Background
COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a global health threat and remains a challenge for modern medicine. Rapid and accurate diagnosis of COVID-19 is vital for proper disease and outbreak management. Our review aimed to analyze scientific articles published in the literature addressing the rapid tests available for COVID-19 diagnosis at the first year of the pandemic. Methods: A systematic review was performed from October 22 to 27, 2020, searching data published in PubMed and Google Scholar databases, using subject headings or keywords related to point of care and rapid test diagnostic for SARS-CoV-2 and COVID-19. Results: The first survey identified 403 articles, but only 23 met the defined criteria for the systematic analysis. The sensitivity and specificity parameters were assessed in 19 studies, and the data suggested that there was lower sensitivity in the period 1 to 7 days after the emergence of symptoms (∼38%) higher sensitivity at 8 to 14 days (∼90%), and the highest at 15 to 39 days (∼98%). Accuracy was reported in six studies, reporting values above 50%. Only three studies reported a possible cross-reaction. Conclusions: Our findings indicate that the rapid tests used in the first year of the pandemic were tested with a small number of samples and not adequately validated. And the studies that described them were conducted with little scientific rigor.
Keywords: SARS-CoV-2, COVID-19, Quick test, Serological test, Rapid Diagnostic test, Point-of-care
1. Introduction
The Coronavirus Disease 2019 (COVID-19) outbreak, caused by the new Severe Acute Respiratory Syndrome Coronavirus type 2 (SARS-CoV-2), has rapidly progressed worldwide after its origin in Wuhan, Hubei Province, China [1], [2]. Since the World Health Organization (WHO) declared COVID-19 as a pandemic [3], the number of cases has increased exponentially, with more than 102 million cases reported up to February 2021, with a death toll exceeding 2.2 million [4].
The rapid and accurate diagnosis of COVID-19 is vital for proper disease and outbreak management, allowing immediate and precise public health surveillance, prevention and control measures [5]. At present, diagnostic tests for COVID-19 rely on two main categories: 1) methods in which the clinical specimen is examined directly for the presence of virus particles, virus antigen, or viral nucleic acids; and 2) serological tests that detect anti-SARS-CoV-2 antibodies.
Real-time reverse transcriptase polymerase chain reaction (RT-PCR), a molecular test of virus nucleic acid found in respiratory samples [6], is currently used as the reference standard for diagnosing COVID-19. However, this method has limitations that include possible false-negative results if the amount of viral genome is insufficient or if the correct time window of viral replication is missed [7]. Hence, patients suspected of having COVID-19 may receive an incorrect diagnosis, leading to greater disease spread or inappropriate treatment in the case of false positives and greater spread of the disease mistaken clinical conduct [8], [9].
Serological tests, called immunoassays, are rapid and simple techniques based on the qualitative or quantitative detection of anti-SARS-CoV-2 immunoglobulins. There are several types of serological tests available, including ELISA (enzyme-linked immunosorbent assay), chemiluminescence immunoassay (CLIA) fluorescence, and qualitative immunochromatography [10], [11], [12], [13], [14]. Serological tests for antibodies against SARS-CoV-2 are an important tool for epidemiological studies, to determinate the immunization status of a population [15], [16], [17], [18], [19] and identify asymptomatic patients [15], [16], [17].
Combined RT-PCR and serological tests increase both the sensitivity and accuracy of COVID-19 diagnosis [7]. Thus, the complementary use of IgM-IgG antibody tests may guarantee a correct diagnosis, allow early intervention, and play a critical role in fighting outbreaks [20], [21]. Due to their advantages, many serological tests for COVID-19 have become available in a short period, including some marketed for use as rapid, point-of-care (POC) tests. These tests provide results within minutes of administering the test and may extend testing to communities and populations that cannot readily access care [22]. In addition, POCs contribute to the rapid screening of sick patients and containment of the spread of Sars-CoV-2 [23].
However, the pace of development of these rapid diagnostic tests (RDT) exceeded that of rigorous evaluation, and many questions about their accuracy remain [24]. It is not clear whether the scientific evidence supports the continued use of these tests for COVID-19. Therefore, in this review, we searched for studies related to POC tests and RDT for SARS-CoV-2 infection.
2. Methods
For this systematic review, we searched the PubMed and Google Scholar databases from November/2019 to November/2020, with no restrictions on language. These databases are large enough to find all relevant articles on the topic of the article. We used subject headings/sub-headings (when applicable) or keywords for the concepts of point-of-care diagnostics or rapid test diagnostic for COVID-19 or SARS-CoV-2. The inclusion criteria were randomized trials, cohort, or case-control studies reporting experimental data on POC tests or RDT for SARS-CoV-2 infection. the WHO and UNICEF mention that rapid diagnostic test (RDT) provides results in 15–30 min. Because of this, we considered POC and RDT quick tests providing results within 30 min after administration [25], [26]. We excluded review articles, editorials, case reports, modeling studies, duplicate articles, preprints, and studies that did not perform POC or RDT for COVID-19. Three investigators independently screened the titles and keywords, and two independently screened the full-text papers. A third reviewer resolved any disagreements between the reviewers at the full-text stage. The authors used PICO to describe all the components related to the identified problem and to structure the research question. Then, the bibliographic search for evidence was carried out by the selection of the search terms identification of terms (descriptors) related to each component of the PICO strategy. The descriptors are classified as controlled: known as “medical subject headings” or “subject descriptors”, which are used for the indexation of articles in the databases. The most known vocabularies of controlled descriptors are: MeSH (MEDLINE/PubMed), DeCS (BIREME) and EMTREE (EMBASE). Not controlled: represent the textual words and their synonyms, orthographic variations, acronyms and correlates. Moreover, the PICO strategy also used Boolean operators which was represented by the connector terms AND, OR and NOT. These terms allow for combinations of descriptors that will be used in the search, with AND for a restrictive combination, OR for an additive combination and NOT for an excluding combination. Therefore, the combination of components of the PICO strategy for the finalization of the search strategy: after the selection of the search terms and use of Booleans operators for each of the four components of the PICO strategy, these must be inter-related in the following final strategy: (P) AND (I) AND (C) AND (O). Such final strategy must be inserted in the search box existent in the databases, so that evidence is located by means of a bibliographic search [27].
3. Results
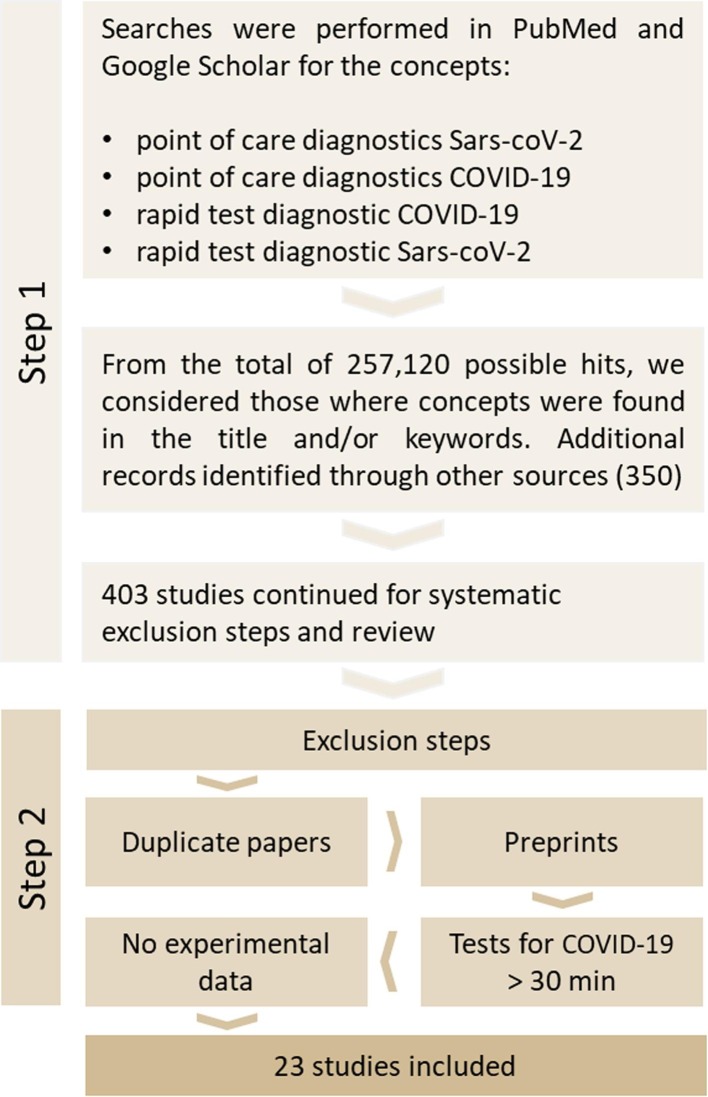
Fig. 1 shows the selection process of the studies. Initially, 257,120, we identified possible results through the database searches. Then, after screening the titles and keywords, we selected 403 studies for inclusion in the whole selection process. Twenty-three studies met the inclusion criteria. Table 1, Table 2 summarize the studies by test method.
Fig. 1.
Study selection.
Table 1.
Serological POC tests and RDT for detection of antibodies against SARS-CoV-2.
| AUTHOR / METHODS USED | SARS-CoV-2 Test |
SENSITIVITY |
SPECIFICITY / ACCURACY | N | ||
|---|---|---|---|---|---|---|
|
Day after symptom onset | ||||||
| 01 – 07 | 08 – 14 | 15 – 39 | ||||
| Yang et al. / Immunochromatography for the qualitative detection of the antibody IgM / IgG to novel Coronavirus (COVID-19) in human whole blood / serum / plasma (USA) | IgM | ND | 82% | ND | 94% / 92% | - |
| IgG | ND | ND | 94% | 97% / 96% | ||
| Abreu et al. / The LFI, SGTi-flex COVID-19 IgM / IgG (Sugentech, Republic of Korea), which is a nanoparticle-based immunochromatographic test kit for the qualitative determination of IgM and IgG COVID-19 antibodies in human whole blood (finger pricked or venous), serum or plasma. | IgM/IgG | ND | 75% | ND | 80,30% / ND | 164 = 27 positives samples and 137 negatives (rt-PCR) |
| Spicuzza et al. / LFA 2019 IgG / IgM Antibody Rapid Test Kit (Beijing Diagreat Biotechnologies Co., Ltd) | IgM/IgG | ND | ND | 83% | 93% / ND | 37 (30 patients admitted to the Infectious Disease Covid 19 or Pneumology Units and 7 healthy controls) |
| Cassaniti et al. / VivaDiag COVID‐19 IgM/IgG Rapid Test lateral flow immunoassay (LFIA) with serum or whole blood sample | IgM/IgG | 18.4% | ND | ND | 91.7% / ND | 50 patients confirmed positive for SARS-CoV-2 by rRT-PCR |
| Li et al. / Lateral flow qualitative immunoassay (LFQI) for the rapid determination of the presence or absence of both anti‐SARS‐CoV‐2‐IgM and anti‐SARS‐CoV‐2‐IgG in human serum, and plasma), manufactured by Jiangsu Medomics Medical Technologies. Specimens (whole blood) | SARS‐CoV‐2 IgG‐IgM combined antibody | 88.66% | ND | ND | 90.63% / ND | A total of 525 cases were tested: 397 (positive) clinically confirmed (including rRT-PCR test) SARS‐CoV‐2‐infected patients and 128 non‐SARS‐CoV‐2‐infected patients (128 negative). |
| Lee et al. / SARS-CoV-2 was detected in a throat swab sample by rRT-PCR. | IgG | 100.0% | 98% / 98.6% | 1 case of COVID-19 confirmed by rRT-PCR | ||
| Lyu et al / LFA (COVI040, GenBody Inc., Cheonan, Republic of Korea) to detect IgM or IgG and ELISA for IgG (RAI010R, BioVendor Laboratories) against SARS-CoV-2 | IgM/IgG | ND | ND | ND | ND / ND | 400 all samples from 2015 - 2017 |
| Capello et al. / VivaDiag ™ COVID-19 IgM / IgG Rapid Test is an in vitro diagnostic test for the qualitative determination of COVID-19 IgM and IgG antibodies | IgM/IgG | ND | ND | ND | ND | 30 (56% men, 44% women; general age 60.9 + 2.71 years; x ± SEM) |
| Pan et al. / Blood samples were collected, and blood serum, plasma or whole blood were subjected to ICG assay in accordance with the manufacturer’s protocol (Zhuhai Livzon Diagnositic Inc.). | IgM | 11.1% | 78.6% | 74.2% | ND | A total of 86 samples from 67 cases of real-time rRT-PCR confirmed SARS-CoV-2 positive |
| IgG | 3.6% | 57.1% | 96.8% | |||
| IgM | 22.2% | 33.3% | 57.1% | ND | Antibody detection in nucleic acid-negative “clinically diagnosed” patients | |
| IgG | 44.4% | 66.7% | 71.4% | |||
| Zhong et al. / ELISA methods for serum samples to detect IgM and IgG antibodies | rN-based IgM | 97.9% | 99.7% / ND | 47 COVID-19 patients ho were nucleic acid-positive (Table S1) and 300 healthy controls were analyzed | ||
| rN-based IgG | 97.9% | 99.7% / ND | ||||
| rS-based IgM | 89.1% | 97.0% / ND | ||||
| rS-based IgG | 95.7% | 85.7% / ND | ||||
| Chemiluminescence methods for serum samples to detect IgM and IgG antibodies | IgM | 97.7% | 95.2% / ND | |||
| IgG | 95.6% | 96.6% / ND | ||||
| Liu et al. / Two Enzyme-Linked Immunosorbent Assay (ELISA) kits based on recombinant SARS-CoV-2 nucleocapsid protein (rN) and spike protein (rS) were used for detecting IgM and IgG antibodies. | rN-based IgM | 68.2% | ND | ND | ND | 214 patients diagnosed with COVID-19. All laboratory confirmed positive for SARS-CoV-2 by rRT-PCR |
| rN-based IgG | 70.1% | ND | ND | ND | ||
| rN-based IgM and-or IgG | 80.4% | ND | ND | ND | ||
| rS-based IgM | 77.1% | ND | ND | ND | ||
| rS-based IgG | 74.3% | ND | ND | ND | ||
| rS-based IgM and-or IgG | 82.2% | ND | ND | ND | ||
| To et al. / Antibody levels against the SARS-CoV-2 internal nucleoprotein (NP) and surface spike protein receptor binding domain (RBD) were measured using EIA (EIA of IgG and IgM against internal viral NP and RBD). | IgM and IgG (EIA) | ND | ND | 108 serum specimens were obtained from 23 patients confirmed positive for SARS-CoV-2 by rRT-PCR | ||
| Zhang et al. / anti-SARSr-CoV IgG and IgM ELISA kits (MaxiSorp Nunc-immuno). | - | Day 0 | Day 5 | Day 15 | ND | 16 patients were viral nucleotide detection positive |
| IgM | ND | 50.0% | 81.0% | ND | ||
| IgG | 81.0% | 100.0% | ND | |||
| Okba et al. / Serologic assays for detection of SARS-CoV-2 neutralizing, spike protein–specific, and nucleocapsid-specific antibodies. Performed anti-SARS-CoV-2 S1 IgG and IgA ELISAs by using β-versions of 2 commercial kits (EUROIMMUN Medizinische Labordiagnostika AG | IgG and IgA (ELISA) | ND | ND | Serum samples (n = 10) collected from 3 patients + serum samples (n = 31) All serum samples from patients with rRT-PCR-confirmed SARS-CoV-2 | ||
| Jin et al. / SARS-CoV-2 IgM and IgG chemiluminescence immunoassay (CLIA). All serum antibody tests were performed by iFlash3000 fully automatic chemiluminescence immunoassay analyzer from Shenzhen YHLO Biotech Co., Ltd (China). | IgM | 48.1% | ND | ND | 100.0% / ND | 27 patients with laboratory confirmed infection and at least one viral serological test (Laboratory confirmation of the virus was based on the result of real time rRT-PCR). |
| IgG | 88.9% | ND | ND | 90.9% / ND | ||
| Zhao et al. / Test for total antibodies, IgM and IgG against SARS-CoV-2 in plasma samples using enzyme linked immunosorbent assay (ELISA) kits supplied Beijing Wantai Biological Pharmacy Enterprise Co., Ltda. | Total antibodies | ND | 89.6% | 100.0% | 99.1% / ND | 173 cases of COVID-19 confirmed by rRT-PCR |
| IgM | ND | 73.3% | 94.3% | 98.6% / ND | ||
| IgG | ND | 54.1% | 78.9% | 99.0% / ND | ||
| RNA by rRT-PCR | ND | 66.7% | 45.5% | ND | ||
| Guo et al. / Western blot and ELISA assays to evaluate the potential cross-reactivities between SARS-CoV-2 and other human coronavirus. | rRT-PCR CONFIRMED CASES | IgM | 75.6% | ND | ND | 208 blood samples from 2 cohorts: 39 patients confirmed positive for SARS-CoV-2; 101 inpatients; 135 plasma samples collected in 2018; 150 plasma samples from healthy adults in 2018-2019 (control); plasma samples positive for human CoV-229E, -NL63, -OC43, -NKU1 and SARS-CoV |
| rRT-PCR NO CONFIRMED CASES IgM |
IgM | 93.1% | ND | ND | ||
| 85.4% | ND | ND | ND | |||
| IgA | 92.7% | ND | ND | ND | ||
| IgG | ND | 77.9% | ND | |||
| rRT-PCR | ND | ND | ||||
| rRT-PCR + Igs |
98.6% | ND | ||||
Table 2.
POC tests and RDT for direct detection of SARS-CoV-2.
| AUTHOR / METHODS USED | SARS-CoV-2 Test |
SENSITIVITY |
SPECIFICITY |
N | ||
|---|---|---|---|---|---|---|
|
Days after symptom onset | ||||||
| 01 – 07 | 08 – 14 | 15 – 39 | ||||
| Huang et al. / (RT‐LAMP) to achieve the detection of SARS‐CoV‐2 in nasopharyngeal secretions in 30 minutes (China) | Direct Test RNA | 100% | ND | ND | 100% / ND | 16 clinic samples; with 8 positives (rt-PCR) and 8 negatives |
| Wang et al. / SARS-CoV-2 nucleic acid was detected using rRT-PCR (kit provided by Shanghai Zhijiang Biotechnology Co., Shanghai, China; detection instrument provided by Hongshi Biotechnology Co., Shanghai, China). | RNA by rRT-PCR | ND | ND | ND | ND | 86 patients: 72 with IgM-positive sera to different diseases (Influenza A and B, Mycoplasma pneumoniae, Legionella pneumophila, HIV infection, hypertensive and diabetes mellitus) without symptoms of COVID-19, 14 COVID-19 positive patients. and 36 RF-IgM-positive serum samples |
| Wang et al. / GICA and ELISA were used for SARS-CoV-2 IgM detection (kit provided by Beijing Hotgen Biotechnology Co., Beijing, China, for GICA for ELISA). Optical density in ELISA plates using a Microplate Reader (PHOMO, Autobio Diagnostics Co., Zhengzhou, China. | ||||||
| Mohanty et al / STANDARD Q COVID-19 do SD Biosensor LFIA for the qualitative detection of SARS-CoV-2 specific antigens present in the human nasopharynx (South Korea) |
Direct Test Structural antigens | 67% | ND | ND | 96% / ND | - |
| Diao et al. / Detection of NP antigen with double antibody sandwich fluorescence immunochromatographic analysis technology | Direct Test Structural antigens | ND | 75% | ND | 100% / 80.5% | 251= 122 men (48.6%) and 129 women (51.4%), aged 16-75 (average: 40.2 years) |
| Porte et al. / Fluorescence antigen rapid test kit Bioeasy 2019-Novel (2019-nCoV) with anti-SARS-CoV-2 IgY antibodies (fluorescence immunochromatographic assay) (Bioeasy Biotechnology Co., Shenzhen, China) | Direct Test Structural antigens | 93,90% | ND | ND | 100% / 96,1% | 127 = 82 rt-PCR positives and 45 negatives. 53.5% were male and the median age was 38 years |
| Scohy et al. / COVID-19 Ag Respi-Strip is an immunochromatographic test with reagent tape designed to detect the SARS-CoV-2 antigen in nasopharyngeal secretions in 15 minutes (Coris BioConcept, Gembloux, Belgium) |
Direct Test Structural antigens | 30% | ND | ND | 100% / 50% | 148 nasopharyngeal samples (RT-Qpcr: 42 negative and 106 positive). Median age of the study population was 57.5 (range: 0-94) with a sex ratio of 0.8 (64 men and 84 women) |
We identified 14 studies evaluating the levels of IgG, and IgM antibodies [5], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], one assessing the levels of IgG, IgM and IgA antibodies [7], one the levels of IgG and IgA antibodies [40], one the levels of IgG antibodies only [41], and one the levels of IgM antibodies [42]. Also, we identified four rapid tests directed to the detection of antigenic structures (44–47) and two for the detection of RNA [42], [47].
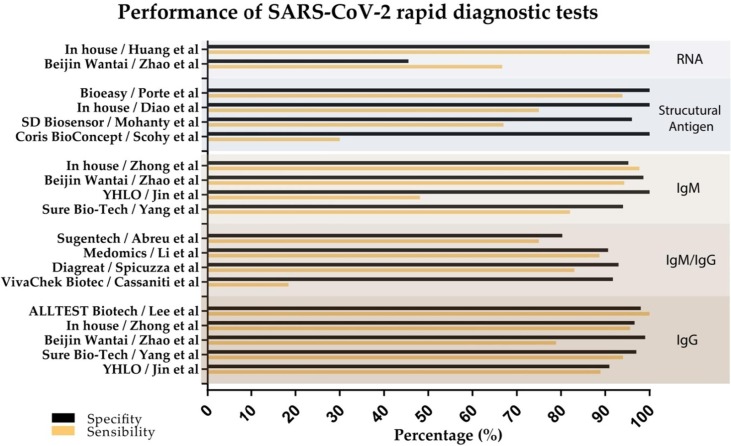
Among the included studies, 18 reported the sensitivity of the test [5], [7], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [42], [44], [45], [46], [47], [48], and 14 reported the specificity [5], [28], [29], [33], [34], [35], [36], [37], [41], [43], [44], [45], [46], [47]. Fig. 2 shows a graphical representation of the sensitivity and specificity of the analyzed tests. Only five studies provided information about the accuracy of the test [36], [42], [45], [46], [47]. Four studies [32], [39], [40], [41] performed tests to detect antibodies but did not reveal any of these parameters. Cross-reaction was assessed in three studies [7], [29], [43], and different diseases were considered, such as mycoplasma, influenza, legionella, pneumophila, HIV, and chronic diseases. Only two studies evaluated cross-reaction for other coronaviruses [7], [29].
Fig. 2.
Sensitivity and specificity of tests.
4. Discussion
Serological tests are an essential tool for diagnosing of COVID-19; however, there is uncertainty about the accuracy of the point-of-care (POC) and rapid diagnostic tests (RDT), that are currently available. We, therefore, aimed to analyze the scientific literature published in this area. Here, we overview of the rapid POC tests used to diagnose SARS-CoV-2 infection from 2,739 samples.
We identified 23 studies reporting on the use of POC tests and RDT for the diagnosis of COVID-19. We divided these studies into serological POC tests and RDT for detecting IgM and/or IgG antibodies against SARS-CoV-2 (Table 1), and POC tests and RDT for direct detection SARS-CoV-2 (Table 2). The reviewed studies used different quick diagnosis methods such as Lateral flow immunoassay (LFIA), chemiluminescence, enzyme-linked immunosorbent assay (ELISA), enzyme immunoassay (EIA), RT‐LAMP, rRT-PCR, colloidal gold immunochromatography assay (GICA), and fluorescence assays.
Despite the methodology applied, diagnostic tests should be requested at different stages of infection considering both its sensitivity and specificity [10], [11], [12], [13], [14], [48] and its accuracy and chance of cross-reactions [35], [36]. However, four serological studies included in this review [31], [38], [39], [40] did not display any of the abovementioned parameters.
4.1. Specificity and sensitivity
The analytical specificity of a test is its ability to detect the intended target while not being affected by interfering substances under well-controlled laboratory conditions. The analytical sensitivity often describes the lowest amount of analyte that can be accurately measured through an assay. Adequate analytical specificity and sensitivity will, in the end, lead to optimal clinical performance.
Evidence on the diagnostic sensitivity and specificity for COVID-19 POC tests and RDTs was found, respectively, in 18 and 14 studies (Table 1, Table 2). Therefore, the sensitivity and specificity parameters together were assessed in 19 studies. Pooled sensitivities and specificities are shown in Fig. 2.
In respect of SARS-CoV-2 contagion test criteria, Zhao et al. [33] reported that in the period from 1 to 7 days after the advent of symptoms, the RT-PCR method has a sensitivity of 67%, while between 8 and 14 days, the sensitivity decreases to 54%, and from 15 to 39 days, it drops to 45%.
In comparison, IgM and IgG levels have a sensitivity of 29% and 19%, respectively, within 7 days of symptoms; 73% and 54%, from 8 to 14 days; and 94% and 89%, from 15 and 39 days. Thus, RT-PCR should be used as a diagnostic tool from 1 to 7 days of symptoms to obtain the best results [49], [50], while serological assays for combined or isolated antibodies (IgG and IgM) present higher sensitivity after 15 days of symptoms [51].
From the 18 studies reporting POC/RDT sensitivity, only seven studies (two reporting direct detection and five identifying antibodies) reported good sensitivity, ranging from 90% to 100% (Fig. 2). This observation shows that although the POC/RDT has the advantage of producing results quickly, the sensitivity values vary depending on the type of test, the brand, and the lot; and for most tests found, the values are low.
Regarding the different methods, RT-LAMP exhibited 100% sensitivity from 1 to 7 days, and ELISA showed a sensitivity increase for both IgG and IgM after 15 days of contagion [34]. The chemiluminescence technique seems to be more sensitive from day 8 to 14 of contagion, with a 97.7% sensitivity for IgM and 95.6% for IgG [34]. The enzyme-linked fluorescent assay (ELFA) showed a sensitivity of 18.4% on the first day of symptoms [34]; these data agree with Rong et al. [14], who suggested that this methodology is not capable of detecting antibodies early.
As for specificity, most of the studies presented values higher than 90% (Fig. 2).
4.2. Accuracy and Cross-reaction
Accuracy was reported in six studies, describing values above 50%, frighteningly low, and reinforces the lack of reliable information. Indeed, there is some uncertainty about the accuracy of COVID-19 POC tests. Most of them are produced in China and do not have Food and Drug Administration (FDA) approval. In addition, these tests have frequently been found to be inaccurate and nonspecific, with the occurrence of cross-reactivity with other viruses, such as influenza [52]. Only three studies described cross-reactions. They considered various diseases [7], [28], [42] such as mycoplasma, influenza, legionella, pneumophila, HIV and chronic diseases. Of these, only two evaluated cross-reaction for other coronaviruses [7], [28].
Many tests can cross-react with other microorganisms, resulting in false-positive results [53]. A false-positive test may disrupt individuals’ lives and contribute to a false impression about the disease with the consequent quarantine requirements [43], [14]. Both false-positive and false-negative results can have devastating consequences in respect of this epidemic. A false-negative result will lead to undetected cases that propagate the epidemic.
5. Study limitations
In general, the limitations of the studies identified in this review were: low sample numbers (varying between 16 and 525 individuals); a lack of validation; and diverse clinical parameters [19], [48]. Thus, we found significant variability and little scientific rigor in the data, and there is a clear need for more studies about this type of COVID-19 test to determine their reliability and safety for use in clinical practice.
5. Final Considerations
Block serial evaluations for symptom days seem to be urgently needed to confirm the sensitivity and specificity of rapid point-of-care tests, being subsequently validated by techniques such as immunochromatography, ELISA, chemiluminescence, and fluorescence [55,57]. Overall, the studies suggest that direct diagnostic tests are more sensitive and specific during the earlier phase of symptom onset (with best results between 1 and 3 days), and serological tests that detect IgM and IgG antibodies are more accurate after 15 days of contagion, with titers increasing according to symptoms evolution [35], [43].
Moreover, due to the urgent need imposed by the COVID-19 pandemic, validation of POC tests/RDT should not be delayed, as they are a cornerstone for the extensive use of rapid tests. Nevertheless, questions on the quality of rapid COVID-19 tests remain, as there is a real lack of indisputable and conclusive evidence. Therefore, increasing the number of samples tested and comparing them with molecular techniques should be the key strategy for a better diagnosis, validation of rapid tests, and, consequently, technical decision-making by healthcare workers. However, for excellent diagnostic reliability, a larger number of samples should be evaluated to validate rapid tests such as POC and RDTs.
To develop more sensitive and specific tests, companies must adopt criteria in selecting antigens used in their fabrication. These antigens must be specific and immunogenic and can be selected by bioinformatics tools. Similarly, the production of glycosylated antigens, such as the Spike protein, must be done in eukaryotic cells to preserve their antigenic characteristics.
For the development of direct test tests that detect viral components, it is fundamental to use monoclonal or polyclonal antibodies that have been tested for cross-reactivity to other pathogens. Especially in the case of tests for COVID-19, these antibodies and antigens should be tested for reactivity to antigens and antibodies of alphacoronavirus, as they are ubiquitous in the population and can generate false-positive results in not validated tests. Finally, this information should be present in the test inserts. The fact that they are not shown raises suspicions that these criteria were not used to manufacture the tests.
Regarding the ability of these tests to recognize new virus variants, there are already some works showing differences in the binding of monoclonal antibodies due to the existence of variants. However, even kits that use monoclonal antibodies to detect antigens will probably detect new variants because they accumulate mutations in various parts of the genome and not necessarily in the same target proteins of the test.
And if the kit uses polyclonal antibodies, the mutations present in some variants should not prevent binding to these antibodies either. In these kits, however, the use of polyclonal antibodies can impair the accuracy of the test. It occurs because the antibodies will bind to antigens that present epitopes of related viruses, such as Alpha coronavirus 229E and NL63, Beta coronavirus OC43, and HKU1, which share many epitopes in the protein spike with sarscov2.
In conclusion, our findings corroborate the need for further studies to determine the best rapid testing method. These studies are pivotal to establishing rapid tests as an essential surveillance tool against the SARS-CoV-2 epidemic. We also found that the citizen tests in this article were released to the market without being adequately validated and did not show information about cross-reactivity with other pathogens, especially against other coronaviruses. All of these tests and the technological advances employed in them, and the pandemic lessons could be very useful in developing new tests for diagnosing other diseases and pathogens.
Author contributions
All authors have read and agreed to the published version of the manuscript. LP was responsible for the conception and design of the manuscript; AL, BM, BP, RL, DR, MG and PC analyzed and interpreted the data and completed the draft; MR, JS and LJ carried out a critical review of intellectual content; and HDMC and NM contributed with the final draft of the manuscript and with resources for the research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;15:395. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu H., Wang L., Fang C., Peng S., Zhang L., Chang G., et al. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Translational Pediatrics. 2020;9:51. doi: 10.21037/tp.2020.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Pneumonia of unknown cause – China (2020). https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/ [Acessed April 20, 2020].
- 4.Kang S., Peng W., Zhu Y., Lu S., Zhou M., Lin W., et al. Recent progress in understanding 2019 novel coronavirus (SARS-CoV-2) associated with human respiratory disease: detection, mechanisms and treatment. International Journal of Antimicrobial Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105950. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Y., Wang M., Zuo Z., Fan C., Ye F., Cai Z., et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. International Journal of Infectious Diseases. 2020;94:49. doi: 10.1016/j.ijid.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou L.R., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L., Ren L., Yang S., Xiao M., Chang Yang F., et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin. Infect. Dis. 2020 21 Mar doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai T.H.T., Tang E.W.H., Chau S.K.Y., Fung K.T.S.C., Li K.K.W. Stepping up infection controlmeasures in ophthalmology during the novel coronavirus outbreak: anexperience from Hong Kong. Graefes Arch Clin Exp Ophthalmol. 2020;258:1049. doi: 10.1007/s00417-020-04641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J., Ding N., Cehn H., Liu X.J., He W.J., Dai W.C., et al. Infection Control against COVID-19 in Departments of Radiology. Academic Radiology. 2020;27:614. doi: 10.1016/j.acra.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yongchen Z., Shen H., Wang X., Shi X., Li Y., Yan J., et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerging Microbes & Infections. 2020;9:833. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., et al. Antibody Detection and Dynamic Characteristics in Patients with COVID-19. Clinical Infectious Diseases. 2020 doi: 10.1093/cid/ciaa461. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., et al. SARS-CoV-2 seroconversion inhumans: A detailed protocol for a serological assay, antigenproduction, and test setup. Current Protocols in Microbiology. 2020;57:100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim E. Drawing on Israel’s Experience Organizing Volunteers to Operationalize Drive-Through Coronavirus Testing Centers. Disaster Medicine and Public Health Preparedness (202), 1-3. doi.org/10.1017/dmp.2020.104. [DOI] [PMC free article] [PubMed]
- 14.Rong X., Liu Y., Chu H., Fan M. Effect of delay in diagnosis on transmission of COVID-19. Mathematical Biosciences and Engineering. 2020:17:2725. doi: 10.3934/mbe.2020149. [DOI] [PubMed] [Google Scholar]
- 15.Sung H, Yoo CK, Han MG, Lee SW, Lee H, Chun S, et al. Preparedness and Rapid Implementation of External Quality Assessment Helped Quickly Increase COVID-19 Testing Capacity in the Republic of Korea. Clinical Chemistry 2020, in press. doi.org/10.1093/clinchem/hvaa097. [DOI] [PMC free article] [PubMed]
- 16.Maxmen A. Untapped potential: More US labs could be providing tests for coronavirus. Nature. 2020 doi: 10.1038/d41586-020-01154-6. in press. [DOI] [PubMed] [Google Scholar]
- 17.Bassi LL, Hwenda L. COVID-19: time to plan for prompt universal access to diagnostics and treatments. Lancet Glob Health (2020) in press. doi.org/10.1016/S2214-109X(20)30137-6. [DOI] [PMC free article] [PubMed]
- 18.Zitek T. The Appropriate Use of Testing for COVID-19. The Western Journal of Emergency Medicine: Integrating Emergency Care with Population Health (2020) in press. doi.org/10.5811/westjem.2020.4.47370.
- 19.Gareth I. Covid-19: Antibody tests will not be rolled out in UK until at least May. MPs hear. BMJ. 2020;369:1449. doi: 10.1136/bmj.m1449. [DOI] [PubMed] [Google Scholar]
- 20.Broadhurst M.J., Brooks T.J.G., Pollock N.R. Diagnosis of Ebola Virus Disease: Past, Present, and Future. Clin. Microbiol. Rev. 2016;29:773. doi: 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kluge H., Martin-Moreno J.M., Emiroglu N., Rodier G., Kelley E., Vujnovic M., et al. Strengthening global health security by embedding the International Health Regulations requirements into national health systems. BMJ Global Health. 2018;3:1. doi: 10.1136/bmjgh-2017-000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CDC. Centers for Disease Control and Prevention. Guidance for SARS-CoV-2 Point-of-Care Testing, 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/point-of-care-testing.html. [accessed on January 8, 2021].
- 23.Rai P., Kumar B.K., Deekshit V.K., Karunasagar I., Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021;105:441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization. WHO, Global partnership to make available 120 million affordable, quality COVID-19 rapid tests for low- and middle-income countries, (2020). https://www.who.int/news/item/28-09-2020-global-partnership-to-make-available-120-million-affordable-quality-covid-19-rapid-tests-for-low--and-middle-income-countries (accessed August 27, 2021).
- 25.UNICEF Supply Division, Most affordable COVID-19 rapid diagnostic test now available |, (2021). https://www.unicef.org/supply/stories/most-affordable-covid-19-rapid-diagnostic-test-now-available (accessed August 27, 2021).
- 26.da Santos C.M., Pimenta C., de C.A., Nobre M.R.C M. The PICO strategy for the research question construction and evidence search. Rev. Lat. Am. Enfermagem. 2007;15:508–511. doi: 10.1590/S0104-11692007000300023. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. Journal of Infection. 2020 doi: 10.1016/j.jinf.2020.03.051. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., et al. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020 30 Mar doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 27 Feb doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W., Chang S., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L., et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. Journal of Infection. 2020;1:48. doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet Infectious Diseases. 2020;20:565. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong L., Chuan J., Gong B., Shuai P., Zhou Y., Yi Zhang, et al. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci. China Life Sci. 2020:63:777. doi: 10.1101/2020.02.11.20021493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang T., Gentile M., Shen C.F., Cheng C.M. Combining point-of-care diagnostics and internet of medical things (IOMT) to combat the Covid-19 pandemic. Diagnostics. 2020;10:4–6. doi: 10.3390/diagnostics10040224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Abreu M.C., Choquet C., Petit H., Bouzid D., Damond F., et al. SARS-CoV-g IGM and IGG rapid serologic test for the diagnosis of COVID-19 in the emergency department. Journal of Infection. 2020;81(5):816–846. doi: 10.1016/j.jinf.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spicuzza L., Montineri A., Manuele R., Crimi C., Pistorio M.P., Campisi R., et al. Reliability and usefulness of a rapid IgM-IgG antibody test for the diagnosis of SARS-CoV-2 infection: A preliminary report. J. Infect. 2020;81:e53–e54. doi: 10.1016/j.jinf.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyu Z., Harada Sassa M., Fujitani T., Harada K.H. Serological Tests for SARS-CoV-2 Coronavirus by Commercially Available Point-of-Care and Laboratory Diagnostics in Pre-COVID-19 Samples in Japan. Diseases. 2020;8:36. doi: 10.3390/diseases8040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capello F., Cipolla M., Cosco L., Gnasso A., et al. The VivaDiag COVID-19 lgM/IgG Rapid Test for the Screening and Early Diagnosis of COVID-19 in Patients with No Clinical Signs of the Disease. Int. J. Endocrinol. Metab. Disord. 2020;6 doi: 10.16966/2380-548x.167. [DOI] [Google Scholar]
- 40.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee K.H., Zeng H.Q. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017;89:12743. doi: 10.1021/acs.analchem.7b02862. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q., Du Q., Guo B., Mu D., Lu X., Ma Q., et al. A method to prevent SARS-CoV-2 IgM false positives in gold immunochromatography and enzyme-linked immunosorbent assays. Journal of Clinical Microbiology. Apr 2020 doi: 10.1128/JCM.00375-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohanty A., Kabi A., Kumar S., Hada V. Role of Rapid Antigen Test in the Diagnosis of COVID-19 in India. J. Adv. Med. Med. Res. 2020;32:77–80. doi: 10.9734/jammr/2020/v32i1830657. [DOI] [Google Scholar]
- 44.Huang W.E., Lim B., Hsu C.C., et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maxmen A. Coronavirus tests go unused in their thousands. Nature. 2020;580:312. doi: 10.1038/d41586-020-01068-3. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Chen S, Yang Z, Guan W, Liu D, Lin Z. SARS-CoV-2 Viral Load in Clinical Samples of Critically Ill Patients. AJRCCM (2020) in press. doi.org/10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed]
- 47.Xiao S.Y., Wu Y., Liu H. Evolving status of the 2019 novel coronavirus infection: proposal of conventional serologic assays for disease diagnosis and infection monitoring. J Med Virol. 2020;92:464. doi: 10.1002/jmv.25702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y., Xiao M., Liu X., Xu S., Du T., Xu J. Significance of Serology Testing to Assist Timely Diagnosis of SARS-CoV-2 infections: Implication from a Family Cluster. Emerging Microbes & Infections. 2020;9:924. doi: 10.1080/22221751.2020.1752610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nickbakhsh S., Ho A., Marques D.F.P., McMenamin J., Gunson R.N., Murcia P.R. Epidemiology of seasonal coronaviruses: Establishing the context for COVID-19 emergence. MedRxiv. 2020 doi: 10.1101/2020.03.18.20037101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J., et al. Limits of Detection of Six Approved RT–PCR Kits for the Novel SARS-coronavirus-2 (SARS-CoV-2) Clinical Chemistry. 2020 doi: 10.1093/clinchem/hvaa099. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Padula W.V. Why Only Test Symptomatic Patients? Consider Random Screening for COVID-19. Appl. Health Econ. Health Policy. 2020 doi: 10.1007/s40258-020-00579-4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Surkova E, Nikolayevskyy V, Drobniewski F. False-positive COVID-19 results: hidden problems and costs. Lancet Respir Med. 2020 Dec;8(12):1167-1168. doi: 10.1016/S2213-2600(20)30453-7. Epub 2020 Sep 29. PMID: 33007240; PMCID: PMC7524437. [DOI] [PMC free article] [PubMed]
- 53.Du Z., Zhu F., Guo F., Yang B., Wang T. Detection of antibodies against SARS‐CoV‐2 in patients with COVID‐19. Journal of Medical Virology. 2020 doi: 10.1002/jmv.25820. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]