Abstract
Dengue is hyperendemic in most Southeast Asian countries including Thailand, where all four dengue virus serotypes (DENV-1 to -4) have circulated over different periods and regions. Despite dengue cases being annually reported in all regions of Thailand, there is limited data on the relationship of epidemic DENV infection between humans and mosquitoes, and about the dynamics of DENV during outbreaks in the northeastern region. The present study was conducted in this region to investigate the molecular epidemiology of DENV and explore the relationships of DENV infection in humans and in mosquitoes during 2016–2018. A total of 292 dengue suspected patients from 11 hospitals and 902 individual mosquitoes (at patient’s houses and neighboring houses) were recruited and investigated for DENV serotypes infection using PCR. A total of 103 patients and 149 individual mosquitoes were DENV -positive. Among patients, the predominant DENV serotypes in 2016 and 2018 were DENV-4 (74%) and DENV-3 (53%) respectively, whereas in 2017, DENV-1, -3 and -4 had similar prevalence (38%). Additionally, only 19% of DENV infections in humans and mosquitoes at surrounding houses were serotypically matched, while 81% of infections were serotypically mismatched, suggesting that mosquitoes outside the residence may be an important factor of endemic dengue transmission. Phylogenetic analyses based on envelope gene sequences showed the genotype I of both DENV-1 and DENV-4, and co-circulation of the Cosmopolitan and Asian I genotypes of DENV-2. These strains were closely related to concurrent strains in other parts of Thailand and also similar to strains in previous epidemiological profiles in Thailand and elsewhere in Southeast Asia. These findings highlight genomic data of DENV in this region and suggest that people’s movement in urban environments may result in mosquitoes far away from the residential area being key determinants of DENV epidemic dynamics.
Introduction
Dengue, caused by the dengue virus (DENV), is one of the most important re-emerging arboviral diseases in the tropical and subtropical regions of the world, including Southeast Asia. The virus is transmitted between humans primarily by the mosquitoes Aedes aegypti, and Ae. albopictus being a secondary vector [1]. The disease is endemic in more than 100 countries, with more than 2.5 billion people at risk [2]. There are an estimated 390 million DENV infections annually, of which 96 million are symptomatic, ranging from mild dengue fever, with or without warning signs, to severe dengue with plasma leakage that may lead to shock, bleeding, and/or organ impairment [3]. Increasing numbers of dengue cases, and the propagation of all four serotypes of DENV, are facilitated by uncontrolled urbanization, suboptimal management of water and solid waste, gaps in vector control, and rapid population movement, especially travel [4, 5]. In the absence of specific antiviral drugs and an effective vaccine, virus surveillance for early warning and integrated vector management are the primary options for the prevention and control of dengue outbreaks [6, 7]. Dengue prevention and control programs in Thailand are mainly based on hospital case reporting, conducted jointly by the hospital and the Offices of Disease Prevention and Control (ODPC), Ministry of Public Health. The dengue surveillance team responds when a case is reported by a hospital within 24 hours of notice in order to prevent transmission by spraying insecticides within 100 meters of the patient’s house [8].
DENV is a single-stranded, positive-sense RNA virus within the Flaviviridae family, and has four distinct serotypes: DENV-1 to -4 [9]. The viral genome has length approximately 11 kb which contains a single open reading frame (ORF) encoding three structural proteins (C, prM/M, E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) [10]. Within each serotype, genotypes can be phylogenetically classified on the basis of their E gene; DENV-1 includes six genotypes (I, II, III (sylvatic), IV, V, and VI); DENV 2 includes six genotypes (Asian I, Asian II, Asian/American, American, Cosmopolitan, and sylvatic); DENV-3 includes five genotypes (I, II, III, IV, and V); DENV-4 includes five genotypes (I, IIA, IIB, III, and sylvatic) [2, 11]. Previous studies have suggested that the transmission of the various serotypes is cyclic, with distinct serotypes periodically re-emerging to dominate, and the introduction of new serotypes or genotypes leading to new epidemics or outbreaks [11–15]. However, the circulating patterns of viruses during infection in human hosts and mosquito vectors are still unclear.
In Thailand, the first case of dengue disease was reported in 1949 and the first major outbreak of severe dengue (dengue hemorrhagic fever, DHF) was documented in Bangkok (central region) in 1958 [2, 11]. Dengue affects inhabitants throughout the country and has become a major public health problem annually [2, 16]. Although the dengue situation in northeastern Thailand has become critical in recent years, with both rising case numbers and increasing disease severity, there is no information available on the DENV genotype in this region.
In the present study, we performed molecular characterization of DENV detected in humans and Aedes mosquitoes in order to explore, for the first time, the epidemiological relationship between humans and mosquitoes and molecular characteristics of circulating DENV strains in northeastern Thailand.
Materials and methods
Study site, recruitment, and sample collection
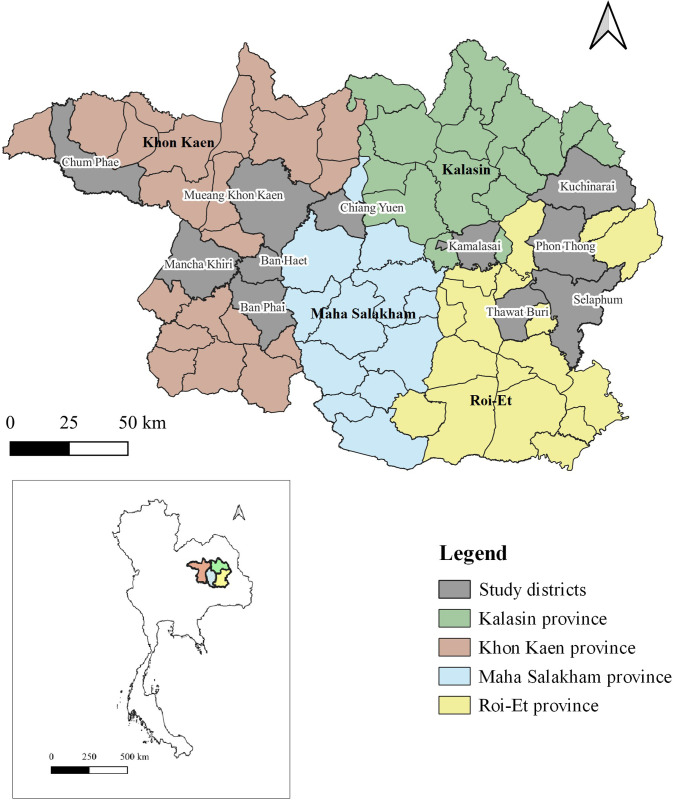
Plasma and mosquito samples obtained from a prospective, hospital-based, case-control study in northeastern Thailand [8] were used. The study was conducted in four provinces in northeastern Thailand from June 2016 to December 2018 (Fig 1). Eleven district hospitals in these provinces were included: Mancha Khiri (16° 12’ N 102° 29’ E), Chum Phae (16° 34’ N 102° 5’ E), Ban Phai (16° 3’ N 102° 43’ E), Ban Haet (16° 12’ N 102° 46’ E), and Mueang Khon Kaen (16° 25’ N 102° 49’ E) district hospitals in Khon Kaen Province; Selaphum (16° 2’ N 103° 59’ E), Phon Thong (16° 18’ N 103° 57’ E), and Thawat Buri (16° 1’ N 103° 45’ E) district hospitals in Roi Et Province; Kamalasai (16° 18’ N 103° 36’ E) and Kuchinarai (16° 30’ N 104° 1’ E) district hospitals in Kalasin Province; and Chiang Yuen (16° 25’ N 103° 3’ E) district hospital in Maha Sarakham Province.
Fig 1. Map of the study area.
Sample collections were done in four provinces of northeastern Thailand which are highlighted in green, brown, blue, and yellow. The locations of study districts are marked on the map in gray. The map was created using QGIS 3.16 software.
The methodology of sample collection was previously described [8]. Briefly, plasma samples were collected from patients presenting with dengue-like symptoms, with potential dengue infections based on the presence of fever (> 38°C), no recent travel history during the previous 7 days, and being older than five years of age.
For the entomological investigation, adult Aedes mosquitoes were collected from the patient’s household and four additional neighboring houses within a 100-meter radius of the patient’s house using portable Prokopack aspirators [17]. The collection was conducted for 15 minutes indoors (mainly in bedrooms and living rooms) and 15 minutes outdoors near the house (primarily around human-made articles, backyard/patio, vegetation, etc). Mosquito sex and species (Ae. aegypti or Ae. albopictus) were identified under a stereomicroscope using morphological keys [18, 19]. Aedes mosquitoes from the patient’s house and neighboring houses were designated as being from one combined collection cluster.
Confirmation of dengue virus infection in humans
During the study period, we obtained 292 plasma samples from suspected dengue patients (S1 Fig). The preliminary screening for DENV in the patient’s plasma was performed using the SD BIOLINE Dengue Duo kit (Standard Diagnostics, Suwon, Korea), according to the manufacturer’s instructions. Laboratory-confirmed DENV detection and serotyping were carried out in the laboratory at Khon Kaen University, Khon Kaen province, Thailand. Briefly, viral RNA was extracted from 140 μl of each plasma sample using QIAamp Viral RNA Mini Kits following the manufacturer’s instructions (Qiagen, Germany), and DENV detection and serotyping were performed using qRT-PCR as described by Shu et al. [20].
DENV detection in adult mosquitoes
A total of 192 female Aedes mosquito pools containing 902 Aedes mosquitoes (1–32 mosquitoes per pool) were processed for DENV identification by qRT-PCR (S1 Fig). These 192 pools originated from different collection clusters. Mosquito abdomens were separated from the head-thorax and pooled. These pooled and individual head-thorax samples were triturated, using sterile pestles and 1.5 ml Eppendorf tubes (Eppendorf AG, Hamburg, Germany) in 500 μl of Leibovitz’s L-15 medium (Gibco, Thermo Fisher Scientific, USA). The resulting suspension was clarified by centrifugation 800×g at 4°C for 5 minutes. Next, 140 μl of each sample was transferred to Eppendorf tubes for RNA extraction, and the remaining suspensions were stored at −80°C. Viral RNA extraction was performed by using QIAamp Viral RNA Mini Kits following the manufacturer’s instructions (Qiagen, Germany). Mosquito samples were subjected to two rounds of PCR for DENV detection and serotyping using the primer set of Lanciotti et al. [21] with minor modifications to perform it on real-time PCR. The viral RNA was reverse-transcribed into cDNA by specific D2 primer synthesized using SuperScript III first-strand synthesis system (Invitrogen, USA) according to the manufacturer’s instructions and cDNA was stored at -20°C until used. The first round of PCR aimed to screen DENV infection from mosquito pooled samples using SYBR green-based real-time and the primer set of Lanciotti et al. [21]. All individual mosquito specimens from each PCR-positive pool were subjected to the second round of PCR for DENV serotyping by SYBR green-based real-time PCR using the primer set of Lanciotti et al. [21]. The PCR reaction comprised pre-denaturation at 95°C for 2 minutes followed by 35 cycles at 95°C for 15 seconds, 55°C for 30 seconds, and a final period of 72°C for 42 seconds. The PCR amplification used an Applied Biosystems® 7500 Real-Time PCR machine (Applied Biosystems, CA, USA). Positive control and negative control (without cDNA template) were included with each amplification reaction. The resulting data was analyzed using the software provided by Applied Biosystems based on the Tm and Ct amplification plot values. The positive results were re-confirmed by visualization on 2% agarose gel electrophoresis staining with ethidium bromide.
Serotype matching between dengue patients and mosquitoes
To evaluate the relationship of DENV infection between patients and mosquitoes in the same residential area, DENV serotypes from 75 patients’ plasma were matched with those from mosquitoes collected from the collection clusters around each patient’s residence (S1 Fig). Where the patient’s plasma had at least one serotype in common with the mosquito pool, a match was assigned. On the other hand, if there was no DENV serotype in common between the patient’s plasma sample and the mosquito pool, or if the latter was negative for DENV, this was assigned as a serotype mismatch.
DENV envelope gene sequencing
Full-length E gene PCR products were directly amplified from patient plasma and mosquito pooled samples to avoid genetic changes due to the culture process. The viral RNA was extracted from the samples using QIAamp Viral RNA Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. cDNA fragments covering the E gene of DENV were synthesized by specific primers of each serotype as previously described [22] using SuperScript III first-strand synthesis system (Invitrogen, USA) according to the manufacturer’s instructions and stored at -20°C until use. Two overlapping fragments (F and L fragments) of the E gene were amplified primers as previously described [22], using PrimeStar GXL DNA polymerase (Takara, Japan) according to the manufacturer’s instructions. The PCR amplification cycles were performed with a step of initial DNA denaturation at 98°C for 5 minutes followed by 35 cycles of denaturation at 98°C for 10 seconds and annealing at 55°C for 15 seconds and extension at 68°C for 90 seconds. The specific amplicons were purified from 0.8% agarose gel by gel cutting and extraction using the QIAquick gel extraction kit (Qiagen, Germany). Purified amplicons of each sample were subjected to Sanger sequencing by using cycle sequencing reactions and dye terminator methodologies of Macrogen company (Macrogen, Seoul, Korea) using four overlapping primers for each serotype as previously described [22].
Phylogenetic analysis
The phylogenetic analysis based on the nucleotide sequence of the envelope gene-encoding region of the DENV was constructed and used to elucidate the origins of disease outbreaks. The E gene sequences obtained in this study were aligned with other DENV sequences from those previously isolated in Thailand and neighboring countries and from other regions of the world available in the GenBank database (www.ncbi.nlm.nih.gov) using ClustalW in Bioedit. Maximum-likelihood trees were constructed in MEGA X using 35 reference sequences for DENV-1, 65 sequences for DENV-2, and 40 sequences for DENV-4 (S2 Table) with the best-fit nucleotide substitution model (TN93+G). Bootstrap values were done with 1000 replications. Genotypic classification of DENV-1, -2, and -4 followed Goncalvez et al. [23], Twiddy et al. [24] and AbuBakar et al. [25] respectively.
Accession numbers
The full-length E gene sequences obtained in this study were deposited in GenBank and granted accession numbers MT524489 to MT524513 (Table 1).
Table 1. Information on dengue virus strains in this study.
| Sample code | Host | Location (District, Province) | Collection date | Serotype | Genotype | Accession No. |
|---|---|---|---|---|---|---|
| D1H/4024-01/16 | Human | Ban Haet, Khon Kaen | Jun-2016 | 1 | I | MT524490 |
| D1H/4024-02/16 | Human | Ban Haet, Khon Kaen | Jun-2016 | 1 | I | MT524493 |
| D1H/4024-07/16 | Human | Ban Haet, Khon Kaen | Jun-2016 | 1 | I | MT524492 |
| D1M/4024-07/16 | Mosquito | Ban Haet, Khon Kaen | Jun-2016 | 1 | I | MT524491 |
| D1M/4024-08/16 | Mosquito | Ban Haet, Khon Kaen | Jun-2016 | 1 | I | MT524494 |
| D1H/4010-02/16 | Human | Ban Phai, Khon Kaen | Sep-2016 | 1 | I | MT524489 |
| D1H/4405-08/17 | Human | Chiang Yuen, Maha Salakham | Jul-2017 | 1 | I | MT524496 |
| D1H/4603-24/18 | Human | Kamalasai, Kalasin | Aug-2018 | 1 | I | MT524495 |
| D2H/4005-02/16 | Human | Chum Phae, Khon Kaen | Jun-2016 | 2 | Asian I | MT524502 |
| D2H/9918-02/18 | Human | Mueang khon kaen, Khon Kaen | May-2018 | 2 | Cosmopolitan | MT524498 |
| D2H/4603-10/18 | Human | Kamalasai, Kalasin | Jun-2018 | 2 | Cosmopolitan | MT524499 |
| D2H/4603-11/18 | Human | Kamalasai, Kalasin | Jun-2018 | 2 | Asian I | MT524501 |
| D2H/4405-26/18 | Human | Chiang Yuen, Khon Kaen | Jun-2018 | 2 | Cosmopolitan | MT524500 |
| D2H/9918-09/18 | Human | Mueang khon kaen, Khon Kaen | Aug-2018 | 2 | Cosmopolitan | MT524497 |
| D4H/4005-06/16 | Human | Chum Phae, Khon Kaen | Jun-2016 | 4 | I | MT524505 |
| D4M/4005-07/16 | Mosquito | Chum Phae, Khon Kaen | Jun-2016 | 4 | I | MT524506 |
| D4H/4507-09/16 | Human | Phon Thong, Roi Et | Jul-2016 | 4 | I | MT524504 |
| D4M/4510-08/16 | Mosquito | Selaphum, Roi-Et | Aug-2016 | 4 | I | MT524503 |
| D4H/4005-11/16 | Human | Chum Phae, Khon Kaen | Aug-2016 | 4 | I | MT524508 |
| D4H/4005-15/16 | Human | Chum Phae, Khon Kaen | Aug-2016 | 4 | I | MT524507 |
| D4H/4507-15/16 | Human | Phon Thong, Roi Et | Aug-2016 | 4 | I | MT524510 |
| D4H/4507-18/16 | Human | Phon Thong, Roi-Et | Sep-2016 | 4 | I | MT524509 |
| D4H/4507-19/16 | Human | Phon Thong, Roi-Et | Sep-2016 | 4 | I | MT524512 |
| D4H/4005-29/16 | Human | Chum Phae, Khon Kaen | Oct-2016 | 4 | I | MT524513 |
| D4H/4605-01/17 | Human | Kuchinarai, Kalasin | Jan-2017 | 4 | I | MT524511 |
Samples were named in a format consisting of “DENV serotype host/sample ID/Year of sample collection”.
Ethics statement
The human samples in the present study were obtained from a dengue case-control study [8]. The present study was approved by the Khon Kaen University Ethics Committee for Human Research (KKUEC, no. HE611454). Informed consent was obtained in writing from all participants before sample collection.
Results
DENV serotype distribution in human samples
A total of 103 of 292 plasma samples were positive for DENV (S1 Fig). Serotype identification revealed all four DENV serotypes circulated in these samples. Of the 103 DENV positive samples, single DENV serotypes indicating mono-infection were detected in 79 (77%) samples, while multiple serotypes indicating co-infection were found in 24 (23%) samples. The predominant DENV serotypes in 2016 and 2018 were DENV-4 (74%) and DENV-3 (53%), respectively, whereas in 2017 DENV-1, -3, and DENV-4 were found with equal prevalence (Table 2).
Table 2. Dengue virus infection and serotype distribution in human blood and individual mosquitoes per year.
| 2016 | ||||||
| Sample | No. of samples | DENV positive | DENV-1 | DENV-2 | DENV-3 | DENV-4 |
| Human blood | 133 | 42 | 9 (21%) | 3 (7%) | 9 (21%) | 31 (74%) |
| Ae. aegypti | 337 | 77 | 8 | 18 | 56 | 7 |
| Ae. albopictus | 9 | 4 | 0 | 1 | 3 | 1 |
| Total Aedes | 346 | 81 | 8 (10%) | 19 (23%) | 59 (73%) | 8 (10%) |
| 2017 | ||||||
| Sample | No. of samples | DENV positive | DENV-1 | DENV-2 | DENV-3 | DENV-4 |
| Human blood | 35 | 8 | 3 (38%) | 0 | 3 (38%) | 3 (38%) |
| Ae. aegypti | 137 | 4 | 1 | 3 | 3 | 0 |
| Ae. albopictus | 0 | 0 | 0 | 0 | 0 | 0 |
| Total Aedes | 137 | 4 | 1 (13%) | 3 (38%) | 3 (38%) | 0 |
| 2018 | ||||||
| Sample | No. of samples | DENV positive | DENV-1 | DENV-2 | DENV-3 | DENV-4 |
| Human blood | 124 | 53 | 15 (28%) | 22 (42%) | 28 (53%) | 4 (8%) |
| Ae. aegypti | 417 | 64 | 9 | 50 | 37 | 2 |
| Ae. albopictus | 2 | 0 | 0 | 0 | 0 | 0 |
| Total Aedes | 419 | 64 | 9 (17%) | 50 (94%) | 37 (70%) | 2 (4%) |
Note: The number of DENV serotypes distribution reported in the table consists of mono-infected and co-infected cases.
DENV serotype distribution in mosquito samples
A total of 902 female Aedes individual mosquitoes (contained in 192 pools) were captured during the study period (Table 2). The predominant mosquito species was Ae. aegypti (98.8%) followed by Ae. albopictus (1.2%) (Table 2). One hundred forty-nine (16.5%) of the 902 female mosquitoes were positive for DENV. Serotyping of individual mosquito head-thoraces revealed that the predominant DENV serotypes were DENV-3 (73%) in 2016, DENV-2 and DENV-3 (38% of each serotype) in 2017, and DENV-2 (94%) in 2018 (Table 2). Of the 149 DENV positive individual mosquito samples, single DENV serotypes indicating mono-infection were detected in 105 (70%) samples, while multiple serotypes indicating co-infection were found in 44 (30%) samples.
Relationships of DENV infection between dengue patients and mosquitoes from the same residence area
Out of 103 DENV-positive patients, mosquito collection was done only in 75 patient’s residential areas. DENV serotypes in plasma of those patients were compared with DENV serotypes from the infected mosquitoes from their residences. Of the 75 plasma samples, 14 (19%) matched a serotype from the DENV positive mosquitoes samples (Fig 2). Among 61 (81%) mismatched samples, 10 (13%) had a different DENV serotype in the corresponding mosquito samples, while 51 (68%) corresponded to DENV negative mosquito samples. The molecular characteristics of 24 pairs out of 75 paired samples are presented in S3 Table.
Fig 2. Serotypes in 75 DENV-positive patients from households where corresponding DENV-positive mosquitoes were collected.
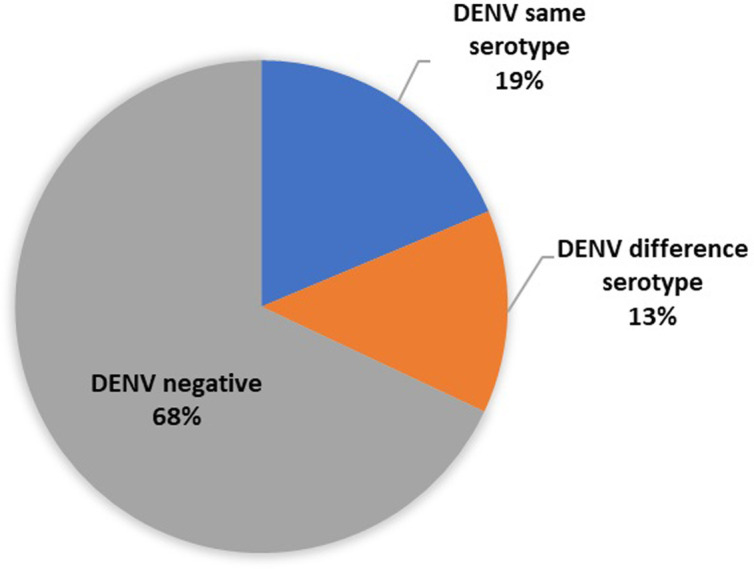
In 14 households (19%), the serotype in patients and mosquitoes matched each other, i.e. were the same (blue); in 10 households (13%), the serotypes in patients and mosquitoes were the different (orange); and in 51 households (68%) patients were DENV-positive whereas mosquitoes were DENV-negative (grey). More details of the molecular characteristics of DENV-positive patients and mosquitoes are provided in S3 Table.
Phylogenetic analysis
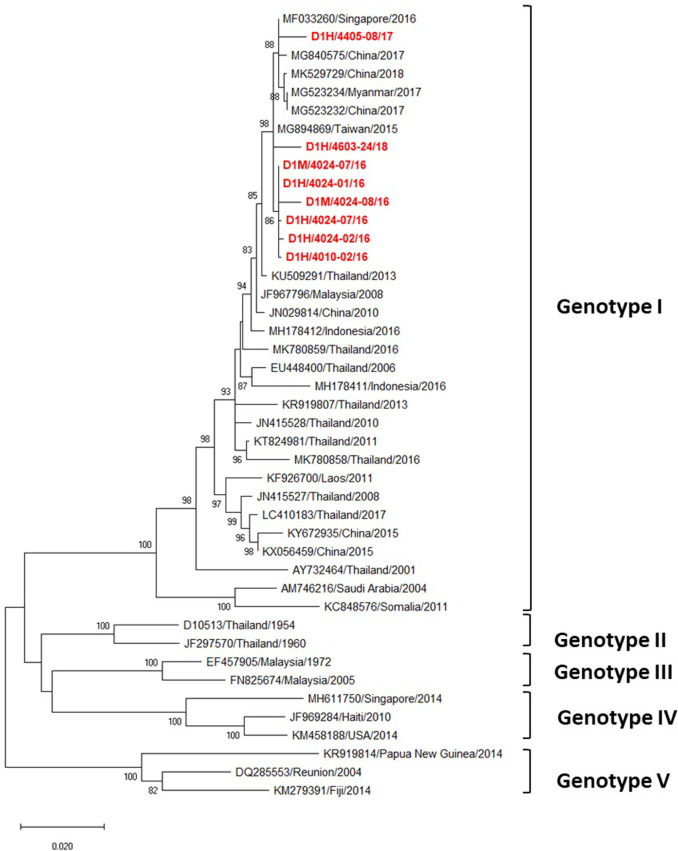
Sequencing of the E gene was successful for 25 samples, 21 from patients, and 4 from mosquito pools. They contained eight DENV-1, six DENV-2, and 11 DENV-4 sequences (Table 1).
Phylogenetic analysis of DENV-1 following Goncavez’s classification [23] showed that all eight DENV-1 strains were genotype I (Fig 3), and that all eight were from the same clade. Seven strains were closely related to the DENV-1 genotype I isolated from Taiwan in 2015, the other (D1H/4405-08/17) being closely related to DENV-1 isolated between 2016 and 2018 from other Asian countries including Singapore in 2016, China in 2017–18 and Myanmar in 2017 (Fig 3).
Fig 3. Phylogenetic analysis of DENV-1.
Maximum-likelihood tree of the E gene sequences of DENV-1 was generated in the MEGA X program using the TN93 + G model with 1000 bootstrap replications. Bootstrap support values exceeding 80% are shown on branch nodes. The virus names of each sequence retrieved from the NCBI database are labeled as follows: GenBank accession number/country/isolated year. Samples collected in the present study are in red bold font. Annotation on the right denotes DENV genotype.
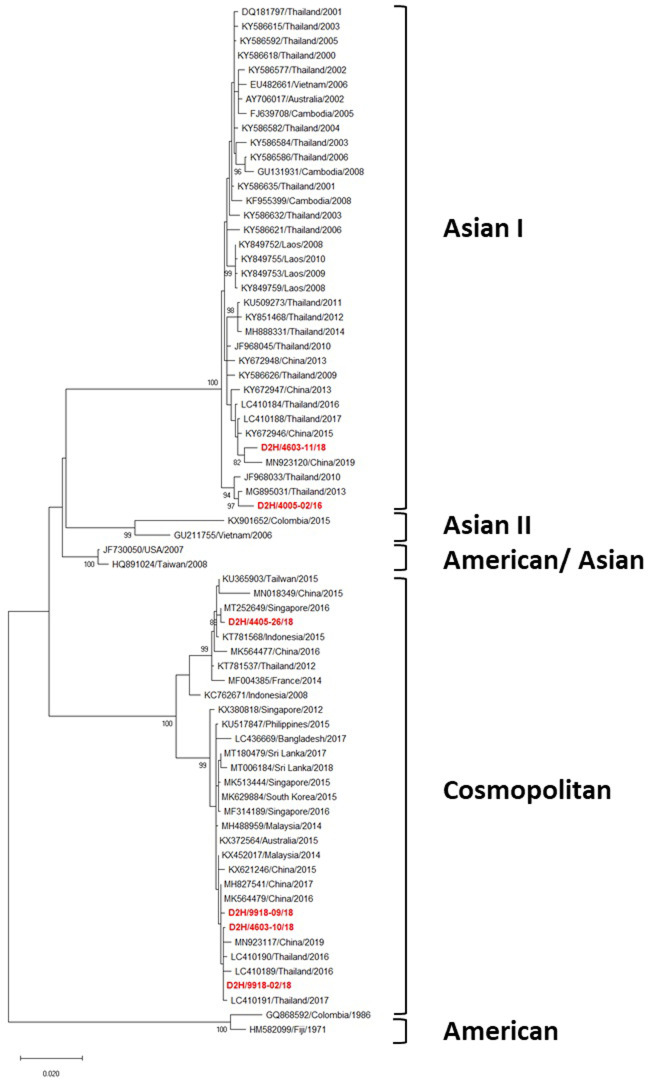
Of the six DENV-2 strains, two were classified as Asian I and four as Cosmopolitan genotypes [24] (Fig 4). In turn, the Asian I genotype was clustered into two clades. The Asian I strain D2H/4005-02/16 was closely related to the DENV-2 Thai strain isolated in 2013, while the other (D2H/4603-11/18) was closely related to one isolated in Thailand in 2017, and in China in 2015 (Fig 4). The four Cosmopolitan strains also clustered into two distinct clades. Three Cosmopolitan strains (D2H/4603-10/18, D2H/9918-02/18, and D2H/9918-09/18) were closely related to ones isolated in Thailand and China during 2016–2017, whereas the remaining strain (D2H/4405-26/18) was closely related to DENV-2 isolated from Singapore in 2016 (Fig 4).
Fig 4. Phylogenetic analysis of DENV-2.
Maximum-likelihood tree of the E gene sequences of DENV-2 was generated in the MEGA X program using the TN93 + G model with 1000 bootstrap replications. Bootstrap support values exceeding 80% are shown on branch nodes. The virus names of each sequence retrieved from the NCBI database are labeled as follows: GenBank accession number/country/isolated year. Samples collected in the present study are in bold red font. Annotation on the right denotes DENV genotype.
Phylogenetic analysis of DENV-4 following AbuBakar et al. [25] revealed that all 11 DENV 4 strains obtained in this study belonged to genotype I (Fig 5). However, they were clustered into four distinct clades (Fig 5). Seven strains from 2016 were closely related to one isolated in Taiwan in 2015 and in Thailand in 2017, whereas two strains collected during 2016 (D4H/4507-15/16 and D4H/4507-18/16) were closely related to one isolated in Taiwan in 2013. The two remaining strains were closely related to ones found in Thailand and neighboring countries, e.g. Singapore in 2014 and China in 2015 to 2016 (Fig 5).
Fig 5. Phylogenetic analysis of DENV-4.
Maximum-likelihood tree of the E gene sequences of DENV-4 was generated in the MEGA X program using the TN93 + G model with 1000 bootstrap replications. Bootstrap support values exceeding 80% are shown on branch nodes. The virus names of each sequence retrieved from the NCBI database are labeled as follows: GenBank accession number/country/isolated year. Samples collected in the present study are in red bold font. Annotation on the right denotes DENV genotype.
Discussion
Recent evidence indicates that the dengue outbreak situation in northeastern Thailand has become critical, causing annually peaks of the disease associated with increasing morbidity and mortality humans [2]. So far, there is limited information on the molecular characteristics of DENV in this region. The present study is the first to describe the relationship of DENV infection between humans and mosquitoes, and molecular characteristics of DENV in northeastern Thailand.
Approximately one third (35.3%) of dengue-suspected patients recruited in this study had a confirmed DENV infection, indicating a high burden of dengue in 2016 and 2018 (Table 2). All four DENV serotypes were found co-circulating during the three years of the study. These findings are similar to those from other regions of Thailand during the same period [11, 16]. The most prevalent serotypes in 2016 and 2018 were DENV-4 and DENV-3 respectively, whereas, DENV-1, -3, -4 were equally prevalent in 2017. With the lack of molecular data on DENV in northeastern Thailand, we were not able to compare this serotype profile to those of previous years. During the three years (June 2016 to December 2018) of our study, Thailand faced dengue outbreaks in 2016 and 2018 in several provinces [26, 27]. Fewer dengue cases in 2017 were also reported by national surveillance (The Office of Disease Prevention and Control Region, ODPC). In addition, a previous report also showed that, prior to our study, during 2013–2015, the yearly distribution of DENV serotypes in Thailand included a high prevalence of DENV-2 [27]. This could have resulted in fewer DENV-2 human cases in 2017 due to recently acquired prior immunity. However, DENV-2 is still present in the collected mosquitoes at the same time, it is common to find all serotypes circulating in vectors or hosts, given the hyperendemicity of DENV in Thailand. The virus can also be directly transmitted to the next vector generation by vertical transmission. Similarly, a study conducted in India observed that the DENV-2 disappeared in 2016 after predominating between 2011–2015 [28].
Dengue virus detection in mosquitoes during an outbreak has been suggested as a potential tool for early warnin and designing effective vector control strategies [29, 30]. We collected Aedes mosquitoes from houses of suspected dengue patients in northeastern Thailand, the most prevalent species was Ae. aegypti (98.8%) followed by Ae.albopictus (1.2%) (Table 2). Our study found 16.5% (149/902) of mosquitoes were DENV-positive. Similarly, in southern Thailand, 16% of field-caught Aedes aegypti and 36.2% of Aedes albopictus were infected with DENV during the early rainy season of 2005 [31]. Several studies have described DENV infection in field-caught Aedes mosquitoes, with various infection rate including 10.8% in Brazil [32], 12.7% and 62% in Colombia [33, 34], 2.8% in Philippines [35]. These findings suggested that the infection rate of virus in field-caught mosquitoes is highly variable, possibly depending on the design and conditions of each study [36, 37]. Serotyping results illustrated that all four serotypes were circulating in the mosquitoes in this region. DENV-3 and DENV-2 were predominant in 2016 and 2018 respectively, while both serotypes were most prevalent in 2017.
Moreover, we found co-infection of different DENV serotypes: 23% and 30% in human and mosquito samples respectively. This finding is consistent with several studies which reported the concurrence of DENV infection in the same patient with multiple serotypes at different percentages ranging from 3% to 43% [38–41]. Similar observations of co-infection with multiple DENV serotypes have been reported in other countries and also in Thailand [31, 42, 43]. All four serotypes circulating in the region have been found in combinations within single patients [41]. Several factors might be involved in co-infection including: i) the multiple feeding behavior of Ae. aegypti, which feeds more than once during its genotropic cycle [31, 41, 42], ii) the high attack rates of cases during dengue epidemics which may result in many infections with multiple serotypes in humans, and also provide opportunities for mosquitoes to become infected with multiple serotypes [42], iii) transovarial transmission of dengue viruses in mosquitoes [31], iv) asymptomatic cases as a source of infection in mosquitoes [42].
Results from the present study, show the difference of serotype distribution in hosts and vectors by year, and suggest that mosquitoes may serve as a maintenance mode for the virus in the environment. When the immunity level in human population to a certain serotype decreases, a burst of infections may follow [32]. Moreover, our results are consistent with previous studies which showed specific DENV serotypes building up in mosquitoes, then becoming predominant in humans in the subsequent year [32, 33].
Although several studies have evaluated the presence of DENV-infected Aedes mosquitoes in all four regions of Thailand including central [44–49], southern [31], northern [47, 50], and northeastern [51], our study is the first to target the residential area of DENV-infected patients within the onset of symptoms in northeastern Thailand. The DENV serotypes of patient samples were compared with mosquito samples collected from the patient’s residential area within a 100-meter radius. About one fifth (19%) of the patients had a serotype matching the serotype in the mosquitoes collected, while remaining 81% were serotype mismatched, consistsing of 13% with a different serotype and another 68% whose corresponding mosquitoes were DENV negative (Fig 2). This result suggests that transmission outside the residence may be important. Since most of the cases were children and adults (S4 Table) who have spent day time outside their residence (e.g., school, worksite/office), it is possible that they were exposed there to infectious mosquitoes. This finding is consistent with previous studies conducted in Brazil and Philippines that found differences in DENV serotypes between patients and mosquitoes in the same home [32, 35]. Previous reports suggest that DENV infection often occurs elsewhere, other than the home, e.g. workplace, school or college, temple or farm field [9, 35, 52]. Numerous factors may influence these results including: i) asymptomatic infections with different DENV serotypes in members of the same household, ii) transovarial transmission of DENV in the household involving serotypes to which the residents have recent prior immunity [35]. It is essential to note that spending time in an area or work environment during the day that is infested with Aedes mosquitoes may increase the risk of dengue infection. This supports the idea that DENV transmission is probably driven by the movement of infected humans [35].
The phylogenetic analysis revealed that all eight DENV-1 strains were classified as genotype I (Fig 3), which mainly circulates in Asian countries especially Thailand [11, 16, 53–56]. Interestingly, these DENV-1 strains were not grouped within the same clade as Thai strains isolated during the same period in northern [11] and southern [16] regions. Rather, those other strains were closely related to strains from other Asian countries during 2015–2018, such as a Taiwanese strain isolated from a traveler who returned from Thailand in 2015, a Singaporean strain in 2016, a Myanmar strain in 2017, and Chinese strains in 2017–2018. This indicates that DENV-1 circulating strains in northeastern Thailand are likely to have been imported from other Asian countries (Fig 3).
We observed DENV-2 Asian I and Cosmopolitan genotypes co-circulating in this region (Fig 4), as they commonly do elsewhere in Asia, especially Southeast Asia (SEA) [11, 14, 16, 57–59]. The Asian I strains were clustered into different 2 clades. Interestingly, the strain D2H/4005-02/16 was closely related to the Taiwanese strain isolated from two travelers returning from Thailand in 2010 [60] and 2013 [61], suggesting they were likely to have circulated for at least 3–4 years in northeastern Thailand. Moreover, the remaining strain D2H/4603-11/18 was closely related to one isolated in Guangzhou (China) during 2019, suggesting they have a common ancestor. Therefore, northeastern Thailand seems to be a potential source of dengue transmission to other parts of Asian countries (Fig 4). The four Cosmopolitan strains were clustered into two clades. Three strains (D2H/4603-10/18, D2H/9918-02/18, and D2H/9918-09/18) were closely related to ones isolated during the same period in the central and northern regions of the country, and in other Asian countries such as China and Singapore (Fig 4). This demonstrates that the DENV-2 northeastern strains were not only transferred from other parts of Thailand in the same period but also were persistent in this region (Fig 4).
All 11 DENV-4 strains were clustered into genotype I, which commonly circulates and was recently found in Thailand [11, 16, 62] (Fig 5). Although all strains were grouped into this single genotype, they were clustered into 3 different clades (Fig 5). Nine strains were closely related to ones isolated in Thailand, Taiwan, and China during 2015–2016, with the remaining two (D4H/4507-15 and D4H/4507-18) being closely related to one isolated in Taiwan in 2013 from a traveler who returned from Thailand (Fig 5). This demonstrates that DENV-4 strains circulating in this region were not only transferred from other regions of Thailand in the same period but also imported from Southeast Asian countries (Fig 5). Interestingly, all seven DENV-4 northeastern strains collected during 2016 were closely related to the LC410202-03/Thailand/2017 strain which was isolated from the central region of Thailand in 2017 [11]. These results suggest that the northeastern region is a locus of active circulation of DENV genotypes, with the potential to export them not only to other regions of the country but also other countries around the world especially SEA through the movement of infected persons and mosquitoes [10].
Our study has some limitations, in particular in terms of generalizability (extrapolation). First, we defined inconsistent serotyping results as a mismatch, which included both the occurrence of different genotypes, and an infection linked to negative sample. Since negative was assigned as mismatched, it could increase the chance of mismatch due to the low titer of DENV in mosquitoes. Second, our mosquito collection focused on households, but transmission could occur in other locations where patients spend their daytime such as schools, work offices, and community centers [8]. Last, we were unable to sequence any DENV-3 viruses. Since our study performed E gene sequencing directly from the sample without virus isolation, to avoid working on passaged viruses, this gap might be attributed to low viral titers which were insufficient for conventional PCR amplification. Shorter fragments of PCR products have been suggested to be more suitable for sequencing [10], and this is being considered for our future studies.
In conclusion, our study provides the first data on molecular characteristics of DENV in northeastern Thailand during 2016 to 2018. Our data confirmed the hyperendemicity of all four DENV serotypes in northeastern Thailand. The low proportion of matches between DENV serotypes in patients and mosquitoes from the patients’ residence areas suggests that non-residential transmission may be important. Our phylogenetic analysis revealed that the virus circulating in this region shared high homology with the virus from other regions of Thailand and Southeast Asian countries in the same period, as well as with persisting strains that originated in Thailand and other southeast Asian countries. These findings indicated that human movement has an important role in infection and disease transmission in the region. Taken together, our findings highlight that vector control, and early warning based on DENV detection in vectors, are important strategies for the prevention of dengue epidemics in northeastern Thailand. Continuous molecular surveillance of DENV in northeastern Thailand to observe the virus replacement and also better understand dengue transmission dynamics in this region is required.
Supporting information
(TIF)
(DOCX)
(accessed from GenBank, the National Centre for Biotechnology Information https://www.ncbi.nlm.nih.gov/genbank/).
(DOCX)
The molecular characteristics of 24 pairs of DENV positive samples were shown in this table. Gray row label represents serotype matched between patients and mosquitoes from their resident area.
(DOCX)
(DOCX)
Acknowledgments
We would like to thank the attending physicians, nurses, and clinical laboratory staff at eight district hospitals in Khon Kaen, Roi Et, Kalasin, and Mahasarakham provinces for their kind assistance in recruiting potential participants and logistic support. We would like to acknowledge the entomology team, Office of Disease Prevention and Control, Region 7, Khon Kaen for mosquito collections. The officers of Mahidol-Osaka Center for Infectious Diseases (MOCID), Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand are thanked for their technical support. We are grateful to all members of Assoc. Prof. Sunchai Payungporn lab group (SP lab), Chulalongkorn University for their kind assistance and support in data analysis of DENV sequence. Our special thanks to Thawaree Nakpook, Le Thi Bao Chi and Benedicte Fustec for their valuable suggestions and assistance in the laboratory.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Research Council of Norway (project no. 250443); the Invitation Research Grant, Faculty of Medicine, Khon Kaen University, Thailand, grant number IN62113; the Japan Agency for Medical Research and Development (AMED) JP19fm0108003 and 20wm0225010h0101.
References
- 1.Sim S, Aw PPK, Wilm A, Teoh G, Hue KDT, Nguyen NM, et al. Tracking Dengue Virus Intra-host Genetic Diversity during Human-to-Mosquito Transmission. Pimenta PF, editor. PLoS Negl Trop Dis. 2015;9: e0004052. doi: 10.1371/journal.pntd.0004052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Limkittikul K, Brett J, L’Azou M. Epidemiological Trends of Dengue Disease in Thailand (2000–2011): A Systematic Literature Review. Halstead SB, editor. PLoS Negl Trop Dis. 2014;8: e3241. doi: 10.1371/journal.pntd.0003241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Special Programme for Research and Training in Tropical Diseases, World Health Organization, editors. Dengue: guidelines for diagnosis, treatment, prevention, and control. New ed. Geneva: TDR: World Health Organization; 2009.
- 4.Gubler DJ. Dengue, Urbanization and Globalization: The Unholy Trinity of the 21st Century. Trop Med Health. 2011;39: S3–S11. doi: 10.2149/tmh.2011-S05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harapan H, Michie A, Sasmono RT, Imrie A. Dengue: A Minireview. Viruses. 2020;829: 1–35. doi: 10.3390/v12080829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Achee NL, Grieco JP, Vatandoost H, Seixas G, Pinto J, Ching-NG L, et al. Alternative strategies for mosquito-borne arbovirus control. Kittayapong P, editor. PLoS Negl Trop Dis. 2019;13: e0006822. doi: 10.1371/journal.pntd.0006822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee K-S, Lai Y-L, Lo S, Barkham T, Aw P, Ooi P-L, et al. Dengue Virus Surveillance for Early Warning, Singapore. Emerg Infect Dis. 2010;16: 847–849. doi: 10.3201/eid1605.091006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fustec B, Phanitchat T, Hoq MI, Aromseree S, Pientong C, Thaewnongiew K, et al. Complex relationships between Aedes vectors, socio-economics and dengue transmission—Lessons learned from a case-control study in northeastern Thailand. Carvalho MS, editor. PLoS Negl Trop Dis. 2020;14: e0008703. doi: 10.1371/journal.pntd.0008703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu T-S, Zhang H-L, Feng Y, Fan J-H, Tang T, Liu Y-H, et al. Epidemiological and molecular characteristics of emergent dengue virus in Yunnan Province near the China-Myanmar- Laos border, 2013–2015. BMC Infect Dis. 2017;17: 12. doi: 10.1186/s12879-016-2102-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahri S, Yohan B, Trimarsanto H, Sayono S, Hadisaputro S, Dharmana E, et al. Molecular Surveillance of Dengue in Semarang, Indonesia Revealed the Circulation of an Old Genotype of Dengue Virus Serotype-1. Charrel R, editor. PLoS Negl Trop Dis. 2013;7: e2354. doi: 10.1371/journal.pntd.0002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phadungsombat J, Lin MY-C, Srimark N, Yamanaka A, Nakayama EE, Moolasart V, et al. Emergence of genotype Cosmopolitan of dengue virus type 2 and genotype III of dengue virus type 3 in Thailand. Samy AM, editor. PLOS ONE. 2018;13: e0207220. doi: 10.1371/journal.pone.0207220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz Y, Chen-Germán M, Quiroz E, Carrera J-P, Cisneros J, Moreno B, et al. Molecular Epidemiology of Dengue in Panama: 25 Years of Circulation. Viruses. 2019;11: 764. doi: 10.3390/v11080764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvo MA, Aliota MT, Moncla LH, Velez ID, Trujillo I, Friedrich TC. Tracking dengue virus type 1 genetic diversity during lineage replacement in an hyperendemic area in Colombia. PLoS ONE. 2019;14: 22. doi: 10.1371/journal.pone.0212947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harapan H, Michie A, Yohan B, Shu P-Y, Mudatsir M, Sasmono RT, et al. Dengue viruses circulating in Indonesia: A systematic review and phylogenetic analysis of data from five decades. Rev Med Virol. 2018; 17. doi: 10.1002/rmv.2037 [DOI] [PubMed] [Google Scholar]
- 15.Harapan H, Michie A, Mudatsir M, Sasmono RT, Imrie A. Epidemiology of dengue hemorrhagic fever in Indonesia: analysis of five decades data from the National Disease Surveillance. BMC Res Notes. 2019;350: 1–6. doi: 10.1186/s13104-019-4379-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamel R, Surasombatpattana P, Wichit S, Dauvé A, Donato C, Pompon J, et al. Phylogenetic analysis revealed the co-circulation of four dengue virus serotypes in Southern Thailand. Roques P, editor. PLOS ONE. 2019;14: e0221179. doi: 10.1371/journal.pone.0221179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazquez-Prokopec GM, Galvin WA, Kelly R, Kitron U. A New, Cost-Effective, Battery-Powered Aspirator for Adult Mosquito Collections. J Med Entomol. 2010;46: 8. doi: 10.1603/033.046.0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rattanarithikul R, Harbach RE, Harrison BA, Panthusiri P, Coleman RE, Richardson JH. Illustrated keys to the mosquitoes of Thailand VI. Tribe Aedini. Southeast Asian J Trop Med Public Health. 2010;41: 38. [PubMed] [Google Scholar]
- 19.Rueda LM. Pictorial keys for the identification of mosquitoes (Diptera: Culicidae) associated with Dengue Virus Transmission. Zootaxa. 2004;589: 1. doi: 10.11646/zootaxa.589.1.1 [DOI] [Google Scholar]
- 20.Shu P-Y, Chang S-F, Kuo Y-C, Yueh Y-Y, Chien L-J, Sue C-L, et al. Development of Group- and Serotype-Specific One-Step SYBR Green I-Based Real-Time Reverse Transcription-PCR Assay for Dengue Virus. J Clin Microbiol. 2003;41: 2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol. 1992;30: 545–551. doi: 10.1128/jcm.30.3.545-551.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warrilow D, Northill JA, Pyke AT. Sources of Dengue Viruses Imported into Queensland, Australia, 2002–2010. Emerg Infect Dis. 2012;18: 1850–1857. doi: 10.3201/eid1811.120014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, et al. Diversity and Evolution of the Envelope Gene of Dengue Virus Type 1. Virology. 2002;303: 110–119. doi: 10.1006/viro.2002.1686 [DOI] [PubMed] [Google Scholar]
- 24.Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, et al. Phylogenetic Relationships and Differential Selection Pressures among Genotypes of Dengue-2 Virus. Virology. 2002;298: 63–72. doi: 10.1006/viro.2002.1447 [DOI] [PubMed] [Google Scholar]
- 25.AbuBakar S, Wong P-F, Chan Y-F. Emergence of dengue virus type 4 genotype IIA in Malaysia. J Gen Virol. 2002;83: 2437–2442. doi: 10.1099/0022-1317-83-10-2437 [DOI] [PubMed] [Google Scholar]
- 26.Suphanchaimat R, Thammavijaya P, Taweewigyakarn P, Boonchalermvichien T, Pensuk P, Niyamrattanakulsiri P, et al. Systemic Investigation of Dengue Incidence and Control Measures in Surin, Thailand, 2018. Outbreak Surveill Investig Response OSIR J. 2019;12: 9. [Google Scholar]
- 27.Kerdpanich P, Kongkiatngam S, Buddhari D, Simasathien S, Klungthong C, Rodpradit P, et al. Comparative Analyses of Historical Trends in Confirmed Dengue Illnesses Detected at Public Hospitals in Bangkok and Northern Thailand, 2002–2018. Am J Trop Med Hyg. 2020. [cited 8 Jul 2021]. doi: 10.4269/ajtmh.20-0396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Islam A, Mohd Abdullah, Tazeen A, Naqvi IH, Kazim SN, Ahmed A, et al. Circulation of dengue virus serotypes in hyperendemic region of NewDelhi, India during 2011–2017. J Infect Public Health. 2020;13: 1912–1919. doi: 10.1016/j.jiph.2020.10.009 [DOI] [PubMed] [Google Scholar]
- 29.Khan J, Ghaffar A, Khan SA. The changing epidemiological pattern of Dengue in Swat, Khyber Pakhtunkhwa. Sekaran SD, editor. PLOS ONE. 2018;13: e0195706. doi: 10.1371/journal.pone.0195706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aranda C, Martínez MJ, Montalvo T, Eritja R, Navero-Castillejos J, Herreros E, et al. Arbovirus surveillance: first dengue virus detection in local Aedes albopictus mosquitoes in Europe, Catalonia, Spain, 2015.: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thavara U, Siriyasatien P, Tawatsin A, Asavadachanukorn P, Anantapreecha S, Wongwanich R, et al. Double infection of heteroserotypes of dengue viruses in field populations of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) and serological features of dengue viruses found in patients in Southern Thailand. Southeast Asian J Trop Med Public Health. 2006;37: 10. [PubMed] [Google Scholar]
- 32.Guedes DRD, Cordeiro MT, Melo-Santos MAV, Magalhaes T, Marques E, Regis L, et al. Patient-based dengue virus surveillance in Aedes aegypti from Recife, Brazil. J Vector Borne Dis. 2010;47: 67–75. [PubMed] [Google Scholar]
- 33.Parra B, Rengifo G, Méndez F, Burbano ME, Arias JF, Muñoz J, et al. Human and mosquito infections by dengue viruses during and after epidemics in a dengue–endemic region of Colombia. Am J Trop Med Hyg. 2006;74: 678–683. doi: 10.4269/ajtmh.2006.74.678 [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Castro R, Castellanos JE, Olano VA, Matiz MI, Jaramillo JF, Vargas SL, et al. Detection of all four dengue serotypes in Aedes aegypti female mosquitoes collected in a rural area in Colombia. Mem Inst Oswaldo Cruz. 2016;111: 233–240. doi: 10.1590/0074-02760150363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balingit JC, Carvajal TM, Saito-Obata M, Gamboa M, Nicolasora AD, Sy AK, et al. Surveillance of dengue virus in individual Aedes aegypti mosquitoes collected concurrently with suspected human cases in Tarlac City, Philippines. Parasit Vectors. 2020;13: 594. doi: 10.1186/s13071-020-04470-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu W, Novak RJ. Short report: detection probability of arbovirus infection in mosquito populations. Am J Trop Med Hyg. 2004;71: 636–638. doi: 10.4269/ajtmh.2004.71.636 [DOI] [PubMed] [Google Scholar]
- 37.Lord CC, Bustamante DM. Sources of Error in the Estimation of Mosquito Infection Rates Used to Assess Risk of Arbovirus Transmission. Am J Trop Med Hyg. 2010;82: 1172–1184. doi: 10.4269/ajtmh.2010.09-0323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, et al. Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol J. 2008;5: 1. doi: 10.1186/1743-422X-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colombo TE, Vedovello D, Mondini A, Reis AFN, Cury AAF, Oliveira FH de, et al. Co-infection of dengue virus by serotypes 1 and 4 in patient from medium sized city from Brazil. Rev Inst Med Trop São Paulo. 2013;55: 275–281. doi: 10.1590/S0036-46652013000400009 [DOI] [Google Scholar]
- 40.Reddy MN, Dungdung R, Valliyott L, Pilankatta R. Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013–2015 in northern Kerala, India. PeerJ. 2017;5: e2970. doi: 10.7717/peerj.2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senaratne UTN, Murugananthan K, Sirisena PDNN, Carr JM, Noordeen F. Dengue virus co-infections with multiple serotypes do not result in a different clinical outcome compared to mono-infections. Epidemiol Infect. 2020;148: e119. doi: 10.1017/S0950268820000229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beaty BJ, Farfán JA, Rosado-Paredes EP, Loroño-Pino MA, Flores-Flores LF, Vorndam AV, et al. Common occurrence of concurrent infections by multiple dengue virus serotypes. Am J Trop Med Hyg. 1999;61: 725–730. doi: 10.4269/ajtmh.1999.61.725 [DOI] [PubMed] [Google Scholar]
- 43.Vazeille M, Gaborit P, Mousson L, Girod R, Failloux A-B. Competitive advantage of a dengue 4 virus when co-infecting the mosquito Aedes aegypti with a dengue 1 virus. BMC Infect Dis. 2016;16: 318. doi: 10.1186/s12879-016-1666-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thongrungkiat S, Maneekan P, Wasinpiyamongkol L, Prummongkol S. Prospective field study of transovarial dengue-virus transmission by two different forms of Aedes aegypti in an urban area of Bangkok, Thailand. J Vector Ecol. 2011;36: 147–152. doi: 10.1111/j.1948-7134.2011.00151.x [DOI] [PubMed] [Google Scholar]
- 45.Scott TW, Amerasinghe PH, Morrison AC, Lorenz LH, Clark GG, Strickman D, et al. Longitudinal Studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: Blood Feeding Frequency. J Med Entomol. 2000;37: 89–101. doi: 10.1603/0022-2585-37.1.89 [DOI] [PubMed] [Google Scholar]
- 46.Yoon I-K, Getis A, Aldstadt J, Rothman AL, Tannitisupawong D, Koenraadt CJM, et al. Fine Scale Spatiotemporal Clustering of Dengue Virus Transmission in Children and Aedes aegypti in Rural Thai Villages. Barrera R, editor. PLoS Negl Trop Dis. 2012;6: e1730. doi: 10.1371/journal.pntd.0001730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mammen MP, Pimgate C, Koenraadt CJM, Rothman AL, Aldstadt J, Nisalak A, et al. Spatial and Temporal Clustering of Dengue Virus Transmission in Thai Villages. Riley S, editor. PLoS Med. 2008;5: e205. doi: 10.1371/journal.pmed.0050205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sithiprasasna R, Patpoparn S, Attatippaholkun W, Suvannadabba S, Srisuphanunt M. The geographic information system as an epidemiological tool in the surveillance of dengue virus-infected Aedes mosquitos. Southeast Asian J Trop Med Public Health. 2004;35: 9. [PubMed] [Google Scholar]
- 49.Siriyasatien P, Pengsakul T, Kittichai V, Phumee A, Kaewsaitiam S, Thavara U, et al. Identification of blood meal of field caught Aedes aegypti (L.) by multiplex PCR. Southeast Asian J Trop Med Public Health. 2010;41: 6. [PubMed] [Google Scholar]
- 50.Hutamai S, Suwonkerd W, Suwannchote N, Somboon P, Prapanthadara L. A survey of dengue viral infection in Aedes aegypti and Aedes albopictus from re-epidemic areas in the North of Thailand using nucleic acid sequence based amplification assay. Southeast Asian J Trop Med Public Health. 2007;38: 8. [PubMed] [Google Scholar]
- 51.Teerasut C, Petphuwadee U, Thammapalo S, Jampangern W, Limkittikul K. Identification of dengue virus in Aedes mosquitoes and patients’ sera from Si Sa Ket province, Thailand. Southeast Asian J Trop Med Public Health. 2012;43: 5. [PubMed] [Google Scholar]
- 52.Sun J, Wu D, Zhou H, Zhang H, Guan D, He X, et al. The epidemiological characteristics and genetic diversity of dengue virus during the third largest historical outbreak of dengue in Guangdong, China, in 2014. J Infect. 2016;72: 80–90. doi: 10.1016/j.jinf.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, Mammen MP, Chinnawirotpisan P, Klungthong C, Rodpradit P, Monkongdee P, et al. Clade Replacements in Dengue Virus Serotypes 1 and 3 Are Associated with Changing Serotype Prevalence. J Virol. 2005;79: 15123–15130. doi: 10.1128/JVI.79.24.15123-15130.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jarman RG, Holmes EC, Rodpradit P, Klungthong C, Gibbons RV, Nisalak A, et al. Microevolution of Dengue Viruses Circulating among Primary School Children in Kamphaeng Phet, Thailand. J Virol. 2008;82: 5494–5500. doi: 10.1128/JVI.02728-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su C-L, Huang J-H, Shu P-Y, Chang M-C, Hu H-C, Chang S-F, et al. Molecular Characterization of Dengue Viruses Imported Into Taiwan during 2003–2007: Geographic Distribution and Genotype Shift. Am J Trop Med Hyg. 2009;80: 1039–1046. doi: 10.4269/ajtmh.2009.80.1039 [DOI] [PubMed] [Google Scholar]
- 56.Huang L, Luo X, Shao J, Yan H, Qiu Y, Ke P, et al. Epidemiology and characteristics of the dengue outbreak in Guangdong, Southern China, in 2014. Eur J Clin Microbiol Infect Dis. 2016;35: 269–277. doi: 10.1007/s10096-015-2540-5 [DOI] [PubMed] [Google Scholar]
- 57.Haryanto S, Hayati RF, Yohan B, Sijabat L, Sihite IF, Fahri S, et al. The molecular and clinical features of dengue during outbreak in Jambi, Indonesia in 2015. Pathog Glob Health. 2016;110: 119–129. doi: 10.1080/20477724.2016.1184864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wardhani P, Aryati A, Yohan B, Trimarsanto H, Setianingsih TY, Puspitasari D, et al. Clinical and virological characteristics of dengue in Surabaya, Indonesia. Munderloh UG, editor. PLOS ONE. 2017;12: e0178443. doi: 10.1371/journal.pone.0178443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Megawati D, Masyeni S, Yohan B, Lestarini A, Hayati RF, Meutiawati F, et al. Dengue in Bali: Clinical characteristics and genetic diversity of circulating dengue viruses. Gubler DJ, editor. PLoS Negl Trop Dis. 2017;11: e0005483. doi: 10.1371/journal.pntd.0005483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang C-F, Su C-L, Hsu T-C, Lin C-C, Shu P-Y, Liao T-L, et al. Molecular Characterization and Phylogenetic Analysis of Dengue Viruses Imported into Taiwan during 2008–2010. Am J Trop Med Hyg. 2012;87: 349–358. doi: 10.4269/ajtmh.2012.11-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang C-F, Chang S-F, Hsu T-C, Su C-L, Wang T-C, Lin S-H, et al. Molecular characterization and phylogenetic analysis of dengue viruses imported into Taiwan during 2011–2016. Blacksell SD, editor. PLoS Negl Trop Dis. 2018;12: e0006773. doi: 10.1371/journal.pntd.0006773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klungthong C, Zhang C, Mammen MP, Ubol S, Holmes EC. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology. 2004;329: 168–179. doi: 10.1016/j.virol.2004.08.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)
(accessed from GenBank, the National Centre for Biotechnology Information https://www.ncbi.nlm.nih.gov/genbank/).
(DOCX)
The molecular characteristics of 24 pairs of DENV positive samples were shown in this table. Gray row label represents serotype matched between patients and mosquitoes from their resident area.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.