Abstract
Genetic screens for modifiers of activated Ras phenotypes have identified a novel protein, kinase suppressor of Ras (KSR), which shares significant sequence homology with Raf family protein kinases. Studies using Drosophila melanogaster and Caenorhabditis elegans predict that KSR positively regulates Ras signaling; however, the function of mammalian KSR is not well understood. We show here that two predicted kinase-dead mutants of KSR retain the ability to complement ksr-1 loss-of-function alleles in C. elegans, suggesting that KSR may have physiological, kinase-independent functions. Furthermore, we observe that murine KSR forms a multimolecular signaling complex in human embryonic kidney 293T cells composed of HSP90, HSP70, HSP68, p50CDC37, MEK1, MEK2, 14-3-3, and several other, unidentified proteins. Treatment of cells with geldanamycin, an inhibitor of HSP90, decreases the half-life of KSR, suggesting that HSPs may serve to stabilize KSR. Both nematode and mammalian KSRs are capable of binding to MEKs, and three-point mutants of KSR, corresponding to C. elegans loss-of-function alleles, are specifically compromised in MEK binding. KSR did not alter MEK activity or activation. However, KSR-MEK binding shifts the apparent molecular mass of MEK from 44 to >700 kDa, and this results in the appearance of MEK in membrane-associated fractions. Together, these results suggest that KSR may act as a scaffolding protein for the Ras-mitogen-activated protein kinase pathway.
Ras activation is an essential step in signaling in response to a variety of extracellular signals, including receptor tyrosine kinase ligands which bind and activate their corresponding tyrosine kinase receptors. Activation of receptor tyrosine kinases leads to activation of Ras via the action of specific guanine nucleotide exchange factors. Activated Ras can physically interact with numerous downstream targets and activate several different signaling pathways (15).
One of the best-characterized Ras signaling pathways is the Raf-MEK-ERK pathway, also known as the mitogen-activated protein (MAP) kinase cascade (20, 25). Ras directly binds Raf in a GTP-dependent manner, and this interaction appears to be critical for recruitment of Raf to the membrane, where it undergoes activation. Activated Raf directly phosphorylates and activates MEK, also known as MAP kinase kinase, which in turn directly phosphorylates and activates ERK (25). Activation of ERK is critical for numerous Ras-induced cellular responses, including transcriptional activation of a number of genes (11). Ternary complex factors (TCFs) are among the best-characterized physiological substrates of ERK. Activated ERK directly phosphorylates and thereby activates the transcription activation potential of TCFs (8, 12, 18). It appears that TCFs, in association with the serum response factor, play an essential role in the activation of many mitogen-inducible genes (11).
The Ras-MAP kinase pathway is highly conserved in eukaryotes. Genetic studies of Caenorhabditis elegans and Drosophila melanogaster indicate that this pathway is involved in many developmental programs (15). Genetic screens for mutations that suppress constitutively active Ras mutants have identified numerous components of the MAP kinase pathway. Such screens identified KSR (for kinase suppressor of Ras), a putative protein kinase with significant sequence identity to Raf (17, 30, 31). Genetic data indicated that KSR plays a positive role in Ras signaling and functions parallel to, or downstream of, Ras. Microinjection experiments using Xenopus oocytes showed that KSR is able to enhance Ras-induced germinal vesicle breakdown and MAP kinase activation, indicating that KSR has a positive role in Ras signaling in vertebrates (32). However, recent reports suggest that KSR may have a negative role in certain aspects of Ras signaling (6, 14, 29, 39). KSR was found to inhibit Ras-induced cellular transformation in NIH 3T3 cells and to inhibit MEK and/or ERK activation (6, 14, 39). However, the mechanism of this inhibition remains to be elucidated. We have previously reported that KSR selectively inhibits TCF phosphorylation and transcription activity, without significantly affecting ERK activation in COS1 cells (29). Therefore, the biochemical function of KSR in mammalian cells is rather perplexing, though KSR clearly plays a role in Ras-MAP kinase signal transduction.
The deduced amino acid sequence of KSR predicts it to be a protein kinase, and one report has suggested that KSR is a ceramide-activated protein kinase (40). However, whether KSR has intrinsic kinase activity remains to be confirmed. It has been proposed that KSR may function as a scaffold protein to assemble a signaling complex in mammalian cells (14, 39). Some limited data exist to support this notion. Previous studies have suggested that KSR can interact directly with 14-3-3 proteins, MEK, and possibly ERK (6, 14, 37, 39). In addition, KSR translocates to membrane fractions and associates with Raf (perhaps indirectly) in a Ras-dependent manner (22, 37).
We tested the hypothesis that KSR may function independently of its presumed protein kinase activity. Surprisingly, we observed that upon microinjection, kinase domain mutants of KSR complement KSR loss-of-function alleles in C. elegans vulval induction. With this in mind, we turned our attention to the mouse homolog of KSR (mKSR), testing whether kinase-independent functions of mKSR exist in mammalian cells and whether they are sensitive to mutations analogous to genetically isolated loss-of-function alleles. Consistent with this possibility, we observed that KSR is a component of a multimolecular complex in human embryonic kidney (HEK) 293T cells consisting of MEK1, MEK2, HSP90, HSP70, HSP68, p50CDC37, and 14-3-3 in addition to several other, unidentified proteins. ERK also associates with KSR, though this binding appears to be much weaker than KSR-MEK interactions. A loss-of-function mutant, KSR C809Y, specifically lacks the ability to interact with MEK yet retains the ability to bind other KSR-associated proteins. Interestingly, ectopic KSR expression alters the apparent molecular mass of MEK from 44 kDa to approximately 700 kDa and results in the translocation of MEK from a soluble to a membrane-associated fraction. Treatment of PC12 cells with nerve growth factor (NGF) resulted in induction of KSR protein levels and a concomitant increase in KSR-MEK association, suggesting that KSR may play a role in vertebrate differentiation. These data suggest that KSR may function as a scaffolding protein in vivo.
MATERIALS AND METHODS
C. elegans strains and plasmids.
Standard methods were used for the handling and culture of animals (2). Mutations used were let-60(n1046) (G13E) (1, 9), ksr-1(ku68) (R531H) (29), and ksr-1(n2526) (W255STOP) (17). The ksr-1 transgenes pMS44 (wild type), pMS77 (K503M), pMS108 (D618A), and pMS153 (R531H) consist of genomic positions 1809 (BamHI) to 15401 (BamHI) from cosmid F13B9 (GenBank accession no. U39853), followed by cDNA positions 520 (BamHI) to 2394 (ClaI) from ksr-1 (GenBank accession no. U38820), followed by genomic positions 19965 (ClaI) to 21454 (SacII) from cosmid F13B9, in pBluescript SK(+). Point mutations were introduced into the cDNA fragment by PCR. For in vitro translation, C. elegans KSR-1 cDNAs were cloned into the BamHI site pcDNA3-HA (29) to create either pMS82 or pMS83. C. elegans KSR-1b (CeKSR-1b; pMS183) consists of the entire cDNA; CeKSR-1a (pMS182) begins from an alternate initiation methionine at position 189 and may be a naturally occurring variant (29a).
Cell culture.
HEK 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS; Life Technologies). Transfections were performed by using Lipofectamine (Life Technologies) as directed by the manufacturer. Rat PC12 cells were cultured in DMEM containing 10% horse serum and 5% FBS and were induced to differentiate by incubating in DMEM containing 2% horse serum, 1% FBS, and 50 ng of NGF (Calbiochem) per ml for 5 days, with a change of medium every 48 h.
Plasmids and protein expression.
To construct a yeast expression vector for two-hybrid screening, the entire open reading frame of mKSR was subcloned into the bait vector plex-Ade (kindly provided by Anne Vojtek), creating an in-frame fusion with LexA. To construct Lex-Ade-KSR KD and Lex-Ade-KSR KD C809Y fusions, a 1,188-bp EcoRV fragment from pcDNA3-HA-KSR or pcDNA3-HA-KSR C809Y (see below) was ligated into plex-Ade that had been digested with BamHI and blunted with Klenow polymerase. MEK1-Gal4 activation domain fusions were constructed by ligating the entire open reading frame of human MEK1 or MEK1Δproline rich (kindly provided by Melanie Cobb) into pGAD10 (Clontech). Expression vectors encoding point mutants of murine KSR were constructed by site-directed mutagenesis of pcDNA3-HA-KSR (29), using a Quick Change kit (Stratagene). pcDNA3-HA-KSR 1-301 was constructed by amplifying amino acids 1 to 301 of mKSR via PCR and subcloning into the XbaI-EcoRI sites of pcDNA3-HA (29). HA-KSR KD was constructed by amplifying the region coding for amino acids 500 to the stop codon of mKSR and subcloning as an XbaI-EcoRI fragment into pcDNA3-HA. pcDNA3-HA-MEK2 was constructed by subcloning a BamHI fragment containing the entire open reading frame of human MEK2 (41) into the BamHI site of pcDNA3-HA. Expression vectors for Elk-1 have been described elsewhere (28).
Glutathione S-transferase (GST)–MEK1, GST-ERK1, and a kinase-dead mutant, GST-ERK1 KR, were expressed in Escherichia coli and purified as described previously (28, 41). GST-CeMEK2 was expressed and purified as described elsewhere (36b).
Metabolic labeling, immunoprecipitation, gel filtration chromatography, and subcellular fractionation.
Subconfluent 293T cells in 10-cm-diameter dishes were transfected with 10 μg of the indicated expression vector; 48 h posttransfection, cells were labeled for 4 h in cysteine- and methionine-free DMEM–10% dialyzed FBS (Life Technologies) containing [35S]methionine/cysteine at 200 μCi/ml (Tran35S-label; ICN). Cells were lysed in buffer (10 mM Tris-Cl [pH 7.5], 150 mM NaCl, 50 mM NaF, 1 mM EDTA, 1% Nonidet P-40 [NP-40]) containing 1 mM dithiothreitol [DTT], 1 mM NaVO4, and a protease inhibitor cocktail (Complete; Boehringer Mannheim) for 20 min at 4°C. Precleared lysates were incubated for 3 h at 4°C with 10 μg of antibody precoupled to protein G-Sepharose (Pharmacia) or protein A-agarose (Pierce). For peptide competition, antibody was preincubated with hemagglutinin (HA) peptide (YPYDVPDYA) before addition to lysates. Immunocomplexes were collected by gentle centrifugation, washed five times in lysis buffer, boiled in sodium dodecyl sulfate (SDS) sample, and separated by SDS-polyacrylamide gel electrophoresis (PAGE). For KSR immunoprecipitation experiments using PC12 cells, 1 mg of extract from PC12 cells treated with or without NGF for 5 days was immunoprecipitated with 10 μg of KSR-specific antibody (α-KSR; Santa Cruz Biotechnology) in 1% NP-40–150 mM NaCl lysis buffer. Where indicated, antibody was incubated with a twofold excess of immunizing peptide prior to immunoprecipitation.
To determine the effects of geldanamycin on KSR protein levels, 293T cells transfected with HA-KSR were labeled for 30 min with 100 μCi of [35S]methionine/cysteine per ml with or without 10 μM geldanamycin (Sigma), followed by a chase with complete medium for the indicated times with methionine/cysteine-containing medium. Lysates were prepared and immunoprecipitated as described above with α-HA followed by SDS-PAGE and autoradiography.
For GST pull-down experiments, full-length CeKSR and mKSR cDNAs were translated in vitro (Promega) in the presence of [35S]methionine. A portion of the translation reaction mixture was incubated with 5 μg of either GST, GST-human MEK1, or GST-CeMEK2 bound to glutathione-Sepharose (Pharmacia) for 1 h at 4°C in 1% NP-40–150 mM NaCl buffer followed by four washes, SDS-PAGE, and autoradiography.
For gel filtration experiments, two 10-cm-diameter dishes of 293T cells, transfected as described above, were lysed by homogenization in phosphate-buffered saline (PBS) plus protease and phosphatase inhibitors. Cellular debris was pelleted by centrifugation in a microcentrifuge at 6,000 × g for 15 min at 4°C. The resulting supernatant (about 1 mg of protein) was fractionated on a Superose 6-HR column (Pharmacia) equilibrated in PBS at a flow rate of 0.5 ml/min. Fractions of 333 μl, 15 μl of which was subjected to SDS-PAGE and immunoblot analysis with the indicated antibodies, were collected.
For subcellular fractionation, transfected 293T cells were lysed by homogenization in PBS containing protease and phosphatase inhibitors. Cellular debris was removed by centrifugation at 6,000 × g for 15 min at 4°C. The resulting supernatant was centrifuged for 1 h at 100,000 × g at 4°C. The microsomal pellet (P100) was resuspended in buffer containing 0.5% NP-40 followed by centrifugation at 13,000 × g to fully remove remaining insoluble material. Equal volumes of each fraction were examined by SDS-PAGE and immunoblotting with the indicated antibodies.
Peptide sequencing.
α-HA immunocomplexes from approximately 5 × 108 293T cells transfected with HA-KSR were subjected to SDS-PAGE and Coomassie blue staining. Bands were excised, digested with lysyl endopeptidase, purified via reversed-phase high-performance liquid chromatography (HPLC), and sequenced as described previously (38).
Kinase assays.
KSR or MEK immunoprecipitates were equilibrated in buffer (25 mM HEPES [pH 8.0], 0.5 mM EDTA, 0.25% β-mercaptoethanol) before addition of 1 μg of GST-ERK1 or GST-ERK1 KR in 10 mM HEPES (pH 8.0)–10 mM MgCl2–1 mM DTT–50 μM ATP in a volume of 30 μl. GST-ERK1 or GST-ERK1 KR activation was allowed to proceed for 15 min at 30°C, after which 5-μl aliquots were removed and assayed for ERK activity for 15 min at 30°C in 10 mM HEPES (pH 8.0)–10 mM MgCl2–1 mM DTT–50 μM ATP–10 μCi of [γ-32PO4]ATP (ICN) and 10 μg of myelin basic protein (MBP; Sigma). Reactions were terminated by adding EDTA (pH 8.0) to 50 mM, and reaction mixtures were spotted on P81 phosphocellulose paper (Whatman), washed, and counted in a scintillation counter.
Two-hybrid interactions and immunoblotting.
Two-hybrid screening using mKSR as a bait was carried out essentially as described elsewhere (35). α-14-3-3 and α-KSR were from Santa Cruz Biotechnology; α-HA was from Babco; α-phospho-MEK antibody was from New England Biolabs; α-MEK and α-ERK have been described elsewhere (42); and α-HSP90 was from Transduction Laboratories. Blots were developed by using enhanced chemiluminescence (Amersham).
RESULTS
Integrity of the KSR kinase domain is not required for its biological function in C. elegans vulval induction.
Mutations in the C. elegans ksr-1 gene were isolated as suppressors of the Multivulva (Muv) phenotype caused by activated Ras (17, 30), suggesting that in C. elegans, the KSR-1 protein normally plays a positive role in Ras-mediated signaling. Of the 12 ksr-1 mutations originally described (17, 30), 8 are missense mutations affecting the putative kinase domain; this finding suggested that kinase activity could be important for KSR-1 function. To test directly the importance of kinase activity, we constructed ksr-1 transgenes bearing substitutions predicted to eliminate kinase activity and tested these transgenes for the ability to complement the suppressor phenotype caused by ksr-1 loss-of-function mutations. In these assays, complementation is observed as a restoration of the activated Ras Muv phenotype. We created a mutant in the Mg2+-ATP binding motif (K503M) as well as a mutant lacking the catalytic nucleophile aspartic acid (D618A). These residues are highly conserved in protein kinases, and mutations analogous to these in other protein kinases have been used to create kinase-inactive molecules. Surprisingly, both of the kinase-dead transgenes, KSR-1 K503M and KSR-1 D618A, retained complementing activity comparable to that of wild-type KSR-1 (Table 1). However, a KSR-1 R531H transgene (bearing the same substitution caused by the endogenous ksr-1 allele ku68) lacked complementing activity. These data argue that KSR-1 does not require kinase activity to promote Ras signaling and are inconsistent with models in which KSR acts as a protein kinase. They do, however, raise the possibility that KSR has other biochemical functions.
TABLE 1.
The putative kinase activity of KSR is not required for its ability to function in C. elegans vulval inductiona
| Chromosomal genotype | KSR-1 protein encoded by transgene | % Muv (n)
|
No. of independent transgenic lines | Complementation | |
|---|---|---|---|---|---|
| F1 | F2 | ||||
| let-60(n1046gf); ksr-1(ku68) | None | 9 (107) | 7 (153) | 5 | − |
| let-60(n1046gf); ksr-1(ku68) | WT | 39 (72) | 89 (182) | 8 | + |
| let-60(n1046gf); ksr-1(ku68) | K503M | 52 (65) | 70 (276) | 9 | + |
| let-60(n1046gf); ksr-1(ku68) | D618A | 54 (93) | 59 (160) | 7 | + |
| let-60(n1046gf); ksr-1(ku68) | R531H | 1 (150) | 0 (57)b | 3 | − |
| let-60(n1046gf); ksr-1(n2526) | None | 12 (73) | 17 (167)b | 3 | − |
| let-60(n1046gf); ksr-1(n2526) | WT | 70 (130) | 87 (250) | 8 | + |
| let-60(n1046gf); ksr-1(n2526) | D618A | 66 (79) | 66 (181) | 8 | + |
| let-60(n1046gf); ksr-1(n2526) | K503M | 67 (43) | 63 (269) | 8 | + |
Each ksr-1 transgene (5 to 10 ng/μl) was coinjected with the marker plasmid pRF4 (65 to 100 ng/μl), which causes a dominant Roller phenotype (21). F1 and F2 Roller progeny were scored for the Muv phenotype by using a dissecting microscope as described previously (9). F2 data from multiple independent transgenic lines were pooled. Mutations used were let-60(n1046) (G13E) (1, 9), ksr-1(ku68) (R531H) (29), and ksr-1(n2526) (W255STOP) (17). The ksr-1 transgenes (encoding either wild-type [WT], K503M, D618A, or R531H KSR-1) consist of genomic positions 1809 (BamHI) to 15401 (BamHI) from cosmid F13B9 (GenBank accession no. U39853), followed by cDNA positions 520 (BamHI) to 2394 (ClaI) from ksr-1 (GenBank accession no. U38820), followed by genomic positions 19965 (ClaI) to 21454 (SacII) from cosmid F13B9, in pBluescript SK(+).
Includes F2 and F3 animals.
KSR is a component of a high-molecular-weight complex in vivo.
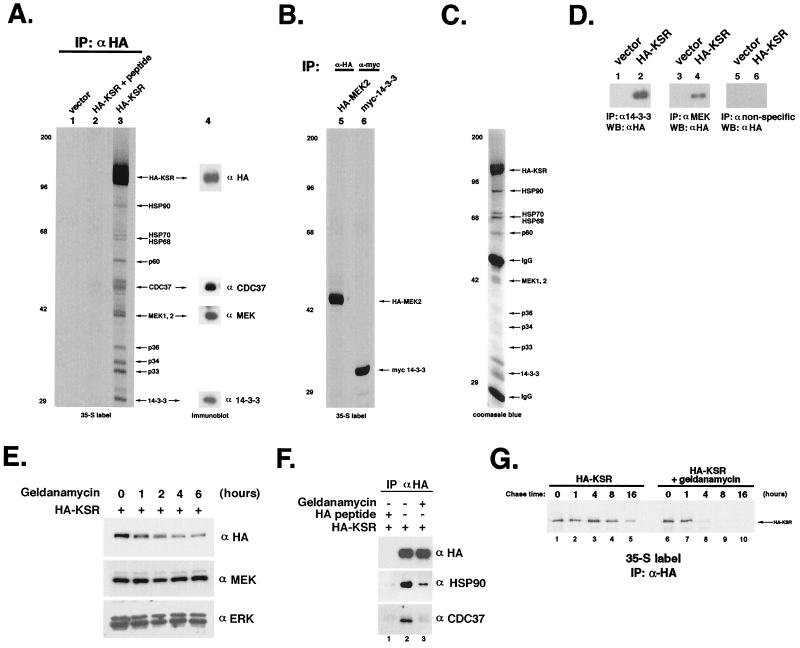
The above results, in addition to repeated failed attempts to detect intrinsic kinase activity in vitro, led us to test whether any other biochemical function could be attributed to KSR. We therefore performed immunoprecipitation experiments using transfected HEK 293T cells expressing HA-tagged mKSR to identify KSR-associated proteins. When 35S-labeled α-HA immunocomplexes were subjected to SDS-PAGE and autoradiography, we observed that several proteins associated specifically with HA-KSR (Fig. 1A, lane 3). Inclusion of HA peptide prior to immunoprecipitation competed all coprecipitated proteins, demonstrating the specificity of the immunoprecipitation (Fig. 1A, lane 2). These coimmunoprecipitated bands were not due to C-terminal proteolytic degradation of HA-KSR, as immunoblot analysis with α-HA detected only KSR. In fact, several of the KSR-associated bands have been identified as distinct cellular proteins, as opposed to fragments of KSR (see below). In addition, certain KSR-associated proteins can be coprecipitated by N- or C-terminal truncations of KSR, which themselves migrate faster than some of the KSR-associated proteins (Fig. 2A). We obtained similar results using COS1 cells or with a Myc-tagged KSR expression vector. These results demonstrate that KSR specifically associates with numerous cellular proteins.
FIG. 1.
KSR specifically associates with a number of proteins in vivo. (A) Metabolic labeling and immunoprecipitation (IP) of transfected mouse KSR. 293T cells were transfected with HA-KSR (lanes 2 and 3) or vector (lane 1), labeled with [35S]Met/Cys, and immunoprecipitated with α-HA (lanes 1 and 3) or with α-HA that had been preincubated with competitor peptide (lane 2). Immunocomplexes were separated by SDS-PAGE and visualized by autoradiography. Lane 4 represents an immunoblot of lane 3 probed with α-HA, α-p50CDC37, α-MEK, and α-14-3-3 as indicated. Positions of size markers are indicated in kilodaltons on the left. (B) Metabolic labeling and immunoprecipitation of transfected MEK and 14-3-3. 293T cells, transfected with HA-MEK2 or Myc-14-3-3 β, were processed as for panel A, separated by SDS-PAGE, and visualized by autoradiography. (C) Coomassie blue staining of HA-KSR complex affinity purified from 293T cells as for panel A. (D) HA-KSR is present in MEK1/2 and 14-3-3 immunoprecipitates. 293T cells transfected with HA-KSR were subjected to IP with α-14-3-3 (lanes 1 and 2), α-MEK1/2 (lanes 3 and 4), or control antibody (lanes 5 and 6) as described above, followed by Western blotting (WB) with α-HA to detect HA-KSR. (E) Reduction of KSR protein levels in response to geldanamycin treatment. HEK 293T cells transfected with HA-KSR were treated with 10 μM geldanamycin for the indicated times before harvesting and immunoblotting of whole-cell lysates with α-HA (top), α-MEK (middle), and α-ERK (bottom). (F) Geldanamycin treatment selectively disrupts the association of KSR with HSP90 and p50CDC37. HEK 293T cells transfected with HA-KSR were treated with 10 μM geldanamycin for 1 h (lane 3) before harvesting and immunoprecipitation with α-HA followed by immunoblotting with α-HA (top), α-HSP90 (middle), and α-p50CDC37 (bottom). HA peptide was included (lane 1) to demonstrate the specificity of the IP. (G) Geldanamycin destabilizes KSR. HA-KSR-transfected 293T cells were labeled for 30 min with [35S]Met/Cys with or without geldanamycin. Cells were chased for the indicated times and then immunoprecipitated with α-HA. Cells that had been treated with geldanamycin (lanes 6 to 10) showed a rapid loss of 35S-labeled HA-KSR. All results in this and subsequent figures are representative examples of at least three independent experiments except for Fig. 3A (performed twice).
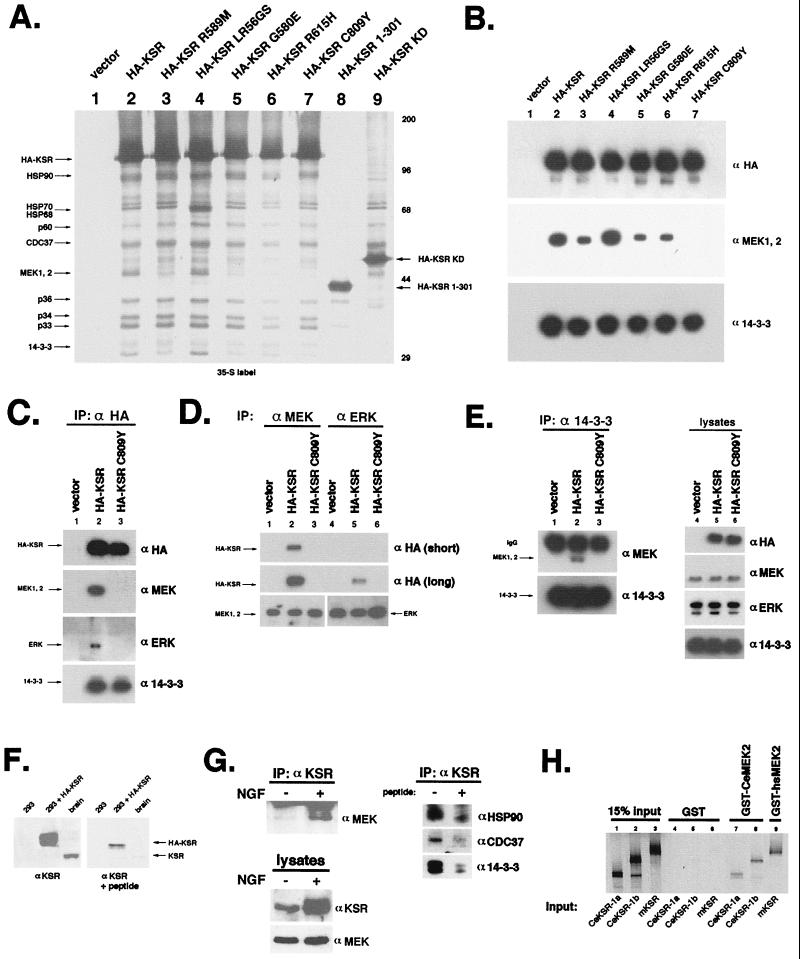
FIG. 2.
Mutant KSRs, corresponding to loss-of-function alleles in C. elegans, are compromised in MEK1/2 binding. (A) Metabolic labeling and immunoprecipitation (IP) of mutant KSRs. 293T cells were transfected with vector (lane 1), wild-type KSR (lane 2), or mutant HA-tagged KSR (lanes 3 to 9). Metabolically labeled lysates were subjected to IP with α-HA as for Fig. 1A prior to SDS-PAGE and autoradiography. Positions of size markers are indicated in kilodaltons on the right. (B) Loss-of-function mutant KSR C809Y binds 14-3-3 but not MEK1/2. 293T cells were transfected as for panel A and then immunoprecipitated with α-HA. Immunocomplexes were separated by SDS-PAGE and immunoblotted with α-HA (to detect HA-KSR; top) α-MEK1/2 (middle), and α-14-3-3 (bottom). KSR C809Y showed no detectable MEK binding, while KSR R589M, G580E, and R615H displayed reduced MEK binding. (C) KSR forms a signaling complex in vivo with ERK and MEK. 293T cells were transfected with vector (lane 1), wild-type KSR (lane 2), or HA-KSR C809Y (lane 3) and then immunoprecipitated with α-HA prior to SDS-PAGE and immunoblotting with α-HA, α-MEK, α-ERK, and α-14-3-3 as indicated. Note that we reproducibly detect far more MEK1/2 (>10-fold) than ERK1/2 associated with HA-KSR in these cells. (D) Comparison of MEK-KSR and ERK-KSR association in vivo. 293T cells, transfected as for panel A, were lysed, immunoprecipitated with α-MEK (lanes 1 to 3) or α-ERK (lanes 4 to 6), and then immunoblotted with α-HA (to detect KSR; upper two panels), α-ERK (lower panel, lanes 4 to 6), and α-MEK (lower panel, lanes 1 to 3). Two exposures of the α-HA blot are presented to emphasize the relative amounts of KSR in MEK and ERK immunoprecipitates. (E) MEK-14-3-3 association is mediated by KSR and dependent on cysteine 809. Lysates were immunoprecipitated with α-14-3-3 and then immunoblotted with α-MEK (upper panel, lanes 1 to 3) and α-14-3-3 (lower panel). Representative immunoblot of whole-cell extracts shows expression of transfected HA-KSRs and endogenous MEK, ERK, and 14-3-3 (lanes 4 to 6). IgG, immunoglobulin G. (F) Detection of endogenous KSR in brain and PC12 cells. Extracts from vector- or HA-KSR-transfected 293T cells or mouse brain were probed with affinity-purified α-KSR. Reactive bands were effectively competed by competition by peptide antigen. (G) Association of endogenous KSR with MEK, HSP90, p50CDC37, and 14-3-3 in PC12 cells. Lysates from undifferentiated or day 5 differentiated PC12 cells were blotted with α-KSR or α-MEK as indicated. KSR is induced by NGF treatment. Lysates from differentiated PC12 cells were immunoprecipitated with α-KSR and then immunoblotted with α-MEK, α-HSP90, α-p50CDC37, and α-14-3-3. Where indicated, immunizing peptide was included in the IP. (H) MEK binding is a conserved function of KSR. 35S-labeled in vitro-translated CeKSR-1a and CeKSR-1b (see Materials and Methods) were incubated with either GST- or GST-CeMEK2 coupled to glutathione-Sepharose. After mixing for 1 h, beads were collected by centrifugation and washed several times in buffer before elution with glutathione. Eluted proteins were separated by SDS-PAGE and subjected to autoradiography. Mammalian KSR and GST-MEK were included for comparison. Note that CeMEK2 and CeKSR-1 can associate as effectively as their mammalian counterparts.
We reproducibly detect several major HA-KSR-associated proteins in immunoprecipitation experiments; a number of these proteins (p90, p70, p68, p60, p50, p46, p44, and p30) are present in near-stoichiometric amounts with respect to one another and can be detected by Coomassie blue staining of as few as 107 transfected cells (Fig. 1C). Size exclusion chromatography demonstrated that transfected KSR has a molecular mass of about 106 Da (Fig. 3A), suggesting the possibility that KSR is a constituent of a multiprotein complex in vivo. These observations are supported by the fact that a significant portion of KSR-associated proteins remains bound even under high-salt conditions (1 M NaCl) or in the presence of 0.1% SDS (data not shown).
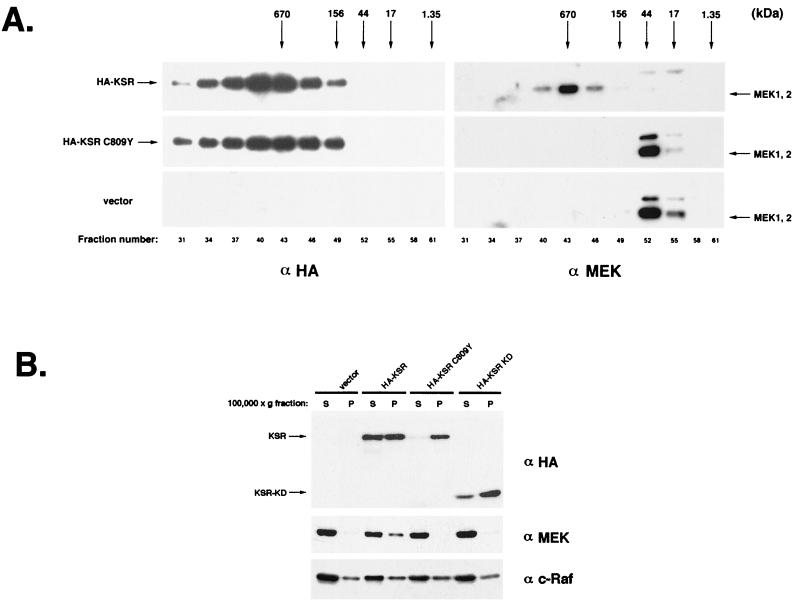
FIG. 3.
KSR exists in a high-molecular-weight complex in vivo. (A) HA-KSR recruits MEK to a large complex. HA-KSR-, HA-KSR C809Y-, or vector-transfected 293T cells were homogenized, and the resulting lysates were fractionated on a Superose 6 column (Pharmacia). Equal volumes of each fraction were subjected to SDS-PAGE followed by immunoblotting with α-HA (left) and α-MEK (right). Elution profiles of molecular weight standards are indicated (thyroglobulin, 670 kDa; gamma globulin, 156 kDa; ovalbumin, 44 kDa; myoglobin, 17 kDa; and cobalamin, 1.35 kDa; Bio-Rad Laboratories). Endogenous MEK1/2 exhibits an apparent molecular mass of 700 kDa only in the presence of HA-KSR (top). In contrast, endogenous MEK elutes at approximately 44 kDa in cells transfected with vector or KSR C809Y (bottom and middle). (B) KSR alters the cellular distribution of MEK. HA-KSR-, HA-KSR C809Y-, or HA-KSR KD-transfected 293T cells were Dounce homogenized and then centrifuged at 6,000 × g for 15 min. The resulting supernatant was spun at 100,000 × g for 60 min to separate it into soluble (S) and membrane (P) fractions. Equal volumes of each fraction were subjected to SDS-PAGE and immunoblot analysis with α-HA, α-MEK, and α-c-Raf.
MEK1, MEK2, 14-3-3, HSP90, HSP70, HSP68, and p50CDC37 are components of the KSR complex.
Previous reports have indicated that KSR is capable of associating with MEK1 and -2 as well as 14-3-3 proteins (6, 37, 39). We have also isolated 14-3-3 β and MEK2 in yeast two-hybrid screens using as baits full-length mKSR and the kinase domain of KSR, respectively (data not shown). We therefore tested whether MEK1, MEK2, or 14-3-3 proteins were present in our KSR immunoprecipitates. Immunoblot analysis using α-MEK1/2 clearly detected the presence of a tightly spaced pair of immunoreactive bands corresponding to MEK1 and MEK2, respectively (Fig. 1A, lane 4). The presence of MEK in the KSR complex was also demonstrated by direct peptide sequencing (Table 2). The specificity of the KSR-MEK interaction was confirmed by reciprocal coimmunoprecipitation. α-MEK specifically precipitated HA-KSR, which was not precipitated by a nonspecific antibody (Fig. 1D, lanes 3 to 6).
TABLE 2.
Peptide sequence analysis of KSR-associated proteinsa
| Molecular weight (106) | Peptide sequence | Identity |
|---|---|---|
| 90 | HNDDEQYADE | HSP90 |
| VILHLK | ||
| 70 | KRQAVTNPNNTF | HSP70 |
| L?QDFFNGRDLNK | ||
| KEDIERMVQEA | ||
| 68 | KEEIERMVQEA | HSP68 |
| 44/43 | KNPAERADLK | MEK1/2 |
| KRLERFLTQK | ||
| KKLEELELDEQ | ||
| KGLTYREK | ||
| 30 | KNVIGARRASW | 14-3-3 |
α-HA immunoprecipitates from 5 × 108 transfected cells were subjected to SDS-PAGE followed by silver staining. Bands corresponding to KSR-associated proteins were excised, digested with lysyl endopeptidase, and purified via reversed-phase HPLC (38), and the amino acid sequence was determined via Edman degradation. Peptide sequence identities were determined by using the Blast-P database search program (National Center for Biotechnology Information).
Similarly, we also probed HA-KSR immunoprecipitates with α-14-3-3. We detected immunoreactive bands that correspond well with the predicted molecular mass of 14-3-3 proteins, approximately 30 kDa (Fig. 1A, lane 4). Immunoprecipitation with α-14-3-3 also demonstrated the associated HA-KSR (Fig. 1D, lane 2). By peptide sequence analysis, we demonstrated that the 30-kDa KSR-associated protein is a 14-3-3 protein (Table 2).
In marked contrast to HA-KSR-transfected cells, no specific proteins were associated with HA-MEK2 in immunocomplexes in the absence of KSR transfection under identical conditions (Fig. 1B, lane 1). Likewise, Myc-14-3-3 β immunocomplexes consisted primarily of 14-3-3 (Fig. 1B, lane 2). The fact that no endogenous KSR was visible in HA-MEK2 and Myc-14-3-3 β immunocomplexes is consistent with our observation that 293T cells express little or no endogenous KSR (Fig. 2F). Thus, we conclude that the large number of proteins copurifying with HA-KSR are not MEK- and/or 14-3-3-associated proteins but rather appear to be highly specific and dependent on the presence of HA-KSR.
To determine the identity of KSR-associated proteins, we attempted direct protein sequencing from α-HA immunoprecipitates. We were successful in obtaining amino acid sequences from peptides corresponding to p90, p70, p68, p44/43, and p30. The sequences of two peptides from p90 were perfect matches to the human heat shock protein HSP90 (Table 2). Peptide sequences of p70 and p68 were identical to those of human HSP70 and HSP68, respectively (Table 2). We also tested whether p50CDC37 (4, 7, 16, 27), a protein kinase-targeting subunit of the HSP90 complex, was present in KSR immunocomplexes. Immunoblot analysis revealed the presence of a 50-kDa protein using α-p50CDC37, suggesting that p50CDC37 is capable of associating with KSR (Fig. 1A).
The functions of HSP90 and p50CDC37 have been implicated in signal transduction and cell cycle regulation through assembly of protein kinase complexes (4, 7, 16, 23, 24, 26, 27, 34). Therefore, we tested the effects of the HSP90 inhibitor geldanamycin on KSR protein levels since it has been observed that geldanamycin treatment reduces levels of Raf, which also associates with the HSP90 complex (26). Treatment of HA-KSR-transfected cells with geldanamycin significantly reduced steady-state levels of KSR yet had no effect on either MEK or ERK (Fig. 1E). Consistent with this finding brief treatment of HA-KSR-transfected 293T cells with geldanamycin nearly abolished the association between KSR and HSP90 and p50CDC37, as determined by coimmunoprecipitation experiments (Fig. 1F). To test whether geldanamycin destabilizes KSR, we performed pulse-chase labeling experiments in the presence or absence of geldanamycin. Treatment of HA-KSR-transfected cells with geldanamycin significantly reduced (>50%) the half-life of HA-KSR (Fig. 1G), suggesting that the HSP90 complex may act to stabilize KSR in vivo.
Loss-of-function alleles of KSR are defective in MEK1/2 binding.
Several missense mutations in CeKSR-1 were identified as loss-of-function alleles in genetic screens (17, 30). Some of these are mutations at positions absolutely conserved in all protein kinases and would therefore be likely to affect the overall kinase structure of KSR (10). Other mutations, however, are found in residues conserved only within the Raf and KSR subfamily of protein kinases. Since our data indicated that kinase activity was not required for KSR function in C. elegans, we tested whether mutants analogous to genetically derived loss-of-function alleles would alter formation of the multimolecular KSR complex. We constructed HA-KSR expression vectors bearing the substitution G580E, R615H, or C809Y in the kinase domain of mKSR corresponding to C. elegans mutant alleles ku83, ku68, and ku148, respectively (30). 35S metabolic labeling and immunoprecipitation experiments were performed as described above. These mutant proteins were efficiently expressed and generally associated with all KSR-associated proteins exception MEK1 and MEK2 (Fig. 2A). No detectable MEK1 or MEK2 was observed in HA-KSR C809Y immunoprecipitates, while the amount of MEK1/2 associated with HA-KSR G580E and R615H mutants was significantly reduced (Fig. 2A and B). Thus, in these cases, loss of ksr-1 function in C. elegans correlates with reduced MEK binding of the corresponding murine KSR mutant protein. We also tested HA-KSR-L56G,R57S, containing a substitution in the CA1 domain corresponding to the Drosophila weak loss-of-function allele S-548 (31). HA-KSR L56G,R57S displayed wild-type binding to all KSR-associated proteins.
All protein kinases contain a conserved lysine residue in the ATP binding domain (10). Surprisingly, both mouse and human KSRs contain an arginine residue in place of the conserved lysine at this position, whereas C. elegans and Drosophila KSRs contain a lysine. HA-KSR R589M, which contains a methionine in place of this arginine and is presumably kinase dead, was also tested for its ability to associate with MEK proteins. HA-KSR R589M also displayed a decreased association with MEK1 and MEK2 (roughly 50% of wild-type level) but still interacted with other KSR-associated proteins (Fig. 2A and B, lanes 3). This appears to contradict our observation that a ksr-1 transgene encoding a K503M substitution can still function in vulva induction. However, it is not clear whether the two mutant proteins in question, mKSR R589M and CeKSR-1 K503M, are biochemically equivalent.
These observations were expanded upon by immunoblotting the same immunoprecipitates with α-MEK1/2 and α-14-3-3. Our results demonstrate that the KSR C809Y mutant is completely defective in MEK1/2 association yet retains wild-type affinity for 14-3-3 proteins (Fig. 2B). Similarly, HA-KSR G580E, R615H, and R589M showed a significant decrease in MEK1/2 association, while the association with 14-3-3 proteins was not changed (Fig. 2B).
We tested which regions of KSR mediate binding and complex formation. To accomplish this, we expressed either the N-terminal amino acids 1 to 301 of KSR (HA-KSR 1-301), a region which contains the N-terminal CA1 and CA2 domains (31), or the kinase domain (HA-KSR KD; amino acid 500 to the stop codon) of KSR. In vivo labeling and immunoprecipitation experiments demonstrated that the isolated kinase domain of KSR was sufficient for binding to most of the proteins detected in full-length HA-KSR immunoprecipitates (Fig. 2A, lane 9). Specifically, HSP70, HSP68, p60, and p50CDC37 were capable of associating with the isolated kinase domain of KSR. Immunoblot analysis with αHSP90 revealed that HSP90 is also easily detectable in HA-KSR KD immunocomplexes, though it does not appear particularly dramatic on this gel (data not shown). In contrast, the only proteins that were able to associate with the amino-terminal 301 amino acids were p36, p33, and in a reduced amount, p34 (Fig. 2A, lane 8). These results suggest that different domains of KSR can bind different KSR-associated proteins in vivo.
We performed experiments in the yeast two-hybrid system to determine if KSR directly interacts with MEK. We observed that LexA-mKSR construct showed little interaction with MEK1 in the yeast two-hybrid assay (Table 3). In contrast, the isolated kinase domain of KSR fused to LexA interacts with MEK1 strongly in yeast. Mutation of cysteine 809 to tyrosine completely eliminated the interaction with MEK (Table 3). These results indicate that cysteine 809 of KSR may be directly involved in its interaction with MEK1 and MEK2. MEK1 and MEK2 contain a proline-rich region between the kinase subdomains IX and X (5, 10, 13). This proline-rich domain has been implicated in interaction between Raf and MEK and has a role in the biological function of MEK (5, 13). Our results indicate that the proline-rich region of MEK1 is not required, however, for interaction with KSR, as deletion of it had no effect on KSR KD-MEK1 interaction (Table 3).
TABLE 3.
KSR interacts with 14-3-3 and MEK1 in the yeast two-hybrid system
| Bait | Interaction with indicated targeta
|
|||
|---|---|---|---|---|
| Vector | 14-3-3 | MEK1 | MEK1ΔP | |
| Lamin | − | − | − | − |
| KSR | − | + | − | − |
| KSR KD | − | − | + | + |
| KSR KD C809Y | − | − | − | − |
A positive interaction denotes both a positive signal by His3+ and positive staining of β-galactosidase. KSR KD is the C-terminal kinase domain of the KSR. KSR KD C809Y contains a point mutation of Cys809 replaced by Tyr; this mutant corresponds to a loss-of-function KSR allele, ku148, isolated from C. elegans (30). MEK1ΔP is a MEK1 construct lacking the proline-rich domain between kinase subdomains X and XI (10).
KSR mediates protein-protein interactions in vivo.
The presence of ERK was reproducibly detected in the KSR complex (Fig. 2C), consistent with previous observations (39). However, the amount of ERK associated with KSR appears to be far less than the amounts of MEK1 and MEK2, since we cannot detect ERK by Coomassie blue staining, although MEK1 and MEK2 are easily detectable (Fig. 1D). The association between ERK and KSR was confirmed by reciprocal immunoprecipitation. α-ERK consistently precipitated far less KSR than α-MEK (Fig. 2D; compare long and short α-HA exposures). These results indicate that KSR may weakly or indirectly interact with ERK. Interestingly, the KSR C809Y mutant failed to precipitate detectable amounts of ERK (Fig. 2C, D), suggesting that MEK1/2 binding may affect this interaction.
The results shown in Fig. 2A and B indicate that KSR interacts with MEK and 14-3-3 proteins via different domains. We tested whether KSR could mediate interactions between MEK and 14-3-3 proteins. α-14-3-3 precipitated MEK1 and MEK2 only upon transfection of HA-KSR (Fig. 2E). Furthermore, MEK–14-3-3 interaction was not observed in the absence of KSR expression or when the MEK binding-defective KSR C809Y mutant was expressed (Fig. 2E, lane 3). The above results support the hypothesis that the interaction between MEK and 14-3-3 is facilitated by KSR and suggest that KSR can serve as a link between MEK and 14-3-3 proteins.
Induction of KSR in differentiated PC12 cells.
We also tested whether endogenous KSR was associated with MEK, HSP90, p50CDC37, and 14-3-3. Immunoblot analysis with αKSR revealed that mouse brain and PC12 cells express detectable amounts of KSR (Fig. 2F, left panel). α-KSR-reactive bands were competed by preincubation with peptide antigen, demonstrating the specificity of the antibody (Fig. 2F, right panel). We also observed that the level of KSR was significantly increased during NGF-induced differentiation of PC12 cells. Immunoprecipitation experiments from differentiated PC12 cell extracts revealed that KSR antibody coprecipitated endogenous MEK1 and MEK2, in addition to HSP90, p50CDC37, and 14-3-3 protein (Fig. 2G). KSR immunoprecipitates from differentiated PC12 cells contain considerably more MEK than those from undifferentiated cells, presumably due to increased KSR protein levels in differentiated cells (Fig. 2G). We reasoned that C. elegans MEK and KSR homologs should be capable of direct interaction. Therefore, both C. elegans and murine KSRs were in vitro translated and tested for MEK binding in a GST pull-down assay using either GST, GST-CeMEK2 (36b), or GST-human MEK2. Our data clearly indicate that both nematode and mammalian KSRs can specifically associate with MEKs in vitro (Fig. 2H) and appear to bind with comparable affinities. These results demonstrate that MEK-KSR binding is an evolutionarily conserved function of KSR.
KSR recruits MEK to a high-molecular-weight complex and alters its subcellular distribution.
If KSR and MEK form a large, stable complex, this may be reflected in their molecular weights and/or subcellular distribution. In 293T cells, MEK1 and MEK2 exhibit an apparent molecular mass of 44 kDa based on Superose 6 size-exclusion chromatography (Fig. 3A). This is in good agreement with the predicted molecular weights of monomeric MEK1 and MEK2. However, in KSR-transfected cells, the apparent molecular mass of MEK shifts to approximately 700 kDa (Fig. 3A). In fact, endogenous MEK quantitatively resides in a high-molecular-weight complex in KSR-transfected cells. We tested whether this alteration in the apparent molecular mass of MEK requires its interaction with KSR. Identical experiments performed with the KSR C809Y mutant did not alter the apparent molecular mass of MEK (Fig. 3A). We also tested the possibility that the large KSR complex that we observed was due to KSR self-oligomerization. HA-KSR and a Myc-tagged variant of mKSR were coexpressed in 293T cells. Lysates were immunoprecipitated with either α-HA or α-Myc, and the extent of KSR self-oligomerization was examined by immunoblotting with α-Myc or α-HA. No detectable KSR oligomerization was observed with either HA- or Myc-tagged KSR (data not shown). These results support our hypothesis that the KSR complex contains KSR and other KSR-associated proteins, including MEK.
MEK normally exists as a soluble cytoplasmic protein (42). KSR has been reported to exist in both cytoplasmic and membrane-associated forms (22, 37). To test whether association with KSR altered the subcellular distribution of MEK, KSR-transfected cells were fractionated into soluble and particulate fractions by ultracentrifugation. Immunoblot analysis indicated that KSR exists in both soluble (S100) and particulate (P100) fractions (Fig. 3B). Immunoblotting with α-MEK indicated that a significant portion of MEK existed in the P100 fraction in KSR-transfected cells (Fig. 3B). Membrane association of MEK required KSR binding, as the KSR C809Y mutant did not noticeably alter the subcellular distribution of MEK. As with KSR C809Y, KSR KD had no detectable effect on MEK localization. The above observation suggests that a potential physiological role of KSR is to alter the subcellular distribution of MEK. It is interesting that the KSR C809Y mutant exists almost exclusively in the P100 fraction whereas a significant portion of wild-type KSR remains in the S100 fraction (Fig. 3B). This observation suggests that MEK binding can, in turn, affect the subcellular distribution of KSR. Thus, it appears that the localization of MEK and KSR is a cooperative process. Immunoblotting with α-c-Raf revealed that c-Raf distribution between S100 and P100 fractions was not significantly altered by expression KSR (Fig. 3B), which is consistent with our observation that little endogenous c-Raf is associated with KSR (data not shown).
KSR-associated MEK is phosphorylated and active.
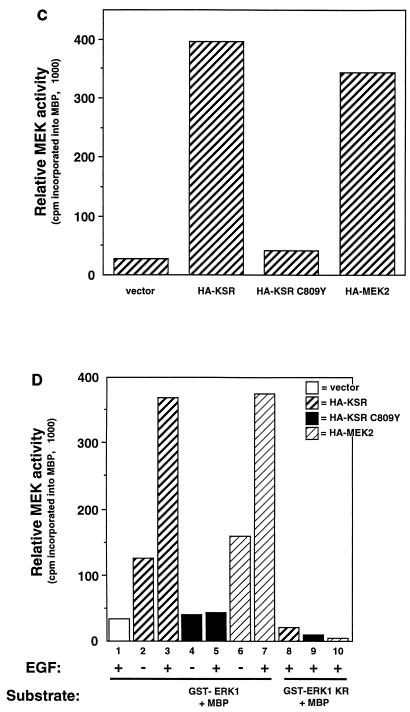
We tested the effects of expressing wild-type KSR or KSR C809Y on the ability of MEK1 and MEK2 to undergo phosphorylation and activation in response to growth factors. Epidermal growth factor (EGF) stimulated a dramatic increase of active, phosphorylated MEK1 and MEK2, which can be detected by α-phospho-MEK. Cotransfection of either HA-KSR or KSR C809Y had no effect on the ability of EGF to stimulate MEK1 and MEK2 phosphorylation (Fig. 4A).
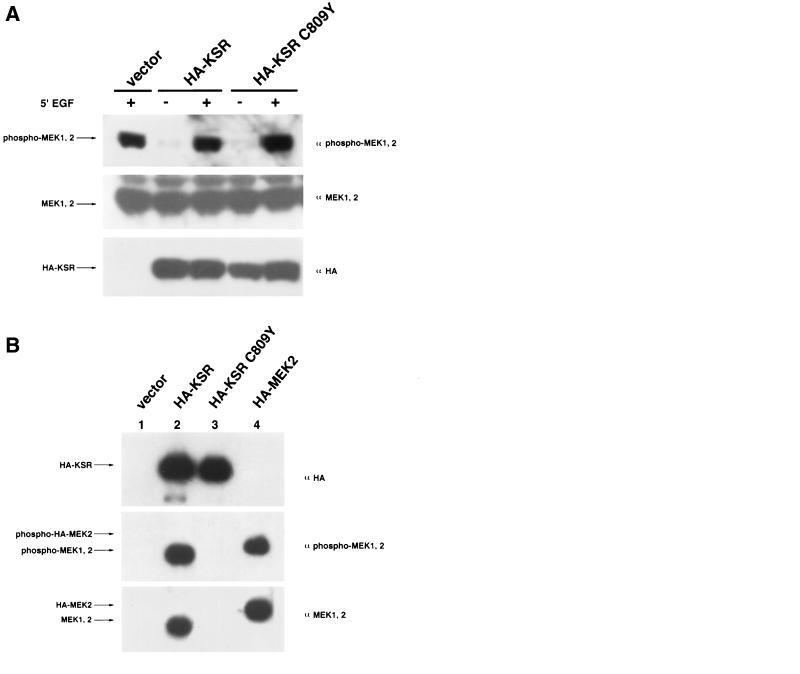
FIG. 4.
KSR-associated MEK is phosphorylated and can activate ERK in response to growth factors. (A) KSR expression does not effect MEK1/2 phosphorylation at serine 218/222. 293T cells were transfected with vector, HA-KSR, or HA-KSR C809Y (1 μg of each). After 24 h of serum deprivation, cells were stimulated for 5 min with EGF (50 ng/ml) as indicated. Whole-cell extracts were prepared and separated by SDS-PAGE followed by immunoblot analysis with α-phospho-MEK1/2, α-MEK, and α-HA, as indicated (lower panel). (B) MEK in the KSR complex is phosphorylated. α-HA immunoprecipitates from vector-, HA-KSR-, HA-KSR C809Y-, or HA-MEK2-transfected 293T cells were subjected to SDS-PAGE and immunoblot analysis with α-phospho-MEK1/2, α-MEK, and α-HA, as indicated. Note that the electrophoretic mobility of HA-MEK2 is slower than that of endogenous MEK due to the presence of two HA epitopes at its amino terminus. (C) KSR-associated MEK is active. Equal amounts of MEK from each of the immunoprecipitates shown in panel B were used to activate recombinant GST-ERK1 in a coupled kinase assay using MBP as a substrate (see Materials and Methods). (D) Binding of KSR to MEK does not alter the ability of MEK to activate ERK in response to growth factor stimulation. 293T cells were transfected with the indicated KSR or MEK expression vector. After 24 h of serum starvation, cells were stimulated with EGF for 5 min as indicated and then immunoprecipitated with α-HA. Equal amounts of MEK from all immunoprecipitates were used to activate recombinant GST-ERK1 or GST-ERK1 KR (kinase-inactive control) in a coupled kinase assay.
To directly examine the phosphorylation state of MEK1 and MEK2 in the KSR complex, we transfected cells with either HA-KSR, HA-KSR C809Y, or HA-MEK2; then lysates from cells grown in the presence of serum were precipitated with α-HA and blotted with α-MEK and α-phospho-MEK (Fig. 4B). Our results clearly indicate that the MEK1/2 in the KSR complex is phosphorylated. In fact, the relative phosphorylation of MEK1/2 in the KSR complex is no less than that in HA-MEK2 immunoprecipitates without KSR transfection (Fig. 4B). We also directly examined the ability of growth factors to induce MEK activity in the KSR complex by a coupled kinase assay. Our results indicate that coprecipitated MEK1/2 in the KSR complex is fully capable of activating recombinant ERK1 (Fig. 4C and D). Furthermore, the specific activity of MEK in the KSR complex was no different than that of HA-MEK2 alone (Fig. 4C and D). HA-KSR C809Y immunocomplexes contained virtually no detectable MEK activity. This finding is consistent with our observation that the C809Y mutant is unable to associate with MEK and also demonstrates the high specificity of this kinase assay (Fig. 4D). These results demonstrate that KSR-bound MEK1/2 can be phosphorylated and activated by Raf, and, in turn, can phosphorylate and activate GST-ERK1 in vitro. Our results support the hypothesis that KSR inhibits neither the activity nor the activation of MEK but rather modulates its localization in the cell. However, it remains possible that the effect of KSR on MEK activation is cell type dependent or dependent on relative expression levels.
KSR C809Y is defective Elk-1 regulation.
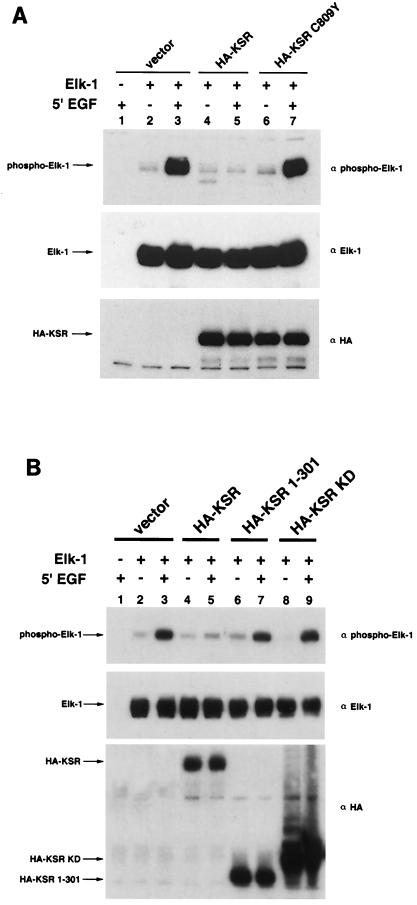
We have observed that KSR can specifically block phosphorylation and activation of Elk-1, a physiological substrate of MAP kinases (29). To test the relationship between the ability of KSR to block Elk-1 phosphorylation and its ability to interact with MEK, KSR C809Y was used. Although C809Y may affect other functions of KSR, a plausible explanation is that KSR-MEK interaction is needed for its ability to block Elk-1 activation in these cells. We tested this hypothesis. KSR effectively blocked EGF-stimulated Elk-1 phosphorylation, while KSR C809Y had no effect on Elk-1 phosphorylation (Fig. 5A). This result suggests that the ability of KSR to bind MEK1 and MEK2 may correlate with its ability to block Elk-1 activation by MAP kinases. We also tested truncated mutants of KSR in the same assay to determine which regions of KSR are necessary and sufficient to inhibit Elk-1. Our results show that the full-length KSR protein is required to inhibit Elk-1 activation (Fig. 5B).
FIG. 5.
Effects of KSR mutants on Elk-1 phosphorylation. (A) KSR C809Y is deficient in Elk-1 inhibition. 293T cells were transfected with Elk-1 (250 ng) and either vector, HA-KSR, or HA-KSR C809Y (1 μg). After 24 h of serum starvation, cells were stimulated with EGF (50 ng/ml) for 5 min prior to lysis and immunoblotting with the indicated antibodies. α-Phospho-Elk-1 recognizes only phosphorylated, active Elk-1. (B) Neither the isolated amino terminus nor the kinase domain of KSR is sufficient to block Elk-1 phosphorylation. 293T cells were transfected with Elk-1 and the indicated KSR expression vector. After 24 h of serum starvation, cells were stimulated with EGF (50 ng/ml) for 5 min prior to lysis and immunoblotting with α-phospho-Elk-1 (top), α-Elk-1 (middle), and α-HA (bottom).
DISCUSSION
Kinase-independent function of KSR.
KSR was originally identified by genetic means to be a regulator of Ras-MAP kinase signaling pathways controlling Drosophila photoreceptor differentiation and C. elegans vulval induction (17, 30, 31). Subsequent studies confirmed that KSR indeed plays a role in Ras-MAP kinase signaling in Xenopus and mammalian cells as well (6, 14, 29, 32, 37, 39). However, the mechanism of KSR function is not well understood in any system. Although the sequence of KSR predicts it to be a protein kinase, to date we are unable to demonstrate any kinase activity intrinsic to KSR whether it is expressed in bacteria, insect cells, or mammalian cells as either a full-length protein or the isolated C-terminal kinase domain. Our C. elegans data argue that if KSR is a protein kinase, this activity is not required for its positive signaling function during vulval induction.
We observed that mutation of the Mg2+-ATP-coordinating lysine residue (KSR-1 K503M) or the catalytic nucleophile aspartic acid residue (KSR-1 D618A) did not compromise the function of ksr-1 in C. elegans vulval induction, although these mutations are frequently used to render a protein kinase dead. Since these complementation assays rely on transgenes, which are likely overexpressed, it remains possible that endogenous levels of a KSR-1 (kinase-dead) protein are signaling deficient. Nevertheless, our data clearly indicate that a kinase-independent function of KSR-1 can promote vulval induction. Our data are consistent with previous reports that a kinase-independent function of KSR can enhance Ras-induced germinal vesicle breakdown in Xenopus oocytes (22, 31) but are inconsistent with a report that KSR acts as a ceramide-activated protein kinase (40). One likely kinase-independent function of KSR is suggested by our finding that murine KSR forms a multimolecular complex in vivo.
Components of the KSR complex.
The KSR complex displays an apparent molecular mass of roughly 106 Da, as determined by size-exclusion chromatography. Immuneprecipitation of HA-KSR revealed numerous cellular proteins that specifically coprecipitate with KSR. We consistently observed the following proteins in the KSR complex affinity purified from HEK 293T cells: HSP90, HSP70, HSP68, p50CDC37, p60, MEK1, MEK2, p36, p34, p33, and 14-3-3. Data derived from experiments using deletion and point mutants of KSR suggest that different regions of KSR may mediate interactions with distinct signaling proteins. For example, the N-terminal domain of KSR (amino acids 1 to 301) interacts with p36, p34, and p33. Similarly, the C-terminal kinase domain of KSR interacts with a unique and distinct set of proteins, notably, HSP90, HSP70, HSP68, and p50CDC37. It is not clear whether all KSR-associated proteins directly interact with KSR, nor do we know that each of the associated proteins exists in the same complex in vivo. However, our data strongly suggest that MEK directly binds KSR, as this interaction is observed in yeast and in vitro binding assays. Furthermore, KSR C809Y, which corresponds to a loss-of-function allele in C. elegans, cannot interact with MEK yet can still associate with other KSR-associated molecules. Similarly, KSR is likely to directly interact with 14-3-3. 14-3-3 proteins have been shown to interact with numerous cellular proteins, including Raf, KSR’s closest relative.
Previous reports have demonstrated interactions between KSR and other proteins, including Raf, MEK, ERK, and 14-3-3 (6, 14, 22, 37, 39). These studies were based on yeast two-hybrid and/or coimmunoprecipitation experiments and have revealed little information regarding the relative strength or functional significance of these interactions. Similarly, it has not been demonstrated whether KSR is capable of simultaneous interactions with different signaling molecules. In this report, we have identified several novel KSR-associated proteins (HSP90, HSP70, HSP68, p50CDC37, p36, p34, and p32) in addition to those previously reported. Furthermore, we demonstrated that KSR is capable of forming a multimolecular complex, although binding affinities among known KSR-associated proteins appear to vary considerably. Specifically, the three kinases of the MAP kinase cascade, Raf, MEK, and ERK, display markedly different abilities to associate with KSR in HEK 293T cells. We consistently observe that MEK stoichiometrically associates with KSR, where as a small fraction of ERK it is detected in KSR immunocomplexes. The fraction of Raf that associates with KSR appears to be smaller still. The data presented here significantly advance the concept of KSR as a scaffolding protein in the Ras-MAP kinase pathway.
Involvement of the HSP90 complex in KSR and Ras signaling.
It is noteworthy that HSP90, HSP70, HSP68, and p50CDC37 are components of the KSR complex. HSP90 has been observed to interact with numerous proteins, including steroid hormone receptors and protein kinases (24). Protein-protein interactions with HSP90 appear to be important for maintaining both protein stability and biological function. In addition, p50CDC37 has been proposed to be a protein kinase-targeting subunit of HSP90 (27). Consistent with this, inhibition of HSP90 with geldanamycin abolishes association between KSR and p50CDC37. In addition, geldanamycin destabilizes KSR in vivo, suggesting a role for these proteins in maintaining levels of KSR. HSPs are also known to facilitate protein folding, and it remains possible that KSR exists in both folded and unfolded forms in our experimental setting. Therefore, it may be difficult to determine whether KSR-HSP90 interactions are biologically significant. Our data do support the hypothesis that KSR-MEK interactions are physiological in nature, as KSR and MEK appear to associate at endogenous protein levels in PC12 cells (Fig. 2G and reference 39). It is noteworthy that KSR protein levels are induced during PC12 cell differentiation. The Ras-MAP kinase pathway plays a pivotal role in PC12 cell differentiation, and further experiments seem warranted to determine whether KSR is involved in this system. Based on several lines of evidence, our data support the model in which HSP90-KSR interactions are also physiologically relevant. First, almost equal amounts of HSPs, MEK, and 14-3-3 proteins are found associated with KSR. Second, distinct regions of KSR mediate protein-protein interactions with HSPs. Third, low-level expression of KSR yields a similar set of coprecipitated molecules. Furthermore, we show here that HSP90 plays a positive role in maintaining KSR protein levels. Finally, mutations in both HSP90 and p50CDC37 have been identified by using genetic screens very similar to those in which KSR was isolated, suggesting that these molecules may function in the same pathway (4, 34).
A model for KSR function as a scaffolding protein.
Scaffolding proteins of other MAP kinase pathways have been shown to maintain specificity and to enhance signaling efficiency (3, 19, 36a). The best example is the Ste5 protein, which tethers components of the Fus3 MAP kinase pathway in the mating pheromone response in budding yeast. Ste5p is essential for mating and maintains specificity of the Fus3p MAP kinase cascade (3, 19). Recently, a scaffolding protein of the mammalian stress-activated MAP kinase cascade has been reported (36a). Although the molecular structure of the KSR complex requires further biochemical characterization, we favor the model in which KSR can simultaneously and directly interact with multiple cellular proteins to form a large signaling complex. Some of the components, such as MEK, HSPs, and 14-3-3, associate with KSR with high stoichiometry. Other components, such as ERK, may transiently (or in a regulated manner) associate with KSR with low stoichiometry. KSR has been reported to weakly interact with Raf in a Ras-dependent manner (22, 32, 37), and we detect Raf-KSR association only upon overexpression of both proteins (data not shown).
We propose that KSR functions in vivo to recruit numerous molecules to form a large signaling complex. Consistent with this model, several loss-of-function mutants exhibited reduced MEK association in vivo. Furthermore, MEK binding is a conserved function of both murine and C. elegans KSRs, though it is not known whether the two are functionally interchangeable. A plausible explanation is that KSR’s interaction with MEK is important for its physiological functions. KSR appears to be a core component of this complex and may function as a scaffold protein to maintain specificity and to increase or restrict the signaling kinase cascade. Other components of the MAP kinase cascade may transiently interact with the complex, depending on intracellular signaling conditions. For example, Raf might transiently interact with the complex and activate the associated MEK. Similarly, ERK may temporally associate with the KSR complex, where it can undergo activation by the tightly bound MEK. Finally, our data suggest that the ability of KSR to form a signaling complex may be more important than its putative kinase activity for its physiological function. The precise role of the KSR signaling complex should be clarified by the identification of the remaining KSR-associated proteins and by the identification of loci that interact genetically with ksr-1 in C. elegans and Drosophila.
ACKNOWLEDGMENTS
We thank Anne Vojtek for reagents and advice on two-hybrid screening; Gerald Rubin and Melanie Cobb for mKSR and MEK plasmids, respectively; Kim Orth and Haris W. Vikis for advice and discussion; and Tianquin Zhu for technical assistance.
This work was supported by the Cancer Biology Training Program, NIH (grant 5T32 CA09676 to S.S.), a Life Sciences Research Foundation/Boehringer Mannheim postdoctoral fellowship (M.S.), American Cancer Society (M.H.), and Public Health Service grant GM51586 and a MacArthur Foundation fellowship (K.-L.G.).
REFERENCES
- 1.Beitel G J, Clark S G, Horvitz H R. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- 2.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi K Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 4.Cutforth T, Rubin G M. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 5.Dang A, Frost J A, Cobb M H. The MEK1 proline-rich insert is required for efficient activation of the mitogen-activated protein kinases ERK1 and ERK2 in mammalian cells. J Biol Chem. 1998;273:19909–19913. doi: 10.1074/jbc.273.31.19909. [DOI] [PubMed] [Google Scholar]
- 6.Denouel-Galy A, Douville E M, Warne P H, Papin C, Laugier D, Calothy G, Downward J, Echène A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1997;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- 7.Gerber M R, Farrell A, Deshaies R J, Herskowitz I, Morgan D O. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc Natl Acad Sci USA. 1995;92:4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gille H, Sharrocks A D, Shaw P E. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 9.Han M, Aroian R V, Sternberg P W. The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics. 1990;126:899–913. doi: 10.1093/genetics/126.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanks S K, Quinn A M. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 11.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 12.Janknecht R, Ernst W H, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jelinek T, Catling A D, Reuter C W, Moodie S A, Wolfman A, Weber M J. RAS and RAF-1 form a signaling complex with MEK-1 but not MEK-2. Mol Cell Biol. 1994;14:8212–8218. doi: 10.1128/mcb.14.12.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joneson T, Fulton J A, Volle D J, Chaika O V, Bar-Sagi D, Lewis R E. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J Biol Chem. 1998;273:7743–7748. doi: 10.1074/jbc.273.13.7743. [DOI] [PubMed] [Google Scholar]
- 15.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Cell Biol. 1997;9:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 16.Kimura Y, Rutherford S L, Miyata Y, Yahara I, Freeman B C, Yue L, Morimoto R I, Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 17.Kornfeld K, Hom D B, Horvitz H R. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 18.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 19.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 21.Mello C C, Kramer J M, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans after microinjection of DNA into germline cytoplasm: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaud N R, Therrien M, Cacace A, Edsall L C, Spiegel S, Rubin G M, Morrison D K. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathan D F, Vos M H, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratt W B. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 25.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 26.Stancato L F, Silverstein A M, Owens-Grillo J K, Chow Y H, Jove R, Pratt W B. The hsp90-binding antibiotic geldanamycin decreases Raf levels and epidermal growth factor signaling without disrupting formation of signaling complexes or reducing the specific enzymatic activity of Raf kinase. J Biol Chem. 1997;272:4013–4020. doi: 10.1074/jbc.272.7.4013. [DOI] [PubMed] [Google Scholar]
- 27.Stepanova L, Leng X, Parker S B, Harper J W. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 28.Sugimoto T, Stewart S, Guan K L. The calcium/calmodulin dependent protein phosphatase calcineurin is the major Elk-1 phosphatase. J Biol Chem. 1997;272:29415–29418. doi: 10.1074/jbc.272.47.29415. [DOI] [PubMed] [Google Scholar]
- 29.Sugimoto T, Stewart S, Han M, Guan K L. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 1998;17:1717–1727. doi: 10.1093/emboj/17.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Sundaram, M. Unpublished observation.
- 30.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 31.Therrien M, Chang H C, Solomon N M, Karim F D, Wassarman D A, Rubin G M. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 32.Therrien M, Michaud N R, Rubin G M, Morrison D K. KSR modulates signal propagation within the MAPK cascade. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 33.van der Geer P, Hunter T, Lindberg R A. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 34.van der Straten A, Rommel C, Dickson B, Hafen E. The heat shock protein 83 (Hsp83) is required for Raf-mediated signalling in Drosophila. EMBO J. 1997;16:1961–1969. doi: 10.1093/emboj/16.8.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 36.Wartmann M, Davis R J. The native structure of the Raf protein kinase is a membrane-bound multi-subunit complex. J Biol Chem. 1994;269:6695–6701. [PubMed] [Google Scholar]
- 36a.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1761–1764. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 36b.Wu Y, Han M, Guan K L. MEK-2, a Caenorhabditis elegans MAP kinase kinase, functions in Ras-mediated vulval induction and other developmental events. Genes Dev. 1995;9:742–755. doi: 10.1101/gad.9.6.742. [DOI] [PubMed] [Google Scholar]
- 37.Xing H, Kornfeld K, Muslin A J. The protein kinase KSR interacts with 14-3-3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 38.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 39.Yu W, Fantl W J, Harrowe G, Williams L T. Regulation of the MAP kinase pathway by mammalian KSR through direct interaction with MEK and ERK. Curr Biol. 1997;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin X H, Basu S, McGinley M, Chan-Hui P Y, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 41.Zheng C F, Guan K L. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J Biol Chem. 1993;268:23933–23939. [PubMed] [Google Scholar]
- 42.Zheng C F, Guan K L. Cytoplasmic localization of the mitogen-activated protein kinase activator MEK. J Biol Chem. 1994;269:19947–19952. [PubMed] [Google Scholar]