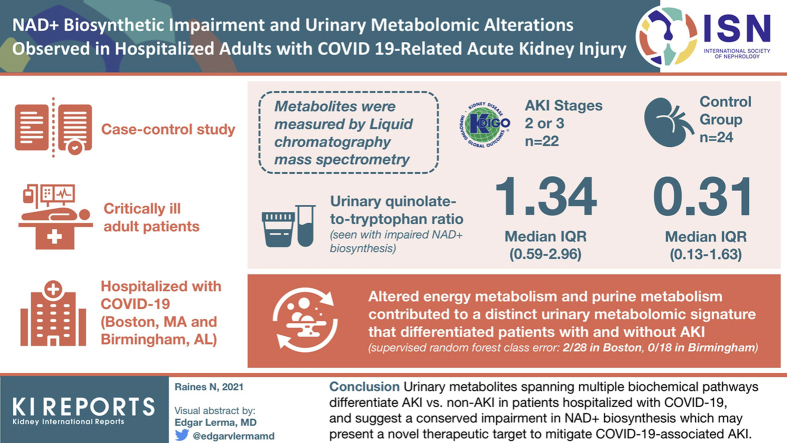
Abstract
Introduction
Acute kidney injury (AKI) is common in COVID-19 and associated with increased morbidity and mortality. We investigated alterations in the urine metabolome to test the hypothesis that impaired nicotinamide adenine dinucleotide (NAD+) biosynthesis and other deficiencies in energy metabolism in the kidney, previously characterized in ischemic, toxic, and inflammatory etiologies of AKI, will be present in COVID-19–associated AKI.
Methods
This is a case-control study among the following 2 independent populations of adults hospitalized with COVID-19: a critically ill population in Boston, Massachusetts, and a general population in Birmingham, Alabama. The cases had AKI stages 2 or 3 by Kidney Disease Improving Global Outcomes (KDIGO) criteria; the controls had no AKI. Metabolites were measured by liquid chromatography–mass spectrometry.
Results
A total of 14 cases and 14 controls were included from Boston and 8 cases and 10 controls from Birmingham. Increased urinary quinolinate-to-tryptophan ratio (Q/T), found with impaired NAD+ biosynthesis, was present in the cases at each location and pooled across locations (median [interquartile range]: 1.34 [0.59–2.96] in cases, 0.31 [0.13–1.63] in controls, P = 0.0013). Altered energy metabolism and purine metabolism contributed to a distinct urinary metabolomic signature that differentiated patients with and without AKI (supervised random forest class error: 2 of 28 in Boston, 0 of 18 in Birmingham).
Conclusion
Urinary metabolites spanning multiple biochemical pathways differentiate AKI versus non-AKI in patients hospitalized with COVID-19 and suggest a conserved impairment in NAD+ biosynthesis, which may present a novel therapeutic target to mitigate COVID-19–associated AKI.
Keywords: acute kidney injury, COVID-19, metabolism, metabolomics, nicotinamide adenine dinucleotide (NAD+)
Graphical abstract
On August 4, 2021, the COVID-19 pandemic caused by SARS-CoV-2 has led to more than 199 million confirmed cases worldwide, with more than 4.2 million deaths.1 COVID-19 is a disease of multiple organ systems beyond the respiratory tract. AKI is one of the more frequent extrapulmonary manifestations, with observational studies in the United States reporting AKI in up to 50% of hospitalized patients.2, 3, 4 AKI may develop through a combination of injury mechanisms, including ischemia, inflammation, endothelial damage, hypercoagulability, direct or indirect viral tropism, and toxicity arising from medications used to treat COVID-19.4 Given the ongoing high global burden of COVID-19, it remains vitally important to further understand physiological alterations that exist in this disease.
Investigations of AKI applying RNA sequencing and metabolomics implicate a consistent deficit in energy metabolism centered on NAD+ during ischemic and inflammatory injuries.5, 6, 7 Production of NAD+ from dietary sources becomes suppressed, whereas enzymes responsible for NAD+ consumption are up-regulated.6,8, 9, 10 Experimental models further suggest that NAD+ biosynthetic impairment may arise as a direct result of SARS-CoV-2 infection.11 Clinical data support a potential role of NAD+ biosynthetic impairment in COVID-19–related AKI: A prospective interventional study administering nicotinamide—an analog of vitamin B3 that may be the most orally bioavailable NAD+ precursor12—to hospitalized patients with COVID-19 with AKI revealed reduction of a composite outcome of kidney replacement therapy or death and prevention of further rise in serum creatinine (sCr) among those receiving supplementation.13 Other studies exploring the urine metabolome in AKI have revealed alterations in metabolites associated with oxidative stress, uremia, and energy metabolism.14,15
To investigate the underlying metabolic features of AKI arising among individuals hospitalized with COVID-19, we used a hypothesis-based approach focused on metabolites related to NAD+ as a primary analysis, complemented by an unbiased approach exploring the broader urine metabolome as a secondary analysis. Given the variability in clinical severity of COVID-19, we evaluated features associated with AKI in both a critically ill hospitalized cohort and in a general hospital population to compare metabolic changes found across the spectrum of the disease. We hypothesized that elevation of urinary Q/T, a marker of NAD+ biosynthetic impairment associated with AKI and subsequent adverse sequelae,5 would be greater in individuals with AKI compared with those with no impairment in kidney function across both groups.
Methods
Study Design and Setting
This is a nested case-control study using urine specimens from a convenience sample of patients hospitalized with COVID-19 at Massachusetts General Hospital in Boston and included in the Massachusetts Consortium on Pathogen Readiness biobank and urine specimens from a convenience sample of patients hospitalized with COVID-19 within the University of Alabama at Birmingham health system. Participants were hospitalized between April 2020 and November 2020. Specimens were not collected explicitly for this study, but for COVID-19 biobanks established through each institution.
This study was approved by the institutional review boards at the Massachusetts General Hospital and the University of Alabama at Birmingham, and collection of specimens for the Massachusetts Consortium on Pathogen Readiness biobank and the University of Alabama at Birmingham biobank was approved by the institutional review boards of the participating institutions. A modified Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement is provided in the Supplementary Materials.
Participants
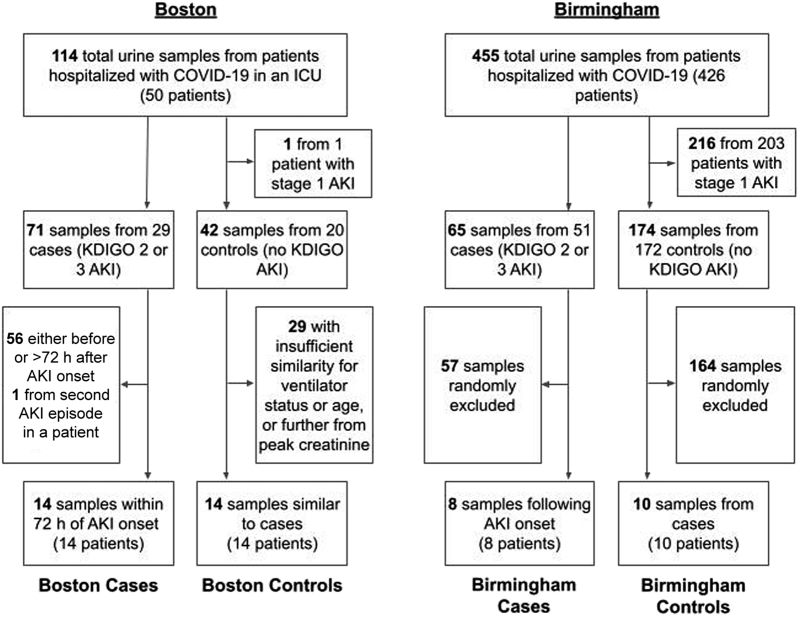
Adult patients aged 18 years and older were eligible for participation. Cases were individuals with AKI stages 2 or 3 by KDIGO sCr criteria.16 Controls were individuals who never met the KDIGO sCr criteria for any stage of AKI during their hospitalization. The Boston cohort included only critically ill patients, with cases selected as individuals receiving mechanical ventilation when they developed AKI, and controls selected to approximate cases preferentially for (i) mechanical ventilation, (ii) sex, and (iii) age without using 1:1 matching. Individuals with stages 2 and 3 AKI were chosen given they were those who derived the greatest benefit from nicotinamide supplementation in our previous work.13 Specimens in the cases were collected within 72 hours of AKI onset. Specimens in the controls were collected while they were mechanically ventilated, and as close as possible to their highest sCr while hospitalized. The Birmingham cohort was selected to represent a more general hospitalized patient population not limited to the critically ill, with cases randomly selected from patients meeting the KDIGO stage 2/3 AKI criteria and controls randomly selected from patients never meeting the KDIGO AKI criteria during their hospitalization (Figure 1). Sample size for this pilot study was determined by the availability of appropriate samples from existing biobanks.
Figure 1.
Participant flowsheet. AKI, acute kidney injury; h, hour; ICU, intensive care unit; KDIGO, Kidney Disease Improving Global Outcomes.
Variables
Information on demographics, medical history, clinical characteristics, and laboratory values was extracted by manual chart review from the medical records of the participating institutions. Baseline sCr used to determine AKI status was defined using the most recent sCr level 7 to 365 days before hospital admission (36% in the Boston group, 28% in the Birmingham group) or back-calculated with the Modification of Diet in Renal Disease equation using a glomerular filtration rate of 75 ml/min per 1.73 m2 when previous sCr values were not available.17 AKI stage 2 or 3 was defined by the standard KDIGO criteria as a doubling of sCr from baseline, sCr of ≥4.0 mg/dl, or kidney replacement therapy initiation, and did not include urine output criteria.16
The primary outcome variable was the ratio of peak area under the curve (AUC) by liquid chromatography-tandem mass spectrometry of urinary Q/T. Secondary outcomes were urinary peak AUC values of other metabolites involved in NAD+ metabolism, each corrected for peak AUC of urinary creatinine, and differences in other urine creatinine-corrected metabolite peak AUCs between the cases and controls investigated in a non–hypothesis-based exploratory fashion.
Specimen and Data Analyses
Details of specimen processing and liquid chromatography-tandem mass spectrometry procedures are provided in the Supplementary Methods. Boston and Birmingham patients were analyzed separately except where indicated. Urinary Q/T using the cationic metabolite curves was compared between the cases and controls using the Mann-Whitney U test and multivariable linear regression models which included age and sex. As a post hoc analysis, we evaluated urine Q/T comparing the cases and controls using multivariable linear regression models, including AKI status and ventilator requirement. Peak AUCs of additional metabolites were corrected for urine creatinine (Supplementary Methods). Those related to NAD+ metabolism were compared between AKI and non-AKI groups using t test for variables with a Gaussian distribution and the Mann-Whitney U test for metabolites without a Gaussian distribution. Peak AUCs for all metabolites were compared between the AKI and non-AKI groups in univariate analysis using volcano plots; in multivariable analysis using supervised random forest with features ranked by mean decrease in classification accuracy and supervised partial least squares-discriminant analysis with variable importance in projection scoring; and through development of unsupervised heatmaps. Analyses were performed using GraphPad Prism 9.1.0 (GraphPad Software, Inc., San Diego, California, USA) and MetaboAnalyst 5.0 (http://www.metaboanalyst.ca).
Results
Baseline clinical and demographic characteristics for the 14 cases and 14 controls from Boston, the 8 cases and 10 controls from Birmingham, and pooled across groups are presented in Table 1. Individuals with AKI tended to be older at both institutions, and with a greater number of comorbidities. Baseline creatinine was similar among individuals who developed AKI and those who did not in the Boston group, but it was higher in patients who developed AKI than those who did not in the Birmingham group. Individuals in Boston were universally on mechanical ventilation regardless of the presence of AKI, whereas in Birmingham, only the AKI group included patients requiring mechanical ventilation.
Table 1.
Baseline characteristics
| Variables | Boston |
Birmingham |
Overall |
|||
|---|---|---|---|---|---|---|
| AKI (n = 14) | No AKI (n = 14) | AKI (n = 8) | No AKI (n = 10) | AKI (n = 22) | No AKI (n = 24) | |
| Age, yr, med (IQR) | 65 (60–72) | 49 (44–67) | 64 (36–69) | 54 (39–58) | 65 (59–70) | 52 (43–65) |
| Female, n (%) | 3 (21.4) | 6 (42.9) | 3 (37.5) | 5 (50.0) | 6 (27.3) | 11 (45.8) |
| Race, n (%) | ||||||
| Asian | 1 (7.1) | 0 (0.0) | 1 (12.5) | 0 (0.0) | 2 (9.1) | 0 (0.0) |
| Black | 3 (21.4) | 2 (14.3) | 5 (62.5) | 4 (40.0) | 8 (36.4) | 6 (25.0) |
| White | 6 (42.9) | 6 (42.9) | 2 (25.0) | 6 (60.0) | 8 (36.4) | 12 (50.0) |
| Other/not known | 4 (28.6) | 6 (42.9) | 0 (0.0) | 0 (0.0) | 4 (18.2) | 6 (25.0) |
| Hispanic, n (%) | 4 (28.6) | 6 (42.9) | 0 (0.0) | 0 (0.0) | 4 (18.2) | 6 (25.0) |
| BMI, med (IQR) | 26 (24–30) | 32 (25–41) | 29 (28–43) | 32 (28–38) | 28 (25–31) | 32 (27–40) |
| sCr (mg/dl), mean (SD) | ||||||
| At baseline | 0.96 (0.14) | 0.97 (0.17) | 1.80 (1.28) | 0.98 (0.85) | 1.26 (0.85) | 0.97 (0.15) |
| Day of urine sample collection | 3.60 (1.54) | 0.74 (0.30) | 4.21 (2.57) | 0.85 (0.21) | 3.82 (1.94) | 0.78 (0.26) |
| Fold change sCr, mean (SD) | 3.76 (1.59) | 0.77 (0.28) | 2.56 (1.11) | 0.87 (1.53) | 3.33 (1.53) | 0.81 (0.25) |
| Comorbidities, n (%) | ||||||
| Diabetes | 7 (50.0) | 4 (28.6) | 3 (37.5) | 2 (20.0) | 10 (45.5) | 6 (25.0) |
| Hypertension | 5 (35.7) | 8 (57.1) | 3 (37.5) | 4 (40.0) | 8 (36.4) | 12 (50.0) |
| Hyperlipidemia | 1 (7.1) | 4 (28.6) | 2 (25.0) | 3 (30.0) | 3 (13.6) | 7 (29.2) |
| CAD | 1 (7.1) | 0 (0.0) | 3 (37.5) | 0 (0.0) | 4 (18.2) | 0 (0.0) |
| CHF | 1 (7.1) | 0 (0.0) | 4 (50.0) | 0 (0.0) | 5 (22.7) | 0 (0.0) |
| Cancera | 3 (21.4) | 1 (7.1) | 1 (12.5) | 0 (0.0) | 4 (18.2) | 1 (4.2) |
| COPD or asthma | 3 (21.4) | 2 (14.3) | 1 (12.5) | 1 (10.0) | 4 (18.2) | 3 (12.5) |
| Baseline GFR <60 | 1 (7.1) | 0 (0.0) | 6 (75.0) | 0 (0.0) | 7 (31.8) | 0 (0.0) |
| Mechanical ventilation, n (%) | 14 (100.0) | 14 (100.0) | 2 (25.0) | 0 (0.0) | 16 (72.7) | 14 (58.3) |
| Death, n (%) | 6 (42.9) | 3 (21.4) | 4 (50.0) | 0 (0.0) | 10 (45.5) | 3 (12.5) |
AKI, acute kidney injury; BMI, body mass index; CAD, atherosclerotic coronary artery disease; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; GFR, glomerular filtration rate; IQR, interquartile range; KDIGO, Kidney Disease Improving Global Outcomes; med, median; sCr, serum creatinine.
AKI was defined as meeting stage 2 or 3 criteria by KDIGO guidelines. GFR measured in ml/min per 1.73 m2.
Excludes nonmetastatic skin cancer.
Results of liquid chromatography-tandem mass spectrometry revealed 66 unique metabolites in the urine of the patients in Boston and 74 unique metabolites among the patients in Birmingham. Of these, 57 metabolites were found in the patients in both Boston and Birmingham, with a total of 88 unique metabolites across both institutions. A total of 21 metabolites had both cationic and anionic forms observed (Supplementary Table S1).
Primary Outcome
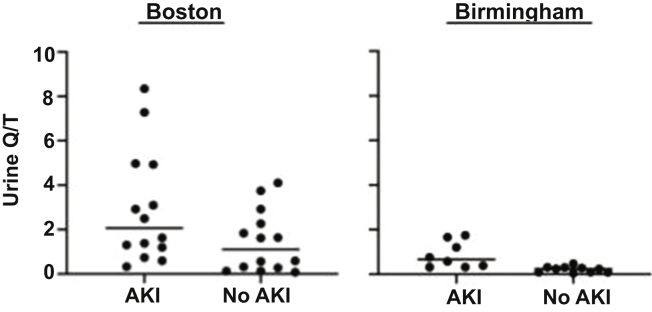
In unadjusted analyses, urinary Q/T was only marginally significantly increased among individuals with AKI in the Boston group (median [interquartile range]: 2.06 [1.08–4.94] in the AKI group, 1.11 [0.24–2.43] in the control group, P = 0.07) but was significantly increased in the Birmingham group (median [interquartile range]: 0.66 [0.34–1.55] in the AKI group, 0.24 [0.09–0.31] in the control group, P = 0.0003). In analyses adjusted for age and sex, AKI status but not age or sex was significantly associated with urinary Q/T in both the Boston (P = 0.03 for AKI status, 0.43 for age, 0.27 for sex) and Birmingham (P = 0.003 for AKI status, 0.06 for age, 0.09 for sex) cohorts (Figure 2). Models incorporating both AKI and mechanical ventilation status at the time of collection pooled across the Boston and Birmingham cohorts revealed that both variables were significant predictors of urine Q/T (P = 0.04 for AKI, 0.006 for mechanical ventilation).
Figure 2.
Ratio of the AUC of urinary Q/T by LC-MS-MS among hospitalized COVID-19 patients with and without AKI at institutions in Boston and Birmingham. Positive ionic forms were used for both quinolinate and tryptophan. In analyses adjusted for age and sex, Q/T was higher in the AKI group at both institutions (multiple regression, P = 0.03 in Boston patients, P = 0.003 in Birmingham patients). AKI, acute kidney injury; AUC, area under the curve; LC-MS-MS, liquid chromatography-tandem mass spectrometry; Q/T, quinolinate-to-tryptophan ratio.
Secondary Outcomes
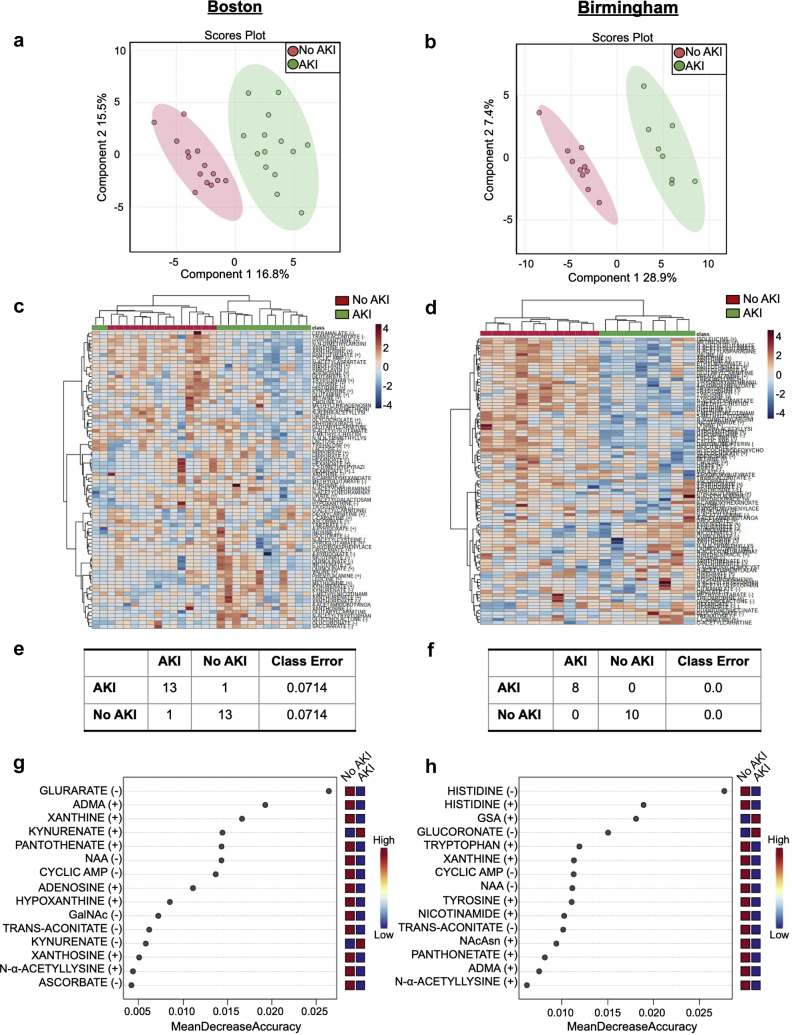
Metabolites that differed significantly between the patients with and without AKI in each of the Boston and Birmingham cohorts, including pooled across the cohorts, are illustrated in Table 2. These included decreases in purine metabolites and in metabolites adjacent to or directly involved in the tricarboxylic acid cycle, including alterations in NAD+-related metabolism. Partial least squares-discriminant analysis revealed complete separation of the 95% confidence limits between the individuals with and without AKI when evaluated in either location, and unsupervised hierarchical clustering analysis and random forest classification also revealed good separation of the 2 groups (Figure 3a–f). In addition, Figure 3g and h reveals the metabolites that were important in distinguishing groups in random forest classification; variable importance in projection scores revealing the metabolites that most contributed to the separation found in partial least squares-discriminant analysis are illustrated in Supplementary Figure S1 and demonstrate considerable overlap with the metabolites identified by random forest analysis. Analysis of pooled Boston and Birmingham data is found in Supplementary Figure S2.
Table 2.
Urinary metabolites significantly differing between groups with and without AKI
| Metabolites | Metabolic pathways involved | Boston |
Birmingham |
Overall |
|||
|---|---|---|---|---|---|---|---|
| FC | Raw P | FC | Raw P | FC | Raw P | ||
| Decreased | |||||||
| Xanthine (+) | Purine, caffeine | 0.26 | 4.00e−6 | 0.30 | 1.52e−6 | 0.28 | 8.14e−10 |
| N,N-dimethylarginine (+) | Arginine, urea | 0.36 | 5.45e−6 | 0.48 | 0.00054 | 0.40 | 1.61e−8 |
| Cyclic AMP (−) | Purine, cellular signaling | 0.42 | 5.80e−6 | 0.43 | 0.00017 | 0.41 | 4.87e−7 |
| N-acetylaspartate (−) | Aspartate/glutamate, cellular signaling, acetyl-CoA | 0.32 | 2.92e−5 | 0.42 | 0.00022 | 0.35 | 2.94e−8 |
| Hypoxanthine (+) | Purine | 0.25 | 4.91e−5 | 0.27 | 0.0016 | 0.27 | 1.32e−7 |
| Pantothenate (+) | Beta-alanine, pantothenate, and CoA | 0.27 | 5.98e−5 | 0.29 | 0.0016 | 0.26 | 3.17e−7 |
| Xanthosine (+) | Purine | 0.43 | 7.11e−5 | 0.80 | 0.20 | 0.59 | 0.0023 |
| N-α-acetyl-lysine (+) | Lysine | 0.53 | 0.0026 | 0.36 | 0.0057 | 0.44 | 4.22e−5 |
| Adenosine (+) | Purine, cell signaling | 0.25 | 0.010 | 0.38 | 0.026 | 0.30 | 0.00043 |
| Transaconitate (−) | TCA cycle | 0.21 | 0.015 | 0.23 | 0.00046 | 0.21 | 7.95e−5 |
| Urate (+) | Purine | 0.75 | 0.023 | 0.77 | 0.20 | 0.77 | 0.080 |
| Glycocholate (+) | Bile acid, lipid metabolism | 0.38 | 0.027 | 0.71 | 0.085 | 0.47 | 0.0040 |
| Histidine (+) | Histidine, β-alanine, ammonia recycling | 0.25 | 0.033 | 0.15 | 9.38e−6 | 0.24 | 0.00012 |
| Methylglutarate (−) | Lipid, fatty acid metabolism | 0.46 | 0.039 | 0.15 | 0.24 | 0.19 | 0.023 |
| 1-Methyl-L-histidine (+) | Histidine | 0.68 | 0.042 | 0.47 | 0.00036 | 0.60 | 0.00031 |
| Tryptophan (+) | Tryptophan | 0.37 | 0.077 | 0.31 | 8.45e−5 | 0.33 | 0.0020 |
| Isocitrate (−) | TCA cycle | 0.92 | 0.34 | 0.42 | 0.00016 | 0.38 | 0.23 |
| Tyrosine (+) | Tyrosine, phenylalanine, catecholamine | 0.67 | 0.47 | 0.31 | 0.00020 | 0.50 | 0.014 |
| Tryptophan (−) | (see above) | 0.55 | 0.42 | 0.30 | 0.0044 | 0.27 | 0.14 |
| Hypoxanthine (−) | (see above) | 0.37 | 0.20 | 0.41 | 0.0047 | 0.36 | 0.13 |
| Tyrosine (−) | (see above) | 0.67 | 0.47 | 0.33 | 0.0080 | 0.30 | 0.64 |
| Betaine (+) | Betaine, glycine/serine, methionine | 0.51 | 0.19 | 0.23 | 0.030 | 0.35 | 0.012 |
| N-acetylglutamate (−) | Glycine/serine, glutamate | 1.01 | 0.62 | 0.28 | 0.00069 | 0.25 | 0.23 |
| Glutarate (−) | Tryptophan, lysine | 0.15 | 0.00011 | — | — | — | — |
| S-adenosylmethionine (+) | Tryptophan, purine, nicotinamide, methionine, glycine/serine, arginine, proline, betaine, carnitine, histidine, tyrosine, spermine/spermidine, catecholamine | 0.49 | 0.0026 | — | — | — | — |
| N-acetylgalactosamine (−) | Carbohydrates | 0.38 | 0.0058 | — | — | — | — |
| Riboflavin (−) | Riboflavin | 0.65 | 0.022 | — | — | — | — |
| Kynurenine (+) | Tryptophan | 0.66 | 0.041 | — | — | — | — |
| Histidine (−) | (see above) | — | — | 0.10 | 1.94e−6 | — | — |
| Ethylmalonate (−) | Lipids | — | — | 0.17 | 4.04e−5 | — | — |
| N-acetylasparagine (+) | Asparagine | — | — | 0.25 | 0.00015 | — | — |
| Nicotinamide (+) | Nicotinamide | — | — | 0.32 | 0.00031 | — | — |
| Dihydrobiopterin (+) | Pterine, catecholamine | — | — | 0.32 | 0.00050 | — | — |
| Trigonelline (+) | Nicotinamide | — | — | 0.27 | 0.0080 | — | — |
| 2-Hydroxybutyrate (−) | Threonine, methionine, cysteine | — | — | 0.49 | 0.011 | — | — |
| Lysine (+) | Lysine | — | — | 0.048 | 0.016 | — | — |
| 4-Hydroxybenzoate (+) | Exogenous | — | — | 0.48 | 0.024 | — | — |
| Theobromine (+) | Caffeine | — | — | 0.082 | 0.025 | — | — |
| 3-Hydroxyanthranilate (+) | Tryptophan | — | — | 0.22 | 0.026 | — | — |
| Increased | |||||||
| Kynurenate (+) | Tryptophan | 3.26 | 0.00021 | 1.43 | 0.36 | 2.21 | 0.00056 |
| Kynurenate (−) | (see above) | 2.78 | 0.0018 | 1.97 | 0.071 | 2.53 | 0.00056 |
| Glucuronate (−) | Inositol, carbohydrate | 1.29 | 0.24 | 4.05 | 0.0020 | 1.66 | 0.014 |
| Leucine (+) | Valine/leucine/isoleucine | 3.28 | 0.012 | — | — | — | — |
| Lactose (+) | Exogenous | 2.35 | 0.049 | — | — | — | — |
| Guanidinosuccinate (+) | Uremia | — | — | 8.72 | 5.53e−7 | — | — |
| Mixed | |||||||
| Glutarylcarnitine (+) | Lipids, fatty acids | 1.93 | 0.0024 | 0.51 | 0.0081 | 0.57 | 0.94 |
| Riboflavin (+) | (see above) | 0.64 | 0.039 | 1.16 | 0.56 | 0.80 | 0.033 |
| Citramalate (−) | Lipids, fatty acids | 0.29 | 0.014 | 1.02 | 0.58 | 0.77 | 0.14 |
| Methionine (+) | Betaine, glycine/serine, methionine, spermine/spermidine | 2.12 | 0.13 | 0.41 | 0.0011 | 1.33 | 0.90 |
| Valine (+) | Propanoate, valine/leucine/isoleucine | 1.66 | 0.22 | 0.42 | 0.0015 | 0.95 | 0.53 |
| Phenylalanine (+) | Phenylalanine, tyrosine, catecholamine | 1.73 | 0.13 | 0.55 | 0.0081 | 1.15 | 0.90 |
AKI, acute kidney injury; AMP, adenosine monophosphate; AUC, area under the curve; CoA, coenzyme A; FC, fold change; TCA, tricarboxylic acid.
Metabolite data used for analysis are peak AUC by liquid chromatography-tandem mass spectrometry for each metabolite, with values for each participant divided by their respective peak AUC of creatinine. FC compares the log2-transformed ratio (AKI/no AKI) between each metabolite in each group. P values are derived from t test without multiple comparison corrections after transformation to achieve Gaussian distributions.
Figure 3.
Urine metabolomic features distinguishing AKI among patients with COVID-19 in a critically ill population in Boston and a general hospital population in Birmingham. (a,b) Supervised PLS-DA plot comparing patients with (green) and without (red) AKI. (c,d) Unsupervised hierarchical clustering analysis comparing urine metabolomic features in patients with (green) and without (red) AKI and relative increases (red) or decreases (blue) in metabolite levels. (e,f) Supervised random forest predictions and class error comparing AKI and no AKI groups. (g,h) Key features identified by random forest comparing patients with and without AKI; red and blue boxes denote relatively high or low concentrations of metabolites important in classification, respectively. ADMA, asymmetric dimethylarginine; AKI, acute kidney injury; AMP, adenosine monophosphate; GalNAc, N-acetylgalactosamine; GSA, guanidosuccinic acid; NAA, n-acetylaspartate; NAcAsn, N-acetylasparagine; PLS-DA, partial least squares-discriminant analysis.
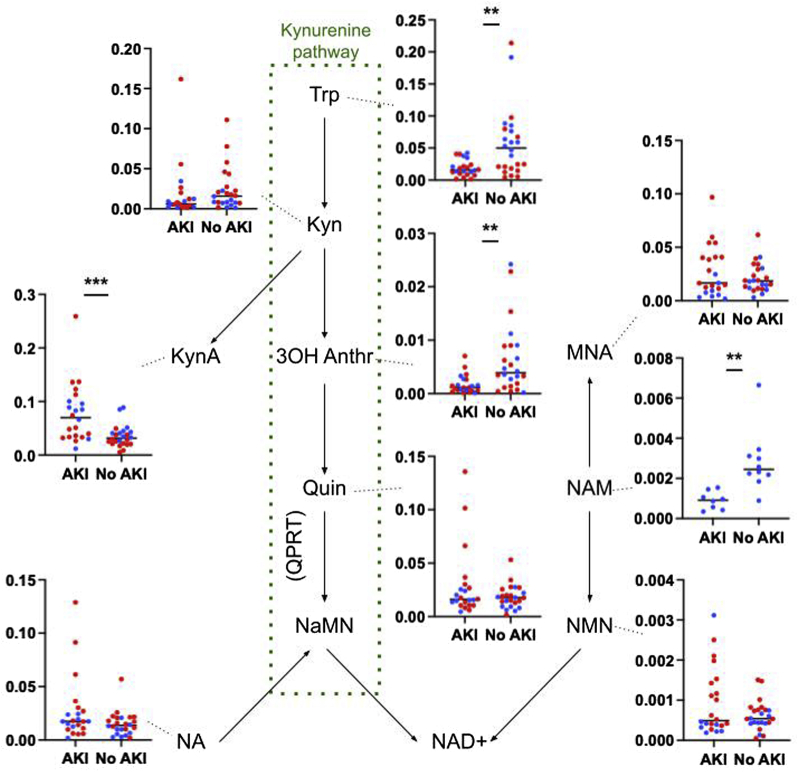
Differences in other metabolites related to NAD+ biosynthesis between the individuals with and without AKI are found in Figure 4. These included increased kynurenic acid and decreased nicotinamide, tryptophan, and 3-hydroxyanthranilic acid in the setting of AKI.
Figure 4.
Differences in metabolites related to NAD+ synthesis between groups of hospitalized patients with COVID-19 with and without AKI. Metabolite data used for analysis are peak AUC by liquid chromatography-tandem MS for each metabolite, with values for each participant divided by their respective peak AUC of creatinine.  are patients from Birmingham,
are patients from Birmingham,  are patients from Boston. Statistical significance between groups with metabolites pooled is designated on each plot: ∗∗P < 0.001; ∗∗∗P < 0.0001; analyses with Boston and Birmingham patients considered separately are reported in Supplementary Table S2. Positive ion forms are used where both positive and negative ions were identifiable by MS. Nicotinamide was not detected in the urine of patients either with or without AKI in the Boston cohort. 3OH Anthr, 3-hydroxyanthranilic acid; AKI, acute kidney injury; AUC, area under the curve; Kyn, kynurenine; KynA, kynurenic acid; MNA, 1-methyl nicotinamide; MS, mass spectrometry; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; NaMN, nicotinic acid mononucleotide; NMN, nicotinamide mononucleotide; QPRT, quinolinate phosphoribosyltransferase; Quin, quinolinate; Trp, tryptophan.
are patients from Boston. Statistical significance between groups with metabolites pooled is designated on each plot: ∗∗P < 0.001; ∗∗∗P < 0.0001; analyses with Boston and Birmingham patients considered separately are reported in Supplementary Table S2. Positive ion forms are used where both positive and negative ions were identifiable by MS. Nicotinamide was not detected in the urine of patients either with or without AKI in the Boston cohort. 3OH Anthr, 3-hydroxyanthranilic acid; AKI, acute kidney injury; AUC, area under the curve; Kyn, kynurenine; KynA, kynurenic acid; MNA, 1-methyl nicotinamide; MS, mass spectrometry; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; NaMN, nicotinic acid mononucleotide; NMN, nicotinamide mononucleotide; QPRT, quinolinate phosphoribosyltransferase; Quin, quinolinate; Trp, tryptophan.
Discussion
In this case-control study of patients hospitalized with COVID-19 in 2 locations in the United States, individuals with AKI had significantly different urine metabolomes when compared with individuals without AKI. These differences were present in an analysis of cases and controls selected to have similarly severe COVID-19 save AKI status (Boston) and in an analysis of a general hospitalized population (Birmingham). In both critically ill and general hospitalized patient populations, AKI was associated with an increase in urinary Q/T, an in vivo marker previously found to be a characteristic of NAD+ biosynthetic impairment, and a profound change in the urinary metabolome of patients with COVID-19 revolving on energy metabolism—including tryptophan/NAD+ metabolism—and purine metabolism. This is the first study to investigate the urine metabolome in patients with COVID-19 with AKI.
Evidence of NAD+ biosynthetic impairment in patients with COVID-19 and KDIGO stage 2 or 3 AKI is consistent with injury mechanisms found in ischemic and inflammatory processes.18 Studies in non-COVID-19 AKI have revealed similar derangements in tryptophan and NAD+ salvage pathway metabolism in both human disease5,19,20 and experimental models.20, 21, 22 Further indirect evidence of the role of NAD+ in COVID-19–associated AKI comes from our previous study of nicotinamide supplementation revealing patients with stage 2 or 3 AKI treated with nicotinamide had stabilization of sCr and decreased risk of a composite of death or need for kidney replacement therapy when compared with those not receiving nicotinamide.13 An important future step will be the evaluation of urine metabolic features of individuals with COVID-19 and AKI treated or not treated with nicotinamide.
Increased Q/T may also reflect metabolic changes related to the immune response to COVID-19. Alterations in the kynurenine pathway of tryptophan metabolism (Figure 3) have been consistently reported across metabolomic studies in the sera of individuals with COVID-19.23, 24, 25 The kynurenine pathway is important in the immune response to viral infection, with greater conversion of tryptophan to kynurenine by indoleamine 2,3-dioxygenase-1 occurring in response to interferons and cytokines.26, 27, 28 In addition, down-regulation of the enzyme quinolinate phosphoribosyltransferase integral to the conversion of quinolinate to NAD+ in macrophages has been implicated in response to an immune challenge, leading to a proinflammatory state.29 In our study, those individuals requiring mechanical ventilation had increased Q/T independent of AKI status, implying a correlation with overall disease severity rather than an association limited to altered metabolism with kidney injury.
Although the urinary metabolome of AKI could reflect altered metabolism originating in the immune system, there may also be an inverse connection. Our previous work in experimental AKI implicates a specific down-regulation of quinolinate phosphoribosyltransferase in the renal tubule.5,30 Because NAD+ precursors may be shuttled between organs,12 effects of COVID-19 on renal metabolism could secondarily affect NAD+ homeostasis in other compartments of the body. For example, blockage of renal NAD+ biosynthesis at quinolinate phosphoribosyltransferase during AKI may promote whole-body accumulation of kynurenate, which in turn modulates immune function.23,28
Metabolites associated with central energy metabolism but which are not directly related to NAD+ metabolism differed between AKI and non-AKI states, reinforcing the role of altered energy metabolism in AKI arising in the setting of COVID-19. Pantothenate is involved in the synthesis of coenzyme A and is therefore critical in the metabolism of carbohydrates, fats, and proteins, including the metabolism of drugs. Impaired pantothenate metabolism has previously been reported in experimental AKI.21 Histidine has antioxidant and anti-inflammatory properties31; it has been found to be decreased in the serum and urine of patients with chronic kidney disease and may be mechanistically related to protein-energy wasting, inflammation, and oxidative stress.32,33 Other studies have similarly revealed disruption of renal mitochondrial energetics and a generally energy-depleted state arising with kidney injury.15,22,34,35
Purine metabolism was markedly altered in AKI, which may relate partially to energy metabolism. For example, N-acetylaspartate may accelerate lipidogenesis36 and adenosine forms the backbone of adenosine triphosphate. Nevertheless, adenosine is also an important signaling molecule with protective effects in AKI, serving both an anti-inflammatory function and as an important mediator of tubuloglomerular feedback.37,38 Adenosine levels are typically increased extracellularly in response to an ischemic injury, so the decreased urinary values may reflect an overall depletion in the context of the severe AKI, or a loss of a protective effect leading to impaired adaptation to hypoxia.
Some metabolite patterns observed likely reflect physiological alterations arising directly from loss of renal function. Glutarate, besides being a tryptophan derivative, is a dicarboxylic acid excreted into the urine by the renal proximal tubular organic anion transporter 1.39,40 Decreased urinary levels may reflect loss of proximal tubular function in the context of kidney injury. Guanidosuccinate, an important feature in the Birmingham cohort, is a uremic toxin.41
A meta-analysis of serum metabolomic data from patients with COVID-19 revealed up-regulation of the propanoate pathway and down-regulation of glyoxylate and dicarboxylate metabolism and tricarboxylic acid metabolites.42 Although we did not observe alterations in propanoate metabolism, we found similar decreases in these other classes of metabolites. Other studies have revealed that disease severity could be predicted using machine learning techniques from metabolomic and proteomic signatures in the sera of patients with COVID-19, similar to the accuracy in predicting AKI from metabolomic signatures in this study.43
An important limitation of this study is the convenience sampling of participants based on the availability of biospecimens contained in the University of Alabama at Birmingham and Massachusetts Consortium on Pathogen Readiness COVID-19 biobanks. Controls in the Boston cohort were carefully selected to mirror the illness severity of cases save the presence of AKI, but controls in both Boston and Birmingham tended to be younger and have fewer preexisting comorbidities than the cases. Analysis of our primary outcome controlling for age nevertheless revealed persistent differences by AKI status in both Boston and Birmingham cohorts. AKI was defined based on sCr criteria alone and did not incorporate urine output criteria. We used urine metabolites only, which may reflect not only intracellular metabolism but also movement between intracellular/extracellular compartments and blood/urine and metabolic activity in both the blood and urine. Owing to the cross-sectional nature of this study, we are unable to determine whether metabolic alterations are a cause or a consequence of AKI.
We were able to compare and contrast findings in our Boston cohort, which was carefully curated to include only individuals with critical illness, against those in our Birmingham cohort which included a randomized selection from individuals with and without AKI from a hospital population. That similar metabolic differences were observed in each adds to the overall generalizability of our findings. The use of untargeted mass spectrometry analyses allowed for discovery of unexpected metabolic differences between the groups but permits less precision with respect to metabolite identification and quantification than a targeted approach. The methodology used in our study provides a high degree of validation through comparison with the molecular metabolite standards database, with incorporation of retention time as a confirming parameter in addition to the mass of the metabolite ion and its tandem mass spectrometry product ion spectrum, which mitigates some of the decreased precision in our untargeted approach. Nevertheless, further study confirming the findings here using targeted metabolomics along the metabolic pathways of interest is merited. For instance, nicotinamide was not identified in the Boston cohort, which may reflect depletion to below the threshold for detection using our methodology. Future studies should include the analysis of changes in the urinary metabolome during sequential measurements in the course of illness of a patient and incorporate transcriptomics to further analyze processes underlying the changes present in the metabolome. Our findings provide further basis for additional studies of NAD+ augmentation in both COVID-19–related AKI and other types of ischemic, inflammatory, and/or toxic AKI.
In summary, we found that COVID-19–related AKI was associated with an increase in urinary Q/T, consistent with NAD+ biosynthetic impairment arising in the context of ischemic, inflammatory, or toxic kidney injury. This result supports ongoing studies investigating the therapeutic potential of NAD+ augmentation as a means to mitigate this injury. The urine metabolome as a whole differed markedly between individuals with and without AKI, particularly with respect to energy metabolism and purine metabolism, and could be used to distinguish patients who had AKI with a high degree of accuracy. Additional studies are merited to further explore pathophysiological and therapeutic implications of our findings.
Disclosure
AA reports serving as a consultant for DynaMed and being on the advisory boards of Goldilocks Therapeutics, Alpha Young, and Angion for work outside the scope of this manuscript. SMP reports receiving consulting fees from Janssen, Pfizer, Mission Therapeutics, and Flagship Pioneering and being on the scientific advisory boards of Cytokinetics and Aerpio. All the other authors declared no competing interests.
Acknowledgments
Please see the Supplementary Acknowledgments for individual members of the Massachusetts General Hospital COVID-19 Collection & Processing Team, the Massachusetts Consortium on Pathogen Readiness members, and the University of Alabama at Birmingham COVID-19 Collection & Processing Team. Dr. Amar Pandit from the University of Massachusetts provided helpful insights into physiological interpretations. This work was supported by an Anderson Innovation Award (Stephen Barnes, Principal Investigator) and from the National Institutes of Health / National Institute of Diabetes, Digestive, and Kidney to the University of Alabama at Birmingham–University of California—San Diego O’Brien Center for Acute Kidney Injury (P30 DK079337, Anupam Agarwal, Principal Investigator). The mass spectrometer was purchased from a National Institutes of Health Shared Instrumentation Grant (S10 RR220872, Stephen Barnes, Principal Investigator). The Massachusetts General Hospital/Massachusetts Consortium on Pathogen Readiness COVID-19 biorepository was supported by a gift from Ms. Enid Schwartz, by the Mark and Lisa Schwartz Foundation, the Massachusetts Consortium for Pathogen Readiness and the Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard. Nathan H. Raines was supported by the National Institutes of Health / National Institute of Diabetes, Digestive, and Kidney T32 DK007199 (Pollak) and is now supported by the Doris Duke Physician Scientist Fellowship Grant 202182. Matthew Cheung is supported by the National Institutes of Health T32-GM008361. Jeffrey C. Edberg is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR003096 (Edberg). Samir M. Parikh is supported by grants R35HL139424, R01DK095072, and R01AG027002 from the National Heart Lung Blood Institute, the National Institute of Diabetes, Digestive, and Kidney Diseases, and the National Institute on Aging.
Footnotes
Supplementary Methods.
Supplementary Acknowledgments.
Table S1. List of metabolites identified in patient urine using liquid chromatography-tandem mass spectrometry.
Table S2. Differences in metabolites related to NAD+ synthesis between groups of hospitalized patients with COVID-19 with and without AKI.
Figure S1. Important features identified by variable importance in projection scores in supervised partial least squares analysis.
Figure S2. Urine metabolomic features distinguishing AKI among patients with COVID-19 pooled across Boston and Birmingham cohorts.
Supplementary Modified STROBE Statement.
STROBE Statement (PDF).
Contributor Information
Stephen Barnes, Email: sbarnes@uab.edu.
Samir M. Parikh, Email: sparikh1@bidmc.harvard.edu.
Supplementary Material
Supplementary Methods.
Supplementary Acknowledgments.
Table S1. List of metabolites identified in patient urine using liquid chromatography-tandem mass spectrometry.
Table S2. Differences in metabolites related to NAD+ synthesis between groups of hospitalized patients with COVID-19 with and without AKI.
Figure S1. Important features identified by variable importance in projection scores in supervised partial least squares analysis.
Figure S2. Urine metabolomic features distinguishing AKI among patients with COVID-19 pooled across Boston and Birmingham cohorts.
Supplementary Modified STROBE Statement.
STROBE Statement (PDF)
References
- 1.WHO coronavirus (COVID-19) dashboard. World Health Organization. https://covid19.who.int Updated August 4, 2021. Accessed August 7, 2021.
- 2.Chan L., Chaudhary K., Saha A., et al. AKI in hospitalized patients with COVID-19. J Am Soc Nephrol. 2021;32:151–161. doi: 10.1681/ASN.2020050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson S., Hirsch J.S., Narasimhan M., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area [published correction appears in JAMA. 2020;323:2098] JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruchfeld A. The COVID-19 pandemic: consequences for nephrology. Nat Rev Nephrol. 2021;17:81–82. doi: 10.1038/s41581-020-00381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poyan Mehr A., Tran M.T., Ralto K.M., et al. De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat Med. 2018;24:1351–1359. doi: 10.1038/s41591-018-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran M.T., Zsengeller Z.K., Berg A.H., et al. PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531:528–532. doi: 10.1038/nature17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsyuba E., Mottis A., Zietak M., et al. De novo NAD+ synthesis enhances mitochondrial function and improves health. Nature. 2018;563:354–359. doi: 10.1038/s41586-018-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ralto K.M., Rhee E.P., Parikh S.M. NAD(+) homeostasis in renal health and disease. Nat Rev Nephrol. 2020;16:99–111. doi: 10.1038/s41581-019-0216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin D.R., Lewington A.J.P., Hammerman M.R., Padanilam B.J. Inhibition of poly(ADP-ribose) polymerase attenuates ischemic renal injury in rats. Am J Physiol Regul Integr Comp Physiol. 2000;279:R1834–R1840. doi: 10.1152/ajpregu.2000.279.5.R1834. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J., Devalaraja-Narashimha K., Singaravelu K., Padanilam B.J. Poly(ADP-ribose) polymerase-1 gene ablation protects mice from ischemic renal injury. Am J Physiol Ren Physiol. 2005;288:F387–F398. doi: 10.1152/ajprenal.00436.2003. [DOI] [PubMed] [Google Scholar]
- 11.Heer C.D., Sanderson D.J., Voth L.S., et al. Coronavirus infection and PARP expression dysregulate the NAD metabolome: an actionable component of innate immunity. J Biol Chem. 2020;295:17986–17996. doi: 10.1074/jbc.RA120.015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Su X., Quinn W.J., 3rd, et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 2018;27:1067–1080 e5. doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raines N.H., Ganatra S., Nissaisorakarn P., et al. Niacinamide may be associated with improved outcomes in COVID-19-related acute kidney injury: an observational study. Kidney360. 2021;2:33–41. doi: 10.34067/KID.0006452020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C., Zhang P., Bao G., Fang Y., Chen W. Discovery of potential biomarkers in acute kidney injury by ultra-high-performance liquid chromatography-tandem quadrupole time-of-flight mass spectrometry (UPLC-Q/TOF-MS). Int Urol Nephrol. Published online March 8, 2021. https://doi.org/10.1007/s11255-021-02829-3 [DOI] [PubMed]
- 15.Martin-Lorenzo M., Gonzalez-Calero L., Ramos-Barron A., et al. Urine metabolomics insight into acute kidney injury point to oxidative stress disruptions in energy generation and H2S availability. J Mol Med (Berl) 2017;95:1399–1409. doi: 10.1007/s00109-017-1594-5. [DOI] [PubMed] [Google Scholar]
- 16.Kellum J.A., Lameire N., Aspelin P., et al. Kidney disease: improving global outcomes (KDIGO) acute kidney injury work group. KDIGO clinical practice guideline for acute kidney injury. Kidney Int Suppl. 2012;2:1–138. doi: 10.1038/kisup.2012.1. [DOI] [Google Scholar]
- 17.Levey A.S., Bosch J.P., Lewis J.B., Greene T., Rogers N., Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 18.Nadim M.K., Forni L.G., Mehta R.L., et al. COVID-19-associated acute kidney injury: consensus report of the 25th Acute Disease Quality Initiative (ADQI) Workgroup [published correction appears in Nat Rev Nephrol. 2020;16:765] Nat Rev Nephrol. 2020;16:747–764. doi: 10.1038/s41581-020-00356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gisewhite S., Stewart I.J., Beilman G., Lusczek E. Urinary metabolites predict mortality or need for renal replacement therapy after combat injury. Crit Care. 2021;25:119. doi: 10.1186/s13054-021-03544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwaki T., Bennion B.G., Stenson E.K., et al. PPARα contributes to protection against metabolic and inflammatory derangements associated with acute kidney injury in experimental sepsis. Physiol Rep. 2019;7 doi: 10.14814/phy2.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Standage S.W., Xu S., Brown L., et al. NMR-based serum and urine metabolomic profile reveals suppression of mitochondrial pathways in experimental sepsis-associated acute kidney injury. Am J Physiol Ren Physiol. 2021;320:F984–F1000. doi: 10.1152/ajprenal.00582.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu X., Gao H., Sun J., et al. Identification of key metabolites during cisplatin-induced acute kidney injury using an HPLC-TOF/MS-based non-targeted urine and kidney metabolomics approach in rats. Toxicology. 2020;431:152366. doi: 10.1016/j.tox.2020.152366. [DOI] [PubMed] [Google Scholar]
- 23.Thomas T., Stefanoni D., Reisz J.A., et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimhofer T., Lodge S., Whiley L., et al. Integrative modeling of quantitative plasma lipoprotein, metabolic, and amino acid data reveals a multiorgan pathological signature of SARS-CoV-2 infection [published correction appears in J Proteome Res. 2021;20:3400] J Proteome Res. 2020;19:4442–4454. doi: 10.1021/acs.jproteome.0c00519. [DOI] [PubMed] [Google Scholar]
- 25.Cai Y., Kim D.J., Takahashi T., et al. Kynurenic acid may underlie sex-specific immune responses to COVID-19. Sci Signal. 2021;14 doi: 10.1126/scisignal.abf8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narui K., Noguchi N., Saito A., et al. Anti-infectious activity of tryptophan metabolites in the L-tryptophan-L-kynurenine pathway. Biol Pharm Bull. 2009;32:41–44. doi: 10.1248/bpb.32.41. [DOI] [PubMed] [Google Scholar]
- 27.Wang J., Simonavicius N., Wu X., et al. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J Biol Chem. 2006;281:22021–22028. doi: 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 28.Sorgdrager F.J.H., Naudé P.J.W., Kema I.P., Nollen E.A., Deyn P.P.D. Tryptophan metabolism in inflammaging: from biomarker to therapeutic target. Front Immunol. 2019;10:2565. doi: 10.3389/fimmu.2019.02565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minhas P.S., Liu L., Moon P.K., et al. Macrophage de novo NAD+ synthesis specifies immune function in aging and inflammation. Nat Immunol. 2019;20:50–63. doi: 10.1038/s41590-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cervenka I., Agudelo L.Z., Ruas J.L. Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science. 2017;357 doi: 10.1126/science.aaf9794. [DOI] [PubMed] [Google Scholar]
- 31.Pisarenko O.I. Mechanisms of myocardial protection by amino acids: facts and hypotheses. Clin Exp Pharmacol Physiol. 1996;23:627–633. doi: 10.1111/j.1440-1681.1996.tb01748.x. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z.H., Wei F., Vaziri N.D., et al. Metabolomics insights into chronic kidney disease and modulatory effect of rhubarb against tubulointerstitial fibrosis. Sci Rep. 2015;5:14472. doi: 10.1038/srep14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe M., Suliman M.E., Qureshi A.R., et al. Consequences of low plasma histidine in chronic kidney disease patients: associations with inflammation, oxidative stress, and mortality. Am J Clin Nutr. 2008;87:1860–1866. doi: 10.1093/ajcn/87.6.1860. [DOI] [PubMed] [Google Scholar]
- 34.Kessler R.H. Effects of ischemia on the concentration of adenine nucleotides in the kidney of anesthetized dogs. Proc Soc Exp Biol Med. 1970;134:1091–1095. doi: 10.3181/00379727-134-34950. [DOI] [PubMed] [Google Scholar]
- 35.Chihanga T., Ruby H.N., Ma Q., Bashir S., Devarajan P., Kennedy M.A. NMR-based urine metabolic profiling and immunohistochemistry analysis of nephron changes in a mouse model of hypoxia-induced acute kidney injury. Am J Physiol Renal Physiol. 2018;315:F1159–F1173. doi: 10.1152/ajprenal.00500.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber K., Hofer D.C., Trefely S., et al. N-acetylaspartate pathway is nutrient responsive and coordinates lipid and energy metabolism in brown adipocytes. Biochim Biophys Acta Mol Cell Res. 2019;1866(3):337–348. doi: 10.1016/j.bbamcr.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vallon V., Osswald H. Adenosine receptors and the kidney. Handb Exp Pharmacol. 2009;193:443–470. doi: 10.1007/978-3-540-89615-9_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauerle J.D., Grenz A., Kim J.H., Lee H.T., Eltzschig H.K. Adenosine generation and signaling during acute kidney injury. J Am Soc Nephrol. 2011;22:14–20. doi: 10.1681/ASN.2009121217. [DOI] [PubMed] [Google Scholar]
- 39.Cihlar T., Ho E.S. Fluorescence-based assay for the interaction of small molecules with the human renal organic anion transporter 1. Anal Biochem. 2000;283:49–55. doi: 10.1006/abio.2000.4633. [DOI] [PubMed] [Google Scholar]
- 40.Aslamkhan A., Han Y.H., Walden R., Sweet D.H., Pritchard J.B. Stoichiometry of organic anion/dicarboxylate exchange in membrane vesicles from rat renal cortex and hOAT1-expressing cells. Am J Physiol Renal Physiol. 2003;285:F775–F783. doi: 10.1152/ajprenal.00140.2003. [DOI] [PubMed] [Google Scholar]
- 41.De Deyn P.P., Vanholder R., Eloot S., Glorieux G. Guanidino compounds as uremic (neuro)toxins. Semin Dial. 2009;22:340–345. doi: 10.1111/j.1525-139X.2009.00577.x. [DOI] [PubMed] [Google Scholar]
- 42.Pang Z., Zhou G., Chong J., Xia J. Comprehensive meta-analysis of COVID-19 global metabolomics datasets. Metabolites. 2021;11:44. doi: 10.3390/metabo11010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen B., Yi X., Sun Y., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.