Abstract
Background:
Airway eosinophilia is a prominent feature of asthma and chronic rhinosinusitis (CRS), and the endothelium plays a key role in eosinophil trafficking. To date, microRNA-1 (miR-1) is the only microRNA known to be regulated in the lung endothelium in asthma models.
Objective:
We sought to determine the role of endothelial miR-1 in allergic airway inflammation.
Methods:
We measured microRNA and mRNA expression using quantitative RT-PCR. We used ovalbumin and house dust mite models of asthma. Endothelium-specific overexpression of miR-1 was achieved through lentiviral vector delivery or induction of a transgene. Tissue eosinophilia was quantified by using Congo red and anti-eosinophil peroxidase staining. We measured eosinophil binding with a Sykes-Moore adhesion chamber. Target recruitment to RNA-induced silencing complex was assessed by using anti-Argonaute2 RNA immunoprecipitation. Surface P-selectin levels were measured by using flow cytometry.
Results:
Serum miR-1 levels had inverse correlations with sputum eosinophilia, airway obstruction, and number of hospitalizations in asthmatic patients and sinonasal tissue eosinophilia in patients with CRS. IL-13 stimulation decreased miR-1 levels in human lung endothelium. Endothelium-specific overexpression of miR-1 reduced airway eosinophilia and asthma phenotypes in murine models and inhibited IL-13–induced eosinophil binding to endothelial cells. miR-1 recruited P-selectin, thymic stromal lymphopoietin, eotaxin-3, and thrombopoietin receptor to the RNA-induced silencing complex; downregulated these genes in the lung endothelium; and reduced surface P-selectin levels in IL-13–stimulated endothelial cells. In our asthma and CRS cohorts, miR-1 levels correlated inversely with its target genes.
Conclusion:
Endothelial miR-1 regulates eosinophil trafficking in the setting of allergic airway inflammation. miR-1 has therapeutic potential in asthmatic patients and patients with CRS.
Keywords: Eosinophil trafficking, vascular endothelium, microRNA, asthma, chronic rhinosinusitis, P-selectin
GRAPHICAL ABSTRACT
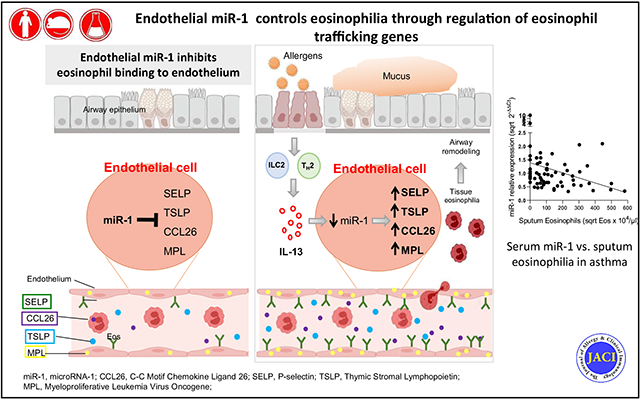
More than half of asthmatic patients and patients with chronic rhinosinusitis (CRS) have high numbers of eosinophils in their blood, sputum, or airway tissues.1,2 Eosinophils play a distinctive role in the pathophysiology of airway allergic disorders, and the presence of eosinophilia has been used as a biomarker of type 2 (T2) inflammation.2-14 However, the discordance between the levels of eosinophilia and T2 cytokines among patients suggests that downstream pathways in inflamed tissues also play a critical role in the eosinophilic response.
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by recruiting mRNAs to the RNA-induced silencing complex (RISC).15-17 Animal and human studies have shown the importance of miRNAs in T2 inflammatory cells.18-28 However, the functions of miRNAs in tissue structural cells, specifically endothelial cells, have not been extensively studied.
To date, microRNA-1 (miR-1) is the only miRNA reported to be altered in the endothelium of murine asthma models.29
In the present study we evaluated the clinical correlations of miR-1 and found that miR-1 levels in asthmatic patients and patients with CRS had significant inverse correlations with eosinophilia. Additionally, stimulation with the T2 cytokine IL-13 caused a dose-dependent decrease in miR-1 levels in both human lung tissue and isolated endothelial cultures, all suggesting that low miR-1 levels play a critical role in the T2-mediated eosinophilic response. Selective overexpression of miR-1 in endothelial cells inhibited airway eosinophilia and major phenotypes of asthma, including airway hyperreactivity and mucus metaplasia in murine asthma models. Mechanistically, miR-1 recruited several eosinophil-trafficking genes to RISC and directly inhibited IL-13–induced eosinophil binding to the endothelial surface. In accord with these findings, miR-1 targets had inverse correlations with miR-1 and direct correlations with eosinophilia in clinical samples. We conclude that miR-1 controls tissue eosinophilia by regulating a network of trafficking genes in the endothelium.
METHODS
Asthma cohort
Serum and sputum from asthmatic patients and healthy subjects were retrieved from the Yale Center for Asthma and Airway Diseases repository under HIC 0102012268. The severity of disease for each patient was assigned based on Global Initiative for Asthma 2018/National Asthma Education and Prevention Program guidelines and was consistent with the previous definitions used for severity.30 The details of sample retrieval and processing were described previously31 and are available in the Methods section in this article’s Online Repository at www.jacionline.org.
CRS cohort
Sinonasal tissue samples from 40 patients undergoing sinonasal surgery at the Yale Department of Otolaryngology were retrieved under Yale Institutional Review Board protocol #0304025173 as deidentified excess tissue. All patients provided consent. The details of tissue processing, miR-1 measurements, and histopathology are described in the Methods section in this article’s Online Repository.
Ex vivo lung culture
Detailed procedures for ex vivo culture are described in the Methods section in this article’s Online Repository. Briefly, histologically normal lung tissue retrieved from patients who underwent surgical resection for lung masses at the Yale Cancer Center were cut into small pieces (2-3 mm) and cultured in M199 and 20% FBS (Gibco, Life Technologies, Waltham, Mass) at 37°C.
miRNA and mRNA measurements
miR-1 levels were measured by using a real-time stem-loop quantitative RT-PCR assay, as described previously.29 Gene expression studies were performed with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, Calif) on a ViiA 7 Real-Time PCR machine (Thermo Fisher Scientific, Waltham, Mass) and according to the manufacturer’s instructions.
Murine asthma models and analyses of lung tissue, bronchoalveolar lavage fluid, and airway hyperreactivity
Murine experiments were performed, as described previously,29 and all the protocols have been reviewed and approved by the Institutional Animal Care and Use Committee at Yale University. Details are available in the Methods section in this article’s Online Repository.
Lentiviral vectors
Vascular-specific miR-1 (V-miR-1) and vascular-specific control (V-ctrl) vectors were prepared and used, as described previously.32
miRNA-induced silencing complex analysis
Argonaute2 co-immunoprecipitation studies were performed, as described previously,29 and details are available in the Methods section in this article’s Online Repository.
MACS for isolation of endothelial cells
Cells were isolated by using Miltenyi Biotech (Bergisch Gladbach, Germany) magnetic beads and columns, as described previously.29,32 Details are available in the Methods section in this article’s Online Repository.
Eosinophil isolation and adhesion assays
Human eosinophils were isolated from blood drawn from healthy human volunteers under approved Yale Institutional Review Board protocol #0902004786. Detailed descriptions of eosinophil isolation and adhesion assays are available in the Methods section in this article’s Online Repository.
Statistical analysis
Normality (Gaussian distribution) of the data sets were tested with D’Agostino-Pearson tests. Normally distributed parametric data were assessed by using t tests, and nonnormally distributed data were analyzed by using the Mann-Whitney test. Adequate sample size (α = 0.05, power = 0.8) for clinical experiments was assessed by using the Power Analysis calculator (http://www.biomath.info/power/).
The Spearman correlation coefficient or Mann-Whitney test was used to assess the association between miR-1 levels and eosinophils in all clinical samples. GraphPad Prism 7 (GraphPad Software, La Jolla, Calif) and SAS software (SAS Institute, Cary, NC) were used for these statistical analyses. Detailed analyses for histopathology and mRNA measurements in the asthma cohort were previously described.33
RESULTS
miR-1 levels have inverse correlations with airway eosinophilia in asthmatic patients and patients with CRS
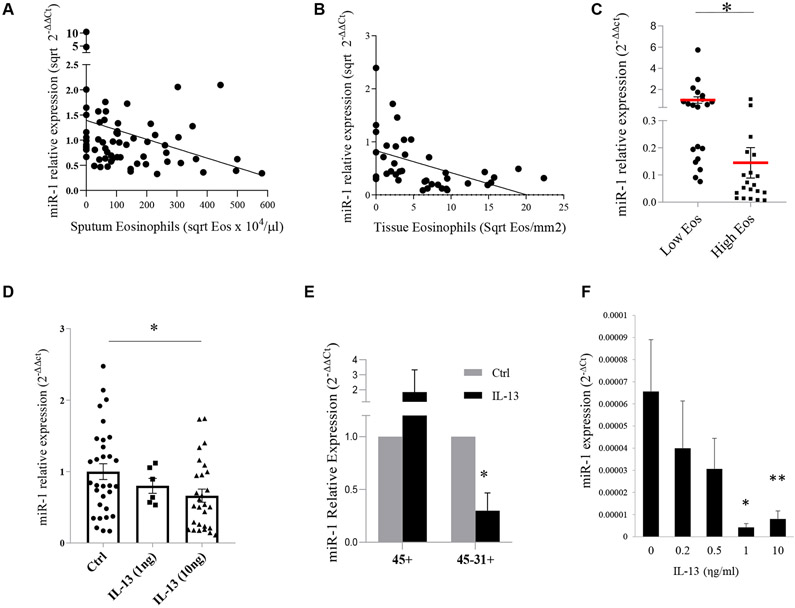
We have previously shown that miR-1 is altered in the lung endothelium in murine asthma models.29 We investigated whether miR-1 has clinical significance in human allergic airway disorders. We first evaluated the correlations of serum miR-1 levels in a 70-patient asthma cohort (see Table E1, A, in this article’s Online Repository at www.jacionline.org) and found that miR-1 levels correlate inversely with sputum eosinophilia and frequency of hospitalizations and directly with FEV1 percent predicted, forced vital capacity (FVC) percent predicted, and Asthma Control Test scores (Fig 1, A, and Table I).
FIG 1.
The effect of T2 inflammation on miR-1 levels in human subjects. A, Association between serum miR-1 levels (normalized to the mean of the control group) and sputum eosinophilia in asthma patients and healthy control subjects (n = 66, R = −0.3037, P = .013). B, Association between miR-1 levels and eosinophil counts (counted after anti-eosinophil peroxidase [EPX] staining) in sinonasal tissue samples from patients with CRS (n = 40, r = −5784, P < .00001). C, Comparison of miR-1 levels in sinonasal tissue samples from patients with CRS divided into high-eosinophil-count (>15 cells/high-power field [hpf]) and low-eosinophil-count (≤15 cells/hpf) groups (n = 40). *P < .0001. Eos, Eosinophils. D, miR-1 levels in cultured human lung tissue samples after stimulation with recombinant IL-13 and normalized to the mean of the control group (ctrl, PBS; control group: n = 8 subjects with 3 or more replicates; 1 ng/mL group, n = 3 subjects with 2 replicates; 10 ng/mL group, n = 7 subjects with 3 or more replicates). *P = .0106. E, miR-1 levels in immune (CD45+) and endothelial (CD31+CD45−) cells isolated from cultured human lungs after treatment with recombinant IL-13 or PBS controls (n = 3 subjects). *P = .026. F, miR-1 levels in HUVECs stimulated with recombinant IL-13 (n = 6 or more in each group from 4 experiments). *P < .01 and **P < .05, Student unpaired t test. R, Spearman correlation coefficient for Fig 1, A and B. Error bars represent SEMs.
TABLE I.
Clinical correlations of miR-1 levels in asthmatic patients (n = 70)
| Variable | |
|---|---|
| Age | P = .4158, r = −0.0988 |
| Sex | P = .351 |
| Atopy | P = .4711 |
| Hospitalization for asthma (lifetime) | P = .0047, r = −0.3339 |
| Hospitalization for asthma (past year) | P = .0163, r = −0.2862 |
| ACT score | P = .0158, r = 0.2875 |
| FEV1 (% predicted)* | P = .0007, r = 0.3996 |
| FVC (% predicted)* | P = .0008, r = 0.3961 |
| OCS use | P = .2082, r = −0.1523 |
| ICS use | P = .0039, r = −0.3409 |
| Sputum cytology† | |
| Eosinophils | P = .0132, r = −0.3037 |
| Neutrophils | P = .0778, r = −0.2187 |
| Macrophages | P = .8875, r = −0.01774 |
| Lymphocytes | P = .9132, r = 0.01368 |
ACT, Asthma Control Test; FVC, forced vital capacity; ICS, inhaled corticosteroid; OCS, oral corticosteroid.
Data are available for 68 subjects.
Data are available for 66 subjects.
Because airway eosinophilia is a significant pathophysiologic determinant, we next tested the miR-1 association with eosinophil infiltration in sinonasal tissues from 40 patients with CRS (see Table E1, B) and found that, consistent with our findings in the asthma cohort, tissue miR-1 levels had an inverse correlation with eosinophilia (Fig 1, B, and see Fig E1 in this article’s Online Repository at www.jacionline.org) and were significantly lower in the high-eosinophil-count group compared with the low-eosinophil-count group (Fig 1, C).
IL-13 decreases miR-1 levels in human lung and endothelial cells
T2 cytokines are one of the main mediators of the eosinophilic response. To determine whether T2 cytokines regulate miR-1, we tested the effect of IL-13, a prototypic T2 cytokine,12,13,34 on miR-1 levels in an ex vivo human lung tissue culture system. IL-13 stimulation caused a dose-dependent reduction of miR-1 levels in the lung (Fig 1, D), showing that T2 inflammation down-regulates miR-1.
To determine whether, similar to the murine lung,29 this down-regulation occurs primarily in the endothelium, we first isolated the immune (CD45+), endothelial (CD31+ CD45−), and epithelial (CD326+ CD45−) fractions from one subject and found that miR-1 is most notably decreased in the endothelial cells and showed a smaller degree of downregulation in the immune cells (see Fig E2 in this article’s Online Repository at www.jacionline.org). We next compared miR-1 levels in stimulated (and control) endothelial and immune cells from 3 subjects and found that miR-1 levels are consistently downregulated in the endothelium, whereas in immune cells they are variable (Fig 1, E). We then asked whether this downregulation will also occur in cultured endothelial cells. We used human umbilical vein endothelial cells (HUVECs), primary human endothelial cells of systemic circulation origin (similar to the bronchial circulation) that are commonly used to characterize T2 responses in the endothelium.35-42 As shown in Fig 1, F, and in accord with the findings in our fractionation experiment, IL-13 decreased miR-1 levels in HUVECs in a dose-dependent manner. Together with our clinical data, these findings suggested that miR-1 downregulation is a downstream inflammatory mechanism and controls the eosinophilic response.
Endothelium-specific miR-1 regulates aeroallergen-induced mouse airway inflammation
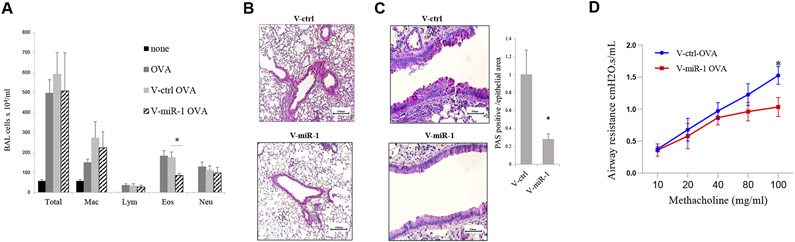
To specifically assess the importance of endothelial miR-1 downregulation, we used 2 strategies to overexpress it in the endothelium in murine asthma models. We first used V-miR-1 vector, in which a VE-cadherin promoter drives cell-specific overexpression of miR-1 in the endothelium.32 We delivered V-miR-1 (or the control vector V-ctrl) intranasally to mice that were previously sensitized to ovalbumin (OVA). After OVA inhalational challenge, the mice that had received V-miR-1 exhibited a significant (approximately 50%) decrease in eosinophil counts in their bronchoalveolar lavage (BAL) fluid compared with control mice. They also showed reduced airway inflammation, mucus metaplasia, and bronchial hyperresponsiveness (Fig 2). The comparison of T2 cytokine levels in BAL fluid from these mice showed a 30% reduction in IL-5 levels and a trend toward lower levels of IL-4 (see Fig E3, A, in this article’s Online Repository at www.jacionline.org) in V-miR-1–treated mice but no significant changes in IL-13, IL-10, or eotaxin levels. Interestingly, mRNA analyses in the lungs of these mice did not show any significant differences in levels of IL-4, IL-5, or IL-13 between the experimental groups (see Fig E3, B), suggesting that the miR-1 effect on eosinophilia is not mediated through a significant alteration of T2 cytokines.
FIG 2.
Effect of vascular-specific lentiviral miR-1 vector on T2 inflammation. Wild-type C57BL/6 mice were sensitized with OVA and received intranasal V-miR-1 or V-ctrl vector 2 weeks before OVA aerosol challenge. A, BAL cytology from PBS-challenged (none), OVA-challenged, OVA-challenged with V-ctrl treatment, and OVA-challenged with V-miR-1 treatment groups (n = 7 and 11 for the none and OVA groups and n = 15 per group for the V-ctrl-OVA and V-miR-1-OVA groups from 3 experiments). *P = .00049. Eos, Eosinophils; Lym, lymphocytes; Mac, macrophages; Neu, neutrophils. B and C, Lung sections from these mice were stained with hematoxylin and eosin for airway inflammation (Fig 2, B) and periodic acid–Schiff (PAS) stain for mucus metaplasia (Fig 2, C). PAS-positive/total airway epithelial areas were counted and presented as a graph in the left panel (n ≥ 6 from 2 experiments). *P = .01909. D, Airway responses were measured by using the forced oscillation technique. Mean airway resistance (in cm H20.s/mL) was measured after exposing mice to increasing concentrations of methacholine (in milligrams per milliliter, n = 8 from 2 experiments). *P = .02. Error bars represent SEMs. Data were assessed by using the Student unpaired t test.
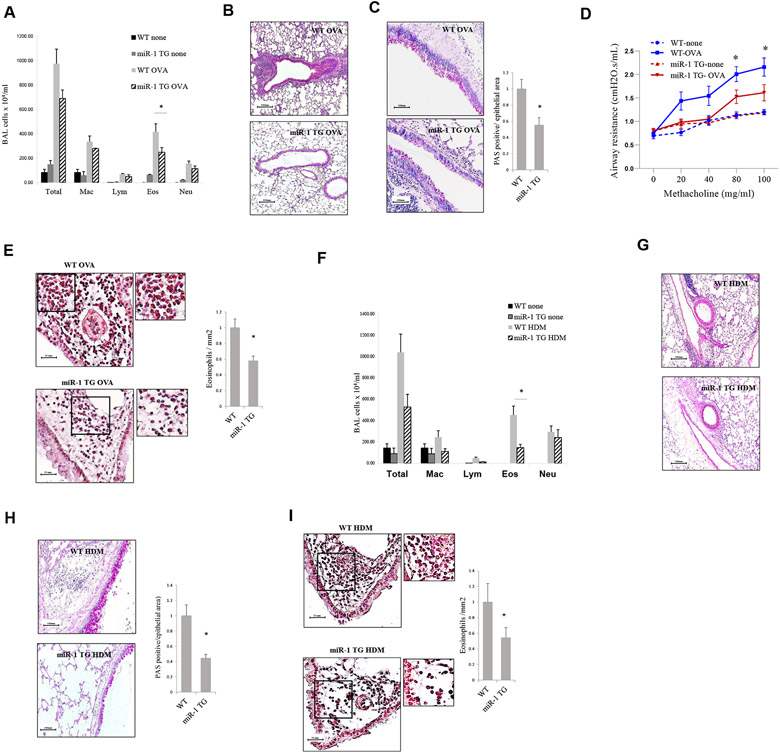
We next tested our hypothesis in V-miR-1 transgenic mice sensitized and challenged with OVA. In the miR-1 transgenic mouse vascular-specific overexpression of miR-1 is induced by adding doxycycline to the drinking water.32 Similar to the vector delivery experiments, transgenic overexpression of miR-1 in the endothelium reduced BAL eosinophilia and airway inflammation, mucus metaplasia, and hyperreactivity compared with those seen in control mice (Fig 3, A-D). Quantification of peribronchial eosinophil infiltration in the lungs of these mice confirmed the observed difference in BAL fluid eosinophil counts (Fig 3, E). Also, like the vector delivery experiments, cytokine measurements in these mice showed only trends toward lower levels of IL-4 and IL-5 in the BAL fluid of transgenic mice and no significant differences in expression of T2 cytokines in the lung (see Fig E4 in this article’s Online Repository at www.jacionline.org).
FIG 3.
Effect of the inducible miR-1 transgene in asthma models. A-E, miR-1 transgenic mice (miR-1 TG) and their wild-type (WT) littermates in an OVA model. Fig 3, A, BAL fluid cytology (n = 4 for the “none” group and n = 12 for the OVA groups from 2 experiments). *P = .00048. Fig 3, B and C, Representative lung sections from these mice stained with hematoxylin and eosin for inflammation (Fig 3, B) and periodic acid-Schiff (PAS) for mucus metaplasia (Fig 3, C). Right panel shows PAS-positive areas measured and presented as in Fig 2, C (n = 4-7 per group). *P = .0018. Fig 3, D, Airway responses to methacholine challenge were measured and presented, as described in Fig 2, D (n = 4-7 per group from 2 experiments). *P < .03 for the OVA group. Fig 3, E, Congo red staining for tissue eosinophilia. Left panel shows representative images. Insets show 2 × magnification of the corresponding area. Right panel shows quantification results (n = 3-5 per group from 1 experiment). *P = .0017. F-I, miR-1 TG mice and their WT litters in an HDM model. Fig 3, F, BAL fluid cytology (n = 4 for the “none” group and n = 8 or 19 for the HDM groups from 2 experiments). *P = .00524. Fig 3, G-I, Lung sections were stained with H&E (Fig 3, G), PAS (n = 3-6 per group from 1 experiment, *P = .0165; Fig 3, H), and Congo red (n = 13 from 2 experiments, *P = .000603; Fig 3, I). Error bars represent SEMs. Data were assessed by using the Student unpaired t test. Eos, Eosinophils; Lym, lymphocytes; Mac, macrophages; Neu, neutrophils.
To confirm our findings with a clinically relevant human allergen, we repeated our experiment with the house dust mite (HDM) model. Our results with the HDM model were similar to those with the OVA model and showed reduction of eosinophilia and inflammation in transgenic mice compared with control mice. Interestingly, however, in this case none of the measured T2 cytokines or chemokines showed a significant difference or trend between the 2 groups (see Fig E5 in this article’s Online Repository at www.jacionline.org). Cumulatively, these data show that endothelial miR-1 plays a significant role in regulating the severity of eosinophilia in the T2 response. They also suggest that miR-1 acts downstream of the T2 cytokines and mediates their effects on the inflamed endothelium.
miR-1 inhibits binding and movement of eosinophils on human endothelial cells
One of the main mechanisms through which the endothelium controls tissue eosinophilia is regulation of eosinophil trafficking.8,13,43 To evaluate the effect of miR-1 on eosinophil trafficking, we tested the interaction of eosinophils with the endothelium after miR-1 overexpression. We used fresh human eosinophils isolated by using a rapid isolation method with chemotactic peptide for these experiments. HUVECs were transduced with V-miR-1 (or V-ctrl) and exposed to eosinophils in an adhesion chamber. miR-1 overexpression significantly reduced binding of eosinophils to HUVECs at baseline (Fig 4, A, and see Fig E6 in this article’s Online Repository at www.jacionline.org) and after IL-13 stimulation (by approximately 50%). Importantly, the method of eosinophil isolation did not make a difference, and human eosinophils isolated through magnetic sorting also showed lower affinity for miR-1–transduced endothelium. Also, interestingly, miR-1 overexpression decreased the postbinding movement of eosinophils on the endothelial surface by more than 50% (Fig 4, B, and see Videos E1 and E2 in this article’s Online Repository at www.jacionline.org), suggesting that miR-1 also inhibits the postcapture activation and crawling of the eosinophils.
FIG 4.
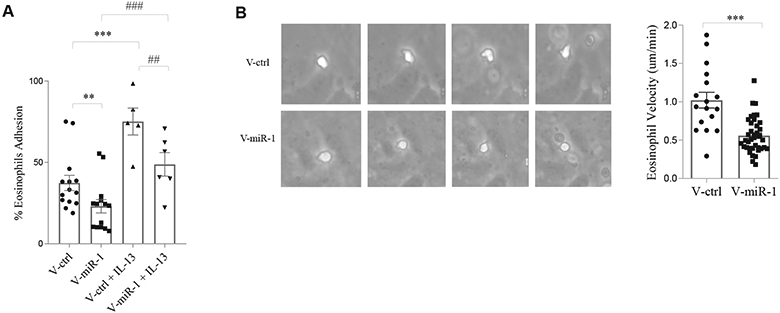
Effect of miR-1 on eosinophil-endothelium interaction. HUVECs were transduced with V-miR-1 or V-ctrl, stimulated with recombinant IL-13 (1 ng/mL) for 24 hours, and then exposed to eosinophils freshly isolated from healthy human subjects. Eosinophils were isolated by using a rapid method with chemotactic peptide. A, Eosinophil adhesion assay in a Sykes-Moore adhesion chamber. Percentages of eosinophils bound to the HUVEC surface were plotted for each group (n = 14 for the v-miR-1 and v-ctrl groups and n = 5 for the IL-13 groups, data from 4 experiments). **P = .0014, ***P = .0006, ##P = .0303, and ###P = .0041, Mann-Whitney test). B, Velocities of eosinophil movement on the surfaces of transduced HUVECs (in micrometers per minute) after binding were recorded by using video microscopy. Left panel shows representative images depicting sequential images from the movement of a human eosinophil (white cell) on the surfaces of transduced HUVECs. Right panel shows the quantification plot (n = 17 for the V-ctrl and n = 39 for the V-miR-1 group, 2 experiments). ***P < .0001, Mann-Whitney test. Error bars represent SEMs.
miR-1 recruits eosinophil-trafficking genes to RISC and reduces their expression
Because overexpression of miR-1 in the endothelium inhibited airway eosinophilia and specifically eosinophil adhesion and activation, we asked whether miR-1 targets eosinophil trafficking genes. We selected a number of candidate genes based on their known role in trafficking and the presence of a predicted miR-1 binding site in their 3′ untranslated region. We then measured the enrichment of these mRNAs on the Argonaute2 RISC versus total cell lysate and compared V-miR-1–transduced cells to V-ctrl–transduced cells. As shown in Fig 5, A, miR-1 overexpression (compared with control values) increased selective enrichment of 3 trafficking genes, eotaxin-3 (CCL26), P-selectin (SELP), and thymic stromal lymphopoietin (TSLP), and its previously known miR-1 targets myeloproliferative leukemia virus oncogene or thrombopoietin receptor (MPL), although the enrichment values for TSLP did not reach statistical significance (P = .111).
FIG 5.
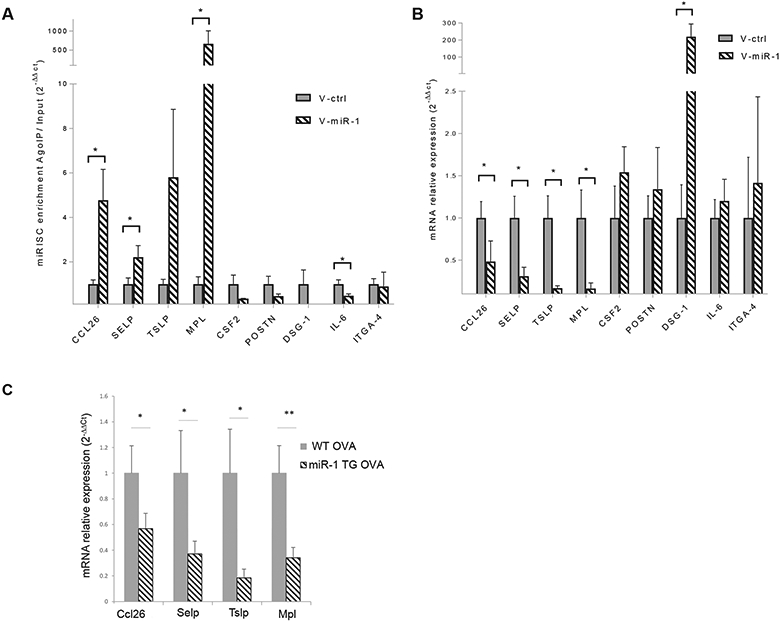
miR-1 targets controlling eosinophil trafficking. A, HUVECs were transduced with V-miR-1 or V-ctrl vectors. Cell lysates were immunoprecipitated with anti–Argonaute2 (Ago2) antibody, and levels of mRNAs bound to Ago2 were measured by using quantitative RT-PCR in the whole lysate (input) and Ago2 immuno-precipitates (AgoIP). RISC recruitment was calculated as mRNA levels in AgoIP/input (2−ΔΔCt, n = 4 per group). *P < .05. B, Expression levels of the target gene mRNAs in V-miR-1– and V-ctrl-transduced HUVECs were normalized to their means in the control (V-ctrl) group and presented as 2−ΔΔCt (n = 4). *P < .05. C, Endothelial cells were isolated from OVA-challenged mice by using magnetic immune sorting, as described in Fig 1, E, and mRNA expression levels were measured, normalized, and presented as described in Fig 5, B (CCL26, SELP, and TSLP: n = 7WT and 8 miR-1 TG; MPL: n = 10 per group; from 2 experiments). *P < .05 and **P = .005331. CSF2, Colony-stimulating factor 2; DSG1, desmoglein 1; ITGA4, integrin subunit α4; POSTN, periostin. Error bars represent SEMs. Data were assessed by using the Student unpaired t test.
We next validated the proposed targets by measuring their levels in both HUVECs and mouse lung endothelium (isolated from the transgenic mice). In either case miR-1 overexpression decreased expression of all 4 genes (Fig 5, B and C).
miR-1 regulates SELP expression at the endothelial surface
To explore the pathway of the miR-1 effect on eosinophil binding, we next tested SELP levels at the endothelial surface. SELP is an endothelial surface protein known to increase with T2 stimulation and to regulate the selective binding of eosinophils to the inflamed airways.44-51 We hypothesized that miR-1 regulates SELP levels at the endothelial surface. To test this hypothesis, we measured surface SELP levels using flow cytometry. We first validated the sensitivity of this method by showing its ability to detect the dose-dependent increase of surface SELP in response to IL-13 (see Fig E7 in this article’s Online Repository at www.jacionline.org). We next tested the miR-1 effect and found that miR-1 overexpression causes a distinct alteration of surface SELP levels (Fig 6, A).
FIG 6.
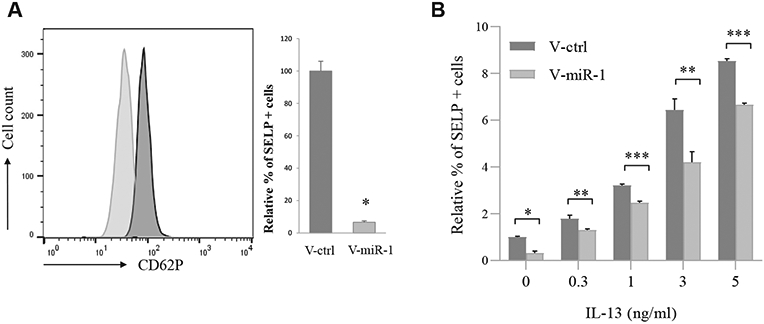
Effect of the miR-1 axis on SELP expression on the endothelial surface. HUVECs were transduced with V-miR-1 (or V-ctrl), and SELP expression on the cell surface was measured by using flow cytometry. A, Left panel shows a typical histogram plot. Right panel shows cumulative data from 2 experiments with percentage SELP expression normalized to the V-ctrl group (n = 9 from 2 experiments). Light gray, V-miR-1; dark gray, V-ctrl. *P = 2.1 × 10−7. B, Transduced HUVECs were treated with increasing concentrations of human recombinant IL-13. SELP expression levels in each group were measured and normalized to V-ctrl in PBS (n = 6 or more from 2 experiments). *P < .000001, **P < .01, and ***P < .05.
We next asked whether, similar to its effect on eosinophil binding, the miR-1–induced inhibition is strong enough to overcome the stimulatory effect of IL-13. As shown in Fig 6, B, miR-1 retained its inhibitory effect, even at higher doses of IL-13 and consistently reduced surface SELP levels.
Correlations between miR-1, its targets, and eosinophilia in clinical cohorts
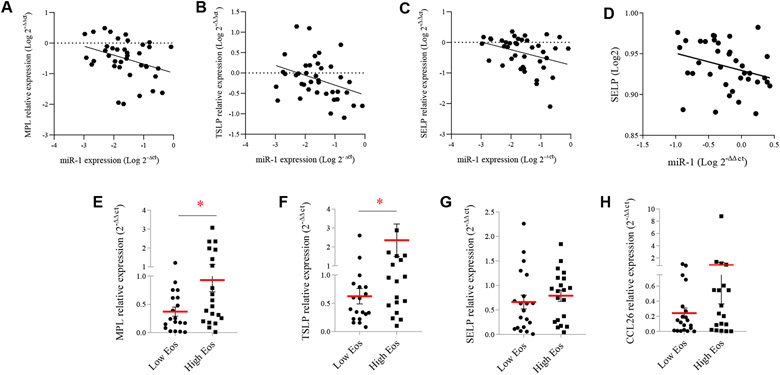
To test the validity of the proposed miR-1–targeting events in clinical samples, we next assessed the correlation between miR-1 levels and its endothelial targets in our clinical cohorts. As shown in Fig 7, A-C, we found a statistically significant inverse correlation between miR-1 levels and SELP, TSLP, and MPL expression in the sinonasal tissues of our CRS cohort and between miR-1 and SELP expression in serum samples of our asthma cohort (Fig 7, D), strongly suggesting that the proposed targeting also occurs in human subjects.
FIG 7.
Associations between miR-1, its target genes, and eosinophilia in clinical samples. A-C, Correlation between miR-1 and its target genes in the CRS cohort described in Fig 1, A (n = 40, except for MPL [n = 39]). Fig 7, A, MPL: R = −0.3356, P = .0367. Fig 7, B, TSLP: R = −0.3461, P = .0309. Fig 7, C, SELP: R = −0.3361, P = .034. D, Correlation between miR-1 and SELP expression in serum samples from the asthma cohort described in Fig 1 (n = 37, R = −0.3589, P = .0291. E-H, mRNA expression levels in the high- and low-eosinophil-count groups described in Fig 1, A. Fig 7, E, MPL: *P = .0468. Fig 7, F, TSLP. *P = .0262. Fig 7, G, SELP: P = .2534. Fig 7, H, CCL26: P = .2829. In each graph means and SEMs are represented as red lines and error bars, respectively. R, Spearman correlation coefficient.
We next asked whether, similar to miR-1 itself, miR-1 target genes also show associations with tissue eosinophilia. We compared levels of miR-1 targets (and several other T2-related genes) between patients with CRS with high and those with low eosinophil counts (Fig 7, E-H, and see Fig E8 in this article’s Online Repository at www.jacionline.org) and found that, consistent with their targeting by miR-1, MPL and TSLP are among the only genes that were expressed at significantly higher levels in the highly eosinophilic samples.
DISCUSSION
In this article we show for the first time that miR-1 is a critical regulator of an endothelial gene network that controls eosinophil trafficking. We show that miR-1 levels have inverse correlations with eosinophilia in asthmatic patients and patients with CRS and are downregulated by IL-13. We then demonstrate the critical role and cell specificity of this downregulation by showing that an isolated increase in endothelial miR-1 levels in murine asthma models inhibits airway eosinophil infiltration and hyperreactivity without imparting a consistent effect on T2 cytokines.
Three lines of evidence support the regulatory effect of miR-1 on eosinophil trafficking. First, in cell-culture experiments miR-1 overexpression directly inhibited IL-13–induced binding of eosinophils to the endothelium. Second, miR-1 targeted a group of genes known to be involved in eosinophil trafficking, including an eosinophil chemokine, an adhesion molecule, and an alarmin. Third, miR-1 targets had inverse correlations with miR-1 and direct correlations with eosinophilia in clinical samples. These findings are consistent with the role of endothelium as an “orchestrator” of the inflammatory response52-55 and suggest that miR-1 plays critical roles in the initiation (through release of alarmins, such as TSLP) and sustenance (through SELP, MPL, and CCL26) of the eosinophilic response.
Eosinophilia is a ubiquitous feature of allergic airway disorders and has been used as a biomarker of T2 inflammation.8,56-60 Eosinophils participate in the T2 inflammatory response through release of enzymes43,61 and secretion of cytokines and chemokines.62,63 The associations between miR-1 and clinically relevant parameters in our asthma cohort might reflect the multifaceted contributions of eosinophils to T2 inflammation. However, eosinophilia also occurs in T2-low patients,61 and levels of eosinophilia are not significantly different between the atopic and nonatopic clusters in the large asthma cohorts.31,64,65 The consistent association between miR-1 and eosinophilia in our clinical cohorts thus suggests that miR-1 acts downstream from T2 cytokines and might be one of the mediators used by these cytokines to regulate airway eosinophilia.
miRNAs play unique roles within specific pathophysiologic contexts, and recent studies have shown their importance in T2 inflammatory cells.18,66-76 However, the role of miRNAs in structural cells has not been studied as extensively, and past studies have mainly focused on epithelial and smooth muscle cells.66,77-85 Our findings in murine asthma models suggested that miR-1 regulates endothelial genes.29 miR-1 regulates a range of molecular mechanisms in myoblasts and cardiomyocytes86-88 and modulates tumor progression and angiogenesis. 32,89-92 Our present findings assign a completely novel function to endothelial miR-1.
We observed miR-1 downregulation and its endothelial selectivity in IL-13–stimulated ex vivo–cultured human lung tissues. Thus we used several strategies to confirm the endothelial origin of the miR-1 response, its effectiveness as a postsensitization intervention, and its applicability to human disease. We used a vascular promoter–driven lentiviral vector and a vascular-specific transgenic mouse to ensure the cell specificity of the miR-1 effect in our murine models and induced miR-1 overexpression only after the sensitization step. We also used 2 different allergens: OVA and HDM. The OVA model is extensively used in mechanistic murine studies. However, it is not a human allergen and lacks properties, such as intrinsic enzymatic activity and direct activation of immune cells, that are characteristics of HDM.93,94 Furthermore, recognizing the cross-species differences of eosinophil recruitment in mice and human subjects,95,96 we validated our gene-targeting events in both murine and human models. These experiments established that miR-1 regulates the eosinophilic response through an endothelium-intrinsic pathway, its effects are not specific to the OVA allergen or the mouse model, and its effects can modify the inflammatory response after sensitization.
Active roles of the endothelium in T2 inflammation have been well recognized.54,97,98 Increased inflammatory cell trafficking is one of these roles99-103 and is initiated by tethering (capture) and rolling of the inflammatory cells on the endothelial surface,104,105 followed by chemokine-mediated activation and directional crawling, integrin-mediated arrest and firm adhesion, and subsequent polarization and transmigration.104-106 In our experiments miR-1 affected both the initial binding and subsequent movement of eosinophils on the endothelial surface, suggesting that it inhibits both capture and activation (crawling) of eosinophils.
miR-1 targeted the SELP, CCL26, TSLP, and MPL genes in the endothelium. SELP is expressed on the surfaces of endothelial cells and specifically regulates the selective attachment of human eosinophils on the endothelium by binding to the P-selectin glycoprotein ligand 1, a homodimeric protein on the surface of eosinophils.44-51,107,108 Eosinophil binding to nasal polyp endothelium is primarily dependent on SELP expression at the surfaces endothelial cells.40,45 Our findings on the effects of miR-1 on surface SELP levels are consistent with its inhibitory effect on eosinophil binding.
Other miR-1 targets also have known roles in eosinophil trafficking. CCL26 is upregulated in asthmatic patients and recruits eosinophils to inflamed tissues through activation of CCR3 on their surfaces.109-111
TSLP is an alarmin expressed in both the epithelium and endothelium and recruits and activates eosinophils.112-114 TSLP levels correlate with disease severity in asthmatic patients and patients with CRS,115,116 and treatment of patients with mild asthma with an anti-TSLP antibody reduced their allergic responses and eosinophilia.117
MPL is a potent inducer of SELP expression in platelets118 and endothelial cells.29 MPL activation leads to signaling through Janus kinase 2/signal transducer and activator of transcription 3 and 5 axes, phosphoinositide 3–kinase, extracellular signal-regulated kinase 1/2, and p38 mitogen-activated protein kinases.119 All these signaling pathways have been implicated in asthma and mediate critical functions within the airway structural cells.120-125 It is of note that even though the elucidated targets of miR-1 provide a coherent molecular mechanism for its effect on tissue eosinophilia, these findings do not rule out a direct effect of miR-1 on the intrinsic signaling events within eosinophils.
There are limitations to our investigation. Pure human bronchial endothelial cells are not readily available, and we have instead used well-characterized HUVECs in our mechanistic studies. HUVECs are commonly used to characterize the effect of cytokines on the endothelium35-39 and resemble bronchial endothelial cells in being part of the systemic circulation. In our experiments both miR-1 regulation and its targets in HUVECs resembled those seen with the immune-selected lung endothelial cells (Figs 1 and 5). However, our findings in HUVECs suggest that miR-1 regulation is a universal feature of endothelial cells and might be applicable to eosinophilia in organs other than the lungs.
Our clinical studies on miR-1 target genes did not show the expected correlations in all instances. For instance, in our CRS expression analysis, we did not find a significant difference in SELP expression between patients with high and those with low eosinophil counts. We have only tested mRNA expression levels in our study, and measurements of the respective proteins might improve these associations. Furthermore, whole-tissue measurements might not capture the cell-specific targeting events, and expression of genes in other cell types (eg, P-selectin in platelets) might obscure the results.
Finally, we have only tested miR-1 correlations in a single cohort of asthmatic patients. Confirmation of the observed miR-1 associations, such as its inverse correlation with airway obstruction or number of hospitalizations, in larger cohorts is necessary for its validation as a biomarker of severe asthma.
In summary, we show for the first time that miR-1 has clinical significance in human airway allergic disorders and that endothelial miR-1 directly regulates eosinophil trafficking. Specifically, we show that an isolated increase in miR-1 levels in the endothelium is adequate to inhibit airway eosinophilia and the hallmarks of allergic airway inflammation. We also propose a gene network for the latter inhibitory effect that involves SELP, CCL26, TSLP, and MPL. This study is the first report of an endothelial miRNA-mediated regulation of eosinophil trafficking and shows the potential therapeutic value of miR-1 as a direct inhibitor of the eosinophilic response in asthmatic patients and patients with CRS.
Supplementary Material
Key messages.
miRNA-1 levels have inverse correlations with airway eosinophilia in asthma and CRS patients.
miRNA-1 controls eosinophil recruitment to the airways through regulation of an adhesion molecule (P-selectin), an alarmin (TSLP), a chemokine (eotaxin-3), and a cytokine receptor (MPL) in the endothelium.
Acknowledgments
We thank Rita Matta for her generous help with adhesion experiments, Jonas Schupp for his help with immunohistochemistry, Ying Sun for her help in airway hyperactivity experiments, and the Yale Pathology–Tissue Procurement and Distribution Facility (TPD) for providing CRS and normal human lung tissues.
This work was supported by National Institutes of Health (NIH)/National Heart, Lung, and Blood Institute (NHLBI) grant R00 HL098695; NIH/National Institute of Allergy and Infectious Diseases grant R56 AI125411-01; and the American Thoracic Society Foundation Recognition Award for Outstanding Established Investigators (R to R; to S.S.T.).
Abbreviations used
- BAL
Bronchoalveolar lavage
- CRS
Chronic rhinosinusitis
- HDM
House dust mite
- HUVEC
Human umbilical vein endothelial cell
- miR-1
MicroRNA-1
- miRNA
MicroRNA
- MPL
Myeloproliferative leukemia virus oncogene or thrombopoietin receptor
- OVA
Ovalbumin
- RISC
RNA-induced silencing complex
- SELP
P-selectin
- T2
Type 2
- TSLP
Thymic stromal lymphopoietin
- V-ctrl
Vascular-specific control
- V-miR-1
Vascular-specific miR-1
Footnotes
Disclosure of potential conflict of interest: G. Chupp has been a speakers’ bureau member, consultant, and clinical trial principal investigator for Genentech, GlaxoSmithKline, AstraZeneca, and BSCI; has been a speakers’ bureau member and clinical trial principal investigator for Genzyme; on serves on the advisory board of TEVA unrelated to this work. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.McGrath KW, Icitovic N, Boushey HA, Lazarus SC, Sutherland ER, Chinchilli VM, et al. A large subgroup of mild-to-moderate asthma is persistently noneosinophilic. Am J Respir Crit Care Med 2012;185:612–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinke JW, Borish L. Chronic rhinosinusitis phenotypes. Ann Allergy Asthma Immunol 2016;117:234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holgate ST. Pathogenesis of asthma. Clin Exp Allergy 2008;38:872–97. [DOI] [PubMed] [Google Scholar]
- 4.Pelaia G, Vatrella A, Maselli R. The potential of biologics for the treatment of asthma. Nat Rev Drug Discov 2012;11:958. [DOI] [PubMed] [Google Scholar]
- 5.Shah SA, Ishinaga H, Takeuchi K. Pathogenesis of eosinophilic chronic rhinosinusitis. J Inflamm (Lond) 2016;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agache I, Sugita K, Morita H, Akdis M, Akdis CA. The complex type 2 endotype in allergy and asthma: from laboratory to bedside. Curr Allergy Asthma Rep 2015;15:29. [DOI] [PubMed] [Google Scholar]
- 7.Kato A Immunopathology of chronic rhinosinusitis. Allergol Int 2015;64: 121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol 2017;12:331–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol 2015;16: 45–56. [DOI] [PubMed] [Google Scholar]
- 10.Fahy JV. Type 2 inflammation in asthma—present in most, absent in many. Nat Rev Immunol 2015;15:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnes PJ. Pathophysiology of allergic inflammation. Immunol Rev 2011;242: 31–50. [DOI] [PubMed] [Google Scholar]
- 12.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, et al. Interleukin-13: central mediator of allergic asthma. Science 1998;282:2258–61. [DOI] [PubMed] [Google Scholar]
- 13.Becerra-Diaz M, Wills-Karp M, Heller NM. New perspectives on the regulation of type II inflammation in asthma. F1000Res 2017;6:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr TF, Zeki AA, Kraft M. Eosinophilic and noneosinophilic asthma. Am J Respir Crit Care Med 2018;197:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–66. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005;353:1793–801. [DOI] [PubMed] [Google Scholar]
- 18.Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol 2013;132:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebane A, Akdis CA. MicroRNAs in allergy and asthma. Curr Allergy Asthma Rep 2014;14:424. [DOI] [PubMed] [Google Scholar]
- 20.Liu F, Qin HB, Xu B, Zhou H, Zhao DY. Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Mol Med Rep 2012;6:1178–82. [DOI] [PubMed] [Google Scholar]
- 21.Qin HB, Xu B, Mei JJ, Li D, Liu JJ, Zhao DY, et al. Inhibition of miRNA-221 suppresses the airway inflammation in asthma. Inflammation 2012;35:1595–9. [DOI] [PubMed] [Google Scholar]
- 22.Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, et al. Transcriptome analysis shows activation of circulating CD81 T cells in patients with severe asthma. J Allergy Clin Immunol 2012;129:95–103. [DOI] [PubMed] [Google Scholar]
- 23.Chen RF, Huang HC, Ou CY, Hsu TY, Chuang H, Chang JC, et al. MicroRNA-21 expression in neonatal blood associated with antenatal immunoglobulin E production and development of allergic rhinitis. Clin Exp Allergy 2010;40:1482–90. [DOI] [PubMed] [Google Scholar]
- 24.Jardim MJ, Dailey L, Silbajoris R, Diaz-Sanchez D. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am J Respir Cell Mol Biol 2012;47:536–42. [DOI] [PubMed] [Google Scholar]
- 25.Wu XB, Wang MY, Zhu HY, Tang SQ, You YD, Xie YQ. Overexpression of microRNA-21 and microRNA-126 in the patients of bronchial asthma. Int J Clin Exp Med 2014;7:1307–12. [PMC free article] [PubMed] [Google Scholar]
- 26.Ma ZX, Tan X, Shen Y, Ke X, Yang YC, He XB, et al. MicroRNA expression profile of mature dendritic cell in chronic rhinosinusitis. Inflamm Res 2015;64: 885–93. [DOI] [PubMed] [Google Scholar]
- 27.Xia G, Bao L, Gao W, Liu S, Ji K, Li J. Differentially expressed miRNA in inflammatory mucosa of chronic rhinosinusitis. J Nanosci Nanotechnol 2015;15: 2132–9. [DOI] [PubMed] [Google Scholar]
- 28.Zhang YN, Cao PP, Zhang XH, Lu X, Liu Z. Expression of microRNA machinery proteins in different types of chronic rhinosinusitis. Laryngoscope 2012;122: 2621–7. [DOI] [PubMed] [Google Scholar]
- 29.Takyar S, Vasavada H, Zhang JG, Ahangari F, Niu N, Liu Q, et al. VEGF controls lung Th2 inflammation via the miR-1-Mpl (myeloproliferative leukemia virus oncogene)-P-selectin axis. J Exp Med 2013;210:1993–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med 2007;357:2016–27. [DOI] [PubMed] [Google Scholar]
- 31.Yan X, Chu JH, Gomez J, Koenigs M, Holm C, He X, et al. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Am J Respir Crit Care Med 2015;191:1116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korde A, Jin L, Zhang JG, Ramaswamy A, Hu B, Kolahian S, et al. Lung endothelial microRNA-1 regulates tumor growth and angiogenesis. Am J Respir Crit Care Med 2017;196:1443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan X, Chu JH, Gomez J, Koenigs M, Holm C, He X, et al. Noninvasive analysis of the sputum transcriptome discriminates clinical phenotypes of asthma. Ann Am Thorac Soc 2016;13(suppl 1):S104–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science 1998;282:2261–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corrigan CJ, Wang W, Meng Q, Fang C, Wu H, Reay V, et al. T-helper cell type 2 (Th2) memory T cell-potentiating cytokine IL-25 has the potential to promote angiogenesis in asthma. Proc Natl Acad Sci U S A 2011;108:1579–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eubank TD, Galloway M, Montague CM, Waldman WJ, Marsh CB. M-CSF induces vascular endothelial growth factor production and angiogenic activity from human monocytes. J Immunol 2003;171:2637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotowicz K, Callard RE, Friedrich K, Matthews DJ, Klein N. Biological activity of IL-4 and IL-13 on human endothelial cells: functional evidence that both cytokines act through the same receptor. Int Immunol 1996;8:1915–25. [DOI] [PubMed] [Google Scholar]
- 38.Wierzbicki T, Iqbal SM, Cuvelier SL, Awong G, Tibbles LA, Patel KD. IL-4 primes human endothelial cells for secondary responses to histamine. J Leukoc Biol 2003;74:420–7. [DOI] [PubMed] [Google Scholar]
- 39.Wu LW, Mayo LD, Dunbar JD, Kessler KM, Ozes ON, Warren RS, et al. VRAP is an adaptor protein that binds KDR, a receptor for vascular endothelial cell growth factor. J Biol Chem 2000;275:6059–62. [DOI] [PubMed] [Google Scholar]
- 40.Woltmann G, McNulty CA, Dewson G, Symon FA, Wardlaw AJ. Interleukin-13 induces PSGL-1/P-selectin-dependent adhesion of eosinophils, but not neutrophils, to human umbilical vein endothelial cells under flow. Blood 2000;95: 3146–52. [PubMed] [Google Scholar]
- 41.Goebeler M, Schnarr B, Toksoy A, Kunz M, Brocker EB, Duschl A, et al. Interleukin-13 selectively induces monocyte chemoattractant protein-1 synthesis and secretion by human endothelial cells. Involvement of IL-4R alpha and Stat6 phosphorylation. Immunology 1997;91:450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinkai A, Yoshisue H, Koike M, Shoji E, Nakagawa S, Saito A, et al. A novel human CC chemokine, eotaxin-3, which is expressed in IL-4-stimulated vascular endothelial cells, exhibits potent activity toward eosinophils. J Immunol 1999; 163:1602–10. [PubMed] [Google Scholar]
- 43.Fulkerson PC, Rothenberg ME. Targeting eosinophils in allergy, inflammation and beyond. Nat Rev Drug Discov 2013;12:117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larbi KY, Dangerfield JP, Culley FJ, Marshall D, Haskard DO, Jose PJ, et al. P-selectin mediates IL-13-induced eosinophil transmigration but not eotaxin generation in vivo: a comparative study with IL-4-elicited responses. J Leukoc Biol 2003;73:65–73. [DOI] [PubMed] [Google Scholar]
- 45.Symon FA, Walsh GM, Watson SR, Wardlaw AJ. Eosinophil adhesion to nasal polyp endothelium is P-selectin-dependent. J Exp Med 1994;180:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulfman LH, Joosten DP, van Aalst CW, Lammers JW, van de Graaf EA, Koenderman L, et al. Platelets promote eosinophil adhesion of patients with asthma to endothelium under flow conditions. Am J Respir Cell Mol Biol 2003;28:512–9. [DOI] [PubMed] [Google Scholar]
- 47.Broide DH, Humber D, Sullivan S, Sriramarao P. Inhibition of eosinophil rolling and recruitment in P-selectin- and intracellular adhesion molecule-1-deficient mice. Blood 1998;91:2847–56. [PubMed] [Google Scholar]
- 48.Broide DH, Sullivan S, Gifford T, Sriramarao P. Inhibition of pulmonary eosinophilia in P-selectin- and ICAM-1-deficient mice. Am J Respir Cell Mol Biol 1998;18:218–25. [DOI] [PubMed] [Google Scholar]
- 49.Lukacs NW, John A, Berlin A, Bullard DC, Knibbs R, Stoolman LM. E- and P-selectins are essential for the development of cockroach allergen-induced airway responses. J Immunol 2002;169:2120–5. [DOI] [PubMed] [Google Scholar]
- 50.Johansson MW, Han ST, Gunderson KA, Busse WW, Jarjour NN, Mosher DF. Platelet activation, P-selectin, and eosinophil beta1-integrin activation in asthma. Am J Respir Crit Care Med 2012;185:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denis CV, André P, Saffaripour S, Wagner DD. Defect in regulated secretion of P-selectin affects leukocyte recruitment in von Willebrand factor-deficient mice. Proc Natil Acad Sci U S A 2001;98:4072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 2011;146:980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pober JS, Tellides G. Participation of blood vessel cells in human adaptive immune responses. Trends Immunol 2012;33:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asosingh K, Cheng G, Xu W, Savasky BM, Aronica MA, Li X, et al. Nascent endothelium initiates Th2 polarization of asthma. J Immunol 2013; 190:3458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asosingh K, Weiss K, Queisser K, Wanner N, Yin M, Aronica M, et al. Endothelial cells in the innate response to allergens and initiation of atopic asthma. J Clin Invest 2018;128:3116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med 2013;368: 2455–66. [DOI] [PubMed] [Google Scholar]
- 57.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, et al. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med 2009;360:973–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, et al. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med 2009;360:985–93. [DOI] [PubMed] [Google Scholar]
- 59.Hamilos DL, Leung DY, Wood R, Cunningham L, Bean DK, Yasruel Z, et al. Evidence for distinct cytokine expression in allergic versus nonallergic chronic sinusitis. J Allergy Clin Immunol 1995;96:537–44. [DOI] [PubMed] [Google Scholar]
- 60.Szczeklik A, Stevenson DD. Aspirin-induced asthma: advances in pathogenesis, diagnosis, and management. J Allergy Clin Immunol 2003;111:913–22. [DOI] [PubMed] [Google Scholar]
- 61.Carr TF, Berdnikovs S, Simon HU, Bochner BS, Rosenwasser LJ. Eosinophilic bioactivities in severe asthma. World Allergy Organ J 2016;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobsen EA, Ochkur SI, Doyle AD, LeSuer WE, Li W, Protheroe CA, et al. Lung pathologies in a chronic inflammation mouse model are independent of eosinophil degranulation. Am J Respir Crit Care Med 2017;195:1321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ying S, Humbert M, Barkans J, Corrigan CJ, Pfister R, Menz G, et al. Expression of IL-4 and IL-5 mRNA and protein product by CD41 and CD81 T cells, eosinophils, and mast cells in bronchial biopsies obtained from atopic and nonatopic (intrinsic) asthmatics. J Immunol 1997;158:3539–44. [PubMed] [Google Scholar]
- 64.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med 2012;18:716–25. [DOI] [PubMed] [Google Scholar]
- 65.Lotvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol 2011;127:355–60. [DOI] [PubMed] [Google Scholar]
- 66.Rebane A microRNA and allergy. Adv Exp Med Biol 2015;888:331–52. [DOI] [PubMed] [Google Scholar]
- 67.Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med 2012;186:965–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levanen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol 2013;131:894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol 2011;128:1077–1085, e1–10. [DOI] [PubMed] [Google Scholar]
- 70.Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, et al. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell-associated cytokine production. Immunity 2016; 44:821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 2009;182: 4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng MJ, Shi F, Qiu C, Peng WK. MicroRNA-181a, −146a and −146b in spleen CD4+ T lymphocytes play proinflammatory roles in a murine model of asthma. Int Immunopharmacol 2012;13:347–53. [DOI] [PubMed] [Google Scholar]
- 73.Malmhall C, Alawieh S, Lu Y, Sjostrand M, Bossios A, Eldh M, et al. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol 2014;133:1429–1438, e1–7. [DOI] [PubMed] [Google Scholar]
- 74.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem 2010;285:30139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu TX, Lim EJ, Itskovich S, Besse JA, Plassard AJ, Mingler MK, et al. Targeted ablation of miR-21 decreases murine eosinophil progenitor cell growth. PLoS One 2013;8:e59397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu TX, Lim EJ, Besse JA, Itskovich S, Plassard AJ, Fulkerson PC, et al. MiR-223 deficiency increases eosinophil progenitor proliferation. J Immunol 2013;190: 1576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pua HH, Ansel KM. MicroRNA regulation of allergic inflammation and asthma. Curr Opin Immunol 2015;36:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perry MM, Baker JE, Gibeon DS, Adcock IM, Chung KF. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am J Respir Cell Mol Biol 2014;50:7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haj-Salem I, Fakhfakh R, Berube JC, Jacques E, Plante S, Simard MJ, et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFbetaR2 gene in severe asthma. Allergy 2015;70:212–9. [DOI] [PubMed] [Google Scholar]
- 80.Hu R, Pan W, Fedulov AV, Jester W, Jones MR, Weiss ST, et al. MicroRNA-10a controls airway smooth muscle cell proliferation via direct targeting of the PI3 kinase pathway. FASEB J 2014;28:2347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jude JA, Dileepan M, Subramanian S, Solway J, Panettieri RA Jr, Walseth TF, et al. miR-140-3p regulation of TNF-alpha-induced CD38 expression in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2012;303: L460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med 2009;180:713–9. [DOI] [PubMed] [Google Scholar]
- 83.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A 2009;106:18704–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, De Preter K, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol 2016;137:1433–46. [DOI] [PubMed] [Google Scholar]
- 85.Bartel S, Schulz N, Alessandrini F, Schamberger AC, Pagel P, Theis FJ, et al. Pulmonary microRNA profiles identify involvement of Creb1 and Sec14l3 in bronchial epithelial changes in allergic asthma. Sci Rep 2017;7:46026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wust S, Drose S, Heidler J, Wittig I, Klockner I, Franko A, et al. Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab 2018;27: 1026–1039, e6. [DOI] [PubMed] [Google Scholar]
- 87.Zaglia T, Ceriotti P, Campo A, Borile G, Armani A, Carullo P, et al. Content of mitochondrial calcium uniporter (MCU) in cardiomyocytes is regulated by microRNA-1 in physiologic and pathologic hypertrophy. Proc Natl Acad Sci U S A 2017;114:E9006–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214–20. [DOI] [PubMed] [Google Scholar]
- 89.Stahlhut C, Suarez Y, Lu J, Mishima Y, Giraldez AJ. miR-1 and miR-206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development 2012; 139:4356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Butz H, Ding Q, Nofech-Mozes R, Lichner Z, Ni H, Yousef GM. Elucidating mechanisms of sunitinib resistance in renal cancer: an integrated pathological-molecular analysis. Oncotarget 2018;9:4661–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin CY, Lee HC, Fu CY, Ding YY, Chen JS, Lee MH, et al. MiR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat Commun 2013;4:2829. [DOI] [PubMed] [Google Scholar]
- 92.Lu J, Zhao FP, Peng Z, Zhang MW, Lin SX, Liang BJ, et al. EZH2 promotes angiogenesis through inhibition of miR-1/endothelin-1 axis in nasopharyngeal carcinoma. Oncotarget 2014;5:11319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barrett NA, Maekawa A, Rahman OM, Austen KF, Kanaoka Y. Dectin-2 recognition of house dust mite triggers cysteinyl leukotriene generation by dendritic cells. J Immunol 2009;182:1119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim CH, Ahn JH, Kim SJ, Lee SY, Kim YK, Kim KH, et al. Co-administration of vaccination with DNA encoding T cell epitope on the Der p and BCG inhibited airway remodeling in a murine model of chronic asthma. J Asthma 2006;43:345–53. [DOI] [PubMed] [Google Scholar]
- 95.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol 2004;172:2731–8. [DOI] [PubMed] [Google Scholar]
- 96.Borchers MT, Ansay T, DeSalle R, Daugherty BL, Shen H, Metzger M, et al. In vitro assessment of chemokine receptor-ligand interactions mediating mouse eosinophil migration. J Leukoc Biol 2002;71:1033–41. [PubMed] [Google Scholar]
- 97.Shoda T, Futamura K, Orihara K, Emi-Sugie M, Saito H, Matsumoto K, et al. Recent advances in understanding the roles of vascular endothelial cells in allergic inflammation. Allergol Int 2016;65:21–9. [DOI] [PubMed] [Google Scholar]
- 98.Asosingh K, Erzurum SC. Angioplasticity in asthma. Biochem Soc Trans 2009; 37:805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson JW, Hii S. The importance of the airway microvasculature in asthma. Curr Opin Allergy Clin Immunol 2006;6:51–5. [DOI] [PubMed] [Google Scholar]
- 100.Vrugt B, Wilson S, Bron A, Holgate ST, Djukanovic R, Aalbers R. Bronchial angiogenesis in severe glucocorticoid-dependent asthma. Eur Respir J 2000;15:1014–21. [DOI] [PubMed] [Google Scholar]
- 101.Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just T(H)2 cells. Nat Rev Immunol 2010;10:838–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Walker JA, McKenzie A. Innate lymphoid cells in the airways. Eur J Immunol 2012;42:1368–74. [DOI] [PubMed] [Google Scholar]
- 103.Islam SA, Luster AD. T cell homing to epithelial barriers in allergic disease. Nat Med 2012;18:705–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol 2005;6:1182–90. [DOI] [PubMed] [Google Scholar]
- 105.Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol 2007;7:678–89. [DOI] [PubMed] [Google Scholar]
- 106.Heit B, Colarusso P, Kubes P. Fundamentally different roles for LFA-1, Mac-1 and alpha4-integrin in neutrophil chemotaxis. J Cell Sci 2005;118:5205–20. [DOI] [PubMed] [Google Scholar]
- 107.Symon FA, Lawrence MB, Williamson ML, Walsh GM, Watson SR, Wardlaw AJ. Functional and structural characterization of the eosinophil P-selectin ligand. J Immunol 1996;157:1711–9. [PubMed] [Google Scholar]
- 108.Patel KD, McEver RP. Comparison of tethering and rolling of eosinophils and neutrophils through selectins and P-selectin glycoprotein ligand-1. J Immunol 1997;159:4555–65. [PubMed] [Google Scholar]
- 109.Cuvelier SL, Patel KD. Shear-dependent eosinophil transmigration on interleukin 4—stimulated endothelial cells: a role for endothelium-associated eotaxin-3. J Exp Med 2001;194:1699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Larose M-C, Chakir J, Archambault A-S, Joubert P, Provost V, Laviolette M, et al. Correlation between CCL26 production by human bronchial epithelial cells and airway eosinophils: involvement in patients with severe eosinophilic asthma. J Allergy Clin Immunol 2015;136:904–13. [DOI] [PubMed] [Google Scholar]
- 111.Errahali YJ, Taka E, Abonyo BO, Heiman AS. CCL26-targeted siRNA treatment of alveolar type II cells decreases expression of CCR3-binding chemokines and reduces eosinophil migration: implications in asthma therapy. J Interferon Cytokine Res 2009;29:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Allakhverdi Z, Comeau MR, Jessup HK, Yoon BR, Brewer A, Chartier S, et al. Thymic stromal lymphopoietin is released by human epithelial cells in response to microbes, trauma, or inflammation and potently activates mast cells. J Exp Med 2007;204:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takai T TSLP expression: cellular sources, triggers, and regulatory mechanisms. Allergol Int 2012;61:3–17. [DOI] [PubMed] [Google Scholar]
- 114.Watson B, Gauvreau GM. Thymic stromal lymphopoietin: a central regulator of allergic asthma. Expert Opin Ther Targets 2014;18:771–85. [DOI] [PubMed] [Google Scholar]
- 115.Ying S, O’Connor B, Ratoff J, Meng Q, Mallett K, Cousins D, et al. Thymic stromal lymphopoietin expression is increased in asthmatic airways and correlates with expression of Th2-attracting chemokines and disease severity. J Immunol 2005;174:8183–90. [DOI] [PubMed] [Google Scholar]
- 116.Nagarkar DR, Poposki JA, Tan BK, Comeau MR, Peters AT, Hulse KE, et al. Thymic stromal lymphopoietin activity is increased in nasal polyps of patients with chronic rhinosinusitis. J Allergy Clin Immunol 2013;132:593–600, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gauvreau GM, O’Byrne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J, et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med 2014;370:2102–10. [DOI] [PubMed] [Google Scholar]
- 118.Tibbles HE, Navara CS, Hupke MA, Vassilev AO, Uckun FM. Thrombopoietin induces p-selectin expression on platelets and subsequent platelet/leukocyte interactions. Biochem Biophys Res Commun 2002;292:987–91. [DOI] [PubMed] [Google Scholar]
- 119.Kaushansky K The molecular mechanisms that control thrombopoiesis. J Clin Invest 2005;115:3339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Southworth T, Mason S, Bell A, Ramis I, Calbet M, Domenech A, et al. PI3K, p38 and JAK/STAT signalling in bronchial tissue from patients with asthma following allergen challenge. Biomark Res 2018;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoo EJ, Ojiaku CA, Sunder K, Panettieri RA Jr. Phosphoinositide 3-kinase in asthma: novel roles and therapeutic approaches. Am J Respir Cell Mol Biol 2017;56:700–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simeone-Penney MC, Severgnini M, Tu P, Homer RJ, Mariani TJ, Cohn L, et al. Airway epithelial STAT3 is required for allergic inflammation in a murine model of asthma. J Immunol 2007;178:6191–9. [DOI] [PubMed] [Google Scholar]
- 123.Nashed BF, Zhang T, Al-Alwan M, Srinivasan G, Halayko AJ, Okkenhaug K, et al. Role of the phosphoinositide 3-kinase p110delta in generation of type 2 cytokine responses and allergic airway inflammation. Eur J Immunol 2007;37: 416–24. [DOI] [PubMed] [Google Scholar]
- 124.Liu W, Liang Q, Balzar S, Wenzel S, Gorska M, Alam R. Cell-specific activation profile of extracellular signal-regulated kinase 1/2, Jun N-terminal kinase, and p38 mitogen-activated protein kinases in asthmatic airways. J Allergy Clin Immunol 2008;121:893–902, e2. [DOI] [PubMed] [Google Scholar]
- 125.Alam R, Gorska MM. Mitogen-activated protein kinase signalling and ERK1/2 bistability in asthma. Clin Exp Allergy 2011;41:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.