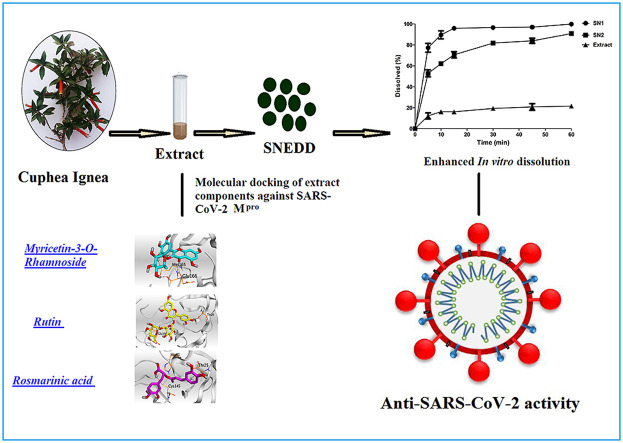
Abstract
The outbreak of coronavirus disease-2019, caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a worldwide emerging crisis. Polyphenols are a class of herbal metabolites with a broad-spectrum antiviral activity. However, most polyphenols encounter limited efficacy due to their poor solubility and degradation in neutral and basic environments. Thus, the effectiveness of their pharmaceutical application is critically dependent on the delivery systems to overcome the aforementioned drawbacks. Herein, Polyphenols-rich Cuphea ignea extract was prepared and its constituents were identified and quantified. Molecular docking was conducted for 15 compounds in the extract against SARS-CoV-2 main protease, among which rutin, myricetin-3-O-rhamnoside and rosmarinic acid depicted the most promising antiviral activity. Further, a self-nanoemulsifying formulation, composed of 10% oleic acid, 40% tween 20 and propylene glycol 50%, was prepared to improve the solubility of the extract components and enable its concurrent delivery permitting combined potency. Upon dilution with aqueous phases, the formulation rapidly Formsnanoemulsion of good stability and excellent dissolution profile in acidic pH when compared to the crude extract. It inhibited SARS-CoV-2 completely in vitro at a concentration as low as 5.87 μg/mL presenting a promising antiviral remedy for SARS-CoV-2, which may be attributed to the possible synergism between the extract components.
Keywords: Antiviral, Covid-19, Cuphea ignea, Molecular docking, Nanoemulsion, Polyphenols, SARS-CoV-2
Graphical abstract
1. Introduction
The latest outbreak of coronavirus disease 2019 (COVID-19) was primarily recognized in Wuhan, China [1] by the end of 2019. This disease is caused by a novel Betacoronavirus that has been given the name of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses [2]. SARS-CoV-2 belongs to the enveloped positive-sense single-stranded RNA viruses and it can be transmitted from human to human [3]. In October 10, 2020, the World Health Organization (WHO) recorded 37.1 million confirmed cases of COVID-19 and 1.07 million confirmed deaths worldwide [4], signifying that COVID-19 is a life-threatening disease that affects public health. Thus, substantial efforts that are currently being exerted to discover effective vaccines or drugs. In numerous countries, positively tested cases are subjected to off-label and repurposed therapies such as chloroquine, hydroxychloroquine, lopinavir-ritonavir, azithromycin, remdesivir, favipiravir, nitazoxanide, convalescent plasma and interleukin-6 inhibitors [5,6]; yet there is a critical and urgent necessity to discover other effective compounds as potential therapies for COVID-19 [7].
Computational studies are very promising tools in drug discovery processes. Several computational methods like molecular docking studies help scientists in the discovery of new drug candidates [8]. The SARS-CoV-2 main protease (Mpro) is responsible for its replication and transcription as well, through the conversion of polypeptides into functional proteins [9,10]. Accordingly, targeting Mpro of this virus can be a very promising target to examine the anti-viral efficacy of any compound [11,12].
Numerous herbal metabolites possess many health benefits. However, the pharmaceutical use of the herbal extracts and ingredients or phytopharmaceuticals is restricted because of their low solubility and bioavailability which is considered the biggest obstacle in the application of herbal products for the management of innumerable health complications. Thus, the effectiveness of phytopharmaceuticals is critically dependent on the availability of suitable drug delivery systems which can overcome the aforementioned drawbacks.
Cuphea ignea A. DC. Which is known as firecracker or cigar flower, is often grown as an ornamental plant in the tropics and as a summer bedding in the temperate zone [13]. Cuphea ignea A. DC. belongs to Lythraceae family that is considered an important source of unique natural materials for development of drugs to treat many diseases. Recent reports revealed that Cuphea ignea A. DC. possesses significant antiulcerogenic, antitumor, antioxidant and antihypertensive activities [[13], [14], [15]]. Thirteen compounds were identified in Cuphea ignea A. DC. ethanolic extract including flavonoids, phenolic acids, hydrolysable tannin and coumarin compound [13]. Different extracts of the plant possess high content of polyphenols comprising phenolic acids and flavonoids especially the ethanolic extract of the leaves [14].
Polyphenols are the most prevalent class of bioactive compounds available in plants. Regarding the structure, polyphenols are the compounds that contain one or more phenolic rings substituted with hydroxyl groups. Accordingly, they can be categorized into phenolic acids, polyphenolic amides, flavonoid and other polyphenolic compounds (including stilbenes or lignans) [5]. Reports have signified that polyphenols exerted an antiviral activity against many viruses with particular emphasis on the influenza [16] and other respiratory tract infecting viruses [[17], [18], [19]]. Nevertheless, the hydrophobic nature of most polyphenolic compounds limits their water solubility and hence they exhibit poor bioavailability which greatly affect their efficacy [20]. Furthermore, the stability of polyphenolic compounds in gastrointestinal tract is an important issue that needs to be considered while designing a successful drug delivery system, as they are stable at acidic pH of stomach while they degrade in neutral or basic pH environments resulting in limited bioavailability [21]. Consequently, a formulation with rapid dissolution which is able to rapidly dissolve the polyphenolic compounds allowing their rapid absorption in the stomach before reaching the intestine is highly demanded.
Nanoemulsion formulations have been investigated for the delivery of various medicines such as phytopharmaceuticals, dietary and nutraceutical as a result of their capability of solving many delivery problems, such as the limited aqueous solubility, instability, poor permeability and bioavailability, by encapsulating the drugs into the oily core and hydrophilic region of the droplets. Herbal extracts contain multi-bioactives which are either hydrophilic or lipophilic in nature; extremely hydrophilic components exhibit poor absorption through the lipid membranes while lipophilic drugs show poor aqueous solubility and these reasons can limit their biological efficacy and pharmacokinetics of herbal extracts. Since nanoemulsion can dissolve both hydrophilic and lipophilic components, they are considered an attractive formulation strategy for herbal extracts.
Self-nanoemulsifying drug delivery systems (SNEDDSs) are isotropic mixtures in which a drug is dissolved, these mixtures composed of oily component, surfactant, and cosurfactant. The conceptual principle of these systems is their capability of forming fine oil-in-water nanoemulsion by means of mild agitation after diluting them with aqueous phases. Such formulations when administered orally, they form nanoemulsions in the gastrointestinal tract by the aid of the gastric and intestinal digestive motility that provide the agitation needed for self-emulsification and spontaneous formation of nanoemulsion [22]. The small droplet size of the obtained nanoemulsions permits the availability of a large interfacial area for solubilization and absorption of the drugs [23]. These properties make SNEDDSs an attractive formulation strategy to enhance the solubility of poorly aqueous soluble drugs.
Therefore, our aims were to explore the phenolic metabolites of Cuphea ignea A. DC. ethanolic extract, study their ability to target SARS-CoV-2 Mpro using molecular docking and propose a rapidly dissolving self-nanoemulsifying formulation to overcome the poor solubility and pH dependent instability of polyphenols, hence their bioavailability can be improved accordingly. Moreover, our aim was to assess the viability of utilizing the developed formulation as an inhibition therapy for SARS-CoV-2. We envisioned that this formulation will be a promising tool to improve the delivery of Cuphea ignea polyphenols and offers a combined antiviral activity against SARS CoV-2 through simultaneous delivery of multi-bioactives. To the best of our knowledge there is no currently published data reporting the use of Cuphea ignea A. DC. extract for inhibition of SARS-CoV-2 and this is the first presented drug delivery system for the extract.
2. Material and methods
2.1. Plant material
Cuphea ignea A. DC. leaves used in this study were obtained in March 2020 from 6th of October city, Egypt and were authenticated by Dr. Mohamed El-Gebali, Senior Botanist at El-Orman Botanic Garden, Egypt. A voucher specimen (No. 4-5-2016-1) of the plant was kept in the Herbarium of Pharmacognosy Department, Faculty of Pharmacy, Cairo University, Egypt.
2.2. Chemicals
Oleic acid and propylene glycol were obtained from Fisher Scientific UK, Loughborough, UK, tween 20 was purchased from Sigma–Aldrich, Germany. HPLC grade Ethanol, HPLC grade methanol, HPLC grade acetonitrile, dimethyl sulfoxide (DMSO) and phosphoric acid were purchased from El-Nasr Chemical Company (Cairo, Egypt).
2.3. Cells
Vero E6 cells were cultured in Dulbecco's modified Eagle's medium (Lonza, Verviers, Belgium) supplemented with 10% fetal bovine serum (Gibco, New York, NY, USA), and 1% antibiotic antimycotic mixture (Lonza). Incubation of the cells were done at a temperature of 37 °C in a humidified 5% CO2 atmosphere. An hCoV-19/Egypt/NRC-3/2020 SARS-CoV-2 virus (Accession Number on GSAID: EPI_ISL_430820) was propagated in Vero E6 cells and harvested after cytopathic effects (CPE) appearance. Viral stock was titred using plaque infectivity assay and stored at −80 °C.
2.4. Preparation of the ethanolic extract of Cuphea ignea A. DC. Leaves
The air-dried powdered leaves (40 g) were extracted with 95% ethanol by cold maceration till exhaustion (5 × 200 mL). The ethanolic extract was dried through evaporation under reduced pressure till dryness at 50 °C to give 5 g of dry residue of the leaves.
2.5. Identification and quantitative estimation of the phenolic compounds in Cuphea ignea A. DC
The 95% ethanolic extract of the leaves was dissolved in methanol using sonication for 10 min followed by filtration through a microfilter (0.4 μm pore size) prior to injection. The standards (Sigma-Aldrich, USA) mixed stock solution was prepared by solubilizing 10 mg of each reference standard in 10 mL methanol. The calibration curve of each standard was attained by further diluting the mixed stock solution with methanol and the mixed standard solutions were injected (20 μL). HPLC analysis was carried out by means of an Agilent 1260 infinity series (Agilent Technologies, USA) equipped with Quaternary pump, a Kinetix EVO C18 column (5 μm, 4.6 × 100 mm) at 30 °C. Mobile phase was composed of a mixture of (A) 0.2% phosphoric acid solution in water (v/v), (B) methanol and (C) acetonitrile in a gradient elution mode. Separation was achieved at a solvent flow rate 1 mL/min, ultraviolet detection was set at 284 nm using variable wavelength detector.
2.6. Docking studies
Molecular docking studies using MOE 2019.012 suite [24] were carried out to propose and evaluate the binding scores and interaction modes of the 15 compounds constituting the ethanolic extract of Cuphea ignea A. DC leaves against SARS-CoV-2 Mpro. The N3 co-crystallized inhibitor was involved in our studies as a reference standard.
2.7. Preparation of the extract compounds
The examined compounds were obtained from the PubChem and prepared for docking using the steps described earlier [25]. A single database containing the co-crystallized N3 inhibitor together with the prepared compounds of the extract was built and saved as an MDB file for the docking process.
2.8. Preparation of the target SARS-CoV-2 Mpro
The SARS-CoV-2 main protease X-ray structure (code: 6LU7) [26] was obtained from the Protein Data Bank and it was subjected for the detailed preparation steps described previously [27].
Docking of the extract compounds to the binding site of SARS-CoV-2 Mpro
The aforementioned database was docked according to the general docking methodology applied [28]. At the end, we selected the best poses based on their scores, binding interactions, and RMSD values. Furthermore, a program validation process was carried out at the start for the target Mpro, and the validation was confirmed by low RMSD values (<1) between the docked and native forms [29].
2.9. Formulation of Cuphea ignea A. DC. SNEDDS and assessment of dispersibility
Six formulations were prepared with changing the concentrations of oleic acid, Tween 20 and propylene glycol as an oily phase, a surfactant and a co-solvent, respectively; as shown in Table 1 . The excipients were weighed into glass vials of 10 mL capacity and stirred at 300 rpm and 50 °C, to ensure the complete mixing of the components. The mixtures were allowed to cool to 37 °C then 1 g of each mixture was transferred to a glass beaker containing 500 mL of distilled water and kept at 37 °C, the mixtures are gently stirred at 50 rpm and subjected to visual inspection. A good SNEDDS is obtained if spontaneous emulsification occurred and a clear transparent nanoemulsion formed after dispersion of the mixture in water at 37 °C [30].
Table 1.
The composition of the prepared formulations.
| Formulation Code | % Oleic acid | % Tween 20 | % Propylene glycol |
|---|---|---|---|
| SN1 | 10 | 40 | 50 |
| SN2 | 10 | 80 | 10 |
| SN3 | 20 | 30 | 50 |
| SN4 | 20 | 70 | 10 |
| SN5 | 30 | 20 | 50 |
| SN6 | 30 | 60 | 10 |
The effectiveness of the self-emulsification process is judged by visual examination utilizing the subsequent grading system: Grade A: for nanoemulsion that formed rapidly within only 1 min depicting a clear transparent or bluish appearance. Grade B: for rapidly developed emulsions but appearing as slightly less clear or bluish white systems. Grade C: for the fine milky emulsions that obtained in 2 min. Grade D: for the slowly formed dull, greyish white and slightly oily emulsions (take more than 2 min to be formed). Grade E: systems showing poor emulsification with large oil droplets floating on the surface. Both grade A and B systems are expected to form nanoemulsions after dispersion in gastrointestinal tract [30]. Thus, only the formulations that could form either grade A or B nanoemulsions were selected and incorporated with Cuphea ignea extract (30 mg/mL) and kept for additional evaluation.
2.10. Evaluation of thermodynamic stability
In order to assess the ability of SNEDDS to solubilize Cuphea ignea extract with the acceptable stability. SNEDDSs were subjected to centrifugation at 3500 rpm for 30 min. The SNEDDS formulations which exhibited stability without any precipitation of the extract or phase separation of the SNEDDSs components were exposed to 6 heating cooling cycles (45 °C and 4 °C). Additionally, passed formulations were further exposed to 6 freeze thaw cycles (between temperatures −21 °C and +25 °C); the formulations were stored not less than 48 h at every single temperature [31].
2.11. Assessment of robustness to dilution and phase separation
With the purpose of simulating the physiological dilution processing of the formulated Cuphea ignea SNEDDS post oral ingestion, the formulations were exposed to 50, 100 and 1000 fold dilution with diverse aqueous phases comprising distilled water, 0.1 N HCl and phosphate buffer pH 6.8. The diluted formulations were stirred on magnetic stirrer till formation of homogenous nanoemulsions. The formed nanoemulsions were kept for 2 h, before being subjected to visual inspection for the transparency of the nanoemulsions and the presence of any separation of the SNEDDS components or precipitation of the extract [32].
2.12. Determination of emulsification time and % transmittance
The emulsification time of SNEDDS is a significant indicator to assess the efficiency of the emulsification. The test was conducted in the dissolution equipment (Hanson Research, USA) operated with USP type II apparatus. A constant amount of each formulation (1 g) was put into the dissolution vessels filled with distilled water (500 mL), the temperature was previously adjusted to 37 °C and the paddle was adjusted to rotate at a speed of 50 rpm to permit mild agitation. The time taken till obtaining a clear homogenous solution was recorded [30].
The transparency of the obtained nanoemulsions was investigated by determining the percentage of transmittance at wavelength of 650 nm utilizing U.V spectrophotometer and distilled water was used as a blank [33].
2.13. In vitro dissolution study
Dissolution study of Cuphea ignea loaded SNEDDSs was done in 0.1 N HCl as a discriminative dissolution medium (900 mL maintained at 37 °C) of using paddle apparatus equipped dissolution instrument (Hanson Research, USA). Samples of SNEDDSs (1 mL) and the suspension of the extract that contains 30 mg of the extract, were dropped into the dissolution media. Samples of dissolution media (5 mL) were withdrawn at predetermined time intervals (5, 10, 15, 30, 45 and 60 min) and the volume replacement was done by fresh dissolution media. The percent dissolved of the phenolic compounds in the extract was analyzed by measuring the absorbance of the samples at 260 nm using UV spectrophotometer, using 0.1 N HCl as a blank. UV spectroscopy was selected as a method for determination of the total phenolic compounds as it was reported as a suitable quantification method of the phenolic compounds as a result of the ability of the phenolic components to strongly absorb the UV light [34]. It was reported that the majority of phenolic compounds show maximum UV absorption at 260 nm [35].
2.14. Droplet size, polydispersity index (PDI) and zeta potential analysis
The formulated Cuphea ignea SNEDDSs were diluted 100-fold using deionized water and vortexed for 30 s to permit the formation of nanoemulsion. Dynamic light scattering technique was used to record the droplet size and PDI of the obtained nanoemulsions employing Malvern Zetasizer 3000, Malvern, U.K. In order to judge the physical stability of the attained nanoemulsions, zeta potential values of the samples (after dilution with water) were also determined. The results were presented as the mean values of 3 measurements ± standard deviation (SD).
2.15. Transmission electron microscope (TEM)
The morphology of the optimum Cuphea ignea loaded nanoemulsion was examined by transmission electron microscope, (H-7650, Hitachi, Japan), after diluting the optimum formulation with distilled water. One drop of the nanoemulsion was put on a film-coated copper grid before negatively stained with 2% (w/v) phosphotungstic acid solution. The sample was photographed after drying by TEM.
2.16. In vitro cytotoxicity and antiviral activity
2.16.1. CC50 and IC50 calculation using crystal violet assay
The assay was performed according to the procedure that was previously described [36] with minor modifications. Vero E6 cells were seeded into 96-well plates in 100 μL of DMEM Complete Medium containing DMEM high glucose medium with 2 mM l-glutamine, 1 mM sodium pyruvate, and 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin. After 24 h (90–100% confluence monolayer of Vero E6), each compound was diluted using infection DMEM into varying concentrations in a separate U shape 96 well plate (with a range of concentration from 10 μg/mL to 1 ng/mL). An aliquot of 100 μL of each dilution was transferred into new U shape 96 well plate and supplemented with 100 TCID50 in 100 μL infection media. In parallel the wells dedicated for CC50 calculation were supplemented with 100 μL infection media without virus. Aliquots of 100 μL infection media containing 100 TCID50 were used as virus control. After 1 h of incubation, 100 μL of each well were transferred to their corresponding wells into the 96-well plates containing Vero E6 cultures. The plates were incubated for 72 h, the cell monolayers were washed with PBS and subjected to cell fixation using 100 μL of 10% formaldehyde for 1 h. Subsequently, the plates are washed well for 3 times with 1x PBS and dried well before staining with 50 μL (0.5%) crystal violet to each well (0.5 g) crystal violet powder, 80 mL distilled H2O and 20 mL methanol)) for 30 min. The plates were then washed well with rinsed water and air-dried at room temperature for 2–24 h. To distain crystal violet, 200 μL methanol were added to each well, and the plate was incubated with its lid on a bench rocker (20 oscillations/minute) for 20 min at room temperature. Finally, the optical density of each well at 590 nm (OD590) was measured with a plate reader. 100% percent was assigned to non-treated control cells, the average OD of each dilution without or with virus was compared to control cells and control virus wells to calculate % toxicity and % reduction in virus replication (respectively). The concentration that is toxic for 50% of the cells (CC50) and the concentration that cause 50% viral inhibition (IC50) values were calculated using nonlinear regression (three parameters) in GraphPad Prism 5.01.
3. Results and discussion
3.1. Identification and quantitative estimation of the phenolic compounds content
Fifteen phenolic compounds were detected and quantified using HPLC/UV detector as shown in Table 2 . Nine phenolic acids were identified such as p-coumaric acid (5.434 mg/g), gallic acid (1.671 mg/g), syringic acid (0.850 mg/g) and rosmarinic acid (0.107 mg/g). Concerning flavonoids, myricetin 3-O-rhamnoside (4.27 mg/g), rutin (1.399 mg/g), quercetin (0.281 mg/g), naringenin (0.195 mg/g) and catechin (0.024 mg/g) were identified in the ethanolic extract of the leaves. The stilbenoid resveratrol (0.371 mg/g) was also detected.
Table 2.
Phenolic compounds identified in the ethanolic extract of Cuphea ignea Leaves using HPLC/UV analysis.
| No. | Identified compounds | (mg/g) |
|---|---|---|
| 1 | p- Coumaric acid | 5.43 |
| 2 | Myricetin 3-O-rhamnoside | 4.27 |
| 3 | Gallic acid | 1.67 |
| 4 | Rutin | 1.40 |
| 5 | Syringic acid | 0.85 |
| 6 | O- Coumaric acid | 0.44 |
| 7 | Resveratrol | 0.37 |
| 8 | Quercetin | 0.28 |
| 9 | Naringenin | 0.19 |
| 10 | Rosmarinic acid | 0.11 |
| 11 | Cinnamic acid | 0.07 |
| 12 | Caffeic acid | 0.06 |
| 13 | Chlorogenic acid | 0.03 |
| 14 | Vanillic acid | 0.03 |
| 15 | Catechin | 0.02 |
Phenolics acids and flavonoids are considered the most abundant bioactive groups of secondary metabolites produced in the plant kingdom. Both classes show different biological activities including antioxidant, anticancer, antiulcer, anti-inflammatory, antispasmodic and antimicrobial activities [37]. Phenolic acids were reported to accomplish potent inhibitory action on different viruses for instance influenza virus, hepatitis C virus, human immunodeficiency virus and herpes simplex virus [38].
3.2. Docking studies
The binding site of SARS-CoV-2 Mpro contains a catalytic dyad (Cys–His) and the N3 inhibitor fitted inside its pocket asymmetrically. Molecular docking of p-Coumaric acid (1), Myricetin-3-O-rhamnoside (2), Gallic acid (3), Rutin (4), Syringic acid (5), O-Coumaric acid (6), Resveratrol (7), Quercetin (8), Naringenin (9), Rosmarinic acid (10), Cinnamic acid (11), Caffeic acid (12), Chlorogenic acid (13), Vanillic acid (14), Catechin (15), and N3 inhibitor (16) into the Mpro binding pocket was performed. They got stabilized inside the binding site by promising score values and several interactions with the receptor amino acids. The descending order of the tested compounds according to their binding scores was as follow: N3 natural inhibitor (docked, 16) ˃ Rutin (4) ˃ Myricetin-3-O-rhamnoside (2) ˃ Rosmarinic acid (10) ˃ Chlorogenic acid (13) ˃ Quercetin (8) ˃ Catechin (15) ˃ Naringenin (9) ˃ Resveratrol (7) ˃ Syringic acid (5) ˃ Caffeic acid (12) ˃ O-Coumaric acid (6), p- Coumaric acid (1) ˃ Vanillic acid (14) ˃ Cinnamic acid (11) ˃ Gallic acid (3). The binding scores and binding modes with the amino acids of SARS-CoV-2 Mpro pocket are depicted in Table 3 and supplementary data.
Table 3.
The binding scores and binding modes of the examined compounds from Cuphea ignea A. DC leaves extract (1-15) and N3 (docked, 16) inside the binding site of the SARS-CoV-2 main protease (Mpro).
| No. | Compound | Sa | RMSDb | Amino acid bond | Distance (A֯) |
|---|---|---|---|---|---|
| 1 | p-Coumaric acid | −4.75 | 1.81 | Glu166/H-donor His41/H-pi |
2.91 3.35 |
| 2 | Myricetin 3-O-rhamnoside | −7.86 | 1.11 |
Glu166/H-donor Met165/H-donor |
3.01 3.23 |
| 3 | Gallic acid | −4.52 | 0.85 | His164/H-donor | 3.04 |
| 4 | Rutin | −9.28 | 1.19 |
Glu166/H-donor Thr26/H-donor |
3.01 3.05 |
| 5 | Syringic acid | −5.18 | 1.52 | His164/H-donor | 3.03 |
| 6 | O-Coumaric acid | −4.78 | 1.02 | Glu166/H-donor | 2.92 |
| 7 | Resveratol | −5.79 | 1.44 | Glu 166/pi-H | 4.61 |
| 8 | Quercetin | −6.33 | 1.43 | Leu141/H-donor Gly143/H-acceptor Met165/H-donor Met165/pi-H |
2.69 3.46 3.78 4.57 |
| 9 | Naringenin | −6.15 | 1.05 | Glu166/H-acceptor Asn187/H-donor |
3.03 3.20 |
| 10 | Rosmarinic acid | −7.05 | 1.56 |
His41/H-acceptor Cys145/H-donor Thr25/pi-H |
3.12 4.38 4.08 |
| 11 | Cinnamic acid | −4.59 | 1.26 | Met166/pi-H Glu166/pi-H |
3.45 4.53 |
| 12 | Caffeic acid | −4.84 | 0.92 | Glu166/H-donor | 2.88 |
| 13 | Chlorogenic acid | −6.58 | 1.26 | Leu141/H-donor Glu166/H-acceptor Gln192/H-acceptor |
2.92 3.05 3.13 |
| 14 | Vanillic acid | −4.73 | 1.85 | – | – |
| 15 | Catechin | −6.23 | 1.64 | Arg188/H-donor Phe140/H-donor Glu166/H-donor |
2.91 3.05 3.12 |
| 16 | N3 | −10.11 | 1.86 | Gln189/H-donor Glu166/H-acceptor His41/H-pi Asn142/pi-H |
2.99 3.04 3.63 4.47 |
S: the score of a compound inside the protein binding pocket (Kcal/mol).
RMSD: The Root Mean Squared Deviation between the predicted pose and the crystal structure.
The results of docking simulation showed that Rutin (4), Myricetin-3-O-rhamnoside (2), and Rosmarinic acid (10) have the best binding scores towards SARS-CoV-2 main protease equal to −9.28, −7.86, and −7.05 kcal/mol, respectively (Table 3). These values were close to the binding energy of the docked N3 inhibitor (S = −10.11 kcal/mol).
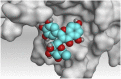
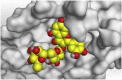
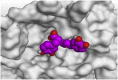
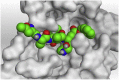
The detailed binding mode of N3 was; the docked N3 inhibitor fitted the asymmetric pocket of SARS-CoV-2 Mpro through the formation of two hydrogen bonds with Gln189 and Glu166 at 2.99 and 3.04 A֯, respectively. Besides, it formed one H-pi interaction with His41 and one pi-H interaction with Asn142 at 3.63 and 4.47 A֯, respectively. On the other hand, Rutin (4) occupied the Mpro pocket by forming two hydrogen bonds with Glu166 and Thr26 at 3.01 and 3.05 A֯, respectively. However, Myricetin-3-O-rhamnoside (2) showed two hydrogen bonds with Glu166 and Met165 at 3.01 and 3.23 A֯, respectively. Moreover, Rosmarinic acid (10) formed two H-bonds with His41 and Met145, and a pi-H interaction with Thr25 at 3.12, 4.38, and 4.08 A֯, respectively (Table 4 ).
Table 4.
3 D pictures of the binding interactions and the Mpro positioning between the most promising tested compounds of Cuphea ignea A. DC leaves extract (Myricetin-3-O-rhamnoside (2), Rutin (4), and Rosmarinic acid (10)) and N3-binding site compared to the N3 (docked, 16). Red dash represents H-bonds and black dash represent H-pi interactions.
| Compound | 3 D binding interactions | 3 D Mpro positioning |
|---|---|---|
| Myricetin-3-O-rhamnoside (2) | 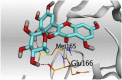 |
 |
| Rutin (4) | 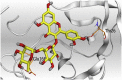 |
 |
| Rosmarinic acid (10) | 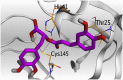 |
 |
| N3 (docked, 16) | 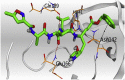 |
 |
In general, the docking results of the tested compounds of Cuphea ignea extract, compared to N3, presented a good idea about their binding affinities towards SARS-CoV-2 main protease. Some compounds of the extract showed ideal and promising binding, which corresponds to high affinities and predicted intrinsic activities as well.
3.3. Preparation of Cuphea ignea A. DC. SNEDDS and assessment of dispersibility
Results revealed that SN1 and SN2 formed clear, isotropic, transparent grade A nanoemulsions of low viscosity judged by visual inspection. The ability of these formulations to form nanoemulsions can be attributed to the presence of low oil concentration (10%) along with sufficient amount of surfactant (40% and 80% in case of SN1 and SN2, respectively). On the other hand, SN4 and SN6 produced emulsions of grade C while SN3 and SN5 yielded grade D coarse emulsions. This can be justified by the presence of higher surfactant content of SN4 and SN6 (70% and 60%, respectively), while the surfactant concentrations in both SN5 and SN3 were not high enough to solubilize the relatively high concentrations of oleic acid (20% and 30%, respectively). Based on the results, only SN1 and SN2 were selected to be loaded with Cuphea ignea extract; both formulations were prepared and loaded with the same amount of the extract (30 mg/mL).
3.4. Evaluation of thermodynamic stability
Both SN1 and SN2 exhibited good thermodynamic stability and the ability to tolerate the centrifugation, storage at the various stress cycles. The prepared systems did not depict any phase separation of the SNEDDS components or precipitation of the extract.
3.5. Assessment of robustness to dilution and phase separation
It is vital to guarantee that uniform size nanoemulsions are obtained through self-emulsification upon dilution of the SNEDDS with different media at various concentrations. The developed Cuphea ignea extract loaded SNEDDS formulations were subjected to dilution with various media with the purpose of simulating the in vivo status and predicting the possible phase separation of SNEDDS components or precipitation of the extract in the gastrointestinal tract where SNEDDSs are exposed to gradual dilution [39]. Visual assessment revealed that both SN1 and SN2 upon dilution with all the investigated media including distilled water, 0.1 N HCl and phosphate buffer (pH 6.8) at 50, 100 and 1000 fold dilutions exhibited formation of transparent nanoemulsions without precipitation of the extract. These observations confirmed that both SN1 and SN2 were robust to dilutions.
3.6. Determination of emulsification time and % transmittance
It was noticed that emulsification rate of SN1 was significantly faster than that of SN2, as SN1 took only 5 ± 1 min to disperse and emulsify in water while SN2 took 12.5 ± 1.5 min (p < 0.05; student t-test). This may be justified by the presence of higher surfactant (tween 20) concentration in SN2 than SN1 (80% and 40%, respectively) which impart SN2 higher viscosity than SN1. Recording % transmittance is important to ensure the formation of clear systems with high transparency upon dilution of SNEDDS. % transmittance of SN1 and SN2 were 88.9% ± 0.6 and 95.8% ± 0.66, respectively; this result indicates that clear nanoemulsion was formed upon dispersion of SNEDDSs in water. The statistically significant higher transmittance of SN2 than SN1 (p < 0.05; student t-test) is attributed to its higher content of tween 20 than SN1 which may lead to the formation of a more packed film of the surfactant at the oil–water interface.
3.7. Droplet size, polydispersity index and zeta potential analysis
Droplet size is an important tool in self-emulsification process, as it judges the rate and extent of dissolution. Moreover, the smaller the droplet size, the greater the interfacial area that is available for drug absorption [40]. Moreover, reducing the droplet size permits the formation of a more packed surfactant film at the oil–water interface, thus impart stability to the oil droplets [41]. The polydispersity index (PDI) is a dimensionless measure of the width of size distribution attained through the cumulative analysis, it is calculated by dividing the standard deviation by the mean droplet size. The values of PDI are usually within the range of 0–1. A smaller PDI value signifies a monodispersed and homogenous droplet size distribution while a greater PDI values represents a broader size distribution and less uniform samples [42]. It is observable from the results that the mean droplet size of SN1 was 119.65 ± 8.7 nm and PDI value was 0.247 ± 0.02 indicating monodispersed droplet size distribution within the formulation; while SN2 exhibited statistically significant larger droplet size of 164 ± 10.4 nm and larger PDI value of 0.62 ± 0.12 (p < 0.05; student t-test).
Measuring zeta potential of the formulations were important to judge the stability of the formulations since larger values of zeta potential refer to higher protection against coalescence and aggregate formation which can be maintained by the repulsive forces present on the surfaces of the droplets. The values of zeta potential of SN1 and SN2 were −28.30 ± 0.52 and −16.26 ± 2.5 mV, respectively. The presence of negatively charged droplets is justified by the incorporation of oleic acid in the formulated SNEDDSs.
3.8. In vitro dissolution study
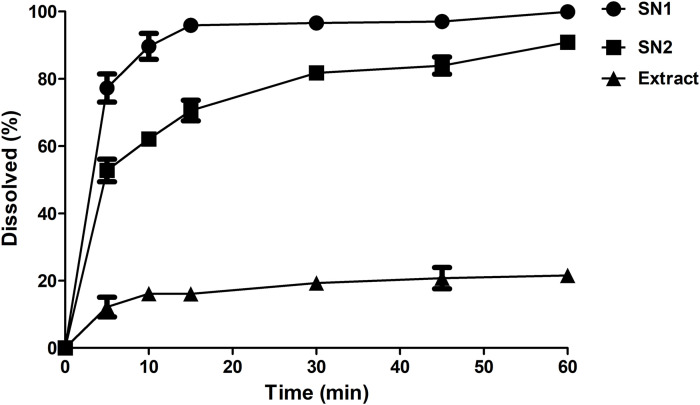
The in vivo bioavailability data for herbal medicines is not readily attainable as a result of the complexity of their composition such as the diversity of the components present in a single extract; and since the potency of a given herbal extract is most probably attributed to the synergism between several compounds [43]. Thus, a discriminative in vitro dissolution study that can estimate the solubility of the extract components and judge the superiority of the developed formulation over the crude extract is beneficial in that case as an indicative and alternative measure to predict the in vivo behavior. Dissolution studies were performed selecting 0.1 N HCl as a discriminating dissolution medium. Dissolution profiles of Cuphea ignea extract from SNEDDSs (SN1 and SN2) were found to be greater as compared with that of the suspension of the crude extract powder. Results are shown in Fig. 1 . It was observed that after 5 min, the highest % extract dissolved was achieved in case of SN1 as it could dissolve 77.25 ± 4.1% of the extract, followed by SN2 that dissolved 52.76 ± 3.3%, while only 12.17 ± 2.9% dissolved in case of the crude extract. Moreover, complete dissolution of the extract was observed after 60 min from SN1 (100 ± 0.1%) while the maximum % dissolved from the crude extract was as low as 21.56 ± 2.2% at the end of the experiment (60 min).
Fig. 1.
In vitro dissolution profile of Cuphea ignea extract from the prepared SNEDDSs and the extract suspension in 0.1 N HCl as a discriminative dissolution medium.
By applying ANOVA statistical analysis, results proved that there is a statistically significant enhancement of the dissolution profile of Cuphea ignea extract (P < 0.05) from the formulated SNEDDSs as a result of the spontaneously formed nanoemulsion that possesses a fine droplet size facilitating a faster and enhanced dissolution profile of the extract in the aqueous phases. The faster dissolution rate of SN1 than SN2 can be attributed to the faster emulsification rate of SN1 than SN2 and the smaller droplet size of SN1 that provides larger interfacial area for drug dissolution. Based the abovementioned emulsification rate, droplet size, PDI, zeta potential and dissolution results, SN1 was selected as the optimum formulation for further investigation. Generally, the rapid dissolution profile that was offered by the SNEDDSs formulations is beneficial because polyphenolic compounds exhibited degradation in the neutral or basic pH environments of the gastrointestinal tract; consequently a successful drug delivery system should be amenable to exhibit rapid and complete dissolution in the acidic pH in the stomach [21].
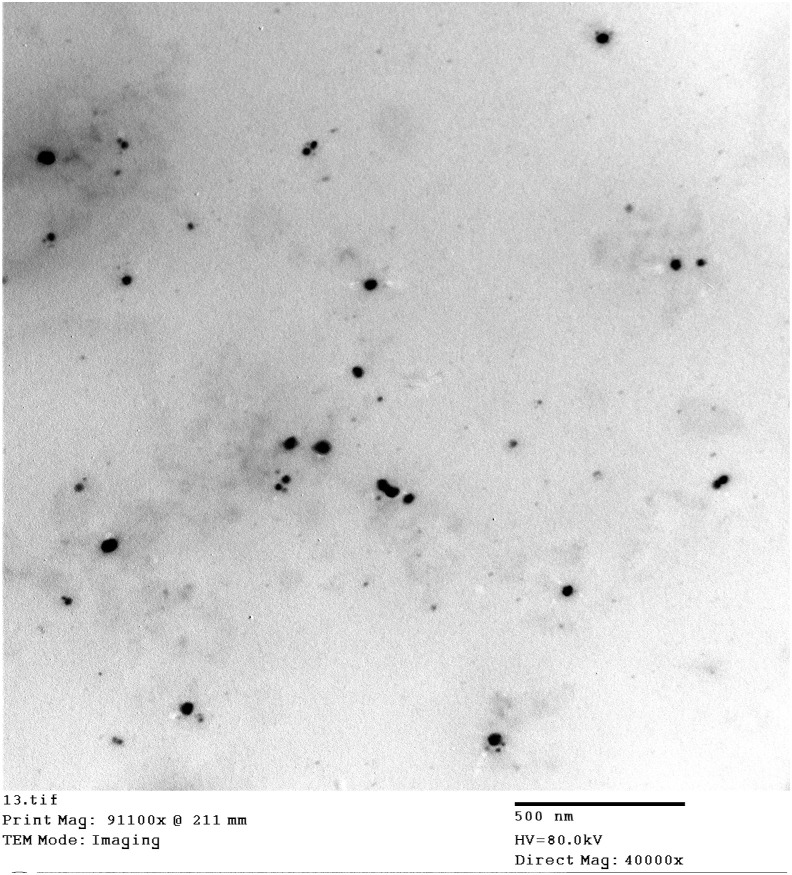
3.9. Transmission electron microscope
Based on the results of the in vitro dissolution study, droplet size and zeta potential analysis, SN1 was selected as the optimum formulation for the further evaluation. The photograph of the transmission electron microscopy of the optimum SNEDD formulation (SN1) was illustrated in Fig. 2 . It could be noticed that small spherical and homogeneous nanoemulsion droplets were obtained as a result of dilution of SNEDDS with water. The observed droplet sizes were smaller than the size obtained with zetasizer analysis as the dynamic light scattering technique is a multiangle measuring method that determines the hydrodynamic radius of the droplet dependent on the intensity of the light scattering. On the other hand, Transmission electron microscopy measures the actual droplet size [44,45].
Fig. 2.
TEM photographs of SN1 depict the small spherical and homogeneous nanoemulsion droplets that formed upon dilution with water.
3.10. In vitro cytotoxicity and antiviral activity
3.10.1. CC50 and IC50 calculation using crystal violet assay
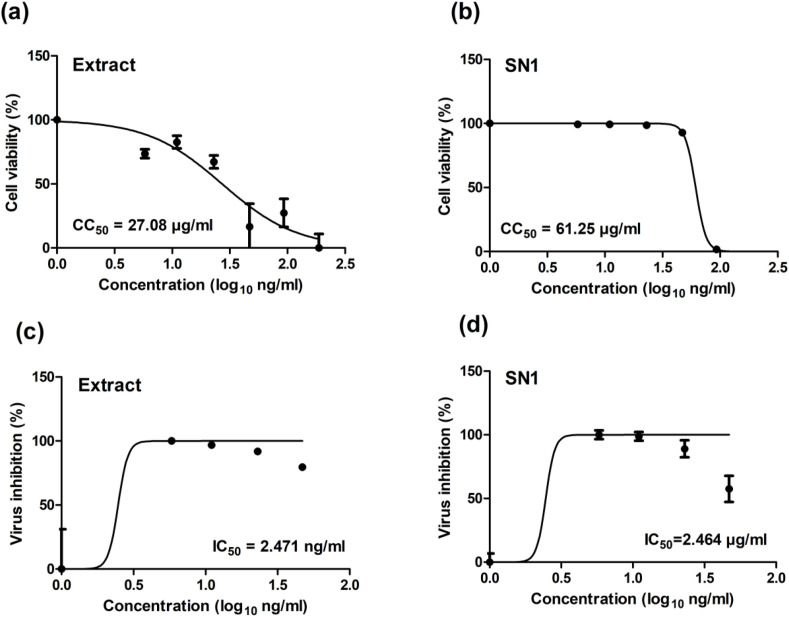
The cytotoxicity of Cuphea ignea extract solution in DMSO and the selected Cuphea ignea self-nanoemulsifying formulation (SN1) in Vero-E6 cells were determined by MTT assay and the results revealed that the CC50 values of DMSO solution of the extract and SN1 were 27.08 and 61.25 μg/ml, respectively (Fig. 3 a and b).
Fig. 3.
Graphs of the determination of (a) CC50.of Cuphea ignea extract in DMSO (b) CC50 of the formulated Cuphea ignea self-nanoemulsifying system (SN1) (c) IC50 of Cuphea ignea extract in DMSO (d) IC50 of the formulated Cuphea ignea self-nanoemulsifying system (SN1).
The antiviral activities of Cuphea ignea extract solution in DMSO and Cuphea ignea self-nanoemulsifying formulation (SN1) were investigated based on the dose–response. Analysis of cytotoxicity was conducted to guarantee that the cell death was due to the inhibitory action of the tested samples of Cuphea ignea extracts on SARS-CoV-2 virus and that the reduction in viral titre was not related to cell death and to determine the safe concentrations for IC50 calculations. The results of antiviral action of Cuphea ignea formulations are shown in Fig. 3c and d. It is observable that both Cuphea ignea extract solution in DMSO and Cuphea ignea self-nanoemulsifying formulation (SN1) exhibited almost statistically insignificant values of the IC50 of 2.47 and 2.46 μg/ml, respectively. Accordingly, the selectivity indexes were calculated as the ratio of CC50 to IC50 of each sample; it was noticed that the SI of SN1 was 25 while the SI of thee extract solution in DMSO was 11 signifying a statistically significant augmentation in the therapeutic activity of SN1 (p < 0.05; student t-test). These results demonstrated that formulation of Cuphea ignea extract in the developed self-nanoemulsifying was capable of retaining the antiviral activity and enhancing the therapeutic index of Cuphea ignea extract; and it could inhibit the virus completely (100% reduction in virus replication) at a concentration as low as 5.87 μg/ml obtained from dose-response measurements. It is noteworthy to mention that there are safety concerns regarding the use of DMSO as a vehicle for solubilization of drugs to be administered in vivo and other solubilizing methods are highly recommended due to the reported toxic effects of DMSO [46] including ocular and renal toxicities [47]. Additionally, other adverse reactions, such as abdominal cramps, dyspnea, flushing and cardiovascular reactions, were linked to the induction of histamine release by DMSO [48].
Thus, proposing a formulation that could retain the same antiviral activity of Cuphea ignea extract that is obtained by dissolving the extract in DMSO was of great significance.
The significant inhibition of SARS-CoV-2 by Cuphea ignea extract can be attributed to the phenolic compounds content that were identified in the ethanolic extract of the leaves and these findings are in accordance with our results of molecular docking studies that proposed the promising affinity of the tested components of Cuphea ignea extract against SARS-CoV-2 main protease, especially rutin, myricetin-3-O-rhamnoside, and rosmarinic acid.
Additionally, it was reported in literature that gallic and caffeic acids showed antiviral properties by different mechanisms of action [38]. Furthermore, recent in silico studies showed the efficient ability of gallic acid, quercetin, resveratrol and naringenin in inhibition of the internalization of SARS-CoV-2 by binding to its cellular receptor angiotensin-converting enzyme 2 (ACE 2) [49]. Other in silico study reported that quercetin showed better inhibition compared to hydroxychloroquine against SAR-CoV-2 main protease active site and the receptor ACE 2 [50]. Additionally, molecular docking has recently depicted that naringenin possess the ability to significantly inhibit SARS-CoV-2 via suppressing its main protease and reducing ACE 2 receptors activity [51]. Although each polyphenolic compound has dissimilar structure, polyphenolic compounds generally share the same chemical characters due to the existence of phenolic rings substituted with hydroxyl groups. Consequently, it was hypothesized that, various categories of polyphenolic compounds may exert comparable antiviral activities while involving different mechanisms of action. Based on this hypothesis, it was of great importance to scrutinize the antiviral activity of the natural polyphenolic Cuphea ignea extract, instead of the single purified compounds. According to these results, it is suggested that the intriguing antiviral activity of the extract and the formulation may be attributed to a possible synergism between the extract components.
4. Conclusion
To summarize, fifteen compounds of the Cuphea ignea A. DC leaves were studied using molecular docking against SARS-CoV-2 Mpro. The tested compounds showed different affinities compared to N3 inhibitor towards COVID-19 main protease as follows: N3 natural inhibitor (docked, 16) ˃ rutin (4) ˃ myricetin-3-o-rhamnoside (2) ˃ rosmarinic acid (10) ˃ chlorogenic acid (13) ˃ quercetin (8) ˃ catechin (15) ˃ naringenin (9) ˃ resveratrol (7) ˃ syringic acid (5) ˃ caffeic acid (12) ˃ o-coumaric acid (6), p-coumaric acid (1) ˃ vanillic acid (14) ˃ cinnamic acid (11) ˃ gallic acid (3). However, rutin (4) is recommended for more preclinical and clinical studies against COVID-19. It may be tested either alone or in combination with Myricetin-3-O-rhamnoside (2) or Rosmarinic acid (10).
Moreover, a self-nanoemulsifying formulation of Cuphea ignea extract was successfully designed. The optimum SNEDDS was formulated utilizing 10% oleic acid, 40% tween 20, 50% propylene glycol and loaded with the extract at a concentration of 30 mg/ml; it exhibited a good thermodynamic stability. The optimum SNEDDS could rapidly form grade A nanoemulsion upon dilution with water or aqueous phases. The obtained nanoemulsion was of small droplet size, narrow size distribution and optimal zeta potential. The dissolution profile of the SNEDDS was excellent when compared to the crude extract as polyphenolic compounds could dissolve completely in 0.1 N HCl in short time, this is expected to ameliorate the absorption in acidic pH before degradation in the intestine. The developed SNEDDS could inhibit SARS-CoV-2 completely at a concentration as low as 5.87 μg/ml, this potent antiviral activity may suggest the possible occurrence of a synergism between the extract components. In the light of these results, it was of great importance to scrutinize the antiviral activity of Cuphea ignea total ethanolic extract where the significant inhibition activity can be attributed to the phenolic compounds identified by the HPLC technique. Finally, SNEDDS offers an attractive formulation strategy to enable pharmaceutical application of Cuphea ignea extract in future clinical studies as a promising natural therapy for Covid-19.
Authors' contributions
Conceptualization: D. B. Mahmoud, W. M. Ismail; methodology: formulation and in vitro characterization:: D. B. Mahmoud, Plant extraction and standardization: W. M. Ismail, virology in vitro studies: Y. Moatasim, O.Kutkut, M. R. Gomaa, A. Mostafa, A.Kandeil; Docking studies: A. Karmalawy writing—original draft preparation: D. B. Mahmoud, W. M. Ismail, Y. Moatasim, O.Kutkut, M. R. Gomaa, A. Karmalawy, A. Mostafa, A.Kandeil; review and editing:, D. B. Mahmoud, A. N. ElMeshad, W. M. Ismail, A. S. M. Ezzat, K. S. El Deeb, A. M. El-Fishawy, Karmalawy, A. Mostafa, A.Kandeil, M. A. Ali; supervision, A. N. ElMeshad, S. M. Ezzat, K. S. El Deeb, A. M. El-Fishawy, M. A. Ali.
Funding
This research was funded by NRC under contract number MP120801 and by Egyptian Academy of Scientific Research & Technology (ASRT) within the “Ideation Fund” program under contract number 7303. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
This work has been submitted for patent application to patent office Academy of Scientific Research and Technology, Egypt under number 2020/1698.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jddst.2021.102845.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wang M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J.F.-W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Coronavirus disease ( COVID-19): global epidemiological situation report. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20201012-weekly-epi-update-9.pdf Available from:
- 5.Annunziata G., et al. May polyphenols have a role against coronavirus infection? An overview of in vitro evidence. Front. Med. 2020;7 doi: 10.3389/fmed.2020.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahmoud D.B., Shitu Z., Mostafa A. Drug repurposing of nitazoxanide: can it be an effective therapy for COVID-19? J. Genetic Eng. Biotechnol. 2020;18(1):1–10. doi: 10.1186/s43141-020-00055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X., et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brogi S. Multidisciplinary Digital Publishing Institute; 2019. Computational Approaches for Drug Discovery. [Google Scholar]
- 9.Lai M., Perlman S., Anderson L. In: Coronaviridae in Fields Virology. Knipe H.P., editor. Williams Lippincott, & Wilkins; Philadelphia, USA: 2007. pp. 1305–1336. DM. [Google Scholar]
- 10.Voss A., et al. Publishing in face of the COVID-19 pandemic. Int. J. Antimicrob. Agents. 2020;56(1) doi: 10.1016/j.ijantimicag.2020.106081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alnajjar R., et al. Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6(12) doi: 10.1016/j.heliyon.2020.e05641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaki A.A., et al. Molecular docking reveals the potential of Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J. Chem. 2020;44(39):16752–16758. [Google Scholar]
- 13.Hassan S.K., et al. Antitumor activity of Cuphea ignea extract against benzo (a) pyrene-induced lung tumorigenesis in Swiss Albino mice. Toxicol. Reports. 2019;6:1071–1085. doi: 10.1016/j.toxrep.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ismail W.M., et al. Angiotensin-converting enzyme and renin inhibition activities, antioxidant properties, phenolic and flavonoid contents of Cuphea ignea A. DC. J. Rep. Pharm. Sci. 2020;9(1):92. [Google Scholar]
- 15.Mousa A.M., et al. Antiulcerogenic effect of Cuphea ignea extract against ethanol-induced gastric ulcer in rats. BMC Compl. Alternative Med. 2019;19(1):1–13. doi: 10.1186/s12906-019-2760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C.-j., et al. Polygonum cuspidatum and its active components inhibit replication of the influenza virus through toll-like receptor 9-induced interferon beta expression. PloS One. 2015;10(2) doi: 10.1371/journal.pone.0117602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T., et al. Resveratrol inhibits the TRIF-dependent pathway by upregulating sterile alpha and armadillo motif protein, contributing to anti-inflammatory effects after respiratory syncytial virus infection. J. Virol. 2014;88(8):4229–4236. doi: 10.1128/JVI.03637-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mastromarino P., et al. Resveratrol inhibits rhinovirus replication and expression of inflammatory mediators in nasal epithelia. Antivir. Res. 2015;123:15–21. doi: 10.1016/j.antiviral.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Zang N., et al. Resveratrol-mediated gamma interferon reduction prevents airway inflammation and airway hyperresponsiveness in respiratory syncytial virus-infected immunocompromised mice. J. Virol. 2011;85(24):13061–13068. doi: 10.1128/JVI.05869-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta S., Moses J.A., Anandharamakrishnan C. Encapsulation of nutraceutical ingredients in liposomes and their potential for cancer treatment. Nutr. Canc. 2018;70(8):1184–1198. doi: 10.1080/01635581.2018.1557212. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes I., et al. Bioavailability of anthocyanins and derivatives. J. Funct. Foods. 2014;7:54–66. [Google Scholar]
- 22.Shah N., et al. Self-emulsifying drug delivery systems (SEDDS) with polyglycolyzed glycerides for improving in vitro dissolution and oral absorption of lipophilic drugs. Int. J. Pharm. 1994;106(1):15–23. [Google Scholar]
- 23.Charman S.A., et al. Self-emulsifying drug delivery systems: formulation and biopharmaceutic evaluation of an investigational lipophilic compound. Pharmaceut. Res. 1992;9(1):87–93. doi: 10.1023/a:1018987928936. [DOI] [PubMed] [Google Scholar]
- 24.Inc C. Chemical Computing Group Inc; Montreal, QC, Canada: 2016. Molecular Operating Environment (MOE), 2013.08. [Google Scholar]
- 25.Al-Karmalawy A.A., Khattab M. Molecular modelling of mebendazole polymorphs as a potential colchicine binding site inhibitor. New J. Chem. 2020;44(33):13990–13996. [Google Scholar]
- 26.Jin Z., et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020:1–5. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 27.Ghanem A., et al. Tanshinone IIA synergistically enhances the antitumor activity of doxorubicin by interfering with the PI3K/AKT/mTOR pathway and inhibition of topoisomerase II: in vitro and molecular docking studies. New J. Chem. 2020;44(40):17374–17381. [Google Scholar]
- 28.Eliaa S.G., et al. Empagliflozin and doxorubicin synergistically inhibit the survival of triple-negative breast cancer cells via interfering with the mTOR pathway and inhibition of calmodulin: in vitro and molecular docking studies. ACS Pharmacol. Transl. Sci. 2020;3(6):1330–1338. doi: 10.1021/acsptsci.0c00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McConkey B.J., Sobolev V., Edelman M. The performance of current methods in ligand–protein docking. Curr. Sci. 2002:845–856. [Google Scholar]
- 30.Mahmoud D.B., Shukr M.H., Bendas E.R. In vitro and in vivo evaluation of self-nanoemulsifying drug delivery systems of cilostazol for oral and parenteral administration. Int. J. Pharm. 2014;476(1–2):60–69. doi: 10.1016/j.ijpharm.2014.09.045. [DOI] [PubMed] [Google Scholar]
- 31.Abouhussein D.M., Bahaa El Din Mahmoud D., Mohammad F E. Design of a liquid nano-sized drug delivery system with enhanced solubility of rivaroxaban for venous thromboembolism management in paediatric patients and emergency cases. J. Liposome Res. 2019;29(4):399–412. doi: 10.1080/08982104.2019.1576732. [DOI] [PubMed] [Google Scholar]
- 32.Ofokansi K., Chukwu K., Ugwuanyi S. The use of liquid self-microemulsifying drug delivery systems based on peanut oil/tween 80 in the delivery of griseofulvin. Drug Dev. Ind. Pharm. 2009;35(2):185–191. doi: 10.1080/03639040802244292. [DOI] [PubMed] [Google Scholar]
- 33.Date A.A., Nagarsenker M.S. Design and evaluation of microemulsions for improved parenteral delivery of propofol. AAPS PharmSciTech. 2008;9(1):138–145. doi: 10.1208/s12249-007-9023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aleixandre-Tudo J.L., du Toit W. Cold maceration application in red wine production and its effects on phenolic compounds: a review. LWT. 2018;95:200–208. [Google Scholar]
- 35.Aaby K., Ekeberg D., Skrede G. Characterization of phenolic compounds in strawberry (Fragaria× ananassa) fruits by different HPLC detectors and contribution of individual compounds to total antioxidant capacity. J. Agric. Food Chem. 2007;55(11):4395–4406. doi: 10.1021/jf0702592. [DOI] [PubMed] [Google Scholar]
- 36.Feoktistova M., Geserick P., Leverkus M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016 doi: 10.1101/pdb.prot087379. 2016, pdb.prot087379. [DOI] [PubMed] [Google Scholar]
- 37.Ghasemzadeh A., Ghasemzadeh N. Flavonoids and phenolic acids: role and biochemical activity in plants and human. J. Med. Plants Res. 2011;5(31):6697–6703. [Google Scholar]
- 38.Wu Y.-H., et al. Structure properties and mechanisms of action of naturally originated phenolic acids and their derivatives against human viral infections. Curr. Med. Chem. 2017;24(38):4279–4302. doi: 10.2174/0929867324666170815102917. [DOI] [PubMed] [Google Scholar]
- 39.Elnaggar Y.S., El-Massik M.A., Abdallah O.Y. Self-nanoemulsifying drug delivery systems of tamoxifen citrate: design and optimization. Int. J. Pharm. 2009;380(1–2):133–141. doi: 10.1016/j.ijpharm.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 40.Gershanik T., Benita S. Self-dispersing lipid formulations for improving oral absorption of lipophilic drugs. Eur. J. Pharm. Biopharm. 2000;50(1):179–188. doi: 10.1016/s0939-6411(00)00089-8. [DOI] [PubMed] [Google Scholar]
- 41.Cui J., et al. Enhancement of oral absorption of curcumin by self-microemulsifying drug delivery systems. Int. J. Pharm. 2009;371(1–2):148–155. doi: 10.1016/j.ijpharm.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 42.Mahmoud D.B., Afifi S.A., El Sayed N.S. Crown ether nanovesicles (crownsomes) repositioned phenytoin for healing of corneal ulcers. Mol. Pharm. 2020 doi: 10.1021/acs.molpharmaceut.0c00742. [DOI] [PubMed] [Google Scholar]
- 43.Pade D., Stavchansky S. Selection of bioavailability markers for herbal extracts based on in silico descriptors and their correlation to in vitro permeability. Mol. Pharm. 2008;5(4):665–671. doi: 10.1021/mp700153h. [DOI] [PubMed] [Google Scholar]
- 44.Delan W.K., et al. Formulation of simvastatin chitosan nanoparticles for controlled delivery in bone regeneration: optimization using Box-Behnken design, stability and in vivo study. Int. J. Pharm. 2020;577 doi: 10.1016/j.ijpharm.2020.119038. [DOI] [PubMed] [Google Scholar]
- 45.Kaasalainen M., et al. Size, stability, and porosity of mesoporous nanoparticles characterized with light scattering. Nanoscale Res. Lett. 2017;12(1):74. doi: 10.1186/s11671-017-1853-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galvao J., et al. Unexpected low‐dose toxicity of the universal solvent DMSO. Faseb. J. 2014;28(3):1317–1330. doi: 10.1096/fj.13-235440. [DOI] [PubMed] [Google Scholar]
- 47.Brayton C.F. Dimethyl sulfoxide (DMSO): a review. Cornell Vet. 1986;76(1):61–90. [PubMed] [Google Scholar]
- 48.Madsen B.K., et al. Adverse reactions of dimethyl sulfoxide in humans: a systematic review. F1000Research. 2018;7(1746) doi: 10.12688/f1000research.16642.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurya V.K., et al. VirusDisease; 2020. Structure-based Drug Designing for Potential Antiviral Activity of Selected Natural Products from Ayurveda against SARS-CoV-2 Spike Glycoprotein and its Cellular Receptor; pp. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekiou O., et al. 2020. In-silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin, Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine against COVID-19 Main Protease Active Site and ACE2. [Google Scholar]
- 51.Tutunchi H., et al. Naringenin, a flavanone with antiviral and anti‐inflammatory effects: a promising treatment strategy against COVID‐19. Phytother Res. 2020 doi: 10.1002/ptr.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.