Abstract
Beyond their rapid rewarding effects, drugs of abuse can durably alter an individual’s response to their environment as illustrated by the compulsive drug seeking and risk of relapse triggered by drug-associated stimuli. The persistence of these associations even long after cessation of drug use demonstrates the enduring mark left by drugs on brain reward circuits. However, within these circuits, neuronal populations are differently affected by drug exposure and growing evidence indicates that relatively small subsets of neurons might be involved in the encoding and expression of drug-mediated associations. The identification of sparse neuronal populations recruited in response to drug exposure has benefited greatly from the study of immediate early genes (IEGs) whose induction is critical in initiating plasticity programs in recently activated neurons. In particular, the development of technologies to manipulate IEG-expressing cells has been fundamental to implicate broadly distributed neuronal ensembles coincidently activated by either drugs or drug-associated stimuli and to then causally establish their involvement in drug responses. In this review, we summarize the literature regarding IEG regulation in different learning paradigms and addiction models to highlight their role as a marker of activity and plasticity. As the exploration of neuronal ensembles in addiction improves our understanding of drug-associated memory encoding, it also raises several questions regarding the cellular and molecular characteristics of these discrete neuronal populations as they become incorporated in drug-associated neuronal ensembles. We review recent efforts towards this goal and discuss how they will offer a more comprehensive understanding of addiction pathophysiology.
1. Introduction
The integration in the central nervous system of external sensory inputs together with internal information reflecting our physiological state is critical for individuals to produce appropriate behavioral responses and to quickly adapt to a changing environment. This continual adjustment of brain circuits through life experiences paradoxically requires both a highly dynamic remodeling of neuronal networks and the capacity to stabilize these adaptations to support the encoding and storage of relevant information into memory (Kandel, Dudai, & Mayford, 2014). Addiction can be studied through this lens of memory formation, maintenance and updating: the high risk of relapse to drug seeking in abstinent individuals even long after cessation of drug taking is proof of such highly stable alterations (Dong, Taylor, Wolf, & Shaham, 2017; Nestler, 2001; Wolf, 2016). The neuronal networks whose activity is dramatically and durably affected by drug exposure are mostly comprised of the interconnected cortical, striatal and limbic regions of the brain’s reward circuitry (Lüscher & Malenka, 2011), which normally integrates midbrain dopamine signals to guide reward-related learning (Hyman, Malenka, & Nestler, 2006; Kelley, 2004).
The persistent imprint of drug exposure onto brain function relies on the ability of neuronal plasticity mechanisms normally engaged in non-pathological processes like reward learning to adjust neuronal physiology in order to modulate the response of brain networks to a given stimulus. The mechanisms that translate rapid and transient electrical and neuro-chemical changes into persistent cellular adaptations require a coordinated response across neuronal compartments from the synapse to the nucleus (Kandel, 2001; Nestler & Lschüer, 2019). Among the myriad of cellular mechanisms orchestrating neuronal plasticity, changes in gene expression are pivotal in reshaping neuronal networks during learning and memory formation (Alberini & Kandel, 2015). Addiction is similarly associated with complex transcriptional adaptations following drug exposure, which underlie the long-term behavioral alterations observed in this pathology (Nestler, 2013; Robison & Nestler, 2011; Walker et al., 2018).
Gene expression is tightly coupled to synaptic activity with signaling mechanisms transducing and propagating information from the membrane to different cell compartments including the nucleus. Activity-dependent transcription thus links rapid environmental changes to long-term neuronal adaptations via specific transcriptional programs (West & Greenberg, 2011; Yap & Greenberg, 2018). The first wave of these activity-regulated genes are called immediate early genes (IEGs) because of their rapid but transient up-regulation in response to a wide variety of cellular stimuli (Sheng & Greenberg, 1990). Bulk analysis of IEG induction provided the first evidence in favor of a brain region- and task-specific recruitment of activity-regulated transcription (Tischmeyer & Grimm, 1999). Because they are supposedly strictly coupled to synaptic activity, these mechanisms seem likely to be restricted to electrophysiologically activated cells, suggesting that changes detected at the bulk level could be driven by the activity of a discrete subpopulation of neurons specifically recruited in memory encoding (Miyashita, Kubik, Lewandowski, & Guzowski, 2008). This hypothesis was further corroborated by the histological mapping of IEG induction in sparse subsets of neurons during learning and memory retrieval (Guzowski et al., 2005; Miyashita et al., 2008). As a result, activity-dependent transcription progressively emerged as a highly reliable marker of behaviorally relevant activated neuronal populations, later termed neuronal ensembles (Minatohara, Akiyoshi, & Okuno, 2016). Activity-dependent mechanisms, including IEG induction, are similarly recruited by drugs of abuse throughout the brain (Chandra & Lobo, 2017; Manning, Williams, & Robison, 2017). As the identification and manipulation of these drug-activated cells in multiple addiction models markedly improved our understanding of the biology behind neuronal ensembles, it also provided new insights into the encoding of drug-associated memories at different stages of addiction (Bobadilla et al., 2017; Cruz, Rubio, & Hope, 2015; Whitaker & Hope, 2018).
Here we review the literature published in the last three decades that has supported the concept that drug-related pathological memories are encoded through plasticity changes in discrete neuronal ensembles in key regions of the brain reward circuitry. First, we discuss the up-regulation of IEGs by drugs of abuse in light of their role in general learning and memory processes, and how these markers of transcriptional plasticity can identify drug-activated cells. Second, we examine the various genetic tools that have more recently been developed to track and manipulate activated cells, and how they have been used to causally demonstrate the importance of IEG-expressing neuronal ensembles in mediating memory- and addiction-related learned behaviors. Third, we summarize the emerging and still relatively scarce data describing the circuit, physiological, cellular, and molecular features that specifically characterize the neuronal ensembles recruited by drug exposure. We finally conclude by arguing that the ongoing investigation of the unique signature of drug-activated cells has the potential to greatly deepen our understanding of basic neuronal biology, as well as to yield preclinical data with strong translational potential to better combat drug addiction.
It is very fitting that this review appears in a special issue devoted to the life and career of Paul Greengard, PhD, in whose laboratory one of us (EJN) completed his dissertation research close to 40 years ago. No one better exemplified the ideals of a superb mentor, and provided much of the field’s basic understanding of how the molecular constituents and circuit activities of neurons are inter-related, than Paul Greengard (Nestler & Huganir, 2019).
2. Immediate early genes to visualize drug-activated cells: toward neuronal ensembles
2.1. IEG Expression as a proxy for neuronal activity and plasticity
The first evidence that neurons can trigger rapid transcriptional activity in response to environmental changes came from the study of the Fos gene, with both the transcript and protein induced within minutes following cell stimulation (Cole, Saffen, Baraban, & Worley, 1989; Greenberg, Ziff, & Greene, 1986; Morgan, Cohen, Hempstead, & Curran, 1987). The idea of a dynamic coupling between extracellular events and transcription was further confirmed through the identification of multiple genes regulated by neuronal activity and referred to as IEGs (Morgan & Curran, 1989; Sheng & Greenberg, 1990). These genes, many of which are expressed at very low levels in basal conditions, are remarkable by their rapid and transient induction in response to a wide range of stimuli. In neurons, IEGs are induced within minutes following synaptic activation and most of them return to their basal levels of expression one to two hours later (Sheng & Greenberg, 1990). Because they encode either transcription factors (e.g., Fos, FosB, JunD, Egr1 [also known as Zif268 or NGFI-A]) or effector proteins involved in activity-dependent plasticity (e.g., Arc, Homer1A) (Lanahan & Worley, 1998; Guzowski, McNaughton, Barnes, & Worley, 1999), these genes were logically proposed to be essential linking blocks that ensure the continuity between the rapid synaptic changes and long-lasting adaptations necessary for neuronal plasticity (Flavell & Greenberg, 2008; Sheng & Greenberg, 1990) that underlie memory formation (Tischmeyer & Grimm, 1999).
The bulk expression of several IEGs such as Fos and Egr-1 is increased within brain regions involved in memory encoding after several behavioral learning tasks (Gallo, Katche, Morici, Medina, & Weisstaub, 2018; Kubik, Miyashita, & Guzowski, 2007). They are induced in the hippocampus following novel environment exploration, spatial memory tasks, object recognition or fear conditioning (Montag-Sallaz, Welzl, Kuhl, Montag, & Schachner, 1999; Guzowski et al., 1999a; Vann, Brown, Erichsen, & Aggleton, 2000; Guzowski, Setlow, Wagner, & McGaugh, 2001; Ramírez-Amaya et al., 2005; Barbosa et al., 2013) and following fear conditioning in the cortex and amygdala as well (Hall, Thomas, & Everitt, 2001; Rosen, Fanselow, Young, Sitcoske, & Maren, 1998). Across these different types of tasks, the spatiotemporal regulation of Fos and Zif268 expression matches the known role and recruitment of specific brain regions during given phases of training and memory encoding (Minatohara et al., 2016). For instance, Zif268 expression is increased in the amygdala after retrieval of both contextual and cued fear associations, and in the hippocampus during contextual fear retrieval only (Hall et al., 2001). Similarly, Arc expression is increased in relevant brain regions during several behavioral tasks, from the exploration of a novel environment to the acquisition of operant behavior (Montag-Sallaz et al., 1999; Guzowski et al., 1999; Kelly & Deadwyler, 2002, 2003). Moreover, the level of Arc expression in the hippocampus positively correlates with novelty (Kelly & Deadwyler, 2002), decreases in trained animals (Kelly & Deadwyler, 2002), and is higher in animals showing higher performance in hippocampus- (Guzowski, McNaughton, Barnes, & Worley, 2001) or striatal- (Daberkow, Riedy, Kesner, & Keefe, 2007) dependent learning paradigms, together suggesting a role in the early phases of learning.
Preliminary evidence showing that patterns of IEG induction reflect the activity of not only given brain regions studied in bulk but also of specific neuronal networks within and across these brain regions came from studies of Arc (Miyashita et al., 2008). Unlike many other IEGs, Arc is localized in the dendritic compartment (Link et al., 1995; Lyford et al., 1995), where it directly regulates synaptic plasticity (Korb & Finkbeiner, 2011). While Arc expression is broadly increased in the brain following the massive and synchronous neuronal activity surge induced by a seizure (Lyford et al., 1995), it also appears to be induced selectively in dendrites exhibiting long-term potentiation (LTP) after electrical stimulation (Moga et al., 2004; Steward, Wallace, Lyford, & Worley, 1998), suggesting that Arc induction could be restricted to recently activated neurons that develop synaptic plasticity. Consistently, more precise histological mapping of IEG-expressing cells demonstrated and confirmed that only subsets of neurons engage these mechanisms in several learning paradigms (Guzowski et al., 2005; Miyashita et al., 2008). In particular, a mRNA labeling method called cellular compartment analysis of temporal activity by fluorescence in situ hybridization (catFISH) has been instrumental in correlating IEG expression with neuronal network activity (Guzowski et al., 1999; Guzowski, McNaughton, et al., 2001). This approach takes advantage of the rapid nuclear induction (within 2min) and export (within 20min) of IEG mRNAs such as Arc, Homer1A and others to distinguish cells activated at two different time points. Using this method, one can backtrack the activity of a given neuron or population of neurons during two different behavioral tasks, such as the exploration of two distinct environments. Exploration of a novel environment increased Arc expression in the hippocampus (Guzowski et al., 1999) and cortex (Vazdarjanova, McNaughton, Barnes, Worley, & Guzowski, 2002). In the hippocampus, the sequential exploration of two different environments induced Arc mRNA in two separate populations, whereas exploring twice the same environment induced Arc expression within the same neuronal population (Guzowski et al., 1999). These studies were fundamental in linking Arc induction with the activation by behavioral experience of precise brain networks and were further used to dissect the activation dynamics of distinct hippocampal subfields (Guzowski, Knierim, & Moser, 2004; Vazdarjanova & Guzowski, 2004). Together, this seminal data strongly supported the idea that IEGs not only serve as a reliable marker to detect global activation of given brain regions but can also help define discrete neuronal ensembles specifically recruited during learning processes, in which activity-dependent plasticity programs are likely to be engaged (Guzowski et al., 2004; Kubik et al., 2007; Minatohara et al., 2016).
2.2. IEG induction and function in drug-activated neuronal ensembles
Both acute and chronic consumption of drugs of abuse dramatically alter an individual’s behavioral, psychological, and cognitive responses in a wide range of situations, and these changes can linger for a very long time after cessation of use (Wolf, 2016). Such enduring effects reflect the development of a pathological form of neuronal and behavioral plasticity likely initiated during the early stages of drug exposure and whose persistence sustains the maladaptations and resulting malfunction of the brain reward circuitry. While decades of work documented the various ways these circuits are altered in addiction (Lüscher & Malenka, 2011), the concept that sparsely activated neuronal populations within those circuits are key in driving drug-induced alterations and network remodeling has emerged more recently and stands as a very promising perspective to refine our understanding of drug-associated memory encoding. IEG expression has been instrumental in identifying these drug-activated cells.
Several IEGs, including Fos, Arc and Zif268, are induced by drugs of abuse in a variety of drug exposure paradigms and across multiple regions of the reward circuitry (Table 1). Fos is induced in the nucleus accumbens (NAc), dorsal striatum and prefrontal cortex (PFC) following treatment with cocaine, amphetamine or methamphetamine (Crombag et al., 2002; Graybiel et al., 1990; Hope et al., 1992; Moratalla et al., 1993; Norman et al., 1993; Robertson, Paul, Moratalla, & Graybiel, 1991; Rosen et al., 1994; Torres et al., 2015; Young et al., 1991). Like Fos, Arc expression is increased in the same three brain regions after both acute and chronic cocaine (Fosnaugh et al., 1995; Freeman et al., 2010; Fumagalli et al., 2006; Salery et al., 2017; Tan et al., 2000; Yuferov et al., 2003), methamphetamine (Fujiyama et al., 2003; Kodama et al., 1998) and amphetamine (Tan et al., 2000). Arc expression is also increased in the CA1 region of the hippocampus after chronic treatment with psychostimulants (Fumagalli et al., 2006; Kodama et al., 1998). Similar patterns of induction are observed for Egr1 in response to psychostimulants (Bhat et al., 1992a, 1992b; Hope et al., 1992; Moratalla et al., 1992; Thomas, Arroyo, & Everitt, 2003; Torres et al., 2015). Consistent with their characteristic rapid and transient induction kinetics, IEGs are induced within minutes following psychostimulants exposure and return to their basal level of expression after minutes to hours (Fumagalli et al., 2006; Salery et al., 2017). The expression of these genes is also regulated by acute or chronic exposure to opioids (Ammon et al., 2003; Bisagno & Cadet, 2019; Browne, Godino, Salery, & Nestler, 2020; Hayward et al., 1990; Lv et al., 2011; Marie-Claire et al., 2004; Ziółkowska et al., 2012), albeit, with a slower time-course (Ziółkowska et al., 2012), likely due to different pharmacokinetic properties of the drugs and possibly different signaling mechanisms. In addition to these three well-described IEGs, several others including FosB (Alibhai, Green, Potashkin, & Nestler, 2007; Hope et al., 1992), Homer1A (Zhang et al., 2007), Jun and JunB (Hope et al., 1992; Moratalla et al., 1993), Egr3 (Chandra et al., 2015), Npas4 (Taniguchi et al., 2017) and Nr4a1 (Carpenter et al., 2020) are induced in diverse paradigms of drug exposure. However, whether all of these IEGs are induced in the exact same cells has yet to be fully substantiated, even if it seems to be the case at least for Fos and Arc following acute cocaine (Guez-Barber et al., 2011; Liu et al., 2014; Salery et al., 2017).
Table 1.
Summary of IEG regulation in rodent brain in response to psychostimulants or opioids.
Amph, amphetamine; CC, cerebral cortex; Cg, cingulate cortex; dStri, dorsal striatum; dmLC, dorsomedial limbic cortex; vmLC, ventromedial limbic cortex; HPC, Hippocampus; LC, locus coeruleus; LS, locomotor sensitization; Meth, methamphetamine; NAc, nucleus accumbens; NeoCx, neocortex: OFC, orbitofrontal cortex; PFC, prefrontal cortex; PC, parietal cortex; vStri, ventral striatum; WD, withdrawal.
In turn, IEG induction in reward-processing circuits contributes to establishing the persistent neuronal and behavioral adaptations characterizing the addicted phenotype in many different ways. Up-regulation of Fos is implicated in downstream changes in gene expression: its selective knock-out in Drd1-containing medium-sized spiny neurons (D1-MSNs) of the NAc alters drug-induced transcriptional programs along with cocaine-induced dendritic remodeling and locomotor sensitization (Xu, 2008; Zhang et al., 2007). The related IEG, FosB, has been studied widely in the context of addiction. FosB regulation is particularly interesting in the context of chronic drug exposure as a spliced product of the gene called ΔFosB resists degradation and thus accumulates in neurons following chronic treatment with psychostimulants and virtually any other class of abused drug (Hope et al., 1994; Chen et al., 1995; Jingshan Chen, Kelz, Hope, Nakabeppu, & Nestler, 1997; Lee et al., 2006; Robison & Nestler, 2011; Lobo et al., 2013). The long-lasting expression of this transcription factor in multiple brain regions following drug exposure (Perrotti et al., 2008) supports the idea that ΔFosB could act as a key “switch” for the drug-induced maladaptations underlying the transition from acute to compulsive drug taking (Brenhouse & Stellar, 2006; Nestler, 2013; Nestler, Kelz, & Chen, 1999). Consistently, ΔFosB overexpression in D1-MSNs of NAc increases cocaine-induced locomotor sensitization and conditioned place preference (CPP) (Kelz et al., 1999), facilitates acquisition of self-administration at low doses and enhances responses in a progressive ratio schedule of reinforcement (Colby, Whisler, Steffen, Nestler, & Self, 2003). Similar effects are seen with opioids (Zachariou et al., 2006). At the cellular level, ΔFosB overexpression in the NAc affects structural (Robison et al., 2013) and synaptic (Grueter, Robison, Neve, Nestler, & Malenka, 2013) plasticity. ΔFosB also regulates transcriptional programs in the NAc (McClung & Nestler, 2003) and has been mechanistically involved in the epigenetic desensitization of the Fos promoter via the recruitment of HDAC1 after chronic amphetamine (Renthal et al., 2008).
The role of Egr1 in drug models appears more subtle than that of Fos family members. Genetic suppression of Egr1 impairs locomotor sensitization but does not affect animals’ responses to cocaine CPP (Valjent et al., 2006), advocating for more comprehensive studies of this transcription factor in drug models, which remains surprisingly scarce despite its pivotal role in synaptic plasticity (Duclot & Kabbaj, 2017).
Unlike Fos, FosB/ΔFosB and Egr1, Arc protein does not interact directly with DNA to regulate transcription but rather exerts its function through the interaction with other proteins involved in plasticity. Due to its peculiar dendritic localization, most studies have focused on Arc’s canonical role in dendrites, where it regulates – among others – synaptic strength through AMPA glutamate receptor endocytosis (Chowdhury et al., 2006; Shepherd et al., 2006) and long-term neuronal plasticity (Guzowski et al., 2000; Shepherd & Bear, 2011; Waung, Pfeiffer, Nosyreva, Ronesi, & Huber, 2008). Recent evidence demonstrates that a nuclear pool of Arc protein might also contribute to neuronal plasticity in a non-canonical manner via indirect transcriptional regulation (Bloomer, VanDongen, & VanDongen, 2007; Korb, Wilkinson, Delgado, Lovero, & Finkbeiner, 2013; Salery et al., 2017). In drug models, cocaine increases Arc expression in the nucleus, and its suppression is associated with increased RNA-PolII activity and Fos expression (Salery et al., 2017). At the behavioral level, Arc loss of function is associated with increased sensitivity to low doses of cocaine in locomotor sensitization and CPP paradigms (Salery et al., 2017) and the local inhibition of Arc in the dorsal striatum attenuates the extinction of cocaine seeking but not seeking itself (Hearing, Schwendt, & McGinty, 2011). Together, cellular and behavioral studies of Arc in cocaine models argue for a homeostatic role of Arc in drug-induced plasticity, a unique property diametrically opposed to the proposed roles of other IEGs like Fos, FosB and Egr1, which seem to be overall positive regulators of drug-evoked maladaptations.
2.3. Plasticity in IEG expression, a substrate for behavioral plasticity in addiction models
While most IEGs are up-regulated by acute drug treatment independent of their molecular function, their response to prolonged exposure is different. The unique induction of ΔFosB protein after chronic drug exposure was discussed above. Arc too shows different responses to acute vs. chronic drug exposure. Chronic psychostimulant administration is for instance associated with a higher induction of Arc protein levels in animals with a drug history in comparison with those exposed to the drug for the first time (Fumagalli et al., 2006). This sensitization of Arc induction is associated with persistent accumulation of its protein and mRNA over days following the last injection. Arc expression is also chronically up-regulated in the absence of the drug during abstinence from cocaine self-administration (Freeman et al., 2010, 2008). After withdrawal, Arc mRNA expression is increased upon re-exposure to a drug-paired context (Hearing et al., 2011) or drug-paired discrete cues (Zavala, Osredkar, Joyce, & Neisewander, 2008), indicating that, through plastic changes, Arc expression tracks not only activation triggered by direct drug exposure but also the retrieval of previously learned drug-related memories.
In an opposite manner, bulk Fos expression is desensitized when cocaine or amphetamine is given in the home cage (Hope et al., 1992, 1994; Rosen et al., 1994), an effect mediated via ΔFosB as noted earlier (Renthal et al., 2008). By contrast, the number of Fos+cells is sensitized when animals are treated in a new environment and exhibit locomotor sensitization (Crombag et al., 2002; Nordquist et al., 2008) and not in the home cage (Mattson et al., 2008). Nonetheless, the absence of Fos sensitization to repeated cocaine or amphetamine when animals are injected in a familiar environment strongly suggests that the Fos+population is involved in the specific encoding of drug-context associations more than in simply tracking drug-evoked neuronal activity in chronically treated animals. Consistent with the persistence of such associations, context-specific Fos sensitization was maintained over 6months after drug exposure along with locomotor sensitization (Hope, Simmons, Mitchell, Kreuter, & Mattson, 2006). In operant models, Fos expression is chronically up-regulated during abstinence from cocaine self-administration (Freeman et al., 2010, 2008) and can be further induced by re-exposure to contextual or discrete drug-paired cues (Bastle et al., 2012; Bossert et al., 2011; Fanous et al., 2012; Hamlin, Clemens, & McNally, 2008; Kufahl et al., 2009; Neisewander et al., 2000; Rubio et al., 2019; Zavala, Biswas, Harlan, & Neisewander, 2007). Similarly, chronic non-contingent cocaine treatment desensitizes Egr1 expression, with reductions in its basal levels in multiple forebrain areas (Bhat et al., 1992a, 1992b; Daunais & McGinty, 1995; Unal, Beverley, Willuhn, & Steiner, 2009). However, similar to Arc and Fos, in a chronic self-administration model, Egr1 expression was induced by exposure to discrete cues previously associated with cocaine (Thomas et al., 2003). This response was not observed in yoked controls, suggesting that Egr1 may participate in the encoding of stimulus-drug conditioned associations.
Together, these findings argue for complex gene-specific, drug-specific and paradigm-specific patterns of IEG expression and plasticity, which could be explained by numerous factors. First, different signaling cascades might converge onto different epigenetic regulation of a given IEG promoter—which has been described in detail only in the specific case of Fos desensitization after chronic amphetamine (Renthal et al., 2008). Second, the precise pharmacodynamics of different drugs and drug administration regimens can greatly influence IEG induction and sensitization, as for example Arc and Fos induction and locomotor sensitization have all been shown to depend on the rate of intravenous cocaine administration (Samaha, Mallet, Ferguson, Gonon, & Robinson, 2004). Third, given that not all cells are activated by drugs, one cannot exclude drug-induced adaptations in non-activated cells or that the global response might be driven by relatively smaller or larger neuronal ensembles, especially as these methods do not allow the tracking of the initially activated population at subsequent stages of drug-related behaviors. Nevertheless, the one consistent finding is that multiple IEGs are induced following the presentation of previously neutral contextual information now associated with the drug. This could indicate a coordinated transcriptional response within a subset of neurons specifically involved in encoding drug-context associations if it were that those IEGs are induced in the same cells—which has not yet been confirmed. This also strengthens the idea that IEG-expressing cells not only track neuronal activity per se but neuronal activity relevant for drug-related learning and plasticity as well. Overall, the fact that the IEG response itself can change over time parallels behavioral plasticity, and that these responses seem to occur in ensembles encoding drug-cue associations reinforces the argument that IEG induction is more than just a good marker of neuronal activity: it can also be used as a proxy for the neuronal plasticity mechanisms engaged in those activated ensembles.
3. Beyond correlation: Causal role of activated cells in controlling behavior
The mapping of endogenous IEG expression across multiple brain regions and learning paradigms has been instrumental in identifying the neuronal populations recruited at different stages of memory formation and expression. We have seen that their activation not only reflects the rapid response of distributed neuronal networks but also contribute to initiate plasticity mechanisms within these neuronal ensembles. While a crucial first step, these early studies remained correlative and did not provide insight into the causal role of IEG-expressing cells in memory processing. The recent advent of optogenetic and chemogenetic approaches has made it possible to assess causality of neuronal ensembles (DeNardo & Luo, 2017; Denny, Lebois, & Ramirez, 2017; Koya, Margetts-Smith, & Hope, 2016; Whitaker & Hope, 2018).
3.1. Genetic tools to tag and manipulate neuronal ensembles
Because of their very transient induction, endogenous IEG expression can only be correlated to performance in recently executed behavioral tasks. Nonetheless, their role in neuronal plasticity suggests that cells where IEGs were initially expressed are likely to be reactivated by further stimulation, and that subsequent and repeated reactivation could be decisive in evoking persistent adaptations in network activity. As a first basis for that argument, the initial peak of Arc expression in the hippocampus after the exploration of a novel environment is followed a few hours later by a second wave of Arc induction in a subset of the originally activated neurons in the absence of any new stimulation (Ramírez-Amaya et al., 2005). Such reactivation—solely driven by network activity—has been proposed to reflect the stabilization of recently activated neuronal networks during memory consolidation, which is corroborated by the fact that Arc is induced by a second exploration of the same environment in the same neurons that were activated during the first exploration (Guzowski et al., 1999a; Vazdarjanova et al., 2002). Yet, studies based on the detection of endogenous IEG expression remain considerably limited in their ability to track the fate of initially activated cells over longer periods of time to decipher their role at later stages of memory encoding and retrieval, which would confirm that they truly represent the neuronal substrates of the corresponding memories. Past and current efforts to establish a causal role for these populations in memory encoding have benefited from the development of research tools allowing for both the permanent labeling (tagging) of activated cells to monitor their reactivation as well as the transient manipulation of their activity during key phases of learning processes (Denny et al., 2017; Garner & Mayford, 2012; Liu, Ramirez, Redondo, & Tonegawa, 2014; Ramirez, Tonegawa, & Liu, 2014).
These transgenic approaches mostly used the promoters of Arc or Fos to drive the expression of artificial tags, which can be either fluorescent reporters to simply visualize IEG-expressing cells, or optogenetic/chemogenetic constructs to manipulate IEG-expressing cells’ activity. The key factor has been to render those tags activity-dependent yet stable, and the persistence of this tagging depends on the system and strategy used. The Arc-GFP knock-in mouse—in which the Arc promoter drives the transient expression of a destabilized form of GFP instead of Arc itself—allowed for the visualization of Arc-expressing neuronal ensembles in different cortical areas during visual information processing (Gao et al., 2010; Wang et al., 2006) and motor learning (Cao et al., 2015; Gao et al., 2010). While this method is useful for in vivo real-time visualization and characterization of Arc-inducing cells, the destabilized GFP does not provide a permanent tagging of activated cells. Studying those cells over longer periods of time is feasible using the TetTag mouse line, another genetic model in which the tag expression can last days to weeks (Reijmers & Mayford, 2009; Reijmers, Perkins, Matsuo, & Mayford, 2007). In these mice, the expression of a longer-lasting tag in Fos-expressing cells is triggered by doxycycline treatment and can be used to both track and manipulate cells that were Fos+during the time window of doxycycline action, which is usually synchronized with a behavior or experience of interest (Garner et al., 2012; X. Liu et al., 2012; Ramirez et al., 2013, 2015). While this tag persists over days, it fails to offer an indelible labeling of activated neurons over weeks to months. A truly permanent tagging of activated cells was made possible via the development of the Arc-Cre/Arc-TRAP and Fos-Cre/Fos-TRAP transgenic mice (Guenthner, Miyamichi, Yang, Heller, & Luo, 2013), as well as the Arc-CreERT2 bacterial artificial chromosome (BAC) mice (Denny et al., 2014) (Fig. 1). Based on the use of a tamoxifen-inducible CreERT2 recombination system, these mouse lines offer improved temporal control over the tagging window to achieve specific and permanent labeling of neuronal populations activated during specific behavioral tasks through recombination of a second Cre-dependent tag-expressing transgene (Allen et al., 2017; Denny et al., 2014; Wall et al., 2019).
Fig. 1.
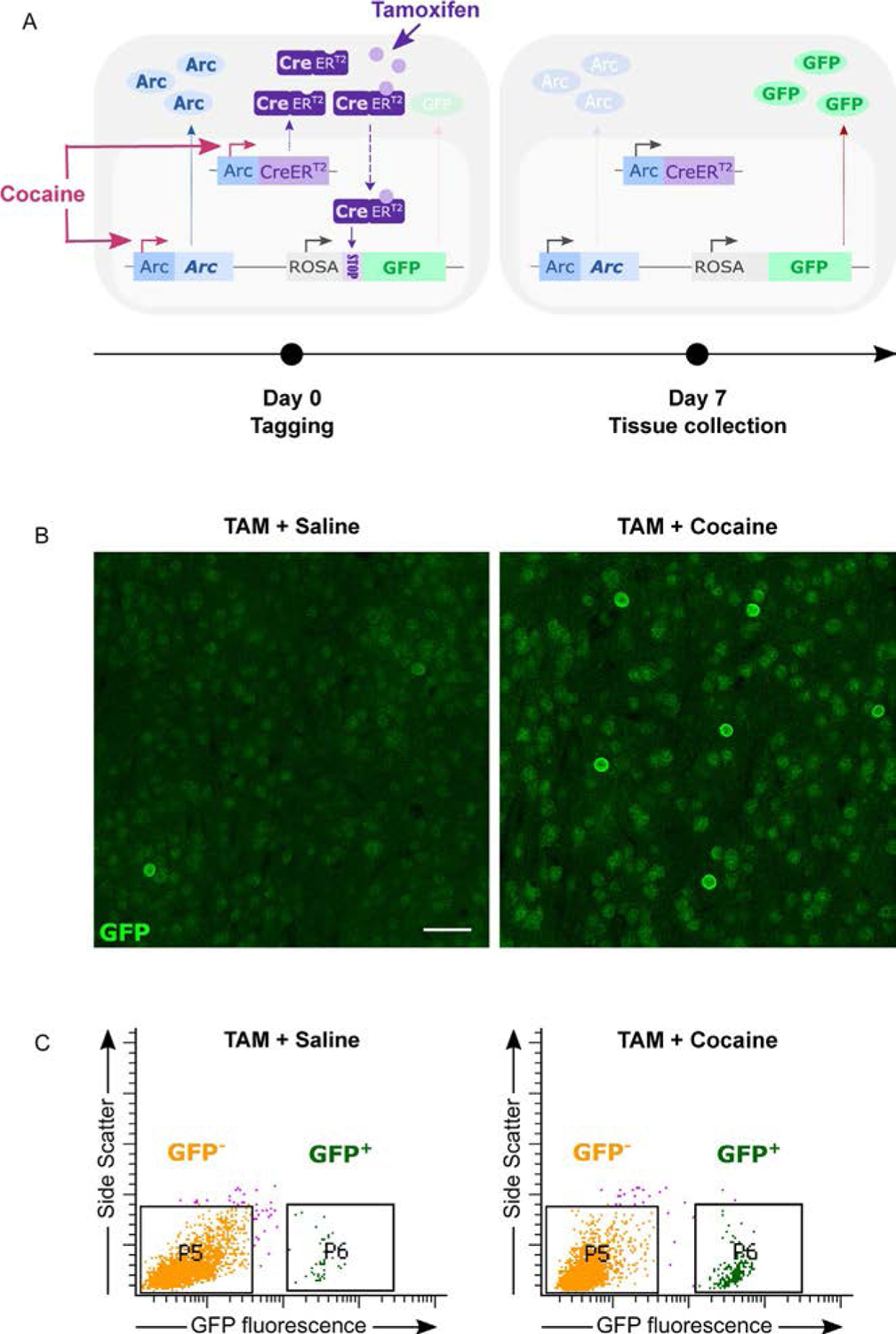
Permanent tagging of cocaine-activated cells in NAc of Arc-CreERT2 mice. Arc-CreERT2 mice were crossed with a GFP reporter line to induce stable GFP expression in activated Arc+ neurons. (A) Schematic representation of the Arc-CreERT2 strategy applied to cocaine-activated cell tagging. On the day of tagging (day 0), animals are injected concomitantly with cocaine and tamoxifen. Cocaine triggers the activation of both the endogenous and artificially-expressed Arc promoters controlling, respectively, endogenous Arc and CreERT2 transcription. This leads to increased expression of both Arc and CreERT2 proteins in activated Arc+ cells. Upon tamoxifen binding, CreERT2 enters the nucleus where the Cre-dependent removal of a floxed-STOP cassette triggers GFP expression. The persistent expression of GFP in cells activated initially by cocaine allows for their visualization at any future time (e.g., 7days). (B–C) On day 0, Arc-CreERT2 mice were treated concomitantly with tamoxifen + cocaine or tamoxifen + saline as a control. Tissue was collected on day 7 post-tagging to visualize cells previously activated by cocaine. TAM = Tamoxifen. (B) Representative confocal images of GFP immunostaining in the NAc at day 7 post-tagging showing a higher number of activated cells in the cocaine-treated group. Scalebar = 50μm. (C) Fluorescence-activated nuclei sorting (FANS) strategy to isolate GFP+ nuclei from NAc punches at day 7 post-tagging. Cocaine-treated group shows a larger population of activated (GFP+) cells in comparison to saline-treated group.
These tools have been utilized to tag neuronal ensembles activated during memory formation and further track and manipulate their activity at later stages of memory expression, with most of the focus being on the acquisition and retrieval of contextual fear memory (Minatohara et al., 2016). Using TetTag mice, (Reijmers et al., 2007) first demonstrated that an amygdala Fos+ensemble activated during the learning phase of fear conditioning is reactivated during the retrieval of that fear memory. The level of reactivation correlated with the freezing fear response, indicating that this reactivated ensemble might represent the neural trace of the associative memory (Reijmers & Mayford, 2009). To evaluate their causal role, these activated and reactivated cells were later manipulated by expressing optogenetic (Channelrhodopsin ChR2, Archaerhodopsin ArchT) or chemogenetic (designer receptors exclusively activated by designer drugs or DREADDs) tags. In the dentate gyrus (DG), the optogenetic reactivation of a ChR2-tagged Fos+ensemble recruited during contextual fear conditioning training was sufficient for the later expression of fear memory (Liu et al., 2012). Reciprocally, the inactivation of hippocampal CA1 Fos+ensemble activated during the conditioning session altered the retrieval of conditioned fear memory (Tanaka et al., 2014). The manipulation of Arc+ ensembles in the DG also controlled fear memory retrieval (Denny et al., 2014). These seminal studies were fundamental in demonstrating the causal role of IEG ensembles in several brain regions in memory retrieval and were followed by many others well reviewed elsewhere (Frankland, Josselyn, & Köhler, 2019; Liu, Ramirez, et al., 2014; Tonegawa, Morrissey, & Kitamura, 2018).
Together, this work provides direct evidence for specific reactivation during memory expression of neuronal ensembles that are initially activated during memory encoding and that such reactivation is both necessary and sufficient for memory expression. The sparse neuronal populations that meet these requirements—activated during memory encoding, reactivated during memory retrieval and both necessary and sufficient for memory expression—have been referred to as memory engram cells, and to date are thought to represent the neuronal substrate of the memory engram (Denny et al., 2017; Josselyn & Tonegawa, 2020; Tonegawa, Liu, Ramirez, & Redondo, 2015).
3.2. Tagging and manipulating neuronal ensembles in addiction
The robustness and persistence of associations formed between contextual information and the rewarding properties of drugs of abuse is proof of the strength of associative learning processes at play in the development of addiction (Robbins, Ersche, & Everitt, 2008). These memories are thought to be driven by a potent and enduring remodeling of reward-processing brain circuits during and after drug exposure (Lüscher & Malenka, 2011). As mentioned, these persistent adaptations are likely to occur in the sparse neuronal ensembles first activated during the early stages of addiction and whose later reactivation could account for the expression of drug-associated memories.
Most efforts towards characterizing neuronal ensembles in addiction have focused on the causal role of drug-activated Fos+ensembles in behavioral alterations across several addiction models (Cruz et al., 2015; Whitaker & Hope, 2018). Correlative studies described above indicate that Fos+ensembles respond to drugs or their associated cues (Cruz et al., 2015). However, the fact that Fos is induced at both early and late phases of drug-related paradigms raises the question of whether the same subset of neurons is recruited at different time points. From data on contextual fear conditioning, one can hypothesize that ensemble reactivation mediates the consolidation of the drug-associated engram and subsequent response to the drug itself or contextual cues. The earliest supporting evidence came from a study that took advantage of ΔFosB accumulation after chronic cocaine treatment to infer the past activation of those neurons (Mattson et al., 2008). Through co-labeling for recently induced Fos and remotely induced ΔFosB, the authors visualized the two waves of activation simultaneously and showed that 80% of the neurons recruited by chronic cocaine exposure (ΔFosB+) in a locomotor sensitization protocol are reactivated (Fos+) in response to a challenge injection in the cocaine-associated context. The causal role of these chronically activated Fos+ensembles in drug memory encoding has been assessed using a transgenic rat line expressing LacZ—coding for ß-galactosidase (ß-gal)—under the control of the Fos promoter. In Fos-LacZ transgenic rats, selective inactivation of Fos expressing neurons can be achieved via the local injection of the prodrug Daun02 that inactivates ß-gal-expressing cells, i.e., Fos+cells (Koya et al., 2009, 2016). In a locomotor sensitization paradigm, the selective inactivation of NAc Fos+neuronal ensembles activated in a cocaine-paired context severely impaired the conditioned locomotor response during re-exposure to the same context. Importantly, the behavioral response was not altered when the authors inactivated the cells activated by a second environment different from the drug-paired one (Koya et al., 2009). These findings support the idea that the drug-context associations are indeed encoded in a sparse and context-specific population of NAc neurons, whose activity is required for the expression of this associative memory, akin to the way contextual fear memories have been proposed to be encoded in the hippocampus and amygdala (Josselyn & Tonegawa, 2020). This scheme was further confirmed in an operant paradigm: selective inactivation of NAc shell neuronal ensembles activated by context-induced drug-seeking prevents the subsequent seeking during another re-exposure to the context (Cruz et al., 2014). Again, inactivation of a non-specific subset of neurons activated in a control context failed to alter context-induced drug seeking, and so did the inactivation of NAc core—not shell—ensembles, suggesting that context-induced drug-seeking is mediated by a context- and subregion-specific ensemble. The same approach was then used to identify medial PFC and orbitofrontal cortex (OFC) ensembles involved in heroin seeking following context (Bossert et al., 2011) or cue (Fanous et al., 2012) re-exposure, and dorsomedial striatum, central nucleus of amygdala and infralimbic cortex ensembles in the incubation of methamphetamine craving (Caprioli et al., 2017), alcohol dependence (de Guglielmo et al., 2016) and alcohol seeking (Pfarr et al., 2015), respectively. It is interesting to note that, across these studies, the proportion of neurons activated and subsequently manipulated is very low (5–10%), suggesting that drug memories are encoded in sparse neuronal populations, a hint at how precise and selective engram allocation mechanisms might be.
3.3. From a drug engram to a drug engram circuit?
Besides the Fos-LacZ mouse, other tools have been occasionally applied to addiction models. The TetTag mouse was used to analyze the encoding of a cocaine-associated spatial memory in hippocampal CA1 ensembles (Trouche et al., 2016). Through the optogenetic silencing of a CA1 Fos+neuronal ensemble associated with a specific environment, the authors were able to shift the representation of this spatial context to a different hippocampal ensemble. They recoded the engram representing the cocaine-paired environment and thereby reversed cocaine-induced CPP (Trouche et al., 2016). A cocaine-context memory engram was also found in the lateral nucleus of amygdala (LA) through the manipulation of a CREB-expressing neuronal population (Hsiang et al., 2014). This approach, previously used to characterize LA neuronal ensembles involved in fear memory (Han et al., 2007, 2009), showed that the artificial overexpression of CREB in a sparse population of LA neurons during contextual conditioning is sufficient to increase their incorporation into an engram encoding the drug memory. While this approach does not directly manipulate the cells activated during encoding of drug memory, it demonstrates that activation of a given population of neurons during memory encoding is sufficient for it to become incorporated into the engram and to drive the expression of the corresponding memory.
Given that neuronal ensembles activated by drug exposure appear to emerge in multiple brain regions, one key question is whether these distributed engram cells are preferentially interconnected across brain regions. Recent but still limited data seem to support that statement. First, anatomically connected ventral CA1 (vCA1) and NAc core ensembles were shown to be concomitantly activated during cocaine CPP training and exhibit preferential synaptic strengthening with conditioning (Zhou et al., 2019). Second, a retrograde tracing study found a similar preferential strengthening of synaptic connections between cocaine-activated neurons from the OFC projecting to cocaine-activated neurons in the dorsal striatum (Wall et al., 2019). In both cases, the manipulation of the coordinated activity of engram cells across regions modulated drug-induced behavioral plasticity. This has led to the proposal that the “cocaine engram” might actually be a “cocaine engram circuit” (Zhou et al., 2019) with preferential connectivity between engram cells of each region.
The broad distribution of sparse activated networks associated with a specific memory also suggests the co-existence within brain regions of neuronal ensembles associated with different experiences. Recently, this was confirmed by the identification of two distinct PFC ensembles either activated by events of positive (cocaine exposure) or negative (foot shock) valence, respectively. The cells also differed in their molecular phenotype and long-range wiring—with positive valence-encoding and negative valence-encoding PFC cells preferentially projecting to the NAc and lateral habenula, respectively—confirming that different experiences can be represented in divergent and specific networks of activated neurons within a given brain region (Ye et al., 2016). Functionally, the reactivation of these cocaine- or shock-associated ensembles in a real-time preference assay increased preference or aversion, respectively (Ye et al., 2016). Similarly, the two different memory traces related to two successive phases of a given paradigm—acquisition and then extinction of food (Warren et al., 2016) or cocaine (Warren et al., 2019) self-administration - were shown to be encoded in distinct non-overlapping neuronal ensembles of the ventral medial PFC. These distinct ensembles also have different outputs, with the self-administration ensembles projecting to the NAc Core and the extinction ensembles projecting to the NAc Shell, putatively reflecting the role of these two ensembles in, respectively, promoting or inhibiting cocaine-seeking (Warren et al., 2019). These studies (Wall et al., 2019; Warren et al., 2016; Warren et al., 2019; Ye et al., 2016; Zhou et al., 2019), all based on activity-dependent and permanent cell tagging methods, emphasize once again the importance of monitoring and modulating the activity of cells recruited at the early stages of drug exposure, i.e., during memory encoding. It is of course also true that the manipulation of ensembles associated with the expression of an already sensitized/learned response has greatly improved our understanding of the encoding of drug associations. Still, our ability to track and eventually modulate the fate of the originally activated ensemble remains a critical step in finding and characterizing a drug memory engram.
4. Cellular and molecular characteristics of neuronal ensembles
Drugs of abuse affect neuronal physiology in part by increasing dopamine neurotransmission and thereby activating dopamine-evoked signaling cascades throughout the brain reward circuitry (Di Chiara & Imperato, 1988). The resulting rapid cellular changes, including activity-dependent transcription of IEGs, are thought to then set the stage for more persistent adaptations within activity-defined neuronal networks (Nestler & Lüscher, 2019). However, the different time courses of these mechanisms limits our ability to assess whether lasting changes actually happen as a consequence of the initial activation of a given neuron. The permanent tagging of IEG expressing cells hence stands as a powerful tool to address these short-comings, as it offers the possibility to subsequently question the neuronal properties and causal role of the populations targeted initially. This approach is particularly relevant in addiction models where the persistence of behavioral alterations is proposed to depend on the strengthening of neuronal alterations built up since the earliest stages of drug exposure. Given the possibility that at least part if not a majority of the several plasticity mechanisms implicated in the pathophysiology of addiction might be restricted to sparse neuronal ensembles, the cellular and molecular characterization of the ensemble cells becomes especially critical to reconcile with the decades of evidence for plastic changes detected in bulk tissue analyses. Efforts toward this goal are twofold: first, the identification of intrinsic properties shared by neurons from the same ensemble that would reflect their preferential recruitment based on their cell type or expression of specific molecular markers and, second, the dissection of acquired properties that would reflect coordinated adaptations developed in cells as they become part of the drug engram.
4.1. Cell type specificity of drug-activated neuronal ensembles
Much evidence regarding the preferential recruitment of specific cell types in drug responses comes from the cellular mapping of IEGs, which highlight a dramatic dichotomy between D1- and D2-MSNs of the NAc (Chandra & Lobo, 2017). These two striatal subpopulations, which have distinct or even opposite effects on drug-related behaviors, also diverge in their cellular and transcriptional profiles in both basal and drug-exposed conditions (Calipari et al., 2016; Heiman et al., 2008; Lobo et al., 2010; Lobo, Karsten, Gray, Geschwind, & Yang, 2006; Salery, Trifilieff, Caboche, & Vanhoutte, 2020). Several IEGs, including Fos, FosB, Egr1 and Arc, are induced in D1-MSNs specifically in response to drugs of abuse (Bertran-Gonzalez et al., 2008; Chandra et al., 2015; Chandra & Lobo, 2017; Heiman et al., 2008; Lobo et al., 2013). Consistently, Fos+ensembles activated by acute and chronic cocaine isolated via fluorescence-activated cell sorting (FACS) were shown to express higher levels of the D1-MSN marker prodynorphin and lower levels of the D2 receptor itself as well as the D2-MSN marker Adora2a (Guez-Barber et al., 2011), indicating a predominant activation of D1-MSNs. Likewise, all drugs of abuse after chronic exposure induce ΔFosB protein in D1-MSNs only, except for opioids which induce it equally in D1- and D2-MSNs (Lobo et al., 2013). This D1-D2 MSN dichotomy holds for ΔFosB in operant (i.e., self-administration) paradigms (Lobo et al., 2013), although less so for Fos: both D1- and D2-MSNs appear to be recruited in Fos+ensembles in the dorsomedial striatum, NAc shell, ventromedial PFC and OFC associated with incubation of methamphetamine craving (Caprioli et al., 2017), context-induced reinstatement of cocaine seeking (Cruz et al., 2014) and heroin seeking (Bossert et al., 2011) and incubation of heroin craving (Fanous et al., 2012), respectively. A contribution of both D1- and D2-MSNs to contextually guided drug seeking is consistent with previous reports of context-specific regulation of Fos expression in D1- and D2-MSNs (Badiani et al., 1999). Preferential recruitment of D1-MSNs in drug memory encoding and retrieval was further confirmed in a study showing that Fos+ensembles activated in the NAc core during both the acquisition and expression of cocaine CPP were mainly composed of D1-MSNs (Zhou et al., 2019). The D1 engram neurons are preferentially targeted by afferents from vCA1 cocaine-activated neuronal ensembles, which are thought to provide contextual information for cocaine CPP memory (Zhou et al., 2019). This D1-MSN engram appears critical in integrating and encoding drug-context memory since manipulation of vCA1 ensembles connecting to the D1-MSN engram cells was further shown to mediate the retrieval of cocaine CPP memory (Zhou et al., 2019). Consistent with the idea of D1-expressing NAc engram cells encoding drug associations, silencing D1-MSNs suppresses cocaine CPP, while activation of D1- or D2-MSNs promoted and impaired retrieval of this memory, respectively (Lobo et al., 2010; Zhou et al., 2019). However, these studies manipulated D1- and D2-MSNs populations as a whole and did not specifically target the activated D1- or D2-MSNs ensembles. While much indirect evidence supports the existence of a predominant D1 engram (Guez-Barber et al., 2011; Zhou et al., 2019), we still lack a comprehensive understanding of the specific contribution of D1- vs. D2-expressing engram cells. For instance, a similarly sparse D2-MSNs population could take part in the encoding of drug-associated memories through opposite dopamine-dependent regulatory mechanisms, and could thereby modulate the recruitment of the drug engram by regulating the signal to noise ratio in these circuits.
4.2. Synaptic adaptations in drug engram cells
The strengthened connectivity between broadly distributed activated neurons discussed above is mediated by synaptic adaptations. Upon cocaine CPP, the preferential recruitment of D1-MSNs is associated with synaptic structural (dendritic spine morphology) and electrophysiological (AMPA transmission, frequency of synaptic inputs and paired-pulse ratio) changes restricted to NAc D1-MSN engram cells compared to neighboring D1-MSN non-engram cells (Zhou et al., 2019). This was associated with a specific synaptic strengthening of vCA1 inputs onto NAc core D1-MSN engram cells compared to neighboring D1-MSN non-engram cells which was further shown to mediate the retrieval of cocaine CPP memory (Zhou et al., 2019). The investigation of similar engram-specific synaptic changes in cortico-striatal networks revealed a preferential connectivity between cocaine-activated cortical and striatal neurons which was associated with enhanced synaptic strength between these co-activated ensembles (Wall et al., 2019). In a cocaine locomotor sensitization paradigm, chronic cocaine specifically enhanced drug-activated OFC projections onto coactive dorsal striatal neurons as evidenced by increased AMPA/NMDA receptor ratios and smaller paired-pulse ratio in activated dorsal striatal neurons compared to non-activated neurons (Wall et al., 2019). Likewise, the silencing of the coactivated OFC-to-dorsal striatum network impaired the expression of cocaine locomotor sensitization. Plastic synaptic alterations were also reported for chronically activated NAc Fos+ensembles, as they formed more silent synapses—proposed to be the substrates of drug-induced plasticity (Dong & Nestler, 2014)—than surrounding non-activated ensembles in animals expressing cocaine locomotor sensitization (Koya et al., 2012). This phenomenon was only observed in context-dependent cocaine sensitization indicating that it could also be restricted to neurons involved in the encoding of drug-context associations (Whitaker et al., 2016).
4.3. Molecular characteristics of drug engram cells
These synaptic adaptations presumably result from drug-evoked molecular and transcriptional adaptations. Genes involved in synaptic plasticity associated with glutamatergic transmission (Homer1, Grin2A, Grin2B) are for instance selectively increased in FACS-isolated Fos+ensembles of the dorsal striatum activated during context-induced reinstatement of cocaine seeking (Li et al., 2015; Rubio et al., 2015). Glutamatergic transmission plays an important role in drug-induced cue seeking, and high levels of glutamate release might expand the neuronal ensembles activated during cue-induced drug-seeking and precipitate relapse-like responses (Bobadilla et al., 2017). The transcriptional profiling of FACS-isolated NAc Fos+ensembles also revealed an increase in the expression of several IEGs including Arc, FosB, and Nr4a3 in comparison with non-activated Fos- neurons (Guez-Barber et al., 2011). Similarly, PFC neuronal ensembles associated with cue-induced heroin seeking expressed higher levels of Arc, Egr1 and Egr2 (Fanous et al., 2013). Importantly, the magnitude of the increase compared to control groups was higher than previously reported in bulk tissue for the same genes in the same behavioral task (Koya et al., 2006; Kuntz, Patel, Grigson, Freeman, & Vrana, 2008), a strong argument defending the idea that changes detected in bulk are mostly driven by the changes engaged in activated cells. This was also demonstrated in methamphetamine-activated dorsostriatal Fos+ensembles, which exhibited a greater induction of FosB and ΔFosB than seen for total homogenates (Liu, Rubio, et al., 2014). Consistent with its major role in addiction (Lobo et al., 2010) and in cue-induced cocaine seeking (Graham et al., 2007; Grimm et al., 2003), the plasticity marker and neurotrophic factor Bdnf along with its receptor TrkB were also selectively increased in Fos+neurons during the incubation of methamphetamine (Li et al., 2015). In this same paradigm, the expression of genes encoding several epigenetic enzymes (e.g., Hdac3, Hdac4, Hdac5, Ehmt1, Dnmt3a, Kdm1a) was also affected, suggesting here again that various epigenetic mechanisms implicated in drug-induced plasticity at the bulk tissue level (LaPlant et al., 2010; Maze & Nestler, 2011; Renthal et al., 2008; Walker & Nestler, 2018) might occur specifically in drug-activated ensembles (Li et al., 2015). While the exploration of activity-regulated changes can inform cellular adaptations following drug exposure, recent evidence also suggests the existence of pre-existing molecular markers that define a neuronal population more likely to be allocated to the encoding of a specific memory (Ye et al., 2016). For instance, PFC neuronal ensembles encoding a positive valence experience preferentially expressing Npas4 at baseline in comparison with negative valence-associated ensembles.
Across cell types, the segregation of neuronal populations based on their level of activation during specific behavioral tasks reveals the existence of complex patterns of gene expression specifically associated with the recruitment of a given cell in drug-activated neuronal ensembles. While D1-MSNs seem pivotal in encoding drug-context associations, the concomitant activation of D1- and D2 MSNs during re-exposure to the drug-paired context indicates that the definition of the cocaine engram could reside beyond cell type specificity. The co-induction of multiple IEGs in drug-activated ensembles suggests that they could be characterized by a transcriptional signature composed of the combination of specific activity-regulated genes. This hypothesis is corroborated by recent findings showing a tight correlation between the expression of different IEGs at the level of individual MSN neurons in the dorsal striatum after acute cocaine (Gonzales, Mukherjee, Ashwal-Fluss, Loewenstein, & Citri, 2020). The spatial segregation of IEG-expressing clusters was proposed by the authors to represent “super-ensembles” of neurons characterized by a coherent and robust expression of a specific set of IEGs. A more comprehensive molecular characterization of NAc populations after acute cocaine exposure was recently achieved using single-nucleus RNA sequencing (Savell et al., 2020). This approach identified a combinatorial cocaine-specific transcriptional program composed of distinct IEGs, and the coordinated manipulation of this set of genes using large-scale multiplexed CRISPR-dCAS9 potentiated cocaine locomotor sensitization (Savell et al., 2020). The implementation of transcriptional profiling of activated populations with single-cell resolution thus appears as a critical step to unmask subtle yet specific transcriptional alterations restricted to sparse neuronal populations involved in drug memory encoding, as well as intrinsic molecular markers that could determine a neuron’s allocation to a drug memory engram. Applying these approaches to chronic drug exposure, in particular, to self-administration and relapse paradigms, is a high priority for the future.
5. Conclusion
From the study of bulk IEG regulation to the visualization and manipulation of IEG-expressing cells across several brain regions and learning paradigms, the exploration of activity-dependent transcription is a major tool to define behaviorally relevant networks at the molecular level. The recent finding that cocaine-activated IEG-expressing cells can form spatially defined clusters whose size is correlated with their level of activation (Gonzales et al., 2020) suggests that their pattern of induction could reflect complex regulation within brain regions and circuits during the encoding of specific information. At the single cell level, the consistent and coordinated induction of an IEG program in cocaine-activated cells provides an experience-specific transcriptional signature that is sufficient to recapitulate drug responses (Savell et al., 2020). While these studies confirm the relevance of studying IEG regulation, they also demonstrate the importance of more comprehensive approaches beyond focusing on a restricted set of genes to reveal experience-specific molecular signatures.
The mapping of neuronal ensembles recruited throughout the brain at different stages of addiction reveals a sparse but broad distribution of activated cells across brain regions. This suggests that different components of a given experience can be encoded in distinct but interconnected neuronal ensembles in brain regions involved in the processing of specific aspects of this experience. Reciprocally, the coexistence within a given brain region of neuronal ensembles encoding distinct experiences suggests complex patterns of intermingled ensembles and subtle mechanisms of cellular allocation to an ensemble, which highlights the importance of coupling permanent tagging of activated cells with wide-range tracing to achieve an extensive characterization of broadly distributed engrams.
Finally, while the study of neuronal ensembles in the addiction field has been fundamental for the identification of the small and specific neuronal populations involved in complex behavioral responses, we still lack a broad description of their cellular and molecular properties. Most molecular studies focusing on rapid changes in gene expression within activated ensembles indicate alterations in their transcriptional responses (Cruz et al., 2015; Koya et al., 2016; Whitaker & Hope, 2018). The exploration of stable molecular alterations in neuronal ensembles over long periods of withdrawal and relapse would also be extremely informative in regard to the persistence of addictive behaviors. Improving our ability to physically isolate neuronal ensembles using sorting techniques (Fig. 1) will facilitate the implementation of cutting edge epigenomic and transcriptomic techniques to this field and greatly expand our understanding of the complex transcriptional signature of activated cells at different stages of addiction.
Acknowledgments
This work was supported by the Boehringer Ingelheim Fonds (doctoral fellowship to AG) and National Institute of Health (Grant No. R01DA014133 [to EJN]).
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- Arc
activity regulated cytoskeleton associated protein
- ArchT
archaerhodopsin
- BAC
bacterial artificial chromosome
- CA1
cornus ammonis subfield 1
- Cas9
CRISPR associated protein 9
- ChR2
channelrhodopsin 2
- CPP
conditioned place preference
- Cre
Cre recombinase
- CREB
cAMP response element-binding protein
- CreERT2
Cre recombinase fused to a mutant estrogen ligand-binding domain
- CRISPR
clustered regularly interspaced short palindromic repeat
- DG
dentate gyrus
- Dnmt3A
DNA methyltransferase 3 Alpha
- Drd1/D1
dopamine receptor D1
- D1-MSN
dopamine receptor D1-expressing medium spiny neuron
- Drd2/D2
dopamine receptor D2
- D2-MSN
dopamine receptor D2-expressing medium spiny neuron
- DREADD
designer receptors exclusively activated by designer drugs
- Egr1
early growth response 1
- Egr2
early growth response 2
- Egr3
early growth response 3
- Ehmt1
euchromatic histone lysine methyltransferase 1
- FACS
fluorescence-activated cell sorting
- Fos
FBJ murine osteosarcoma viral oncogene homolog
- FosB
FBJ murine osteosarcoma viral oncogene homolog B
- GFP
green fluorescent protein
- Grin2A
glutamate Ionotropic receptor N-methyl-d-aspartate type subunit 2A
- Grin2B
glutamate Ionotropic receptor N-methyl-d-aspartate type subunit 2B
- Hdac1
histone deacetylase 1
- Hdac3
histone deacetylase 3
- Hdac4
histone deacetylase 4
- Hdac5
histone deacetylase 5
- Homer1A
homer scaffold protein 1A
- IEGs
immediate early genes
- JunD
JunD proto-oncogene, AP-1 transcription factor subunit
- Kdm1a
Lysine demethylase 1A
- LA
lateral nucleus of the amygdala
- LacZ
bacterial beta-galactosidase gene
- mRNA
messenger ribonucleic acid
- MSN
medium spiny neuron
- NAc
nucleus accumbens
- NGFI-A
nerve growth factor-induced protein A
- Npas4
neuronal PAS domain-containing protein 4
- Nr4a1
nuclear receptor subfamily 4 group A member 1
- Nr4a3
nuclear receptor subfamily 4 group A member 3
- OFC
orbitofrontal cortex
- PFC
prefrontal cortex
- TRAP
targeted recombination in active populations
- vCA1
ventral cornus ammonis subfield 1
- Zif268
zinc finger protein 268
Footnotes
Conflict of interest statement
The authors report no biomedical financial interests or potential conflicts of interest.
References
- Alberini CM, & Kandel ER (2015). The regulation of transcription in memory consolidation. Cold Spring Harbor Perspectives in Biology, 7(1), a021741. 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibhai IN, Green TA, Potashkin JA, & Nestler EJ (2007). Regulation of fosB and ΔfosB mRNA expression: In vivo and in vitro studies. Brain Research, 1143, 22–33. 10.1016/j.brainres.2007.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WE, DeNardo LA, Chen MZ, Liu CD, Loh KM, Fenno LE, et al. (2017). Thirst-associated preoptic neurons encode an aversive motivational drive. Science, 357(6356), 1149–1155. 10.1126/science.aan6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammon S, Mayer P, Riechert U, Tischmeyer H, & Höllt V (2003). Microarray analysis of genes expressed in the frontal cortex of rats chronically treated with morphine and after naloxone precipitated withdrawal. Brain Research. Molecular Brain Research, 112(1–2), 113–125. 10.1016/s0169-328x(03)00057-3. [DOI] [PubMed] [Google Scholar]
- Badiani A, Oates MM, Day HEW, Watson SJ, Akil H, & Robinson TE (1999). Environmental modulation of amphetamine-induced c-fos expression in D1 versus D2 striatal neurons. Behavioural Brain Research, 103(2), 203–209. 10.1016/S0166-4328(99)00041-8. [DOI] [PubMed] [Google Scholar]
- Barbosa FF, Santos JR, Meurer YSR, Macêdo PT, Ferreira LMS, Pontes IMO, et al. (2013). Differential cortical c-Fos and Zif-268 expression after object and spatial memory processing in a standard or episodic-like object recognition task. Frontiers in Behavioral Neuroscience, 7, 112. 10.3389/fnbeh.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastle RM, Kufahl PR, Turk MN, Weber SM, Pentkowski NS, Thiel KJ, et al. (2012). Novel cues reinstate cocaine-seeking behavior and induce fos protein expression as effectively as conditioned cues. Neuropsychopharmacology, 37(9), 2109–2120. 10.1038/npp.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran-Gonzalez J, Bosch C, Maroteaux M, Matamales M, Hervé D, Valjent E, et al. (2008). Opposing patterns of signaling activation in dopamine D1 and D2 receptor-expressing striatal neurons in response to cocaine and haloperidol. Journal of Neuroscience, 28(22), 5671–5685. 10.1523/JNEUROSCI.1039-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RV, Cole AJ, & Baraban JM (1992a). Chronic cocaine treatment suppresses basal expression of zif268 in rat forebrain: In situ hybridization studies. Journal of Pharmacology and Experimental Therapeutics, 263(1), 343–349. [PubMed] [Google Scholar]
- Bhat RV, Cole AJ, & Baraban JM (1992b). Role of monoamine systems in activation of zif268 by cocaine. Journal of Psychiatry and Neuroscience, 17(3), 94–102. [PMC free article] [PubMed] [Google Scholar]
- Bisagno V, & Cadet JL (2019). Expression of immediate early genes in brain reward circuitries: Differential regulation by psychostimulant and opioid drugs. Neurochemistry International, 124, 10–18. 10.1016/j.neuint.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Bloomer WAC, VanDongen HMA, & VanDongen AMJ (2007). Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia (PML) bodies. Brain Research, 1153, 20–33. 10.1016/j.brainres.2007.03.079. [DOI] [PubMed] [Google Scholar]
- Bobadilla A-C, Heinsbroek JA, Gipson CD, Griffin WC, Fowler CD, Kenny PJ, et al. (2017). Chapter 4—Corticostriatal plasticity, neuronal ensembles, and regulation of drug-seeking behavior. In Calvey T, & Daniels WMU (Eds.), Progress in brain research (pp. 93–112). 10.1016/bs.pbr.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Stern AL, Theberge FRM, Cifani C, Koya E, Hope BT, et al. (2011). Ventral medial prefrontal cortex neuronal ensembles mediate context-induced relapse to heroin. Nature Neuroscience, 14(4), 420–422. 10.1038/nn.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenhouse HC, & Stellar JR (2006). C-Fos and deltaFosB expression are differentially altered in distinct subregions of the nucleus accumbens shell in cocaine-sensitized rats. Neuroscience, 137(3), 773–780. 10.1016/j.neuroscience.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Browne CJ, Godino A, Salery M, & Nestler EJ (2020). Epigenetic mechanisms of opioid addiction. Biological Psychiatry, 87(1), 22–33. 10.1016/j.biopsych.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calipari ES, Bagot RC, Purushothaman I, Davidson TJ, Yorgason JT, Peña CJ, et al. (2016). In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proceedings of the National Academy of Sciences of the United States of America, 113(10), 2726–2731. 10.1073/pnas.1521238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao VY, Ye Y, Mastwal S, Ren M, Coon M, Liu Q, et al. (2015). Motor learning consolidates arc-expressing neuronal ensembles in secondary motor cortex. Neuron, 86(6), 1385–1392. 10.1016/j.neuron.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli D, Venniro M, Zhang M, Bossert JM, Warren BL, Hope BT, et al. (2017). Role of dorsomedial striatum neuronal ensembles in incubation of methamphetamine craving after voluntary abstinence. The Journal of Neuroscience, 37(4), 1014–1027. 10.1523/JNEUROSCI.3091-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MD, Hu Q, Bond AM, Lombroso SI, Czarnecki KS, Lim CJ, et al. (2020). Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nature Communications, 11(1), 504. 10.1038/s41467-020-14331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, Francis TC, Konkalmatt P, Amgalan A, Gancarz AM, Dietz DM, et al. (2015). Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. The Journal of Neuroscience, 35(20), 7927–7937. 10.1523/JNEUROSCI.0548-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra R, & Lobo MK (2017). Beyond neuronal activity markers: Select immediate early genes in striatal neuron subtypes functionally mediate psychostimulant addiction. Frontiers in Behavioral Neuroscience, 11, 112. 10.3389/fnbeh.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nye HE, Kelz MB, Hiroi N, Nakabeppu Y, Hope BT, et al. (1995). Regulation of delta FosB and FosB-like proteins by electroconvulsive seizure and cocaine treatments. Molecular Pharmacology, 48(5), 880–889. [PubMed] [Google Scholar]
- Chen J, Kelz MB, Hope BT, Nakabeppu Y, & Nestler EJ (1997). Chronic Fos-related antigens: Stable variants of ΔFosB induced in brain by chronic treatments. Journal of Neuroscience, 17(13), 4933–4941. 10.1523/JNEUROSCI.17-13-04933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, et al. (2006). Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron, 52(3), 445–459. 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby CR, Whisler K, Steffen C, Nestler EJ, & Self DW (2003). Striatal cell type-specific overexpression of ΔFosB enhances incentive for cocaine. Journal of Neuroscience, 23(6), 2488–2493. 10.1523/JNEUROSCI.23-06-02488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AJ, Saffen DW, Baraban JM, & Worley PF (1989). Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature, 340(6233), 474–476. 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- Crombag HS, Jedynak JP, Redmond K, Robinson TE, & Hope BT (2002). Locomotor sensitization to cocaine is associated with increased Fos expression in the accumbens, but not in the caudate. Behavioural Brain Research, 136(2), 455–462. 10.1016/S0166-4328(02)00196-1. [DOI] [PubMed] [Google Scholar]
- Cruz FC, Babin KR, Leao RM, Goldart EM, Bossert JM, Shaham Y, et al. (2014). Role of nucleus accumbens shell neuronal ensembles in context-induced reinstatement of cocaine-seeking. Journal of Neuroscience, 34(22), 7437–7446. 10.1523/JNEUROSCI.0238-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz FC, Rubio FJ, & Hope BT (2015). Using c-fos to study neuronal ensembles in corticostriatal circuitry of addiction. Brain Research, 1628, 157–173. 10.1016/j.brainres.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daberkow DP, Riedy MD, Kesner RP, & Keefe KA (2007). Arc mRNA induction in striatal efferent neurons associated with response learning. European Journal of Neuroscience, 26(1), 228–241. 10.1111/j.1460-9568.2007.05630.x. [DOI] [PubMed] [Google Scholar]
- Daunais JB, & McGinty JF (1995). Cocaine binges differentially alter striatal preprodynorphin and zif/268 mRNAs. Molecular Brain Research, 29(2), 201–210. 10.1016/0169-328X(94)00246-B. [DOI] [PubMed] [Google Scholar]
- DeNardo L, & Luo L (2017). Genetic strategies to access activated neurons. Current Opinion in Neurobiology, 45, 121–129. 10.1016/j.conb.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, et al. (2014). Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron, 83(1), 189–201. 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Lebois E, & Ramirez S (2017). From engrams to pathologies of the brain. Frontiers in Neural Circuits, 11, 23. 10.3389/fncir.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, & Imperato A (1988). Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proceedings of the National Academy of Sciences, 85(14), 5274–5278. 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y,& Nestler EJ(2014). Theneural rejuvenationhypothesisof cocaineaddiction. Trends in Pharmacological Sciences, 35(8), 374–383. 10.1016/j.tips.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Taylor JR, Wolf ME, & Shaham Y (2017). Circuit and synaptic plasticity mechanisms of drug relapse. Journal of Neuroscience, 37(45), 10867–10876. 10.1523/JNEUROSCI.1821-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclot F, & Kabbaj M (2017). The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Frontiers in Behavioral Neuroscience, 11, 35. 10.3389/fnbeh.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Goldart EM, Theberge FRM, Bossert JM, Shaham Y, & Hope BT (2012). Role of orbitofrontal cortex neuronal ensembles in the expression of incubation of heroin craving. Journal of Neuroscience, 32(34), 11600–11609. 10.1523/JNEUROSCI.1914-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous S, Guez-Barber DH, Goldart EM, Schrama R, Theberge FRM, Shaham Y, et al. (2013). Unique gene alterations are induced in FACS-purified Fos-positive neurons activated during cue-induced relapse to heroin seeking. Journal of Neurochemistry, 124(1), 100–108. 10.1111/jnc.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, & Greenberg ME (2008). Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annual Review of Neuroscience, 31, 563–590. 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosnaugh JS, Bhat RV, Yamagata K, Worley PF, & Baraban JM (1995). Activation of arc, a putative “effector” immediate early gene, by cocaine in rat brain. Journal of Neurochemistry, 64(5), 2377–2380. 10.1046/j.1471-4159.1995.64052377.x. [DOI] [PubMed] [Google Scholar]
- Frankland PW, Josselyn SA, & Köhler S (2019). The neurobiological foundation of memory retrieval. Nature Neuroscience, 22(10), 1576–1585. 10.1038/s41593-019-0493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Lull ME, Patel KM, Brucklacher RM, Morgan D, Roberts DC, et al. (2010). Gene expression changes in the medial prefrontal cortex and nucleus accumbens following abstinence from cocaine self-administration. BMC Neuroscience, 11(1), 29. 10.1186/1471-2202-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Patel KM, Brucklacher RM, Lull ME, Erwin M, Morgan D, et al. (2008). Persistent alterations in mesolimbic gene expression with abstinence from cocaine self-administration. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology, 33(8), 1807–1817. 10.1038/sj.npp.1301577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama K, Kajii Y, Hiraoka S, & Nishikawa T (2003). Differential regulation by stimulants of neocortical expression of mrt1, arc, and homer1a mRNA in the rats treated with repeated methamphetamine. Synapse (New York, N.Y.), 49(3), 143–149. 10.1002/syn.10220. [DOI] [PubMed] [Google Scholar]
- Fumagalli F, Bedogni F, Frasca A, Pasquale LD, Racagni G, & Riva MA (2006). Corticostriatal up-regulation of activity-regulated cytoskeletal-associated protein expression after repeated exposure to cocaine. Molecular Pharmacology, 70(5), 1726–1734. 10.1124/mol.106.026302. [DOI] [PubMed] [Google Scholar]
- Gallo FT, Katche C, Morici JF, Medina JH, & Weisstaub NV (2018). Immediate early genes, memory and psychiatric disorders: Focus on c-Fos, Egr1 and Arc. Frontiers in Behavioral Neuroscience, 12. 10.3389/fnbeh.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Sossa K, Song L, Errington L, Cummings L, Hwang H, et al. (2010). A specific requirement of Arc/Arg3.1 for visual experience-induced homeostatic synaptic plasticity in mouse primary visual cortex. Journal of Neuroscience, 30(21), 7168–7178. 10.1523/JNEUROSCI.1067-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner AR, & Mayford M (2012). New approaches to neural circuits in behavior. Learning & Memory (Cold Spring Harbor, N.Y.), 19(9), 385–390. 10.1101/lm.025049.111. [DOI] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, et al. (2012). Generation of a synthetic memory trace. Science (New York, N.Y.), 335(6075), 1513–1516. 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales BJ, Mukherjee D, Ashwal-Fluss R, Loewenstein Y, & Citri A (2020). Subregion-specific rules govern the distribution of neuronal immediate-early gene induction. Proceedings of the National Academy of Sciences, 117(38), 23304–23310. 10.1073/pnas.1913658116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, & Self DW (2007). Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nature Neuroscience, 10(8), 1029–1037. 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Moratalla R, & Robertson HA (1990). Amphetamine and cocaine induce drug-specific activation of the c-fos gene in striosome-matrix compartments and limbic subdivisions of the striatum. Proceedings of the National Academy of Sciences, 87(17), 6912–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M, Ziff E, & Greene L (1986). Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science, 234(4772), 80–83. 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Grimm JW, Lu L, Hayashi T, Hope BT, Su T-P, & Shaham Y (2003). Time-dependent increases in brain-derived neurotrophic factor protein levels within the mesolimbic dopamine system after withdrawal from cocaine: Implications for incubation of cocaine craving. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 23(3), 742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter BA, Robison AJ, Neve RL, Nestler EJ, & Malenka RC (2013). ΔFosB differentially modulates nucleus accumbens direct and indirect pathway function. Proceedings of the National Academy of Sciences of the United States of America, 110(5), 1923–1928. 10.1073/pnas.1221742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, & Luo L (2013). Permanent genetic access to transiently active neurons via trap: targeted recombination in active populations. Neuron, 78(5), 773–784. 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez-Barber D, Fanous S, Golden SA, Schrama R, Koya E, Stern AL, et al. (2011). FACS identifies unique cocaine-induced gene regulation in selectively activated adult striatal neurons. Journal of Neuroscience, 31(11), 4251–4259. 10.1523/JNEUROSCI.6195-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guglielmo G, Crawford E, Kim S, Vendruscolo LF, Hope BT, Brennan M, et al. (2016). Recruitment of a neuronal ensemble in the central nucleus of the amygdala is required for alcohol dependence. Journal of Neuroscience, 36(36), 9446–9453. 10.1523/JNEUROSCI.1395-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Knierim JJ, & Moser EI (2004). Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron, 44(4), 581–584. 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, et al. (2000). Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. Journal of Neuroscience, 20(11), 3993–4001. 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, & Worley PF (1999a). Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nature Neuroscience, 2(12), 1120–1124. 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, & Worley PF (2001). Imaging neural activity with temporal and cellular resolution using FISH. Current Opinion in Neurobiology, 11(5), 579–584. 10.1016/S0959-4388(00)00252-X. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, & McGaugh JL (2001). Experience-dependent gene expression in the rat hippocampus after spatial learning: A comparison of the immediate-early genes Arc, c-fos, and zif268. Journal of Neuroscience, 21(14), 5089–5098. 10.1523/JNEUROSCI.21-14-05089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, & Barnes CA (2005). Mapping behaviorally relevant neural circuits with immediate-early gene expression. Current Opinion in Neurobiology, 15(5), 599–606. 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Hall J, Thomas KL, & Everitt BJ (2001). Cellular imaging of zif268 expression in the hippocampus and amygdala during contextual and cued fear memory retrieval: Selective activation of hippocampal CA1 neurons during the recall of contextual memories. Journal of Neuroscience, 21(6), 2186–2193. 10.1523/JNEUROSCI.21-06-02186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin AS, Clemens KJ, & McNally GP (2008). Renewal of extinguished cocaine-seeking. Neuroscience, 151(3), 659–670. 10.1016/j.neuroscience.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Han J-H, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, et al. (2007). Neuronal competition and selection during memory formation. Science (New York, N.Y.), 316(5823), 457–460. 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- Han J-H, Kushner SA, Yiu AP, Hsiang H-L(L), Buch T, Waisman A, et al. (2009). Selective erasure of a fear memory. Science, 323(5920), 1492–1496. 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- Hayward MD, Duman RS, & Nestler EJ (1990). Induction of the c-fos proto-oncogene during opiate withdrawal in the locus coeruleus and other regions of rat brain. Brain Research, 525(2), 256–266. 10.1016/0006-8993(90)90872-9. [DOI] [PubMed] [Google Scholar]
- Hearing MC, Schwendt M, & McGinty JF (2011). Suppression of activity-regulated cytoskeleton-associated gene expression in the dorsal striatum attenuates extinction of cocaine-seeking. The International Journal of Neuropsychopharmacology/Official Scientific Journal of the Collegium Internationale Neuropsychopharmacologicum (CINP), 14(6), 784–795. 10.1017/S1461145710001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. (2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell, 135(4), 738–748. 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Kosofsky B, Hyman SE, & Nestler EJ (1992). Regulation of immediate early gene expression and AP-1 binding in the rat nucleus accumbens by chronic cocaine. Proceedings of the National Academy of Sciences, 89(13), 5764–5768. 10.1073/pnas.89.13.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, et al. (1994). Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron, 13(5), 1235–1244. 10.1016/0896-6273(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Hope BT, Simmons DE, Mitchell TB, Kreuter JD, & Mattson BJ (2006). Cocaine-induced locomotor activity and Fos expression in nucleus accumbens are sensitized for 6 months after repeated cocaine administration outside the home cage. European Journal of Neuroscience, 24(3), 867–875. 10.1111/j.1460-9568.2006.04969.x. [DOI] [PubMed] [Google Scholar]
- Hsiang H-L(L), Epp JR, van den Oever MC, Yan C, Rashid AJ, Insel N, et al. (2014). Manipulating a “cocaine engram” in mice. The Journal of Neuroscience, 34(42), 14115–14127. 10.1523/JNEUROSCI.3327-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, & Nestler EJ (2006). Neural mechanisms of addiction: The role of reward-related learning and memory. Annual Review of Neuroscience, 29, 565–598. 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Josselyn SA, & Tonegawa S (2020). Memory engrams: Recalling the past and imagining the future. Science, 367(6473), eaaw4325. 10.1126/science.aaw4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER (2001). The molecular biology of memory storage: A dialogue between genes and synapses. Science, 294(5544), 1030–1038. 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, & Mayford MR (2014). The molecular and systems biology of memory. Cell, 157(1), 163–186. 10.1016/j.cell.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Kelley AE (2004). Memory and addiction: Shared neural circuitry and molecular mechanisms. Neuron, 44(1), 161–179. 10.1016/j.neuron.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Kelly MP, & Deadwyler SA (2002). Acquisition of a novel behavior induces higher levels of Arc mRNA than does overtrained performance. Neuroscience, 110(4), 617–626. 10.1016/S0306-4522(01)00605-4. [DOI] [PubMed] [Google Scholar]
- Kelly MP, & Deadwyler SA (2003). Experience-dependent regulation of the immediate-early gene arc differs across brain regions. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 23(16), 6443–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelz MB, Chen J, Carlezon WA, Whisler K, Gilden L, Beckmann AM, et al. (1999). Expression of the transcription factor ΔFosB in the brain controls sensitivity to cocaine. Nature, 401(6750), 272–276. 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- Kodama M, Akiyama K, Ujike H, Shimizu Y, Tanaka Y, & Kuroda S (1998). A robust increase in expression of arc gene, an effector immediate early gene, in the rat brain after acute and chronic methamphetamine administration. Brain Research, 796(1), 273–283. 10.1016/S0006-8993(98)00349-7. [DOI] [PubMed] [Google Scholar]
- Korb E, & Finkbeiner S (2011). Arc in synaptic plasticity: From gene to behavior. Trends in Neurosciences, 34(11), 591–598. 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Wilkinson CL, Delgado RN, Lovero KL, & Finkbeiner S (2013). Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nature Neuroscience, 16(7), 874–883. 10.1038/nn.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya E, Cruz FC, Ator R, Golden SA, Hoffman AF, Lupica CR, et al. (2012). Silent synapses in selectively activated nucleus accumbens neurons following cocaine sensitization. Nature Neuroscience, 15(11), 1556–1562. 10.1038/nn.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya E, Golden SA, Harvey BK, Guez DH, Berkow A, Simmons DE, et al. (2009). Targeted disruption of cocaine-activated accumbens neurons prevents context-specific sensitization. Nature Neuroscience, 12(8), 1069–1073. 10.1038/nn.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya E, Margetts-Smith G, & Hope BT (2016). Daun02 inactivation of behaviorally activated Fos-expressing neuronal ensembles. Current Protocols in Neuroscience, 76, 8.36.1–8.36.17. 10.1002/cpns.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya E, Spijker S, Voorn P, Binnekade R, Schmidt ED, Schoffelmeer ANM, et al. (2006). Enhanced cortical and accumbal molecular reactivity associated with conditioned heroin, but not sucrose-seeking behaviour. Journal of Neurochemistry, 98(3), 905–915. 10.1111/j.1471-4159.2006.03917.x. [DOI] [PubMed] [Google Scholar]
- Kubik S, Miyashita T, & Guzowski JF (2007). Using immediate-early genes to map hippocampal subregional functions. Learning & Memory, 14(11), 758–770. 10.1101/lm.698107. [DOI] [PubMed] [Google Scholar]
- Kufahl PR, Zavala AR, Singh A, Thiel KJ, Dickey ED, Joyce JN, et al. (2009). C-Fos expression associated with reinstatement of cocaine-seeking behavior by response-contingent conditioned cues. Synapse (New York, N.Y.), 63(10), 823–835. 10.1002/syn.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz KL, Patel KM, Grigson PS, Freeman WM, & Vrana KE (2008). Heroin self-administration: II. CNS gene expression following withdrawal and cue-induced drug-seeking behavior. Pharmacology Biochemistry and Behavior, 90(3), 349–356. 10.1016/j.pbb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan A, & Worley P (1998). Immediate-early genes and synaptic function. Neurobiology of Learning and Memory, 70(1–2), 37–43. 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J, Warren BL, et al. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nature Neuroscience, 13(9), 1137–1143. 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-W, Kim Y, Kim AM, Helmin K, Nairn AC, & Greengard P (2006). Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proceedings of the National Academy of Sciences, 103(9), 3399–3404. 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rubio FJ, Zeric T, Bossert JM, Kambhampati S, Cates HM, et al. (2015). Incubation of methamphetamine craving is associated with selective increases in expression of Bdnf and Trkb, glutamate receptors, and epigenetic enzymes in Cue-activated Fos-expressing dorsal striatal neurons. Journal of Neuroscience, 35(21), 8232–8244. 10.1523/JNEUROSCI.1022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, et al. (1995). Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proceedings of the National Academy of Sciences, 92(12), 5734–5738. 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q-R, Rubio FJ, Bossert JM, Marchant NJ, Fanous S, Hou X, et al. (2014). Detection of molecular alterations in methamphetamine-activated Fos-expressing neurons from a single rat dorsal striatum using fluorescence-activated cell sorting (FACS). Journal of Neurochemistry, 128(1), 173–185. 10.1111/jnc.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, et al. (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature, 484(7394), 381–385. 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Redondo RL, & Tonegawa S (2014). Identification and Manipulation of Memory Engram Cells. Cold Spring Harbor Symposia on Quantitative Biology, 79, 59–65. 10.1101/sqb.2014.79.024901. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. (2010). Cell type–specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science, 330(6002), 385–390. 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, & Yang XW (2006). FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nature Neuroscience, 9(3), 443–452. 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Zaman S, Damez-Werno DM, Koo JW, Bagot RC, DiNieri JA, et al. (2013). ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(47), 18381–18395. 10.1523/JNEUROSCI.1875-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, & Malenka RC (2011). Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron, 69(4), 650–663. 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X-F, Xu Y, Han J-S, & Cui C-L (2011). Expression of activity-regulated cytoskeleton-associated protein (Arc/Arg3.1) in the nucleus accumbens is critical for the acquisition, expression and reinstatement of morphine-induced conditioned place preference. Behavioural Brain Research, 223(1), 182–191. 10.1016/j.bbr.2011.04.029. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, et al. (1995). Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron, 14(2), 433–445. 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Manning CE, Williams ES, & Robison AJ (2017). Reward network immediate early gene expression in mood disorders. Frontiers in Behavioral Neuroscience, 11, 77. 10.3389/fnbeh.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Claire C, Courtin C, Roques BP, & Noble F (2004). Cytoskeletal genes regulation by chronic morphine treatment in rat striatum. Neuropsychopharmacology, 29(12), 2208–2215. 10.1038/sj.npp.1300513. [DOI] [PubMed] [Google Scholar]
- Mattson BJ, Koya E, Simmons DE, Mitchell TB, Berkow A, Crombag HS, et al. (2008). Context-specific sensitization of cocaine-induced locomotor activity and associated neuronal ensembles in rat nucleus accumbens. European Journal of Neuroscience, 27(1), 202–212. 10.1111/j.1460-9568.2007.05984.x. [DOI] [PubMed] [Google Scholar]
- Maze I, & Nestler EJ (2011). The epigenetic landscape of addiction. Annals of the New York Academy of Sciences, 1216, 99–113. 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, & Nestler EJ (2003). Regulation of gene expression and cocaine reward by CREB and ΔFosB. Nature Neuroscience, 6(11), 1208–1215. 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- Minatohara K, Akiyoshi M, & Okuno H (2016). Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Frontiers in Molecular Neuroscience, 8, 78. 10.3389/fnmol.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Kubik S, Lewandowski G, & Guzowski JF (2008). Networks of neurons, networks of genes: An integrated view of memory consolidation. Neurobiology of Learning and Memory, 89(3), 269–284. 10.1016/j.nlm.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moga DE, Calhoun ME, Chowdhury A, Worley P, Morrison JH, & Shapiro ML (2004). Activity-regulated cytoskeletal-associated protein is localized to recently activated excitatory synapses. Neuroscience, 125(1), 7–11. 10.1016/j.neuroscience.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M, Welzl H, Kuhl D, Montag D, & Schachner M (1999). Novelty-induced increased expression of immediate-early genes c-fos and arg 3.1 in the mouse brain. Journal of Neurobiology, 38(2), 234–246. [PubMed] [Google Scholar]
- Moratalla R, Robertson HA, & Graybiel AM (1992). Dynamic regulation of NGFI-A (zif268, egr1) gene expression in the striatum. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 12(7), 2609–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratalla R, Vickers EA, Robertson HA, Cochran BH, & Graybiel AM (1993). Coordinate expression of c-fos and jun B is induced in the rat striatum by cocaine. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 13(2), 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, & Curran T (1987). Mapping patterns of c-fos expression in the central nervous system after seizure. Science, 237(4811), 192–197. 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Morgan JI, & Curran T (1989). Stimulus-transcription coupling in neurons: Role of cellular immediate-early genes. Trends in Neurosciences, 12(11), 459–462. 10.1016/0166-2236(89)90096-9. [DOI] [PubMed] [Google Scholar]
- Neisewander JL, Baker DA, Fuchs RA, Tran-Nguyen LTL, Palmer A, & Marshall JF (2000). Fos protein expression and cocaine-seeking behavior in rats after exposure to a cocaine self-administration environment. Journal of Neuroscience, 20(2), 798–805. 10.1523/JNEUROSCI.20-02-00798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ (2001). Molecular basis of long-term plasticity underlying addiction. Nature Reviews Neuroscience, 2(2), 119–128. 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nestler EJ (2013). Cellular basis of memory for addiction. Dialogues in Clinical Neuroscience, 15(4), 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, & Huganir RL (2019). Paul Greengard (1925–2019). Cell, 177(6), 1365–1366. 10.1016/j.cell.2019.05.020. [DOI] [Google Scholar]
- Nestler EJ, Kelz MB, & Chen J (1999). ΔFosB: A molecular mediator of long-term neural and behavioral plasticity1Published on the World Wide Web on 27 November 1998.1. Brain Research, 835(1), 10–17. 10.1016/S0006-8993(98)01191-3. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, & Lüscher C (2019). The molecular basis of drug addiction: Linking epigenetic to synaptic and circuit mechanisms. Neuron, 102(1), 48–59. 10.1016/j.neuron.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordquist RE, Vanderschuren LJMJ, Jonker AJ, Bergsma M, de Vries TJ, Pennartz CMA, et al. (2008). Expression of amphetamine sensitization is associated with recruitment of a reactive neuronal population in the nucleus accumbens core. Psychopharmacology, 198(1), 113–126. 10.1007/s00213-008-1100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman AB, Lu SY, Klug JM, & Norgren RB (1993). Sensitization of c-fos expression in rat striatum following multiple challenges with D-amphetamine. Brain Research, 603(1), 125–128. 10.1016/0006-8993(93)91308-f. [DOI] [PubMed] [Google Scholar]
- Perrotti LI, Weaver RR, Robison B, Renthal W, Maze I, Yazdani S, et al. (2008). Distinct patterns of ΔFosB induction in brain by drugs of abuse. Synapse (New York, N.Y.), 62(5), 358–369. 10.1002/syn.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr S, Meinhardt MW, Klee ML, Hansson AC, Vengeliene V, Schönig K, et al. (2015). Losing control: Excessive alcohol seeking after selective inactivation of cue-responsive neurons in the infralimbic cortex. Journal of Neuroscience, 35(30), 10750–10761. 10.1523/JNEUROSCI.0684-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S, Liu X, Lin P-A, Suh J, Pignatelli M, Redondo RL, et al. (2013). Creating a false memory in the hippocampus. Science, 341(6144), 387–391. 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Liu X, MacDonald CJ, Moffa A, Zhou J, Redondo RL, et al. (2015). Activating positive memory engrams suppresses depression-like behaviour. Nature, 522(7556), 335–339. 10.1038/nature14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S, Tonegawa S, & Liu X (2014). Identification and optogenetic manipulation of memory engrams in the hippocampus. Frontiers in Behavioral Neuroscience, 7, 226. 10.3389/fnbeh.2013.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, & Barnes CA (2005). Spatial exploration-induced arc mRNA and protein expression: evidence for selective, network-specific reactivation. Journal of Neuroscience, 25(7), 1761–1768. 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, & Mayford M (2009). Genetic control of active neural circuits. Frontiers in Molecular Neuroscience, 2, 27. 10.3389/neuro.02.027.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, & Mayford M (2007). Localization of a stable neural correlate of associative memory. Science (New York, N.Y.), 317(5842), 1230–1233. 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, Truong H-T, Alibhai I, et al. (2008). Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 28(29), 7344–7349. 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins TW, Ersche KD, & Everitt BJ (2008). Drug addiction and the memory systems of the brain. Annals of the New York Academy of Sciences, 1141(1), 1–21. 10.1196/annals.1441.020. [DOI] [PubMed] [Google Scholar]
- Robertson HA, Paul ML, Moratalla R, & Graybiel AM (1991). Expression of the immediate early gene c-fos in basal ganglia: Induction by dopaminergic drugs. The Canadian Journal of Neurological Sciences. Le Journal Canadien Des Sciences Neurologiques, 18(3 Suppl), 380–383. 10.1017/s0317167100032480. [DOI] [PubMed] [Google Scholar]
- Robison AJ, & Nestler EJ (2011). Transcriptional and epigenetic mechanisms of addiction. Nature Reviews Neuroscience, 12(11), 623–637. 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Vialou V, Mazei-Robison M, Feng J, Kourrich S, Collins M, et al. (2013). Behavioral and structural responses to chronic cocaine require a feedforward loop involving ΔFosB and calcium/calmodulin-dependent protein kinase II in the nucleus accumbens shell. The Journal of Neuroscience, 33(10), 4295–4307. 10.1523/JNEUROSCI.5192-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen JB, Chuang E, & Iadarola MJ (1994). Differential induction of Fos protein and a Fos-related antigen following acute and repeated cocaine administration. Molecular Brain Research, 25(1), 168–172. 10.1016/0169-328X(94)90295-X. [DOI] [PubMed] [Google Scholar]
- Rosen JB, Fanselow MS, Young SL, Sitcoske M, & Maren S (1998). Immediate-early gene expression in the amygdala following footshock stress and contextual fear conditioning. Brain Research, 796(1–2), 132–142. 10.1016/S0006-8993(98)00294-7. [DOI] [PubMed] [Google Scholar]
- Rubio FJ, Liu Q-R, Li X, Cruz FC, Leão RM, Warren BL, et al. (2015). Context-induced reinstatement of methamphetamine seeking is associated with unique molecular alterations in fos-expressing dorsolateral striatum neurons. Journal of Neuroscience, 35(14), 5625–5639. 10.1523/JNEUROSCI.4997-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio FJ, Quintana-Feliciano R, Warren BL, Li X, Witonsky KFR, del Valle FS, et al. (2019). Prelimbic cortex is a common brain area activated during cue-induced reinstatement of cocaine and heroin seeking in a polydrug self-administration rat model. European Journal of Neuroscience, 49(2), 165–178. 10.1111/ejn.14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salery M, Dos Santos M, Saint-Jour E, Moumné L, Pagès C, Kappès V, et al. (2017). Activity-regulated cytoskeleton-associated protein accumulates in the nucleus in response to cocaine and acts as a brake on chromatin remodeling and long-term behavioral alterations. Biological Psychiatry, 81(7), 573–584. 10.1016/j.biopsych.2016.05.025. [DOI] [PubMed] [Google Scholar]
- Salery M, Trifilieff P, Caboche J, & Vanhoutte P (2020). From signaling molecules to circuits and behaviors: Cell-type–specific adaptations to psychostimulant exposure in the striatum. Biological Psychiatry, 87(11), 944–953. 10.1016/j.biopsych.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Samaha A-N, Mallet N, Ferguson SM, Gonon F, & Robinson TE (2004). The rate of cocaine administration alters gene regulation and behavioral plasticity: Implications for addiction. Journal of Neuroscience, 24(28), 6362–6370. 10.1523/JNEUROSCI.1205-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savell KE, Tuscher JJ, Zipperly ME, Duke CG, Phillips RA, Bauman AJ, et al. (2020). A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Science Advances, 6(26), eaba4221. 10.1126/sciadv.aba4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, & Greenberg ME (1990). The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron, 4(4), 477–485. 10.1016/0896-6273(90)90106-P. [DOI] [PubMed] [Google Scholar]
- Shepherd JD, & Bear MF (2011). New views of Arc, a master regulator of synaptic plasticity. Nature Neuroscience, 14(3), 279–284. 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, et al. (2006). Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron, 52(3), 475–484. 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Wallace CS, Lyford GL, & Worley PF (1998). Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron, 21(4), 741–751. 10.1016/S0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Tan A, Moratalla R, Lyford GL, Worley P, & Graybiel AM (2000). The activity-regulated cytoskeletal-associated protein arc is expressed in different striosome-matrix patterns following exposure to amphetamine and cocaine. Journal of Neurochemistry, 74(5), 2074–2078. 10.1046/j.1471-4159.2000.0742074.x. [DOI] [PubMed] [Google Scholar]
- Tanaka KZ, Pevzner A, Hamidi AB, Nakazawa Y, Graham J, & Wiltgen BJ (2014). Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron, 84(2), 347–354. 10.1016/j.neuron.2014.09.037. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Carreira MB, Cooper YA, Bobadilla A-C, Heinsbroek JA, Koike N, et al. (2017). HDAC5 and its target gene, Npas4, function in the nucleus accumbens to regulate cocaine-conditioned behaviors. Neuron, 96(1), 130–144.e6. 10.1016/j.neuron.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KL, Arroyo M, & Everitt BJ (2003). Induction of the learning and plasticity-associated gene Zif268 following exposure to a discrete cocaine-associated stimulus. European Journal of Neuroscience, 17(9), 1964–1972. 10.1046/j.1460-9568.2003.02617.x. [DOI] [PubMed] [Google Scholar]
- Tischmeyer W, & Grimm R (1999). Activation of immediate early genes and memory formation. Cellular and Molecular Life Sciences: CMLS, 55(4), 564–574. 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S, Liu X, Ramirez S, & Redondo R (2015). Memory engram cells have come of age. Neuron, 87(5), 918–931. 10.1016/j.neuron.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Tonegawa S, Morrissey MD, & Kitamura T (2018). The role of engram cells in the systems consolidation of memory. Nature Reviews Neuroscience, 19(8), 485–498. 10.1038/s41583-018-0031-2. [DOI] [PubMed] [Google Scholar]
- Torres OV, McCoy MT, Ladenheim B, Jayanthi S, Brannock C, Tulloch I, et al. (2015). CAMKII-conditional deletion of histone deacetylase 2 potentiates acute methamphetamine-induced expression of immediate early genes in the mouse nucleus accumbens. Scientific Reports, 5(1), 13396. 10.1038/srep13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche S, Perestenko PV, van de Ven GM, Bratley CT, McNamara CG, Campo-Urriza N, et al. (2016). Recoding a cocaine-place memory engram to a neutral engram in the hippocampus. Nature Neuroscience, 19(4), 564–567. 10.1038/nn.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal CT, Beverley JA, Willuhn I, & Steiner H (2009). Long-lasting dysregulation of gene expression in corticostriatal circuits after repeated cocaine treatment in adult rats: Effects on zif 268 and homer 1a. The European Journal of Neuroscience, 29(8), 1615–1626. 10.1111/j.1460-9568.2009.06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Aubier B, Corbillé A-G, Brami-Cherrier K, Caboche J, Topilko P, et al. (2006). Plasticity-associated gene Krox24/Zif268 is required for long-lasting behavioral effects of cocaine. Journal of Neuroscience, 26(18), 4956–4960. 10.1523/JNEUROSCI.4601-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann SD, Brown MW, Erichsen JT, & Aggleton JP (2000). Fos imaging reveals differential patterns of hippocampal and parahippocampal subfield activation in rats in response to different spatial memory tests. Journal of Neuroscience, 20(7), 2711–2718. 10.1523/JNEUROSCI.20-07-02711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, & Guzowski JF (2004). Differences in hippocampal neuronal population responses to modifications of an environmental context: Evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. Journal of Neuroscience, 24(29), 6489–6496. 10.1523/JNEUROSCI.0350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdarjanova A, McNaughton BL, Barnes CA, Worley PF, & Guzowski JF (2002). Experience-dependent coincident expression of the effector immediate-early genes arc and Homer 1a in hippocampal and neocortical neuronal networks. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 22(23), 10067–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, et al. (2018). Cocaine self-administration alters transcriptome-wide responses in the brain’s reward circuitry. Biological Psychiatry, 84(12), 867–880. 10.1016/j.biopsych.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, & Nestler EJ (2018). Neuroepigenetics and addiction. Handbook of Clinical Neurology, 148, 747–765. 10.1016/B978-0-444-64076-5.00048-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, Neumann PA, Beier KT, Mokhtari AK, Luo L, & Malenka RC (2019). Complementary genetic targeting and monosynaptic input mapping reveal recruitment and refinement of distributed corticostriatal ensembles by cocaine. Neuron, 104(5), 916–930.e5. 10.1016/j.neuron.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, et al. (2006). In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell, 126(2), 389–402. 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Warren BL, Kane L, Venniro M, Selvam P, Quintana-Feliciano R, Mendoza MP, et al. (2019). Separate vmPFC ensembles control cocaine self-administration versus extinction in rats. Journal of Neuroscience, 39(37), 7394–7407. 10.1523/JNEUROSCI.0918-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren BL, Mendoza MP, Cruz FC, Leao RM, Caprioli D, Rubio FJ, et al. (2016). Distinct Fos-expressing neuronal ensembles in the ventromedial prefrontal cortex mediate food reward and extinction memories. Journal of Neuroscience, 36(25), 6691–6703. 10.1523/JNEUROSCI.0140-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, & Huber KM (2008). Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron, 59(1), 84–97. 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, & Greenberg ME (2011). Neuronal activity–regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor Perspectives in Biology, 3(6), a005744. 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker LR, Carneiro de Oliveira PE, McPherson KB, Fallon RV, Planeta CS, Bonci A, et al. (2016). Associative learning drives the formation of silent synapses in neuronal ensembles of the nucleus accumbens. Biological Psychiatry, 80(3), 246–256. 10.1016/j.biopsych.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker LR, & Hope BT (2018). Chasing the addicted engram: Identifying functional alterations in Fos-expressing neuronal ensembles that mediate drug-related learned behavior. Learning & Memory, 25(9), 455–460. 10.1101/lm.046698.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf ME (2016). Synaptic mechanisms underlying persistent cocaine craving. Nature Reviews. Neuroscience, 17(6), 351–365. 10.1038/nrn.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M (2008). C-Fos is an intracellular regulator of cocaine-induced long-term changes. Annals of the New York Academy of Sciences, 1139, 1–9. 10.1196/annals.1432.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap E-L, & Greenberg ME (2018). Activity-regulated transcription: bridging the gap between neural activity and behavior. Neuron, 100(2), 330–348. 10.1016/j.neuron.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, et al. (2016). Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell, 165(7), 1776–1788. 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ST, Porrino LJ, & Iadarola MJ (1991). Cocaine induces striatal c-fos-immunoreactive proteins via dopaminergic D1 receptors. Proceedings of the National Academy of Sciences of the United States of America, 88(4), 1291–1295. 10.1073/pnas.88.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuferov V, Kroslak T, Laforge KS, Zhou Y, Ho A, & Kreek MJ (2003). Differential gene expression in the rat caudate putamen after “binge” cocaine administration: Advantage of triplicate microarray analysis. Synapse (New York, N.Y.), 48(4), 157–169. 10.1002/syn.10198. [DOI] [PubMed] [Google Scholar]
- Zachariou V, Bolanos CA, Selley DE, Theobald D, Cassidy MP, Kelz MB, et al. (2006). An essential role for ΔFosB in the nucleus accumbens in morphine action. Nature Neuroscience, 9(2), 205–211. 10.1038/nn1636. [DOI] [PubMed] [Google Scholar]
- Zavala AR, Biswas S, Harlan RE, & Neisewander JL (2007). Fos and glutamate AMPA receptor subunit coexpression associated with cue-elicited cocaine-seeking behavior in abstinent rats. Neuroscience, 145(2), 438–452. 10.1016/j.neuroscience.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala AR, Osredkar T, Joyce JN, & Neisewander JL (2008). Upregulation of Arc mRNA expression in the prefrontal cortex following cue-induced reinstatement of extinguished cocaine-seeking behavior. Synapse (New York, N.Y.), 62(6), 421–431. 10.1002/syn.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G-C, Mao L-M, Liu X-Y, Parelkar NK, Arora A, Yang L, et al. (2007). In vivo regulation of homer1a expression in the striatum by cocaine. Molecular Pharmacology, 71(4), 1148–1158. 10.1124/mol.106.028399. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu H, Liu Z, Chen X, Su X, Ma C, et al. (2019). A ventral CA1 to nucleus accumbens core engram circuit mediates conditioned place preference for cocaine. Nature Neuroscience, 22(12), 1986–1999. 10.1038/s41593-019-0524-y. [DOI] [PubMed] [Google Scholar]
- Ziółkowska B, Korostyński M, Piechota M, Kubik J, & Przewłocki R (2012). Effects of morphine on immediate-early gene expression in the striatum of C57BL/6J and DBA/2J mice. Pharmacological Reports: PR, 64(5), 1091–1104. 10.1016/s1734-1140(12)70906-4. [DOI] [PubMed] [Google Scholar]


