Abstract
The members of the Sp1 transcription factor family can act as both negative and positive regulators of gene expression. Here we show that Sp1 can be a target for histone deacetylase 1 (HDAC1)-mediated transcriptional repression. The histone deacetylase inhibitor trichostatin A activates the chromosomally integrated murine thymidine kinase promoter in an Sp1-dependent manner. Coimmunoprecipitation experiments with Swiss 3T3 fibroblasts and 293 cells demonstrate that Sp1 and HDAC1 can be part of the same complex. The interaction between Sp1 and HDAC1 is direct and requires the carboxy-terminal domain of Sp1. Previously we have shown that the C terminus of Sp1 is necessary for the interaction with the transcription factor E2F1 (J. Karlseder, H. Rotheneder, and E. Wintersberger, Mol. Cell. Biol. 16:1659–1667, 1996). Coexpression of E2F1 interferes with HDAC1 binding to Sp1 and abolishes Sp1-mediated transcriptional repression. Our results indicate that one component of Sp1-dependent gene regulation involves competition between the transcriptional repressor HDAC1 and the transactivating factor E2F1.
The chromatin of eukaryotic cells is organized in nucleosomes. This organization allows the efficient packaging of chromosomal DNA into the nucleus but limits the access of high-molecular-weight protein complexes of the transcription machinery. At least two different mechanisms enable the eukaryotic cell to relieve nucleosomal repression: the chromatin-remodeling complexes (reviewed in references 55 and 57) and reversible histone acetylation. Two recent reports indicate a direct link between these two activities (60, 67). Posttranslational acetylation on conserved lysine residues within the N-terminal regions of nucleosomal histones is assumed to lead to a reduced attraction between chromosomal DNA and histone tails and changed interactions with neighboring nucleosomes or other nonhistone proteins. The resulting local chromatin decondensation increases the accessibility of particular DNA regions for RNA polymerase complexes. Consistent with this idea, transcriptionally active chromatin correlates with histone hyperacetylation (reviewed in references 18, 30, 47, 49, 61, and 62). This model predicts that histone acetyltransferases would promote transcription, while histone deacetylases (HDACs) should act as repressors. In accordance with this model, several transcriptional adapters and coactivators, such as GCN5 (8, 31), p300/CBP (4, 46), TAFII250 (40), SRC-1 (54), and ACTR (10), have been classified as histone acetyltransferases. Five HDACs have been identified in mammalian cells (12, 14, 56, 58, 63, 64). Three of them, HDAC1, HDAC2, and HDAC3, have significant homology to yeast Rpd3 (44, 50, 59). HDAC4 and HDAC5 belong to the histone deacetylase A (HDA) family (9, 58). HDAC1 and HDAC2 are found in high-molecular-weight complexes associated with adapter proteins like SIN3, SAP18, and SAP30 and nuclear corepressors like N-CoR, SMRT, and SUN-CoR (2, 24, 32, 42, 65, 66). Recently it was demonstrated that several mammalian transcription factors, such as Mad (21, 24, 32, 52), YY1 (64), hormone-dependent nuclear receptors (24, 42), MeCP2 (26, 43), CBF (27), retinoblastoma protein (Rb) (7, 38, 39), and related pocket proteins (16), can repress transcription by recruiting HDACs to specific promoters. In addition, the aberrant recruitment of HDACs by PLZF, PML, and ETO fusion proteins can interfere with the differentiation of hematopoietic precursor cells in acute promyelocytic leukemia (13, 17, 19, 35).
In this study we investigated the potential function of HDACs as transcriptional repressors during the growth arrest of mammalian cells. Using the S-phase-specific mouse thymidine kinase (TK) promoter as a model system, we show that HDAC1 can mediate transcriptional repression via the Sp1 binding site. HDAC1 is associated with Sp1 and binds directly to the C-terminal part of Sp1 that was previously identified as interacting domain for E2F1 (28). Sp1 and E2F1 cooperate in the activation of S-phase-specific promoters (28, 36). Here we show that E2F1 but not E2F4 can compete with HDAC1 binding to Sp1, thereby relieving HDAC1-mediated repression of the TK promoter. Finally, we present a model of how transcription factors and histone-modifying enzymes could regulate the activity of specific promoters at the G1/S boundary of the cell cycle.
MATERIALS AND METHODS
Cell culture and transfection.
Swiss 3T3 fibroblasts and human 293 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with antibiotics and 10% fetal calf serum. Swiss 3T3 cells were stably transfected by Polybrene-assisted gene transfer (3). Briefly, 3T3 fibroblasts were seeded at 106 cells per 100-mm-diameter dish. The culture medium was replaced with 4 ml of a cocktail consisting of 5 μg of Polybrene per ml and 50 ng of plasmid DNA per ml in fresh medium. After 16 to 20 h, the mixture was removed and the cells were treated with 15% dimethyl sulfoxide (DMSO) in growth medium for 4 to 5 min. The cells were rinsed twice with growth medium and returned to the incubator for 24 h before Geneticin-containing medium was added. Geneticin-resistant clones were pooled for further investigations. The results described in this study are representative of those from experiments with both single clones and mixed populations. Transient transfection of 293 cells was carried out by calcium phosphate coprecipitation as described previously (28). Swiss 3T3 cells were growth arrested by reducing the serum concentration in the culture medium to 0.2% for 72 h and restimulated to enter the cell cycle with fresh medium containing 20% fetal calf serum. Trichostatin A (TSA) (Wako) was dissolved in DMSO and added to the culture medium at a final concentration of 80 to 100 ng/ml. A corresponding volume of DMSO was added to the control cells. Growth arrest and stimulation were routinely controlled by fluorescence-activated cell sorter analysis with a Partec PAS-II sorter.
Coimmunoprecipitations.
Whole-cell extracts were prepared as described previously (1), and equal amounts (500 μg) were incubated in 200 μl of extraction buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, Boehringer Complete Protease Inhibitor Cocktail) with 3 to 5 μl of the respective antibody for 1 h at 4°C. After addition of 20 μl of a protein A-Sepharose bead suspension (10%, vol/vol; Pharmacia), the mixture was further incubated with gentle shaking for 12 h at 4°C. After three washes with extraction buffer, the beads were resuspended in 50 μl of extraction buffer, and 30-μl aliquots were examined for protein expression on Western blots. The remaining 20 μl was assayed for HDAC activity (see below).
The following antibodies were used in this study. Rb was immunoprecipitated and detected with C15 (Santa Cruz). Sp1 was immunoprecipitated and detected with a polyclonal rabbit antiserum raised against the full-length protein, generously provided by G. Suske (20). HDAC1 was immunoprecipitated and visualized on Western blots with a polyclonal rabbit antibody raised against a recombinant mouse HDAC1 polypeptide (5) (Upstate Biotechnology). Sin3A and Sin3B were precipitated with AK-11 (Santa Cruz) and AK-12 (Santa Cruz), respectively. Hemagglutinin (HA)-tagged and Myc-tagged proteins were immunoprecipitated and detected with the monoclonal sera for the HA epitope (12CA5 and 16B12) and for the Myc epitope (9E10) (15). Glutathione S-transferase (GST) fusion proteins were detected with a polyclonal rabbit serum that recognizes specifically the GST epitope (generously provided by K. Kuchler, Institute of Molecular Genetics, University of Vienna).
HDAC assays and luciferase reporter assays.
HDAC assays were done as described previously (5, 33). To measure enzymatic HDAC activity, equal amounts (10 μg of whole-cell extract) of protein or 20 μl of immunoprecipitated proteins was incubated with 10 μl of [3H]acetate-labeled chicken erythrocyte histones in a total volume of 50 μl for 1 h at 30°C. The reaction was stopped by addition of 36 μl of 1 N HCl–0.4 M acetate and 800 μl of ethyl acetate. After centrifugation at 8,400 × g for 5 min, the radioactivity in a 600-μl aliquot of the organic phase was counted in 3 ml of liquid scintillation cocktail. For luciferase reporter assays, cells were grown in six-well plates and lysed 48 h after transfection in luciferase lysis buffer (100 mM K-phosphate [pH 7.8], 0.2% Triton X-100). Luciferase activity and β-galactosidase activity (as a control for transfection efficiency) were assayed in parallel by using the Dual Light Chemoluminiscent Reporter Gene Assay System (TROPIX, Bedford, Mass.). An aliquot of each extract was analyzed on Western blots for the expression levels of cotransfected proteins.
GST pull-down assays.
Recombinant proteins were expressed in and purified from Escherichia coli BL21 as described previously (28). Beads coated with GST fusion proteins (2 μg) were incubated in binding buffer (20 mM HEPES [pH 7.9], 1 mM MgCl2, 40 mM KCl, 0.1 mM EDTA, 0.1% Nonidet P-40) with 500 μg of whole-cell extract, radiochemical amounts of in vitro-translated proteins, or 2 footprint units (FPU) of purified human Sp1 (Promega) for 2 h at 4°C. After three washes with GST wash buffer (100 mM KCl, 20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.5 mM dithiothreitol), bound proteins were eluted by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, resolved by electrophoresis, and visualized by Western blotting. In vitro expression of radiolabeled proteins was performed in reticulocyte extracts (Quick Coupled Transcription/Translation system; Promega) in the presence of [35S]methionine. Labeled proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and gels were dried and exposed to X-ray films at −70°C. The signals of labeled proteins were quantified with a Molecular Dynamics Storm 840 Scanner.
Plasmids.
To generate the selectable luciferase reporter plasmid pGL2neo, the neomycin resistance gene from pSV2neo (Clontech) was inserted into the BamHI site of the pGL2 vector (Promega). The EcoRI/NheI fragment encompassing the murine TK promoter was cloned into the pGL2neo vector (pTK-luc). To obtain pTK-Sp1mut-luc and pTK-E2Fmut-luc, the corresponding EcoRI/NheI fragments were excised from pTKEcoSp1mut-ATG-CAT and pTKEcoE2Fmut-ATG-CAT (28) and cloned into pGL2neo. To clone pCIneoHDAC1, the EcoRI/PstI mouse HDAC1 cDNA fragment (5) containing the entire open reading frame was cloned into Bluescript pKS. The mammalian expression plasmid pCIneoHDAC1myc encoding an epitope-tagged version of HDAC1 was described previously (5). The insert was excised by EcoRI and NotI digestion and ligated into pCIneo. The parental plasmid pCIneo-HA was created by inserting a double-stranded oligodeoxynucleotide encoding the peptide MAYPYDVPDYA into the XhoI-cut vector pCIneo (Promega). To clone pCIneo-HA-Sp1, the Sp1 cDNA was inserted into XbaI-cut pCIneo-HA. Expression vectors encoding HA-Sp1 mutants were generated by cutting pCIneo-HA-Sp1 with PpuMI and SmaI [pCIneo-HA-Sp1(1-293)], with SmaI and BamHI (partially) [pCIneo-HA-Sp1(1-621)], with SmaI and XmnI (partially) [pCIneo-HA-Sp1(1-668)], or with BamHI [pCIneo-HA-Sp1(622-788)] and religating the plasmid. The GST-Sp1 constructs have been described previously (28). GST-HDAC1 was generated by inserting the murine HDAC1-coding sequence into BamHI- and EcoRI-cut pGEX-2TK. The reporter plasmids pSp1-luc and pmtSp1-luc were kindly provided by H. Nomura (53).
RESULTS
TSA-dependent induction of the murine TK promoter in G0-phase cells is linked to the Sp1 site.
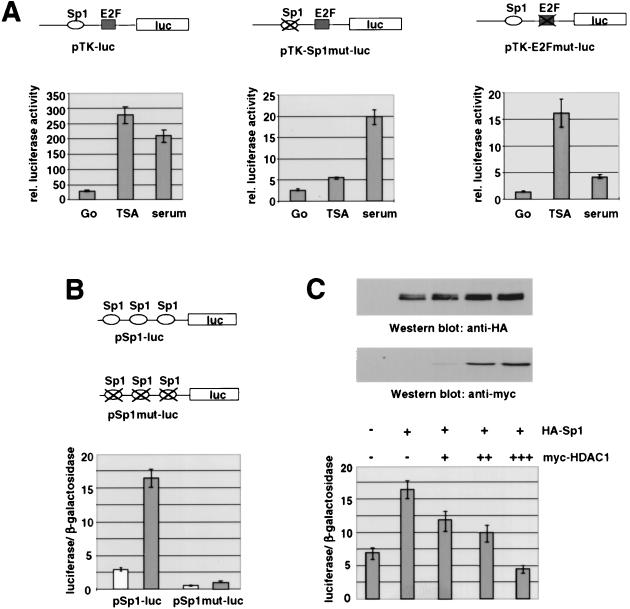
We have previously shown that the mouse TK gene is transcriptionally regulated by E2F and Sp1 (28). Binding of both proteins is essential for activation of the TK promoter during the S phase of the cell cycle. The interaction of the DNA-binding proteins is direct (28) and was shown to be strongly enhanced during the late G1 phase of the cell cycle (36). Given that the TK promoter is inactive in G0-phase cells, we investigated whether HDAC activity is necessary for this repression. Figure 1 shows that a stably integrated TK luciferase reporter gene can be activated in serum-starved Swiss 3T3 cells by the HDAC inhibitor TSA. The activity of the TK promoter was ninefold induced by TSA, compared to a sevenfold stimulation by 20% fetal calf serum. Interestingly, the presence of an intact Sp1 binding site was required for this effect, while the binding of E2F was dispensable for the activating effect of TSA. Both mutated promoters show significantly lower affinity than the wild-type promoter due to loss of the cooperativity between the two transcription factors (28). The promoter construct with a mutated Sp1 site and an intact E2F binding site showed a less-than-twofold response to TSA but was still responsive to serum. Mutation of the E2F site, on the other hand, led to nearly complete loss of the growth factor response but had no effect on the inducibility by the HDAC inhibitor. A reporter construct with mutations in both binding sites was not responsive to TSA (13a).
FIG. 1.
Sp1 binding sites can mediate transcriptional activation by TSA. (A) The Sp1 binding site is required for activation of the murine TK promoter by TSA in resting Swiss 3T3 cells. Serum-deprived cells containing the chromosomally integrated luciferase reporter genes pTK-luc, pTK-E2Fmut-luc, and pTK-Sp1mut-luc were incubated for 20 h either with TSA (80 ng/ml) or fresh medium supplemented with 20% fetal calf serum. Data are means and standard deviations from three independent experiments. rel., relative. (B) 293 cells were transiently transfected with a reporter plasmid containing three Sp1 consensus sites (pSp1-luc) or a construct bearing three mutated Sp1 sites (pmtSp1-luc) together with the control vector pCMVβGal. In each transfection experiment half of the cells were treated with TSA (100 ng/ml) for 20 h. Luciferase activities of untreated cells (white bars) and TSA-treated cells (gray bars) are depicted relative to the respective β-galactosidase activities. (C) pSp1-luc was transfected together with pCIneo, pCIneoHA-Sp1wt, and pCIneoHA-Sp1wt in combination with increasing amounts of pCIneomyc-HDAC1 (0.25, 0.75, and 1.25 μg). Luciferase activities are depicted relative to the respective β-galactosidase activities. Expression levels of epitope-tagged Sp1 and HDAC1 were analyzed on Western blots with HA-specific and Myc-specific antibodies.
To demonstrate that the TSA effect is independent of the presence of other transcription factor binding sites, we examined a promoter that is driven by three Sp1 consensus sites only. A luciferase reporter construct under control of this promoter (pSp1-luc) was previously shown to be strongly induced by TSA in the p53-deficient human cell line MG63 (53). When expressed in 293 cells, the reporter had considerable activity that was more than sixfold enhanced upon treatment with 100 ng of TSA per ml (Fig. 1B). A mutant construct, pmtSp1-luc, with mutated Sp1 consensus sites had significantly reduced luciferase activity that was only weakly responsive (twofold) to TSA. Coexpression of HA-tagged Sp1 with pSp1-luc led to an increase in promoter activity, confirming the role of Sp1 as a transcriptional activator for this promoter. Next, we tested whether HDAC1 can affect the reporter activity in the presence of Sp1. Cotransfection of increasing amounts of mouse HDAC1 (5) abolished the activation by Sp1. Importantly, coexpression of HDAC1 did not reduce the expression levels of Sp1 (Fig. 1C). In contrast, pmtSp1-luc showed no response to coexpression of either Sp1 or HDAC1 (data not shown). These results suggest that Sp1 and/or other members of the Sp1 family not only are transcriptional activators but also are targets for HDACs.
Sp1 interacts with HDAC1 in vivo via its C-terminal domain.
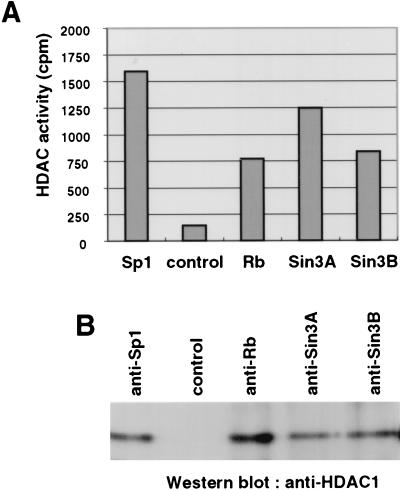
To analyze whether Sp1 interacts with HDACs, we immunoprecipitated Sp1 from extracts of resting Swiss 3T3 cells and measured the associated HDAC activity. As shown in Fig. 2A, Sp1 is associated with significant HDAC activity that is comparable to that found in the coimmunoprecipitation with known HDAC-interacting factors such as Rb, Sin3A, and Sin3B. The immunoprecipitate obtained with the Sp1-specific antibody contained HDAC1, whereas no HDAC1 was observed in an immunoprecipitate obtained with an irrelevant antibody (anti-HA) (Fig. 2B). The differences between associated HDAC activities and amounts of HDAC1 coimmunoprecipitated with Rb and Sp1 may be due to the presence of other HDACs in the respective complexes. For instance, HDAC1 and HDAC2 were recently shown to interact with each other (22), and both deacetylases are present in Sin3-containing complexes (32, 66, 68). In summary, these data indicate that Sp1 is associated with HDAC1 in vivo.
FIG. 2.
Sp1 interacts with HDAC1 in vivo. Whole-cell extracts were immunoprecipitated with anti-Sp1 antibody (Pep2), an irrelevant control antibody (anti-HA), anti-Rb antibody (C-15), anti-Sin3A antibody (AK-11), and anti-Sin3B antibody (AK-12). (A) Sp1 associates with HDAC activity in resting Swiss 3T3 cells similarly to the known HDAC1-binding proteins Rb, Sin3A, and Sin3B. HDAC activity was measured as described previously (5). (B) HDAC1 coimmunoprecipitates with Sp1. HDAC1 was detected by Western blot analysis with the anti-HDAC1 antibody.
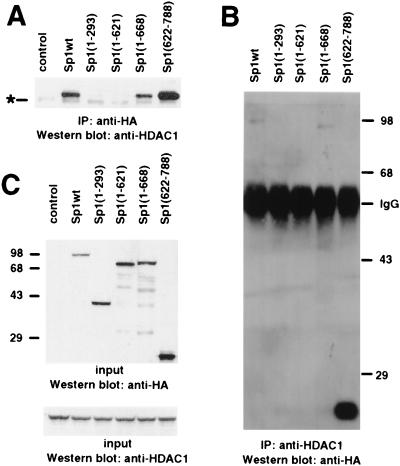
When HDAC1 and HA-tagged Sp1 were coexpressed in mammalian cells, an interaction between the two proteins was observed by immunoprecipitation with the HA-specific antibody and the HDAC1 antiserum (Fig. 3A and B). The association of Sp1 with HDAC1 is dependent on the presence of the carboxy-terminal domain of Sp1, since removal of the first 621 amino acids had no effect on HDAC1 binding, while Sp1 mutants containing amino acids 1 to 293 or 1 to 621 failed to recruit HDAC1 (Fig. 3A and B). An Sp1 protein encompassing amino acids 1 to 668 also had the capacity to bind HDAC1. The strong interaction of Sp1(622-788) with HDAC1 is at least in part due to a significantly higher expression of this protein than of the other Sp1 polypeptides (Fig. 3C). In addition, the HDAC1 binding domain might be more accessible in this deletion mutant.
FIG. 3.
The interaction between Sp1 and HDAC1 requires the C-terminal domain of Sp1. (A) The C-terminal domain (amino acids 622 to 788) of Sp1 is essential and sufficient for HDAC1-Sp1 interaction. Cells were transfected with pCIneoHDAC1 and the Sp1-encoding plasmid pCIneoHA-Sp1wt, pCIneoHA-Sp1(1-293), pCIneoHA-Sp1(1-621), pCIneoHA-Sp1(1-668), or pCIneoHA-Sp1(622-788). Whole-cell extracts were immunoprecipitated (IP) with anti-HA antibody, and HDAC1 was visualized by Western blot analysis with the anti-HDAC1 antibody. An extract from cells transfected with only pCIneoHDAC1 was included as a control. A faint unspecific band from a cross-reacting protein (asterisk) was also visible in the immunoprecipitation from untransfected cells. (B) HA-tagged Sp1 coimmunoprecipitates with HDAC1 from extracts of transfected 293 cells. Cells were transfected with pCIneoHDAC1 and the Sp1-encoding plasmid pCIneoHA-Sp1wt, pCIneoHA-Sp1(1-293), pCIneoHA-Sp1(1-621), or pCIneoHA-Sp1(1-668). Whole-cell extracts were precipitated with the anti-HDAC1 antibody, and HA-Sp1 was visualized by Western blot analysis with the anti-HA antibody. (C) Input extracts were analyzed on a Western blot for expression levels of epitope-tagged Sp1 polypeptides (detected with the anti-HA antibody) and HDAC1 (detected with the anti-HDAC1 antibody).
Direct interaction between HDAC1 and Sp1.
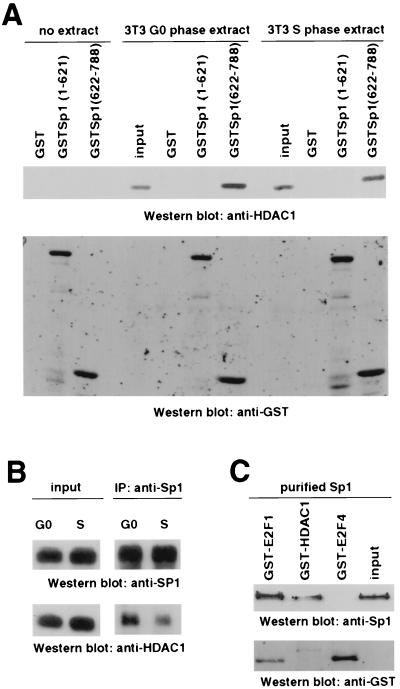
The HDAC1-Sp1 interaction was also observed in GST pull-down experiments with extracts from Swiss 3T3 cells. GST–Sp1(622-788) can complex with HDAC1 in extracts from resting and S-phase 3T3 fibroblasts, while GST or GST–Sp1(1-621) shows no interaction (Fig. 4A). While HDAC1 was expressed at a higher level in replicating cells, the affinity to Sp1 seemed to be slightly reduced. Similarly, a moderate reduction in the in vivo interaction of HDAC1 with Sp1 in serum-stimulated Swiss 3T3 fibroblasts was observed in immunoprecipitation experiments with S-phase extracts prepared from untransfected cells (Fig. 4B). To investigate whether the interaction of Sp1 and HDAC1 is direct, GST-HDAC1 was incubated with purified Sp1. As a negative control, E2F4, which, in contrast to E2F1, lacks the Sp1-interacting domain (28), was included in this experiment. As shown in Fig. 4C, in the absence of additional proteins, GST-HDAC1 could bind to Sp1 to a similar extent as GST-E2F1, while GST-E2F4 showed no interaction.
FIG. 4.
Sp1 interacts directly with HDAC1. (A) GST–Sp1(622-788) precipitates HDAC1 from extracts prepared from resting and serum-stimulated Swiss 3T3 cells. HDAC1 was detected by Western blot analysis with the anti-HDAC1 antibody. GST and GST-Sp1(1-621) failed to interact with HDAC1. Input extracts (8%) and pull-down assays without extract were included as controls. GST fusion proteins were detected on the same blot with a GST-specific antibody. The GST proteins (without the Sp1 portion) running in the front of the gel are not shown. (B) Sp1-HDAC1 interaction is slightly reduced in S-phase cells. Sp1 was immunoprecipitated (IP) with anti-Sp1 antibody (Pep2) from extracts prepared from G0- and S-phase Swiss 3T3 cells. Immunoprecipitated Sp1 and coimmunoprecipitated HDAC1 were visualized on the Western blot with the anti-Sp1 antibody and the anti-HDAC1 antibody, respectively. (C) GST-HDAC1 interacts with purified Sp1 (4 FPU; Promega). Sp1 was visualized by Western blot analysis with the anti-Sp1 antibody. GST-E2F4 failed to interact with recombinant Sp1, while E2F1 binds to Sp1, as previously shown (28). Purified Sp1 (2 FPU) was loaded as a control.
E2F1 can abolish HDAC1 binding to Sp1 and HDAC1-dependent transcriptional repression.
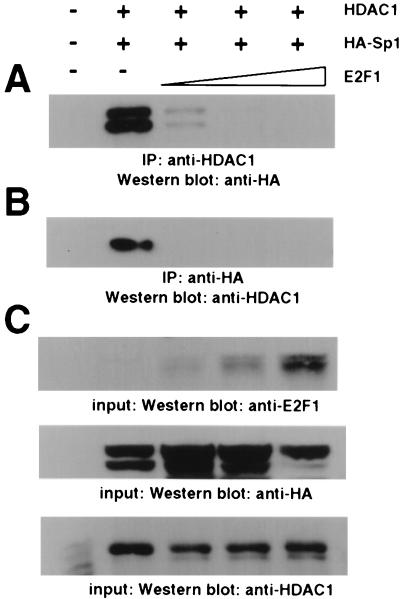
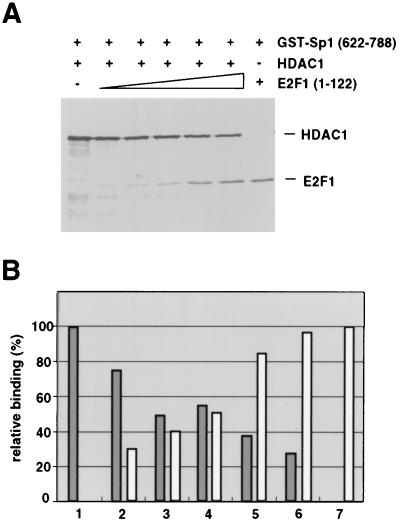
The C-terminal domain of Sp1 that is sufficient to bind HDAC1 was previously shown to interact with the transcription factor E2F1 (28). E2F1 plays an important role in the activation of numerous growth- and cell cycle-regulated promoters. We therefore asked whether E2F1 could influence the interaction between Sp1 and HDAC1. Coexpression of increasing amounts of E2F1 with Sp1 and HDAC1 abolished the interaction between the two proteins as revealed by immunoprecipitations with the HA antibody (for epitope-tagged Sp1) or the HDAC1 antiserum (Fig. 5). Epitope-tagged Sp1 appears on Western blots as a doublet (Fig. 1C and 5) that most probably represents differently modified forms of the transcription factor. Both forms of HA-Sp1 are competed by E2F1. To exclude an indirect effect of E2F1 on the cytomegalovirus promoter that drives the Sp1 and HDAC1 expression constructs, we performed in vitro competition experiments. In pilot experiments the limiting amount of GST–Sp1(622-788) that is still able to bind in vitro-translated radiolabeled HDAC1 was determined (data not shown). Increasing amounts of in vitro-translated E2F1(1-122) were added simultaneously with constant amounts of HDAC1. The E2F1(1-122) protein still contains the Sp1 interaction domain and is easily distinguishable in size from HDAC1. As shown in Fig. 6, addition of E2F1 led to a reduction in the interaction between Sp1 and HDAC1. In contrast, E2F4 that lacks the Sp1 binding domain failed to compete HDAC1 binding to Sp1 (not shown).
FIG. 5.
E2F1 competes Sp1-associated HDAC1. 293 cells were transfected with pCIneoHDAC1, pCIneoHA-Sp1wt, and different amounts of pCIneoE2F1 (0, 1, 3, and 9 μg). Extracts from untransfected cells were included as a control. (A) Concentration-dependent repression of the HDAC1-Sp1 interaction by E2F1. HDAC1 was immunoprecipitated (IP) from whole-cell extracts prepared from transfected 293 cells, and coprecipitated HA-tagged Sp1 was detected by Western blot analysis with the anti-HA antibody. (B) E2F1 abolishes binding of HDAC1 to Sp1. HA-tagged Sp1 was precipitated from whole-cell extracts prepared from transfected 293 cells, and HDAC1 was detected in immunoprecipitates by Western blot analysis with the anti-HDAC1 antibody. (C) Expression levels of HA-Sp1, HDAC1, and E2F1 in input extracts were visualized by Western blot analysis with the anti-E2F1 antibody, the anti-HA antibody, and the anti-HDAC1 antibody.
FIG. 6.
The amino-terminal domain of E2F1 can directly compete HDAC1 binding to Sp1. (A) GST–Sp1(622-788) was incubated with constant amounts of in vitro-translated radiolabeled HDAC1 (3 μl) in the absence or presence of increasing amounts (0.5, 1, 2, 4, and 6 μl) of in vitro-translated radiolabeled E2F1(1-122). As a control, 6 μl of radiolabeled E2F1(1-122) was incubated with GST–Sp1(622-788) in the absence of HDAC1 (lane 7). (B) Amounts of HDAC1 (gray bars) and E2F-1(1-122) (white bars) bound to GST–Sp1(622-788) were quantified on a Molecular Dynamics Storm 840 Scanner and are shown relative to the signal in the absence of the other labeled protein [bar 1 for HDAC1 and bar 7 for E2F-1(1-122)]. Results are from one typical experiment of three.
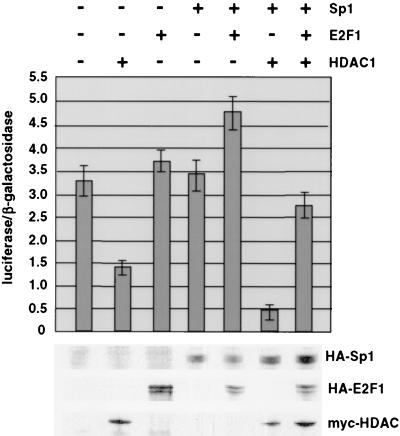
Consistent with the ability of Sp1 to recruit HDAC1, coexpression of both proteins with the TK luciferase reporter resulted in considerable repression of transcriptional activity (Fig. 7, bar 6). HDAC1 alone (bar 2) also had a significant, although less pronounced, repressive effect, while Sp1 expression (bar 4) was without major consequence for the promoter activity. Given that Sp1 can tether HDAC1 to the TK promoter, we examined whether E2F1 affects the transcriptional repression mediated by these proteins. E2F-1 together with Sp1 (Fig. 7, bar 5) slightly stimulated the TK promoter. In combination with Sp1 and HDAC1, E2F1 abrogated the repressing effect of HDAC1 and Sp1 (Fig. 7, bar 7), suggesting that the competition between HDAC1 and E2F1 for binding to Sp1 plays a major role in the regulation of the murine TK promoter.
FIG. 7.
E2F1 abolishes HDAC1-mediated transcriptional repression. Luciferase activity in whole-cell extracts prepared from transfected 293 cells was measured. Cells were transfected with pTK-luc and pCMVβGal (as a control) together with pCIneo (vector control), pCIneoHDAC1myc, pCIneoHA-E2F1, pCIneoHA-Sp1wt, pCIneoHA-Sp1wt plus pCIneoHA-E2F1, pCIneomyc-HDAC1 plus pCIneoHA-Sp1wt, and pCIneomyc-HDAC1 in combination with pCIneoHA-Sp1wt and pCIneoHA-E2F1. Data are means and standard deviations from three independent experiments. The expression levels of HA-Sp1, HA-E2F1, and Myc-tagged HDAC1 were monitored by Western blot analysis with the HA antibody for Sp1 and E2F1 and the Myc-specific antibody for HDAC1.
DISCUSSION
Reversible acetylation of histones and corresponding changes of chromatin structure are substantial elements of gene regulation. Many histone acetyltransferases and HDACs are capable of interacting with constituents of the transcription apparatus, thereby causing promoter-specific alterations of chromatin. We have been interested in the growth control of gene expression, with the S-phase-specific TK gene as a model.
Promoters of growth-regulated genes often carry binding sites for Sp1 and E2F. Depending on the promoter, members of the E2F family can function in one of two ways. First, together with their interacting pocket protein (Rb, p107, or p130), they can inhibit promoter activity; phosphorylation of the pocket protein then causes rapid dissociation of the pocket protein-E2F complexes. This is likely the case for E2F4 and E2F5 and the corresonding pocket proteins p130 and p107. The second type of regulation involves E2F1, -2, or -3. These E2F proteins, together with a pocket protein, can inhibit transcription, but upon release of the phosphorylated pocket protein they can also act as positive transcription factors. The murine TK promoter and the promoter of the dihydrofolate reductase (DHFR) gene are probably regulated in this way. In both cases, there appears to be a strong interaction between Sp transcription factors and E2F in which Sp1, Sp3 and E2F1, -2, or -3 are implicated. Mutation of the binding site for E2F in this case not only leads to deregulation of the promoter but causes nearly complete inactivation. Down-regulation of promoters by the E2F-pocket protein complex is thought to entail deacetylation of histones via pocket protein-HDAC interaction. In contrast to E2F, Sp1 was so far seen primarily as a positive transcription factor. Our study demonstrates that Sp1 can also be targeted by the repressor HDAC1. Binding sites for Sp1 are very common in many promoters, and several Sp1 proteins can bind to these GC-rich motifs, with various consequences. Of these proteins, Sp1 and Sp3 are the most prevalent ones found in mammalian cells. In fact, we observed that Sp3 binds HDAC1 just like Sp1 (49a). The interaction of Sp1 with HDAC1 requires the part of Sp1 which was previously shown to be implicated in the binding to E2F. Accordingly, HDAC1 was found to compete with E2F1 for binding to Sp1. This competition may play an important role in the regulation of promoters, which present closely spaced binding sites for Sp1 and E2F.
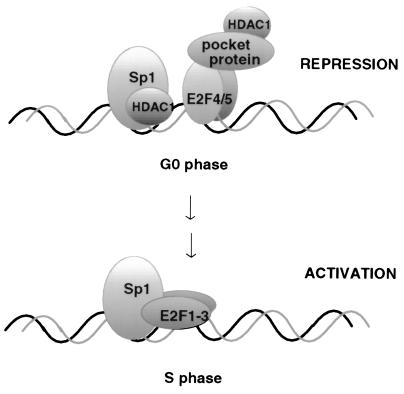
A model for the growth control of the murine TK promoter which incorporates the currently known interactions is shown in Fig. 8. During growth arrest, the E2F complex carries p130 most likely bound to E2F4. As E2F4 lacks the sequence required for interaction with Sp1, the nearby Sp1 is able to bind HDAC1. Thus, both Sp1 and p130 recruit HDAC1, thereby causing full inactivation of the promoter. The fact that a mutant TK promoter lacking the Sp1 binding site is insensitive to TSA is possibly due to the absence of E2F1 in resting fibroblasts. E2F1 was recently shown to be necessary for the effect of TSA on promoters repressed by the Rb-HDAC1 complex (39). After growth stimulation, p130 is phosphorylated, the E2F pocket protein complex dissociates, and E2F4 relocalizes to the cytoplasm (37, 41) or becomes degraded (23). The Sp1-HDAC1 complex keeps the promoter inactive until, in mid-G1, E2F1, -2, and -3 are synthesized, which can bind to the free E2F motif, thereby displacing HDAC1 from the C terminus of Sp1. Notably, E2F1, -2, or -3 itself interacts with RB, which again recruits HDAC1 or HDAC2, thus keeping the promoter inactive until mid-G1, when the pocket protein is phosphorylated. Pocket protein phosphorylation results in its removal from E2F and in the activation of the promoter by the combined activities of Sp1 and E2F. This model could explain the complete shutoff of the mouse TK promoter in growth-arrested cells and would allow for a stepwise reorganization of promoter occupancy during G1, culminating in promoter activation at the G1/S border of the cell cycle. The model is in agreement with recent reports on the regulation of the DHFR-promoter (25, 36, 45, 51). Those studies conclude that Sp1, in addition to E2F, plays an active role in the growth control of the DHFR promoter, although they do not provide a mechanism for such a role.
FIG. 8.
A model for the roles of Sp1, E2F, and HDAC1 in repression and activation of S-phase-specific promoters. (Top) During the G0 phase, heterodimers consisting of E2F4 or E2F5 and DRTF binding protein recruit pocket proteins such as p130 to the E2F binding site of the murine TK promoter. The pocket proteins are associated with HDAC1. Simultaneously, the Sp1 binding site is occupied by Sp1 or Sp3 with HDAC1 bound to its C terminus. (Bottom) At the G1/S boundary, the repressing E2F4/5-p130-HDAC1 complex is replaced by E2F1. HDAC1 is displaced by E2F1 at the C terminus of Sp1, and transcription of the TK gene is subsequently activated.
Since other transcription factors (11, 29, 34, 48) also interact with the C-terminal domain of Sp1, competition between transcriptional regulators and HDACs might be a more general way to regulate gene expression via reversible chromatin modification. The recent finding that the C terminus of Sp1 is phosphorylated during the G0/G1 transition could indicate that the interaction between Sp1 as a target and HDAC1 and the transcription factors as competing binding proteins is in addition modulated by cell cycle phase-specific modification of Sp1 (6).
ACKNOWLEDGMENTS
We thank J. Taplick, E. Ogris, S. Schuechner, K. Kuchler, M. Cotten, and W. Krek for useful discussions, K. Kuchler for the GST antiserum, H. Nomura for the pSp1-luc reporter plasmids, G. Suske for the Sp1 antiserum, and H. Khier for the affinity-purified HDAC1 antiserum.
This work was supported by the Austrian FWF (grants P11179-GEN and P13068-GEN to C.S. and grants P10873-GEN and P13031-MOB to E.W.) and the Austrian National Bank (grant 6123 to C.S.).
REFERENCES
- 1.Adamczewski J P, Gannon J V, Hunt T. Simian virus 40 large T antigen associates with cyclin A and p33cdk2. J Virol. 1993;67:6551–6557. doi: 10.1128/jvi.67.11.6551-6557.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 3.Aubin R J, Weinfeld M, Paterson M C. Factors influencing efficiency and reproducibility of polybrene-assisted gene transfer. Somat Cell Mol Genet. 1988;14:155–167. doi: 10.1007/BF01534401. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Bartl S, Taplick J, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black A R, Jensen D, Lin S-Y, Azizkhan J C. Growth/cell cycle regulation of Sp1 phosphorylation. J Biol Chem. 1999;274:1207–1215. doi: 10.1074/jbc.274.3.1207. [DOI] [PubMed] [Google Scholar]
- 7.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 9.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Lin R J, Schiltz R L, Chakravati D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:596–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 11.Chiang C M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 12.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 13.David G, Alland L, Hong S H, Wong C W, DePinho R A, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 13a.Doetzlhofer, A. Unpublished observation.
- 14.Emiliani S, Fischle W, Van Lint C, Al Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira R, Magnaghi Jaulin L, Robin P, Harel Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, Seiser C, Grignani F, Lazar M A, Minucci S, Pelicci P G. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 18.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 19.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Reduced retinoic acid-sensitivities of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood. 1998;91:2634–2642. [PubMed] [Google Scholar]
- 20.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 22.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 24.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 25.Jensen D E, Black A R, Swick A G, Azizkhan J C. Distinct roles for Sp1 and E2F sites in the growth/cell cycle regulation of the DHFR promoter. J Cell Biochem. 1997;67:24–31. doi: 10.1002/(sici)1097-4644(19971001)67:1<24::aid-jcb3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 26.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 27.Kao H Y, Ordentlich P, Koyano Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi A, Sogawa K, Fujii Kuriyama Y. Cooperative interaction between AhR.Arnt and Sp1 for the drug-inducible expression of CYP1A1 gene. J Biol Chem. 1996;271:12310–12316. doi: 10.1074/jbc.271.21.12310. [DOI] [PubMed] [Google Scholar]
- 30.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 32.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 33.Lechner T, Lusser A, Brosch G, Eberharter A, Goralik Schramel M, Loidl P. A comparative study of histone deacetylases of plant, fungal and vertebrate cells. Biochim Biophys Acta. 1996;1296:181–188. doi: 10.1016/0167-4838(96)00069-6. [DOI] [PubMed] [Google Scholar]
- 34.Lee J S, Galvin K M, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc Natl Acad Sci USA. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 36.Lin S Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindeman G J, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 39.Magnaghi Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, LeVillain J P, Troalen F, Trouche D, Harel Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 40.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 41.Muller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 43.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Nasmyth K, Stillman D, Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987;48:579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 45.Noe V, Chen C, Alemany C, Nicolas M, Caragol I, Chasin L A, Ciudad C J. Cell-growth regulation of the hamster dihydrofolate reductase gene promoter by transcription factor Sp1. Eur J Biochem. 1997;249:13–20. doi: 10.1111/j.1432-1033.1997.00013.x. [DOI] [PubMed] [Google Scholar]
- 46.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 47.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 48.Perkins N D, Agranoff A B, Pascal E, Nabel G J. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth S Y. Something about silencing. Nat Genet. 1996;14:3–4. doi: 10.1038/ng0996-3. [DOI] [PubMed] [Google Scholar]
- 49a.Rotheneder, H. Unpublished observation.
- 50.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slansky J E, Farnham P J. Transcriptional regulation of the dihydrofolate reductase gene. Bioessays. 1996;18:55–62. doi: 10.1002/bies.950180111. [DOI] [PubMed] [Google Scholar]
- 52.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Luescher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 53.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Histone deacetylase inhibitor activates the WAF1/Cip1 gene promoter through the Sp1 sites. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 54.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 55.Steger D J, Workman J L. Remodeling chromatin structures for transcription: what happens to the histones? Bioessays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 56.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 57.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 58.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases—coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 59.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 61.Wade P A, Wolffe A P. Histone acetyltransferases in control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 62.Wolffe A P. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 63.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 64.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W, Bone J R, Edmondson D G, Turner B M, Roth S Y. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Iratni R, Erdjument Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, LeRoy G, Seelig H-P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a component containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Sun Z W, Iratni R, Erdjument Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]