Abstract
In this work, we constructed an exonuclease III cleavage reaction-based isothermal amplification of nucleic acids with CRISPR/Cas12a-mediated pH-induced regenerative Electrochemiluminescence (ECL) biosensor for ultrasensitive and specific detection of SARS-CoV-2 nucleic acids for SARS-CoV-2 diagnosis. The triple-stranded nucleic acid in this biosensor has an extreme dependence on pH, which makes our constructed biosensor reproducible. This is essential for effective large-scale screening of SARS-CoV-2 in areas where resources are currently relatively scarce. Using this pH-induced regenerative biosensor, we detected the SARS-CoV-2 RdRp gene with a detection limit of 43.70 aM. In addition, the detection system has good stability and reproducibility, and we expect that this method may provide a potential platform for the diagnosis of COVID-19.
Keywords: SARS-CoV-2 RdRp gene, CRISPR/Cas12a, ECL, Triple-helix, Regenerative Biosensor, DNA tetrahedron
1. Introduction
Because of the highly infectious nature of the SARS-CoV-2 virus, the COVID-19 outbreak is spreading widely worldwide and is of great global concern [1]. Researchers are currently developing a variety of tests for SARS-CoV-2 [2], [3]. At the same time, the WHO website provides the relevant procedures and steps for conducting virus testing used in several countries such as China, Germany, Japan, and the United States, all of which are based on real-time reverse transcription PCR detection techniques that, despite their high efficiency, still require a high level of technical expertise and expensive [4]. The procedure is based on real-time reverse transcription PCR, which, despite its efficiency, requires a high level of expertise and expensive equipment to operate[5]. Moreover, the inactivation of nucleic acid amplification enzymes and the varying levels of expertise often result in false-positive test results[6]. In this context, the development of any ultrasensitive diagnostic test technology for the evaluation of suspected cases of infection might be helpful for SARS-CoV-2 detection, regardless of the availability of qualified professionals or sophisticated equipment for virus detection.
In the past few years, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology has sparked not only a gene-editing boom but also a revolution in molecular diagnostics[7]. Notably, researchers have recently started to use CRISPR for in vitro detection of nucleic acids, and thus it is expected to be developed as a powerful and accurate molecular diagnostic tool[8] . With the single-stranded DNA Trans-cleavage activity of the Cas12a enzyme, combined with various isothermal nucleic acid amplification techniques, a series of CRISPR-Cas molecular diagnostic platforms based on fluorescent signal output have been developed [9]. Among the CRISPR-Cas effector family, Cas12 is an RNA-directed DNase that belongs to the class II V-A system, which induces the division of arbitrary single-stranded DNA (ssDNA) upon target recognition, which leads to the degradation of the ssDNA reporter and the release of a fluorescent signal at the division site, which can be detected by a portable method[10], [11], [12]. Although optical transduction-based CRISPR-Cas molecular diagnostics have been widely used for nucleic acids[13], small molecules[14] and metal ions[15], considering that these strategies need to be done on expensive and complex devices, there is a need to extend the CRISPR/Cas system to the field of solid-load-free electrochemical biosensing.
For the detection of COVID-19, medical personnel often use real-time reverse transcriptase polymerase chain reaction (RT-PCR) [16] to detect different regions of the SARS-CoV-2 genome, including the nucleocapsid protein gene (N gene) [17], envelope protein gene (E gene), spike protein gene (S gene), and RNA-dependent RNA polymerase gene (RdRp gene). Detection of different regions of the SARS-CoV-2 genome will affect the specificity and accuracy of the assay. the N gene may cross-react with other coronaviruses and its sensitivity is low[17]. The S gene is the most variable region of the genome[18]. the E gene is a conserved region throughout the beta-coronavirus and can be distinguished from other viruses, but not from homologous viruses, such as SARS-CoV-2 from severe acute respiratory syndrome (SARS-CoV), to distinguish them from each other[19]. Notably, the RdRp gene, located in the ORF1ab region, is highly specific and can distinguish SARS-CoV-2 from other viruses [20], [21]. Researchers have widely used the RdRp gene as a detection target, making the screening of COVID-19 with high specificity and accuracy[22].
In addition, SARS-CoV-2 has recently become more transmissible than before. Its latest incarnation, the Delta (or B.1.617.2) variant[23], is the fastest spreading form of the virus to date. Delta was first identified in India, where it swept through this spring (2021), killing hundreds of thousands of people, and it has quickly become the predominant coronavirus variant worldwide[24]. The major mutations in the Delta variant's mutation site on the spike protein are receptor-binding domain mutations (L452R), there is no mutation in the RdRp gene located in the ORF1ab region[24]. So testing for the RdRp gene can also screen for carriers of the Delta variant of the virus.
Electrochemiluminescence (ECL) immunoassay, the most advanced labeling immunoassay technique, is a new generation of labeling immunoassay technique after radioimmunoassay, enzyme immunoassay, fluorescence immunoassay and chemiluminescence immunoassay, which is sensitive, rapid and stable, and is the technological leader in solid-phase labeling immunoassay[25], [26], [27] . Electrochemiluminescence is a specific chemiluminescence reaction triggered by electrochemistry on the electrode surface, which is actually a perfect combination of two processes: electrochemistry and chemiluminescence[2], [28], [29], [30] . The main difference between electrochemiluminescence and ordinary chemiluminescence is that the former is an electrically initiated luminescence reaction, cyclic and multiple luminescence, while the latter is a single instantaneous luminescence reaction initiated by a mixture of compounds[31], [32]. The ECL reaction is therefore easily and precisely controlled and extremely reproducible.
The readout methods for CRISPR/Cas12a systems include the colorimetric method as well as the fluorescence method. In terms of sensor design, universally colorimetric detection methods reflect the content of the target through the mutual reaction between nucleic acid-modified Au nanoparticles caused by the target-participated CRISPR/Cas system, inducing naked-eye visible color changes for analysis of target concentrations[13], [33], [34]. In contrast, the CRISPR/Cas system based on fluorescence readout often causes the separation of the fluorescent moiety from the quenched moiety by severing the molecular probes linking the quenched and fluorescent moieties at each end of ssDNA, finally generating a signal recovery[35]. However, CRISPR/Cas12a-based sensors with electrochemiluminescence readout alter the ECL signal output by introducing a target to cause a change in the energy transfer of the ECL emitting materials on the electrode surface. Since the ECL signal is generated without the stimulation of an external light source, the background signal interference is reduced while possessing a high sensitivity[36].
In addition, with the development of nanotechnology, the development of new ECL materials has become a new hot area[37]. In recent years, two-dimensional (2D) composites, due to their unique metallic nature, tunable nanosheet thickness, and high electron density, which in turn increase the affinity of cations to the surface, have enabled ECL sensors applying 2D composites to have excellent electron selectivity and flux, and greatly facilitate the diffusion of electrons[3], [8]. Therefore, the ECL sensors based on two-dimensional composites show up-and-coming applications.
In this work, we constructed an exonuclease III cleavage reaction-based isothermal amplification of nucleic acids with a CRISPR/Cas12a-mediated ECL biosensor for ultrasensitive and specific detection of SARS-CoV-2 nucleic acids for SARS-CoV-2 diagnosis. The sensor eliminates the need to immobilize the recognition probe on the electrode surface, dramatically simplifies the electrode preparation step, and exhibits a signal-enhanced output pattern. Interestingly, the DNA tetrahedra-based three-stranded nucleic acid in our designed biosensor has an extreme dependence on pH, making the constructed biosensor reproducible[38]. This is crucial for the effective implementation of SARS-CoV-2 detection in areas where resources are currently relatively scarce.
2. Experimental section
2.1. Materials and reagent
The oligonucleotides purified by high-performance liquid chromatography (HPLC) used in our detection strategy were purchased from Genscript Biotech (Nanjing, China). Gold chloride trihydrate (HAuCl4·3H2O), sodium citrate, potassium persulfate (K2S2O8), and sodium borohydride (NaBH4) were obtained from Aladdin Biochemical Technology Co. Ltd. (Shanghai, China). g-C3N4 were obtained from Jiangsu XFNANO Materials Tech, Co. Ltd. (Nanjing, China). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and 6-mercaptohexanol (MCH) were gained from Sigma-Aldrich (St Louis, MO, USA). Exonuclease III and 10 × NEB buffer were obtained from New England Biolabs (USA). The purified DNA fragments (Table S1) were synthesized by Genscript Biotechnology Co. Ltd. (Nanjing, China)
2.2. Instrumentation
Transmission electron microscopy (TEM) images were acquired from JEM-2100F, JEOL (Japan). ECL signals were acquired from an ECL-6B acquisition gifted by the State Key Laboratory of Analytical Chemistry for Life Sciences, Nanjing University. UV–Vis absorption spectra were measured by a Microplate Reader (Spectra Max M5e) in the range of 250 nm to 700 nm (Molecular Devices Co. Ltd, USA).
2.3. Preparation of the Au-g-C3N4
First, 50 μL of HAuCl4 (0.01 M) solution and 4 mL of g-C3N4 (0.15 mg mL−1) were mixed and sonicated for 10 min and stirred at room temperature for 1 h. Then, 100 μL of the just-configured NaBH4 (0.01 M) solution was injected into the mixed solution and the reaction was continued in an ice bath for 20 min. Then 50 μL of sodium citrate solution was added to the above mixed solution and the mixed solution was stirred continuously for 20 min at 25 °C. Finally, to further remove the excess AuNPs, sodium citrate and NaBH4, the above mixed solution was washed and sonicated three times to obtain pure Au-g-C3N4, which was redispersed in 1 mL of ddH2O and stored at 4 °C for the next experiment.
2.4. Preparation of DNA tetrahedron
The construction of DNA tetrahedron was based on previous literature with slight modifications[39]. Briefly, four single-stranded DNAs (T1, T2, T3, T4) of 10 μL each were first dissolved in PBS buffer solution (10 mM TCEP and 50 mM MgCl2, pH = 7.4). Then, the four single-stranded DNAs were mixed together, incubated at 95 °C for 5 min, and slowly cooled to room temperature. In this way, the DNA tetrahedron structure was formed.
2.5. Fabrication of the pH-induced regenerative ECL biosensor
First, the glassy carbon electrode (GCE) was polished with 0.3 and 0.05 μm alumina powder and rinsed with deionized water for the following experiments. Then 10 μL of Au-g-C3N4 was added dropwise to the GCE to obtain Au-g-C3N4/GCE and dried at room temperature. Subsequently, the obtained Au-g-C3N4/GCE was immersed in 100 μL of DNA tetrahedron (1 μM) solution containing 10 mM TCEP for 8 h, allowing the DNA tetrahedron to be modified on the electrode surface. Next, the above-modified GCE was immersed in 60 μL of MCH (100 μM) solution for 1 h to seal the gold nanoparticle surface to prevent non-specific adsorption. In this way, the pH-induced regenerative biosensor was prepared successfully.
2.6. Exo III-assisted targeted nucleic acid cycling reaction
To eliminate the digestion of H1a/H2a duplex by exonuclease III, cleavage of H1/RdRp gene duplex and cleavage of H1/RdRp gene duplex were performed in different solutions. First, both DNA strands, H1 and H2, were annealed at 95 °C for 5 min and then gradually cooled to room temperature over 4 h for the formation of stable hairpin-type structures in different EP tubes, respectively. Then, RdRp gene at different concentrations was introduced in H1 and H2 (1 μM, respectively) and incubated for 2 h at 37 °C and exonuclease III (0.5 U/μL) was added to each tubes for 1 h at 37 °C. After that, the above solutions were kept at 65 °C for 10 min to stop the enzymatic reaction. Finally, the two solutions were mixed and heated at 65 °C for 10 min and then cooled naturally to room temperature to form the H1a/H2a duplex.
For clinical samples assay, voluntary volunteer-donated pharyngeal swabs (ethically compliant) were used to validate the resistance to interference and clinical application potential of this biosensor. Volunteer pharyngeal swabs were spiked with different concentrations of the nucleic acid to be tested for the next experiments.
2.7. Polyacrylamide gel electrophoresis (PAGE) analysis
First, freshly prepared 20% polyacrylamide gels were run at 80 V for 40 min. Next, 10 μL of various DNA samples (1 μM) and loading buffer were mixed and injected into the gel sample bath and run at 110 V for 2.5 h. Immediately thereafter, the gel electrophoresis was immersed in Gel-Red dilution for 30 min. Finally, the stained gels were photographed with a ChemiDoc MP Imager instrument.
2.8. CRISPR/Cas12a cleavage reaction and ECL signal obtain
Next, 50 μL of the above reaction solution was added to lysis buffer (20 mM Tris HCl, 100 mM KCl, 5 mM MgCl2, 5% glycerol and 1 mM DTT) containing 100 nM CRISPR/Cas12a-gRNA and 1 μM DNA1. The above solutions were then mixed and reacted for 30 min, allowing DNA1 to be sufficiently cleaved. Then, the prepared pH-induced regenerative ECL biosensor was submerged in the above solution for 30 min, and then the ECL signal was measured.
The above-mentioned electrodes forming triple-stranded DNA were immersed in 5 mL of PBS (pH = 7.4) containing S2O8 2- (0.1 M) for measurements with a scan range of 0–1.5 V. The tests were performed using a three-electrode system with a 3 mm diameter GCE as the working electrode, a platinum wire electrode as the counter electrode and Ag/AgCl as the reference electrode.
3. Results and discussion
3.1. The working principle
An isothermal amplification reaction based on the targeting molecule-Exo III assisted with tandem CRISPR/Cas12a controlled signal amplification was used to construct an electrochemiluminescent (ECL) sensor for the specific and ultrasensitive detection of RdRp gene nucleic acid fragments. The commonly attributed detection method for commercially available COVID-19 is the conversion of SARS-CoV-2 RNA gene into complementary DNA (cDNA) using RT-qPCR amplification and subsequent signal output by fluorescence method. In this study, a CRISPR/Cas-based signal amplification reaction design using SARS-CoV-2 RNA gene converted cDNA sequences (RdRp gene)[3], [40] were used as a study object for investigating the feasibility of pH-induced regenerative ECL biosensor. Scheme 1 demonstrates the corresponding principle and construction process of the biosensor. The ECL biosensor employs a DNA tetrahedron structure modified on the electrode surface, and the DNA tetrahedron prongs can form DNA triple-stranded complexes with DNA1. In the initial state of the nucleic acid amplification reaction, two hairpin DNAs (H1 and H2) are introduced, and in the presence of exonuclease III (Exo III), different parts of the target are cut synchronously to release the two output DNAs (H1a and H2a). The H1a/H2a duplex can only be formed if both output H1a and H2a are present at the same time, which activates the activity of CRISPR/Cas12a and allows CRISPR/Cas12a to cut DNA1 (Scheme 1B). If there is no target substance present in the system, the H1a/H2a duplex cannot be formed, which in turn cannot activate the activity of CRISPR/Cas12a and cleave DNA1. DNA1 in the system can bind to the DNA tetrahedra on the electrode surface, thus causing a change in the ECL signal. Notably, the biosensor can be regenerated at pH = 10.0 (Scheme 1A), enabling continuous long-term use of this sensing platform for accurate monitoring of the concentration of nucleic acids to be measured and providing a new reference for the design of regenerative DNA tetrahedron ECL biosensors.
Scheme 1.
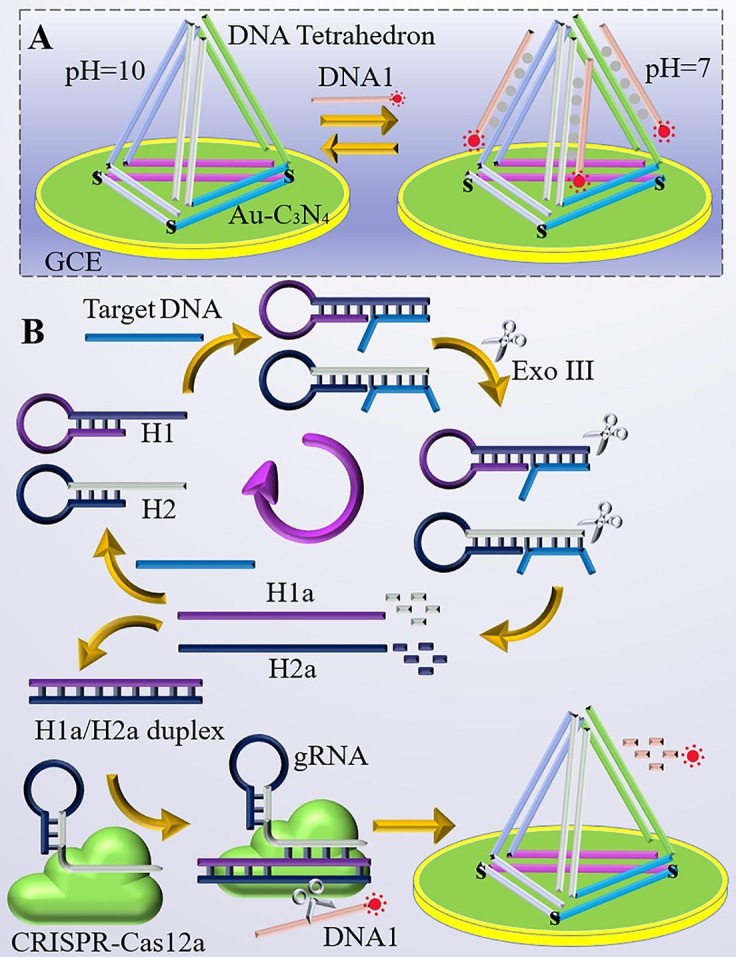
Schematic illustration of the pH-induced regenerative biosensor for the assay of SARS-CoV-2 RdRp gene based on the CRISPR/Cas12a Trans-activity.
In comparison with previous strategies of Electrochemical or Electrochemiluminescence biosensor based on CRISPR technology[8], [10], the previous CRISPR/Cas 12a for single-stranded nucleic acid cleavage needs to be performed on the electrode surface, which will induce a s steric effect of the interface on CRISPR/Cas 12a. In this pH-induced regenerative ECL biosensor, the conventional CRISPR/Cas 12a-based Electrochemical or Electrochemiluminescence biosensor target detection system is cleverly transferred to a homogeneous reaction system with less steric effect and more complete reaction. The method ensures the accuracy of the assay. This method is a novel attempt to ensure the accuracy and high sensitivity of the assay, but also has the flexibility of design.
It is worth noting that the cost of the 2D composite used in the biosensor is within $0.2, the cost of the nucleic acid is within $0.01, the price of the amplification enzymes used (Exo III and CRISPR/Cas12a) is approximately $4.00, the cost of additional chemical reagents for sensor pretreatment is approximately $0.2. Thus, the cost of the sensor is approximately $4.41. It is worth noting that the biosensor can be recycled, thus making the pH-induced regenerative ECL biosensor more affordable.
3.2. Characterization of Au-g-C3N4 and DNA tetrahedron
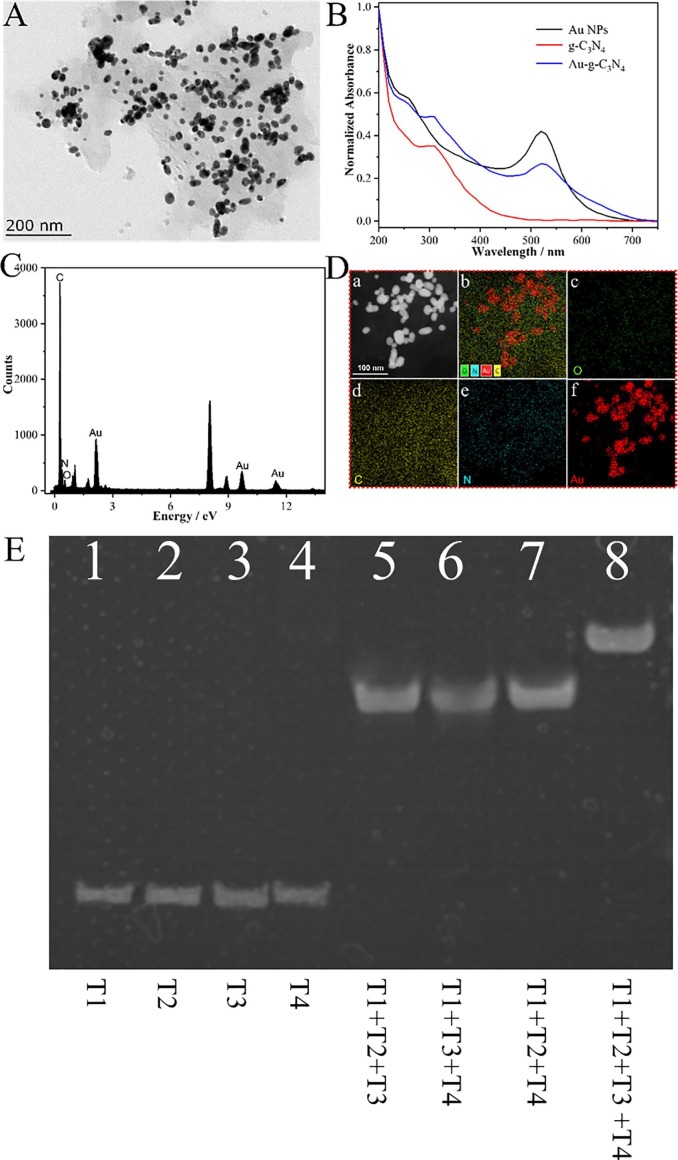
We first characterize the morphology of Au-g-C3N4 as shown in Fig. 1 A, where we can see that gold nanoparticles are dispersed on the surface of C3N4 nanosheets and provide a wide surface area for the linkage of nucleic acid probes. Moreover, the particle size of gold nanoparticles is very uniform, with a diameter of about 17 nm. UV–Vis absorption spectra were also made to characterize the synthesis of Au-g-C3N4, as shown in Fig. 1B, where g-C3N4 has a unique peak at 360 nm and AuNPs has a characteristic peak at 520 nm. If Au-g-C3N4 is successfully synthesized, it should have characteristic peaks at 360 nm and 520 nm. The above results demonstrate the successful synthesis of Au-g-C3N4. Next, we further performed elemental analysis using energy dispersive X-rays (EDX) as depicted in Fig. 1C. The results showed that elemental peaks of C, N and Au were present in the EDX spectrum of Au-g-C3N4. Besides, the elemental mapping data (as shown in Fig. 1D) successfully characterize the elemental distribution of Au-g-C3N4. All these morphological, spectroscopic, and elemental characterizations together verify the successful synthesis of Au-g-C3N4.
Fig. 1.
(A) TEM image of Au-g-C3N4 and (B) UV–vis absorption spectra to illustrate the synthesis of Au-g-C3N4. (C) EDX elemental mapping to show the elemental composites and distribution of the Au-g-C3N4. (D) Elemental mapping data of the Au-g-C3N4. (E) Polyacrylamide gel electrophoresis analysis of DNA tetrahedron: Lane 1 (T1), Lane 2 (T2), Lane 3 (T3), Lane 4 (T4), Lane 5 (T1 + T2 + T3), Lane 6 (T1 + T3 + T4), Lane 7 (T1 + T2 + T4), and Lane 8 (T1 + T2 + T3 + T4).
Polyacrylamide gel electrophoresis (PAGE) was used to characterize the fabrication of DNA tetrahedron. In Fig. 1E, the bands of T1, T2, T3, and T4 were shown in Lane 1, Lane 2, Lane 3 and Lane 4, respectivity. Also, the construction step of DNA tetrahedron were also shown in Fig. 1E: Lane 5 (T1 + T2 + T3), Lane 6 (T1 + T3 + T4), Lane 7 (T1 + T2 + T4), and Lane 8 (T1 + T2 + T3 + T4, namely DNA tetrahedron).
3.3. Possible ECL mechanism about pH-induced regenerative biosensor
In this study, the luminescence signal is initially derived from g-C3N4, whose luminescence mechanism in PBS solution of 0.1 M S2O8 2- enters with a scanning voltage from 0 to −1.5 V. The possible luminescence mechanism of g-C3N4 is as follows Eqs. (1)-(4). First, g-C3N4 acquires an electron under the applied voltage and transforms into g-C3N4 - (Eq. 1). At the same time, S2O8 2- gains one electron under the applied voltage and is reduced to SO4 2- and SO4 - (Equation 2). Further, an electron transfer occurs between SO4 - and g-C3N4 -, and g-C3N4 - becomes g-C3N4* in the excited state (Equation 3). Eventually, the excited state of g-C3N4* completes the electronic leap and transforms into the ground state g-C3N4 while producing light out at 460 nm.
| (1) |
| (2) |
| (3) |
| (4) |
3.4. Characterization of the pH-induced regenerative biosensor stepwise modification process
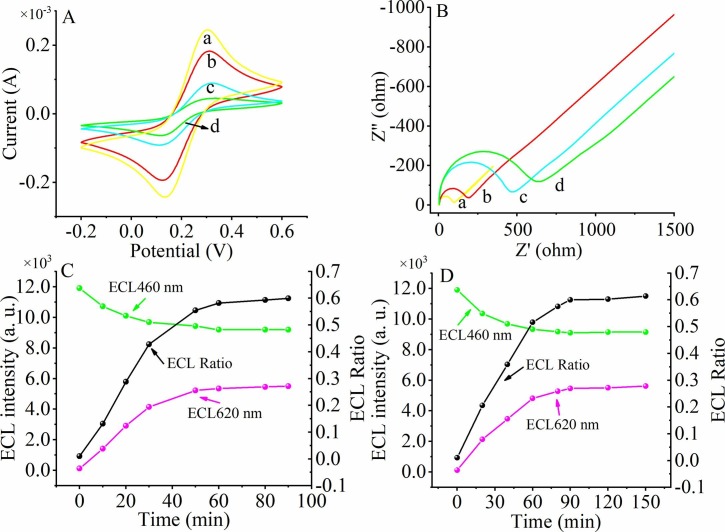
The electrochemical response of the pH-induced regenerative biosensor stepwise modification process is described by cyclic voltammetry (CV). The CV curve of the bare GCE electrode (curve a) shows the maximum current peak. When Au-g-C3N4 (curve b) is deposited on the electrode surface, the current peak decreases significantly. When further modification of DNA tetrahedrons and MCH (curve c) continued on the electrode surface, the current peak continued to decrease, indicating that poorly conductive substances were adsorbed on the electrode surface, which is consistent with the insulating properties of DNA tetrahedrons and MCH. After DNA1 continued to adsorb on the electrode surface through the tetrahedrons, the current continued to decrease (curve d), resulting in a similar decrease in the current peak.
Meanwhile, the stepwise construction process of the pH-induced regenerative biosensor was characterized by the Electrochemical impedance spectroscopy (EIS) shown in Fig. 2 B. The impedance value of the bare GCE electrode (curve a) is the smallest. Continued modification of Au-g- C3N4 (curve b), DNA tetrahedrons and MCH (curve c), and DNA1 (curve d) gradually increases the impedance value. Thus, both electrochemical CV and EIS characterization demonstrated the successful synthesis of a pH-induced regenerative biosensor.
Fig. 2.
Characterize the construction of the biosensor by using cyclic voltammetry (A) and EIS (B). Bare GCE electrode (curve a), Au-g-C3N4 modified GCE (curve b), GCE modified with Au-g-C3N4, DNA tetrahedrons and MCH (curve c). GCE modified with Au-g-C3N4, DNA tetrahedrons, MCH and DNA1 (curve d). Optimization of the Exo III-aided amplification reaction time (C) and triple-strand formation at pH = 7.0 based on Hoogsteen.
In order to verify the successful construction of this pH-induced regenerative biosensor, we performed ECL characterization for the construction process of this biosensor. Figure S1 shows the ECL signal changes during the stepwise modification of the pH-induced regenerative biosensor. pH = 7.0, a strong ECL signal, which is the ECL emission peak of Au-g-C3N4 at this position, appears at 460 nm when the DNA tetrahedron/Au-g-C3N4-modified GCE is incubated with DNA1 before, and no ECL signal at 620 nm. This experimental phenomenon indicates that the pH-induced regenerative biosensor was successfully synthesized. In contrast, when the pH-induced regenerative biosensor was incubated with 1500 aM target DNA, the ECL signal at 460 nm decreased, while a strong ECL signal appeared at 620 nm. This phenomenon indicates that DNA1 forms a three-stranded DNA structure with DNA tetrahedron and that ECL-phenomenon occurs. The experimental results further indicate that the ECL-RET process between Au-g-C3N4 donor and DNA-Ru acceptor occurs successfully.
3.5. Optimization of experimental conditions
The experimental conditions of the Exo III-aided amplification reaction time may affect the detection efficiency and the detection performance of the ECL sensor. Therefore, we optimized the Exo III-aided amplification reaction time. As shown in Fig. 2C, the ECL (620 nm) signal intensity gradually increased and ECL (460 nm) signal gradually decreased stabilized both at 1 h with the increase of time, indicating that the Exo III-aided amplification reaction could be completed within 1 h and 1 h was the optimal reaction time. Also, the ratio of ECL (620 nm)/ECL (460 nm) increased simultaneously with the increase in reaction time and reached stability at 60 min. So the Exo III-aided amplification reaction time of 60 min was selected as the optimal condition in this study.
Since the accuracy and sensitivity of this pH-induced regenerative biosensor sensing platform depends heavily on the ability to form triple-stranded nucleic acids via Hoogsteen with tetrahedra modified on the electrode surface, we further optimized the reaction time for triple-strand formation at pH = 7.0 (Fig. 2D). As the reaction time for nucleic acid triple-strand formation increased, the ECL signal at 460 nm decreased, and the ECL signal at 620 nm increased. At 90 min, the ECL signals at both 460 nm and 620 nm stabilized, and the ratio of ECL (620 nm)/ECL (460 nm) increased simultaneously with the increase in reaction time and reached stability at 90 min. Therefore, the reaction time for the formation of triple-stranded nucleic acids was chosen as 90 min.
3.6. Detection of SARS-CoV-2 RdRp gene with the ratiometric biosensor
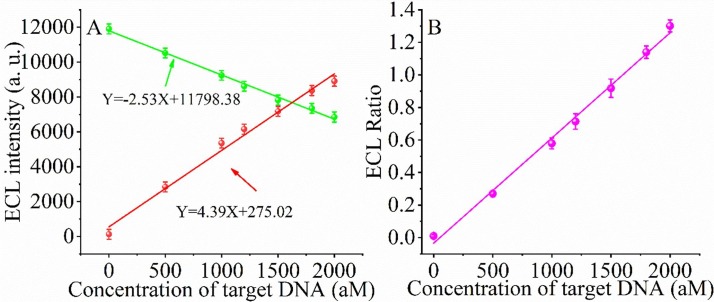
The constructed ratiometric ECL biosensor was used to detect SARS-CoV-2 RdRp gene under optimized conditions. Fig. 3 A depicts the change of ECL signal with increasing concentration of SARS-CoV-2 RdRp gene. As the concentration of SARS-CoV-2 RdRp in the reaction system increases from 10 aM to 10 pM, the ECL signal intensity gradually decreases at 460 nm (Points in green) and increases at 620 nm (Points in red). The decrease of ECL signal intensity at 460 nm is depicted in Fig. 3A. The ECL signal intensity shows an excellent linear relationship with the concentration of SARS-COV-2 RdRp gene: Y = -2.53X + 11798.38, R2 = 0.9956. Similarly, the increase of ECL signal intensity at 620 nm also indicates a good linear relationship with the concentration of SARS-COV-2 RdRp: Y = 4.39X + 275.02, R2 = 0.9887. To further obtain the reliability of the calculated results, we evaluated the correlation between SARS-COV-2 RdRp concentration and ECL (620 nm)/ECL (460 nm) as the concentration of SARS-COV-2 RdRp increased, and the intrinsic correlation. The experimental results are shown in Fig. 3B. It can be clearly found that the variation values of ECL(620 nm)/ECL(420 nm) also showed a linear relationship with ECL(620 nm)/ECL(420 nm): Y = 6.466 × 10-4X-0.034, R2 = 0.9950, where Y is the ratio of ECL(620 nm)/ECL(420 nm) and X is the concentration of RdRp gene. According to the formula of limit of detection (LOD) = 3σ/k, the LOD of the biosensor was 43.70 aM. The reason for the lower detection limit is that we designed the biosensor with a dual amplification effect and used the ratio method to calculate the detection limit.
Fig. 3.
(A) Relationship between the ECL intensity of Au-g-C3N4 at 460 nm (green) and PEI-Ru@Ti3C2@AuNPs at 620 nm (red) and the concentration of SARS-CoV-2 RdRp gene. (B) The relationship between the ECL ratio (ECL460/ECL620) and the concentration of SARS-CoV-2 RdRp gene.
Using this biosensor, the SARS-CoV-2 E gene and N gene were similarly detected after a small change in the nucleic acid sequences of H1 and H2 (The sequences are shown in Table S1, H1E and H2E for E gene assay and H1N and H2N for N gene assay), which recognize the target nucleic acid, and the results were as shown in Figure S2 and Figure S3. By comparing with other SARS-CoV-2 amplification methods, as shown in Table S2, our designed biosensor has comparable or more sensitive detection limits, thus our detection platform has potential applications for highly sensitive detection of SARS-CoV-2, other viruses and even other pathogens by changing the nucleic acid sequences of H1 and H2.
3.7. Stability and selectivity of the pH-induced regenerative biosensor
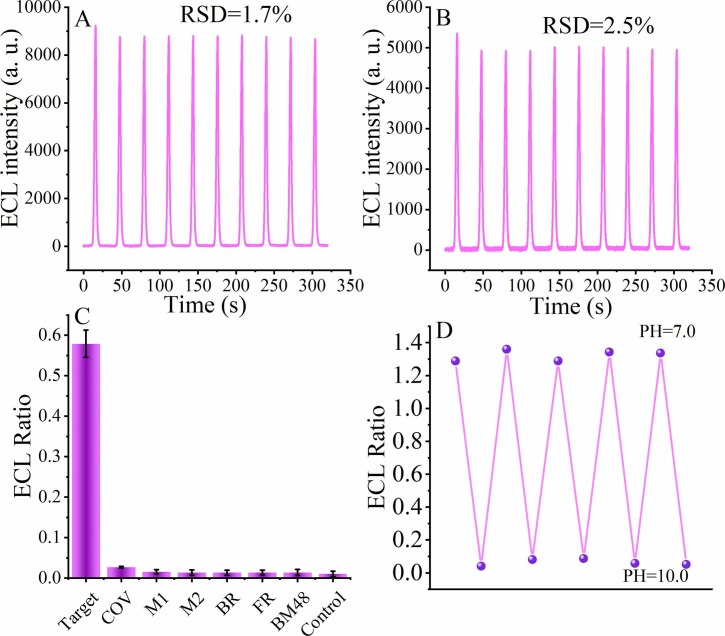
Next, the stability of the biosensor was further evaluated. The modified electrode was immersed in PBS containing 1 pM SARS-CoV-2 RdRp gene and scanned continuously without interruption for 10 cycles. The ECL signal remained stable at 460 nm (Fig. 4 A) with a relative standard deviation (RSD) of 1.7%, and the ECL signal also remained stable at 620 nm (Fig. 4B) with an RSD of 2.5%. These experimental phenomena demonstrate the excellent stability of the biosensor.
Fig. 4.
Stability and selectivity of the pH-induced regenerative biosensor. (A) The ECL signal at 460 nm and (B) the ECL signal at 620 nm by scanning 10 times after treating 1 pM SARS-CoV-2 RdRp gene. (C) Selectivity of the pH-induced regenerative biosensor. (D) Reproducibility of the pH-induced regenerative biosensor.
To further evaluate the selectivity of the pH-induced regenerative biosensor, we introduced some interfering factors to treat the biosensor. When the biosensor was immersed in 1 pM SARS-CoV-2 RdRp gene (Target DNA), 100 pM SARS-CoV RdRp gene (COV), 100 pM random mutant DNA (M1 and M2), 100 pM Bat SARS-related CoV isolate bat-SL-CoVZC45 RdRp gene (BR), 100 pM Frankfurt 1 RdRp gene (FR), 100 pM BM48-31/BGR/2008 RdRp gene (BM48) and blank solution, different ECL signal changes were induced. The ECL (620 nm)/ECL (460 nm) values induced by various analytes are listed in Fig. 4C. It can be seen from the figure that the ECL signal ratio of the SARS-CoV-2-treated biosensor RdRp gene has a significant change compared to the other interferent-treated biosensors, while the other nucleic acids to be tested do not show a strong signal change as a result. The results of this experiment may be due to the fact that only SARS-CoV-2 RdRp can promote the isothermal amplification effect based on EXO III. Therefore, the constructed biosensor has excellent selectivity.
3.8. pH-induced regenerative biosensor validation
We also investigated the pH-induced regenerative of the constructed biosensing platform at different pH values. When the biosensor was incubated in TAE buffer (pH = 10.0), the biosensor forming triple-stranded DNA could be regenerated by nucleic acid dissociation on the electrode surface at that pH value. As shown in Fig. 4D, after the formation of triple-stranded nucleic acids at pH = 7.0, a significant ECL signal change can be observed compared to the initial blank current response of the independent DNA tetrahedra. However, after immersion in TAE buffer at pH = 10.0, the ECL response of the electrode decreases by approximately 98% and reverses to an insignificant ECL response ratio similar to the initial state of the biosensor, indicating successful regeneration of the biosensor. Notably, the pH-induced regenerable biosensor was evaluated five times and still exhibited effective regenerability with regeneration rates ranging from 97.8%, which further demonstrates the good pH-induced regenerability of the biosensor and suggests that the biosensor can be reused many times over.
3.9. Applicability of the biosensor in real samples
To further investigate the resistance of the designed pH-induced regenerative biosensor for SARS-CoV-2 RdRp gene detection to complex environments, we applied the biosensor to detect SARS-CoV-2 RdRp gene in pharyngeal swabs. First, we used the incorporation method to add a graded dose of SARS-CoV-2 RdRp gene in pharyngeal swabs, and then replaced the standard sample with the prepared pharyngeal swabs samples. Then, the concentration of SARS-CoV-2 RdRp gene in different pharyngeal swabs samples was detected by a pH-induced regenerative biosensor and the recovery results for the assay of SARS-CoV-2 RdRp gene were shown in Table 1 . This result indicates that our designed pH-induced regenerative biosensor can be applied to detect the SARS-CoV-2 RdRp gene in human pharyngeal swabs. Therefore, this pH-induced regenerative assay strategy we proposed has the potential to be widely used in the clinic to detect COVID-19-related nucleic acid markers in complex biological samples.
Table 1.
Recovery results for the assay of SARS-CoV-2 RdRp in pharyngeal swabs.
| Sample number | Added | Found | Recovery (%) | RSD (%, n = 3) |
|---|---|---|---|---|
| 1 | 50 aM | 49.63 aM | 99.26 | 1.29 |
| 2 | 100 aM | 99.85 aM | 99.85 | 3.50 |
| 3 | 1 fM | 1.07 fM | 107.0 | 2.07 |
| 4 | 10 fM | 10.07 fM | 100.7 | 1.65 |
| 5 | 100 fM | 102.86 fM | 102.86 | 4.68 |
| 6 | 1 pM | 0.98 pM | 98.00 | 4.49 |
| 7 | 10 pM | 10.19 pM | 101.90 | 2.70 |
4. Conclusions
In summary, we present the first CRISPR/Cas12a-based pH-induced regenerative biosensor for the diagnosis of the SARS-CoV-2 RdRp gene. We carefully designed two segments of hairpin DNA for binding to the nucleic acid of the test subject and cleaved them into two DNA segments in the presence of exonuclease III. The two DNA segments (H1a and H2a) can activate CRISPR/Cas12a after hybridization. If the test subject is not present in the system, the H1a/H2a double strand cannot be formed and thus cannot activate the activity of CRISPR/Cas12a and cleave DNA1. The DNA1 in the system can bind to the DNA tetrahedra on the electrode surface, thus causing a change in the ECL signal. Notably, the biosensor can be regenerated at pH = 10.0, enabling continuous long-term use of this sensing platform for accurate monitoring of the concentration of nucleic acids to be measured and providing a new reference for the design of regenerative DNA tetrahedron ECL biosensors. Our pH-induced regenerative biosensor was used for the detection of SARS-CoV-2 RdRp gene with a detection limit of 43.70 aM, and the pH-induced regenerative biosensor is extremely regenerative as well as resistant to interference. Therefore, it has potential clinical application in the mass screening of the SARS-CoV-2 RdRp gene.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (21705061), the Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017), and the Innovation Capacity Development Plan of Jiangsu Province (BM2018023).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cej.2021.132472.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Abdelhamid H.N., Badr G. Nanobiotechnology as a platform for the diagnosis of COVID-19: a review. Nanotechnology for Environmental Engineering. 2021;6:19. [Google Scholar]
- 2.Fan Z., Yao B., Ding Y., Zhao J., Xie M., Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao B.o., Zhang J., Fan Z., Ding Y., Zhou B., Yang R., Zhao J., Zhang K. Rational Engineering of the DNA Walker Amplification Strategy by Using a Au@Ti3C2@PEI-Ru(dcbpy)32+ Nanocomposite Biosensor for Detection of the SARS-CoV-2 RdRp Gene. ACS Appl. Mater. Inter. 2021;13(17):19816–19824. doi: 10.1021/acsami.1c04453. [DOI] [PubMed] [Google Scholar]
- 4.Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.PM., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12(556):eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji T., Liu Z., Wang G., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B.o., Jia H., Chen Y., Zhou Q. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:23. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng W., Newbigging A.M., Tao J., Cao Y., Peng H., Le C., Wu J., Pang B., Li J., Tyrrell D.L., Zhang H., Le X.C. CRISPR technology incorporating amplification strategies: molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021;12:4683–4698. doi: 10.1039/d0sc06973f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K., Fan Z., Yao B., Ding Y., Zhao J., Xie M., Pan J. Exploring the trans-cleavage activity of CRISPR-Cas12a for the development of a Mxene based electrochemiluminescence biosensor for the detection of Siglec-5. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113019. [DOI] [PubMed] [Google Scholar]
- 9.Zeng R., Wang W., Chen M., Wan Q., Wang C., Knopp D., Tang D. CRISPR-Cas12a-driven MXene-PEDOT:PSS piezoresistive wireless biosensor. Nano Energy. 2021;82 [Google Scholar]
- 10.Dai Y., Somoza R.A., Wang L., Welter J.F., Li Y., Caplan A.I., Liu C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. 2019;58(48):17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng S., Tan Z., Chen S., Lei C., Nie Z. Integrating CRISPR-Cas12a with a DNA circuit as a generic sensing platform for amplified detection of microRNA. Chem. Sci. 2020;11(28):7362–7368. doi: 10.1039/d0sc03084h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan T., Mukama O., Li Z., Chen W., Zhang Y., de Dieu Habimana J., Zhang Y., Zeng R., Nie C., He Z., Zeng L. A rapid and sensitive CRISPR/Cas12a based lateral flow biosensor for the detection of Epstein-Barr virus. The Analyst. 2020;145(19):6388–6394. doi: 10.1039/d0an00663g. [DOI] [PubMed] [Google Scholar]
- 13.Hu M., Yuan C., Tian T., Wang X., Sun J., Xiong E., Zhou X. Single-Step, Salt-Aging-Free, and Thiol-Free Freezing Construction of AuNP-Based Bioprobes for Advancing CRISPR-Based Diagnostics. J. Am. Chem. Soc. 2020;142(16):7506–7513. doi: 10.1021/jacs.0c00217. [DOI] [PubMed] [Google Scholar]
- 14.Xiong Y., Zhang J., Yang Z., Mou Q., Ma Y., Xiong Y., Lu Y.i. Functional DNA Regulated CRISPR-Cas12a Sensors for Point-of-Care Diagnostics of Non-Nucleic-Acid Targets. J. Am. Chem. Soc. 2020;142(1):207–213. doi: 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang M., Li Z., Wang W., Liu J., Liu L., Zhu G., Karthik L., Wang M., Wang K.-F., Wang Z., Yu J., Shuai Y., Yu J., Zhang L., Yang Z., Li C., Zhang Q., Shi T., Zhou L., Xie F., Dai H., Liu X., Zhang J., Liu G., Zhuo Y., Zhang B., Liu C., Li S., Xia X., Tong Y., Liu Y., Alterovitz G., Tan G.-Y., Zhang L.-X. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 2019;10:3672. doi: 10.1038/s41467-019-11648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karami A., Hasani M., Jalilian F.A., Ezati R. Conventional PCR Assisted Single-Component Assembly of Spherical Nucleic Acids for Simple Colorimetric Detection of SARS-CoV-2. Sensor Actuat. B Chem. 2021;328 doi: 10.1016/j.snb.2020.128971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.i., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 21.Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.-B., Cui M., Tao J., Tyrrell D.L., Zhang X.-E., Zhang H., Le X.C. Molecular Diagnosis of COVID-19: Challenges and Research Needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 22.C.B.E.M. Reusken, E.K. Broberg, B. Haagmans, A. Meijer, V.M. Corman, A. Papa, R. Charrel, C. Drosten, M. Koopmans, K. Leitmeyer, o.b.o. EVD-LabNet, ERLI-Net, Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020, 25 (2020) 2000082. [DOI] [PMC free article] [PubMed]
- 23.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C.N.J., Amirthalingam G., Edmunds M., Zambon M., Brown K.E., Hopkins S., Chand M., Ramsay M. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Joshi, M. Kumar, V. Srivastava, D. Kumar, D. Rathore, R. Pandit, C.G. Joshi, First detection of SARS-CoV-2 Delta variant (B.1.617.2) in the wastewater of (Ahmedabad), India, medRxiv (2021) 2021.2007.2007.21260142.
- 25.Zhou X., Zhang W., Wang Z., Han J., Xie G., Chen S. Ultrasensitive aptasensing of insulin based on hollow porous C3N4/S2O82−/AuPtAg ECL ternary system and DNA walker amplification. Biosens. Bioelectron. 2020;148 doi: 10.1016/j.bios.2019.111795. [DOI] [PubMed] [Google Scholar]
- 26.Liu D.i., Zhang X., Zhao J., Chen S., Yuan R. An ultrasensitive sensing platform for microRNA-155 based on H2O2 quenched hydroxide-dependent ECL emission of PFO Pdots. Biosens. Bioelectron. 2020;150:111872. doi: 10.1016/j.bios.2019.111872. [DOI] [PubMed] [Google Scholar]
- 27.Wu M., Xu N., Qiao J., Chen J., Jin L. Bipolar electrode-electrochemiluminescence (ECL) biosensor based on a hybridization chain reaction. Analyst. 2019;144(15):4633–4638. doi: 10.1039/c9an01022j. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y., Jia J., Yang R., Chen D., Wang J., Luo F., Guo L., Qiu B., Lin Z. Ratiometric Immunosensor for GP73 Detection Based on the Ratios of Electrochemiluminescence and Electrochemical Signal Using DNA Tetrahedral Nanostructure as the Carrier of Stable Reference Signal. Anal. Chem. 2019;91(5):3717–3724. doi: 10.1021/acs.analchem.9b00013. [DOI] [PubMed] [Google Scholar]
- 29.Li M.-X., Feng Q.-M., Zhou Z., Zhao W., Xu J.-J., Chen H.-Y. Plasmon-Enhanced Electrochemiluminescence for Nucleic Acid Detection Based on Gold Nanodendrites. Anal. Chem. 2018;90(2):1340–1347. doi: 10.1021/acs.analchem.7b04307. [DOI] [PubMed] [Google Scholar]
- 30.Fan Z., Lin Z., Wang Z., Wang J., Xie M., Zhao J., Zhang K., Huang W. Dual-Wavelength Electrochemiluminescence Ratiometric Biosensor for NF-κB p50 Detection with Dimethylthiodiaminoterephthalate Fluorophore and Self-Assembled DNA Tetrahedron Nanostructures Probe. ACS Appl. Mater. Inter. 2020;12(10):11409–11418. doi: 10.1021/acsami.0c01243. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H.-R., Xu J.-J., Chen H.-Y. Electrochemiluminescence Ratiometry: A New Approach to DNA Biosensing. Anal. Chem. 2013;85:5321–5325. doi: 10.1021/ac400992u. [DOI] [PubMed] [Google Scholar]
- 32.Wang F., Liu X., Willner I. Integration of Photoswitchable Proteins, Photosynthetic Reaction Centers and Semiconductor/Biomolecule Hybrids with Electrode Supports for Optobioelectronic Applications. Adv. Mater. 2013;25(3):349–377. doi: 10.1002/adma.201201772. [DOI] [PubMed] [Google Scholar]
- 33.Yuan C., Tian T., Sun J., Hu M., Wang X., Xiong E., Cheng M., Bao Y., Lin W., Jiang J., Yang C., Chen Q., Zhang H., Wang H., Wang X., Deng X., Liao X., Liu Y., Wang Z., Zhang G., Zhou X. Universal and Naked-Eye Gene Detection Platform Based on the Clustered Regularly Interspaced Short Palindromic Repeats/Cas12a/13a System. Anal. Chem. 2020;92(5):4029–4037. doi: 10.1021/acs.analchem.9b05597. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y., Hu M., Liu A.-A., Lin Y.i., Liu L., Yu B.o., Zhou X., Pang D.-W. Detection of SARS-CoV-2 by CRISPR/Cas12a-Enhanced Colorimetry. ACS Sensors. 2021;6(3):1086–1093. doi: 10.1021/acssensors.0c02365. [DOI] [PubMed] [Google Scholar]
- 35.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K., Fan Z., Yao B., Zhang T., Ding Y., Zhu S., Xie M. Entropy-driven electrochemiluminescence ultra-sensitive detection strategy of NF-κB p50 as the regulator of cytokine storm. Biosens. Bioelectron. 2021;176 doi: 10.1016/j.bios.2020.112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Z., Yao B., Ding Y., Xie M., Zhao J., Zhang K., Huang W. Electrochemiluminescence aptasensor for Siglec-5 detection based on MoS2@Au nanocomposites emitter and exonuclease III-powered DNA walker. Sens. Actuators B Chem. 2021;334 doi: 10.1016/j.snb.2021.129592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X.-L., Yang Z.-H., Chang Y.-Y., Liu D.i., Li Y.-R., Chai Y.-Q., Zhuo Y., Yuan R. Programmable mismatch-fueled high-efficiency DNA signal converter. Chem. Sci. 2020;11(1):148–153. doi: 10.1039/c9sc05084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z., Lei S., Zou L., Li G., Ye B. Grafting homogenous electrochemical biosensing strategy based on reverse proximity ligation and Exo III assisted target circulation for multiplexed communicable disease DNA assay. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112487. [DOI] [PubMed] [Google Scholar]
- 40.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.