Abstract
Key message
Disomic alien chromosome addition Brassica carinata lines with super-high erucic acid content were developed through interspecific hybridization with B. juncea and characterized using molecular, cytological and biochemical techniques.
Abstract
Brassica carinata [A.] Braun (BBCC, 2n = 34) is a climate-resilient oilseed. Its seed oil is high in erucic acid (> 40%), rendering it well suited for the production of biofuel and other bio-based applications. To enhance the competitiveness of B. carinata with high erucic B. napus (HEAR), lines with super-high erucic acid content were developed through interspecific hybridization. To this end, a fad2B null allele from Brassica juncea (AABB, 2n = 36) was introgressed into B. carinata, resulting in a B. carinata fad2B mutant with erucic acid levels of over 50%. Subsequently, the FAE allele from B. rapa spp. yellow sarson (AA, 2n = 20) was transferred to the fad2B B. carinata line, yielding lines with erucic acid contents of up to 57.9%. Molecular analysis using the Brassica 90 K Illumina Infinium™ SNP genotyping array identified these lines as disomic alien chromosome addition lines, with two extra A08 chromosomes containing the BrFAE gene. The alien chromosomes from B. rapa were clearly distinguished by molecular cytogenetics in one of the addition lines. Analysis of microspore-derived offspring and hybrids from crosses with a CMS B. carinata line showed that the transfer rate of the A08 chromosome into male gametes was over 98%, resulting in almost completely stable transmission of an A08 chromosome copy into the progeny. The increase in erucic acid levels was accompanied by changes in the proportions of other fatty acids depending on the genetic changes that were introduced in the interspecific hybrids, providing valuable insights into erucic acid metabolism in Brassica.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00122-021-03883-2.
Introduction
Brassica carinata, also known as Ethiopian or Abyssinian mustard, is a member of the Brassicaceae (mustard) family. It is an allotetraploid (genome BBCC, 2n = 34) formed through the interspecific hybridization between the diploid progenitor species B. nigra (BB, 2n = 16) and B. oleracea (2n = 18) (U 1935; Prakash et al. 2011). Current evidence suggests that it originated in the highland plateaus of Ethiopia and neighboring areas of East Africa and the Mediterranean coast (Gomez-Campo and Prakash 1999; Alemayehu and Becker 2002). While the cultivation of Ethiopian mustard as an oilseed and vegetable crop in East Africa dates back several millennia (Simmonds 1979), interest in this crop in other geographic regions is relatively recent. This interest primarily stems from the well-documented heat and drought tolerance of this crop (Cohen and Knowles 1983; Ferreres et al. 1984; Malik 1990), and its potential to serve as a dedicated feedstock crop for biofuel production and other industrial applications (Taylor et al. 2010, Marillia et al. 2014).
B. carinata seed oil is high in erucic acid, with proportions ranging from 31 to 46% in natural germplasm collections (Röbbelen and Thies 1980; Westphal and Marquard 1980; Becker et al. 1999; Warwick et al. 2006). Plant oils high in erucic acid are of interest for industrial purposes; erucic acid and its derivatives are important renewable raw materials for the oleochemical industry, with more than 1000 potential or patented applications (Scarth and Tang 2006). Erucic acid (cis-13-docosenoic acid, C22:1) is a straight-chained mono-unsaturated very-long-chain fatty acid (VLCFA) with 22 carbon atoms and a double bond at the cis-13 position of the carbon chain. Biosynthesis of this fatty acid involves chain elongation of oleic acid (cis-9-octadecenoic acid, C18:1) to gondoic acid (cis-11-eicosenoic acid, C20:1) and then to C22:1 (Sanyal et al. 2015). This reaction is catalyzed by ß-ketoacyl-CoA synthase (KCS), a membrane-bound enzyme that utilizes malonyl-CoA as the source of the two carbon atoms added in each cycle (Griffiths et al. 1998). KCS is encoded by the gene fatty acid elongase (FAE) (James et al. 1995; Lühs and Friedt 1997) and is the rate-limiting enzyme that plays a key role in determining the levels of erucic acid and other VLCFAs in seed oils (Millar and Kunst 1997). Early work in B. carinata proposed that C22:1 content is controlled by two loci in an additive manner with one allele present in each genome (Getinet et al. 1997). In agreement with this, it was demonstrated that two FAE genes determine the C22:1 content in the closely related species B. napus (Fourmann et al. 1998) and B. juncea (Venkateswari et al. 1999). Several studies, including experiments that involve interspecific hybrids, have demonstrated that the erucic acid content in Brassica seed oils is highly heritable (Getinet et al. 1997; Alemayehu and Becker 2001; Farhatullah et al. 2014).
Results from traditional breeding approaches in B. carinata suggest that C22:1 levels ranging from zero (Alonso et al. 1991; Getinet et al. 1994) to low (< 2%; Fernandez-Escobar et al. 1988; Velasco et al. 1995) to as high as 51% (Alemayehu and Becker 2001) are possible due to natural variation and through selection breeding, compared to <1 to 60% in non-canola types of B. rapa and B. napus (Lühs and Friedt 1994; Lühs et al. 1999) and <1 to 52% in B. juncea (Saikia et al. 2018). Several studies in various Brassica species have manipulated C22:1 levels via mutagenesis or transgenic approaches. Thus, chemical mutagenesis has been used to both reduce and elevate C22:1 levels in B. carinata (Barro et al. 2001). UV treatment has also been used to increase C22:1 levels from 42.8 to 49.5% (Barro et al. 2003). Both sense and antisense FAE constructs were used in B. napus (Zebarjadi et al. 2006) and B. juncea (Kanrar et al. 2006) to increase and decrease the C22:1 content, respectively. In B. napus cultivar CY2, an RNAi approach reduced the C22:1 content from 40 to 3%, with corresponding increases in C18:1 from 20 to 60% (Shi et al. 2015). C22:1 levels were increased in B. carinata by either a co-suppression or antisense construct of omega-6 fatty acid desaturase (FAD2) (Jadhav et al. 2005) through an increase in the amount of the substrate C18:1. Further, silencing of FAD2 via a hairpin construct in combination with a Crambe abyssinica FAE construct in B. carinata resulted in levels of C22:1 as high as 56% (Mietkiewska et al. 2008).
While genetic transformation is a powerful means of effectively transferring genes across reproductive barriers (Cardi et al. 2017), genetically modified crops are facing great regulatory and trade challenges in contrast to crops developed through traditional breeding (Smyth 2016). Serendipitously, in the Brassica genus, several crop species have genomes in common and the ability to perform interspecific hybridizations allows for the transfer of useful adaptive traits between species for targeted crop improvement through conventional breeding (Katche et al. 2019). Interspecific crosses with the compatible allotetraploid species B. napus and B. juncea have resulted in B. carinata lines with decreased glucosinolate content (Márquez-Lema et al. 2008) and increased seed oil content (Sheikh et al. 2009), as well as increased overall genetic diversity (Sheikh et al. 2011). Similarly, C22:1 levels in B. carinata were significantly reduced through crosses to both B. napus (Fernandez-Escobar et al. 1988) and B. juncea (Getinet et al. 1994). On the other hand, B. carinata has been used as a donor for the transfer of a number of traits, such as blackleg (Leptosphaeria maculans) resistance to B. napus (Fredua-Agyeman et al. 2014; Navabi et al. 2010) and resistance to white rust (Albugo candida) and Alternaria blight (Alternaria brassicae) to B. juncea (Gupta et al. 2010). Several groups have utilized B. carinata to introduce yellow seed coat color into B. napus (Rashid et al. 1994; Meng et al. 1998; Rahman 2001). In addition, overall genetic diversity has been enhanced in both B. napus (Chen et al. 2010a, b) and B. juncea (Wei et al. 2016) through crosses with B. carinata. Interspecific crosses of B. carinata with diploid Brassica species have also been explored. The transfer of powdery mildew (Erysiphe polygoni) (Tonguç and Griffiths 2004) and black rot (Xanthomonas campestris) resistance (Sharma et al. 2017) from B. carinata to B. oleracea was examined in early backcross generations; both studies required embryo rescue. Crosses with B. rapa were found to be challenging and highly dependent on the accessions used (Jiang et al. 2007) and the direction of the cross (Choudhary et al. 2000), although successful crosses between B. rapa and B. carinata have been achieved in both directions (FitzJohn et al. 2007; Tian et al. 2010). In particular, successful crosses with diverse B. rapa genotypes including brown sarson, toria and yellow sarson have been achieved (Choudhary et al. 2000; Rahman 2001); yellow sarson is known to have erucic acid levels greater than 55% (Dorrell and Downey 1964).
Industrial applications demand an oil composition predominated by a single fatty acid. Compared to low-erucic Brassica oils, high erucic oils have a higher energy potential (Semenov et al. 2006) and are more desirable substrates for biodiesel (Vicente et al. 2005) and biofuel (Zhao et al. 2016) production. High erucic acid rapeseed (HEAR), currently the main feedstock for erucic acid used in industrial applications, contains about 50% of this fatty acid (Sanyal et al. 2015). For improved technical utility and enhanced marketability of carinata oil, it is desirable to increase the C22:1 content and to simultaneously decrease the content of C20:1, polyunsaturated fatty acids (PUFA, linoleic [C18:2] and linolenic [C18:3] acids) and saturated fatty acids (palmitic [C16:0] and stearic [C18:0] acids).
The purpose of the present study was to create a B. carinata breeding line with super-high C22:1 content through interspecific hybridization with B. juncea and B. rapa. Herein, we report the development of a B. carinata disomic A genome chromosome addition line, carrying the fad2B/FAE gene variants, with erucic acid levels of close to 60%, and its characterization using molecular, cytological and biochemical techniques. Further, we demonstrate the almost completely stable transfer of the chromosome addition through crosses with cytoplasmic male sterile B. carinata and the production of offspring via isolated microspore culture. This study provides the first account of the development of super-high erucic acid Ethiopian mustard through conventional breeding and contributes to the understanding of erucic acid metabolism in Brassica. It is also the first report to describe a B. carinata-rapa disomic chromosome addition line.
Materials and methods
Plant materials and breeding strategy
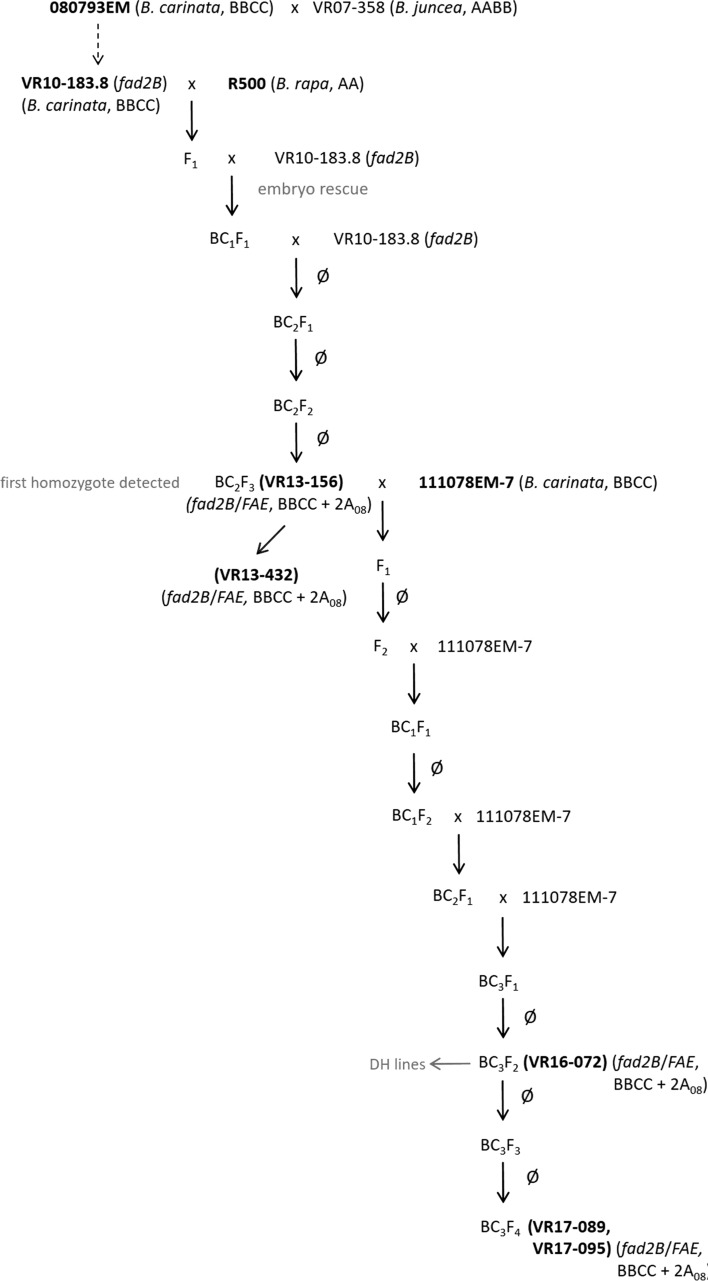
In previous work, the high erucic acid B. carinata breeding line 080793EM was crossed with canola-quality B. juncea line VR07-358 to increase the C18:1 content by introducing a fad2B null allele (Roslinsky et al. 2011). In the present study, the BC3F3 progeny line VR10-183.8 was crossed to the yellow sarson B. rapa line R500 in order to introduce the B. rapa FAE gene (BrFAE) (Fig. 1). The resulting F1 plants were backcrossed to VR10-183.8 and embryos rescued 16 days after pollination (DAP) according to the method described below. A second backcross was completed, followed by two rounds of self-pollination. The BC2F3 line VR13-156 was identified as a stable double mutant for the fad2B null allele and the B. rapa-derived FAE gene via molecular marker analysis, and was subsequently crossed to the doubled haploid (DH) B. carinata breeding line 111078EM-7 to improve total seed oil content and overall agronomics. Improvements to the FAD2-specific marker allowed for F1 lines to be used in the ensuing crossing procedure. Three backcrosses were carried out with 111078EM-7 as the recurrent parent, followed by three rounds of self-pollination. To determine the transfer rate of the B. rapa FAE gene during meiosis, BC3F2 line VR16-072 was used as the donor line for the production of microspore-derived plants. Additionally, individual BC3F4 lines were crossed to a cytoplasmic male sterile (CMS) line to examine pollen viability as well as segregation of the BrFAE allele in the F1 progeny. BC3F4 line VR17-095 was used for the molecular cytogenetic study. BC3F4 lines VR17-089 and VR17-095 were analyzed on a Brassica 90K Illumina Infinium™ SNP genotyping array. Parent material and progeny lines were grown in 2019 in the field in a randomized complete block design with four replicates alongside the two commercial B. carinata cultivars AAC A110 and AAC A120 (Agriculture and Agri-Food Canada—Saskatoon Research and Development Centre, Canada). Harvested seed was subjected to fatty acid analysis.
Fig. 1.
Schematic representation of the breeding strategy for the development of the super-high erucic acid B. carinata lines VR13-156 and VR17-095. Note: Line VR10-183.8 previously developed by (Roslinsky et al. 2011)
Embryo rescue
F1 hybrids resulting from the cross between VR10-183.8 and yellow sarson B. rapa line R500 were used as the female in backcrosses with the recurrent parent VR10-183.8. BC1F1 plants were developed by embryo rescue followed by in vitro culture according to the protocol of Ayotte et al. (1987). Plantlets were transferred to soil and placed in a greenhouse.
FAE and FAD2 gene sequencing and marker development
Genomic DNA was isolated from young leaf tissue of the parents and progeny lines using a sodium dodecyl sulfate protocol as per Somers et al. (1998). The primer pair FAE F and FAE R (Online Resource 1) was designed to amplify all FAE alleles present in a sample, based on Gupta et al. (2004). All PCR reactions were carried out as follows: genomic DNA was used as the template for PCR amplification in a thermocycler with 35 cycles of 45 s for denaturation at 94 °C, primer annealing at 57 °C and 1 minute extension at 72 °C. The PCR products from the parental lines R500 and VR10-183.8 were subsequently cloned and sequenced; the expected bands were cloned into the pGEM-T Vector System I (Promega, Cat. No. A3600) and plasmids were extracted using the QIAprep Spin Miniprep Kit (Qiagen, Cat. No. 27106) following the manufacturer’s recommended protocols. All samples were sequenced at the National Research Council in Saskatoon, Canada. Screening of early generations for the presence of BrFAE was carried out with an HpaI post-amplification digest using the above primer pair. The digest was run on a 2% agarose gel for 3 hours at 120 v to ensure adequate separation of fragments. To improve screening capacity, KASP primers (LGC Genomics) were developed for routine screening utilizing the HpaI SNP position (Fig. 2) and implemented at the first backcross to recurrent parent 111078EM-7 and subsequent generations. The KASP reaction contains 100 ng genomic DNA, 4.0 μL KASP 2x Master Mix (LGC Genomics, Cat. No. KBS-1016-002-US) and 0.11 μL primer assay mix (Online Resource 1) in a total volume of 8 μL. All amplifications were performed in a CFX96 Real-Time Thermal Cycler (Bio-Rad Laboratories) using a touchdown PCR protocol as per the manufacturer’s recommended procedure.
Fig. 2.
KASP primer positions in the FAE gene sequence. The targeted SNP at position 506 in the fragment is highlighted in blue, the sequence targeted by the selective primer in yellow and the sequence targeted by the common primer in green. The bases in red represent variation in the B allele, while bold and underlined bases were utilized for further selective amplification of the BrFAE sequence
Primer pair fad2B F and fad2B R (Online Resource 1) was designed to amplify the FAD2B allele. The amplification followed the same PCR protocol as described above, except that the annealing temperature was 64 °C. With this primer pair, selection against the amplified product was required to select the fad2B null allele. As more sequence information was obtained later in the pedigree, new primers were developed facilitating a co-dominant marker system. This new primer pair (fad2B F2 and fad2B R2) (Online Resource 1) amplified a region tightly linked to FAD2 present in all samples. The PCR protocol is again similar to the one described above, with an annealing temperature of 55 °C. Amplified products for both primer pairs were separated on 2% agarose gels and visualized through ethidium bromide staining. All samples at the BC2F1 × 111078EM-7 stage and beyond were tested with the second FAD2 primer set.
Seed fatty acid analysis
Seed fatty acid composition of field-grown parental and progeny lines was analyzed according to Thies (1971) with the following modifications: methyl esters were separated by gas chromatography using an Agilent INNOWAX fused silica capillary column (7.5 m x 250 µm diameter x 0.5 µm film thickness) at 250 °C with hydrogen as the carrier gas. The methyl esters were prepared by base-catalyzed transesterification using sodium methoxide in methanol and hexane on oil extracted from 3.0 g of crushed seed.
Genotyping
Parental and progeny lines were analyzed using the Brassica 90K Illumina Infinium™ SNP genotyping array as per the supplied protocol. Data analysis was performed utilizing the GenomeStudio software package v 2.0 (https://support.illumina.com/array/array_software/genomestudio/documentation.html) according to suggested best practices (Mason et al. 2017; Clarke et al. 2016).
Isolated microspore culture
To determine the transfer rate of BrFAE during meiosis, BC3F2 line VR16-072, derived from cross VR13-156 × 111078EM-07, was used as the donor line for the production of microspore-derived plants. Isolated microspore culture was performed as described in Zatylny et al. (in press). Germinated embryos with true leaves and roots were transplanted to pots and transferred to the greenhouse.
Molecular cytogenetics
Root tips were collected and chromosome spreads prepared from root tips following the protocol described in Gaebelein et al. (2019), adapted from Snowdon et al. (1997). Cells were observed using a Leica DRME microscope equipped with a Leica EL6000 fluorescent light source. Pictures were taken using a Leica DFC450 C camera and Leica Application Suite X software. Grayscale images showing the red and green fluorescent signals were processed to achieve optimal signal strength and contrast in ImageJ and combined. The combined image was then overlaid with the original grayscale image of the respective cell taken before hybridization using Photoshop CS6.
Labeled probe to detect B genome chromosomes was prepared using Roche Dig-Nick Translation Mix (Cat. No. 11745 816 910) according to the manufacturer’s specifications, using genomic B. nigra DNA as a template. Labeled probe to detect C genome chromosomes was prepared using the Invitrogen BioPrime DNA Labelling System (Cat. No. 18094011) according to the manufacturer’s specifications using DNA from BAC (Bacterial Artificial Chromosome) clone BoB014O06 (Howell et al. 2008) as template DNA, which contains the C genome-specific repetitive element Bot1 (Alix et al. 2008), which is distributed across all C genome chromosomes. Fluorescent in situ hybridization was carried out according to the methods detailed in Gaebelein et al. (2019), as adapted from Leflon et al. (2016). 100 ng of B genome DNA and 200 ng of C genome-specific BAC DNA were used in the hybridization mix, and 2 ng/µl Anti-Digoxigenin-FITC (Roche, Cat. No. 11207741910) and 10 ng/µl Cy3-Streptavidin (GE-Healthcare, Cat. No. PA43001) dissolved in 5% BSA (Bovine Serum Albumin) were applied for visualization of the antibody-labeled DNA probes. DAPI (4′,6-diamidino-2-phenylindole)-containing medium (Vectashield H-1200) was used to visualize the chromosomes under UV-light excitation.
Statistical analysis
Fatty acid profile data were analyzed by ANOVA using the Glimmix Procedure of SAS (SAS for Windows 9.3, SAS Institute, USA). Treatment mean comparisons were made using the Tukey–Kramer method at the P = 0.05 level.
Results
Development and characterization of super-high erucic acid B. carinata
In B. carinata BC3F3 breeding line VR10-183.8 a B1/A5 translocation event had removed a portion of chromosome B1 that contained the FAD2B allele (Roslinsky et al. 2011), as a result of the cross with canola-quality B. juncea (Potts et al. 1999). This translocation caused an increase in the C18:1 content, combined with an increase in C22:1, and reductions in both C18:2 and C18:3 levels. In the present study, crosses were initiated to combine the fad2B null allele with the FAE gene from the yellow sarson B. rapa line R500. The BC1 crosses were embryo rescued to obtain viable interspecific hybrids. All following generations naturally set varying amounts of seed. At the BC2F3 stage, the breeding line VR13-156 was identified to be a double mutant homozygous for both the fad2B null allele and the rapa-derived FAE allele. This line showed a further significant increase in C22:1 content compared to the parental line VR10-183.8. At the same time, this line displayed poor overall growth and seed set, and had a low total seed oil content. Three additional rounds of backcrossing to the elite B. carinata breeding line 111078EM-7 and three cycles of self-pollination were completed to develop true-breeding double mutant lines with improvements in agronomic and seed quality traits. BC3F4 line VR17-095 was selected based on superior seed yield in the greenhouse (unpublished data).
FAE and FAD2 sequence analysis and marker development
A 1254 bp portion of the FAE genes from the breeding lines VR10-183.8 and R500 was amplified and sequenced using primers based on data from Gupta et al. (2004). A SNP was detected in an HpaI digestion site at position 506 of the fragment which is present in both B. carinata endogenous FAE alleles, but absent in the R500 allele. Originally, this SNP was utilized in a post-amplification restriction digest that produced a unique 538 bp band specific for the BrFAE allele (Online Resource 2). To improve marker efficiency, this SNP was converted to a KASP marker. As three homoeologous copies of the FAE gene are present (BcFAE-B, BcFAE-C and BrFAE), both the selective and common primers were designed to preferentially amplify BrFAE to ensure adequate separation of the allele clusters and to capture all rare A genome insertion events (Fig. 2). This marker design is not typical of KASP markers and does not allow for the identification of heterozygous samples (Fig. 3).
Fig. 3.

Bio-Rad CFX Maestro image of the FAE KASP marker. Blue squares represent samples homozygous for the A2 allele (no BrFAE allele), the orange circles represent both the homozygous A1 and heterozygous alleles (BrFAE allele present), and the black diamonds represent no template controls
The fad2B null allele was identified in canola-quality B. juncea and transferred to B. carinata (Roslinsky et al. 2011). The molecular marker (Genbank DQ777854) that was initially developed targets the FAD2B gene itself and amplifies a single 456 bp fragment (Online Resource 3). Because the desired genotype is the homozygous null allele and selection for the absence of amplification was required, selections could only be made in an F2 generation. This resulted in one additional round of selfing after each backcross. In order to save time and avoid the occurrence of false negatives, a second marker was designed based on the sequence of a non-coding fragment associated with the B1/A5 translocation (Figure S2). When analyzed with this marker, the B. carinata lines developed in this study contain a 473 bp fragment associated with the wild-type FAD2B allele, a 615 bp band associated with the A5 translocation and the fad2B null allele and a 440 bp fragment that appears to be associated with the FAD2C allele (Fig. 4). Because of the co-dominant nature of this marker, the selection of lines homozygous for the fad2B null allele is readily feasible.
Fig. 4.
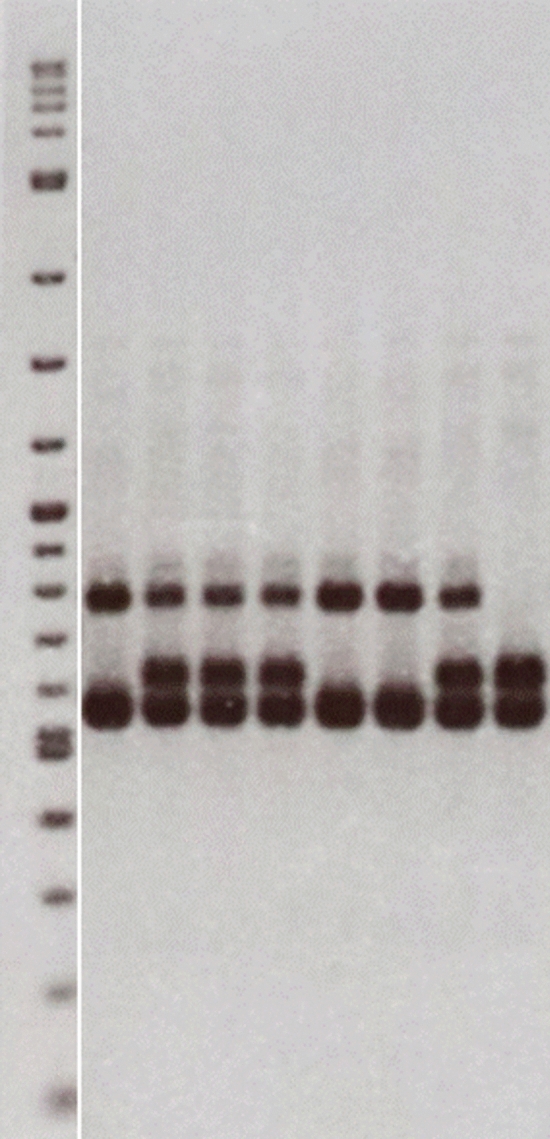
Gel image of samples analyzed with the co-dominant FAD2B marker. Lane 1 contains the 2 log DNA ladder (NEB), the bottom band is 100 bp, with 100 bp increments up to 1.0 kb; lanes 2, 6 and 7 represent samples homozygous for the fad2B null allele, lanes 3, 4, 5 and 8 represent samples heterozygous for the fad2B null allele, and lane 9 represents a sample with a wild-type FAD2B allele. The top band is associated with the fad2B null allele, the middle band with the carinata wild-type FAD2B allele and the bottom band with the FAD2C allele
The FAE and FAD2 markers described herein were used throughout the pedigree to identify stable double mutants homozygous for the juncea-derived fad2B null allele and the rapa-derived FAE gene.
Seed fatty acid analysis
The lines representing important steps in the pedigree alongside samples of two commercial B. carinata cultivars, AAC A110 and AAC A120, were grown in replicate in a field trial at Saskatoon, Saskatchewan, and their seed fatty acid profiles were analyzed by gas chromatography. The relative amount (percentage of total fatty acids) of the major fatty acids C18:1, C18:2, C18:3, C20:1, C22:1 and nervonic acid (C24:1) as well as the percentage of unsaturated fatty acids is presented in Table 1. The fatty acid profiles of cultivars AAC A110 and AAC A120 are representative of generic B. carinata germplasm, while the seed oil of breeding line 080793EM is characterized by elevated C22:1 levels (net increase of 4.9% and 3.9% compared to AAC A110 and AAC A120, respectively) and lower amounts of C18:1, C18:3 and C24:1. The results of the analysis show that fad2B line VR10-183.8 possesses a C18:1 content that is 3.3% higher than that in B. carinata parent line 080793EM. This increase in C18:1 is accompanied by a 4.3% increase in C22:1 and reductions in both C18:2 and C18:3. Both C20:1 and C24:1 levels remain unchanged. BC2F3 line VR13-156, the first line in the pedigree identified as homozygous for both the fad2B null allele and BrFAE, shows a further increase in C22:1 levels with values that are on average 11% above those in line 080793EM (57.9 vs. 46.9%) which corresponds to a proportional increase of 23%. Compared to VR10-183.8, line VR13-156 is characterized by an increased amount of C22:1 (57.9 vs. 51.2%) and proportional reductions of C18:3 and C20:1 levels, by 20.8 and 59.7%, respectively. Line VR13-432, a direct progeny line of VR13-156 derived through selfing, features a fatty acid profile very similar to that of its parent with a statistically significant difference, albeit small, only in C18:1 levels. Line 111078EM-7, used as the recurrent parent in backcrosses to VR13-156, to improve the seed oil content and agronomics of this line, has a fatty acid profile that is very similar to that of the two commercial cultivars included in this study. Interestingly, BC3F4 lines VR17-095 and VR17-89 have C22:1 levels that are 9.8% and 9.4% above those of the recurrent parent and 3.5 and 3.9% below those of donor parent VR13-156. In all fad2B/FAE lines the percentage of saturated fatty acids was slightly but significantly reduced when compared to that of the parent lines 080793EM and 111078EM-7, respectively.
Table 1.
Mean values for fatty acids as percentage of total fatty acids of the seed oil of Brassica carinata parent and progeny lines grown in a field trial at Saskatoon, Saskatchewan, in 2019; n = 4 unless specified otherwise
| Line | Fatty acids (% of total) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Sat. | C18:1 | C18:2 | C18:3 | C20:1 | C22:1 | C24:1 | ||||||||
| AAC A110§§ | 5.4 | c | 7.4 | d | 15.3 | a | 15.7 | a | 7.1 | ab | 42.0 | f | 2.9 | ab |
| AAC A120§§ | 5.7 | b | 7.4 | d | 14.7 | ab | 14.6 | b | 7.7 | a | 43.0 | ef | 2.8 | b |
| 080793EM | 5.9 | a | 6.3 | f | 15.4 | a | 13.0 | c | 6.2 | b | 46.9 | d | 1.9 | c |
| VR10-183.8 | 5.2 | c | 9.6 | a | 10.8 | e | 12.0 | d | 6.7 | b | 51.2 | c | 1.8 | cd |
| VR13-156§ | 4.9 | d | 9.0 | b | 11.7 | cd | 9.5 | e | 2.7 | d | 57.9 | a | 1.6 | e |
| VR13-432 | 4.8 | d | 8.5 | c | 12.0 | c | 9.9 | e | 2.8 | cd | 57.4 | a | 1.7 | de |
| 111078EM-7 | 5.3 | c | 6.7 | ef | 14.2 | b | 15.6 | a | 6.2 | b | 44.6 | e | 3.0 | a |
| VR17-095 | 4.8 | d | 6.8 | ef | 11.4 | d | 12.9 | c | 3.4 | cd | 54.4 | b | 2.7 | b |
| VR17-089 | 4.8 | d | 6.9 | de | 11.9 | cd | 12.5 | cd | 3.5 | c | 54.0 | b | 2.7 | b |
Means within a column with different lowercase letters differ significantly at the p < 0.05 level
AAC A110, AAC A 120, 080793EM and 111078EM-7: generic B. carinata cultivars and breeding lines, respectively; VR10-183.8: fad2B null line derived from B. carinata 080793EM x B. juncea; VR13-156: double mutant line homozygous for fad2B and B. rapa FAE, BC2F3 derived from VR10-183.8 x R500; VR13-432: double mutant line homozygous for fad2B and B. rapa FAE, BC2F4 derived from selfing of VR13-156; VR17-089 and VR17-095: double mutant line homozygous for fad2B and B. rapa FAE, BC3F4 lines derived from VR13-156 x 111078EM-7
Total sat. Total saturated fatty acids include C12:0 (lauric), C14:0 (myristic), C16:0 (palmitic), C18:0 (stearic), C20:0 (arachidic), C22:0 (behenic) and C24:0 (lignoceric)
C18:1 = oleic acid, C18:2 = linoleic acid, C18:3 = linolenic acid, C20:1 = eicosenoic (gondoic) acid, C22:1 = erucic acid, C24:1 = nervonic acid
Fatty acids not listed include C16:1 (palmitoleic), C22:2 (docosadienoic), C22:3 (docosatrienoic), and trace amounts of other unidentified fatty acids
§ Three samples analyzed
§§ Two samples analyzed
Genotyping
In order to determine the exact position and size of the BrFAE introgression, the BC3F4 lines VR17-089 and VR17-095 were analyzed using the Brassica 90K Illumina Infinium™ SNP genotyping array. FAE has previously been mapped to the A08 chromosome in B. napus (Fourmann et al. 1998; Delourme et al. 2006) and it was therefore expected that a portion of the A08 chromosome would be introgressed into the genome of B. carinata. The array data revealed that the entire B. rapa A08 chromosome was present, rendering VR17-089 and VR17-095 disomic alien chromosome addition lines (BBCC+2A08). The array data also showed that the distal portion of chromosome B1 was replaced with a distal portion of chromosome A5 (Figure S2). This re-arrangement led to the loss of the FAD2B allele, making VR17-089 and VR17-095 fad2B/FAE B. carinata disomic A-chromosome addition lines.
Molecular cytogenetics
Unambiguous counts of 36 chromosomes were found in 50 cells from 23 independent plants of line VR17-095, supporting a stable karyotype of 2n = 36 in this line. Three microscope slides were successfully hybridized with probes specific to the Brassica B and C genomes, with sufficient signal strength, probe specificity and limited degradation of chromosomes on the slides. Fifteen chromosome spreads with good chromosome spreading and chromosome condensation were found to show 16 clear signals representing the B genome chromosomes, 18 clear signals representing the C genome chromosomes and two distinctly unstained chromosomes putatively representing the chromosome pair A08 introgressed from B. rapa (Fig. 5).
Fig. 5.
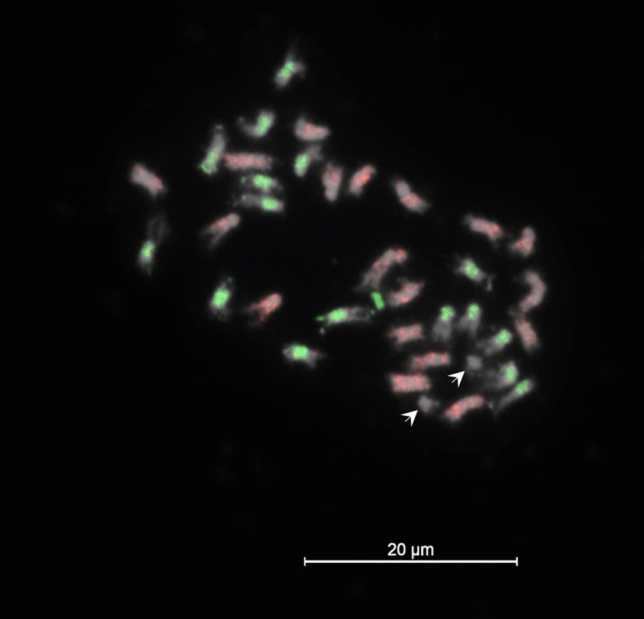
FISH/GISH stained mitotic cell of VR17-095 with a karyotype of 2n = 36. Green (fluorescein) signals represent chromosomes stained by probe derived from genomic DNA of B. nigra, identifying B genome chromosomes. Red signals (Cy3) represent chromosomes stained by probe derived from repetitive DNA sequence of B. oleracea, identifying C genome chromosomes. Two distinct small chromosomes (arrows) remain unstained, identifying them as putative A genome chromosomes
Stability of the B. rapa-derived FAE introgression
To determine the transfer rate of the BrFAE gene during meiosis, BC3F2 line VR16-072 was used as a donor line for the production of microspore-derived plants. Of the 171 plantlets that were tested, 168 contained the BrFAE allele, which corresponds to a transfer rate of 98.25%. Further, in order to confirm that the pollen of the chromosome addition lines is viable and not selected against during pollination, crosses were carried out between 12 BC3F4 plants and a CMS B. carinata line. Seed set after pollination was normal. To confirm the transfer of the BrFAE allele, eight F1 plants from each of the 12 cross combinations were tested for its presence using the above described markers, with 95 of the 96 lines containing the BrFAE gene, equating to a transfer rate of 98.96%.
Discussion
Erucic acid is one of several hundred unusual fatty acids produced by plants (Ohlrogge et al. 2018) and is found almost exclusively in triacylglycerols (TAGs) in the seed oil of the Brassicaceae family and the genus Limnanthes (meadowfoams) (Shi et al. 2015). C22:1 and its derivatives are important industrial feedstocks. Applications include the use of erucamide as a slip-promoting agent in plastic manufacturing and the production of high temperature lubricants, detergents, emulsifiers, photographic materials, as well as cosmetics and even pharmaceuticals (Leonard 1994; Sonntag 1995; Piazza and Foglia 2001; McVetty and Scarth 2002; Impallomeni et al. 2011). According to Li et al. (2012), it is estimated that a 10% increase in C22:1 content of total seed fatty acids results in a 50% reduction of the costs of erucamide production. The plethora of applications and the economics of production in combination with the need for improved technical utility and enhanced marketability of carinata oil have motivated our efforts in the development of B. carinata germplasm with super-high C22:1 levels. Here, we show that the level of C22:1 in B. carinata can be significantly increased through introgression of a fad2B null allele from B. juncea and introduction of an extra copy of FAE from B. rapa.
The enzyme FAD2 catalyzes the desaturation of C18:1 to C18:2, the first committed step in the synthesis of polyunsaturated fatty acids (Beisson et al. 2003). In line VR10-183.8, the loss of FAD2B causes a significant increase of the C18:1 proportion in seed TAGs compared to the seed oil of B. carinata parent line 080793EM, at the expense of the 18-carbon polyunsaturated fatty acids C18:2 and C18:3. Similarly, suppression of FAD2 expression has been used successfully to increase the proportions of C18:1 in transgenic soybean and cotton as well as canola-quality B. napus and B. juncea lines (Stoutjesdijk et al. 2000; Kinney et al. 2002; Liu et al. 2002). It appears that the increased pool of C18:1 in VR10-183.8 was utilized by the endogenous fatty acid elongases (BcFAE-B, BcFAE-C), resulting in a simultaneous increase of the proportion of C22:1 to over 50% of total fatty acids. Thus, the reductive effect of the fad2B null allele on C18:1 desaturation may have enhanced substrate availability for the biosynthesis of C22:1 via elongation. This finding is in agreement with results from earlier work in Arabidopsis thaliana (Okuley 1994), B. rapa (Bao et al. 1998) and B. napus (Domergue et al. 1999). Jadhav et al. (2005) downregulated the expression of FAD2 in B. carinata using co-suppression and antisense silencing approaches. While the resulting transgenic carinata lines had significantly increased C22:1 levels, none reached levels above 46% of total fatty acids. In addition, the effectiveness of these particular transgenic strategies is variable and unpredictable and they require the production of large transgenic populations to obtain an adequate number of lines showing sufficient levels of target gene suppression (Knutzon et al. 1992; Kinney 1996; Liu et al. 2002). Interestingly, in contrast to our results and the above cited studies, downregulation of FAD2 in Crambe abyssinica by RNAi did not increase the C22:1 content, suggesting that oleate availability is not a rate-limiting factor in the biosynthesis of this fatty acid in crambe (Li et al. 2012; Cheng et al. 2013).
Line VR13-156, which in addition to the fad2B null allele possesses a heterologously expressed B. rapa FAE gene, exhibited an additional increase in C22:1 levels to just under 58%. Millar and Kunst (1997) were the first to show that introducing A. thaliana FAE1 in A. thaliana or Nicotiana tabacum (tobacco) tissue void of significant quantities of VLCFAs resulted in their accumulation and that increasing gene dosage through introduction of extra copies of FAE1 resulted in higher levels of these fatty acids. Since then, various FAE genes encoding enzymes with different acyl-CoA specificity have been identified, cloned and overexpressed in plants to increase C22:1 levels in the seed oil. Thus, overexpression of a FAE gene from Tropaeolum majus (garden nasturtium) in A. thaliana led to an increase in C22:1 proportions in the seed oil of up to eight-fold (Mietkiewska et al. 2004). In the present study, the very high proportion of C22:1 in the seed oil of fad2B/FAE line VR13-156 was associated with lower proportions of C18:3 and C20:1 compared to seed oil of VR10-183.8. This indicates that the new heterologous BrFAE acts in concert with the endogenous B. carinata FAE enzymes in elongating C18:1 to C22:1, which would limit the pool of C18:1 available for desaturation, as suggested by Mietkiewska et al. (2007). BrFAE may also be more efficient than the endogenous B. carinata FAE enzymes in elongating C18:1 to C22:1, a hypothesis corroborated by the naturally very high C22:1 proportions in seed oil of yellow sarson B. rapa line R500 (Dorrell and Downey 1964), the source of BrFAE in this study. Further, it is tempting to speculate that the BrFAE enzyme shows stronger elongase activity with gondoyl-CoA than with oleoyl-CoA, explaining the decrease in C20:1 levels in VR13-156 compared to those in VR10-183.8. The co-expression of elongases with different but complementary substrate preferences has been used successfully in a transgenic approach to increase the production of C22:1 in seeds of B. carinata (Mietkiewska et al. 2004), albeit not to the levels reported in this study.
While the introgression of the fad2B null allele alone results in a significant increase in the C18:1 content in VR10-183.8 compared to parent line 080793EM, the simultaneous introduction of this gene and BrFAE into wild-type 111078EM-7 did not yield significant changes in the content of this fatty acid in the lines VR17-089 and VR17-095. This may be explained by the immediate elongation of the increased pool of C18:1 through the heterologous BrFAE in seeds of the double mutant lines. This proposition is substantiated by the observation of Mietkiewska et al. (2007) that the C18:1 elongation pathway has a somewhat stronger ‘metabolic pull’ over C18:1 desaturation in developing seeds.
To the best of our knowledge, the current introgression of the B. juncea fad2B null allele and FAE gene from B. rapa has resulted in the highest level of C22:1 documented thus far for B. carinata seed oil. In comparison, the best result obtained through transgenic manipulations of B. carinata was observed in an RNAi FAD2/Crambe FAE transgenic line with 56% C22:1 (Mietkiewska et al. 2008). The C22:1 levels in double mutant lines VR17-089 and VR17-095 are lower than those in VR13-156 very likely because the recurrent parent 111078EM-7 they originated from also has a lower C22:1 content than line 080793EM which was used in the first cross with B. juncea. This stresses the importance of giving consideration to the level of the target fatty acid in the genetic background to be improved, in this case the recurrent parent, when attempting to develop germplasm with substantially increased proportions of that fatty acid.
Theoretically, the highest level of erucic acid that can be achieved in B. carinata seed oil through conventional breeding is 66.7%. This is because as in most Brassicaceae, in B. carinata the endogenous lysophosphatidic acid acyltransferase (LPAT), the enzyme responsible for placing acyl moieties in the sn-2 or middle position on the glycerol backbone during triacylglycerol biosynthesis via the Kennedy pathway (Kennedy 1961), cannot use erucic acid as a substrate (Frentzen 1993; Kuo and Gardner 2002; McVetty and Scarth 2002). In order to further increase the erucic acid content in our fad2B/FAE carinata lines, one option might be to introduce a diacylglycerol acyltransferase (DGAT) from a closely related Brassica species. DGATs catalyze the final step of triacylglycerol synthesis and it has been shown that specialized DGAT2s play a key role in channeling unusual acyl groups into TAGs (Shockey et al. 2006; Burgal et al. 2008; Li et al. 2010). In this scenario, the addition of an extra Brassica DGAT2 gene with high activity toward erucic acid would allow the plant to more efficiently channel erucic acid into TAG, as discussed by Demski et al. (2019).
In the present study, the introgression of BrFAE into the B. carinata genome was achieved through the transfer of the entire B. rapa A08 chromosome during the process of backcrossing. Almost completely stable fad2B/FAE B. carinata alien chromosome addition lines carrying two copies of chromosome A08 were produced and maintained. Alien chromosome addition lines in general are characterized by the addition of one (monosomic) or two (disomic) copies of a certain chromosome from one species to the genome of another species (Budahn et al. 2006). In Brassica, alien chromosome addition lines have been used extensively to assign linkage groups to chromosomes, study genomic relationships within the genus and to transfer agronomically important traits between species (Budahn et al. 2006; Chen et al. 1997; Fan and Tai 1985; McGrath and Quiros 1990; Quiros et al. 1987; Struss et al. 1991), with numerous studies particularly on the development and characterization of B. napus-nigra monosomic chromosome addition lines (AACC + 1B) (Chevre et al. 1997; Jahier et al. 1989; McGrath and Quiros 1990; Struss et al. 1991, 1992; Zhu et al. 1993). Successful examples of gene introgression from more distantly related species through monosomic addition lines include the transfer of resistance to Leptosphaeria maculans from Sinapis arvensis (wild mustard) (Snowdon et al. 2000) and resistance to the beet cyst nematode from Raphanus sativus (oil radish) (Peterka et al. 2004) into B. napus. While monosomic addition lines have proven to be useful in the dissection of genomes and studying chromosomal homoeologies, their practical utility in plant breeding programs is limited by segregation of the alien chromosome in offspring and potential loss of chromosome integrity by homoeologous recombination (Budahn et al. 2006). Thus, within the Brassica genus, transmission rates of alien monosomic chromosomes vary greatly and are usually below 50% (Budahn et al. 2006; Chen et al. 2010a, b; Chevre et al. 1997).
In contrast, disomic addition lines, which have been reported less frequently, display higher stability and fertility (Chen et al. 2010a, b; Chevre et al. 1991; Quiros et al. 1987; Wang et al. 2006; Zhang et al. 2013). To the best of our knowledge, our report is the first to describe a B. carinata-rapa disomic chromosome addition line. The transfer of the B. rapa A08 chromosome into the carinata genome is clearly demonstrated by the results of the Brassica 90K Illumina Infinium™ SNP genotyping array and further supported by the molecular cytogenetic analysis. Thus, considering the relatively unambiguous chromosome counts based on multiple plants, a stable karyotype of 36 chromosomes can be assumed for the interspecific hybrids, as shown for line VR17-095. The results of the molecular cytogenetic analysis suggest that two of the 36 chromosomes are indeed A genome chromosomes, for a karyotype of 2n = 34 + 2A.
The transfer rate of the BrFAE gene can be used to directly extrapolate the transmission rate of chromosome A08 as FAE is located on this chromosome (Fourmann et al. 1998; Delourme et al. 2006). It is well known that chromosome transmission rates are generally higher through female gametes than via male gametes (Budahn et al. 2006; Multani et al. 1994; Singh et al. 1998; Zhang et al. 2013); therefore, the focus of this study was on determining the transmission rate of chromosome A08 for male gametes by producing microspore-derived progeny and hybrid offspring by conducting crosses with a CMS B. carinata line. Both experimental approaches yielded rates of transmission of over 98%, suggesting virtually stable addition of A08 to the B. carinata genome. In comparison, B. napus-B. nigra disomic chromosome addition lines showed transmission rates for different nigra chromosomes between 50 and 100% (Chevre et al. 1991), indicating that the degree of transmission can vary depending on the species and chromosome under investigation.
A search on Ensembl Plants (https://plants.ensembl.org/index.html) revealed that chromosome A08 from B. rapa harbors additional genes that play a role in lipid metabolism, such as desaturases FAD4 and FAD6 as well as an acyltransferase and LPAT5 (lysophosphatidic acid acyltransferase 5). However, it appears that these genes do not have a major impact on the fatty acid profile in the fad2B/FAE B. carinata disomic A-chromosome addition lines as the seed oil composition phenotype of these lines is consistent with that of FAE1 over expression lines in other plants (Kunst 1997; Mietkiewska et al. 2004). It is also worth noting that FAD2 and FAD3, the major genes involved in the synthesis of polyunsaturated fatty acids (Beisson et al. 2003), are located on chromosomes A03, A04 and A05, and not on A08.
In conclusion, this study has shown that the development of super-high erucic acid Ethiopian mustard is possible through traditional interspecific hybridization. To our knowledge, the results presented herein currently represent the highest accumulation of erucic acid achieved in B. carinata seeds. Although the lines we developed are not yet suitable for introgression into elite B. carinata breeding lines for cultivar development, because of the lack of completely stable introgression of the A08 chromosome, our results nevertheless make B. carinata an increasingly attractive alternative to HEA B. napus as a feedstock crop for industrial applications. In order to achieve a 100% stability of the FAE introgression, an attempt to introgress the FAE gene into the C genome through non-homologous recombination between A08 and a B. carinata C chromosome using intraspecific hybridization between the fad2B/FAE B. carinata disomic A-chromosome addition lines and resynthesized B. carinata is ongoing. Utilizing available B. rapa whole genome sequence information, a collection of A08-specific markers can be developed and, in conjunction with the existing FAE and fad2B markers, will allow for the selection of promising lines while reducing linkage drag.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge the excellent technical support by David Sarich, Ryan Vetter, Annette Zatylny, Lori Bobowski, Leonard Chester, Jillian Hueller and Gerald Serblowski. We would also like to thank Isobel Parkin, Mark Smith and Van Ripley for their critical reviews of the manuscript. This work was supported by funding provided by the Developing Innovative Agri-Products Initiative of the Growing Forward Canadian Agri-Innovation Program, the Growing Forward II Agri-Innovation Program and the Canadian Agricultural Partnership’s Agri-Science Program. The Mason Lab is partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy–EXC 2070–390732324.
Author contributions statement
KCF, VR and CE conceived the research plans; VR designed and performed the experiments; CE designed the field experiment; KCF, VR and CE supervised the experiments; ASM and RG designed and performed the molecular cytogenetics analyses; VR and CE analyzed the data; VR and CE wrote the article with input from ASM; all authors contributed to, read and approved the final version of the article.
Funding
This work was supported by funding provided by the Developing Innovative Agri-Products Initiative of the Growing Forward Canadian Agri-Innovation Program, the Growing Forward II Agri-Innovation Program and the Canadian Agricultural Partnership’s Agri-Science Program. The Mason Lab is partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy–EXC 2070 390732324.
Data availability
All datasets generated for this study are included in the article/supplementary material.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alemayehu N, Becker HC. Variation and inheritance of erucic acid content in Brassica carinata germplasm collections from Ethiopia. Plant Breed. 2001;120:331–335. doi: 10.1046/j.1439-0523.2001.00623.x. [DOI] [Google Scholar]
- Alemayehu N, Becker HC. Genotypic diversity and pattern of variation in a germplasm material of Ethiopian Mustard (Brassica carinata A. Braun) Genet Resour Crop Evol. 2002;49:573–582. doi: 10.1023/A:1021204412404. [DOI] [Google Scholar]
- Alix K, Joets J, Ryder CD, Moore J, Barker GC, Bailey JP, et al. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. Plant J. 2008;56:1030–1044. doi: 10.1111/j.1365-313X.2008.03660.x. [DOI] [PubMed] [Google Scholar]
- Alonso LC, Fernandez-Serrano O, Fernandez-Escobar J (1991) The onset of a new oilseed crop: Brassica carinata with low erucic acid content. In: Proc 8th Intern Rapeseed Congr, pp 170–176.
- Ayotte R, Harney PM, Souza Machado V. The transfer of triazine resistance from Brassica napus L. to B. oleracea L. I. Production of F1 hybrids through embryo rescue. Euphytica. 1987;36:615–624. doi: 10.1007/BF00041511. [DOI] [Google Scholar]
- Bao X, Pollard M, Ohlrogge J. The biosynthesis of erucic acid in developing embryos of Brassica rapa. Plant Physiol. 1998;118:183–190. doi: 10.1104/pp.118.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro F, Fernández-Escobar J, De La Vega M, Martín A. Doubled haploid lines of Brassica carinata with modified erucic acid content through mutagenesis by EMS treatment of isolated microspores. Plant Breed. 2001;120:262–264. doi: 10.1046/j.1439-0523.2001.00602.x. [DOI] [Google Scholar]
- Barro F, Fernández-Escobar J, De la Vega M, Martín A. Modification of glucosinolate and erucic acid contents in doubled haploid lines of Brassica carinata by UV treatment of isolated microspores. Euphytica. 2003;129:1–6. doi: 10.1023/A:1021578318098. [DOI] [Google Scholar]
- Becker HC, Löptien H, Röbbelen G. Breeding: an overview. In: Gomez-Campo C, editor. Biology of Brassica Coenospecies. Amsterdam: Elsevier Science BV; 1999. pp. 413–460. [Google Scholar]
- Beisson F, Koo AJ, Ruuska S, Schwender J, Pollard M, Thelen JJ, et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 2003;132:681–697. doi: 10.1104/pp.103.022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budahn H, Peterka H, Schrader O, Zhang S. Intergeneric transfer of nematode resistance from Raphanus to Brassica using a series of rape-radish chromosome addition lines. Acta Hortic. 2006;706:145–150. doi: 10.17660/ActaHortic.2006.706.16. [DOI] [Google Scholar]
- Burgal J, Shockey J, Lu C, Dyer J, Larson T, Graham I, et al. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J. 2008;6:819–831. doi: 10.1111/j.1467-7652.2008.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardi T, D’Agostino N, Tripodi P. Genetic transformation and genomic resources for next-generation precise genome engineering in vegetable crops. Front Plant Sci. 2017;8:241. doi: 10.3389/fpls.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BY, Cheng BF, Jørgensen RB, Heneen WK. Production and cytogenetics of Brassica campestris-alboglabra chromosome addition lines. Theor. Appl. Genet. 1997;94:633–640. doi: 10.1007/s001220050460. [DOI] [Google Scholar]
- Chen XP, Shen EQ, Zhang CH, Li XF, Xuan SX, Shen SX. Transmission of n + 1 gametes and obtaining two disomic addition lines of flowering Chinese cabbage – Chinese kale. Sci Agric Sin. 2010;43:4871–4876. [Google Scholar]
- Chen S, Zou J, Cowling WA, Meng J. Allelic diversity in a novel gene pool of canola-quality Brassica napus enriched with alleles from B. rapa and B. carinata. Crop Pasture Sci. 2010;61:483–492. doi: 10.1071/CP09327. [DOI] [Google Scholar]
- Cheng J, Salentijn EMJ, Huang B, Krens FA, Dechesne AC, Visser RGF, et al. Isolation and characterization of the omega-6 fatty acid desaturase (FAD2) gene family in the allohexaploid oil seed crop Crambe abyssinica Hochst. Mol. Breed. 2013;32:517–531. doi: 10.1007/s11032-013-9886-0. [DOI] [Google Scholar]
- Chevre AM, This P, Eber F, Deschamps M, Renard M, Delseny M, et al. Characterization of disomic addition lines Brassica napus-Brassica nigra by isozyme, fatty acid, and RFLP markers. Theor Appl Genet. 1991;81:43–49. doi: 10.1007/BF00226110. [DOI] [PubMed] [Google Scholar]
- Chevre AM, Eber F, Barret P. Identification of the different Brassica nigra chromosomes from both sets of B. oleracea – B. nigra and B. napus – B. nigra addition lines with a special emphasis on chromosome transmission and self-incompatibility. Theor Appl Genet. 1997;94:603–611. doi: 10.1007/s001220050457. [DOI] [Google Scholar]
- Choudhary BR, Joshi P, Ramarao S. Interspecific hybridization between Brassica carinata and Brassica rapa. Plant Breed. 2000;119:417–420. doi: 10.1046/j.1439-0523.2000.00503.x. [DOI] [Google Scholar]
- Clarke WE, Higgins EE, Plieske J, Wieseke R, Sidebottom C, Khedikar Y, et al. A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor Appl Genet. 2016;129:1887–1899. doi: 10.1007/s00122-016-2746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BM, Knowles PF (1983) Evaluation of Brassica species in California. In: Proc 6th Intern. Rapeseed Congr, pp 282-285.
- Delourme R, Falentin C, Huteau V, Clouet V, Horvais R, Gandon B, et al. Genetic control of oil content in oilseed rape (Brassica napus L.) Theor Appl Genet. 2006;113:1331–1345. doi: 10.1007/s00122-006-0386-z. [DOI] [PubMed] [Google Scholar]
- Demski K, Jeppson S, Lager I, Misztak A, Jasieniecka-Gazarkiewicz K, Waleron M, et al. Isoforms of Acyl-CoA: Diacylglycerol Acyltransferase2 differ substantially in their specificities toward erucic acid. Plant Physiol. 2019;181:1468–1479. doi: 10.1104/pp.19.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue F, Chevalier S, Santarelli X, Cassagne C, Lessire R. Evidence that oleoyl- CoA and ATP-dependent elongations coexist in rapeseed (Brassica napus L.) Eur J Biochem. 1999;263:464–470. doi: 10.1046/j.1432-1327.1999.00520.x. [DOI] [PubMed] [Google Scholar]
- Dorrell DG, Downey RK. The inheritance of erucic acid content in rapeseed (Brassica campestris) Can J Plant Sci. 1964;44:499–504. doi: 10.4141/cjps64-099. [DOI] [Google Scholar]
- Fan Z, Tai W. A cytogenetic study of monosomics in Brassica napus L. Can J Genet Cytol. 1985;27:683–688. doi: 10.1139/g85-102. [DOI] [Google Scholar]
- Farhatullah S, Nasim A, Kanwal M, Fayyaz L. Heritability studies for seed quality traits in introgressed segregating populations of Brassica. Pak J Bot. 2014;46:239–243. [Google Scholar]
- Fernandez-Escobar J, Dominguez J, Martin A, Fernandez-Martinez JM. Genetics of the erucic acid content in interspecific hybrids of Ethiopian mustard (Brassica carinata Braun) and rapeseed (B. napus L.) Plant Breed. 1988;100:310–315. doi: 10.1111/j.1439-0523.1998.tb00257.x. [DOI] [Google Scholar]
- Ferreres E, Fernandez M, Minguez I, Dominguez J (1983) Productivity of B. juncea and B. carinata in relation to rapeseed B. napus. I. Agronomic studies. In: Proc 6th Intern Rapeseed Congr, pp 293-299
- FitzJohn RG, Armstrong TT, Newstrom-Lloyd LE, Wilton AD, Cochrane M. Hybridisation within Brassica and allied genera: evaluation of potential for transgene escape. Euphytica. 2007;158:209–230. doi: 10.1007/s10681-007-9444-0. [DOI] [Google Scholar]
- Fourmann M, Barret P, Renard M, Pelletier G, Delourme R, Brunel D. The two genes homologous to Arabidopsis FAE1 co-segregate with the two loci governing erucic acid content in Brassica napus. Theor Appl Genet. 1998;96:852–858. doi: 10.1007/s001220050812. [DOI] [Google Scholar]
- Fredua-Agyeman R, Coriton O, Huteau V, Parkin IA, Chèvre AM, Rahman H. Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus× B. carinata interspecific hybrids. Theor Appl Genet. 2014;127:1305–1318. doi: 10.1007/s00122-014-2298-7. [DOI] [PubMed] [Google Scholar]
- Frentzen M. Acyltransferases and triacylglycerols. In: Moore TS, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 195–220. [Google Scholar]
- Gaebelein R, Alnajar D, Koopmann B, Mason AS. Hybrids between Brassica napus and B. nigra show frequent pairing between the B and A/C genomes and resistance to blackleg. Chromosom. Res. 2019;27:221–236. doi: 10.1007/s10577-019-09612-2. [DOI] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gardner Kuo T. M, HW, Lipid biotechnology. New York, Basel: Marcel Dekker Inc; 2002. [Google Scholar]
- Getinet A, Rakow G, Raney JP, Downey RK. Development of zero erucic acid Ethiopian mustard through an interspecific cross with zero erucic acid Oriental mustard. Can J Plant Sci. 1994;74:793–795. doi: 10.4141/cjps94-141. [DOI] [Google Scholar]
- Getinet A, Rakow G, Raney JP, Downey RK. The inheritance of erucic acid content in Ethiopian mustard. Can J Plant Sci. 1997;77:33–41. doi: 10.4141/P96-074. [DOI] [Google Scholar]
- Gomez-Campo C, Prakash S. Origin and domestication of the Brassica. In: Gomez-Campo C, editor. Biology of Brassica Coenospecies. Amsterdam: Elsevier Science BV; 1999. pp. 33–58. [Google Scholar]
- Griffiths DW, Birch ANE, Hillman JR. Antinutritional compounds in the Brassicaceae. Analysis, biosynthesis, chemistry and dietary effects. J Hortic Sci Biotech. 1998;73:1–18. doi: 10.1080/14620316.1998.11510937. [DOI] [Google Scholar]
- Gupta V, Mukhopadhyay A, Arumugam N, Sodhi YS, Pental D, Pradhan AK. Molecular tagging of erucic acid trait in oilseed mustard (Brassica juncea) by QTL mapping and single nucleotide polymorphisms in FAE1 gene. Theor Appl Genet. 2004;108:743–749. doi: 10.1007/s00122-003-1481-z. [DOI] [PubMed] [Google Scholar]
- Gupta K, Prem D, Agnihotri A. Pyramiding white rust resistance and Alternaria blight tolerance in low erucic Brassica juncea using Brassica carinata. J. Oilseed Brassica. 2010;1:55–65. [Google Scholar]
- Howell EC, Kearsey MJ, Jones GH, King GJ, Armstrong SJ. A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics. 2008;180:1849–1857. doi: 10.1534/genetics.108.095893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impallomeni G, Ballistreri A, Carnemolla GM, Guglielmino SPP, Nicolo MS, Cambria MG. Synthesis and characterization of poly (3-hydroxyalkanoates) from Brassica carinata oil with high content of erucic acid and very long chain fatty acids. Int J Biol Macromol. 2011;48:137–145. doi: 10.1016/j.ijbiomac.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Jadhav A, Katavic V, Marillia EF, Giblin EM, Barton DL, Kumar A, et al. Increased levels of erucic acid in Brassica carinata by co-suppression and antisense repression of the endogenous FAD2 gene. Metab Eng. 2005;7:215–220. doi: 10.1016/j.ymben.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Jahier J, Chevre AM, Tanguy AM, Eber F. Extraction of disomic addition lines of Brassica napus-B. nigra. Genome. 1989;32:408–413. doi: 10.1139/g89-463. [DOI] [Google Scholar]
- James DW, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONATION1 (FAE1) gene with maize transposon activator. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Tian E, Li R, Chen L, Meng J. Genetic diversity of Brassica carinata with emphasis on the interspecific crossability with B. rapa. Plant Breed. 2007;126:487–491. doi: 10.1111/j.1439-0523.2007.01393.x. [DOI] [Google Scholar]
- Kanrar S, Venkateswari J, Dureja P, Kirti PB, Chopra VL. Modification of erucic acid content in Indian mustard (Brassica juncea) by up-regulation and down-regulation of the Brassica juncea FATTY ACID ELONGATION1 (BjFAE1) gene. Plant Cell Rep. 2006;25:148–155. doi: 10.1007/s00299-005-0068-3. [DOI] [PubMed] [Google Scholar]
- Katche E, Quezada-Martinez D, Katche EI, Vasquez-Teubner P, Mason AS. Interspecific hybridization for Brassica crop improvement. Crop Breed Genet Genom. 2019 doi: 10.20900/cbgg20190007. [DOI] [Google Scholar]
- Kennedy EP. Biosynthesis of complex lipids. Fed Proc Am Soc Exp Biol. 1961;20:934–940. [PubMed] [Google Scholar]
- Kinney AJ (1996) Improving soybean seed quality (or Designer oils for better nutrition). Nat Biotechnol 14:946 [DOI] [PubMed]
- Kinney AJ, Cahoon EB, Hitz WD. Manipulating desaturase activities in transgenic crop plants. Biochem Soc Trans. 2002;30:1099–1103. doi: 10.1042/bst0301099. [DOI] [PubMed] [Google Scholar]
- Knutzon DS, Thompson GA, Radke SE, Johnson WB, Knauf VC, Kridl JC. Modification of Brassica seed oil by antisense expression of a stearoyl-acyl carrier protein desaturase gene. Proc Natl Acad Sci USA. 1992;89:2624–2628. doi: 10.1073/pnas.89.7.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leflon M, Letanneur Eber F, JC, Chelysheva L, Coriton O, Huteau V, , et al. Pairing and recombination at meiosis of Brassica rapa (AA)× Brassica napus (AACC) hybrids. Theor Appl Genet. 2006;113:1467–1480. doi: 10.1007/s00122-006-0393-0. [DOI] [PubMed] [Google Scholar]
- Leonard C. Sources and commercial applications of high erucic vegetable oils. Lipid Technol. 1994;6:79–83. [Google Scholar]
- Li R, Yu K, Hatanaka T, Hildebrand DF. Vernonia DGATs increase accumulation of epoxy fatty acids in oil. Plant Biotechnol J. 2010;8:184–195. doi: 10.1111/j.1467-7652.2009.00476.x. [DOI] [PubMed] [Google Scholar]
- Li X, van Loo EN, Gruber J, Fan J, Guan R, Frentzen M, et al. Development of ultra-high erucic acid oil in the industrial oil crop Crambe abyssinica. Plant Biotechnol J. 2012;10:862–870. doi: 10.1111/j.1467-7652.2012.00709.x. [DOI] [PubMed] [Google Scholar]
- Lichter R. Anther culture of Brassica napus in a liquid culture medium. Z Pflanzenphysiol. 1981;103:229–237. doi: 10.1016/S044-328X(81)80155-9. [DOI] [Google Scholar]
- Lichter R. Induction of haploid plants from isolated pollen of Brassica napus. Z Pflanzenphysiol. 1982;105:427–434. doi: 10.1016/S044-328X(82)80040-8. [DOI] [Google Scholar]
- Liu Q, Singh SP, Green AG. High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 2002;129:1732–1743. doi: 10.1104/pp.001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lühs W, Friedt W. The major oil crops. In: Murphy DJ, editor. Designer Oil Crops: Breeding, Processing and Biotechnology. Weinheim: VCH Verlagsgesellschaft mbH; 1994. pp. 5–71. [Google Scholar]
- Lühs W, Friedt W (1997) Erucic acid allelism in Brassica napus. In: 10th Crucifer Genetics Workshop, p 229
- Lühs WW, Voss A, Seyis F, Friedt W (1999) Molecular genetics of erucic acid content in the genus Brassica. In: Proc 10th Intern Rapeseed Congr. http://www.regional.org.au/au/gcirc/4/442.htm. Accessed 13 December 2020
- Malik RS. Prospects for Brassica carinata as an oilseed crop in India. Exp Agric. 1990;26:125–130. doi: 10.1017/S0014479700015465. [DOI] [Google Scholar]
- Marillia EF, Francis T, Falk KC, Smith M, Taylor DC. Palliser’s promise: Brassica carinata, an emerging western Canadian crop for delivery of new bio-industrial oil feedstocks. Biocatal Agric Biotechnol. 2014;3:65–74. doi: 10.1016/j.bcab.2013.09.012. [DOI] [Google Scholar]
- Márquez-Lema A, Fernández-Martínez JM, Pérez-Vich B, Velasco L. Development and characterisation of a Brassica carinata inbred line incorporating genes for low glucosinolate content from B. juncea. Euphytica. 2008;164:365–375. doi: 10.1007/s10681-008-9678-5. [DOI] [Google Scholar]
- Mason AS, Higgins EE, Snowdon RJ, Batley J, Stein A, Werner C, et al. A user guide to the Brassica 60K Illumina Infinium TM SNP genotyping array. Theor Appl Genet. 2017;130:621–633. doi: 10.1007/s00122-016-2849-1. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Quiros CF. Generation of alien chromosome addition lines from synthetic Brassica napus: morphology, cytology, fertility, and chromosome transmission. Genome. 1990;33:374–383. doi: 10.1139/G90-057. [DOI] [Google Scholar]
- McVetty PBE, Scarth S. Breeding for improved oil quality in Brassica oilseed species. J Crop Prod. 2002;5:345–369. doi: 10.1300/J144v05n01_14. [DOI] [Google Scholar]
- Meng J, Shi S, Gan L, Li Z, Qu X. The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B. napus. Euphytica. 1998;103:329–333. doi: 10.1023/A:1018646223643. [DOI] [Google Scholar]
- Mietkiewska E, Giblin EM, Wang S, Barton DL, Dirpaul J, Brost JM, et al. Seed-specific heterologous expression of a nasturtium FAE gene in Arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol. 2004;136:2665–2675. doi: 10.1104/pp.104.046839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietkiewska E, Brost JM, Giblin EM, Barton DL, Tayler DC. Cloning and functional characterization of the fatty acid elongase 1 (FAE1) gene from high erucic Crambe abyssinica cv: prophet. Plant Biotechnol. J. 2007;5:636–645. doi: 10.1111/j.1467-7652.2007.00268.x. [DOI] [PubMed] [Google Scholar]
- Mietkiewska E, Hoffman TL, Brost JM, Giblin EM, Barton DL, Francis T, et al. Hairpin-RNA mediated silencing of endogenous FAD2 gene combined with heterologous expression of Crambe abyssinica FAE gene causes an increase in the level of erucic acid in transgenic Brassica carinata seeds. Mol. Breed. 2008;22:619–627. doi: 10.1007/s11032-008-9204-4. [DOI] [Google Scholar]
- Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:101–111. doi: 10.1046/j.1365-313X.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- Monnier M (1973) Croissance et dévelopement des embryons globulaires de Capsella bursa-pastoris cultivés in vitro dans un milieu à basé d'une nouvelle solution minérale. Soc Bot Fr Memoires Colloq Morphologie, pp 179-94
- Multani DS, Jena KK, Brar DS, Reyes BG, Angeles ER, Khush GS. Development of monosomic alien addition lines and introgression of genes from Oryza australiensis Domin. to cultivated rice O. sativa L. Theor Appl Genet. 1994;88:102–109. doi: 10.1007/BF00222401. [DOI] [PubMed] [Google Scholar]
- Navabi ZK, Parkin IAP, Pires JC, Xiong Z, Thiagarajah MR, Good AG, et al. Introgression of B-genome chromosomes in a doubled haploid population of Brassica napus × B. carinata. Genome. 2010;53:619–29. doi: 10.1139/g10-039. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Thrower N, Mhaske V, Stymne S, Baxter M, Yang W, Liu J, Shaw K, Shorr B, Zhang M, Wilkerson C, Matthäus B. PlantFAdb: a resource for exploring hundreds of plant fatty acid structures synthesized by thousands of plants and their phylogenetic relationships. Plant J. 2018;96:1299–1308. doi: 10.1111/tpj.14102. [DOI] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell Online. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka H, Budahn H, Schrader O, Ahne R, Schütze W. Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition lines. Theor Appl Genet. 2004;109:30–41. doi: 10.1007/s00122-004-1611-2. [DOI] [PubMed] [Google Scholar]
- Piazza GJ, Foglia TA. Rapeseed oil for oleochemical uses. Eur J Lipid Sci Technol. 2001;103:405–454. doi: 10.1002/1438-9312(200107)103:7<450::AID-EJLT450>3.0.CO;2-D. [DOI] [Google Scholar]
- Potts DA, Rakow GW, Males DR (1999) Canola-quality Brassica juncea, a new oilseed crop for the Canadian prairies. In: Proc 10th Intern Rapeseed Congr. http://www.regional.org.au/au/gcirc/4/70.htm. Accessed 13 December 2020
- Prakash S, Wu XM, Bhat SR. History, evolution, and domestication of Brassica crops. Plant Breed Rev. 2011;35:19–84. [Google Scholar]
- Quiros CF, Ochoa O, Kianian SF, Douches D. Analysis of the Brassica oleracea genome by the generation of B. campestris-oleracea chromosome addition lines: characterization by isozymes and rDNA genes. Theor Appl Genet. 1987;74:758–766. doi: 10.1007/BF00247554. [DOI] [PubMed] [Google Scholar]
- Rahman MH. Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breed. 2001;120:463–472. doi: 10.1046/j.1439-0523.2001.00640.x. [DOI] [Google Scholar]
- Rashid A, Rakow G, Downey RK. Development of yellow seeded Brassica napus through interspecific crosses. Plant Breed. 1994;112:127–134. doi: 10.1111/j.1439-0523.1994.tb00660.x. [DOI] [Google Scholar]
- Reynolds TL. Pollen embryogenesis. Plant Mol Biol. 1997;33:1–10. doi: 10.1023/A:1005748614261. [DOI] [PubMed] [Google Scholar]
- Röbbelen G, Thies W. Biosynthesis of seed oil and breeding for improved oil quality of rapeseed. In: Tsunoda S, Hinta K, Gomez-Campo C, editors. Brassica Crops and Wild Allies: Biology and Breeding. Tokyo, Japan: Scientific Societies Press; 1980. pp. 253–283. [Google Scholar]
- Roslinsky V, Falk KC, Ripley VL (2011) Analysis of FAD2B alleles in interspecific derived Brassica juncea and Brassica carinata. In: Proc 13th Intern Rapeseed Congr, p 556
- Saikia SL, Rai GK, Salgotra RK, Rai SK, Singh M, Rai PK. Erucic acid and glucosinolate variability in Brassica juncea. L. Int J Chm Stud. 2018;6:1223–1226. [Google Scholar]
- Sanyal A, Pinochet X, Merrien A, Laustriat M, Decocq G, Fine F. Erucic acid rapeseed: 1. Prospects of Improvements OCL. 2015 doi: 10.1051/ocl/2015011. [DOI] [Google Scholar]
- Scarth R, Tang J. Modification of Brassica oil using conventional and transgenic approaches. Crop Sci. 2006;46:1225–1236. doi: 10.2135/cropsci2005.08-0245. [DOI] [Google Scholar]
- Semenov VG, Semenova DU, Slipushenko VP. Calculation of the high heat value of biofuels. Chem Tech Fuels Oil. 2006;42:144–149. doi: 10.1007/s10553-006-0046-7. [DOI] [Google Scholar]
- Sharma BB, Kalia P, Singh D, Sharma TR. Introgression of black rot resistance from Brassica carinata to cauliflower (Brassica oleracea botrytis group) through embryo rescue. Front Plant Sci. 2017;8:1255. doi: 10.3389/fpls.2017.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh FA, Shashi B, Banga SS, Najeeb S, Lone BA, Shikari AB, et al. Development of Ethiopian mustard (Brassica carinata) with improved quality traits through interspecific hybridization with elite lines of Brassica napus and Brassica juncea. J Agric Biol Sci. 2009;4:6–13. [Google Scholar]
- Sheikh FA, Banga S, Banga SS, Najeeb S. Development of Ethiopian mustard (Brassica carinata) with broad genetic base through interspecific hybridization with elite lines of Brassica napus and Brassica juncea. J Agric Biotech Sustantainable Dev. 2011;3:77–84. [Google Scholar]
- Shi J, Lang C, Wu X, Liu R, Zheng T, Zhang D, et al. RNAi knockdown of FATTY ACID ELONGASE1 alters fatty acid composition in Brassica napus. Biochem Bioph Res C. 2015;466:518–522. doi: 10.1016/j.bbrc.2015.09.062. [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan J-C, Dhanoa PK, Bland JM, et al. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell. 2006;18:2294–2313. doi: 10.1105/tpc.106.043695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds NW. Principles of crop improvement. London: Longman; 1979. [Google Scholar]
- Singh RP, Huerta-Espinos J, Rajaram S, Crossa J. Agronomic effects from chromosome translocations 7DL.7Ag and 1BL.1RS in spring wheat. Crop Sci. 1998;38:27–33. doi: 10.2135/cropsci1998.0011183X003800010005x. [DOI] [Google Scholar]
- Smyth SJ. Genetically modified crops, regulatory delays, and international trade. Food Energy Secur. 2017;6:78–86. doi: 10.1002/fes3.100. [DOI] [Google Scholar]
- Snowdon RJ, Köhler W, Friedt W, Köhler A. Genomic in situ hybridization in Brassica amphidiploids and interspecific hybrids. Theor Appl Genet. 1997;95:1320–1324. doi: 10.1007/s001220050699. [DOI] [Google Scholar]
- Snowdon RJ, Winter H, Diestel A, Sacristan MD. Development and characterisation of Brassica napus-Sinapis arvensis addition lines exhibiting resistance to Leptosphaeria maculans. Theor Appl Genet. 2000;1010:1008–1014. doi: 10.1007/s001220051574. [DOI] [Google Scholar]
- Somers DJ, Friesen KRD, Rakow G. Identification of molecular markers associated with linoleic acid desaturation in Brassica napus. Theor Appl Genet. 1998;96:897–903. doi: 10.1007/s001220050817. [DOI] [Google Scholar]
- Sonntag NOV. Industrial utilization of long-chain fatty acids and their derivatives. In: Kimber DS, McGregor DI, editors. Brassica Oilseeds. Oxon, United Kingdom: CAB International; 1995. pp. 339–352. [Google Scholar]
- Stoutjesdijk PA, Hurlestone C, Singh SP, Green AG. High-oleic acid Australian Brassica napus and B. juncea varieties produced by co-suppression of endogenous Delta12-desaturases. Biochem Soc Trans. 2000;28:938–940. doi: 10.1042/bst0280938. [DOI] [PubMed] [Google Scholar]
- Struss U, Bellin U, Röbbelen G. Development of B-genome chromosome addition lines of B. napus using different interspecific Brassica hybrids. Plant Breed. 1991;106:209–214. doi: 10.1111/j.1439-0523.1991.tb00503.x. [DOI] [Google Scholar]
- Struss D, Quiros CF, Röbbelen G. Mapping of molecular markers on Brassica B-genome chromosomes added to Brassica napus. Plant Breed. 1992;108:320–323. doi: 10.1111/j.1439-0523.1992.tb00137.x. [DOI] [Google Scholar]
- Taylor DC, Falk KC, Palmer CD, Hammerlindl J, Babic V, Mietkiewska E, et al. Brassica carinata - a new molecular farming platform for delivering bio-industrial oil feedstocks: case studies of genetic modifications to improve very long-chain fatty acid and oil content in seeds. Biofuel Bioprod Bior. 2010;4:538–561. doi: 10.1002/bbb.231. [DOI] [Google Scholar]
- Thies W. Schnelle und einfache Analyse der Fettsäurezusammensetzung in einzelnen Raps-Kotyledonen. Z. Pflanzenzüchtung. 1971;65:181–202. [Google Scholar]
- Tian E, Jiang Y, Chen L, Zou J, Liu F, Meng J. Synthesis of a Brassica trigenomic allohexaploid (B. carinata x B. rapa) de novo and its stability in subsequent generations. Theor Appl Genet. 2010;121:1431–1440. doi: 10.1007/s00122-010-1399-1. [DOI] [PubMed] [Google Scholar]
- Tonguç M, Griffiths PD. Transfer of powdery mildew resistance from Brassica carinata to Brassica oleracea through embryo rescue. Plant Breed. 2004;123:587–589. doi: 10.1111/j.1439-0523.2004.00987.x. [DOI] [Google Scholar]
- U N, Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot. 1935;7:389–452. [Google Scholar]
- Velasco L, Fernández-Martínez JM, De Haro A. Isolation of induced mutants in Ethiopian mustard (Brassica carinata Braun) with low levels of erucic acid. Plant Breed. 1995;114:454–456. doi: 10.1111/j.1439-0523.1995.tb00832.x. [DOI] [Google Scholar]
- Venkateswari J, Kanrar S, Kirti PB, Malathi VG, Chopra VL. Molecular cloning and characterization of FATTY ACID ELONGATION1 (BjFAE1) gene of Brassica juncea. J Plant Biochem Biot. 1999;8:53–55. doi: 10.1007/BF03263058. [DOI] [Google Scholar]
- Vicente G, Martínez M, Aracil J. Optimization of Brassica carinata oil methanolysis for biodiesel production. J Amer Oil Chem Soc. 2005;82:899–904. doi: 10.1007/s11746-005-1162-6. [DOI] [Google Scholar]
- Wang Y, Sonntag K, Rudloff E, Wehling P, Snowdon RJ. GISH analysis of disomic Brassica napus – Crambe abyssinica chromosome addition lines produced by microspore culture from monosomic addition lines. Plant Cell Rep. 2006;25:35–40. doi: 10.1007/s00299-005-0031-3. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Gugel RK, McDonald T, Falk KC. Genetic variation and agronomic potential of Ethiopian mustard (Brassica carinata A. Braun) in western Canada. Genet Res Crop Evol. 2006;53:297–312. doi: 10.1007/s10722-004-6108-y. [DOI] [Google Scholar]
- Wei Z, Wang M, Chang S, Wu C, Liu P, Meng J et al (2016) Introgressing subgenome components from Brassica rapa and B. carinata to B. juncea for broadening its genetic base and exploring intersubgenomic heterosis. Front Plant Sci 7:1677. doi: 10.3389/fpls.2016.01677 [DOI] [PMC free article] [PubMed]
- Westphal A, Marquard R. Yield and quality of Brassica species in Ethiopia. Plant Res Dev. 1980;13:114–127. [Google Scholar]
- Zatylny A, Catinot V, Bundrock T, Wu SW, Eynck C (In press) Protocol for the production of doubled haploid plants of Brassica carinata. In: Seguí-Simarro JM (ed) Doubled Haploid Technology - Methods and Protocols, series Methods in Molecular Biology. Springer Science + Business Media, New York, USA [DOI] [PubMed]
- Zebarjadi AR, Jalali JM, Karimzadeh G, Moeini A, Salmanian Mousavi A, AH, Transformation of rapeseed (Brassica napus L.) plants with sense and antisense constructs of the fatty acid elongase gene. Iran J Biotechnol. 2006;4:79–87. [Google Scholar]
- Zhang SL, Shen EQ, Liu JF, Dong LJ, Zhang CH, Shen KQ, et al. Identification of disomic alien chromosome addition lines and a substitution line from monosomic alien addition lines of Chinese kale – flowering Chinese cabbage. J Hortic Sci Biotechnol. 2013;88:201–207. doi: 10.1080/14620316.2013.11512957. [DOI] [Google Scholar]
- Zhao X, Weim L, Cheng S, Kadis E, Cao Y, Boakye E, et al. Hydroprocessing of carinata oil for hydrocarbon biofuel over Mo-Zn/Al2O3. Appl Catal B. 2016;196:41–49. doi: 10.1016/j.apcatb.2016.05.020. [DOI] [Google Scholar]
- Zhu JS, Struss D, Röbbelen G. Studies on resistance to Phoma lingam in Brassica napus-Brassica nigra addition lines. Plant Breed. 1993;111:192–197. doi: 10.1111/j.1439-0523.1993.tb00629.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.