the C-terminal regions of Hot1p with Msn1p and Gcr1p are shown. Arrows delineate a region of 64 residues that has the highest similarity. The similarity between Hot1p and Msn1p extends further towards the C-terminal end.
FIG. 2.
Hot1p is a nuclear protein. Logarithmically growing cells of strain VRY140 (HOT1-GFP) were examined by fluorescence or light microscopy under normal growth conditions and 5 min after addition of NaCl to a final concentration of 0.4 M. DIC, differential interference contrast.
Efficient induction of GPD1 expression depends on Hot1p.
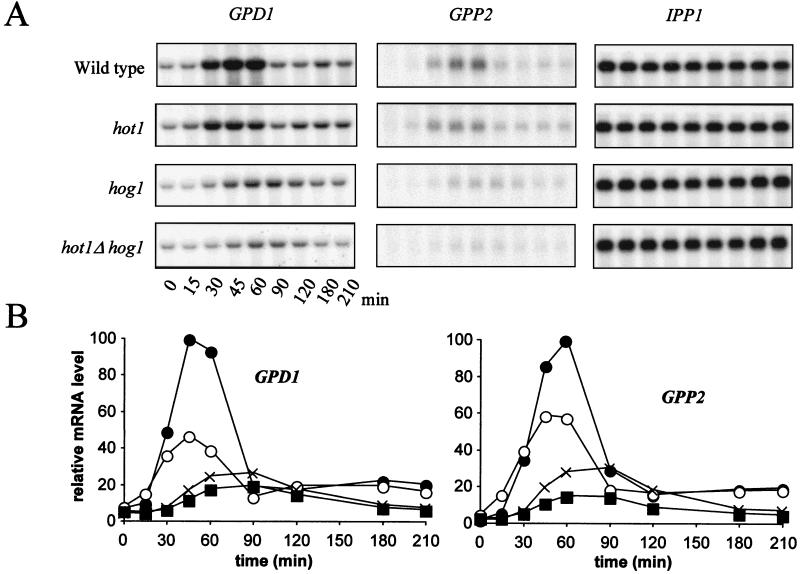
Since Hot1p may interact with Hog1p and exhibits similarity to the transcriptional activators Gcr1p and Msn1p, we tested whether hot1 mutations affect the expression of genes induced by high-osmolarity stress via the HOG pathway. Particularly, we tested whether the expression of the key glycerol biosynthesis genes GPD1 and GPP2, which are known to be partly regulated by the HOG pathway (1, 20, 36, 44), was affected by the mutation. Northern analysis demonstrated that this was indeed the case (Fig. 3). The main effect of a hot1 mutation was a significant decrease of maximal transcript accumulation after a shift to high-salt conditions (0.7 M NaCl). Unlike a hog1 mutant, hot1 cells are not delayed in their transcriptional response. In hog1 hot1 double mutants, the remaining response is kinetically similar to that of a hog1 mutant but is reduced even further. This result indicates that Hot1 could act downstream of Hog1p, perhaps as a mediator of nuclear kinase activity, although it also suggests that the two proteins have independent functions in the control of GPD1 and GPP2 expression. The specific reduction of mRNA levels in the hot1 mutant should be caused at the transcriptional level, since a CYC1-lacZ fusion gene driven by GPD1 UAS elements exhibited a reduction of its mRNA level in a hot1 mutant that was similar to that for the entire GPD1 gene (Fig. 4).
FIG. 3.
Role of Hog1p and Hot1p in the induction of expression of GPD1 and GPP2. Cells of the wild type (●) and of the hot1 (○), hog1 (×), and hot1 hog1 (■) mutants were exposed to 0.7 M NaCl at time zero. (A) Northern blotting; (B) quantification. The highest relative mRNA level in the wild type was set to 100.
FIG. 4.
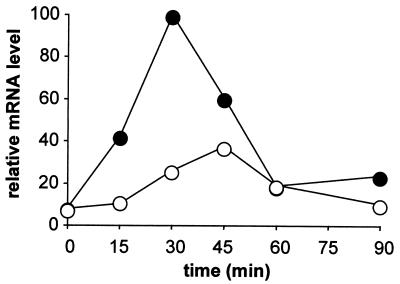
GPD1-lacZ induction is affected by the hot1 mutation. The mutant effect was tested by using a reporter consisting of a CYC1-lacZ construct lacking its own UAS fused to part (position −693 to −177) of the GPD1 promoter. Wild-type (●) and hot1 (○) cells were transformed with the pGPD1-lacZ plasmid and exposed to 0.7 M NaCl, and total RNA was probed for lacZ mRNA. The graph shows lacZ mRNA levels relative to those of IPP1; the highest relative mRNA level in the wild type was set to 100.
Hot1p and Msn1p are necessary for a complete transcriptional response to osmotic stress.
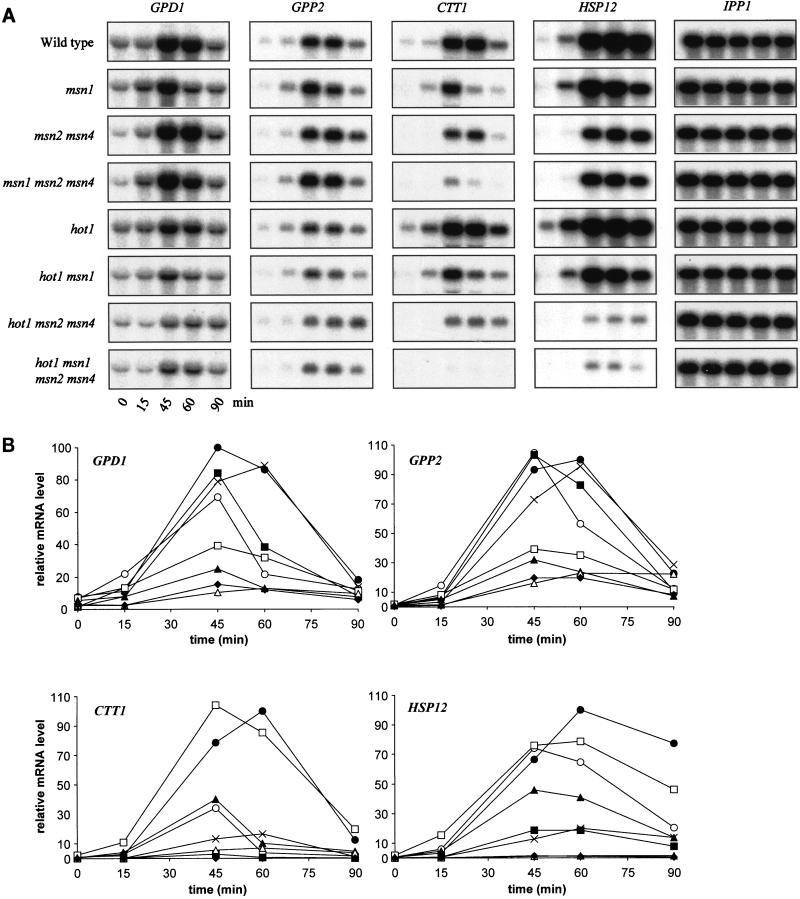
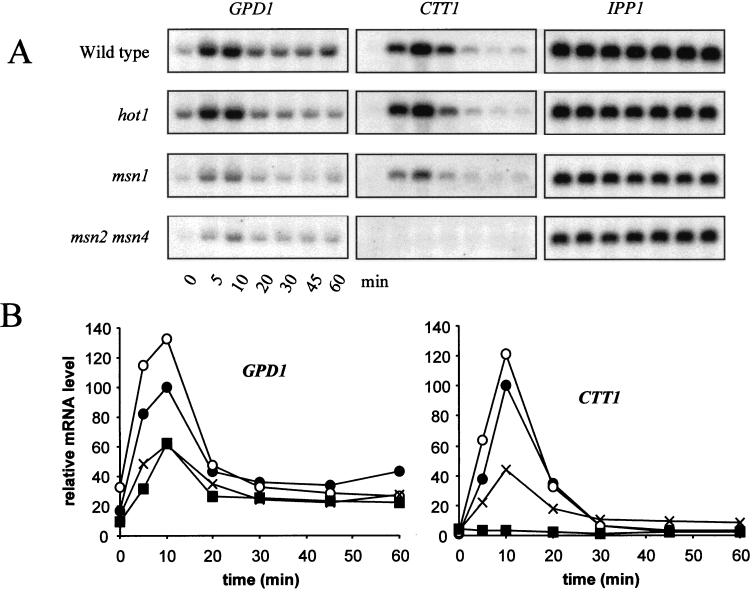
Having obtained evidence that HOT1 controls the promoters of GPD1 and GPP2, we tested the influence of the absence of various combinations of transcription factors that might affect expression of GPD1 and GPP2, as well as of CTT1 and HSP12, two general stress-responsive genes predominantly controlled via STREs (33, 57). While the upstream region of the GPD1 gene contains STRE-like sequences (at positions −330, −286, and −34), no evidence for their functionality has yet been reported. Deletion of MSN2 and MSN4, the two genes encoding two redundant transcription factors binding to STREs, has little effect on osmotic induction of GPD1 and GPP2 beyond a slight delay (Fig. 5). Deletion of the MSN1 gene also has little effect on mRNA induction. On the other hand, an important part of the response still existing in hot1 mutants is lost when MSN1, MSN2, and MSN4 are also deleted, suggesting a complex functional overlap and perhaps cooperation of these factors. Interestingly, there was about a 5-fold induction of the GPD1 mRNA level even in the quadruple mutant (wild type, 13-fold), indicating that additional factors must be involved in the regulation of GPD1 expression.
FIG. 5.
Effects of combinations of hot1, msn1, msn2, and msn4 mutations on osmotic induction of GPD1, GPP2, CTT1, and HSP12. Cells of the wild type (●) and the msn1 (○), msn2 msn4 (×), msn1 msn2 msn4 (■), hot1 (□), hot1 msn1 (▴), hot1 msn2 msn4 (▵), and hot1 msn1 msn2 msn4 (⧫) mutants were exposed to osmotic stress (0.7 M NaCl) at time zero. (A) Northern blotting; (B) quantification. The highest relative mRNA level in the wild type was set to 100.
A similar but not identical pattern of dependencies became apparent in the case of the predominantly STRE-regulated genes. As previously published, osmotic induction of expression of the CTT1 gene is dependent mainly on MSN2 and MSN4 (35). However, an msn1 deletion has a strong negative effect on expression in this case, while a single hot1 mutation has little effect. When various mutations are tested in combination, most of the response is lost in msn1 msn2 msn4 triple mutants, and the slight response still remaining is lost virtually completely in the hot1 msn1 msn2 msn4 quadruple mutant (see particularly Fig. 5A). A somewhat different pattern was observed in the case of HSP12, another STRE-regulated gene (57). Like CTT1, this gene is regulated predominantly by MSN2 and MSN4, as either the single hot1 or single msn1 mutation has only negligible effects, while a combination of both leads to a 50% reduction in maximal expression (Fig. 5B). However, in the msn2 msn4 double mutant the remaining induction (20% of maximal wild-type levels) is strongly dependent on HOT1. Although there is still a visible increase of HSP12 mRNA in the quadruple mutant during the osmotic upshift (Fig. 5A), the maximal level is only 1% of wild type in this mutant. Overall, it is apparent that individual contributions of the two sets of regulatory genes are highly dependent on promoter context and cannot be predicted by the presence or number of STRE elements alone. Nevertheless, since for all genes tested by us the osmotic stress-specific expression was drastically reduced in the absence of Hot1p, Msn1p, Msn2p, and Msn4p, we suggest that this might also hold true for most other osmotic stress-induced genes.
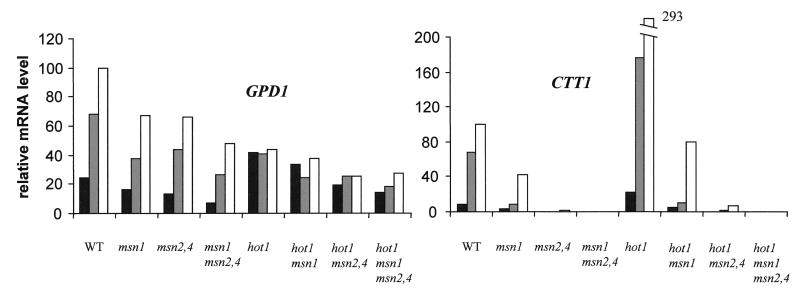
Having established that Hot1p, Msn1p, and Msn2p plus Msn4p affect osmotic induction of glycerol biosynthesis and general stress-responsive gene transcripts differentially, we tested the effect of the respective mutations on the expression of two of the genes affected, GPD1 and CTT1, during continuous growth under normal and high-osmolarity conditions (Fig. 6). These data confirm the differential effects of Msn1p and Hot1p on a glycerol biosynthesis gene and on a general stress-responsive gene. They also demonstrate that the hot1 mutation results in increased CTT1 mRNA levels in cells that are fully adapted to high salt concentrations. This stimulating effect of hot1 is clearly dependent on the presence of Msn1p, Msn2p, and Msn4p, since it is lost in the respective double and triple mutants. A similar effect of hot1 was observed in the case of other general stress-responsive genes, particularly HSP12 (not shown). In the case of GPD1 mRNA, there is a stimulating effect of hot1 only in the absence of NaCl, which is again dependent on MSN1 and MSN2 plus MSN4. Under steady-state growth conditions, Hot1p appears to be required for proper regulation of GPD1 expression in response to the NaCl concentration, while Msn1p and Msn2p plus Msn4p contribute to expression irrespective of the NaCl concentration (Fig. 6).
FIG. 6.
Effects of combinations of hot1, msn1, msn2, and msn4 mutations on GPD1 and CTT1 expression. mRNA levels during continuous growth in rich glucose medium containing no NaCl (black bars), 0.5 M NaCl (grey bars), or 0.85 M NaCl (open bars) are shown. The highest relative mRNA level in the wild type (WT) was set to 100.
Hot1p is specifically required for the osmotic induction of GPD1.
To establish if the effects mediated by Hot1p and Msn1p are osmospecific, the respective mutants and the msn2 msn4 mutant were exposed to heat stress via a shift from 30 to 37°C, and mRNA induction of GPD1 and CTT1 was monitored. In contrast to induction by osmotic stress, induction by heat stress of CTT1 mRNA was completely dependent on Msn2p and Msn4p (Fig. 7). However, deletion of MSN1 also reduced maximal mRNA levels, to about 40%. Heat induction of GPD1 is also affected by loss of MSN1, MSN2, and MSN4 function but is completely independent of HOT1 (Fig. 7). Involvement of Msn2p and Msn4p in GPD1 expression indicates that some or all of the STRE-like elements in its promoter may be functional, at least under heat stress. More importantly, these data indicate that Hot1p is dedicated to the induction by high osmolarity, whereas Msn1p, like Msn2p and Msn4p, contributes to the transcriptional response induced by several kinds of stress.
FIG. 7.
Induction by heat shock of GPD1 and CTT1 expression in hot1, msn1, and msn2 msn4 mutants. Cells of the wild type (●) and the hot1 (○), msn1 (×), and msn2 msn4 (■) mutants were exposed to heat stress (37°C) at time zero. (A) Northern blotting; (B) quantification. The highest relative mRNA level in the wild type was set to 100.
Hot1p is functionally linked to the HOG pathway.
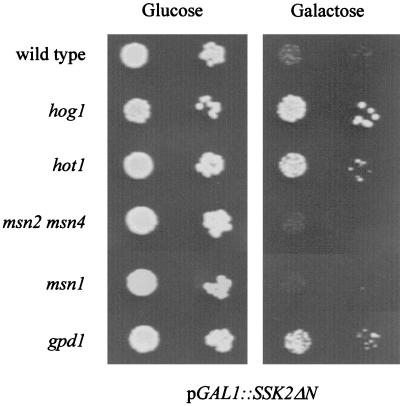
In a wild-type strain, the MAP kinase kinase kinase Ssk2p is kept inactive under basal conditions due to the function of an N-terminal negative regulatory domain (37). Deletion of this domain leads to constitutive activation of the downstream kinase cascade independent of the function of the upstream osmosensors. While expression of this truncated kinase is detrimental for cell growth, this effect can be suppressed by deletions of genes encoding downstream components of the pathway, such as HOG1 (Fig. 8) (30, 37). Here we show that deletion of GPD1 also partially suppresses the growth defect caused by a hyperactive HOG pathway (Fig. 8). Since a plasmid encoding a constitutively open Fps1p glycerol export channel (53) also partially suppressed unregulated Ssk2p (data not shown), overaccumulation of intracellular glycerol probably contributes to growth inhibition by an overactive HOG pathway. If Hot1p was to act at or downstream of the HOG kinase cascade controlling GPD1 induction, then a hot1 mutation might also suppress the negative effect of a constitutively active HOG pathway. Indeed, this appears to be the case (Fig. 8). The partial suppression of negative effects of constitutive HOG pathway activity by hot1 and gpd1 mutations is consistent with a role of Hot1p in GPD1 expression. It is noteworthy that deletion of MSN1 or of MSN2 plus MSN4 does not lead to such a suppression phenotype (Fig. 8).
FIG. 8.
Suppression of the growth defect caused by a constitutively activated HOG pathway. The wild type and hot1, hog1, msn2 msn4, msn1, and gpd1 mutants were transformed with plasmid pGAL1::SSK2ΔN, encoding Ssk2p with its N-terminal inhibitory domain (amino acids 1 to 1172) deleted (30). Expression of this construct is induced by a shift to galactose medium. Transformants were grown to logarithmic phase in selective medium containing glucose, sedimented, washed with water, and diluted to an OD600 of 1.0. Ten microliters of this suspension and of a 1:10 dilution were dropped on plates containing glucose or galactose.
Nuclear factors mediating the transcriptional response to stress contribute to nuclear retention of Hog1p MAP kinase.
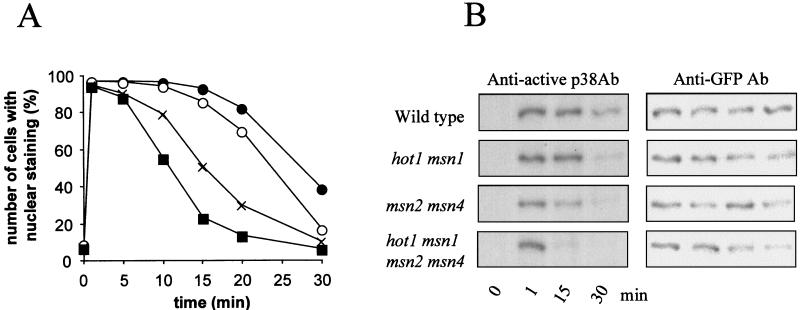
Formally, the effects of the hot1 mutation could be attributed to inefficient activation of the kinase cascade of the HOG pathway. We therefore investigated whether and how hot1 might influence Hog1p kinase activity. Activation of Hog1p in wild-type cells can be correlated with its rapid translocation to the nucleus. Also, during adaptation, the inactivation (dephosphorylation) of the kinase can be correlated with nuclear export (16, 43). Both nuclear accumulation and reexport of the MAP kinase can be visualized by using a functional Hog1-GFP fusion. When msn1 hot1 and msn1 msn2 msn4 hot1 mutants were tested for kinetics of nuclear Hog1-GFP import, it was apparent that Hog1p levels were not significantly affected by the mutations and that nuclear accumulation and Hog1-GFP dual phosphorylation were not delayed compared to those in the wild-type control. In contrast, the period of nuclear residence and dual phosphorylation of Hog1-GFP was clearly shortened in the msn2 msn4 mutant, as previously reported (43), and in the hot1 msn1 and quadruple mutants (Fig. 9). This observation suggests that Hot1p and Msn1p may be part of nuclear structures that help retain the activated Hog1p kinase in the nucleus.
FIG. 9.
Hot1p, Msn1p, Msn2p, and Msn4p affect kinetics of Hog1p phosphorylation and nuclear residence. (A) Logarithmically growing cells of the wild type (●) and the hot1 msn1 (○), msn2 msn4 (×), and msn1 msn2 msn4 hot1 (■) mutants were transformed with pVR65-WT (HOG1-GFP) and examined in time courses by fluorescence microscopy under normal growth conditions and after addition of NaCl to a final concentration of 0.4 M. (B) Determination of the phosphorylation profile of Hog1-GFP. Samples were taken at the time points indicated, and protein extracts were prepared. Western blots were analyzed with anti-active p38 antibody (Ab) or with anti-GFP antibody to determine the protein level of Hog1-GFP.
hot1 mutants are delayed in glycerol accumulation and are hypersensitive to killing by severe osmotic stress.
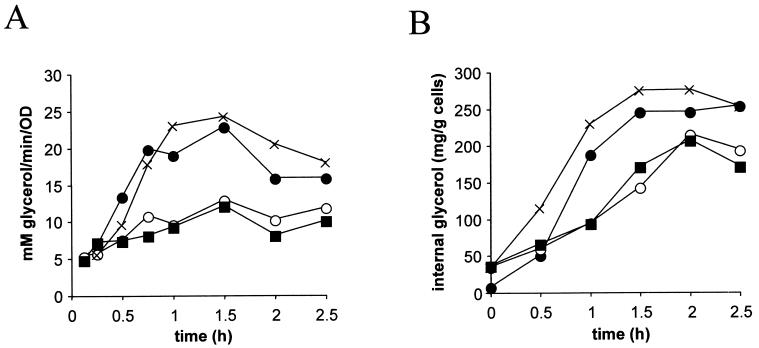
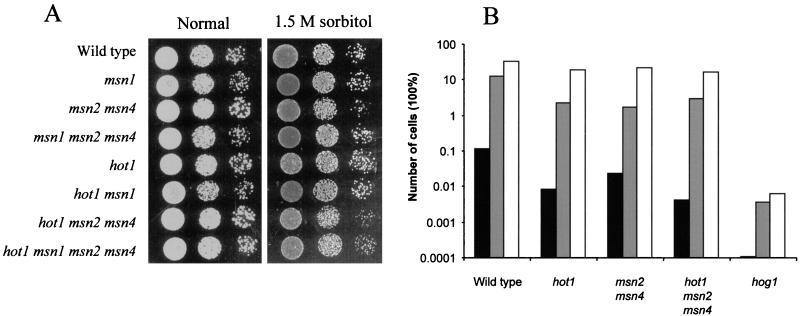
To further clarify whether the effects on gene expression described above are relevant for a functional osmotic stress response, we tested several physiological parameters, including glycerol production and cell survival during or after stress exposure. Consistent with effects on GPD1 transcript levels, the glycerol production rate (Fig. 10A) and internal glycerol accumulation (Fig. 10B) were clearly decreased in hot1 mutants compared to the wild type for a time period of about 2.5 h when the osmolarity of the medium was suddenly increased (0.7 M NaCl). Wild-type and mutant cells were tested for growth on 1.5 M sorbitol and on 0.7 M NaCl, a condition that prevents the growth of a hog1 mutant. Surprisingly, none of the single or multiple mutations tested caused a pronounced defect in growth under these conditions (Fig. 11A and data not shown). Since the defect might be apparent only during the period of adaptation to a changing environment, we shifted cells from normal medium to 1.7 M NaCl and counted the number of survivors after 20 h. Two sets of experiments were performed. In one set cells were preinduced with a 15- or 60-min mild osmotic shock (0.4 M NaCl) before exposure to 1.7 M NaCl, and in the other set cells were directly transferred into 1.7 M NaCl. With the exception of hog1 mutants, all cells showed high survival rates when preinducing conditions were applied. On the other hand, cells not preconditioned by mild stress are dramatically more sensitive to the severe type of stress. Under these conditions, hot1 mutant cells are more sensitive than wild-type cells or even mutants lacking the general stress transcription factors Msn2p and Msn4p (Fig. 11B). Perhaps not surprisingly, hog1 cells are most sensitive in this type of experiment, presumably because important targets of the Hog1p MAP kinase are transcriptional effectors different from Hot1p or do not act at the transcriptional level. Perhaps already under noninducing conditions, Hot1p determines a pattern of gene expression that guards a cell against sudden osmotic insults.
FIG. 10.
Hot1p affects glycerol production rate (A) and glycerol accumulation (B). Cells of the wild type (●) and the hot1 (○), msn2 msn4 (×), and hot1 msn2 msn4 (■) mutants were exposed to 0.7 M NaCl at time zero. The total glycerol production rate and intracellular glycerol accumulation were measured as described in Materials and Methods.
FIG. 11.
(A) Growth of a hot1 strain is unaffected by 1.5 M sorbitol. Cells were grown to logarithmic phase and diluted to an OD600 of 1.0. A 10-fold serial dilution series was plated on rich glucose medium containing either no osmolyte or 1.5M sorbitol. (B) Cells with HOT1 deleted are more sensitive to severe osmotic stress conditions. Logarithmically growing strains were incubated in the presence of 0.4 M NaCl for 0 (black bars), 15 (grey bars), or 60 (open bars) min before the final concentration of NaCl was increased to 1.7 M. After 20 h, the samples were appropriately diluted and plated at about 300 cells per plate. After 3 days, the total number of colonies was counted and expressed relative to that in the corresponding unstressed sample.
DISCUSSION
New factors mediating osmotic stress signaling.
The S. cerevisiae Hog1p kinase is related to the p38 MAP kinase from mammalian cells and to Sty1 from Schizosaccharomyces pombe. While p38 and Sty1 are responsive to different types of stress, Hog1p is activated specifically by osmotic stress, probably reflecting an adaptation to the specific environment of S. cerevisiae, whose osmotic pressure can change dramatically (19, 55). Although the architecture of the HOG pathway of S. cerevisiae is known in more detail than that of the pathway stimulating p38, the actual targets of the Hog1p kinase have remained elusive (19). In other eukaryotic systems the transcription factor substrate(s) of p38/Hog1p appears to be restricted to ATF-related proteins. This does not appear to be the case in S. cerevisiae. Sko1p, which has been described recently as an ATF-like transcriptional repressor, may be a Hog1p target, but it appears to control only a subset of the Hog1p-regulated genes (42, 44, 45). In our aim to identify nuclear regulators involved in the osmotic response, we have focused our attention on two structurally related nuclear proteins, Msn1p and Hot1p. Until now neither factor had been implicated in the osmotic stress response. From the studies presented here, we conclude that in addition to the general stress factors Msn2p and Msn4p, these two proteins are required for most of the short-term transcriptional response to hyperosmotic stress. This raised the question of whether the novel factor Hot1p and Msn1p are direct targets of the HOG pathway.
Hot1p may be a target of the HOG pathway.
The first indication that HOT1 might be implicated in osmotic stress came from the fact that a partial clone was obtained as an interacting partner of Hog1p in the yeast two-hybrid system. Although this suggested a potential physical interaction of Hog1p and Hot1p, we failed to find any corroborating biochemical evidence for direct Hog1p-Hot1p interaction. On the other hand, our data undoubtedly support a physiological interaction of Hot1p with the HOG pathway. First, the fact that a hot1 mutation partially suppresses the negative effects of constitutive hyperactivation of the HOG pathway is consistent with a functional role of Hot1p in responses triggered via the Hog1p MAP kinase. Consistently, we found that deletion of GPD1, a transcriptional target of the HOG pathway, also suppressed the effects conferred by hyperactive Hog1p. On the other hand, deletion of MSN1 and of MSN2 plus MSN4, which encode transcription factors controlling STRE-controlled genes but not the hyperosmotic induction of glycerol biosynthesis genes, did not suppress these effects. This finding further supports a specific role of Hot1p in mediating (part of the) HOG pathway-controlled osmotic regulation of glycerol biosynthesis genes.
Two observations make the connection between Hot1p and Hog1p even closer. The hot1 defect reduces the transient induction of GPD1 and GPP2 mRNAs and, consistent with that, also the rate of cellular glycerol accumulation upon an increase in osmolarity. Finally, a hot1 mutation has a negative effect on survival of yeast cells upon severe high-salt stress. Therefore, Hot1p must somehow affect the effectivity of the response system downstream of the MAP kinase. The following arguments support this notion. The dichotomy of the transcriptional response—namely, Hot1p initiating mainly GPD1 expression and Msn1p contributing mainly to CTT1 induction—indicates that these two factors are unlikely to influence the activation mechanism of the MAP kinase per se. Unless their activity is dedicated to signaling systems that act in parallel with the HOG pathway, the Hog1p kinase must act upstream of these two nuclear factors. This view is consistent with the observations that upon osmotic upshift in a hot1 msn1 mutant or even a quadruple msn1 msn2 msn4 hot1 mutant, dual phosphorylation of Hog1p is unaffected and the kinase is able to accumulate in the nucleus with unimpaired kinetics. What is different from wild-type cells in such mutants, however, is the abbreviated nuclear residence of Hog1p. In the fission yeast S. pombe, a similar phenomenon concerning the Sty1 kinase and the transcription factor Atf1 has been interpreted to indicate that the transcription factor provides nuclear anchorage for the kinase (17). From such a viewpoint, Hot1p and Msn1p should either be nuclear retention factors or at least control the activity of such factors in some way. Another, more remote possibility is that loss of Hot1p and Msn1p function somehow leads to an increase in the activity of stress adaptation factors such as protein phosphatases controlling the Hog1p phosphorylation state (60).
While the data are consistent with a role of Hot1p in mediating osmotic stress signaling downstream of Hog1p, we note that these two proteins may have roles in the control of expression of the glycerol biosynthesis genes GPD1 and GPP2 independent of each other. This notion is based on the observation that the profiles of induction of the two glycerol biosynthesis genes are different in hog1 and hot1 mutants. Moreover, additional deletion of HOT1 in a hog1 mutant results in a hog1-like induction profile, but maximal induction is further diminished. These observations are inconsistent with a simple linear pathway from Hog1p to Hot1p and further to GPD1 or GPP2 expression but rather suggest that at least part of the function of Hot1p (and Msn1p) function is exerted in a pathway parallel to Hog1p.
A number of observations made in this investigation are in line with the assumption that Hot1p acts as a transcriptional activator of the expression of some genes, which are controlled via the HOG pathway. Hot1p is a resident nuclear protein, and its C-terminal region exhibits sequence similarity to the DNA-binding domain of Gcr1p and a similar region in the transcriptional activator Msn1p. The loss-of-function phenotype resulting in the diminished induction of GPD1 and GPP2 mRNAs indicates that Hot1p acts as a positive factor in this situation. Since the function of a GPD1 upstream region driving the expression of a CYC1-lacZ reporter gene is also impaired in hot1 cells, the effect of Hot1p is most likely on transcriptional induction rather than mRNA stability. Despite a number of attempts, we have been unable to demonstrate binding of Hot1p to upstream elements of the genes investigated (GPD1 and CTT1) by gel retardation experiments (data not shown). Gcr1p and Msn1p, which are similar to Hot1p in their (putative) DNA-binding domains, bind DNA only weakly and with low sequence specificity (14, 22). Recently, we have obtained preliminary evidence by use of chromatin precipitation methods that Msn1p indeed binds specifically to stress-responsive promoters, suggesting that the same may hold true for Hot1p in the case of glycerol biosynthesis genes. At least in some Gcr1p-controlled promoters, interaction with another DNA-binding factor, Rap1p, is required for specific DNA binding (11, 56). The GPD1 promoter contains Rap1p binding sites (13), and Hot1p might well bind to promoters only in cooperation with this or other DNA-binding proteins. In conclusion, we presently favor a direct role of Msn1p and Hot1p in the regulation of GPD1 and GPP2 expression. We have observed, however, that the expression of HOT1 is itself about twofold stimulated after an osmotic shift in a time course similar to that observed for GPD1 (data not shown), which could indicate further levels of interdependent regulation of these factors.
Msn1p, Msn2p, Msn4p, and Hot1p mediate a large part of the osmotic induction of several stress-inducible genes.
To what extent do the four factors Msn1p, Msn2p, Msn4p, and Hot1p account for the normally observed transcriptional effects upon an osmotic upshift? According to the present data, all positive factors seem to have been recognized in the case of CTT1, while at least one additional, still-unknown factor appears to contribute to the induction of HSP12 and the glycerol biosynthesis genes GPD1 and GPP2. The Sko1p-Ssn6p-Tup1p repressor system, which has been shown to play an important role in negative control of some genes induced by high osmolarity (34, 42, 45), does not appear to be important at least for regulation of GPD1 (44, 45). Nevertheless, in all cases investigated, the contribution of the four factors is substantial, as the residual expression levels found in the quadruple mutant plunge to a minor fraction of those found in wild-type cells.
With regard to the individual contributions, one can clearly define separate spheres of influence. Hot1p appears to be most important in the case of glycerol biosynthesis gene expression, where Msn2p plus Msn4p and Msn1p have minor importance. In the case of the general stress-responsive genes CTT1 and HSP12, where Msn2p plus Msn4p are much more important for regulation, Msn1p and/or Hot1p makes an additional contribution to the high-osmolarity response.
The described effects of the hot1 mutation on the initial transcriptional response when cells experience an increase of medium osmolarity are also rather different from its long-term effects, i.e., its effects on mRNA levels in cells fully adapted to a given external osmolarity. Although deletion of HOT1 has both a short- and a long-term negative effect on GPD1 transcription in salt-exposed cells, it has a strong stimulating effect on CTT1 and HSP12 mRNA levels in fully adapted cells. This increase in steady-state levels, already seen under basal conditions, is dependent on Msn1p and on Msn2p plus Msn4p. Although this may point to a specific repressor function of Hot1p under some conditions, these effects could also be due to compensating mechanisms acting on general stress response genes, possibly via Msn1p and/or Msn2p plus Msn4p.
Effects on growth, survival, and glycerol accumulation.
It seems surprising that the reduced amount of GPD1 mRNA in the hot1 mutant and, especially, in the hot1 msn1 msn2 msn4 quadruple mutant has no strong effect on growth in media with high solute concentrations. In contrast to the case for hog1 mutants, there is no or only a minor effect of hot1 on the growth rate when assayed on plates or in liquid medium (not shown). The GPD1 mRNA level in adapted cells apparently supports enough enzyme synthesis for glycerol production to sustain the turgor pressure required for growth. This observation also makes it likely that the MAP kinase controls substrates important for growth and proliferation in addition to transcription factors. Nevertheless, the initial response to hyperosmotic stress, i.e., the accumulation of intracellular glycerol, is clearly delayed in a hot1 mutant, leaving it less well prepared for sudden rises in osmolarity. Although this could explain the reduced fitness of hot1 mutants upon sudden exposure to high NaCl concentrations, this effect might still involve as-yet-unrecognized targets of Hot1p.
ACKNOWLEDGMENTS
The first three authors have contributed equally to this publication.
We are grateful to C. Schüller (Vienna, Austria) for helpful discussions and critically reading the manuscript, to A. Marchler-Bauer (Vienna, Austria) for first pointing out to us the similarity between HOT1 and MSN1 sequences, and to H. Saito (Boston, Mass.) and H.-J. Schüller (Greifswald, Germany) for providing plasmids.
We acknowledge support from the Commission of the European Union via contracts BIO4-CT95-0161 to J.M.T. and S.H. and FMRX-CT96-0007 to J.M.T., H.R., and S.H. The Vienna groups have also been supported by grants P10815 and P12478 (to H.R.) and W001 (predoctoral fellowship to V.R.) from the Fonds zur Förderung der wissenschaftlichen Forschung, Vienna, Austria.
REFERENCES
- 1.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansell R, Granath K, Hohmann S, Thevelein J, Adler L. The two isoenzymes for yeast NAD-dependent glycerol 3-phosphate dehydrogenase, encoded by GPD1 and GPD2, have distinct roles in osmoadaption and redox regulation. EMBO J. 1997;16:2179–2187. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker H. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in CTTCC sequence motif. Proc Natl Acad Sci USA. 1991;88:9443–9447. doi: 10.1073/pnas.88.21.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker H. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol Cell Biol. 1986;6:3774–3784. doi: 10.1128/mcb.6.11.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 6.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 7.Chambers A, Packham E A, Graham I R. Control of glycolytic gene expression in the budding yeast (Saccharomyces cerevisiae) Curr Genet. 1995;29:1–9. doi: 10.1007/BF00313187. [DOI] [PubMed] [Google Scholar]
- 8.Clifton D, Frankel D G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981;256:13074–13078. [PubMed] [Google Scholar]
- 9.Cyrcková F, de Virgilio C, Manser E, Pringle J R, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 10.de Winde J H, Crauwels M, Hohmann S, Thevelein J M, Winderickx J. Differential requirement of the yeast sugar kinases for sugar sensing in the establishment of the catabolite repressed state. Eur J Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- 11.Drazinic C M, Smerage J B, Lopez M C, Baker H V. Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p) Mol Cell Biol. 1996;16:3187–3196. doi: 10.1128/mcb.16.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eide D, Guarente L. Increased dosage of a transcriptional activator gene enhances iron-limited growth of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:347–354. doi: 10.1099/00221287-138-2-347. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson P, Adler L, Blomberg A. Ph.D. thesis. Gothenburg, Sweden: Gothenburg University; 1997. [Google Scholar]
- 14.Estruch F, Carlson M. Increased dosage of the MSN1 gene restores invertase expression in yeast mutants defective in the SNF1 protein kinase. Nucleic Acids Res. 1990;11:6959–6964. doi: 10.1093/nar/18.23.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estruch F, Carlson M. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3872–3881. doi: 10.1128/mcb.13.7.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrigno P, Posas F, Koepp D, Saito H, Silver P A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaits F, Degols G, Shiozaki K, Russell P. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 1998;12:1464–1473. doi: 10.1101/gad.12.10.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirayama T, Maeda T, Saito H, Shinozaki K. Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol Gen Genet. 1995;249:127–138. doi: 10.1007/BF00290358. [DOI] [PubMed] [Google Scholar]
- 21.Hohmann S. Shaping up: the response of yeast to osmotic stress. In: Hohmann S, Mager W H, editors. Yeast stress responses. R.G. Austin, Tex: Landes; 1997. pp. 101–145. [Google Scholar]
- 22.Huie M A, Baker H V. DNA-binding properties of the yeast transcriptional activator, Gcr1p. Yeast. 1996;12:307–317. doi: 10.1002/(sici)1097-0061(19960330)12:4<307::aid-yea912>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 23.Huie M A, Scott E W, Drazinic C M, Lopez M C, Hornstra I K, Yang T P, Baker H V. Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2690–2700. doi: 10.1128/mcb.12.6.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby T, Flanagan H, Faykin A, Seto A G, Mattison C, Ota I. Two protein tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen activated protein kinase, Hog1. J Biol Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- 25.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambrechts M G, Bauer F F, Marmur J, Pretorius I S. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci USA. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambrechts M G, Sollitti P, Marmur J, Pretorius I S. A multicopy suppressor gene, MSS10, restores STA2 expression in Saccharomyces cerevisiae strains containing the STA10 repressor gene. Curr Genet. 1996;29:523–529. [PubMed] [Google Scholar]
- 28.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luyten K, Albertyn J, Skibbe W F, Prior B A, Ramos J, Thevelein J, Hohmann S. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 1995;14:1360–1371. doi: 10.1002/j.1460-2075.1995.tb07122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 31.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 32.Manivasakam P, Weber S C, McElver J, Schiestl R H. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:2799–2800. doi: 10.1093/nar/23.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Márquez J A, Pascual-Ahuir A, Proft M, Serrano R. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Pastor M T, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 36.Norbeck J, Påhlman A K, Akhtar N, Blomberg A, Adler L. Purification and characterization of two isoenzymes of dl-glycerol 3-phosphatase from Saccharomyces cerevisiae. Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing MAP kinase signal transduction pathway. J Biol Chem. 1996;271:13875–13881. doi: 10.1074/jbc.271.23.13875. [DOI] [PubMed] [Google Scholar]
- 37.Posas F, Saito H. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 39.Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- 40.Posas F, Witten E A, Saito H. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol. 1998;18:5788–5796. doi: 10.1128/mcb.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multi-step phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 42.Proft M, Serrano R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol. 1999;19:537–546. doi: 10.1128/mcb.19.1.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reiser V, Ruis H, Ammerer G. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1147–1161. doi: 10.1091/mbc.10.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rep M, Albertyn J, Thevelein J M, Prior B A, Hohmann S. Different signalling pathways contribute to the control of GPD1 expression by osmotic stress in Saccharomyces cerevisiae. Microbiology. 1999;145:715–727. doi: 10.1099/13500872-145-3-715. [DOI] [PubMed] [Google Scholar]
- 45.Rep M, Thevelein J M, Hohmann S. The role of ATF-like transcription factors in yeast osmoadaptation. 1999. Unpublished results. [Google Scholar]
- 46.Rose M D, Winston F, Hieter P. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Schmitt A P, McEntee K. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5777–5782. doi: 10.1073/pnas.93.12.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schüller G, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schüller H J, Hahn A, Troster F, Schutz A, Schweizer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992;11:107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siderius M, Rots E, Mager W H. High-osmolarity signalling in Saccharomyces cerevisiae is modulated in a carbon-source-dependent fashion. Microbiology. 1997;143:3241–3250. doi: 10.1099/00221287-143-10-3241. [DOI] [PubMed] [Google Scholar]
- 52.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamás M J, Luyten K, Sutherland F C W, Hernandez A, Albertyn J, Valadi H, Li H, Prior B A, Kilian S G, Ramos J, Gustafsson L, Thevelein J M, Hohmann S. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- 54.Thomas B J, Rothstein R J. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 55.Toone W M, Jones N. Stress-activated signalling pathways in yeast. Genes Cells. 1998;3:485–498. doi: 10.1046/j.1365-2443.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- 56.Tornow J, Zeng X, Gao W, Santangelo G M. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 1993;12:2431–2437. doi: 10.1002/j.1460-2075.1993.tb05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varela J C S, Praekelt U M, Meacock P A, Planta R J, Mager W H. The Saccharomyces cerevisiae HSP12 gene is activated by the high-osmolarity glycerol pathway and negatively regulated by protein kinase A. Mol Cell Biol. 1995;15:6232–6245. doi: 10.1128/mcb.15.11.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 59.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 60.Wurgler-Murphy S M, Maeda T, Witten E A, Saito H. Regulation of the Saccharomyces cerevisiae Hog1 mitogen-activated protein kinase by the Ptp2 and Ptp3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zachariae, W., and K. Nasmyth. Unpublished results.