Abstract
Background:
With the ongoing worldwide coronavirus disease 2019 (COVID-19) pandemic, an increasing number of viral variants are being identified, which poses a challenge for nucleic acid-based diagnostic tests. Rapid tests, such as real-time reverse transcription-polymerase chain reaction (rRT-PCR), play an important role in monitoring COVID-19 infection and controlling its spread. However, the changes in the genotypes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants may result in decreased sensitivity of the rRT-PCR assay and it is necessary to monitor the mutations in primers and probes of SARS-CoV-2 detection over time.
Methods:
We developed two rRT-PCR assays to detect the RNA-dependent RNA polymerase (RdRp) and nucleocapsid (N) genes of SARS-CoV-2. We evaluated these assays together with our previously published assays targeting the ORF1ab and N genes for the detection and confirmation of SARS-CoV-2 and its variants of concern (VOCs). In addition, we also developed two rRT-PCR assays (S484K and S501Y) targeting the spike gene, which when combined with the open reading frames (ORF)1ab assay, respectively, to form duplex rRT-PCR assays, were able to detect SARS-CoV-2 VOCs (lineages B.1.351 and B.1.1.7).
Results:
Using a SARS-CoV-2 stock with predetermined genomic copies as a standard, the detection limit of both assays targeting RdRp and N was five copies/reaction. Furthermore, no cross-reactions with six others human CoVs (229E, OC43, NL63, HKU1, severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus) were observed using these assays. In addition, the S484K and S501Y assays were combined with the ORF1ab assay, respectively.
Conclusions:
Four rRT-PCR assays (RdRp, N, S484K, and S501Y) were used to detect SARS-CoV-2 variants, and these assays were shown to be effective in screening for multiple virus strains.
Keywords: COVID-19, SARS-CoV-2, RT-PCR assay, Variants of concern, RNA polymerase, Nucleocapsid, SARS-CoV-2 B.1.351, SARS-CoV-2 B.1.1.7, SARS-CoV-2 20A S484K variant
Introduction
After the novel coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was identified in patients with pneumonia in December 2019,[1–3] the coronavirus disease 2019 (COVID-19) pandemic became a global phenomenon. As of March 30, 2021, 129,215,179 confirmed cases of COVID-19, including 2,820,098 deaths from 223 countries, areas, or territories, have been reported to the World Health Organization (WHO).[4] With the continuous monitoring of SARS-CoV-2 by researchers globally, new variants have been identified with mutations in key regions, such as the viral receptor-binding domain. The emergence of these variants may result in greater difficulty in controlling COVID-19.
On February 25, 2021, the WHO provided working definitions for SARS-CoV-2 variants of interest (VOIs) and variants of concern (VOCs), with the threshold for determining VOCs being greater than that for VOIs, and recommended that the greatest attention and resources should be directed at the variants with the highest public health implications. SARS-CoV-2 VOCs are associated with (i) increased transmissibility or detrimental changes in COVID-19 epidemiology, (ii) increased virulence or change in clinical disease presentation, or (iii) decreased effectiveness of public health and social measures and available diagnostics, vaccines, and therapeutics.[5] At present, there are six SARS-CoV-2 VOIs and three VOCs assessed by the WHO, including lineages B.1.1.7, B.1.351, and B.1.1.28.1.[6]
High-frequency nucleotide substitutions in the primer or probe binding regions of emerging VOIs and VOCs may result in decreased sensitivity of SARS-CoV-2 detection techniques. Some primer and probe sets have already shown nucleotide mismatches;[7] hence, it is necessary to develop new, sensitive, and specific real-time reverse transcription-polymerase chain reaction (rRT-PCR) assays to support the public health response to SARS-CoV-2 and COVID-19. In this study, we report the development of rRT-PCR assays (RNA-dependent RNA polymerase [RdRp], nucleocapsid [N], S484K, and S501Y) to detect SARS-CoV-2 variants. We provide detailed technical data and a comprehensive comparison of the results with the performance of our published ORF1ab and N assays[8] using multiple viral strains.
Methods
Viruses
SARS-CoV-2 strains (Pango lineage: B, B.1.351, and B.1.1.7) were isolated in our laboratory from clinical specimens obtained from SARS-CoV-2 positive patients. Human coronavirus-229E (229E-CoV), -OC43 (OC43-CoV), -NL63 (NL63-CoV), and -HKU1 (HKU1-CoV) strains were stored in our laboratory. Severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle Eastern respiratory syndrome (MERS)-CoV infectious complementary DNA (cDNA) clones were provided by Dr Jin-Cun Zhao (Guangzhou Medical University, China).
RNA extraction
Viral RNA including SARS-CoV-2 (Pango lineage: B, B.1.351, and B.1.1.7), human coronavirus-229E (229E-CoV), -OC43 (OC43-CoV), -NL63 (NL63-CoV), and -HKU1 (HKU1-CoV) were extracted from 140 μL of processed coronavirus cell culture supernatants using the QIAamp Viral RNA Mini Kit (QIAGEN GmbH, Hiden, Germany) according to the manufacturer's instructions. Approximately 60 μL of the total nucleic acid eluate was recovered into nuclease-free tubes and either tested immediately or stored at −70°C until further use.
Primer and probe design
Using Primer Premier software version 5 (Applied Biosystems, Austin, TX, USA), we designed multiple primer and probe sets targeting the RdRp, N, and Spike (S) gene regions of the SARS-CoV-2 genome (hCoV-19/Wuhan/IVDC-HB-04/2020, Accession ID: EPI_ISL_402120, B lineage; hCoV-19/South Africa/KRISP-K007869/2020, Accession ID: EPI_ISL_860630, B.1.351 lineage; hCoV-19/England/MILK-9E05B3/2020, Accession ID: EPI_ISL_601443, B.1.1.7 lineage; hCoV-19/Brazil/SP-1869/2021, Accession ID: EPI_ISL_1293069, B.1.1.28 lineage; hCoV-19/USA/CA-CZB-14228/2020, Accession ID: EPI_ISL_738815, B.1.429 lineage; hCoV-19/Finland/FinD796H/2021, Accession ID: EPI_ISL_1061414, B.1.1.318 lineage). Hydrolysis probes were labeled at the 5′ end with 6-carboxyfluorescein (6-FAM) and with black hole quencher 1 at the 3′ end [Table 1]. The S484K and S501Y rRT-PCR assays targeting S gene of SARS-CoV-2 variants, the primers and probes were (forward: 5′- CTGAAATCTATCAGGCCGGTAG -3′; reverse: 5′-CTACTCTGTATGGTTGGTAACC-3′; probe: 5′-FAM-TAATGGTGTTAAAGGTT-MGB-3′) and (forward:5′- GTTACTTTCCTTTACAATCATATG-3′; reverse: 5′- TTTAGGTCCACAAACAGTTGC-3′; probe: 5′-FAM- AACCCACTTATGGTG-MGB-3′), respectively.
Table 1.
SARS-CoV-2 rRT-PCR assays primer/probe sequences.
| Target | Gene | Primer | Sequence (5′–3′) | Genome location∗ |
| ORF1ab | nsp10-11 | Forward | CCCTGTGGGTTTTACACTTAA | 13342–13362 |
| Reverse | ACGATTGTGCATCAGCTGA | 13442–13460 | ||
| Probe | FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 | 13377–13404 | ||
| N | N | Forward | GGGGAACTTCTCCTGCTAGAAT | 28881–28902 |
| Reverse | CAGACATTTTGCTCTCAAGCTG | 28958–28979 | ||
| Probe | FAM-TTGCTGCTGCTTGACAGATT-BHQ1 | 28934–28953 | ||
| N9 | N | Forward | TTTGGTGGACCCTCAGATTC | 28322–28341 |
| Reverse | TTGCCATGTTGAGTGAGAGC | 28436–28455 | ||
| Probe | FAM-CAGTAACCAGAATGGAGAACGCAGTGG-BHQ1 | 28348–28374 | ||
| RdRp | RdRp | Forward | CAGGTGGAACCTCATCAGGA | 15470–15489 |
| Reverse | CCGTGACAGCTTGACAAATG | 15525–15544 | ||
| Probe | FAM-ATGCCACAACTGCTTATGCTAATAGTG-BHQ1 | 15491–15517 |
The location on the reference genome, accession ID: EPI_ISL_402119, EPI_ISL_402120. BHQ1: Black hole quencher 1; N: Nucleocapsid; ORF: Open reading frames; RdRp: RNA-dependent RNA polymerase; rRT-PCR: Real-time reverse transcription-polymerase chain reaction; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
rRT-PCR assay
The rRT-PCR assay was performed using the AgPath-IDTM One-Step RT-PCR Kit (Applied Biosystems). Each 25 μL of reaction mixture contained 12.5 μL of 2× RT-PCR buffer, 1 μL of RT-PCR enzyme mix, 5 μL of RNA, 400 nmol/L each of forward and reverse primers, and 200 nmol/L of probe [Table 1]. Thermal cycling was performed at 45°C for 10 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Each run included one SARS-CoV-2 genomic template control and at least two negative or mock controls (for extraction and PCR amplification). Assay specificity was determined using high-titer virus stock and infectious cDNA clones from our laboratory. Identities and viral RNA concentrations were validated by virus-specific RT-PCR before the experiment.[9] The limit of detection was independently assessed using SARS-CoV-2 stock with predetermined genomic copies. The calibration curve for the genomic copy number vs. Ct value was obtained using the Mx3005P Real-Time PCR System (Agilent Technologies, La Jolla, CA, USA).
S484K and S501Y rRT-PCR assays
The primer and probe sets for the S484K and S501Y assays are listed in Table 2. The assays were developed to target the E484K and N501Y mutations in the S gene of the B.1.351 and B.1.1.7 lineage viruses, respectively. These assays were then combined with ORF1ab to form duplex rRT-PCR assays for the preliminary screening of B.1.351 and B.1.1.7 lineage infections. The PCR mix and thermal cycling parameters for the two duplex rRT-PCR assays are as stated above in the rRT-PCR assay subsection of the “Methods” section.
Table 2.
SARS-CoV-2 rRT-PCR assays limits of detection (viral RNA copies/reaction).
| No. of positive tests/No. of test replicates (%) | ||||
| Predicted No. of viral copies/reaction | ORF1ab | N | RdRp | N9 |
| 20 | 24/24 (100.0) | 24/24 (100.0) | 24/24 (100.0) | 24/24 (100.0) |
| 10 | 24/24 (100.0) | 24/24 (100.0)∗ | 24/24 (100.0) | 24/24 (100.0) |
| 5 | 24/24 (100.0)∗ | 23/24 (95.8) | 24/24 (100.0)∗ | 24/24 (100.0)∗ |
| 2.5 | 21/24 (87.5) | 21/24 (87.5) | 20/24 (83.3) | 20/24 (83.3) |
| 1.25 | 18/24 (75.0) | 14/24 (58.3) | 16/24 (66.7) | 17/24 (70.8) |
Limits of detection at which 100% of replicates were positive. ORF: Open reading frames; RdRp: RNA-dependent RNA polymerase; rRT-PCR: Real-time reverse transcription-polymerase chain reaction; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Results
Analysis of primer and probe signatures
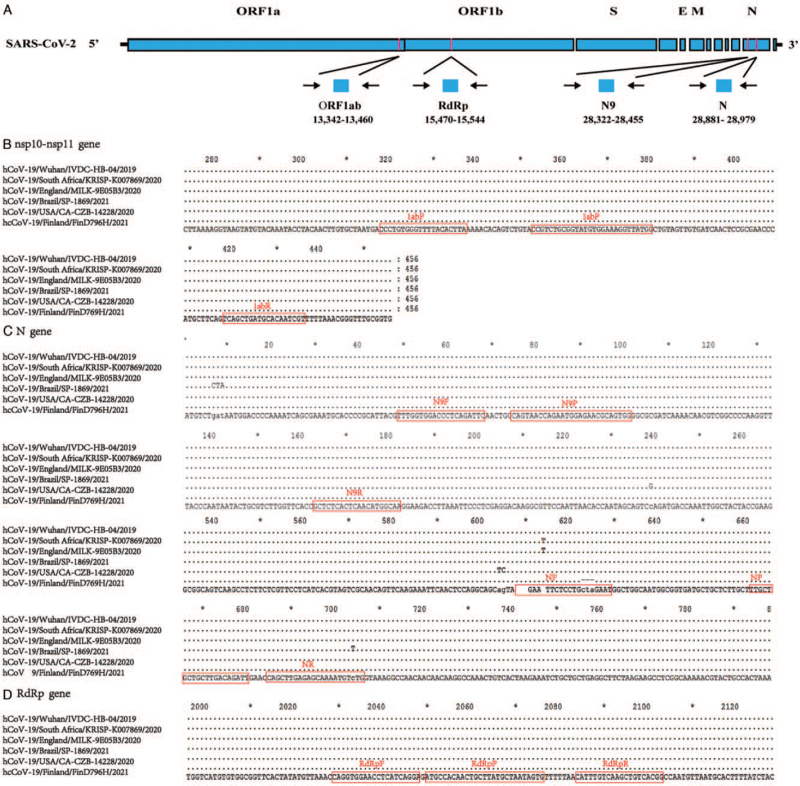
As noted above, we downloaded five SARS-CoV-2 sequences from global initiative on sharing all influenza data (https://www.gisaid.org/) that have recently garnered much attention, including three VOCs and two VOIs. These sequences were aligned to the original strain (HCoV-19/Wuhan/IVDC-HB-04/2020), and the alignment was used for assay design. Two assays (RdRp and N9) were selected based on how well they matched with the SARS-CoV-2 genome. The positions of the amplicons for the two assays, as well as our previously published assays (ORF1ab and N) relative to the SARS-CoV-2 genome, are shown in Figure 1A. The alignments confirmed how well the selected primers matched all sequences for the three assays (ORF1ab, RdRp, and N9). However, there were six nucleotide mismatches in the N forward primer, which predicted assay failure. Alignments of SARS-CoV-2 showing the primer and probe binding domains are presented in Figure 1B–D.
Figure 1.
(A) Relative positions of amplicon targets on SARS-CoV-2 genome. Numbers below amplicons are genome positions (accession ID: EPI_ISL_402120). Partial alignments are shown on (B) nsp10-nsp11 gene, (C) N gene, and (D) RdRp gene. Boxes indicate regions targeted by primers and probes designed in this study. E: Envelope; M: Membrane; N: Nucleocapsid; ORF: Open reading frames; RdRp: RNA-dependent RNA polymerase; S: Spike; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Assay sensitivity based on SARS-CoV-2 strain
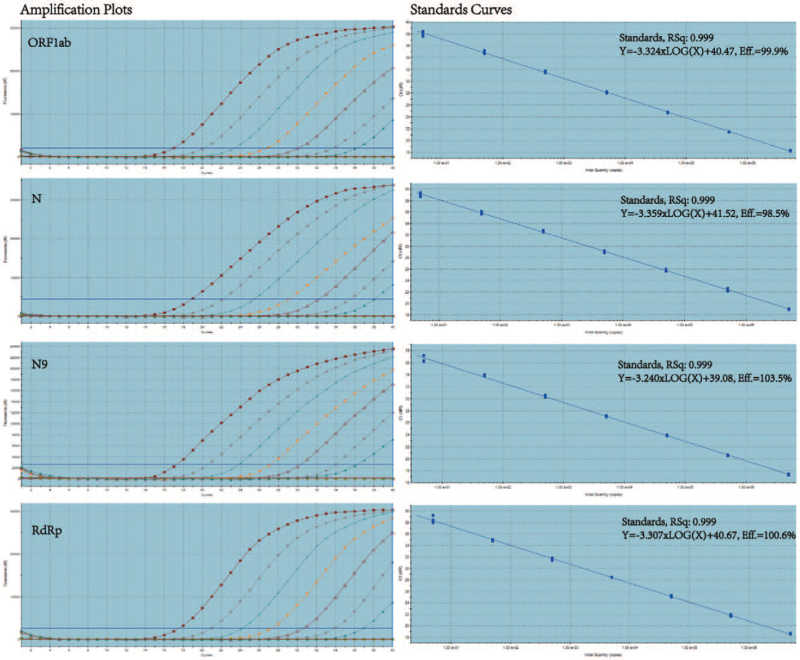
Using SARS-CoV-2 viral RNA with a known copy number, we found that the four assays showed a dynamic range of approximately seven orders of magnitude (5 × 106 copies/reaction). The amplification efficiency of the four assays ranged from 98.5% to 103.5%, R2 > 0.999, in each assay [Figure 2]. To accurately determine the detection limits of these assays, serial 2-fold dilutions of quantified viral RNA (1.2520 copies/reaction) were also tested with each assay in 24-fold replicates. The lowest detection limit was defined as the dilution at which all replicates were positive for each assay. The minimum detection limit was five copies/reaction for the ORF1ab, RdRp, and N9 assays and ten copies/reaction for the N gene assay [Table 2].
Figure 2.
Amplification plots of SARS-CoV-2 RNA serial ten-fold dilutions. Dilutions ranged between 5 and 5 × 106 SARS-CoV-2 RNA copies/reaction for the ORF1ab, N, N9, and RdRp assays. Linear correlation coefficients (R2) and efficiencies for each rRT-PCR assay were determined (3 tests/dilution). N: Nucleocapsid; ORF: open reading frames; RdRp: RNA-dependent RNA polymerase; rRT-PCR: Real-time reverse transcription-polymerase chain reaction; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Assay specificity based on hCoV RNA
The rRT-PCR assays were performed with RNA from six human coronaviruses, including four laboratory strains (229E-CoV, OC43-CoV, NL63-CoV, and HKU1-CoV) and two infectious cDNA clones (SARS-CoV and MERS-CoV). No cross-reaction was observed with any of the primer and probe sets for the ORF1ab, N, RdRp, and N9 assays.
Duplex rRT-PCR assays to detect viruses from the B.1.351 and B.1.1.7 lineages
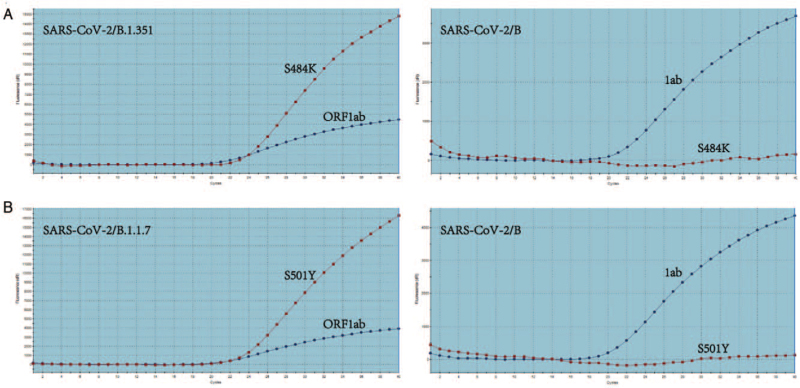
The S484K and ORF1ab duplex rRT-PCR assays were used to detect B (original strain [HCoV-19/Wuhan/IVDC-HB-04/2020]) and B.1.351 lineage viruses, respectively. The results showed that both assays could amplify and detect the B.1.351 lineage viruses; however, only the ORF1ab assay detected the B lineage virus. Similar results were obtained using the S501Y and ORF1ab duplex rRT-PCR assays for detecting B and B.1.1.7 lineage viruses, respectively [Figure 3]. Our preliminary results suggested that the S501Y assay is as sensitive as the ORF1ab assay and the S484K assay is slightly less sensitive than the ORF1ab assay in detecting viral RNA (data not shown).
Figure 3.
SARS-CoV-2 variants detected by each duplex rRT-PCR assay. SARS-CoV-2/B (hCoV-19/Wuhan/IVDC-HB-04/2020) was used as a control. (A) SARS-CoV-2/B.1.351 variants detected by ORF1ab and S484K assays. (B) SARS-CoV-2/B.1.1.7 variants detected by ORF1ab and S501Y assays. ORF: Open reading frames; rRT-PCR: Real-time reverse transcription-polymerase chain reaction; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2.
Discussion
The average evolutionary rate for coronaviruses is approximately 10−4 nucleotide substitutions per site per year,[10] with mutations arising in each replication cycle. As the COVID-19 pandemic became global, variants of SARS-CoV-2 started garnering increasing attention. At present, the WHO has designated three SARS-CoV-2 variants as VOCs and six as VOIs. The B.1.1.7 lineage, one of the most closely monitored SARS-CoV-2 VOCs, was detected in November 2020, and it quickly spread around the world. The genetic mutations identified in this variant cause many changes in the biological characteristics of SARS-CoV-2. The N501Y substitution in the S protein has been shown to increase ACE2 binding,[11] potentially reducing susceptibility to neutralizing antibodies[12] with the result of increased viral transmission advantage.[13] In addition, the S gene variations of the B.1.1.7 lineage have been associated with diagnostic test failure of a commercial S gene rRT-PCR assay,[14] which significantly hinders the tracking and management of the spread of SARS-CoV-2. Thus, it is imperative to align rRT-PCR primer/probe sets and SARS-CoV-2 variant sequences to assess the possible shortcomings of the current testing methods and to widen their detection spectrum.
With the emergence of SARS-CoV-2 variants, it is necessary to monitor mutations in primers and probes and update them over time. In the present study, we found an increasing number of mismatches over time in the forward primer of our previously published rRT-PCR N assay that targets the SARS-CoV-2 variant genome. Therefore, we developed two new rRT-PCR assays (RdRp and N9) that target the RdRp and N genes of SARS-CoV-2, respectively, and used these new assays to detect SARS-CoV-2 VOCs. The RdRp and N9 assay panels were shown to be both sensitive and specific, similar to our previously published rRT-PCR ORF1ab assay. The primer/probe sets targeting the RdRp gene, which has one of the lowest evolutionary rates in the SARS-CoV-2 genome, is to a certain extent resistant to mismatches, and targeting the N gene should offer enhanced sensitivity because of the relative abundance of N gene mRNA produced during virus replication. RdRp and N9 were combined with the ORF1ab assay to enhance the clinical diagnosis of SARS-CoV-2. In addition, we developed S484K and S501Y assays. When combined with the ORF1ab assay to form duplex rRT-PCR assays for preliminary detection of the SARS-CoV-2 B.1.351 and B.1.1.7 lineages, these PCRs showed good detection ability. In addition, there are some other significant mutations in SARS-CoV-2 variants, such as D614G, L452R, and E484Q, which can also be considered for specific detection with rRT-PCR assay; relevant research in our laboratory is in progress.
To conclude, we developed four rRT-PCR assays (RdRp, N, S484K, and S501Y) to detect SARS-CoV-2 variants and applied them to multiple virus strains. These assays provide a material basis for the monitoring, management, and control of SARS-CoV-2 infection.
Funding
This work was supported by grants from the National Key Research and Development Program of China (Nos. 2016YFD0500301, 2021YFC0863300, and 2020YFC0840900).
Conflicts of interest
None.
Footnotes
How to cite this article: Lu RJ, Zhao L, Huang BY, Ye F, Wang WL, Tan WJ. Real-time reverse transcription-polymerase chain reaction assay panel for the detection of severe acute respiratory syndrome coronavirus 2 and its variants. Chin Med J 2021;134:2048–2053. doi: 10.1097/CM9.0000000000001687
References
- 1.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet 2020; 3:470–473. doi: 10.1016/S0140-6736 (20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan WJ, Zhao X, Ma XJ, Wang WL, Niu PH, Xu WB, et al. Notes from the field: a novel coronavirus genome identified in a cluster of pneumonia cases – Wuhan, China 2019−2020. China CDC Wkly 2020; 2:61–62. doi: 10.46234/ccdcw2020.017. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 3:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. WHO Coronavirus (COVID-19) Dashboard; 2021. Available from: https://covid19.who.int/. [Last accessed on April 5, 2021] [Google Scholar]
- 5.World Health Organization. Weekly Epidemiological Update; February 25, 2021. Available from: https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update/. [Last accessed on April 5, 2021] [Google Scholar]
- 6.World Health Organization. Weekly Epidemiological Update on COVID-19; March 30, 2021. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---31-march-2021/. [Last accessed on April 5, 2021] [Google Scholar]
- 7. Publicly Database, GISAID. (GISAID_hCoV19_Analysis_Update_20210618); 2021. Available from: https://www.epicov.org/epi3/entities/tmp/tmp_sd_2021_06_21_20_58_qv1lid_10z93bc9bafd/GISAID_hCoV-19_Analysis_Update_20210618.pdf/. [Last accessed on June 21, 2021] [Google Scholar]
- 8.Niu P, Lu R, Zhao L, Wang H, Huang B, Ye F, et al. Three novel real-time RT-PCR assays for detection of COVID-19. China CDC Wkly 2020; 2:453–457. doi: 10.46234/ccdcw2020.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu P, Shen J, Zhu N, Lu RJ, Tan WJ. Two-tube multiplex real-time reverse transcription PCR to detect six human coronaviruses. Virol Sin 2016; 31:85–88. doi: 10.1007/s12250-015-3653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016; 24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan KK, Tan TJC, Narayanan KK, Procko E. An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci Adv 2021; 7:eabf1738.doi: 10.1126/sciadv.abf1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. ECDC to Assess Risk Associated With Spread of SARS-CoV-2 in Mink Farms. European 148 Centre for Disease Prevention and Control; 2020. Available from: https://www.ecdc.europa.eu/en/news-149events/ecdc-assess-risk-associated-spread-sars-cov-2-mink-farms. [Last accessed on Nov 8, 2020] [Google Scholar]
- 13.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv 2020; doi: 10.1101/2020.12.30.20249034. [Google Scholar]
- 14.Bal A, Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill 2021; 26:2100008.doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]