Abstract
Background:
Long non-coding RNA (lncRNA) actin filament-associated protein 1 antisense RNA 1 (AFAP1-AS1) functions as a competing endogenous RNA to regulate target genes expression by sponging microRNAs (miRs) to play cancer-promoting roles in cancer stem cells. However, the regulatory mechanism of AFAP1-AS1 in cervical cancer (CC) stem cells is unknown. The present study aimed to provide a new therapeutic target for the clinical treatment of CC.
Methods:
Hyaluronic acid receptor cluster of differentiation 44 variant exon 6 (CD44v6)(+) CC cells were isolated by flow cytometry (FCM). Small interfering RNAs of AFAP1-AS1 (siAFAP1-AS1) were transfected into the (CD44v6)(+) cells. The levels of AFAP1-AS1 were measured by quantitative real-time PCR (qRT-PCR). Sphere formation assay, cell cycle analysis, and Western blotting were used to detect the effect of siAFAP1-AS1. RNA pull-down and luciferase reporter assay were used to verify the relationship between miR-27b-3p and AFAP1-AS1 or vascular endothelial growth factor (VEGF)-C.
Results:
CD44v6(+) CC cells had remarkable stemness and a high level of AFAP1-AS1. However, AFAP1-AS1 knockdown with siAFAP1-AS1 suppressed the cell cycle transition of G(1)/S phase and inhibited self-renewal of CD44v6(+) CC cells, the levels of the stemness markers octamer-binding transcription factor 4 (OCT4), osteopontin (OPN), and cluster of differentiation 133 (CD133), and the epithelial-mesenchymal transition (EMT)-related proteins Twist1, matrix metalloprotease (MMP)-9, and VEGF-C. In the mechanism study, miR-27b-3p/VEGF-C signaling was demonstrated to be a key downstream of AFAP1-AS1 in the CD44v6(+) CC cells.
Conclusions:
LncRNA AFAP1-AS1 knockdown inhibits the CC cell stemness by upregulating miR-27b-3p to suppress VEGF-C.
Keywords: Hyaluronic acid receptor cluster of differentiation 44 variant exon 6, Cell stemness, Cervical cancer, Long non-coding RNA actin filament-associated protein 1 antisense RNA 1, MicroRNA-27b-3p
Introduction
Cervical cancer (CC), the fourth most common cancer among women, kills 300,000 people each year worldwide and has a mortality rate of 85% in developing countries.[1] Although the prevention and treatment of early CC had made great progress, statistics showed that, nevertheless, the prognosis of CC patients with local or distant metastases was generally poor.[2] Carcinogenesis of CC cells is a complex biological process, and the mechanism of CC occurrence, metastasis, and even treatment is still unclear, as a result of which these require further exploration.
Mesenchymal transformation-related genes expression was reported to be stimulated by human papillomavirus (HPV) 16 E6 oncoproteins, which combined with fibroblast growth factor (FGF), acts to inhibit E-cadherin and induce epithelial-mesenchymal transition (EMT).[3] EMT is a key cell phenotype transformation for tumor cells to acquire metastasis ability, and can affect the migration and invasion abilities of CC cells. Cancer cells that undergo EMT had significantly weakened adhesion ability and were easy to transfer to other parts of the body with the circulation of fluid.[3] However, the regulatory mechanism of EMT in CC cells is still unclear.
Cancer stem cells (CSCs) are a subset of cancer cells. They have some characteristics that are similar to embryonic stem cells (ESCs) and show stem cell characteristics, including self-renewal, tumorigenesis, tumor growth, and differentiation. In many different types of tumors, CSCs promoted tumorigenesis and drug resistance and were considered to play a key role in the recurrence and metastasis of cancer.[4] Certain cell surface markers, such as cluster of differentiation 133 (CD133), octamer-binding transcription factor 4 (OCT4), and CD44, have been commonly used to isolate CSCs subpopulations. It was showed that CC cells formed tumor spheres when cultured in single cells, and simultaneously express stem cell markers CD133 and OCT4.[4] However, so far, the mechanism of how CSCs promote the progression of CC remains unclear. Enhancing the understanding of the molecular mechanisms of CSCs in CC progression can promote the development of CC therapies and improve the prognosis of patients with CC.
The cluster of differentiation 44 variant exon 6 (CD44v6), a variant exon of the hyaluronic acid receptor CD44, has received extensive attention from scholars in the research of various tumors. CD44v6 was almost not expressed in normal tissues, but it was highly expressed in a variety of cancers, including CC, lung cancer, hepatocellular carcinoma, endometrial cancer, and ovarian cancer.[5–7] Transfecting benign tumor cells with CD44v6 enhanced the malignant characteristics of benign cells, such as invasion and metastasis,[8] and CD44v6(+) cells had stemness in lip, liver, and colon cancer.[6,8,9] However, few studies were investigating the stem cell function and molecular mechanism of CD44v6(+) CC cells. Recently, a growing number of studies found that the dysregulation of long non-coding RNAs (lncRNAs, >200 nucleotides in length and non-translated noncoding RNAs) played an important role not only in carcinogenesis but also in metastasis of CC.[10,11] LncRNA actin filament-associated protein 1 antisense RNA 1 (AFAP1-AS1) was largely found to play a role of an oncogenic gene in many types of cancers.[12] The upregulation of AFAP1-AS1 was closely related to the poor prognosis of breast cancer,[13] non-small cell lung cancer (NSCLC),[14] gastric cancer (GC),[15] and nasopharyngeal carcinoma.[16] Furthermore, AFAP1-AS1 regulated cell stemness and chemoresistance in laryngeal carcinoma cells and promoted the proliferation, migration, and invasion of GC cells.[17] Recently, it was reported that AFAP1-AS1 was associated with invasion of CC.[18] However, the mechanism of AFAP1-AS1 in the regulation of CC stemness is still unknown.
Otherwise, a growing body of evidence indicated that lncRNAs were combined with its target microRNAs (miRs) so that impeded the combination from miRs and the 3′-untranslated region (3′-UTR) of the target gene, thereby controlling the expression of the target gene and regulating cell function.[19] This kind of RNAs is known as competing endogenous RNAs (ceRNA).[20] For example, lncRNA DANCR promoted EMT, migration, and invasion of cholangiocarcinoma cells by transcriptionally activating Twist1 by sponging miR-345-5p.[21] MiR-27b-3p, an anti-tumor gene in many types of tumor,[22] was sponged by lncRNA Gm15290,[23] thereby regulating the expression of peroxisome proliferator-activated receptor gamma (PPARγ) to promote PPARγ-induced proliferation of adipocytes in mice. In CC cells, lncRNA DANCR aggravated CC through miR-34c and miR-613 by targeting matrix metalloprotease (MMP)-9.[24] Importantly, AFAP1-AS functioned as a ceRNA to regulate ras-related protein Rap-1b expression by sponging miR-181a in the Hirschsprung disease.[25] Besides, AFAP1-AS1 increased the RBP-J expression by inhibiting miR-320 in laryngeal carcinoma cells.[17]
In the present study, the CC cell stemness and EMT by inhibiting AFAP1-AS1 were analyzed. We further investigated the molecular mechanism and the potential relationship between AFAP1-AS1 and miR-27b-3p. Intriguingly, our preliminary results found that AFAP1-AS1 plays a role of ceRNA, which had an inhibition response on miR-27b-3p and a positive effect on the expression of vascular endothelial growth factor (VEGF-C), which is a strong promoter of lymphatic vessel formation and serves a crucial role during tumorigenesis and metastasis.[26] The present study aimed to provide a new therapeutic target for the clinical treatment of CC.
Methods
Cell lines and cell cultures
We obtained SiHa and Hela, the human CC cells, and normal human cervical epithelial cell line (HUCEC) from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were identified as free of contamination after Short Tandem Repeat (STR) identification. Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum was used for SiHa and Hela culture. For the culture of HUCEC, RPMI-1640 medium supplemented with 10% fetal bovine serum was used.
Microarray
LncRNA microarrays of SiHa, Hela, and HUCEC were conveyed by KangChen Biotech (Shanghai, China). Differential expression of lncRNA was analyzed by volcano plot. The threshold set for significant differences was log2|fold change|≥1 and P value < 0.05.
Flow cytometer (FCM)
The SiHa and Hela cells (1 × 106 cells/mL), were centrifuged (250×g, 5 min, 4°C); subsequently the supernatant was discarded and then was added 2 mL PBS to wash. The cells were resuspended in 1 mL ALDEFLUOR buffer (STEMCELL Technologies, Vancouver, BC, Canada) and incubated at 37°C for 45 min. 20 μL CD44v6-PE antibody (FAB3660P, R&D Systems, Minneapolis, MN, USA) and 20 μL lgG1-PE, the control, were incubated in a refrigerator at 4°C for 30 min with avoiding light. After centrifuging (250×g for 5 min), and the supernatant was discarded, 500 μL ALDEFLUOR buffer was added to resuspend the cells. The CD44v6(+) cell group and the CD44v6(−) cell group were sorted out with an FCM.
Quantitative real-time PCR (qRT-PCR)
Total RNA in the CC cells and CD44v6(+) CC cells were respectively extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. To synthesize the miR and lncRNA cDNA Synthesis Kit (Takara Biotechnology, Dalian, China) was used. And, a miScript SYBR Green PCR Kit was used for analyzing the expression of miR-27b-3p and AFAP1-AS1. Real-time PCR was performed using the Applied Biosystems 7500 Sequence Detection system (Applied Biosystems, Foster City, CA, USA). The miR-27b-3p (primer: forward, 5′- TTCACAGTGGCTAAGTTCTGC-3′) expression was normalized to U6 (primer: forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′; reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′). And, AFAP1-AS1 (primer: forward, 5′-ACTGAAGAGGAACCAGGGACAG-3′; reverse, 5′- GGGGAAACTGAAATGAATGAAG-3′) were normalized to the geometric mean expression level of GAPDH (primer: forward, 5′-GACTCATGACCACAGTCCATGC-3′; reverse, 5′-AGAGGCAGGGATGATGTTC TG-3′). 2−ΔΔCt method was used for the analysis of quantitative changes in gene expression according to the manufacturer's protocols.
Sphere formation assay
The CD44v6(+) and CD44v6(−) cells were suspended in serum-free DMEM/F12 medium (GIBCO, NY, USA) with 100 IU/mL penicillin (GIBCO), 100 μg/mL streptomycin (GIBCO), 20 ng/mL recombinant human epidermal growth factor (EGF) (GIBCO), 10 ng/mL human recombinant basic fibroblasts growth factor (bFGF) (GIBCO), 2% B27 supplement (GIBCO), and 1% N-2 supplement and 1% methylcellulose (GIBCO). Then the cells were inoculated at a density of 5 × 103 cells/mL in 24-well plates with ultra-low adhesion for 24 h. The spheres with a diameter >100 μm were counted using a microscope. The experiment was repeated three times independently.
CD44v6(+) CC cell transfection
The CD44v6(+) SiHa and CD44v6(+) Hela cells were seeded in 24-well plates 24 h before the experiment. First, small interfering RNAs (siRNAs) (GenePharma, Shanghai, China) were used to knock down the expression of AFAP1-AS1 (1-siAFAP1-AS1 and 2- siAFAP1-AS1) in the CD44v6(+) CC cells. siAFAP1-AS1 and the negative control (si-NC) were transfected into the CD44v6(+) SiHa and CD44v6(+) Hela cells.[43] Second, siAFAP1-AS1 and miR-27b-3p-inhibitor (GenePharma) were co-transfected into CD44v6(+) Hela cells for 24 h by Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Third, miR-27b-3p-mimic (miR-mimic, GenePharma) was transfected into CD44v6(+) SiHa and CD44v6(+) Hela cells for 24 h by Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Fourthly, pcDNA3.1(+)/VEGF-C (Invitrogen), the overexpression vector of VEGF-C or empty vector were transfected into CD44v6(+) Hela cells using liposomes according to a previous study.[44] siAFAP1-AS1 and pcDNA3.1(+)/VEGF-C were co-transfected into CD44v6(+) Hela cells for 24 h by Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Sequences used for silencing in the present study were 1-siAFAP1-AS1 (Forward, 5′-GUCCCAGCUUACACUUGUATT-3′; Reverse, 5′-UACAAGUGUAAGCUGGGACTT-3′), 2-siAFAP1-AS1 (Forward, 5′-GGGCUUCAAUUUACAAGCATT-3′; Reverse: 5′-UGCUUGUAAAUUGAAGCCCTT-3′), and Negative control (siNC, Forward, 5′-GCGACGAUCUGCCUAAGA-3′; Reverse, 5′-AUCUUAGGCAGAUCGUCG-3′).
CD44v6(+) CC cell cycle analysis
The CD44v6(+) CC cells were collected and fixed in ice-cold 70% ethanol overnight. The fixed cells were washed with phosphate-buffered saline and subjected to the cell cycle analysis using the Cell Cycle Detection Kit (KeyGen Biotechnology, Nanjing, China) according to the manufacturer's instructions.
Western blotting
The protein level changes of stemness markers (OCT4, OPN, CD133) and the EMT markers (Twist1, MMP-9, VEGF-A, VEGF-C) were measured by Western blotting. Total protein in CD44v6(+) CC cells was extracted and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto nitrocellulose membranes (Millipore, Temecula, CA, USA). The membranes were incubated with primary antibodies (Abcam, Cambridge, UK) against CD133 (1:550, ab19898), OCT4 (1:700, ab181557), OPN (1:1000, ab228748), Twist1 (1:900, ab50581), MMP-9 (1:600, ab73734), VEGF-A (1:800, ab52917), VEGF-C (1:600, ab135506), and GAPDH (1:2000, ab9485) respectively at 4°C overnight. Subsequently, the membranes were incubated with anti-rabbit horseradish peroxidase secondary antibody (Sigma) for further incubation. The blots were detected using an enhanced chemiluminescence Western blotting detection (Thermo Fisher Scientific). GAPDH was used as an internal control.
RNA pull-down assay
The exact relationship between AFAP1-AS1 and miR-27b-3p was investigated by RNA pull-down assay, as described previously.[45] At first, that biotin-labeled AFAP1-AS1 wild-type (Bio-AFAP1-AS1-WT), biotin-labeled AFAP1-AS1-mutant RNA fragments (Bio-AFAP1-AS1-mut), and biotinylated negative control for biotin-labeled AFAP1-AS1 (NC-1), biotin-labeled miR-27b-3p wild-type (Bio-miR-27b-3p-WT), biotin-labeled miR-27b-3p-mutant (Bio-miR-27b-3p-Mut), and biotinylated negative control for biotin-labeled miR-27b-3p (NC-2) were respectively transfected into Hela for 48 h using Lipofectamine 2000 according to manufacturer procedures. Cells were lysed with RNase-free DNase I (Roche Applied Science, IN, USA) and complete protease inhibitor (Roche). Cell lysates were incubated with streptavidin agarose beads (Invitrogen) coated with RNase-free bovine serum albumin and yeast tRNA (both from Sigma-Aldrich). After an ice bath for 2 h, cell lysates were isolated (10,000 ×g for 10 min), and a pull-down assay was performed as described previously.[46] Levels of miR-27b-3p in the pull-down of wt-Bio-AFAP1-AS1, mut-Bio- AFAP1-AS1, and NC-1, as well as levels of AFAP1-AS1 in the pull-down of wt-Bio-27b-3p, mut-Bio-27b-3p, and NC-2, were quantified by real-time PCR.
Luciferase reporter assay
miR-27b-3p promoter regions containing different wild type AFAP1-AS1 and/or VEGF-C binding sites were inserted into pGL3-Basic reporter gene vector (Promega, Madison, WI, USA). Also, the mutated sequence of AFAP1-AS1 and VEGF-C was inserted into the pGL3-Basic reporter gene vector (Promega). The mutant of AFAP1-AS11 and VEGF-C (-mut) was amplified respectively using AFAP1-AS11 and VEGF-C-3′-UTR as the template. DharmFECT Duo transfection reagent (Thermo Fisher Scientific, Glasgow, UK) was used for the analysis of the luciferase activity analysis. Then, the luciferase assays were performed with the Dual-Glo Luciferase assay system (Promega) according to the manufacturer's instructions.
Statistical analyses
Continuous variables were expressed as the means ± standard deviation (SD). The quantitative data between groups were compared and analyzed by Student's t test (two-tailed) or a one-way analysis of variance (ANOVA) followed by Bonferroni post hoc tests, which were analyzed with SPSS version 19.0 (SPSS Inc., Chicago, USA). Instances of P < 0.05 were considered to indicate statistically significant differences. Each experiment was repeated at least three times.
Results
AFAP1-AS1 is upregulated in CD44v6(+) CC cells
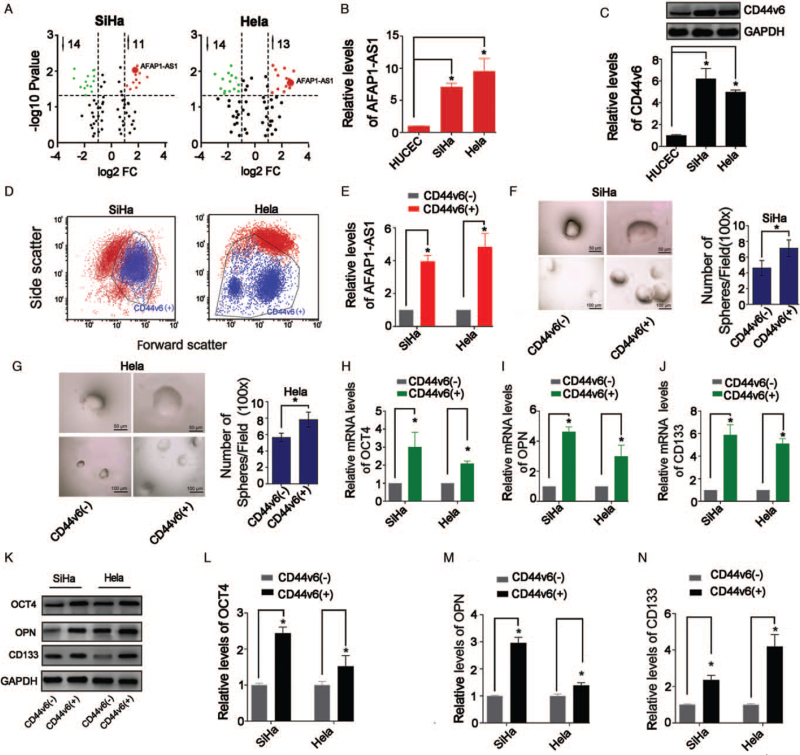
AFAP1-AS1 was remarkably upregulated in both the SiHa and Hela (Figure 1A and 1B; P < 0.05). Furthermore, results showed that the CD44v6 expression levels in the CC cells were all significantly up-regulated compared with HUCEC (P < 0.05, Figure 1C). Furthermore, we separated CD44v6(+) and CD44v6(−) cells from SiHa and Hela cell lines by FCM [Figure 1D]. AFAP1-AS1 was upregulated in the CD44v6(+) cells compared with that in the CD44v6(−) cells (P < 0.05; Figure 1E).
Figure 1.
Level changes of AFAP1-AS1 and stemness in CD44v6(+) CC cells. (A) Volcano plots showing expression profiles of lncRNAs. (B) qRT-PCR detected AFAP1-AS1 levels, ∗P < 0.05; (C) Western blotting detected CD44v6 levels. ∗P < 0.05. (D) CD44v6(+)/(−) cells sorting by FCM. (E) AFAP1-AS1 levels were increased in CD44v6(+) cells, ∗P < 0.05; (F) and (G) Representative images of spheres for CC cells and number of spheres (100×). (H–J) qRT-PCR results showed that OCT4, OPN, CD133 mRNA levels were increased in CD44v6(+) cells. ∗P < 0.05. (K–N) Western blotting results showed that OCT4, OPN, CD133 protein levels were increased in CD44v6(+) cells, ∗P < 0.05. AFAP1-AS1: Actin filament-associated protein 1 antisense RNA 1; CD133: Cluster of differentiation 133; CC: Cervical cancer; CD44v6: Hyaluronic acid receptor cluster of differentiation 44 variant exon 6; HUCEC: Human cervical epithelial cell line; LncRNA: Long non-coding RNA; OCT4: Octamer-binding transcription factor 4; OPN: Osteopontin; qRT-PCR: Quantitative real-time PCR.
CD44v6(+) CC cells have the characteristics of cervical cancer stem cells (CCSCs)
Spheroid formation experiments showed that CD44v6(+) cells formed larger and more spheroids than CD44v6(−) cells, indicating that CD44v6(+) cells have enhanced self-renewal ability than CD44v6(−) cells (all P < 0.05; Figure 1F and 1G). qRT-PCR and Western blotting showed that CD44v6(+) cells expressed more stemness-related genes (OCT4, OPN, CD133) than CD44v6(−) cells (all P < 0.05; Figure 1H–N).
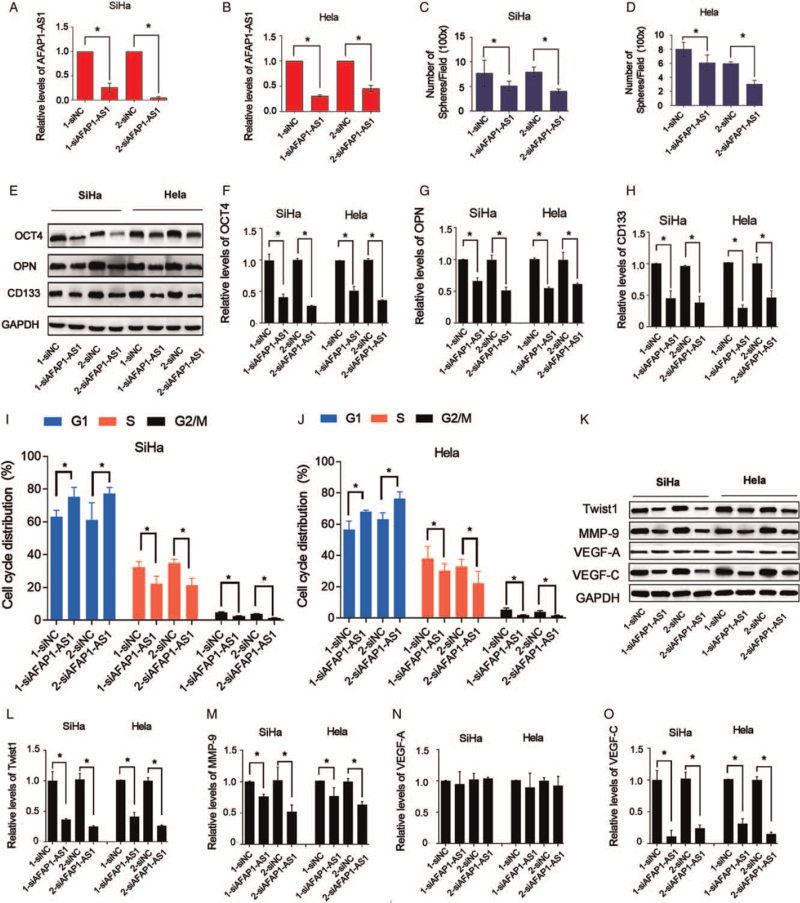
AFAP1-AS1 knockdown inhibits stem cell markers in CD44v6(+) CC cells
The present data confirmed that AFAP1-AS1 knockdown with its siRNAs (1-siAFAP1-AS1 and 2-siAFAP1-AS1) (all P < 0.05; Figure 2A and 2B) significantly suppressed the spheroid formation of CD44v6(+) cells (P < 0.05; Figure 2C and D). Furthermore, AFAP1-AS1 siRNAs remarkably downregulated the expression of OCT4, OPN, and CD133 in the CD44v6(+) cells (P < 0.05; Figure 2E–H).
Figure 2.
Knockdown of AFAP1-AS1 significantly attenuates stemness properties and G1/S cell cycle and EMT markers of CD44v6(+) CC cells. (A) and (B) qRT-PCR is used to verify effect of AFAP1-AS1 knockdown in CD44v6(+) SiHa and CD44v6(+) Hela cells, ∗P < 0.05. (C, D) Numbers of spheres were decreased by AFAP1-AS1 knockdown, ∗P < 0.05. (E–H) Western blotting results showed that OCT4, OPN, CD133 protein levels were decreased in AFAP1-AS1 knocked-down CD44v6(+) SiHa and CD44v6(+) Hela cells, ∗P < 0.05. (I) and (J) Knockdown of AFAP1-AS1 significantly arrests G1/S cell cycle of CD44v6(+) SiHa and CD44v6(+) Hela cells, ∗P < 0.05. (K–O) Western blotting results showed that Twist1, MMP-9, VEGF-A, VEGF-C protein levels were tested in AFAP1-AS1 knocked-down CD44v6(+) SiHa and CD44v6(+) Hela cells, ∗P < 0.05. AFAP1-AS1: Actin filament-associated protein 1 antisense RNA 1; CD133: Cluster of differentiation 133; CC: Cervical cancer; CD44v6: Cluster of differentiation 44 variant exon 6; EMT: Epithelial-mesenchymal transition; LncRNA: Long non-coding RNA; MMP-9: Matrix metalloproteinase 9; OCT4: Octamer-binding transcription factor 4; OPN: Osteopontin; qRT-PCR: Quantitative real-time PCR; siAFAP1-AS1: Small interfering RNA of AFAP1-AS1; siNC: Negative control of AFAP1-AS1; VEGF-A: Vascular endothelial growth factor A; VEGF-C: Vascular endothelial growth factor C.
AFAP1-AS1 knockdown inhibits cell cycle transition of G(1)/S phase and EMT-markers in CD44v6(+) CC cells
Given that inhibition of AFAP1-AS1 inhibited CC cell proliferation and EMT,[8] the effect of AFAP1-AS1 knockdown on the CD44v6(+) CC cells is unclear. Here, AFAP1-AS1 knockdown significantly suppressed the cell cycle transition of the G(1)/S phase of the CD44v6(+) cells (P < 0.05; Figure 2I and 2J). Furthermore, the protein levels of Twist1, the transcription factor of EMT,[27] and MMP-9 and VEGF-C, the key EMT makers[28] in the CD44v6(+) cells, were downregulated by AFAP1-AS1 siRNAs (P < 0.05; Figure 2K–O).
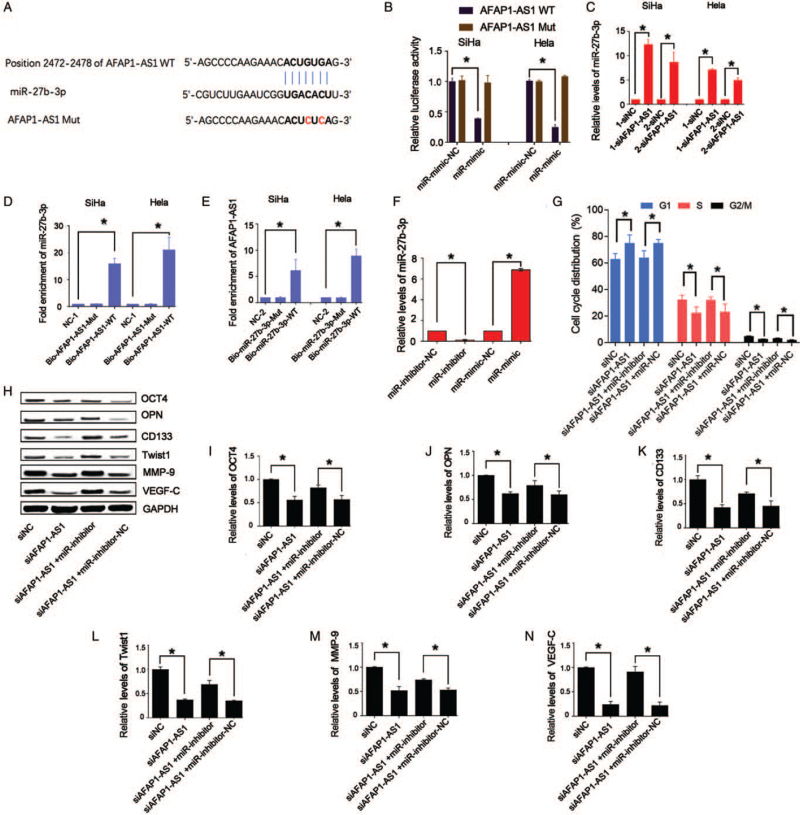
AFAP1-AS1 acts as an RNA sponge of miR-27b-3p
LncRNAs serve as ceRNAs that may positively affect downstream target genes by absorbing miRs.[15,29] Bioinformatic methods predicted several binding sites between AFAP1-AS1 (Position 2472–2478) and miR-27b-3p [Figure 3A]. Intriguingly, we found that miR-27b-3p inhibited the luciferase activity of AFAP1-AS1 (all P < 0.05; Figure 3B) and that knockdown of AFAP1-AS1 increased the levels of miR-27b-3p (Figure 3C; P < 0.05) in CD44v6(+) CC cells. The RNA pull-down assay showed that Bio-AFAP1-AS1-WT could pull down miR-27b-3p (P < 0.05) but that Bio-AFAP1-AS1-mut, which disrupted base pairing between AFAP1-AS1 and miR-27b-3p, could not [Figure 3D]. Additionally, Bio-miR-27b-3p-WT could pull down AFAP1-AS1 (P < 0.05), but Bio-miR-27b-3p-Mut abolished the binding ability between miR-27b-3p and AFAP1-AS1 [Figure 3E], confirming that AFAP1-AS1 was an RNA sponge of miR-27b-3p.
Figure 3.
Knockdown of AFAP1-AS1 in CD44v6(+) SiHa and CD44v6(+) Hela cells increases levels of miR-27b-3p by miR “sponge”. (A) Potential binding site of miR-27b-3p in the sequence of AFAP1-AS1 in humans. (B) miR-27b-3p overexpression decreases luciferase activity of AFAP1-AS1-WT but not AFAP1-AS1-WT, ∗P < 0.05. (C) miR-27b-3p levels were tested in AFAP1-AS1 knocked-down CD44v6(+) cells, ∗P < 0.05. (D) and (E) RNA pull-down assay is used to verify miR “sponge” mechanism in CD44v6(+) cells, ∗P < 0.05. (F) miR-27b-3p levels were downregulated in the miR-inhibitor group and upregulated in miR-mimic group in CD44v6(+) Hela. ∗P < 0.05. (G) G(1)/S phase of CD44v6(+) Hela that is impeded by knockdown of AFAP1-AS1 was significantly upregulated by miR-inhibitor, ∗P < 0.05. (H–N) Western blotting results showed that OCT4, OPN, CD133, Twist1, MMP-9, and VEGF-C protein levels were tested in CD44v6(+) Hela cells with both AFAP1-AS1 knockdown and miR-27b-3p inhibition, ∗P < 0.05. AFAP1-AS1: Actin filament-associated protein 1 antisense RNA 1; Bio-AFAP1-AS1-WT: Biotin-labeled AFAP1-AS1 wild-type; Bio-AFAP1-AS1-mut: AFAP1-AS1-mut RNA fragments; Bio-27b-3p-mut: Biotin-labeled miR-27b-3p mutant; Bio-miR-27b-3p-WT: Biotin-labeled miR-27b-3p wild-type; CD133: Cluster of differentiation 133; CD44v6: Cluster of differentiation 44 variant exon 6; MMP-9: Matrix metalloproteinase 9; miR: MicroRNA; Mut: Mutant; miR-mimic: MiR-27b-3p mimics; miR-mimic-NC: Negative control of miR-27b-3p mimics; miR-inhibitor: MiR-27b-3p inhibitors; miR-inhibitor-NC: Negative control of miR-27b-3p inhibitors; NC-1: Negative control for biotin-labeled AFAP1-AS1; NC-2: Negative control for biotin-labeled miR-27b-3p; OCT4: Octamer-binding transcription factor 4; OPN: Osteopontin; siAFAP1-AS1: Small interfering RNA of AFAP1-AS1; siNC: Negative control of AFAP1-AS1; VEGF-C: Vascular endothelial growth factor C; WT: Wild type.
miR-27b-3p inhibition impeded transition of G(1)/S phase, stemness, and EMT-related markers
We suppressed miR-27b-3p by using miR-27b-3p-inhibitor in the CD44v6(+) Hela cells (all P < 0.05; Figure 3F). The inhibitory effects of AFAP1-AS1 knockdown on cell cycle transition of G(1)/S phase (Figure 3G; P < 0.05), the cell stemness, and EMT-related markers in the CD44v6(+)cells were all recovered by miR-27b-3p suppression (Figure 3H–N; P < 0.05). Taken together, AFAP1-AS1 was confirmed as the RNA sponge of miR-27b-3p, which could be as a functional target of AFAP1-AS1.
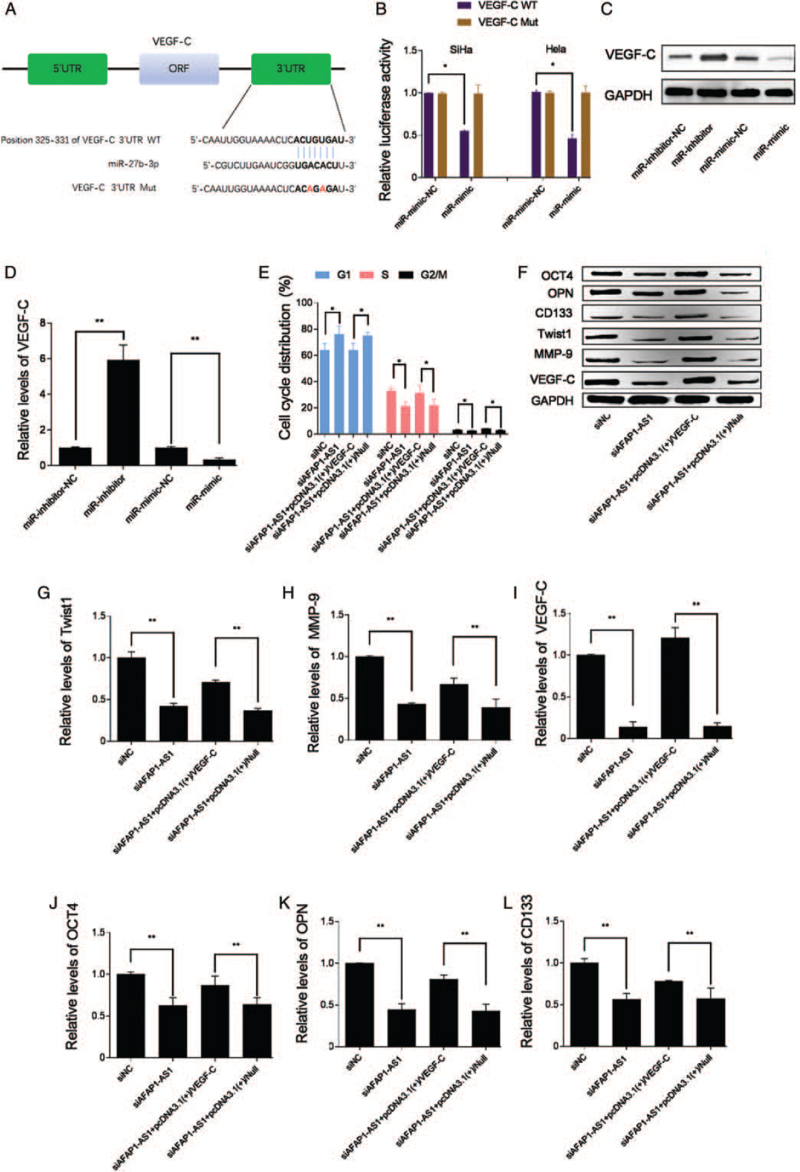
MiR-27b-3p targets VEGF-C in CD44v6(+) CC cells
The relationship between miR-27b-3p and VEGF-C were studied. Bioinformatic prediction implied VEGF-C may be a target gene of miR-27b-3p [Figure 4A]. Furthermore, a dual-luciferase reporter assay confirmed that miR-27b-3p inhibited the luciferase activity of VEGF-C (Figure 4B; P < 0.05), indicating that interaction between miR-27b-3p and VEGF-C 3′-UTR occurs in the CD44v6(+) CC cells. Additionally, miR-27b-3p overexpression by miR-mimic significantly decreased the protein levels of VEGF-C (Figure 4C and 4D; P < 0.05). Thus, VEGF-C had been identified as a target gene of miR-27b-3p in CD44v6(+) CC cells. The present studies above indicated that AFAP1-AS1 played a role of ceRNA to regulate the level of VEGF-C via sponging miR-27b-3p in CD44v6(+) CC cells, which was a key mechanism of AFAP1-AS1 in regulating CD44v6(+) CC cell cycle transition of G(1)/S phase, EMT and stemness.
Figure 4.
VEGF-C is a functional target gene of miR-27b-3p. (A) Potential binding site of miR-27b-3p in 3′UTR sequence of VEGF-C in human. (B) CD44v6(+) cells were co-transfected with miR-mimic-NC, or miR-mimic, and luciferase reporters containing 3′UTR of VEGF-C-WT sequence or 3′UTR of VEGF-C-Mut sequence. miR-27b-3p overexpression decreases luciferase activity of 3′UTR of VEGF-C-WT but not 3′UTR of VEGF-C-Mut, ∗P < 0.05. (C) and (D) VEGF-C levels were tested in miR-27b-3p overexpressed CD44v6(+)Hela cells, ∗P < 0.05. (E) G(1)/S phase of CD44v6(+) Hela that is impeded by knockdown of AFAP1-AS1 were significantly upregulated by VEGF-C overexpression, ∗P < 0.05. (F–L) Western blotting results showed that OCT4, OPN, CD133, Twist1, MMP-9, and VEGF-C protein levels were tested in CD44v6(+) Hela cells with both AFAP1-AS1 knockdown and VEGF-C overexpression, ∗P < 0.05. AFAP1-AS1: Actin filament-associated protein 1 antisense RNA 1; CD133: Cluster of differentiation 133; CD44v6: Cluster of differentiation 44 variant exon 6; MMP-9: Matrix metalloproteinase 9; Mut: Mutant; miR-mimic: MiR-27b-3p mimics; miR-mimic-NC: Negative control of miR-27b-3p mimics; miR-inhibitor: MiR-27b-3p inhibitors; miR-inhibitor-NC: Negative control of miR-27b-3p inhibitors; OCT4: Octamer-binding transcription factor 4; OPN: Osteopontin; siAFAP1-AS1: Small interfering RNA of AFAP1-AS1; siNC: Negative control of AFAP1-AS1; VEGF-C: Vascular endothelial growth factor C; WT: Wild type.
AFAP1-AS1 knockdown attenuated cell cycle transition of G(1)/S phase, the cell stemness, and EMT-related markers by increasing miR-27b-3p to inhibit VEGF-C
To clarify whether VEGF-C was a functional downstream target of AFAP1-AS1, the CD44v6(+) CC cells were respectively co-administrated with siAFAP1-AS1 and pcDNA3.1(+)/VEGF-C. Compared with the siAFAP1-AS1 alone group, the inhibition effects of siAFAP1-AS1 on the cell cycle transition of G(1)/S phase (Figure 4E; P < 0.05), the cell stemness and the EMT-related markers were significantly attenuated in the siAFAP1-AS1+pcDNA3.1(+)/VEGF-C group [Figure 4F–L], confirming that VEGF-C was a functional target of AFAP1-AS1 and that overexpression VEGF-C acts similarly to miR-27b-3p knockdown. This finding further implicates VEGF-C as a functional target of AFAP1-AS1. All these results confirmed that AFAP1-AS1 knockdown attenuated cell cycle transition of G(1)/S phase, the stemness, and EMT-related markers of the CD44v6(+) CC cells by increasing miR-27b-3p to inhibit VEGF-C.
Discussion
An inhibitory role of AFAP1-AS1 knockdown in cell stemness and a ceRNA role of AFAP1-AS1 in miR-27b-3p and target gene VEGF-C in CD44v6(+) CC cells were demonstrated in the present study. Firstly, we provided evidence that CD44v6 and AFAP1-AS1 were all upregulated in the SiHa and Hela cell lines. Secondly, AFAP1-AS1 was upregulated in CD44v6(+)SiHa and CD44v6(+)Hela cells and AFAP1-AS1 knockdown inhibited the cell stemness, cycle transition of G(1)/S phase, and EMT-related proteins. Then, miR-27b-3p had been identified as a target of AFAP1-AS1 and miR-27b-3p inhibitor attenuated the roles of AFAP1-AS1 knockdown in the CD44v6(+) CC cells. Additionally, VEGF-C was demonstrated as a target gene of miR-27b-3p and was also controlled by AFAP1-AS1. Furthermore, VEGF-C overexpression attenuated the role of AFAP1-AS1 knockdown in the CD44v6(+) CC cells. The present findings suggest an important mechanism for AFAP1-AS1 in regulating stemness, cell cycle, and EMT of CD44v6(+)CC cells.
AFAP1-AS1, a large (6.81 kb) lncRNA was first confirmed by Wu et al[30] which was derived from the antisense strand of DNA at the AFAP1 coding gene locus[30] and had become a focus of attention in various tumor researches because of its abnormal expression.[12] In our study, a high level of AFAP1-AS1 expression was found in the CC cell lines, which was consistent with the studies of Bo et al[18] These results suggested that AFAP1-AS1 was a potential tumor-promoting gene in CC. However, it had only been reported to play a role in regulating stemness in pancreatic cancer (PC) cells and laryngeal carcinoma cells.[17]
A number of studies had suggested that AFAP1-AS1 knockdown inhibited oncogenesis in various tumors, including CC. Ma et al[13] found that the high expression of AFAP1-AS1 had a poor prognosis in breast cancer patients, and AFAP1-AS1 knockdown inhibited the proliferation, metastasis, and promotion apoptosis of MCF-7, a breast cancer cell line. Similar roles of AFAP1-AS1 were confirmed in colorectal cancer,[31] NSCLC cells (14), GC,[32] nasopharyngeal carcinoma,[16] and cholangiocarcinoma.[33] The knockdown of AFAP1-AS1 was confirmed to impair PC cell self-renewal ability, tumorigenicity, invasion, migration, and stemness.[34] AFAP1-AS1 silencing also inhibited the stemness and chemoresistance of laryngeal carcinoma cells.[17] In the present study, the ability to form spheres and the stemness markers OCT4, OPN, CD133 in the CD44v6(+) CC cells were inhibited by AFAP1-AS1 knockdown, as well as the cell cycle progression and the expression levels of EMT-related proteins Twist1, MMP-9, and VEGF-C. Bo et al[18] demonstrated that AFAP1-AS1 was elevated and hypomethylated in CC and was associated with a poor prognosis of patients with CC. Furthermore, the antitumor effects induced by the silencing of AFAP1-AS1 were mainly mediated through the regulation of the Rho/Rac signaling pathway and EMT-related genes,[18] which was consistent with the present study. These findings suggested that AFAP1-AS1 was a key modulator involved in maintaining CD44v6(+) CC stem cell self-renewal and tumorigenesis, and may open a new direction for the treatment of CC.
In recent studies, lncRNAs serve as ceRNA which could play a positive role in downstream target genes via absorbing miRs.[33] Our further studies indicated that miR-27b-3p, a key miR, plays a role of tumor suppressor in a variety of cancer cell types,[35] was significantly upregulated by siAFAP1-AS1 though a ceRNA way. Similarly, AFAP1-AS regulated Ras-related Rap-1b protein expression by sponging miR-181a in Hirschsprung disease.[25] In addition, AFAP1-AS1 increased the RBP-J expression by inhibiting miR-320 in laryngeal carcinoma cells, which was involved in the regulation of stemness of laryngeal carcinoma cells.[17] The present findings indicated that miR-27b-3p was a target of AFAP1-AS1 and was sponged by AFAP1-AS1.
MiR-27b-3p was frequently reported as a tumor suppressor.[36] Chen et al[22] have reported miR-27b-3p as an anti-oncogene in breast cancer cells in vitro. A similar role of miR-27b-3p was reported in lung cancer cells.[23] Furthermore, the high expression of miR-27b-3p might be related to the vascular invasion of lung adenocarcinoma,[37] but in the present study, we found that the inhibition of miR-27b-3p in siAFAP1-AS1-transfected CD44v6(+)CC cells upregulated the expression levels of VEGF-C and MMP-9, the two important factors which can promote invasion and EMT of cancer cells.[38] Additionally, inhibition of miR-27b-3p could significantly increase the cell cycle transition of G(1)/S phase, as well as the expression of stemness markers (CD133, OCT4, and OPN) and EMT-related proteins Twist1, in the presence of siAFAP1-AS1 in CD44v6(+)CC cells. It was suggesting that miR-27b-3p was a functional target of AFAP1-AS1 in regulating stemness and EMT of CD44v6(+)CC cells.
More importantly, VEGF-C was confirmed as a target gene of miR-27b-3p in CD44v6(+)CC cells in the present study; additionally, it was also confirmed that miR-27b-3p-inhibitor can significantly upregulate the protein levels of VEGF-C. VEGF-C was a protein that was a member of the VEGF family.[26] Recent studies have revealed that VEGF-targeted treatments suppress tumor angiogenesis and growth significantly in CC.[39] Several studies showed that VEGF inhibition played an anti-malignant effect in CC cells and tissue.[40] In addition, inhibition of VEGF also suppressed migration and invasion of CC cells,[41] and targeting VEGF-C inhibited metastasis and stemness of skin cancer.[42] Importantly, in the present study, we found that the expression of VEGF-C was decreased by AFAP1-AS1 inhibition, as well as by miR-27b-3p overexpression in CD44v6(+) CC cells. Thus, it was indicated that AFAP1-AS1 knockdown inhibited CC cell cycle progression, stemness, and EMT by inhibiting the expression of VEGF-C via upregulating miR-27b-3p.
There were some shortcomings in this study. In vivo experiments on stem cell functions regulated by AFAP1-AS1/miR-27b-3p/VEGF-C need to be further validated in CC animal model, and the correlation between AFAP1-AS1 and tumor stem cell characteristics needs to be analyzed in clinical samples to further determine the therapeutic potential of inhibiting AFAP1-AS1 for malignant CC.
In conclusion, all our results indicate that inhibition of AFAP1-AS1 mitigates stemness, cell cycle progression, and EMT in CD44v6(+)CC cells, and miR-27b-3p /VEGF-C axis is the direct target of AFAP1-AS1, which helps AFAP1-AS1 to play roles in regulating the stemness of CC cells. Our finding suggests that AFAP1-AS1/miR-27b-3p /VEGF-C axis is a potential target for CC treatment.
Conflicts of interest
None.
Footnotes
How to cite this article: Xia M, Duan LJ, Lu BN, Pang YZ, Pang ZR. LncRNA AFAP1-AS1/miR-27b-3p/VEGF-C axis modulates stemness characteristics in cervical cancer cells. Chin Med J 2021;134:2091–2101. doi: 10.1097/CM9.0000000000001665
References
- 1.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet 2019; 393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 2.Shrestha AD, Neupane D, Vedsted P, Kallestrup P. Cervical cancer prevalence, incidence and mortality in low and middle income countries: A systematic review. Asian Pac J Cancer Prev 2018; 19:319–324. doi: 10.22034/APJCP.2018.19.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Q, Qiu MT, Zhu Z, Zhou JH, Chen L, Zhou Y, et al. Twist induces epithelial-mesenchymal transition in cervical carcinogenesis by regulating the TGF-β/Smad3 signaling pathway. Oncol Rep 2015; 34:1787–1794. doi: 10.3892/or.2015.4143. [DOI] [PubMed] [Google Scholar]
- 4.Tyagi A, Vishnoi K, Mahata S, Verma G, Srivastava Y, Masaldan S, et al. Cervical cancer stem cells selectively overexpress HPV oncoprotein E6 that controls stemness and self-renewal through upregulation of HES1. Clin Cancer Res 2016; 22:4170–4184. doi: 10.1158/1078-0432.CCR-15-2574. [DOI] [PubMed] [Google Scholar]
- 5.Porcellini S, Asperti C, Corna S, Cicoria E, Valtolina V, Stornaiuolo A, et al. CAR T cells redirected to CD44v6 control tumor growth in lung and ovary adenocarcinoma bearing mice. Front Immunol 2020; 11:99–109. doi: 10.3389/fimmu.2020.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014; 14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Ni J, Cozzi PJ, Hao JL, Beretov J, Chang L, Duan W, et al. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate 2014; 74:602–617. doi: 10.1002/pros.22775. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Wang R, Bai S, Xiong S, Li Y, Liu M, et al. Musashi2 contributes to the maintenance of CD44v6+ liver cancer stem cells via notch1 signaling pathway. J Exp Clin Cancer Res 2019; 38:505–516. doi: 10.1186/s13046-019-1508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia AS, Assao A, Carvalho AL, Soares FA, Kowalski LP, Oliveira DT. The stem cell markers expression CD44v6 and podoplanin in lip cancer: Clinical significance. Virchows Arch 2019; 474:745–754. doi: 10.1007/s00428-019-02539-3. [DOI] [PubMed] [Google Scholar]
- 10.Sheng L, Wu J, Gong X, Dong D, Sun X. SP1-induced upregulation of lncRNA PANDAR predicts adverse phenotypes in retinoblastoma and regulates cell growth and apoptosis in vitro and in vivo. Gene 2018; 668:140–145. doi: 10.1016/j.gene.2018.05.065. [DOI] [PubMed] [Google Scholar]
- 11.Zhang A, Shang W, Nie Q, Li T, Li S. Long non-coding RNA H19 suppresses retinoblastoma progression via counteracting miR-17-92 cluster. J Cell Biochem 2018; 119:3497–3509. doi: 10.1002/jcb.26521. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, Li J, Xiao H, Zou Y, Liu Y, Huang W. AFAP1-AS1: A novel oncogenic long non-coding RNA in human cancers. Cell Prolif 2018; 51:e12397.doi: 10.1111/cpr.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma D, Chen C, Wu J, Wang H, Wu D. Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis and promotes carcinogenesis of breast cancer. Breast Cancer 2019; 26:74–83. doi: 10.1007/s12282-018-0891-3. [DOI] [PubMed] [Google Scholar]
- 14.Yin D, Lu X, Su J, He X, De W, Yang J, et al. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer 2018; 17:92–99. doi: 10.1186/s12943-018-0836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Zhang K, Wang T, Cui J, Xi H, Wang Y, et al. Long non-coding RNA AFAP1-antisense RNA 1 promotes the proliferation, migration and invasion of gastric cancer cells and is associated with poor patient survival. Oncol Lett 2018; 15:8620–8626. doi: 10.3892/ol.2018.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Zeng J, Chao W, Chen X, Huang Y, Deng K, et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget 2017; 8:41166–41177. doi: 10.18632/oncotarget.17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan Z, Xiu C, Song K, Pei R, Miao S, Mao X, et al. Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. J Cell Mol Med 2018; 22:4253–4262. doi: 10.1111/jcmm.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bo H, Fan L, Gong Z, Liu Z, Shi L, Guo C, et al. Upregulation and hypomethylation of lncRNA AFAP1-AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep 2019; 41:2431–2439. doi: 10.3892/or.2019.7027. [DOI] [PubMed] [Google Scholar]
- 19.Samir N, Matboli M, El-Tayeb H, El-Tawdi A, Hassan MK, Waly A, et al. Competing endogenous RNA network crosstalk reveals novel molecular markers in colorectal cancer. J Cell Biochem 2018; 119:6869–6881. doi: 10.1002/jcb.26884. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Yan J, Yuan X, Yang R, Dan T, Wang X, et al. A computationally constructed ceRNA interaction network based on a comparison of the SHEE and SHEEC cell lines. Cell Mol Biol Lett 2016; 21:21–30. doi: 10.1186/s11658-016-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu CY, Fan CR, Zhang YL, Sun QX, Yan MJ, Wei W, et al. LncRNA DANCR affected cell growth, EMT and angiogenesis by sponging miR-345-5p through modulating Twist1 in cholangiocarcinoma. Eur Rev Med Pharmacol Sci 2020; 24:2321–2334. doi: 10.26355/eurrev_202003_20498. [DOI] [PubMed] [Google Scholar]
- 22.Chen D, Si W, Shen J, Du C, Lou W, Bao C, et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GCC2 in breast cancer cells. Cell Death Dis 2018; 9:188–199. doi: 10.1038/s41419-017-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Ma C, Yang B, Yin C, Zhang B, Xiao Y. LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem Biophys Res Commun 2017; 493:1168–1175. doi: 10.1016/j.bbrc.2017.09.114. [DOI] [PubMed] [Google Scholar]
- 24.Wang JX, Yang Y, Li K. Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J Cell Physiol 2018; 233:6986–6995. doi: 10.1002/jcp.26621. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Peng L, Zhu Z, Du C, Shen Z, Zang R, et al. LncRNA AFAP1-AS functions as a competing endogenous RNA to regulate RAP1B expression by sponging miR-181a in the HSCR. Int J Med Sci 2017; 14:1022–1030. doi: 10.7150/ijms.18392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Eriksson U. Novel VEGF family members: VEGF-B, VEGF-C and VEGF-D. Int J Biochem Cell Biol 2001; 33:421–426. doi: 10.1016/s1357-2725(01)00027-9. [DOI] [PubMed] [Google Scholar]
- 27.Al-Hattab DS, Safi HA, Nagalingam RS, Bagchi RA, Stecy MT, Czubryt MP. Scleraxis regulates Twist1 and Snai1 expression in the epithelial-to-mesenchymal transition. Am J Physiol Heart Circ Physiol 2018; 315:H658–H668. doi: 10.1152/ajpheart.00092.2018. [DOI] [PubMed] [Google Scholar]
- 28.Milone MR, Pucci B, Bruzzese F, Carbone C, Piro G, Costantini S, et al. Acquired resistance to zoledronic acid and the parallel acquisition of an aggressive phenotype are mediated by p38-MAP kinase activation in prostate cancer cells. Cell Death Dis 2014; 5:e1276.doi: 10.1038/cddis.2013.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y, Jiaoming L, Xiang W, Yanhui L, Shu J, Maling G, et al. Analyzing the interactions of mRNAs, miRs, lncRNAs and circRNAs to predict competing endogenous RNA networks in glioblastoma. J Neurooncol 2018; 137:493–502. doi: 10.1007/s11060-018-2757-0. [DOI] [PubMed] [Google Scholar]
- 30.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology 2013; 144:956–966. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J, Zhong G, Wu J, Chen H, Jia Y. Long noncoding RNA AFAP1-AS1 facilitates tumor growth through enhancer of zeste homolog 2 in colorectal cancer. Am J Cancer Res 2018; 8:892–902. [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Y, Zhang Q, Wang J, Liu P. Increased lncRNA AFAP1-AS1 expression predicts poor prognosis and promotes malignant phenotypes in gastric cancer. Eur Rev Med Pharmacol Sci 2017; 21:3842–3849. [PubMed] [Google Scholar]
- 33.Lu X, Zhou C, Li R, Deng Y, Zhao L, Zhai W. Long noncoding RNA AFAP1-AS1 promoted tumor growth and invasion in cholangiocarcinoma. Cell Physiol Biochem 2017; 42:222–230. doi: 10.1159/000477319. [DOI] [PubMed] [Google Scholar]
- 34.Wu XB, Feng X, Chang QM, Zhang CW, Wang ZF, Liu J, et al. Cross-talk among AFAP1-AS1, ACVR1 and microRNA-384 regulates the stemness of pancreatic cancer cells and tumorigenicity in nude mice. J Exp Clin Cancer Res 2019; 38:107–118. doi: 10.1186/s13046-019-1051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B, Li D, Kovalchuk A, Litvinov D, Kovalchuk O. Ionizing radiation-inducible miR-27b suppresses leukemia proliferation via targeting cyclin A2. Int J Radiat Oncol Biol Phys 2014; 90:53–62. doi: 10.1016/j.ijrobp.2014.04.055. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, Zou Z, Nie P, Kou X, Wu B, Wang S, et al. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis 2016; 7:e2454.doi: 10.1038/cddis.2016.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, Yang JM, Jin Y, Jheon S, Kim K, Lee CT, et al. MicroRNA expression profiles and clinicopathological implications in lung adenocarcinoma according to EGFR, KRAS, and ALK status. Oncotarget 2016; 8:8484–8498. doi: 10.18632/oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hawinkels LJ, Zuidwijk K, Verspaget HW, de Jonge-Muller ESM, van Duijn W, Ferreira V, et al. VEGF release by MMP-9 mediated heparan sulphate cleavage induces colorectal cancer angiogenesis. Eur J Cancer 2008; 44:1904–1913. doi: 10.1016/j.ejca.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 39.Salomon-Perzyńska M, Perzyński A, Rembielak-Stawecka B, Michalski B, Skrzypulec-Plinta V. VEGF - targeted therapy for the treatment of cervical cancer - literature review. Ginekol Pol 2014; 85:461–465. doi: 10.17772/gp/1754. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez-Freixinos V, Mackay HJ. Breaking down the evidence for bevacizumab in advanced cervical cancer: past, present and future. Gynecol Oncol Res Pract 2015; 2:8.doi: 10.1186/s40661-015-0015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Wu YY, Liu P, Wang J, Wang G, Qin J, et al. Down-regulation of HPV18 E6, E7, or VEGF expression attenuates malignant biological behavior of human cervical cancer cells. Med Oncol 2011; 28:S528–539. doi: 10.1007/s12032-010-9690-1. [DOI] [PubMed] [Google Scholar]
- 42.Yeh YW, Cheng CC, Yang ST, Tseng CF, Chang TY, Tsai SY, et al. Targeting the VEGF-C/VEGFR3 axis suppresses Slug-mediated cancer metastasis and stemness via inhibition of KRAS/YAP1 signaling. Oncotarget 2017; 8:5603–5618. doi: 10.18632/oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q, Wu JY, et al. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol 2016; 48:1590–1598. doi: 10.3892/ijo.2016.3385. [DOI] [PubMed] [Google Scholar]
- 44.Teifel M, Friedl P. New lipid mixture for efficient lipid-mediated transfection of BHK cells. Biotechniques 1995; 19:79–80. doi: 10.1134/S1063785007100203. [PubMed] [Google Scholar]
- 45.Tang J, Xie Y, Xu X, Yin Y, Jiang R, Deng L, et al. Bidirectional transcription of Linc00441 and CC1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis 2017; 8:e2675.doi: 10.1038/cddis.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, Noh JH, Lee SK, Munk R, Sharov A, Lehrmann E, et al. LncRNA OIP5-AS1/cyrano suppresses GAK expression to control mitosis. Oncotarget 2017; 8:49409–49420. doi: 10.18632/oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]