Abstract
The mechanistic target of rapamycin (mTOR) is a kinase at the center of an evolutionarily conserved signaling pathway that orchestrates cell growth and metabolism. mTOR responds to an array of intra and extracellular stimuli and in turn controls multiple cellular anabolic and catabolic processes. Aberrant mTOR activity is associated with numerous diseases, with particularly profound impact on the nervous system. mTOR is found in two protein complexes, mTOR complex 1 (mTORC1) and 2 (mTORC2), which are governed by different upstream regulators and have distinct cellular actions. Mutations in genes encoding for mTOR regulators result in a collection of neurodevelopmental disorders known as mTORopathies. While these disorders can affect multiple organs, neuropsychiatric conditions such as epilepsy, intellectual disability, and autism spectrum disorder have a major impact on quality of life. The neuropsychiatric aspects of mTORopathies have been particularly challenging to treat in a clinical setting. Current therapeutic approaches center on rapamycin and its analogues, drugs that are administered systemically to inhibit mTOR activity. While these drugs show some clinical efficacy, adverse side effects, incomplete suppression of mTOR targets, and lack of specificity for mTORC1 or mTORC2 may limit their utility. An increased understanding of the neurobiology of mTOR and the underlying molecular, cellular and circuit mechanisms of mTOR-related disorders will facilitate the development of improved therapeutics. Animal models of mTORopathies have helped unravel the consequences of mTOR-pathway mutations in specific brain cell types and developmental stages, revealing an array of disease-related phenotypes. In this review we discuss current progress and potential future directions for the therapeutic treatment of mTORopathies with a focus on findings from genetic mouse models.
Keywords: mTORC1, mTORC2, rapamycin, Tuberous Sclerosis Complex, mTORopathy, epilepsy, neurodevelopmental disorders, Raptor, Rictor, TSC1, TSC2, PTEN
Introduction
In 1964, a scientific expedition on Easter Island led to the discovery of one of the most widely studied compounds today. This compound, named rapamycin after Rapa Nui (Easter Island), exhibited strong immunosuppressant and anti-tumor properties [1]. The target of rapamycin (TOR) was first reported in yeast in 1991 [2, 3] and a few years later followed the discovery of the mammalian TOR homologue [4, 5].
The serine/threonine-protein kinase mTOR (mechanistic target of rapamycin) was discovered just over 25 years ago [4, 5]. Initial studies on mTOR focused on its role in cancer, as it is a key regulator of cell growth and proliferation [6]. However, subsequent studies revealed broader roles for mTOR as a signaling hub, coordinating information between the intra and extracellular environment. mTOR was found to respond to various stimuli including amino acids, trophic factors and energy status and in turn regulate the balance between anabolic (i.e. protein synthesis) and catabolic (i.e. autophagy) processes [7].
mTOR’s role in protein synthesis made this pathway a point of interest for neuroscientists in the early 2000s. De novo protein synthesis had been identified as a requirement for long-term synaptic plasticity in neurons [8, 9] and mTOR was known to control the translational regulators ribosomal protein S6 kinase beta-1 (S6K1 or p70S6K1) and 4E binding proteins (4E-BPs) [10]. Indeed, in 2002, mTOR was shown to be required for the late phase of hippocampal long-term potentiation (LTP), as LTP was inhibited by rapamycin in brain slices [11]. From then on, the list of mTOR’s functions in neurons has steadily expanded. Today, mTOR signaling has been linked to fundamental neural processes such as progenitor proliferation, neuronal migration, cell survival, axon and dendrite development, membrane excitability and synaptic properties [12, 13]. Consistent with its multifaceted roles in the nervous system, deregulation of mTOR signaling is associated with numerous neurological and psychiatric disorders [14–16].
In this review we will discuss 1) how mutations in genes that encode mTOR regulators lead to disorders with shared neurological manifestations (“mTORopathies”), 2) the current status of therapeutic interventions for mTORopathies in patients and animal models, and 3) potential considerations for developing improved treatment strategies for mTOR-related disorders.
The mTOR complexes
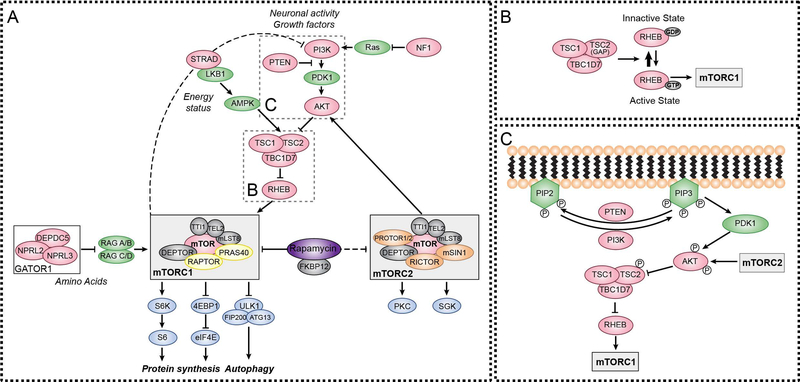
The regulatory-associated protein of mTOR (RAPTOR) was the first mTOR binding partner to be discovered and was identified as an obligatory component of mTOR complex 1 (mTORC1) [17, 18]. A few years later, a second mTOR complex was found (mTOR complex 2, mTORC2), after the discovery of a protein that bound to mTOR independently of RAPTOR, the rapamycin-insensitive companion of mTOR (RICTOR) [19]. mTORC1 and mTORC2 share some of the same protein components, while others are unique to each complex (shown in Fig. 1A). Shared proteins include the mTOR kinase itself, mTOR-associated protein, LST8 homolog (mLST8 also known as GβL), DEP domain-containing mTOR interacting protein (DEPTOR), TELO 2 interacting protein 1 (TTI1) and telomere maintenance 2 (TELO2) [20–22]. Specific to mTORC1 are RAPTOR and proline-rich AKT1 substrate 1 (AKT1S1 or PRAS40) [23]. Besides RICTOR, mTORC2 also contains proline-rich protein 5 or 5-like (PRR5/5L or PROTOR1/2) and target of rapamycin complex 2 subunit MAPKAP1 (MAPKAP1 or mSIN1) [24, 25] (shown in Fig. 1A).
Figure 1. Mutations in regulators of mTOR signaling cause mTORopathies.
(A) mTOR is a protein kinase found in two distinct multiprotein complexes, mTORC1 and mTORC2, which are composed of shared and unique protein components. Several upstream regulators collectively work to control the activity of the two complexes in response to various stimuli including growth factors and nutrients. Mutations in genes that encode for mTOR regulators (denoted in pink) result in neurodevelopmental disorders, collectively termed mTORopathies (see Table 1). Current treatments for mTORopathies include rapalogs, which are derivatives of rapamycin that suppress mTORC1 activity and indirectly inhibit mTORC2 signaling when administered chronically.
(B) The TSC complex functions as a GTPase activating protein (GAP) for the small GTPase Rheb, which is a direct activator of mTORC1.
(C) PI3K converts PIP2 into PIP3 via phosphorylation at the cell membrane. PIP3 recruits the kinase PDK1 that, along with mTORC2, phosphorylates and activates AKT. In turn, AKT phosphorylates TSC2, inhibiting the TSC1/TSC2/TBC1D7 complex, and promoting mTORC1 activity. PTEN is a phosphatase that negatively regulates mTOR signaling by dephosphorylating and converting PIP3 to PIP2.
Both complexes work as integrators of extra and intracellular signals to orchestrate cellular responses [12, 26] (shown in Fig. 1A). mTORC1 is regulated by various inputs including nutrients, growth factors, neuropeptides, and neurotransmitters [12]. In response to these stimuli, mTORC1 controls fundamental cellular processes such as protein synthesis, metabolism and autophagy. In neurons, mTORC1 controls differentiation, migration, cell morphology, physiology and synaptic properties [12, 13]. Our knowledge of the functions of mTORC2 in the central nervous system is still limited. Several studies, however, have shown that mTORC2 responds to growth factors, hormones and neurotransmitters to regulate cytoskeletal organization and thus can impact neuronal morphology and physiology [12].
mTORopathies
The critical importance of balanced mTOR signaling is underscored by the fact that mutations in genes encoding mTOR regulators cause neurodevelopmental disorders collectively termed mTORopathies (Table 1) [27, 28]. Some of the most well characterized genes associated with mTORopathies are TSC1, TSC2, PTEN, AKT, STRADA, and DEPDC5 [29]. While mutations in these genes can affect multiple organ systems, for this review we will focus on their shared neurologic and psychiatric manifestations, which can include cortical malformations, intellectual disability, epilepsy, and autism spectrum disorder (ASD).
Table 1.
mTORopathy genes, diseases and clinical manifestations
| Gene mutations | Associated Diseases and Syndromes | Clinical Manifestations* | Refs | ||
|---|---|---|---|---|---|
| Abbreviation | Neurological | Psychiatric | |||
|
TSC1
TSC2 |
Tuberous Sclerosis Complex | TSC | Tubers, SENs, SEGAs, Epilepsy, Infantile spasms, Altered white matter | ID, ASD, ADHD, other behavioral conditions | [36, 38, 42, 51–58] |
| PTEN | PTEN hamartoma tumor syndrome (incl. Cowden syndrome, Bannayan-Riley-Ruvalcaba syndrome and Lhermitte-Duclos disease) | PHTS | Macrocephaly | ID, ASD | [61, 63–67] |
|
MTOR
PIK3CA RHEB AKT3 TSC1 TSC2 DEPDC5 NPRL2 NPRL3 |
Malformations of cortical development | MCD | FCD, HME, ME, Epilepsy, Infantile spasms | ID | [68–74, 85–87] |
| NF1 | Neurofibromatosis type 1 | NF1 | Macrocephaly, Epilepsy, Ataxia, Altered white matter | ID, ASD, ADHD, Learning disabilities | [91] |
| STRADA | Polyhydramnios, megalencephaly, and symptomatic epilepsy syndrome | PMSE | ME, Epilepsy | ID, Psychomotor retardation | [90] |
| TBC1D7 | - | Macrocephaly/ME | ID | [59–60] | |
(ADHD) Attention deficit hyperactivity disorder, (ASD) Autism spectrum disorder, (FCD) Focal cortical dysplasia, (HME) Hemimegalencephaly, (ID) Intellectual disability, (ME) megalencephaly, (SENs) subependymal nodules, (SEGAs) subependymal giant cell astrocytomas
Listed are the primary neuropsychiatric presentations of these diseases as listed in the references noted, the NIH Genetic and Rare Diseases Information Center, and OMIM database. Other manifestations may be present in these disorders and not all patients may present with all manifestations listed here.
Tuberous Sclerosis Complex (TSC)
TSC is a multisystem developmental disorder with varying symptom severity caused by mutations in either the TSC1 or TSC2 genes that encode for the proteins tuberin and hamartin, respectively [30, 31]. TSC1, TSC2 and TBC1 domain family member 7 (TBC1D7) form a protein complex that acts as an essential negative regulator of mTORC1 [32–34] (shown in Fig. 1B). Loss of either TSC1 or TSC2 results in destabilization of the complex, leading to loss of its GTPase activating protein (GAP) activity towards the GTP-binding protein Rheb (RHEB), a direct activator of mTORC1 [35]. Loss-of-function (LoF) mutations in TSC1 or TSC2 therefore lead to constitutive mTORC1 activity.
TSC neuropathology includes focal malformations called tubers that contain enlarged and dysplastic neurons, astrocytes, and so called “giant” or “balloon” cells [36]. Tubers form during embryonic development and are primarily found in the cortex and occasionally in other regions such as the cerebellum [37, 38]. Tubers, or the peri-tuberal cortex can become seizure foci [39, 40] and may be surgically resected as a treatment for intractable epilepsy in TSC [41]. Approximately 80% of individuals with TSC develop benign growths emanating from the ventricular walls called subependymal nodules (SENs) [42]. In 5–15% of TSC patients, SENs can progress to benign glioneuronal tumors called subependymal giant cell astrocytomas (SEGAs) [42]. A prevailing model is that TSC-associated brain lesions including SEGAs and tubers are caused by somatic second-hit mutations [29, 43]. According to this model, individuals with TSC have a germline heterozygous LoF mutation in TSC1 or TSC2 and during brain development a small number of neural progenitor cells acquire a somatic mutation that disrupts the expression of the functional allele, causing bi-allelic inactivation. In this scenario, progenitor cells with a second-hit mutation give rise to abnormal, dysplastic tuber cells that are surrounded by cells with normal-appearing morphology, which are derived from heterozygous progenitors. This model, in which the secondary mutation occurs stochastically, is consistent with the variable number, size and location of cortical tubers observed in individuals with TSC. Second-hit mutations have been consistently observed in resected SEGAs and TSC-associated tumors (called hamartomas) in peripheral organs [44–46]. However, they have only been identified in a subset of cortical tubers, resulting in some debate over the origins of cortical tuber cells [47–50].
TSC is also associated with several neuropsychiatric conditions, collectively termed “TAND” (TSC-Associated Neuropsychiatric Disorders) [51]. Approximately 80–90% of TSC patients develop seizures that can begin in infancy (infantile spasms) [52]. Earlier onset and increased severity of TSC-related seizures is correlated with greater risk of intellectual disability (ID), ASD, and attention deficit hyperactivity disorder (ADHD) [52–56]. Impaired and disordered myelination has also been observed in individuals with TSC using brain imaging techniques [57, 58]. Interestingly, while mutations in TBC1D7 have not been reported in TSC patients, they have been identified in individuals with macrocephaly/megalencephaly and ID [59, 60], presentations that are shared among several mTORopathies.
Phosphatase and tensin homolog (PTEN) hamartoma tumor syndrome (PHTS)
PHTS is a spectrum of multi-system disorders caused by LoF germline mutations in PTEN [61]. PTEN is a phosphatase that negatively regulates the PI3K/AKT/mTOR pathway by dephosphorylating phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at the cell membrane. LoF mutations in PTEN cause elevated PIP3 that recruits several proteins including 3-phosphoinositide-dependent protein kinase-1 (PDK1) and AKT family members. PDK1 and subsequently mTORC2 phosphorylate and fully activate AKT, which in turn phosphorylates and inhibits TSC2 leading to increased mTORC1 activity (shown in Fig. 1C) [62].
PHTS disorders include Cowden syndrome (CS), Bannayan–Riley–Ruvalcaba syndrome and adult Lhermitte–Duclos disease [61]. These disorders are associated with increased cancer and tumor risk [61]. Neuropsychiatric manifestations associated with LoF mutations in PTEN include macrocephaly, developmental delay, ASD, and intellectual disability [61]. Some studies have also identified patients carrying PTEN mutations who present with cortical malformations and seizures [63–65]. In addition, PTEN mutations have been linked to ASD and PTEN is one of the most prominent risk genes in idiopathic ASD [66, 67].
Malformations of cortical development (MCD)
MCD is a group of disorders characterized by abnormal development of the cerebral cortex, such as focal cortical dysplasia (FCD), megalencephaly (ME), and hemimegalencephaly (HME) [68]. FCD is defined by focal regions of the cortex that contain enlarged, dysplastic, and mislaminated neurons and glia, which can be observable by MRI and vary in size and location [69]. ME is defined by increased head circumference two standard deviations above the age-related mean, which is caused by increased growth of brain structures. HME is the enlargement of an individual hemisphere of the cerebral cortex [70, 71]. MCDs are common causes of pediatric epilepsy, ID and neurological deficits [72]. Epilepsy in MCD patients is often intractable, occasionally life-threatening, and can require surgical resection [73, 74]. Studies of resected tissue from FCD and HME patients have shown increased phosphorylation of the mTORC1 downstream target, ribosomal protein S6 [75–78]. More recent sequencing studies revealed that somatic brain mutations in mTOR regulators including mTOR itself, PIK3CA, RHEB, AKT3, TSC1 and TSC2 [79–82] can result in focal cortical dysplasia (Table 1), which share several features with syndromic disorders including cortical malformations and epilepsy [81, 83]. In addition, mutations in DEPDC5, NPRL2 and NPRL3, which are components of the mTORC1 inhibitor GATOR complex 1 [84], have been linked to FCD, infantile spasms, focal epilepsy and sudden unexpected death in epilepsy (SUDEP) [85–87].
Mutations in STRADA and NF1, which encode negative regulators of mTORC1 [88, 89] have been identified in syndromes that present with MCD, epilepsy and cognitive deficits. Specifically, LoF mutations in STRADA cause polyhydramnios, megalencephaly, and symptomatic epilepsy syndrome (PMSE) [90] and LoF mutations in NF1 cause neurofibromatosis type 1 (NF1) syndrome [91]. Polyhydramnios refers to excess amniotic fluid during pregnancy, which is a hallmark of PMSE. Aside from mTORopathies, aberrant mTOR signaling has been detected in several neurodevelopmental disorders such as Fragile X syndrome, Down syndrome and idiopathic ASD [92–94]. In addition, mTOR dysregulation has been observed in several psychiatric disorders and neurodegenerative diseases [12, 14, 15, 95]. However, a direct causal link between mTOR and these disorders has yet to be defined. In particular, given that mTOR signaling is highly responsive to neuronal activity [96], it is possible that altered mTOR signaling may occur as a secondary phenotype to changes in network activity in many of these diseases.
Current treatment strategies for mTORopathies
Several clinical studies have demonstrated the utility of rapamycin and its analogues (rapalogs), such as sirolimus and everolimus, as treatments for TSC and other mTORopathies including PHTS, NF1 and PMSE [97–99]. Rapamycin is an allosteric mTOR inhibitor that binds to FK506-binding protein 12 (FKBP12). The rapamycin-FKBP12 complex then binds to the FKBP-rapamycin binding domain on mTOR and blocks its catalytic domain [4, 100]. The first clinical evidence of rapalog efficacy for neurological manifestations came from Franz et al. in 2006 who showed SEGA regression in five TSC patients following rapamycin treatment [101]. Several larger follow-up studies corroborated the effectiveness of everolimus treatment in TSC patients with SEGAs [102–104]. Clinical studies have also shown that rapalogs can be useful treatments for other TSC-related phenotypes including seizures [104–106], with one study demonstrating 40% reduction of seizure severity in 40% of patients in a trial of over 300 patients spanning a broad age range [105]. However, not all studies have reported successful outcomes with rapalogs in individuals with TSC-related seizures [107]. Recent clinical studies such as “Stopping TSC Onset and Progression 2: Epilepsy Prevention in TSC Infants (STOP2) (https://clinicaltrials.gov/ct2/show/NCT04595513)” have focused on early treatment or prevention of seizures in infants and children with TSC, as it may be difficult to control seizures with rapalogs or other drugs after prolonged occurrence [108, 109]. Notably, other antiseizure medications, including vigabatrin for infantile spasms, have been successfully used in TSC patients [110].
A limited number of studies have explored the effect of rapalogs on the cognitive and psychiatric conditions associated with TSC. In 2017, Krueger et al. found that a 6-month long everolimus treatment did not significantly improve neurocognitive function or behavioral abnormalities in children with TSC [111]. Similarly, in 2019, Overwater et al, showed that 12-month everolimus treatment in children with TSC did not improve intelligence quotient (IQ) or ASD symptoms [112]. However, a study involving 35 Japanese patients showed promising results of everolimus treatment on both TSC-related ASD behavioral symptoms and seizures [113]. The mixed results of these studies suggest that while rapalogs can be effective at treating neuropsychiatric aspects of TSC in some individuals, further research and development of therapeutic approaches is warranted.
While rapalogs are extensively used in clinical trials as therapeutic interventions for mTORopathies, there are limitations to their usage. For instance, rapamycin does not equally block the phosphorylation of all mTORC1 substrates [114]. Specifically, while rapamycin abolishes phosphorylation of ribosomal protein S6, inhibition of 4E-BP1 phosphorylation is incomplete [115], suggesting that phenotypes arising due to 4E-BP1 deregulation may be largely rapalog-resistant. In addition, while the phenotypes of mTORopathies are generally thought to be due to mTORC1 hyperactivity (although see further discussion below), it is known that chronic rapamycin treatment inhibits both mTORC1 and mTORC2, likely via sequestering mTOR kinase and prohibiting mTORC2 formation [116]. This non-selective inhibition, in addition to on-target off-tissue effects, has been shown to contribute to the adverse effects associated with rapalog treatment such as glucose intolerance, insulin resistance and new-onset diabetes [117–119]. These side-effects are an important consideration given that long-term treatment with rapalogs may be required as the beneficial results can be reversible. For example, upon cessation of rapalog treatment, seizures can resume and SEGAs can regrow [101]. However, studies in mice have shown that intermittent, low-level dosing of rapamycin can maintain the mTOR inhibiting effects while minimizing unwanted systemic side effects [120]. Consistent with this, it was shown that the antiepileptic properties of rapamycin in a mouse model of TSC could be maintained with as long as 24 day inter-treatment intervals [121].
Development of novel drug-based therapeutics
Currently, much research is geared towards the development of new drugs, including novel rapalogs, with more favorable pharmacokinetic characteristics, greater selectivity towards mTORC1, and fewer or less severe side effects. A recently developed rapalog, DL001, showed higher selectivity for mTORC1 when compared to rapamycin and had substantially fewer side effects when tested in mice [122]. In addition to rapalogs, a second generation of ATP-competitive mTOR kinase inhibitors have been under development [123]. These inhibitors target the catalytic site of mTOR and while all downstream mTORC1 targets are equally affected, these inhibitors exhibit no selectivity between the two mTOR complexes, similar to extended rapamycin treatment. Recent studies have attempted to improve selectivity by targeting mTOR regulators. For example, the small molecule NR1 potently binds to Rheb and selectively inhibits mTORC1 activity in mice [124]. More recently, another small molecule, EN6, inhibits mTORC1 by binding to an ATP-proton pump that normally aids mTORC1 recruitment onto the lysosome, which is necessary for its activation [125]. In vitro, EN6 was shown to suppress phosphorylation of both S6 and 4E-BP1, increase autophagy, and did not affect the mTORC2 target AKT [125].
The off-tissue effects of systemically administered small molecules are often a challenge in drug development. The ability to selectively target aberrant mTOR signaling in the brain would be transformative to long-term management of patient care. One recently developed strategy is a dual-molecule approach aiming to restrict mTOR inhibition to the brain. Specifically, Shokat et al. used RapaLink-1, a brain permeable mTOR inhibitor, along with RapaBlock, a brain impermeable FKB12 ligand. RapaBlock inhibits RapaLink-1 function outside the brain and thus prevents mTOR inhibition in peripheral tissues [126]. This approach has the potential to overcome the problem of systemic side effects.
While small molecule mTOR inhibitors are an active area of research, the question of whether we fully understand the functions of the mTOR complexes and the consequences of manipulating these functions remain. The variability of the clinical data underscores the importance of further exploring the mechanisms underlying neuropsychiatric presentations in mTORopathies. In addition, treatment strategies may need to be optimized and tailored toward specific phenotypes. For example, when targeting cells in tumors, complete suppression of mTORC1 activity may be beneficial to inhibit cell growth and eventually induce apoptosis. However, balancing mTOR activity rather than completely inhibiting it, may be a more desirable outcome for treating neuropsychiatric conditions. Thus, it will be important to examine alternative treatment strategies and utilize robust models to study these disorders.
Animal models of mTORopathies
Various animal models have been generated to enable investigations into the underlying mechanisms of mTORopathies. In early studies, drosophila and rat models of TSC were used to uncover the roles of Tsc1 and Tsc2 in the cell cycle, proliferation and cancer [127–131]. Subsequently, numerous genetic mouse models have been developed harboring mutations in genes that encode for mTOR regulators including Strada [132, 133], Nf1 [134] and Depc5 [135] (Table 2). In this review, we will focus on several of the most well-studied mouse models with mutations in the Tsc1, Tsc2 or Pten genes (Table 2). Germline homozygous loss of these genes is embryonically lethal [136–139]. While mice with heterozygous LoF mutations in Tsc1, Tsc2 or Pten can exhibit some synaptic, cellular and behavioral phenotypes [138, 140–147], they often do not fully recapitulate the spectrum and severity of human disease phenotypes. In particular, mice with heterozygous Tsc1 or Tsc2 mutations do not exhibit robust mTORC1 hyperactivity, cortical malformations, or significant spontaneous seizures.
Table 2.
Summary of mouse models discussed in this review
| Disorder | Animal model | Refs |
|---|---|---|
| TSC | Tsc1−/− (germline KO) | [136] |
| Tsc2−/− (germline KO) | [137,138] | |
| Tsc1+/− (germline Het) | [136,140–141], | |
| Tsc2+/− (germline Het) | [137–138, 140,145,147] | |
| Tsc1fl/fl;Emx1Cre | [150] | |
| Tsc1fl/fl;Syn1Cre | [149] | |
| Tsc1fl/fl;Camk2aCre | [151–152] | |
| Tsc1fl/fl;GFAPCreER | [154] | |
| Tsc1fl/fl;L7Cre | [155, 172] | |
| Tsc2-/fl;L7Cre | [156] | |
| Tsc1fl/fl;Slc6a3Cre | [158] | |
| Tsc1fl/fl;Slc6a4Cre | [159] | |
| Tsc1 fl/fl ;Drd1a-Cre | [160[ | |
| Tsc1 fl/fl ;Adora2a-Cre | [160] | |
| Tsc1fl/fl;Gbx2CreER | [161] | |
| Tsc1fl/fl+ Cre in utero electroporation | [173] | |
| Tsc1flox/-+ Cre in utero electroporation | [162] | |
| PHTS | PTEN−/− (germline KO) | [139] |
| PTEN+/− (germline Het) | [139,143–144, 180] | |
| PTEN fl/fl;CamK2aCre | [152–153] | |
| PTEN fl/fl;L7Cre | [157] | |
| PTENfl/fl;Slc6a3Cre | [143] | |
| PTENfl/fl + AAV-Cre | [171] | |
| PMSE | Strada−/− (germline KO) | [133] |
| Strada shRNA in utero electroporation | [132] | |
| MCD | Depdc5fl/fl;Syn1Cre | [135] |
| RhebCAin utero electroporation | [163, 170] | |
| Tsc1 CRISPR/Cas9 editing in utero electroporation | [82] | |
| Tsc2 CRISPR/Cas9 editing in utero electroporation | [82] |
The use of Cre-dependent conditional knock-out mice avoids the confound of embryonic lethality and enables the generation of models with cell type- and developmental stage-specific mutations. Several mouse models with deletion of either Tsc1, Tsc2 or Pten in specific cell types have been generated, which recapitulate various disease phenotypes including dysmorphic neurons, astrogliosis, hypomyelination, mislamination, seizures and behavioral alterations (as reviewed previously [12, 148]). In these models, the onset and severity of phenotypes depend on the developmental stage of the genetic perturbation. For example, studies of Tsc1 conditional knock-out mice (Tsc1-cKO) have shown that Cre expression driven by the Emx1 or Syn1 promoters, which lead to loss of Tsc1 from neurons during mid-embryogenesis, causes spontaneous seizures first observable at two weeks of age with premature mortality and a median survival of ~18–35 days [149, 150]. Loss of Tsc1 or Pten from postmitotic neurons between 2–4 weeks of age using Camk2a-Cre mice results in seizures that begin later, between 5–10 weeks of age, with a median survival of approximately 50 days [151–153]. This demonstrates that seizures can occur independent of early developmental alterations. A study by Zou et al. using a tamoxifen-inducible system showed that GFAP-Cre-driven Tsc1 loss primarily from astrocytes at 2 weeks of age, but not at 6 weeks, was sufficient to cause seizures, signifying that the timing of the perturbation affects the phenotypic severity [154]. These findings are important to consider when interpreting the results of therapeutic interventions in these models. For example, later-onset seizures may arise from distinct cellular or circuit mechanisms and may therefore respond differently to treatment compared to early developmental seizures. In addition, studies that provide treatment prior to or at the onset of seizures (i.e. prevention) may have a better outcome than those in which treatment occurs after seizures have already begun.
Cell type-specific disruption of mTOR regulators has revealed brain regions and cell types that may be key drivers of cognitive and behavioral deficits in TSC and PHTS. For instance, LoF mutations in Tsc1, Tsc2 or Pten in Purkinje cells (PC) of the cerebellum induce ASD-like phenotypes including altered sociability and cognitive inflexibility [155–157]. Selective loss of Tsc1 from dopaminergic (DA) neurons using Slc6a3(DAT)-Cre mice leads to cognitive inflexibility in a reversal learning task [158], while DA-neuron specific loss of Pten impairs social preference and novelty [143]. Selective loss of Tsc1 from serotonergic neurons using Slc6a4(SERT)-Cre mice was also sufficient to cause autism-like behaviors including social behavior deficits and increased repetitive behaviors [159]. Deletion of Tsc1 from one of the primary targets of dopamine neurons, striatal projection neurons (SPNs), results in enhanced motor routine learning, which only occurs when Tsc1 is lost from direct pathway, but not indirect pathway, SPNs [160]. In addition, deletion of Tsc1 from thalamic neurons during mid-embryonic development leads to repetitive grooming [161]. Notably, loss of Tsc1 from cerebellar, dopaminergic, serotonergic or striatal neurons can induce behavioral alternations in the absence of seizures. Therefore, different cell types and circuits may be responsible for the distinct neuropsychiatric manifestations of TSC and related disorders.
Novel approaches have emerged to capture the focal nature of mTOR-driven cortical malformations using in utero electroporation in mice to induce mutations in a small population of cortical cells. In one study, Cre was electroporated at ~E15 into the cortex of Tsc1flox/− mouse embryos to induce a second-hit mutation. This led to the formation of cortical heterotopic nodules with neuronal hypertrophy and abnormal migration together with decreased seizure threshold [162]. Similarly, Lim et al. used in utero electroporation to induce CRISPR/Cas9-based gene disruption of Tsc1 or Tsc2. These mice exhibited spontaneous seizures, neuronal hypertrophy and cortical mislamination [82]. Focal, constitutive activation of Rheb in the developing mouse cortex, which results in robust mTORC1 hyperactivity, has also been shown to result in neuronal hypertrophy, misplacement and spontaneous seizures [163].
As a complement to these animal models, human stem cell-based models of TSC and other mTORopathies have emerged over recent years due to advances in human stem cell and genome engineering [164–168]. These systems can capture human or patient-specific aspects of cell biology, genetics and brain development and thus provide an important complement to mouse models. Together, animal and human cellular models of mTORopathies provide a platform from which investigate key outstanding questions, in particular: 1) what is the neurodevelopmental impact of mutations in mTOR regulators? 2) what are the molecular, cellular and circuit mechanisms that drive pathophysiology? 3) which brain regions and cell types are responsible for different disease manifestations? and 4) what is the best therapeutic approach to maximize improvement and minimize side effects?
Drug-based therapies in animal models
There is a large literature examining the effects of rapalogs in animal models of mTOR-related disorders. These studies have revealed a range of possible outcomes including full reversal of neuropsychiatric phenotypes even after they have been established, prevention of phenotypes by pre-symptomatic mTOR inhibition, rescue within certain critical treatment windows, or a lack of response to rapalog treatment. One of the first studies to show efficacy of rapalogs in treating brain phenotypes was in a conditional mouse model of TSC. Meikle et al. showed that by administering rapamycin every other day starting at P7-P9 in Syn1-Cre;Tsc1-cKO mice they could shift median survival from 33 to over 100 days and reverse neuronal hypertrophy and demyelination phenotypes [169]. In subsequent work, Carson et al began rapamycin treatment at P13 in Emx1-Cre;Tsc1-cKO mice and were able to prevent premature mortality and largely reverse glial pathology. However, abnormal neuronal lamination was rapamycin-resistant [150]. Lin et al. showed that mislamination in a mouse model of hyperactive mTORC1 could, in fact, be prevented through administration of a constitutively active mutant 4E-BP1 if this occurred at the same time as the manipulation that caused mTORC1 hyperactivity [170]. Similarly, a study examining Pten loss from dentate gyrus neurons showed that rapamycin could prevent but not reverse abnormal migration [171]. Together, these studies show that while seizures may be partly treatable with postnatal rapamycin, abnormal lamination and neuronal migration are early developmental phenotypes that may be preventable by mTORC1 suppression but cannot be reversed at later stages of development. For phenotypes such as these it will be key to determine how much they contribute to disease manifestations and whether functional improvement can be achieved in the absence of complete rescue of these abnormalities.
The idea of critical periods for treatment has gained attention in recent studies. Two studies led by Tsai et al. showed that mice which lack Tsc1 from cerebellar Purkinje cell’s exhibit social deficits and repetitive behaviors [155, 172]. The authors demonstrated that treatment starting within the first week of life could rescue both phenotypes [155]. However, treatment starting at 6 weeks of age, after the phenotypes were already established, could only reverse social behavior aberrations, but not behavioral inflexibility or repetitive behaviors [172]. Cox et al. characterized the effects of rapamycin on dendritic arborization and spine deficits caused by embryonic Tsc1 deletion from cortical neurons at two different time points. In the early treatment group (P1-P7) they could rescue abnormal arborization and with later treatment (P15-P27) they could reverse abnormal spine maturation [173]. Critical periods might also exist for treatment of seizures in mTORopathies. Studies reporting the most success have either treated animals prior to symptom onset or have shown rescue in models in which the mutation occurs later in life [148]. It is possible that early on, potentially during epileptogenesis, seizures are sensitive to rapamycin [174] but once epilepsy is established, seizures are less responsive to mTOR inhibition. Together, these data suggest that it is important to identify critical periods for disease phenotypes in order to determine the optimal timing of treatment. Treatment given past the critical window may be responsible for some of the differential drug effectiveness observed in clinical studies.
Rapalogs have been the main pharmacological approach used in animal and cell models to inhibit mTORC1 in the context of genetic mTORopathies. However, as discussed above, chronic rapamycin inhibits both mTOR complexes and may not be effective at targeting all cell types in the brain, for example dopamine neurons [158]. In addition, a study assessing the effect of prenatal rapamycin treatment in wild-type animals showed that a single dose at embryonic day 16 resulted in adverse effects including motor abnormalities and increased anxiety that persisted in adulthood [175]. Thus, strong and non-specific suppression of mTOR activity, particularly in the developing brain, may not be an optimal therapeutic approach. Further work is needed to understand the basic biology of the two mTOR complexes, especially in neurons, the contribution of each complex to specific disease endophenotypes, and whether selective targeting of one complex will yield improved treatment with fewer side effects.
Genetic approaches to manipulating mTORC1 and mTORC2 in disease models
Several studies have attempted to selectively manipulate mTORC1 or mTORC2 in the mouse brain by disrupting their specific components, Raptor or Rictor, respectively. Such studies have revealed that while the two complexes share some common functions such as regulation of somatodendritic morphology [176], they also have distinct functions and differentially impact a range of processes from development to synaptic transmission to behavior [177–179]. The distinct contributions of the two mTOR complexes to neural development and function suggest that therapeutic approaches for modifying disease phenotypes may benefit from an ability to control each complex independently and in a temporally precise manner.
Recent studies have begun disentangling the contribution of mTORC1 and mTORC2 to disease phenotypes in genetic mouse models of mTORopathies. Huang et al. crossed germline Pten+/− mice with mice that had heterozygous loss of Rptor from forebrain neurons (Emx1-Cre;Rptorfl/+) [180]. Pten+/− mice exhibited neuronal hypertrophy in cortical layer V and a deficit in social approach behavior. Partial downregulation of mTORC1 in Pten+/− mice was sufficient to correct both neuronal hypertrophy and social deficits [180]. Chen et al. used CamK2a-Cre mice to induce loss of Pten in postmitotic forebrain neurons [153]. Pten-cKO animals exhibited hyperactivity of both mTORC1 and mTORC2 signaling, macrocephaly, seizures, premature mortality, and behavioral abnormalities. Concomitant deletion of Rictor in Pten-cKO mice prolonged their lifespan by approximately two-fold (i.e. median survival shifted from ~50 to ~110 days postnatal) but did not completely prevent premature mortality as all mice died between ~P90-P130. In this study, mTORC2 downregulation also prevented seizures and corrected behavioral abnormalities. Postnatal intracerebroventricular injection with an antisense oligonucleotide (ASO) targeting Rictor at four weeks of age was sufficient to improve seizures and behavioral deficits in Pten-cKO mice. Rptor deletion in Pten-cKO mice normalized brain size; however, surprisingly, neither seizures nor behavioral impairments were improved, and the median survival was only modestly increased by a few days [153]. These studies show that both mTORC1 and mTORC2 complexes contribute to Pten-related pathology in mice. One discrepancy between the ability to prevent behavioral phenotypes with Raptor (Huang et al.) or Rictor (Chen et al.) manipulation might be because Huang et al. used constitutive Pten+/− animals while Chen et al. studied homozygous loss of Pten from postnatal forebrain neurons. It would be interesting to investigate whether embryonic Pten haploinsufficiency can also be rescued via manipulation of mTORC2.
While TSC phenotypes tend to be ascribed to mTORC1 hyperactivity, the studies described above in Pten mouse models suggest that there could be a potential contribution of mTORC2. It should be noted, however, that while Akt signaling is elevated in response to Pten loss, phosphorylation of Akt at the mTORC2 site (Ser473) is consistently reduced in the context of Tsc1 or Tsc2 mutations [164, 181]. The contribution of mTORC1 and mTORC2 signaling to the neuropsychiatric presentations of TSC has not been comprehensively investigated. However, a recent study demonstrated that heterozygous loss of Rptor restored several TSC-related phenotypes in mice with loss of Tsc1 from dopamine neurons (Slc6a3-Cre;Tsc1-cKO) [158]. These mice exhibited dopamine neuron hypertrophy and impaired striatal dopamine release that led to deficits in cognitive flexibility. Concomitant heterozygous loss of Rptor, while not sufficient to prevent somatic hypertrophy, significantly improved striatal dopamine release and prevented cognitive inflexibility. Notably, the authors showed that homozygous deletion of Rptor in Tsc1-cKO dopamine neurons caused neuronal hypotrophy and was not able to improve dopamine release deficits [158]. This demonstrates that complete suppression of mTORC1 can be just as detrimental to neuronal function as mTORC1 hyperactivation. These findings underscore the idea that rebalancing rather than completely suppressing mTORC1 signaling may be a preferable therapeutic approach for mTOR-related brain disorders.
The importance of considering tissue- and mTOR complex-selectivity of therapeutic approaches was signified by two studies conducted in a mouse model of fragile X syndrome (Fmr1-KO), which exhibits mTORC1 hyperactivity [182]. Yan et al. selectively downregulated mTORC1 by injecting shRNA targeting Rptor directly into the hippocampal CA1 region of Fmr1-KO mice; rescuing, in part, aberrant spine morphology, synaptic function and memory deficits [183]. In contrast, a different study showed that orally administered rapamycin in Fmr1-KO mice did not reverse behavioral deficits and had adverse effects on social behavior and sleep in both control and Fmr1-KO mice [184]. These studies suggest that selective manipulation of a specific mTOR complex within a targeted brain region, as opposed to systemic non-specific inhibition, might be a more beneficial strategy for the treatment of disorders with altered mTOR signaling.
Conclusions
While the mTOR pathway has been extensively studied in many systems, it remains enigmatic due to its significant complexity and breadth of actions. Research using in vitro and in vivo models has revealed a multitude of upstream regulators and downstream targets and shown that these can vary significantly based on cell type and developmental stage. Given this complexity, it is not surprising that results from treatment with rapamycin and its analogues have had mixed success. In both people and in animal models, rapalogs exhibit variability in their efficacy and side effects. While it will be interesting to see how recently developed dual-drug strategies that enable brain-specific targeting of rapalogs affect neuropathophysiology, the development of alternative therapeutic strategies is warranted. One exciting approach currently under development is gene therapy [185]. While this has traditionally meant the delivery of a gene that is lacking via a virus-based carrier, it has expanded to include approaches such as ASOs and CRISPR/Cas9-based systems, including those that modify gene expression without altering the genome sequence [186, 187]. Although there are still several limitations and barriers to the wide-spread implementation of these technologies, gene-based therapies offer potential advantages of selective brain region- and cell type-targeting [188, 189].
In summary, building off the initial success of rapalogs as therapeutic treatments for mTORopathies, new insights into the basic biology of the mTOR complexes and their functions in different neural cell types will facilitate the generation of improved treatments for mTOR-related disorders. Based on our current information, therapeutic strategies that 1) target the most relevant mTOR complex for disease phenotypes, 2) are given at the optimal age and stage of disease progression, 3) act on disease-relevant cellular targets with minimal off-tissue activity, and 4) rebalance signaling to physiologic levels, are likely to be most successful.
Acknowledgements
The authors thank the members of the Bateup lab for helpful discussions on this topic.
Funding Sources
This work was supported by R01 NS105634, R21 NS096415, grants from the Rennie Fund for the Study of Epilepsy, and a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (to H.S.B.). V.K. was supported by an Elizabeth Roboz Einstein graduate fellowship and pre-doctoral NIH/NIGMS training grant T32 GM7232-40. H.S.B. is a Chan Zuckerberg Biohub Investigator.
Footnotes
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
References
- 1.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo). 1975;28(10):721–6. [DOI] [PubMed] [Google Scholar]
- 2.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253(5022):905–9. [DOI] [PubMed] [Google Scholar]
- 3.Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, et al. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol. 1991;11(3):1718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78(1):35–43. [DOI] [PubMed] [Google Scholar]
- 5.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270(2):815–22. [DOI] [PubMed] [Google Scholar]
- 6.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24(1):200–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17(6):596–603. [DOI] [PubMed] [Google Scholar]
- 8.Fifkova E, Anderson CL, Young SJ, Van Harreveld A. Effect of anisomycin on stimulation-induced changes in dendritic spines of the dentate granule cells. J Neurocytol. 1982;11(2):183–210. [DOI] [PubMed] [Google Scholar]
- 9.Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452(1–2):57–65. [DOI] [PubMed] [Google Scholar]
- 10.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15(7):807–26. [DOI] [PubMed] [Google Scholar]
- 11.Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci U S A. 2002;99(1):467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Switon K, Kotulska K, Janusz-Kaminska A, Zmorzynska J, Jaworski J. Molecular neurobiology of mTOR. Neuroscience. 2017;341:112–53. [DOI] [PubMed] [Google Scholar]
- 13.Jaworski J, Sheng M. The growing role of mTOR in neuronal development and plasticity. Mol Neurobiol. 2006;34(3):205–19. [DOI] [PubMed] [Google Scholar]
- 14.Lipton JO, Sahin M. The neurology of mTOR. Neuron. 2014;84(2):275–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16(11):1537–43. [DOI] [PubMed] [Google Scholar]
- 16.Wong M Mammalian target of rapamycin (mTOR) pathways in neurological diseases. Biomed J. 2013;36(2):40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110(2):177–89. [DOI] [PubMed] [Google Scholar]
- 18.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110(2):163–75. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14(14):1296–302. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11(4):895–904. [DOI] [PubMed] [Google Scholar]
- 21.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, et al. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem. 2010;285(26):20109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25(6):903–15. [DOI] [PubMed] [Google Scholar]
- 24.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16(18):1865–70. [DOI] [PubMed] [Google Scholar]
- 25.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405(3):513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21(4):183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crino PB. Focal brain malformations: seizures, signaling, sequencing. Epilepsia. 2009;50 Suppl 9:3–8. [DOI] [PubMed] [Google Scholar]
- 28.Crino PB. mTOR: A pathogenic signaling pathway in developmental brain malformations. Trends Mol Med. 2011;17(12):734–42. [DOI] [PubMed] [Google Scholar]
- 29.Crino PB. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat Rev Neurol. 2016;12(7):379–92. [DOI] [PubMed] [Google Scholar]
- 30.van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277(5327):805–8. [DOI] [PubMed] [Google Scholar]
- 31.European Chromosome 16 Tuberous Sclerosis C. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75(7):1305–15. [DOI] [PubMed] [Google Scholar]
- 32.Nakashima A, Yoshino K, Miyamoto T, Eguchi S, Oshiro N, Kikkawa U, et al. Identification of TBC7 having TBC domain as a novel binding protein to TSC1-TSC2 complex. Biochem Biophys Res Commun. 2007;361(1):218–23. [DOI] [PubMed] [Google Scholar]
- 33.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47(4):535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11(6):1457–66. [DOI] [PubMed] [Google Scholar]
- 35.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13(15):1259–68. [DOI] [PubMed] [Google Scholar]
- 36.Mizuguchi M, Takashima S. Neuropathology of tuberous sclerosis. Brain Dev. 2001;23(7):508–15. [DOI] [PubMed] [Google Scholar]
- 37.Ertan G, Arulrajah S, Tekes A, Jordan L, Huisman TA. Cerebellar abnormality in children and young adults with tuberous sclerosis complex: MR and diffusion weighted imaging findings. J Neuroradiol 2010;37(4):231–8. [DOI] [PubMed] [Google Scholar]
- 38.Curatolo P, Moavero R, de Vries PJ. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol. 2015;14(7):733–45. [DOI] [PubMed] [Google Scholar]
- 39.Ma TS, Elliott RE, Ruppe V, Devinsky O, Kuzniecky R, Weiner HL, et al. Electrocorticographic evidence of perituberal cortex epileptogenicity in tuberous sclerosis complex. J Neurosurg Pediatr. 2012;10(5):376–82. [DOI] [PubMed] [Google Scholar]
- 40.Sosunov AA, McGovern RA, Mikell CB, Wu X, Coughlin DG, Crino PB, et al. Epileptogenic but MRI-normal perituberal tissue in Tuberous Sclerosis Complex contains tuber-specific abnormalities. Acta Neuropathol Commun. 2015;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grayson LE, Peters JM, McPherson T, Krueger DA, Sahin M, Wu JY, et al. Pilot Study of Neurodevelopmental Impact of Early Epilepsy Surgery in Tuberous Sclerosis Complex. Pediatr Neurol. 2020;109:39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Northrup H, Krueger DA, International Tuberous Sclerosis Complex Consensus G. Tuberous sclerosis complex diagnostic criteria update: recommendations of the 2012 Iinternational Tuberous Sclerosis Complex Consensus Conference. Pediatr Neurol. 2013;49(4):243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magri L, Galli R. mTOR signaling in neural stem cells: from basic biology to disease. Cell Mol Life Sci. 2013;70(16):2887–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henske EP, Scheithauer BW, Short MP, Wollmann R, Nahmias J, Hornigold N, et al. Allelic loss is frequent in tuberous sclerosis kidney lesions but rare in brain lesions. Am J Hum Genet. 1996;59(2):400–6. [PMC free article] [PubMed] [Google Scholar]
- 45.Sepp T, Yates JR, Green AJ. Loss of heterozygosity in tuberous sclerosis hamartomas. J Med Genet. 1996;33(11):962–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Au KS, Hebert AA, Roach ES, Northrup H. Complete inactivation of the TSC2 gene leads to formation of hamartomas. Am J Hum Genet. 1999;65(6):1790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crino PB, Aronica E, Baltuch G, Nathanson KL. Biallelic TSC gene inactivation in tuberous sclerosis complex. Neurology. 2010;74(21):1716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin W, Chan JA, Vinters HV, Mathern GW, Franz DN, Taillon BE, et al. Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Pathol. 2010;20(6):1096–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin KR, Zhou W, Bowman MJ, Shih J, Au KS, Dittenhafer-Reed KE, et al. The genomic landscape of tuberous sclerosis complex. Nat Commun. 2017;8:15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan JA, Zhang H, Roberts PS, Jozwiak S, Wieslawa G, Lewin-Kowalik J, et al. Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol. 2004;63(12):1236–42. [DOI] [PubMed] [Google Scholar]
- 51.de Vries PJ, Wilde L, de Vries MC, Moavero R, Pearson DA, Curatolo P. A clinical update on tuberous sclerosis complex-associated neuropsychiatric disorders (TAND). Am J Med Genet C Semin Med Genet. 2018;178(3):309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nabbout R, Belousova E, Benedik MP, Carter T, Cottin V, Curatolo P, et al. Epilepsy in tuberous sclerosis complex: Findings from the TOSCA Study. Epilepsia Open. 2019;4(1):73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–56. [DOI] [PubMed] [Google Scholar]
- 54.Curatolo P, Verdecchia M, Bombardieri R. Tuberous sclerosis complex: a review of neurological aspects. Eur J Paediatr Neurol. 2002;6(1):15–23. [DOI] [PubMed] [Google Scholar]
- 55.Numis AL, Major P, Montenegro MA, Muzykewicz DA, Pulsifer MB, Thiele EA. Identification of risk factors for autism spectrum disorders in tuberous sclerosis complex. Neurology. 2011;76(11):981–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu-Shore CJ, Major P, Camposano S, Muzykewicz D, Thiele EA. The natural history of epilepsy in tuberous sclerosis complex. Epilepsia. 2010;51(7):1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simao G, Raybaud C, Chuang S, Go C, Snead OC, Widjaja E. Diffusion tensor imaging of commissural and projection white matter in tuberous sclerosis complex and correlation with tuber load. AJNR Am J Neuroradiol. 2010;31(7):1273–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arulrajah S, Ertan G, Jordan L, Tekes A, Khaykin E, Izbudak I, et al. Magnetic resonance imaging and diffusion-weighted imaging of normal-appearing white matter in children and young adults with tuberous sclerosis complex. Neuroradiology. 2009;51(11):781–6. [DOI] [PubMed] [Google Scholar]
- 59.Capo-Chichi JM, Tcherkezian J, Hamdan FF, Decarie JC, Dobrzeniecka S, Patry L, et al. Disruption of TBC1D7, a subunit of the TSC1-TSC2 protein complex, in intellectual disability and megalencephaly. J Med Genet. 2013;50(11):740–4. [DOI] [PubMed] [Google Scholar]
- 60.Alfaiz AA, Micale L, Mandriani B, Augello B, Pellico MT, Chrast J, et al. TBC1D7 mutations are associated with intellectual disability, macrocrania, patellar dislocation, and celiac disease. Hum Mutat. 2014;35(4):447–51. [DOI] [PubMed] [Google Scholar]
- 61.Pilarski R PTEN Hamartoma Tumor Syndrome: A Clinical Overview. Cancers (Basel). 2019;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13(5):283–96. [DOI] [PubMed] [Google Scholar]
- 63.Elia M, Amato C, Bottitta M, Grillo L, Calabrese G, Esposito M, et al. An atypical patient with Cowden syndrome and PTEN gene mutation presenting with cortical malformation and focal epilepsy. Brain Dev. 2012;34(10):873–6. [DOI] [PubMed] [Google Scholar]
- 64.Adachi T, Takigawa H, Nomura T, Watanabe Y, Kowa H. Cowden Syndrome with a Novel PTEN Mutation Presenting with Partial Epilepsy Related to Focal Cortical Dysplasia. Intern Med. 2018;57(1):97–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheung KM, Lam CW, Chan YK, Siu WK, Yong L. Atypical focal cortical dysplasia in a patient with Cowden syndrome. Hong Kong Med J. 2014;20(2):165–7. [DOI] [PubMed] [Google Scholar]
- 66.Varga EA, Pastore M, Prior T, Herman GE, McBride KL. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med. 2009;11(2):111–7. [DOI] [PubMed] [Google Scholar]
- 67.Satterstrom FK, Kosmicki JA, Wang J, Breen MS, De Rubeis S, An JY, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180(3):568–84 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pang T, Atefy R, Sheen V. Malformations of cortical development. Neurologist. 2008;14(3):181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blumcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, Palmini A, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pavone P, Pratico AD, Rizzo R, Corsello G, Ruggieri M, Parano E, et al. A clinical review on megalencephaly: A large brain as a possible sign of cerebral impairment. Medicine (Baltimore). 2017;96(26):e6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mirzaa GM, Poduri A. Megalencephaly and hemimegalencephaly: breakthroughs in molecular etiology. Am J Med Genet C Semin Med Genet. 2014;166C(2):156–72. [DOI] [PubMed] [Google Scholar]
- 72.Leventer RJ, Guerrini R, Dobyns WB. Malformations of cortical development and epilepsy. Dialogues Clin Neurosci. 2008;10(1):47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Desbiens R, Berkovic SF, Dubeau F, Andermann F, Laxer KD, Harvey S, et al. Life-threatening focal status epilepticus due to occult cortical dysplasia. Arch Neurol. 1993;50(7):695–700. [DOI] [PubMed] [Google Scholar]
- 74.Barkovich AJ, Dobyns WB, Guerrini R. Malformations of cortical development and epilepsy. Cold Spring Harb Perspect Med. 2015;5(5):a022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baybis M, Yu J, Lee A, Golden JA, Weiner H, McKhann G 2nd, et al. mTOR cascade activation distinguishes tubers from focal cortical dysplasia. Ann Neurol. 2004;56(4):478–87. [DOI] [PubMed] [Google Scholar]
- 76.Miyata H, Chiang AC, Vinters HV. Insulin signaling pathways in cortical dysplasia and TSC-tubers: tissue microarray analysis. Ann Neurol. 2004;56(4):510–9. [DOI] [PubMed] [Google Scholar]
- 77.Ljungberg MC, Bhattacharjee MB, Lu Y, Armstrong DL, Yoshor D, Swann JW, et al. Activation of mammalian target of rapamycin in cytomegalic neurons of human cortical dysplasia. Ann Neurol. 2006;60(4):420–9. [DOI] [PubMed] [Google Scholar]
- 78.Aronica E, Boer K, Baybis M, Yu J, Crino P. Co-expression of cyclin D1 and phosphorylated ribosomal S6 proteins in hemimegalencephaly. Acta Neuropathol. 2007;114(3):287–93. [DOI] [PubMed] [Google Scholar]
- 79.Jansen LA, Mirzaa GM, Ishak GE, O’Roak BJ, Hiatt JB, Roden WH, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138(Pt 6):1613–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao S, Li Z, Zhang M, Zhang L, Zheng H, Ning J, et al. A brain somatic RHEB doublet mutation causes focal cortical dysplasia type II. Exp Mol Med. 2019;51(7):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim JS, Kim WI, Kang HC, Kim SH, Park AH, Park EK, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21(4):395–400. [DOI] [PubMed] [Google Scholar]
- 82.Lim JS, Gopalappa R, Kim SH, Ramakrishna S, Lee M, Kim WI, et al. Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia. Am J Hum Genet. 2017;100(3):454–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baldassari S, Ribierre T, Marsan E, Adle-Biassette H, Ferrand-Sorbets S, Bulteau C, et al. Dissecting the genetic basis of focal cortical dysplasia: a large cohort study. Acta Neuropathol. 2019;138(6):885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baulac S mTOR signaling pathway genes in focal epilepsies. Prog Brain Res. 2016;226:61–79. [DOI] [PubMed] [Google Scholar]
- 86.Ribierre T, Deleuze C, Bacq A, Baldassari S, Marsan E, Chipaux M, et al. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest. 2018;128(6):2452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iffland PH 2nd, Carson V, Bordey A, Crino PB. GATORopathies: The role of amino acid regulatory gene mutations in epilepsy and cortical malformations. Epilepsia. 2019;60(11):2163–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2(4):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lau N, Feldkamp MM, Roncari L, Loehr AH, Shannon P, Gutmann DH, et al. Loss of neurofibromin is associated with activation of RAS/MAPK and PI3-K/AKT signaling in a neurofibromatosis 1 astrocytoma. J Neuropathol Exp Neurol. 2000;59(9):759–67. [DOI] [PubMed] [Google Scholar]
- 90.Puffenberger EG, Strauss KA, Ramsey KE, Craig DW, Stephan DA, Robinson DL, et al. Polyhydramnios, megalencephaly and symptomatic epilepsy caused by a homozygous 7-kilobase deletion in LYK5. Brain. 2007;130(Pt 7):1929–41. [DOI] [PubMed] [Google Scholar]
- 91.Nix JS, Blakeley J, Rodriguez FJ. An update on the central nervous system manifestations of neurofibromatosis type 1. Acta Neuropathol. 2020;139(4):625–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoeffer CA, Sanchez E, Hagerman RJ, Mu Y, Nguyen DV, Wong H, et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 2012;11(3):332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Troca-Marin JA, Casanas JJ, Benito I, Montesinos ML. The Akt-mTOR pathway in Down’s syndrome: the potential use of rapamycin/rapalogs for treating cognitive deficits. CNS Neurol Disord Drug Targets. 2014;13(1):34–40. [DOI] [PubMed] [Google Scholar]
- 94.Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83(5):1131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Olney NT, Alquezar C, Ramos EM, Nana AL, Fong JC, Karydas AM, et al. Linking tuberous sclerosis complex, excessive mTOR signaling, and age-related neurodegeneration: a new association between TSC1 mutation and frontotemporal dementia. Acta Neuropathol. 2017;134(5):813–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bateup HS, Denefrio CL, Johnson CA, Saulnier JL, Sabatini BL. Temporal dynamics of a homeostatic pathway controlling neural network activity. Front Mol Neurosci. 2013;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Komiya T, Blumenthal GM, DeChowdhury R, Fioravanti S, Ballas MS, Morris J, et al. A Pilot Study of Sirolimus in Subjects with Cowden Syndrome or Other Syndromes Characterized by Germline Mutations in PTEN. Oncologist. 2019;24(12):1510–e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weiss B, Widemann BC, Wolters P, Dombi E, Vinks A, Cantor A, et al. Sirolimus for progressive neurofibromatosis type 1-associated plexiform neurofibromas: a neurofibromatosis Clinical Trials Consortium phase II study. Neuro Oncol. 2015;17(4):596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parker WE, Orlova KA, Parker WH, Birnbaum JF, Krymskaya VP, Goncharov DA, et al. Rapamycin prevents seizures after depletion of STRADA in a rare neurodevelopmental disorder. Sci Transl Med. 2013;5(182):182ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273(5272):239–42. [DOI] [PubMed] [Google Scholar]
- 101.Franz DN, Leonard J, Tudor C, Chuck G, Care M, Sethuraman G, et al. Rapamycin causes regression of astrocytomas in tuberous sclerosis complex. Ann Neurol. 2006;59(3):490–8. [DOI] [PubMed] [Google Scholar]
- 102.Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381(9861):125–32. [DOI] [PubMed] [Google Scholar]
- 103.Krueger DA, Care MM, Agricola K, Tudor C, Mays M, Franz DN. Everolimus long-term safety and efficacy in subependymal giant cell astrocytoma. Neurology. 2013;80(6):574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cardamone M, Flanagan D, Mowat D, Kennedy SE, Chopra M, Lawson JA. Mammalian target of rapamycin inhibitors for intractable epilepsy and subependymal giant cell astrocytomas in tuberous sclerosis complex. J Pediatr. 2014;164(5):1195–200. [DOI] [PubMed] [Google Scholar]
- 105.French JA, Lawson JA, Yapici Z, Ikeda H, Polster T, Nabbout R, et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. Lancet. 2016;388(10056):2153–63. [DOI] [PubMed] [Google Scholar]
- 106.Samueli S, Abraham K, Dressler A, Groppel G, Muhlebner-Fahrngruber A, Scholl T, et al. Efficacy and safety of Everolimus in children with TSC - associated epilepsy - Pilot data from an open single-center prospective study. Orphanet J Rare Dis. 2016;11(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Overwater IE, Rietman AB, Bindels-de Heus K, Looman CW, Rizopoulos D, Sibindi TM, et al. Sirolimus for epilepsy in children with tuberous sclerosis complex: A randomized controlled trial. Neurology. 2016;87(10):1011–8. [DOI] [PubMed] [Google Scholar]
- 108.Jozwiak S, Kotulska K, Domanska-Pakiela D, Lojszczyk B, Syczewska M, Chmielewski D, et al. Antiepileptic treatment before the onset of seizures reduces epilepsy severity and risk of mental retardation in infants with tuberous sclerosis complex. Eur J Paediatr Neurol. 2011;15(5):424–31. [DOI] [PubMed] [Google Scholar]
- 109.Cusmai R, Moavero R, Bombardieri R, Vigevano F, Curatolo P. Long-term neurological outcome in children with early-onset epilepsy associated with tuberous sclerosis. Epilepsy Behav. 2011;22(4):735–9. [DOI] [PubMed] [Google Scholar]
- 110.Curatolo P, Verdecchia M, Bombardieri R. Vigabatrin for tuberous sclerosis complex. Brain Dev. 2001;23(7):649–53. [DOI] [PubMed] [Google Scholar]
- 111.Krueger DA, Sadhwani A, Byars AW, de Vries PJ, Franz DN, Whittemore VH, et al. Everolimus for treatment of tuberous sclerosis complex-associated neuropsychiatric disorders. Ann Clin Transl Neurol. 2017;4(12):877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Overwater IE, Rietman AB, Mous SE, Bindels-de Heus K, Rizopoulos D, Ten Hoopen LW, et al. A randomized controlled trial with everolimus for IQ and autism in tuberous sclerosis complex. Neurology. 2019;93(2):e200–e9. [DOI] [PubMed] [Google Scholar]
- 113.Mizuguchi M, Ikeda H, Kagitani-Shimono K, Yoshinaga H, Suzuki Y, Aoki M, et al. Everolimus for epilepsy and autism spectrum disorder in tuberous sclerosis complex: EXIST-3 substudy in Japan. Brain Dev. 2019;41(1):1–10. [DOI] [PubMed] [Google Scholar]
- 114.Thoreen CC, Sabatini DM. Rapamycin inhibits mTORC1, but not completely. Autophagy. 2009;5(5):725–6. [DOI] [PubMed] [Google Scholar]
- 115.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105(45):17414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–68. [DOI] [PubMed] [Google Scholar]
- 117.Kaplan B, Qazi Y, Wellen JR. Strategies for the management of adverse events associated with mTOR inhibitors. Transplant Rev (Orlando). 2014;28(3):126–33. [DOI] [PubMed] [Google Scholar]
- 118.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kleinert M, Sylow L, Fazakerley DJ, Krycer JR, Thomas KC, Oxboll AJ, et al. Acute mTOR inhibition induces insulin resistance and alters substrate utilization in vivo. Mol Metab. 2014;3(6):630–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Arriola Apelo SI, Neuman JC, Baar EL, Syed FA, Cummings NE, Brar HK, et al. Alternative rapamycin treatment regimens mitigate the impact of rapamycin on glucose homeostasis and the immune system. Aging Cell. 2016;15(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rensing N, Han L, Wong M. Intermittent dosing of rapamycin maintains antiepileptogenic effects in a mouse model of tuberous sclerosis complex. Epilepsia. 2015;56(7):1088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schreiber KH, Arriola Apelo SI, Yu D, Brinkman JA, Velarde MC, Syed FA, et al. A novel rapamycin analog is highly selective for mTORC1 in vivo. Nat Commun. 2019;10(1):3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schenone S, Brullo C, Musumeci F, Radi M, Botta M. ATP-competitive inhibitors of mTOR: an update. Curr Med Chem. 2011;18(20):2995–3014. [DOI] [PubMed] [Google Scholar]
- 124.Mahoney SJ, Narayan S, Molz L, Berstler LA, Kang SA, Vlasuk GP, et al. A small molecule inhibitor of Rheb selectively targets mTORC1 signaling. Nat Commun. 2018;9(1):548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chung CY, Shin HR, Berdan CA, Ford B, Ward CC, Olzmann JA, et al. Covalent targeting of the vacuolar H(+)-ATPase activates autophagy via mTORC1 inhibition. Nat Chem Biol. 2019;15(8):776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shokat K, Fan Q, Luo X, Lou K, Weiss W. Brain-Restricted mTOR Inhibition with Binary Pharmacology. bioRxiv 1021203/rs3rs-92256/v1. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ito N, Rubin GM. gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell. 1999;96(4):529–39. [DOI] [PubMed] [Google Scholar]
- 128.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105(3):345–55. [DOI] [PubMed] [Google Scholar]
- 129.Eker R MJ. A dominant gene for renal adenomas in the rat. Nature. 1961;189:858–9.13780066 [Google Scholar]
- 130.Yeung RS, Xiao GH, Jin F, Lee WC, Testa JR, Knudson AG. Predisposition to renal carcinoma in the Eker rat is determined by germ-line mutation of the tuberous sclerosis 2 (TSC2) gene. Proc Natl Acad Sci U S A. 1994;91(24):11413–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hino O, Kobayashi T, Tsuchiya H, Kikuchi Y, Kobayashi E, Mitani H, et al. The predisposing gene of the Eker rat inherited cancer syndrome is tightly linked to the tuberous sclerosis (TSC2) gene. Biochem Biophys Res Commun. 1994;203(2):1302–8. [DOI] [PubMed] [Google Scholar]
- 132.Orlova KA, Parker WE, Heuer GG, Tsai V, Yoon J, Baybis M, et al. STRADalpha deficiency results in aberrant mTORC1 signaling during corticogenesis in humans and mice. J Clin Invest. 2010;120(5):1591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dang LT, Glanowska KM, Iffland Ii PH, Barnes AE, Baybis M, Liu Y, et al. Multimodal Analysis of STRADA Function in Brain Development. Front Cell Neurosci. 2020;14:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gutmann DH, Giovannini M. Mouse models of neurofibromatosis 1 and 2. Neoplasia. 2002;4(4):279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yuskaitis CJ, Jones BM, Wolfson RL, Super CE, Dhamne SC, Rotenberg A, et al. A mouse model of DEPDC5-related epilepsy: Neuronal loss of Depdc5 causes dysplastic and ectopic neurons, increased mTOR signaling, and seizure susceptibility. Neurobiol Dis. 2018;111:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kobayashi T, Minowa O, Sugitani Y, Takai S, Mitani H, Kobayashi E, et al. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc Natl Acad Sci U S A. 2001;98(15):8762–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, Noda T. Renal carcinogenesis, hepatic hemangiomatosis, and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 1999;59(6):1206–11. [PubMed] [Google Scholar]
- 138.Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104(6):687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19(4):348–55. [DOI] [PubMed] [Google Scholar]
- 140.Sato A, Kasai S, Kobayashi T, Takamatsu Y, Hino O, Ikeda K, et al. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat Commun. 2012;3:1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62(6):648–55. [DOI] [PubMed] [Google Scholar]
- 142.Kirschstein T Synaptic plasticity and learning in animal models of tuberous sclerosis complex. Neural Plast. 2012;2012:279834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Clipperton-Allen AE, Page DT. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum Mol Genet. 2014;23(13):3490–505. [DOI] [PubMed] [Google Scholar]
- 144.Li L, Liu F, Salmonsen RA, Turner TK, Litofsky NS, Di Cristofano A, et al. PTEN in neural precursor cells: regulation of migration, apoptosis, and proliferation. Mol Cell Neurosci. 2002;20(1):21–9. [DOI] [PubMed] [Google Scholar]
- 145.Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, et al. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14(8):843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8(12):1727–34. [DOI] [PubMed] [Google Scholar]
- 147.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480(7375):63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jeong A, Wong M. Tuberous Sclerosis and Other mTORopathies. Models of Seizures and Epilepsy (Second edition): Elsevier Inc.; 2017. p. 797–810. [Google Scholar]
- 149.Meikle L, Talos DM, Onda H, Pollizzi K, Rotenberg A, Sahin M, et al. A mouse model of tuberous sclerosis: neuronal loss of Tsc1 causes dysplastic and ectopic neurons, reduced myelination, seizure activity, and limited survival. J Neurosci. 2007;27(21):5546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Carson RP, Van Nielen DL, Winzenburger PA, Ess KC. Neuronal and glia abnormalities in Tsc1-deficient forebrain and partial rescue by rapamycin. Neurobiol Dis. 2012;45(1):369–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78(3):510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McMahon J, Huang X, Yang J, Komatsu M, Yue Z, Qian J, et al. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J Neurosci. 2012;32(45):15704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen CJ, Sgritta M, Mays J, Zhou H, Lucero R, Park J, et al. Therapeutic inhibition of mTORC2 rescues the behavioral and neurophysiological abnormalities associated with Pten-deficiency. Nat Med. 2019;25(11):1684–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zou J, Zhang B, Gutmann DH, Wong M. Postnatal reduction of tuberous sclerosis complex 1 expression in astrocytes and neurons causes seizures in an age-dependent manner. Epilepsia. 2017;58(12):2053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488(7413):647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Reith RM, McKenna J, Wu H, Hashmi SS, Cho SH, Dash PK, et al. Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol Dis. 2013;51:93–103. [DOI] [PubMed] [Google Scholar]
- 157.Cupolillo D, Hoxha E, Faralli A, De Luca A, Rossi F, Tempia F, et al. Autistic-Like Traits and Cerebellar Dysfunction in Purkinje Cell PTEN Knock-Out Mice. Neuropsychopharmacology. 2016;41(6):1457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kosillo P, Doig NM, Ahmed KM, Agopyan-Miu A, Wong CD, Conyers L, et al. Tsc1-mTORC1 signaling controls striatal dopamine release and cognitive flexibility. Nat Commun. 2019;10(1):5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McMahon JJ, Yu W, Yang J, Feng H, Helm M, McMahon E, et al. Seizure-dependent mTOR activation in 5-HT neurons promotes autism-like behaviors in mice. Neurobiol Dis. 2015;73:296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Benthall KNCK, Agopyan-Miu AHCW,Chen EY, Bateup HS. Loss of Tsc1 from striatal direct pathway neurons impairs endocannabinoid-LTD and enhances motor routine learning. bioRxiv 101101/20191215877126. 2019. [Google Scholar]
- 161.Normand EA, Crandall SR, Thorn CA, Murphy EM, Voelcker B, Browning C, et al. Temporal and mosaic Tsc1 deletion in the developing thalamus disrupts thalamocortical circuitry, neural function, and behavior. Neuron. 2013;78(5):895–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Feliciano DM, Su T, Lopez J, Platel JC, Bordey A. Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J Clin Invest. 2011;121(4):1596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nguyen LH, Mahadeo T, Bordey A. mTOR Hyperactivity Levels Influence the Severity of Epilepsy and Associated Neuropathology in an Experimental Model of Tuberous Sclerosis Complex and Focal Cortical Dysplasia. J Neurosci. 2019;39(14):2762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Blair JD, Hockemeyer D, Bateup HS. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med. 2018;24(10):1568–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Blair JD, Bateup HS. New frontiers in modeling tuberous sclerosis with human stem cell-derived neurons and brain organoids. Dev Dyn. 2020;249(1):46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Costa V, Aigner S, Vukcevic M, Sauter E, Behr K, Ebeling M, et al. mTORC1 Inhibition Corrects Neurodevelopmental and Synaptic Alterations in a Human Stem Cell Model of Tuberous Sclerosis. Cell Rep. 2016;15(1):86–95. [DOI] [PubMed] [Google Scholar]
- 167.Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, et al. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell. 2017;20(3):385–96 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Afshar Saber W, Sahin M. Recent advances in human stem cell-based modeling of Tuberous Sclerosis Complex. Mol Autism. 2020;11(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Meikle L, Pollizzi K, Egnor A, Kramvis I, Lane H, Sahin M, et al. Response of a neuronal model of tuberous sclerosis to mammalian target of rapamycin (mTOR) inhibitors: effects on mTORC1 and Akt signaling lead to improved survival and function. J Neurosci. 2008;28(21):5422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lin TV, Hsieh L, Kimura T, Malone TJ, Bordey A. Normalizing translation through 4E-BP prevents mTOR-driven cortical mislamination and ameliorates aberrant neuron integration. Proc Natl Acad Sci U S A. 2016;113(40):11330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Getz SA, DeSpenza T Jr., Li M, Luikart BW. Rapamycin prevents, but does not reverse, aberrant migration in Pten knockout neurons. Neurobiol Dis. 2016;93:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tsai PT, Rudolph S, Guo C, Ellegood J, Gibson JM, Schaeffer SM, et al. Sensitive Periods for Cerebellar-Mediated Autistic-like Behaviors. Cell Rep. 2018;25(2):357–67 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cox RL, Calderon de Anda F, Mangoubi T, Yoshii A. Multiple Critical Periods for Rapamycin Treatment to Correct Structural Defects in Tsc-1-Suppressed Brain. Front Mol Neurosci. 2018;11:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zeng LH, Rensing NR, Wong M. The mammalian target of rapamycin signaling pathway mediates epileptogenesis in a model of temporal lobe epilepsy. J Neurosci. 2009;29(21):6964–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tsai PT, Greene-Colozzi E, Goto J, Anderl S, Kwiatkowski DJ, Sahin M. Prenatal rapamycin results in early and late behavioral abnormalities in wildtype C57BL/6 mice. Behav Genet. 2013;43(1):51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Urbanska M, Gozdz A, Swiech LJ, Jaworski J. Mammalian target of rapamycin complex 1 (mTORC1) and 2 (mTORC2) control the dendritic arbor morphology of hippocampal neurons. J Biol Chem. 2012;287(36):30240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McCabe MP, Cullen ER, Barrows CM, Shore AN, Tooke KI, Laprade KA, et al. Genetic inactivation of mTORC1 or mTORC2 in neurons reveals distinct functions in glutamatergic synaptic transmission. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Angliker N, Burri M, Zaichuk M, Fritschy JM, Ruegg MA. mTORC1 and mTORC2 have largely distinct functions in Purkinje cells. Eur J Neurosci. 2015;42(8):2595–612. [DOI] [PubMed] [Google Scholar]
- 179.Zhu PJ, Chen CJ, Mays J, Stoica L, Costa-Mattioli M. mTORC2, but not mTORC1, is required for hippocampal mGluR-LTD and associated behaviors. Nat Neurosci. 2018;21(6):799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Huang WC, Chen Y, Page DT. Hyperconnectivity of prefrontal cortex to amygdala projections in a mouse model of macrocephaly/autism syndrome. Nat Commun. 2016;7:13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Weston MC, Chen H, Swann JW. Loss of mTOR repressors Tsc1 or Pten has divergent effects on excitatory and inhibitory synaptic transmission in single hippocampal neuron cultures. Front Mol Neurosci. 2014;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30(2):694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yan J, Porch MW, Court-Vazquez B, Bennett MVL, Zukin RS. Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc Natl Acad Sci U S A. 2018;115(41):E9707–E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sare RM, Song A, Loutaev I, Cook A, Maita I, Lemons A, et al. Negative Effects of Chronic Rapamycin Treatment on Behavior in a Mouse Model of Fragile X Syndrome. Front Mol Neurosci. 2017;10:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18(5):358–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fellmann C, Gowen BG, Lin PC, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2017;16(2):89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Adli M The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1):1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ingusci S, Verlengia G, Soukupova M, Zucchini S, Simonato M. Gene Therapy Tools for Brain Diseases. Front Pharmacol. 2019;10:724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Deverman BE, Ravina BM, Bankiewicz KS, Paul SM, Sah DWY. Gene therapy for neurological disorders: progress and prospects. Nat Rev Drug Discov. 2018;17(9):641–59. [DOI] [PubMed] [Google Scholar]