Abstract
Background
Ovarian cancer (OC) is a fatal gynaecological malignancy. The study aimed to conduct a comprehensive study to determine the role of ELF3 in OC through bioinformatic analysis.
Methods
Kruskal–Wallis test, Wilcoxon sign-rank test, and logistic regression were used to evaluate the relationship between clinical characteristics and ELF3 expression. Kaplan–Meier method and Cox regression analysis were used to evaluate the prognostic factors. Gene set enrichment analysis (GSEA) and immuno-infiltration analysis were used to evaluate the significant involvement of ELF3 in function.
Results
High ELF3 expression in OC was associated with age (P< 0.001). High ELF3 expression predicted a poorer overall survival (OS) (HR: 1.37; 95% CI: 1.05–1.78; P=0.019) and disease specific survival (DSS) (HR: 1.43; 95% CI: 1.08–1.89; P=0.013). And ELF3 expression (HR: 1.779; 95% CI: 1.281–2.472; P<0.001) was independently correlated with OS in OC patients. GSEA demonstrated that pathways including GPCR-ligand binding, neuronal system, signaling by WNT, translation, neuroactive ligand-receptor interaction, and TCF dependent signaling in response to WNT were differentially enriched in ELF3 low expression phenotype. Immune infiltration analysis showed that ELF3 expression was correlated with immune infiltrates.
Conclusion
ELF3 expression in OC patients was significantly associated with poor survival and immune infiltration and a promising prognostic biomarker in OC.
Keywords: ovarian cancer, ELF3, prognosis, immune infiltrates, biomarkers
Introduction
Ovarian cancer (OC) is the most common gynaecological tumor, ranking fourth in incidence and third in mortality worldwide.1 In China, OC has the second highest mortality rate among gynaecological tumors and is on the rise, while the incidence is declining.2 High grade serous ovarian cancer (HGSOC) is the most common and fatal type of epithelial ovarian cancer, accounting for 75% of OC cases.3 Non-epithelial ovarian cancer (NEOC) accounts for approximately 10% of all OC cases and includes malignancies of germ cell origin, malignancies of gonadal-stromal cell origin, small cell carcinomas and sarcomas.4 OC has no specific symptoms in its early stages, and over 70% of OC cases are diagnosed when the tumor has progressed to an advanced stage (stage III–IV; International Federation of Gynecology and Obstetrics, FIGO).5 Despite aggressive first-line surgery and adjuvant chemotherapy, the 5-year overall survival (OS) rate is still about 30%.6 The identification of key prognostic factors and predictive biomarkers is important to provide evidence for individualized treatment of OC.
Transcription factor E74-like factor 3 (ELF3) is an epithelial-restricted member of the Ets transcription factor family.7 ELF-1 binds an essential repetitive GGAA cis-acting element at the OAS1 promoter and cooperates with RB1 and SP1 recruitment to contribute to regulation in response to IFN stimulation.8 However, the relevance of ELF3 to immunity is also unclear. ELF3 is a well-documented tumor suppressor in some tumors, but shows oncogenic properties in others.9 ELF3 is an oncogene and putative therapeutic target in Lung adenocarcinoma (LUAD).9 ELF3 is a potential prognostic marker for patients with thyroid cancer (THCA).10 ELF3 is an independent prognostic factor for survival in HR+HER2+ breast cancer (BRCA) patients.11 ELF3 is a key driver of β-catenin signaling in colorectal cancer (CRC) and highlights the potential prognostic and therapeutic significance of ELF3 in CRC.12 ELF3 overexpression is a prognostic biomarker for recurrence of stage II in CRC.13 Although ELF3 has been shown to be a negative regulator of epithelial-mesenchymal transition (EMT) in OC cells, the detailed correlation between ELF3 and OC has not been studied. This study aims to explore the expression of ELF3 in OC, which may provide new directions for the development of diagnostic and therapeutic strategies for OC.
Based on the Cancer Genome Atlas (TCGA) database and OC RNA-seq data in GTEx, this study compared the differences in ELF3 expression between tumor tissues and normal samples, investigated the correlation between ELF3 expression and clinical features of OC, and assessed the prognostic value of ELF3 in OC patients. Genomic enrichment analysis (GSEA) was performed on ELF3 high and low ELF3 expression groups to reveal the possible functions of ELF3. The correlation between ELF3 expression and immune infiltration was analyzed to explore the potential mechanisms by which ELF3 regulates the onset and progression of OC.
Materials and Methods
Differential Expression of ELF3
Baseline information sheet. The analysis was carried out according to the literature.14 Target molecule: ELF3 [ENSG00000163435]. Subgroup: Median.
Unpaired samples. The analysis was carried out according to the literature.14,15 Target molecule: ELF3.
ROC Analysis. The analysis was carried out according to the literature.15,16 Target molecule: ELF3.
The Relationship Between ELF3 and Clinical Characteristics and Prognosis
Correlation of gene expression with clinical characteristics. The analysis was carried out according to the literatures.17 Target molecule: ELF3. Clinical variables: Age.
Logistics analysis. The analysis was carried out according to the literatures.17 Dependent variable: ELF3.
The Relationship Between ELF3 and Clinical Characteristics
Kaplan-Meier method. The analysis was carried out according to the literatures.17,18 Target Molecule: ELF3. Prognosis type: OS and disease-specific survival (DSS). Subgroups: 0–50 vs 50–100.
COX regression. The analysis was carried out according to the literatures.17,18
Forest plot. Software: R (version 3.6.3). R package: ggplot2 package.
Nomogram plot. The analysis was carried out according to the literatures.17,18 R package: rms package and survival package. Prognosis type: Overall Survival. Included variables: FIGO stage; Primary therapy outcome; Race; Age; Tumor residual; ELF3.
Gene Set Enrichment Analysis (GSEA)
Single gene differential analysis. The analysis was carried out according to the literatures.17,19 Target molecule: ELF3. Low expression group: 0–50%. High expression group: 50–100%.
GSEA analysis. The analysis was carried out according to the literatures.17,20,21
Immune Infiltration Analysis by ssGSEA
The analysis was carried out according to the literatures.14,22,23 Target molecule: ELF3.
Results
The Clinical Characteristics of OC Patients
As shown in Table 1, the age range was 51 to 68 years, with a median of 59 years. There were 1 stage I (0.3%), 23 stage II (6.1%), 295 stage III (78.5%), and 57 stage IV (15.2%) in the FIGO stage. There were 27 PD (8.8%), 22 SD (7.1%), 43 PR (14%), and 216 CR (70.1%) in the primary therapy outcome. There were 328 white patients, 12 Asian patients, and 25 Black or African American patients in race. There were 208 patients (≤60, 54.9%) and 171 patients (>60, 45.1%) in the age. There were 1 G1 (1%), 45 G2 (12.2%), 322 G3 (87.3%), and 1 G4 (0.3%) in the histological grade. There were 102 unilateral (28.6%) and 255 bilateral (71.4%) in the anatomic neoplasm subdivision. There were 64 yes (61%) and 41 no (39%) in the venous invasion. There were 48 No (32.2%) and 101 Yes (67.8%) in the lymphatic invasion. There were 67 NRD (20%) and 268 RD (80%) in the tumor residual.
Table 1.
Clinical Characteristics of OC Patients in TCGA
| Characteristic | Levels | Overall |
|---|---|---|
| n | 379 | |
| FIGO stage, n (%) | Stage I | 1 (0.3%) |
| Stage II | 23 (6.1%) | |
| Stage III | 295 (78.5%) | |
| Stage IV | 57 (15.2%) | |
| Primary therapy outcome, n (%) | PD | 27 (8.8%) |
| SD | 22 (7.1%) | |
| PR | 43 (14%) | |
| CR | 216 (70.1%) | |
| Race, n (%) | Asian | 12 (3.3%) |
| Black or African American | 25 (6.8%) | |
| White | 328 (89.9%) | |
| Age, n (%) | ≤60 | 208 (54.9%) |
| >60 | 171 (45.1%) | |
| Histologic grade, n (%) | G1 | 1 (0.3%) |
| G2 | 45 (12.2%) | |
| G3 | 322 (87.3%) | |
| G4 | 1 (0.3%) | |
| Anatomic neoplasm subdivision, n (%) | Unilateral | 102 (28.6%) |
| Bilateral | 255 (71.4%) | |
| Venous invasion, n (%) | No | 41 (39%) |
| Yes | 64 (61%) | |
| Lymphatic invasion, n (%) | No | 48 (32.2%) |
| Yes | 101 (67.8%) | |
| Tumor residual, n (%) | NRD | 67 (20%) |
| RD | 268 (80%) | |
| Age, median (IQR) | 59 (51, 68) |
ELF3 Expression is Correlated with Poor Clinicopathological Characteristics of OC
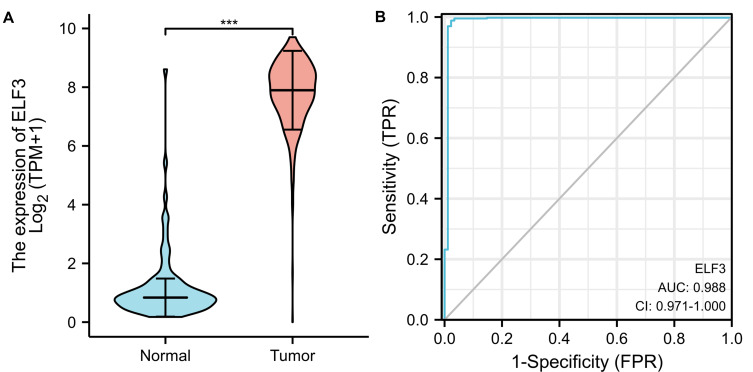
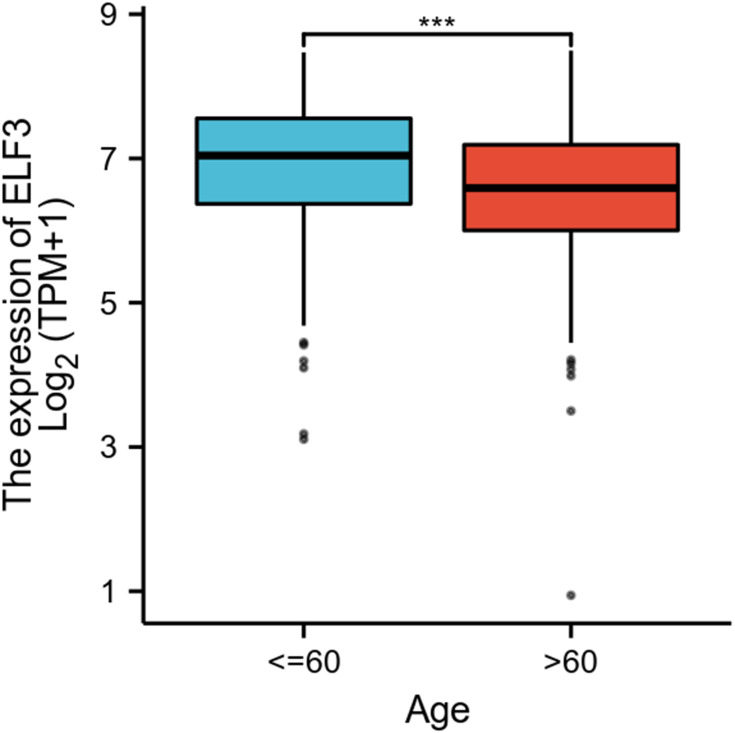
As shown in Figure 1A, ELF3 was highly expressed in OC tissues (1.188 ± 0.129 vs 7.792 ± 0.055, P<0.001). As shown in Figure 1B, the area under curve (AUC) of ELF3 was 0.988, suggesting that ELF3 could be served as an ideal biomarker to distinguish OC from nontumor tissue. As shown in Table 2, ELF3 expression was associated with age (P<0.001). The Logistic regression results in Figure 2 and Table 3 suggested that ELF3 was significantly related to age (HR: 0.465; 95% CI: 0.307–0.701; P<0.001).
Figure 1.
ELF3 is significantly upregulated in OC than normal tissues. (A) The difference expression of ELF3 in OC and normal ovarian tissues. (B) The efficiency of ELF3 expression levels in distinguishing OC from normal ovarian tissues. Significance markers: ***p<0.001.
Table 2.
Correlation of ELF3 Expression with Clinical Characteristics of OC Patients
| Characteristic | Low Expression of ELF3 | High Expression of ELF3 | p |
|---|---|---|---|
| n | 189 | 190 | |
| FIGO stage, n (%) | 1.000 | ||
| Stage I | 1 (0.3%) | 0 (0%) | |
| Stage II | 11 (2.9%) | 12 (3.2%) | |
| Stage III | 147 (39.1%) | 148 (39.4%) | |
| Stage IV | 28 (7.4%) | 29 (7.7%) | |
| Primary therapy outcome, n (%) | 0.274 | ||
| PD | 15 (4.9%) | 12 (3.9%) | |
| SD | 15 (4.9%) | 7 (2.3%) | |
| PR | 19 (6.2%) | 24 (7.8%) | |
| CR | 106 (34.4%) | 110 (35.7%) | |
| Race, n (%) | 0.835 | ||
| Asian | 5 (1.4%) | 7 (1.9%) | |
| Black or African American | 13 (3.6%) | 12 (3.3%) | |
| White | 163 (44.7%) | 165 (45.2%) | |
| Age, n (%) | < 0.001 | ||
| ≤60 | 86 (22.7%) | 122 (32.2%) | |
| >60 | 103 (27.2%) | 68 (17.9%) | |
| Histologic grade, n (%) | 0.722 | ||
| G1 | 1 (0.3%) | 0 (0%) | |
| G2 | 24 (6.5%) | 21 (5.7%) | |
| G3 | 160 (43.4%) | 162 (43.9%) | |
| G4 | 0 (0%) | 1 (0.3%) | |
| Anatomic neoplasm subdivision, n (%) | 0.073 | ||
| Unilateral | 57 (16%) | 45 (12.6%) | |
| Bilateral | 114 (31.9%) | 141 (39.5%) | |
| Venous invasion, n (%) | 0.938 | ||
| No | 21 (20%) | 20 (19%) | |
| Yes | 31 (29.5%) | 33 (31.4%) | |
| Lymphatic invasion, n (%) | 0.250 | ||
| No | 26 (17.4%) | 22 (14.8%) | |
| Yes | 43 (28.9%) | 58 (38.9%) | |
| Tumor residual, n (%) | 0.848 | ||
| NRD | 34 (10.1%) | 33 (9.9%) | |
| RD | 130 (38.8%) | 138 (41.2%) | |
| Age, median (IQR) | 62 (52, 71) | 57 (49.25, 65) | < 0.001 |
Figure 2.
The relationship between ELF3 expression and age of OC patients. Significance markers: ***p<0.001.
Table 3.
Correlation Between ELF3 Expression and Clinical Characteristics (Logistic Analysis)
| Characteristics | Total (N) | Odds Ratio (OR) | P value |
|---|---|---|---|
| FIGO stage (Stage III & Stage IV vs Stage I & Stage II) | 376 | 1.011 (0.438–2.335) | 0.979 |
| Primary therapy outcome (CR vs PD&SD&PR) | 308 | 1.183 (0.726–1.932) | 0.501 |
| Race (White vs Asian & Black or African American) | 365 | 0.959 (0.483–1.899) | 0.904 |
| Age (>60 vs ≤60) | 379 | 0.465 (0.307–0.701) | <0.001 |
| Histologic grade (G3&G4 vs G1&G2) | 369 | 1.213 (0.653–2.272) | 0.542 |
| Anatomic neoplasm subdivision (Bilateral vs Unilateral) | 357 | 1.567 (0.988–2.496) | 0.057 |
| Venous invasion (Yes vs No) | 105 | 1.118 (0.509–2.460) | 0.781 |
| Lymphatic invasion (Yes vs No) | 149 | 1.594 (0.800–3.204) | 0.186 |
| Tumor residual (RD vs NRD) | 335 | 1.094 (0.639–1.873) | 0.743 |
Role of ELF3 in OC Patient Survival
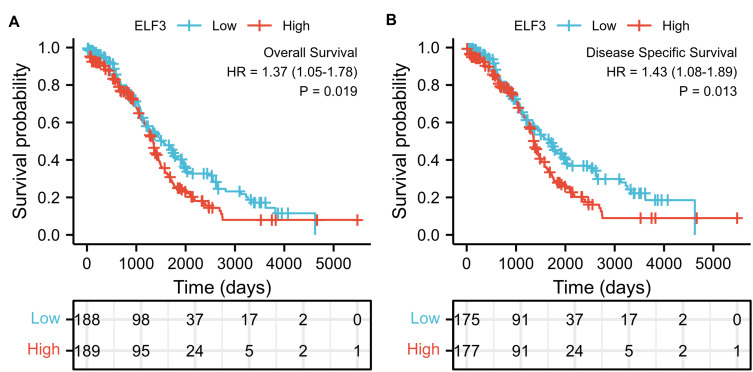
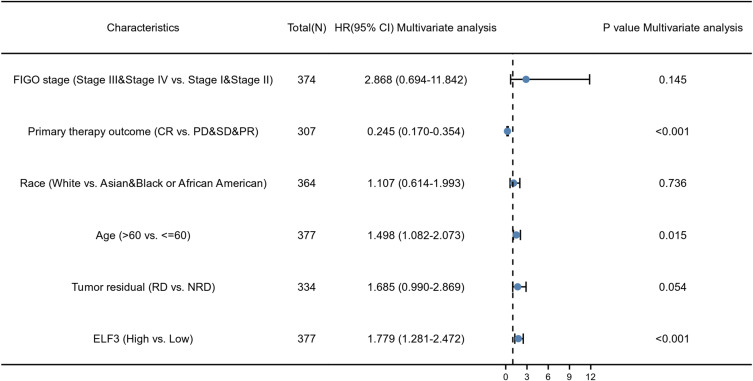
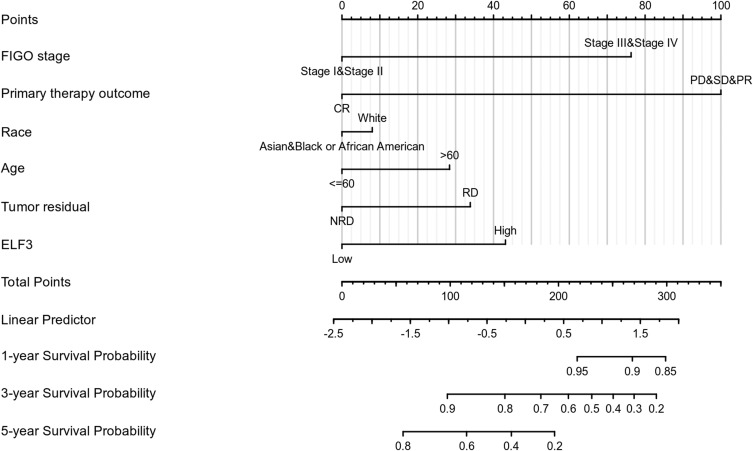
The expression of ELF3 was positively correlated with poor OS (HR: 1.37; 95% CI: 1.05–1.78; P=0.019) and DSS (HR: 1.43; 95% CI: 1.08–1.89; P=0.013) of OC patients (Figure 3). As shown in Table 4, high ELF3 expression levels were associated with worse OS (HR: 1.368, 1.054–1.775, P=0.019), primary therapy outcome (HR: 0.229, 95% CI: 0.166–0.318, P<0.001), age (HR: 1.355, 95% CI: 1.046–1.754, P=0.021), and tumor residual (HR: 2.313, 95% CI: 1.486–3.599, P<0.001). As in Table 4 and Figure 4, ELF3 (HR: 1.779; 95% CI: 1.281–2.472; P<0.001), primary therapy outcome (HR: 0.245; 95% CI: 0.170–0.354; P<0.001), and age (HR: 1.498; 95% CI: 1.082–2.073; P=0.015) were independently correlated with OS in multivariate analysis. The above data indicated ELF3 is a prognostic factor and increased ELF3 level is associated with poor OS. A nomogram was constructed to predict the 1-, 3-, and 5-year survival probability of OC patients by combining the expression level of ELF3 with clinical variables, as shown in Figure 5.
Figure 3.
High expression of ELF3 in OC patients is associated with poor OS and DSS. (A) OS, over survival; (B) DSS, disease-specific survival.
Table 4.
Univariate and Multivariate Analysis (Cox Regression) Between OS and Clinical Characteristics in OC Patients
| Characteristics | Total (N) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| Hazard Ratio (95% CI) | P value | Hazard Ratio (95% CI) | P value | ||
| FIGO stage (Stage III & Stage IV vs Stage I & Stage II) | 374 | 2.115 (0.938–4.766) | 0.071 | 2.868 (0.694–11.842) | 0.145 |
| Primary therapy outcome (CR vs PD&SD&PR) | 307 | 0.229 (0.166–0.318) | <0.001 | 0.245 (0.170–0.354) | <0.001 |
| Race (White vs Asian & Black or African American) | 364 | 0.637 (0.405–1.004) | 0.052 | 1.107 (0.614–1.993) | 0.736 |
| Age (>60 vs ≤60) | 377 | 1.355 (1.046–1.754) | 0.021 | 1.498 (1.082–2.073) | 0.015 |
| Histologic grade (G3&G4 vs G1&G2) | 367 | 1.229 (0.830–1.818) | 0.303 | ||
| Anatomic neoplasm subdivision (Bilateral vs Unilateral) | 356 | 1.049 (0.776–1.418) | 0.757 | ||
| Venous invasion (Yes vs No) | 105 | 0.896 (0.487–1.649) | 0.723 | ||
| Lymphatic invasion (Yes vs No) | 148 | 1.413 (0.833–2.396) | 0.200 | ||
| Tumor residual (RD vs NRD) | 334 | 2.313 (1.486–3.599) | <0.001 | 1.685 (0.990–2.869) | 0.054 |
| ELF3 (High vs Low) | 377 | 1.368 (1.054–1.775) | 0.019 | 1.779 (1.281–2.472) | <0.001 |
Figure 4.
Forest plot of the multivariate Cox regression analysis in OC.
Figure 5.
Nomogram for predicting the probability of patients with 1-, 3- and 5-year overall survival.
ELF3-Related Pathways Based on GSEA
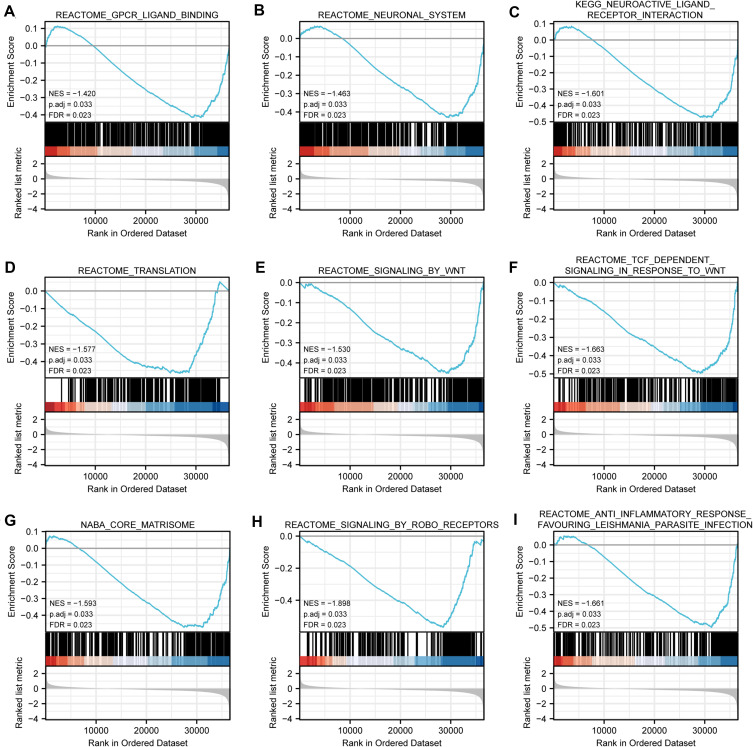
There were 111 data sets which showed significantly differential enrichment in ELF3 low expression phenotype, and we selected the top 9 data sets with high value of normalized enrichment score (NES), in Table 5 and Figure 6, including GPCR-ligand binding, neuronal system, signaling by WNT, translation, neuroactive ligand-receptor interaction, TCF dependent signaling in response to WNT, core matrisome, signaling by ROBO receptors, and anti-inflammatory response favouring Leishmania parasite infection.
Table 5.
Gene Sets Enriched in the ELF3 Low Expression Group
| Description | NES | P Adjust | q values |
|---|---|---|---|
| REACTOME_GPCR_LIGAND_BINDING | −1.420 | 0.033 | 0.023 |
| REACTOME_NEURONAL_SYSTEM | −1.463 | 0.033 | 0.023 |
| REACTOME_SIGNALING_BY_WNT | −1.530 | 0.033 | 0.023 |
| REACTOME_TRANSLATION | −1.577 | 0.033 | 0.023 |
| KEGG_NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | −1.601 | 0.033 | 0.023 |
| NABA_CORE_MATRISOME | −1.593 | 0.033 | 0.023 |
| REACTOME_TCF_DEPENDENT_SIGNALING_IN_RESPONSE_TO_WNT | −1.663 | 0.033 | 0.023 |
| REACTOME_ANTI_INFLAMMATORY_RESPONSE_FAVOURING_LEISHMANIA_PARASITE_INFECTION | −1.661 | 0.033 | 0.023 |
| REACTOME_SIGNALING_BY_ROBO_RECEPTORS | −1.898 | 0.033 | 0.023 |
Figure 6.
Enrichment plots from gene set enrichment analysis (GSEA). (A) GPCR-ligand binding, (B) neuronal system, (C) neuroactive ligand-receptor interaction, (D) translation, (E) signaling by WNT, (F) TCF dependent signaling in response to WNT, (G) core matrisome, (H) signaling by ROBO receptors and (I) anti-inflammatory response favoring Leishmania parasite infection.
Abbreviations: NES, normalized ES; FDR, false discovery rate.
The Correlation Between ELF3 Expression and Immune Infiltration
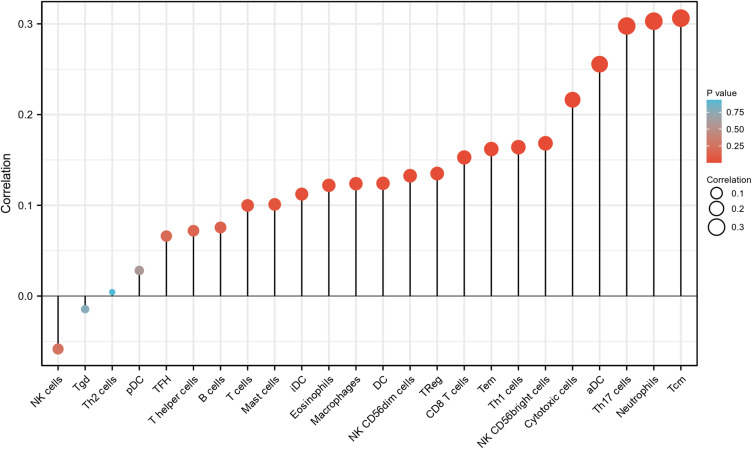
As shown in Figure 7 and Table 6, analysis of the relationship between ELF3 and immune infiltration based on ssGSEA with Spearman r showed that showed that ELF3 expression was positively correlated with that of aDC (P<0.001), CD8 T cells (P=0.003), cytotoxic cells (P<0.001), DC (P=0.016), Eosinophils (P=0.018), iDC (P=0.029), Macrophages (P=0.016), Mast cells (P=0.049), Neutrophils (P<0.001), NK CD56bright cells (P=0.001), NK CD56dim cells (P=0.01), Tcm (P<0.001), Tem (P=0.002), Th1 cells (P=0.001), Th17 cells (P<0.001), and TReg (P=0.008).
Figure 7.
The expression level of ELF3 was related to the immune infiltration in the tumor microenvironment. The forest plot shows the correlation between ELF3 expression level and 24 immune cells. The size of dots indicates the absolute value of Spearman r.
Table 6.
ELF3 Expression Associated with Immune Cells (Spearman Method)
| Gene Name | Cell Type | Correlation Coefficient (Spearman) | P value (Spearman) |
|---|---|---|---|
| ELF3 | aDC | 0.256 | <0.001 |
| ELF3 | B cells | 0.076 | 0.142 |
| ELF3 | CD8 T cells | 0.153 | 0.003 |
| ELF3 | Cytotoxic cells | 0.216 | <0.001 |
| ELF3 | DC | 0.124 | 0.016 |
| ELF3 | Eosinophils | 0.122 | 0.018 |
| ELF3 | iDC | 0.112 | 0.029 |
| ELF3 | Macrophages | 0.124 | 0.016 |
| ELF3 | Mast cells | 0.101 | 0.049 |
| ELF3 | Neutrophils | 0.303 | <0.001 |
| ELF3 | NK CD56bright cells | 0.168 | 0.001 |
| ELF3 | NK CD56dim cells | 0.133 | 0.01 |
| ELF3 | NK cells | −0.058 | 0.257 |
| ELF3 | pDC | 0.028 | 0.585 |
| ELF3 | T cells | 0.1 | 0.052 |
| ELF3 | T helper cells | 0.072 | 0.162 |
| ELF3 | Tcm | 0.306 | <0.001 |
| ELF3 | Tem | 0.162 | 0.002 |
| ELF3 | TFH | 0.066 | 0.199 |
| ELF3 | Tgd | −0.014 | 0.779 |
| ELF3 | Th1 cells | 0.164 | 0.001 |
| ELF3 | Th17 cells | 0.298 | <0.001 |
| ELF3 | Th2 cells | 0.004 | 0.934 |
| ELF3 | TReg | 0.135 | 0.008 |
Discussion
Despite the many advances that have been made in treatment strategies for OC, OS has not improved in these patients and the search for novel biomarkers that can be used to predict the prognosis of these patients is warranted. SLC7A2 is a novel biomarker for the diagnosis and treatment of OC.24 PRDX-1 expression in tumor tissue can be a biomarker for the prognosis of patients with OC.25 Increased expression of TET3 predicts an unfavorable prognosis for OC patients.26 Low expression of BCL7A is an independent risk factor for poor prognosis in patients with OC.27 Overexpression of PRC1 indicates poor prognosis of OC.28 Therefore, it is important to study mRNAs as new OC biomarkers and therapeutic targets in the future.
The high expression of ELF3 in OC patients in this study was significantly associated with age (P<0.001). The expression of ELF3 is high in subjects with age (≤60) and low in subjects with age (>60). The reasons for this are subject to further research. High ELF3 expression predicted a poorer OS (HR: 1.37; 95% CI: 1.05–1.78; P=0.019) and DSS (HR: 1.43; 95% CI: 1.08–1.89; P=0.013). And ELF3 expression (HR: 1.779; 95% CI: 1.281–2.472; P<0.001) was independently correlated with OS in OC patients. Therefore, ELF3 can be used as a promising prognostic marker for patients with OC.
ELF3 forms a positive feedback loop with the MAPK pathway, leading to the progression of BRAF-mutant THCA.10 The miR-1224-5p/ELF3 axis may serve as a novel diagnostic, therapeutic, and prognostic biomarker for pancreatic cancer (PAAD) and the associated PI3K/AKT/Notch/EMT signaling pathway greatly contributes to the progression of PAAD.29 The MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer (NSCLC) through the PI3K/Akt pathway.30 In this study, ELF3 was found to be associated with the pathways GPCR-ligand binding, neuronal system, signaling by WNT, translation, neuroactive ligand-receptor interaction, TCF dependent signaling in response to WNT, core matrisome, signaling by ROBO receptors, and anti-inflammatory response favoring Leishmania parasite infection based on GESA analysis.
Immune infiltration in OC is currently a hot topic and knowledge of immune infiltrating cells is beneficial to the development of immunotherapy for OC. Early efforts in this approach evaluated cytokine therapy for OC, but failed to present convincing Phase III data.31 On the other hand, immune checkpoint inhibitors (ICIs) have emerged as important immune stimulants and the immunological properties of OC provide a basis for their introduction into disease management.31 However, when evaluated in pretreated patients with OC, ICIs have delivered only modest efficacy as monotherapy, necessitating additional approaches to realize the potential.31 Since then, several strategies have aimed to sensitive OC to immunotherapy by combining it with chemotherapy, anti-angiogenics, PARPi, radiotherapy, and dual immune checkpoint blockade.31 The present study showed that ELF3 expression was associated with infiltration of aDC, CD8 T cells, Cytotoxic cells, DC, Eosinophils, iDC, Macrophages, Mast cells, Neutrophils, NK CD56bright cells, NK CD56dim cells, Tcm, Tem, Th1 cells, Th17 cells, and TReg in OC. This means that ELF3 promotes the function of aDC, CD8 T cells, Cytotoxic cells, DC, Eosinophils, iDC, Macrophages, Mast cells, Neutrophils, NK CD56bright cells, NK CD56dim cells, Tcm, Tem, Th1 cells, Th17 cells, and TReg.
This study explored the relationship between ELF3 and OC. However, there are some limitations to this study. This study was based on RNA sequencing from the TCGA database and we were unable to describe the specific molecular mechanisms of ELF3 in OC patients. The specific molecular mechanisms by which ELF3 mediates OC occurrence and development were further investigated.
Conclusion
ELF3 was highly expressed in OC tissues and significantly associated with poor OS and DSS in OC patients. ELF3 is involved in the development and progression of OC through pathways including GPCR-ligand binding, neuronal system, signaling by WNT, translation, neuroactive ligand-receptor interaction, TCF dependent signaling in response to WNT, core matrisome, signaling by ROBO receptors, anti-inflammatory response favoring Leishmania parasite infection. ELF3 was associated with immune infiltrating cells. This study suggested that ELF3 was a promising prognostic biomarker for ovarian cancer.
Acknowledgments
The datasets generated in this study are available from TCGA that provide free resources.
Funding Statement
This work was supported by Natural Science Foundation of Hubei Province (WJ2019H205).
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 3.Chen B, Liu S, Wang H, Li G, Lu X, Xu H. Differential expression profiles and function prediction of transfer RNA-derived fragments in high-grade serous ovarian cancer. Biomed Res Int. 2021;2021:5594081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boussios S, Moschetta M, Zarkavelis G, Papadaki A, Kefas A, Tatsi K. Ovarian sex-cord stromal tumours and small cell tumours: pathological, genetic and management aspects. Crit Rev Oncol Hematol. 2017;120:43–51. doi: 10.1016/j.critrevonc.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 5.Zeppernick F, Meinhold-Heerlein I. The new FIGO staging system for ovarian, fallopian tube, and primary peritoneal cancer. Arch Gynecol Obstet. 2014;290(5):839–842. doi: 10.1007/s00404-014-3364-8 [DOI] [PubMed] [Google Scholar]
- 6.Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Prim. 2016;2:16061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wasylyk B, Hagman J, Gutierrez-Hartmann A. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem Sci. 1998;23(6):213–216. doi: 10.1016/S0968-0004(98)01211-0 [DOI] [PubMed] [Google Scholar]
- 8.Larsen S, Kawamoto S, Tanuma S, Uchiumi F. The hematopoietic regulator, ELF-1, enhances the transcriptional response to Interferon-β of the OAS1 anti-viral gene. Sci Rep. 2015;5(1):17497. doi: 10.1038/srep17497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enfield KSS, Marshall EA, Anderson C, et al. Epithelial tumor suppressor ELF3 is a lineage-specific amplified oncogene in lung adenocarcinoma. Nat Commun. 2019;10(1):5438. doi: 10.1038/s41467-019-13295-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Chen W, Zhang X, et al. E26 transformation (ETS)‑specific related transcription factor‑3 (ELF3) orchestrates a positive feedback loop that constitutively activates the MAPK/Erk pathway to drive thyroid cancer. Oncol Rep. 2019;41(1):570–578. [DOI] [PubMed] [Google Scholar]
- 11.Kar A, Koto K, Walker D, et al. ETS transcription factor ESE-1/Elf3 is an independent prognostic factor of survival in HR(+)HER2(+) breast cancer patients. Breast Cancer Res Treat. 2020;182(3):601–612. doi: 10.1007/s10549-020-05734-y [DOI] [PubMed] [Google Scholar]
- 12.Wang JL, Chen ZF, Chen HM, et al. Elf3 drives β-catenin transactivation and associates with poor prognosis in colorectal cancer. Cell Death Dis. 2014;5(5):e1263. doi: 10.1038/cddis.2014.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaoka A, Ishikawa T, Okazaki S, et al. ELF3 overexpression as prognostic biomarker for recurrence of stage II colorectal cancer. In Vivo (Brooklyn). 2021;35(1):191–201. doi: 10.21873/invivo.12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X, Li G, Liu S, Wang H, Zhang Z, Chen B. Bioinformatics analysis of KIF1A expression and gene regulation network in ovarian carcinoma. Int J Gen Med. 2021;14:3707–3717. doi: 10.2147/IJGM.S323591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314–316. doi: 10.1038/nbt.3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Tang H, Li T, et al. Comprehensive analysis of the expression, prognosis, and biological significance of OVOLs in breast cancer. Int J Gen Med. 2021;14:3951–3960. doi: 10.2147/IJGM.S326402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Zhu C, Wang X, Pan Y. LncRNA ELF3-AS1 is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Can J Gastroenterol Hepatol. 2021;2021:8323487. doi: 10.1155/2021/8323487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14(1):7. doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 24.Sun T, Bi F, Liu Z, Yang Q. SLC7A2 serves as a potential biomarker and therapeutic target for ovarian cancer. Aging. 2020;12(13):13281–13296. doi: 10.18632/aging.103433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sieńko J, Teliga-Czajkowska J, Przytula E, Czajkowski K, Smolarczyk R, Nowis D. Peroxiredoxin-1 as a prognostic factor in patients with ovarian cancer. Ann Agric Environ Med. 2019;26(3):415–419. doi: 10.26444/aaem/105899 [DOI] [PubMed] [Google Scholar]
- 26.Cao T, Pan W, Sun X, Shen H. Increased expression of TET3 predicts unfavorable prognosis in patients with ovarian cancer-A bioinformatics integrative analysis. J Ovarian Res. 2019;12(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Z, Sun L, He M, Pang Y, Yang Z, Wang J. Low BCL7A expression predicts poor prognosis in ovarian cancer. J Ovarian Res. 2019;12(1):41. doi: 10.1186/s13048-019-0518-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bu H, Li Y, Jin C, et al. Overexpression of PRC1 indicates a poor prognosis in ovarian cancer. Int J Oncol. 2020;56(3):685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L, Liu P, Zheng M, et al. The miR-1224-5p/ELF3 axis regulates malignant behaviors of pancreatic cancer via PI3K/AKT/notch signaling pathways. Onco Targets Ther. 2020;13:3449–3466. doi: 10.2147/OTT.S248507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao W, Sun Q, Yu Z, et al. MiR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene. 2018;670:31–37. doi: 10.1016/j.gene.2018.05.100 [DOI] [PubMed] [Google Scholar]
- 31.Demircan NC, Boussios S, Tasci T, Öztürk MA. Current and future immunotherapy approaches in ovarian cancer. Ann Transl Med. 2020;8(24):1714. doi: 10.21037/atm-20-4499 [DOI] [PMC free article] [PubMed] [Google Scholar]