Summary
Streptococcal pyruvate oxidase (SpxB) is a hydrogen peroxide-generating enzyme and plays a critical role in Streptococcus sanguinis interspecies interactions, but little is known about its biochemistry. We examined SpxB subcellular localization using protein fractionation and microscopy and found SpxB to be primarily cytoplasmic, but a small portion is also membrane associated. Potential post-translational modifications of SpxB were determined using coimmunoprecipitation and mass spectrometry. Two mutant strains were constructed to further validate the presence of predicted site-specific post-translational modifications. These site mutated SpxB proteins exhibited reduced solubility in vivo, which likely contributes to the observed phenotypic changes in colony morphology, bacterial growth, and H2O2 production. Overall, our data suggest that SpxB posttranslational modifications likely play a major role to regulate SpxB function in S. sanguinis.
Keywords: Pyruvate oxidase, SpxB, Gram-positive, Streptococcus sanguinis
Introduction
Streptococcus sanguinis is a pioneer colonizer of the tooth surface and its abundance typically correlates with oral health. For example, early ecological studies demonstrated its inverse relationship with the cariogenic species Streptococcus mutans (Becker et al., 2002; Caufield et al., 2000). Similarly, it also antagonizes several other pathobionts of the oral microbiome, such as Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, and Prevotella intermedia (Herrero et al., 2016). While several factors might aid S. sanguinis in the colonization and niche occupation at the early developmental steps of biofilm formation (J. Kreth et al., 2017), its ability to produce and release hydrogen peroxide (H2O2) has been directly associated with the inhibition of S. mutans, A. actinomycetemcomitans, P. gingivalis, and P. intermedia (Herrero et al., 2016; Jens Kreth et al., 2005, 2008). Bacterial H2O2 production and its release into the environment plays important roles in multiple microbial processes, such as extracellular DNA production for horizontal gene exchange (Jens Kreth et al., 2009) as well as biofilm development (Jang et al., 2016; Watson et al., 2018). It also regulates cell death (Li et al., 2016; Sumioka et al., 2017), contributing to intraspecies and interspecies competition (Jens Kreth et al., 2005) and bacterial virulence (Brissac et al., 2018; Erttmann & Gekara, 2019; Orihuela et al., 2004; Pericone et al., 2000; Spellerberg et al., 1996). H2O2 production is typically associated with aerobic metabolism. The pyruvate oxidase SpxB is one of the largest sources of H2O2 production among most oral streptococci, including S. sanguinis (Jan Carlsson et al., 1987; Zheng et al., 2011). SpxB is a decarboxylase that can convert pyruvate, inorganic phosphate, and oxygen into acetyl phosphate, carbon dioxide (CO2), and H2O2 (J. Carlsson & Kujala, 1984; Tittmann, 2009). Pyruvate oxidase is important in several metabolic processes. Besides the production of H2O2, it also affects colony morphologies (Belanger et al., 2004), mediates the lactate-to-acetate pathway (Lorquet et al., 2004), produces additional ATP for stationary phase survival (Goffin et al., 2006), and influences natural competence development (Bättig & Mühlemann, 2008).
SpxB reaches its highest specific activity in the early phase of aerobic exponential growth (Jan Carlsson & Edlund, 1987; Zheng et al., 2011). Under anaerobic conditions, spxB transcription and SpxB abundance both decrease (Zheng et al., 2011). SpxB is not only highly conserved among various S. sanguinis strains (J. Kreth et al., 2017), but is also conserved among the majority of oral streptococcal species with ~96–98% amino acid identity (Redanz et al., 2018). Furthermore, SpxB expression has been detected in oral plaque samples (Lin Zhu et al., 2014), indicating its production in vivo. Our current understanding of SpxB is primarily limited to its ecological function and spxB gene regulation, whereas little is known about SpxB biochemistry. Thus, we examined SpxB subcellular localization and post-translational modification using a combination of fluorescent microscopy, mass spectrometry, and site-specific mutagenesis to better understand its functional role in S. sanguinis.
Results
SpxB is primarily localized in the cytoplasm of S. sanguinis.
Bioinformatic analysis of S. sanguinis SpxB (SSA_0391) indicates that it lacks transmembrane segments, a signal peptide, and a sortase LPXTG motif, which suggests SpxB is likely to be localized in the cytoplasm (data not presented). To verify this prediction, we employed our established fractionation protocol to examine SpxB abundance in both the cytoplasm and cell membrane (Mu et al., 2019). The chromosomal copy of spxB was modified to express a C-terminal FLAG tag, while the wild type SK36 served as a negative control. We also constructed FtsH-FLAGECT, which expresses an N-terminal FLAG-tagged FtsH from vector pVA380 in the wild type background as a positive control for membrane localization (Ito & Akiyama, 2005; Mu et al., 2019). In addition, both mid-logarithmic and stationary phase cells were examined via anti-FLAG western blot (Fig. 1A&1B). In agreement with our predictions, SpxB was mostly localized in the cytoplasmic fraction at both growth stages. However, a small portion of SpxB was detected in the membrane fraction as well. As expected, FtsH was exclusive to the membrane fraction.
Fig. 1. In vivo SpxB protein localization.
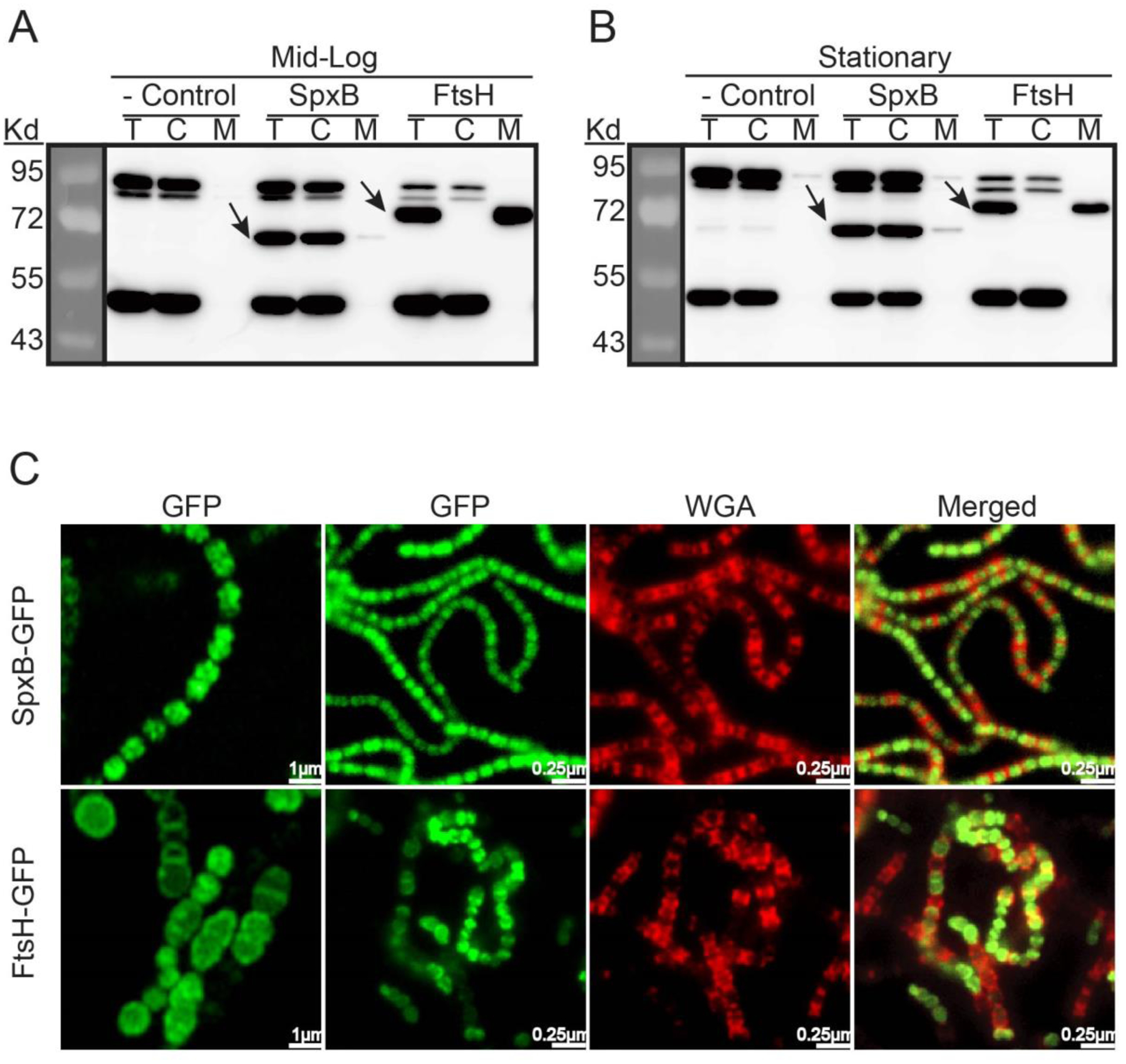
Compartmental protein isolation and Western blot analysis of: A) Cells harvested at exponential phase. B) Cells harvested at early stationary phase. Bacterial cultures were collected at OD600 ~ 0.5 (mid logarithmic phase) and ~ 1.0 (early stationary phase). 40μg protein of each sample from cell lysates (T), cell cytoplasmic (C) and membrane (M) fractions were analyzed by SDS-PAGE. FLAG-tagged SpxB and FtsH were immunoblotted with anti-FLAG antibody. S. sanguinis strain SK36 was used as negative control. Black arrows point to the respective proteins. C) Fluorescent microscopy of live cells. Cells were stained with wheat germ agglutinin (WGA, red) and visualized for green fluorescent protein fluorescence (GFP, green).
As further evidence, we also generated the strain SpxB-GFP, which encodes a C-terminal fusion of sfGFP (Kjos et al., 2015) to SpxB. Furthermore, strain FtsH-GFPECT was also constructed as a microscopy control for membrane localization (Table 1). Fluorescence microscopy revealed a distinct cellular localization of SpxB and FtsH. SpxB was localized in the cytoplasm, sometimes appearing in clusters (Fig. 1C). However, FtsH seemed to be distributed around the cell membrane as well as the division septum (Fig. 1C). Overlay images with the cell surface lectin Wheat Germ Agglutinin (WGA) (Fig.1C), exhibited minimal overlap with SpxB, whereas FtsH overlapped with WGA red fluorescence, especially at the division septum. Taken together, our data suggest SpxB is primarily a cytoplasmic protein (Fig. 1A&1B).
Table 1.
Strains used and generated in this study
| Strains | Description | Reference |
|---|---|---|
| SK36 | WT Streptococcus sanguinis | (Xu et al., 2007) |
| SK36-IFDC-1 | SK36 with IFDC inserted in spxB ORF | this study |
| SK36-IFDC-2 | SK36 with IFDC inserted after spxB ORF | this study |
| SpxB-FLAG | C-terminally FLAG-tagged spxB in SK36 | (Cheng et al., 2018) |
| SpxB-M5 | C-terminally FLAG-tagged spxB with Y212D, Y384D, T409D, T415D and T508D mutations in SK36 | This study |
| SpxB-M2 | C-terminally FLAG-tagged spxB with Y212D and Y384D mutations in SK36 | This study |
| FtsH-FLAGECT | SK36 + pftsHF, Specr | This study |
| FtsH-GFPECT | SK36 + pftsHG, Specr | This study |
| SpxB-GFP | C-terminal sfgfp fused spxB in strain SK36 | This study |
| ΔSpxB | spxB deletion in strain SK36, Ermr | (Cheng et al., 2018) |
| SpxB-FLAGECT | ΔspxB + pspxBF | This study |
| SpxB-M5ECT | ΔspxB + pspxBM5F | This study |
| SpxB-M2ECT | ΔspxB + pspxBM2F | This study |
| Plasmid | Description | Reference |
| pZX9 | pVA380::gyrAP::xylR::ldhP::xylAO::luc, Specr | (Xie et al., 2013b) |
| pftsHF | pVA380::gyrAP::xylR::ldhP::xylAO::N-FLAG-ftsH, Specr | This study |
| pftsHG | pVA380::gyrAP::xylR::ldhP::xylAO::N-sfgfp-ftsH, Specr | This study |
| pDL278 | E.coli-Streptococcus shuttle vector, Specr | (Xie et al., 2011b) |
| pspxBF | pVA380::ldhP::C-spxB-FLAG, Specr | This study |
| pspxBM5F | pVA380::ldhP::C-spxB-M5-FLAG, Specr | This study |
| pspxBM2F | pVA380::ldhP::C-spxB-M2-FLAG, Specr | This study |
Specr, spectinomycin resistance; Emr, erythromycin resistance
Identification of post-translational modifications by coimmunoprecipitation and mass spectrometry.
Our subcellular protein fractionation studies of SpxB showed two bands from the IP sample that exhibited strong signals with different molecular weights (approx. 70 Kd for the SpxB monomer; and a high molecular weight band over 170 kD after SDS-PAGE in reducing conditions) (Fig. 2). Both bands were excised and analyzed via mass spectrometry and confirmed to be SpxB (Table S2), indicating SpxB either forms multimers or a subfraction is post-translationally modified. The MS/MS search program Byonic (Bern et al., 2012) was employed to predict post-translational modification sites based upon observed molecular weight shifts within tryptic peptides. Five modified sites were identified to have 80 Da added to the identified amino acids consistent with phosphorylation (Fig. S1). The SpxB monomer contained four predicted phosphorylation sites (212Y, 409T, 415T and 508T) and the multimer contained two predicted phosphorylation sites (212Y and 384Y), respectively. However, subsequent analysis of SpxB using Phos-tag™ SDS- PAGE (Fig. S2), did not show any differentially migrating bands (Fig. S2).
Fig. 2. Western blot of FLAG-tagged SpxB.
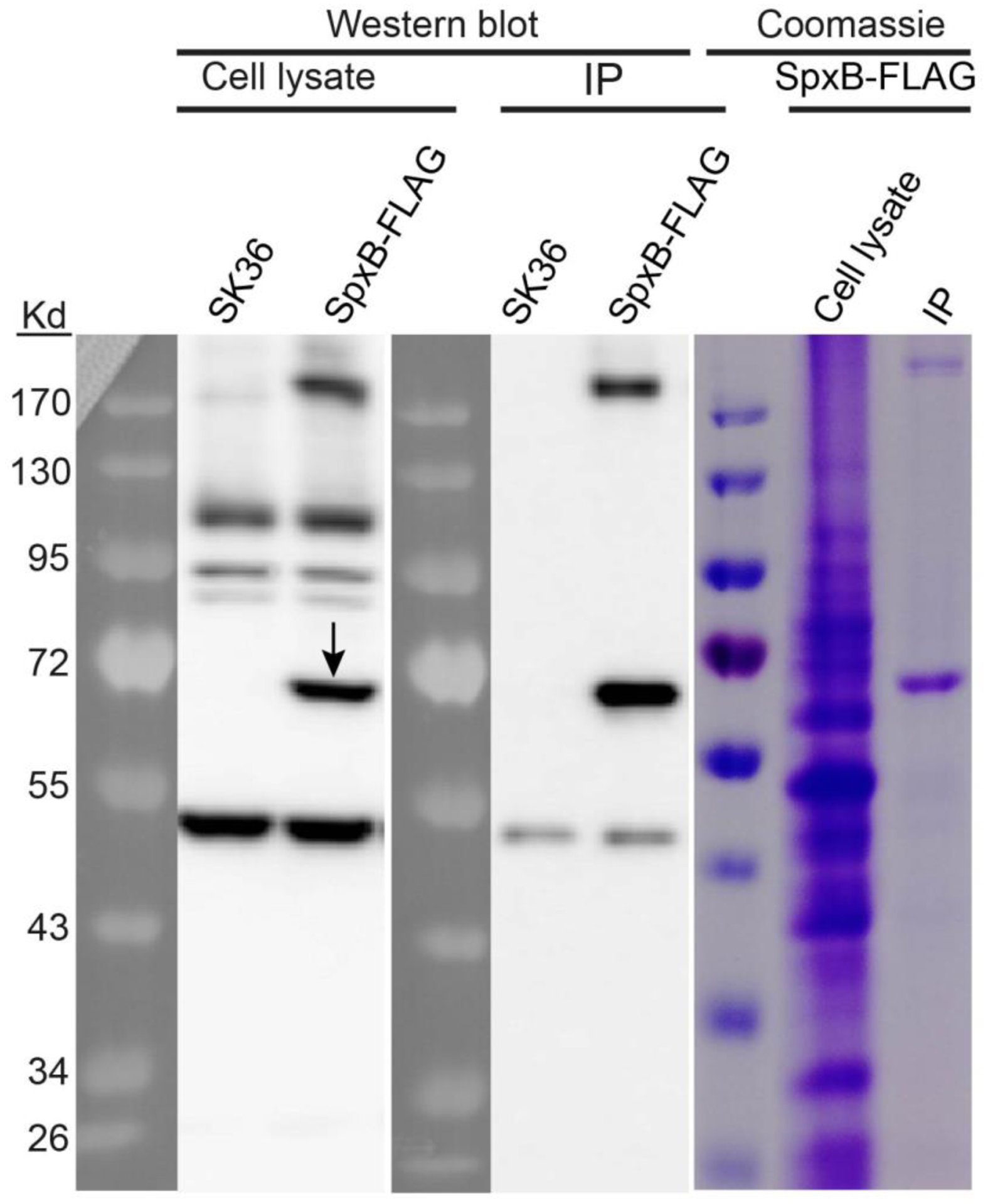
S. sanguinis strain SK36 was used as the negative control. Cell lysates and immunoprecipitation (IP) samples from both SK36 strain and SpxB-FLAG strain were detected by anti-FLAG antibody on blots. The cell lysate and the IP sample from SpxB-FLAG strain were also examined by Coomassie staining. Black arrow indicates SpxB protein.
SpxB site-specific mutations interfere with colony morphology, biofilm formation and cell growth.
To understand whether the predicted post-translational modifications affect SpxB function and S. sanguinis phenotypic traits, two site-specific SpxB mutants were generated (Table 1). Strain SpxB-M5 contains aspartic acid substitutions in each of the predicted post-translational modification sites, while strain SpxB-M2 contains aspartic acid substitutions in two predicted tyrosine phosphorylation sites (Fig.3A). Colony morphology was examined after growth under aerobic conditions in the presence of 5% CO2, which allows for the expression of spxB as we previously demonstrated (Zheng et al., 2011). The strains harboring either wild type SpxB or C-terminal FLAG-tagged SpxB produced small and flat colonies (Fig. 3B&3C), while the SpxB deletion strain ΔSpxB (Cheng et al., 2018) and the other two site-specific mutation strains produced larger and raised colonies (Fig.3B&3C). We next examined the growth of planktonic cells in Brain Heart Infusion (BHI) medium. The growth curve showed that strain SpxB-M2 and ΔSpxB had a statistically significant differences at certain time points during the log phase when compared to the wild type, which was possibly caused by the delayed lag phase, but the overall growth rate seemed to be similar in the exponential phase (Fig. 3D). In addition, biofilm formation was determined and confirmed the previously reported biofilm phenotype for the spxB deletion in streptococci (Blanchette et al., 2016). While SpxB-M2 showed a similar biofilm phenotype as the spxB deletion mutant, SpxB-M5 showed no difference when compared to the wild type (Fig. 3E).
Fig. 3. Colony morphology, biofilm formation and growth of SpxB mutants.
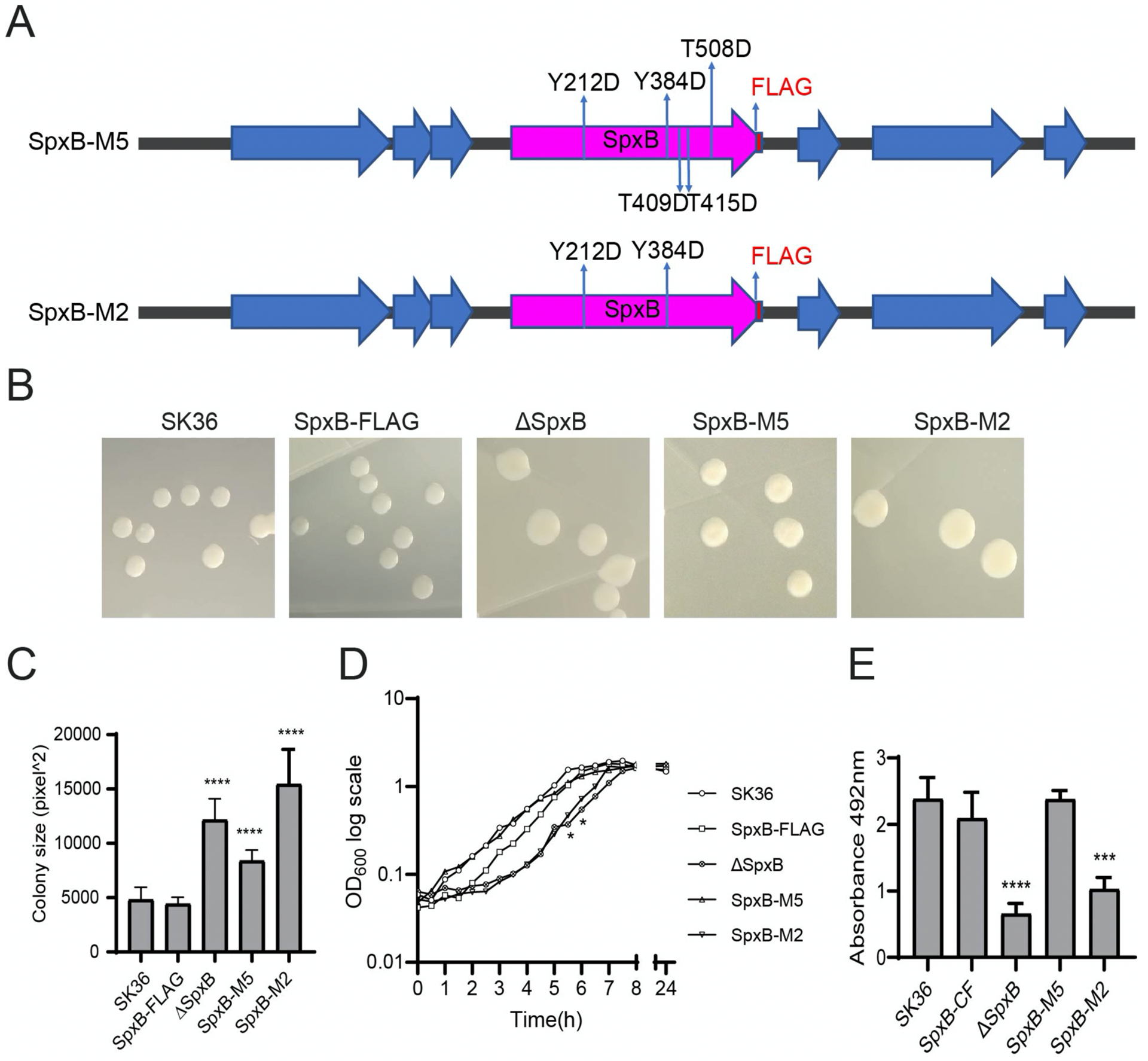
A) Schematic representation of introduced site-specific mutations in spxB-M5 and spxB-M2. B) Representative colony morphologies of each indicated strain. C) Colony size was measured by Image J- software and the average size of colonies is presented in the bar graph (n=6). D) Growth curves were generated by growing bacteria in Brain Heart Infusion (BHI) broth. OD600 values were recorded at 30 min intervals and are presented on a logarithmic scale. The data presented are representative of three independent experiments with similar results. Statistical differences between time points were identified by unpaired T test compared with wild type SK36. (* P < 0.05, ** P< 0.01). E) Biofilm formation of SK36 and mutants. The error bars indicate standard deviation. Differences in biofilm mass among mutants was examined by unpaired T test compared with SK36. (* P<0.05; ** P< 0.01; *** P< 0.001; **** P<0.0001; n=5)
SpxB site-specific mutations also affect H2O2 production.
Prussian blue agar plates were used for the detection of H2O2 production from the spxB point mutant strains. The size of the blue halo around the inoculation spot is proportional to H2O2 abundance. Both SpxB-M5 and SpxB-M2 demonstrated a decreased production of H2O2 compared to SK36 and SpxB-FLAG (Fig. 4A and B). We also examined the expression of spxB in the various strains. While some variation was detected, gene expression differences were not statistically significant (Fig. S4).
Fig. 4. H2O2 production of SpxB mutants.
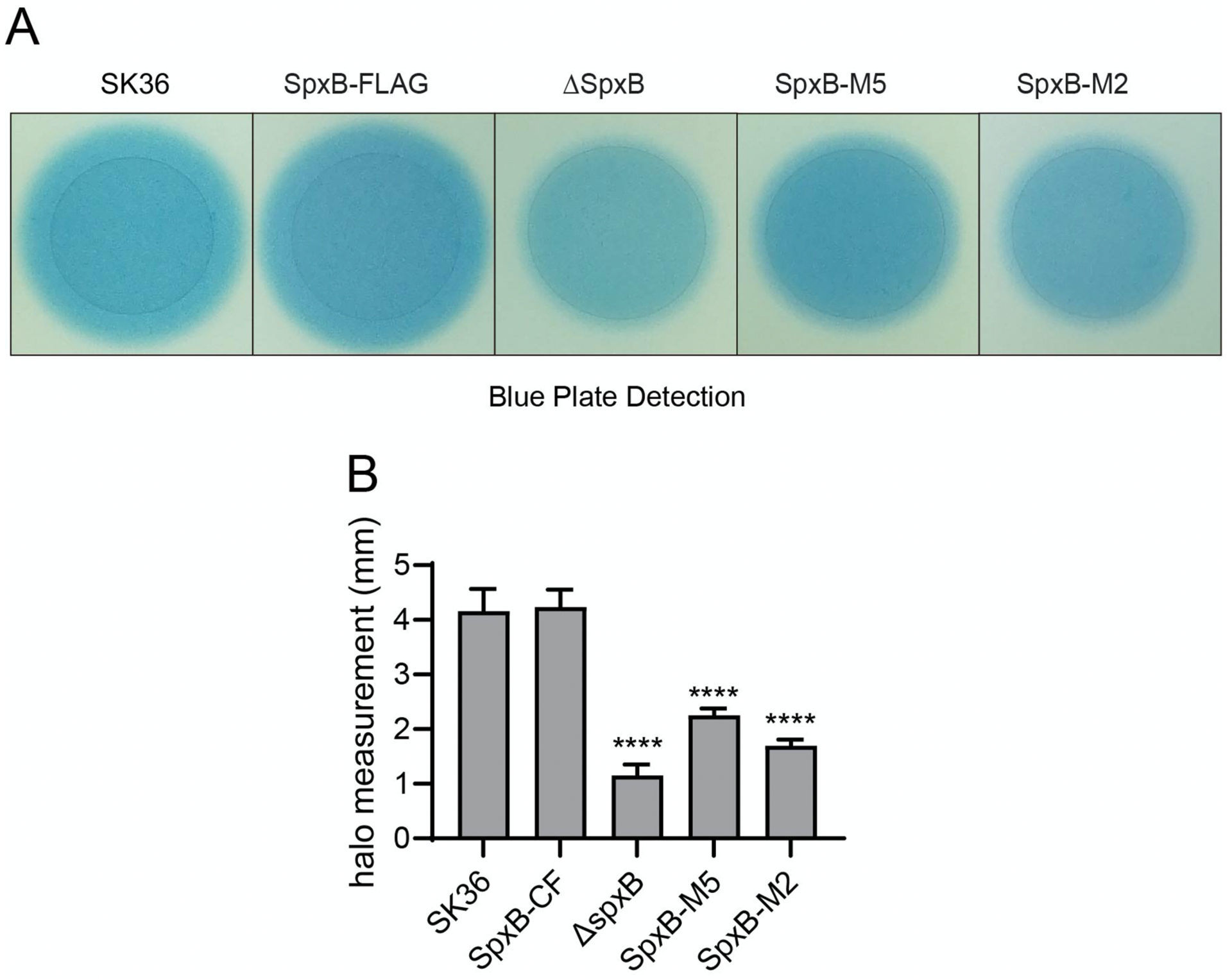
A) Prussian blue agar plates were used to examine the H2O2 production. Overnight cultures were spotted on the plates and incubated for 24 h. The individual inoculation spots from the same plate are presented side by side for easier comparison. The unaltered plate picture is presented in Fig. S3. Shown are representative images of three independent experiments. B) Quantification of H2O2 production. For each strain, the distance from the edge of the colony to the outer circle was randomly measured for four times from different positions. The difference of the distance among mutants was examined by unpaired T test compared with SK36. (* P < 0.05, ** P< 0.01, *** P< 0.001; **** P<0.0001. n = 2).
SpxB protein abundance is changed in site-specific mutant strains.
To learn if the site-specific mutations of the putative post-translational modification sites influence SpxB abundance, we examined the protein expression in wild-type and mutant SpxB from mid-logarithmic phase cultures. As shown in Fig. 5, the amount of mutated SpxB from both SpxB-M5 and SpxB-M2 strains obviously decreased in the protein extracts (Fig. 5A) when compared to the C-terminal FLAG-tagged SpxB wild type control. We also noticed that protein from the SpxB-M5 mutant exhibited slightly slower mobility, presumably due to changes in its overall charge. Immunoprecipitation (Fig. 5B) was also conducted to confirm the weak signals of SpxB-M5 and SpxB-M2 in Fig. 5A are not unspecific bands or background noise.
Fig. 5. SpxB site-specific mutations affect protein abundance.
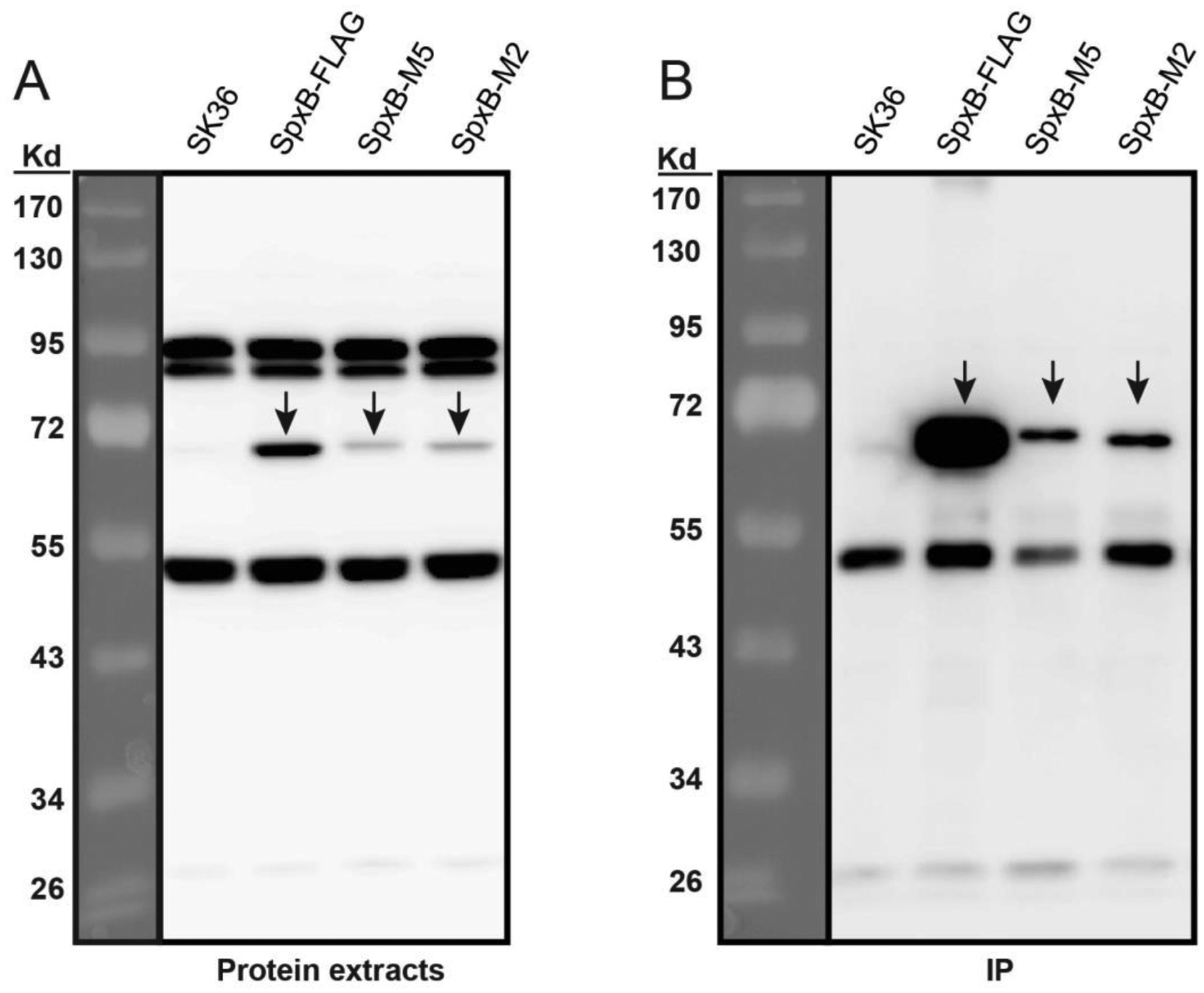
Protein samples were derived from cultures collected at OD600=0.5. A) Western blot of protein extracts. Forty μg of protein extract was loaded for each sample and detected by anti-FLAG antibodies. Wild type SK36 strain was used as a negative control. B) Western blot of protein immunoprecipitation (IP). The IP samples were obtained by immunoprecipitating 1 mg protein extracts through anti-FLAGM2 Affinity Gel and eluted with FLAG peptides. The same volume of IP samples were loaded for comparison among different stains. SpxB protein and its mutated forms are indicated by black arrows.
Site-specific mutations interfere with SpxB protein solubility.
We noticed a difference in protein abundance with the SpxB-M5 and SpxB-M2 mutants compared to the parental strain despite exhibiting no obvious differences in gene expression. Next, we examined the protein stability of SpxB in ectopically expressing strains SpxB-FLAGECT, SpxB-M5ECT, and CspxB-M2ECT. SpxB and its site-specific mutations were ectopically expressed under the control of the lactate dehydrogenase (ldh) promoter in the parental strain ΔSpxB (Cheng et al., 2018) to eliminate any effect of the native SpxB protein. As shown in Fig. S5, all SpxB proteins were quite stable in vivo within 16 hours. Interestingly, when ectopically expressed and analyzed as total cell lysates SpxB-M5 and SpxB-M2 proteins seemed to be as abundant as the WT SpxB protein (Fig. S5), which prompted us to investigate if protein solubility is affected by the site-specific mutations.
Whole cell lysates as well as soluble and insoluble fractions from SpxB-FLAGECT, SpxB-M5ECT, and SpxB-M2ECT were examined for SpxB abundance. As shown in Fig.6, the proportion of soluble protein decreased significantly in strains SpxB-M5ECT and SpxB-M2ECT, indicating the site-specific mutations reduced SpxB solubility. In addition, multimer formation seemed to be affected as well since the soluble portion did not contain any high molecular bands while the insoluble samples had an increased abundance in the site-specific mutations.
Fig. 6. SpxB site-specific mutations changed protein solubility.
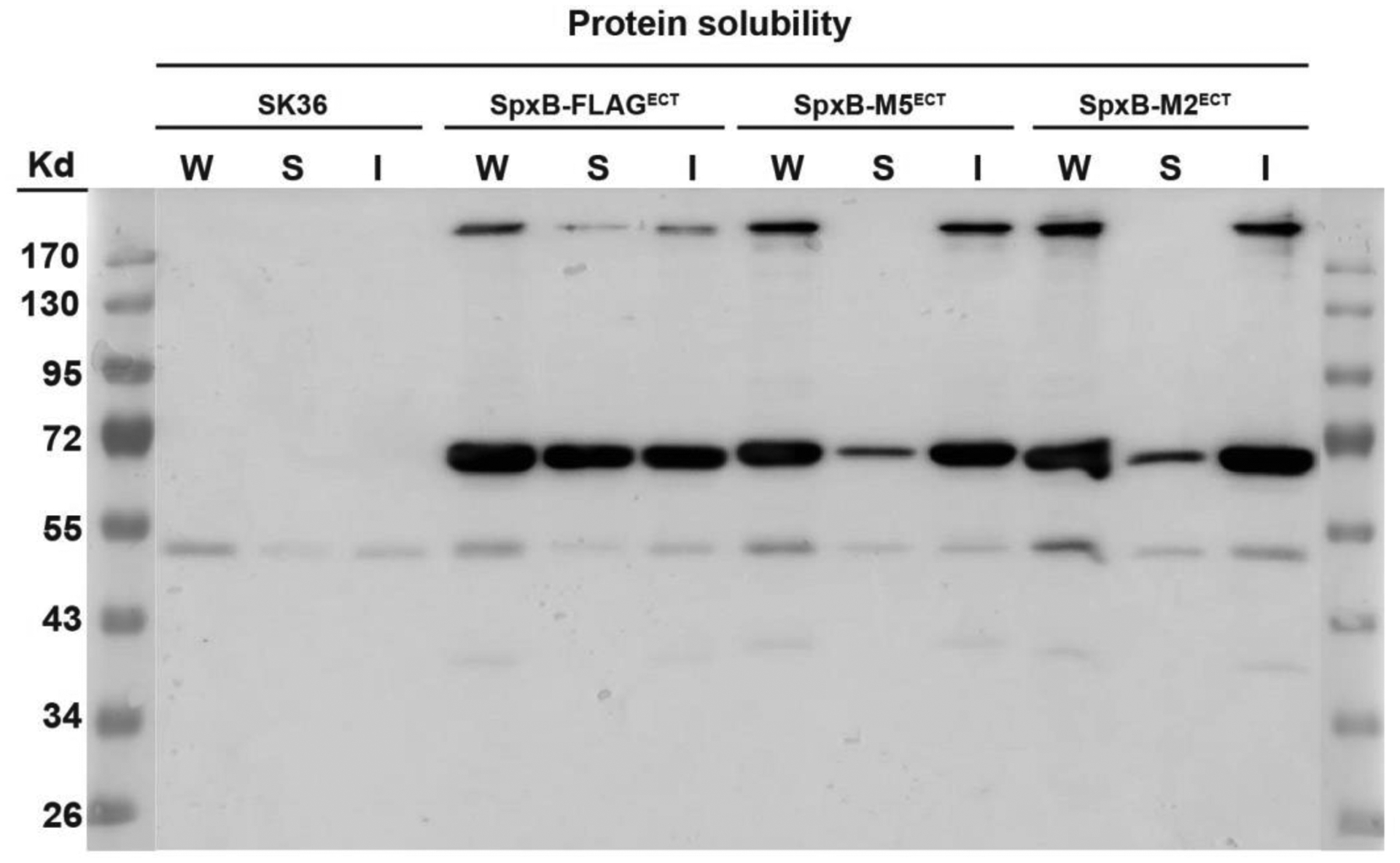
FLAG-tagged SpxB with and without site-specific mutations were ectopically expressed on the plasmid pVA380 under the control of the lactate dehydrogenase promoter (Pldh) in the spxB deletion background. Wild type SK36 was used as a negative control. Cells were grown to mid logarithmic phase in BHI medium for protein extraction. After cell homogenization, the suspension was collected and designated as whole cell lysate (W). The whole cell lysate was centrifuged to separate the soluble proteins (S) and the pellet containing insoluble proteins (I).
Discussion
In the present article, we further investigated S. sanguinis SK36 SpxB cellular localization and post-translational modifications. We confirmed the intracellular localization of SpxB by fractionating cellular lysates combined with western blot analysis and fluorescence microscopy. Furthermore, post-translational modifications of SpxB were predicted through mass spectrometry (Fig.S1). Importantly, we confirmed that SpxB biological functions are affected when the identified sites are mutated. The engineered mutations in SpxB affected the protein electrophoretic mobility, colony morphology, cell growth, and SpxB solubility. As a consequence, the production of H2O2 by SpxB was impaired. Cellular localization studies revealed SpxB to be localized mainly in the cytoplasm. Similarly, fluorescence microscopy analysis of SpxB failed to demonstrate colocalization with the cell surface stain WGA, further supporting cytoplasmic localization. We did, however, observe a faint band in the membrane fraction western blots, suggesting a small portion of the protein could be membrane associated. The functional related pyruvate oxidase (EcPOX, EC 1.2.2.2) of Escherichia coli has the ability to interact with the membrane through a flexible C-terminal membrane anchor (Neumann et al., 2008). Structural modeling of SpxB showed an overall conservation of structural features with EcPOX (Fig. S6A). Further comparison of the respective C-terminal segments of both enzymes demonstrated a similar coiled structure that has been postulated as a linker region (Fig. S6B, arrows) in EcPOX. This is followed by the EcPOX alpha peptide required for membrane interaction (Fig. S6B). However, the alpha peptide seems to be absent or replaced by another coiled structure in SpxB. Although the structural features of the C-terminal end are somewhat divergent, it is conceivable that the SpxB C-terminal end may also mediate membrane interactions. Future studies are planned to investigate this in further detail.
In agreement with published data of the pyruvate oxidase of Lactobacillus plantarum (Sedewitz et al., 1984), which has 49% identity at the protein level and 66% positive conservative exchanges (Redanz et al., 2018), SpxB does form multimers as evident from the high molecular weight bands visible during SDS-PAGE (see also Fig. S7 for un-boiled samples separated by SDS-PAGE). The high-molecular mass bands were confirmed to be SpxB by mass spectrometry. The biologically relevant forms of E. coli and L. plantarum pyruvate oxidases are homo-tetramers (Neumann et al., 2008; Sedewitz et al., 1984), but other multimeric forms (pentamers) have been described in the case of Lactobacillus delbrueckii pyruvate oxidase Pox (Cornacchione & Hu, 2020). Modeling of the protein structure using the Swiss Model server (Waterhouse et al., 2018) predicted a similar homo-tetramer configuration for SpxB (Fig. S6C). While it seems unusual that multimeric forms are present in SDS gels containing the reducing agent 2-mercaptoethanol, it is not unprecedented for pyruvate oxidases. Staphylococcus aureus pyruvate:menaquinone oxidoreductase CidC also shows resistance to 2-mercaptoethanol treatment (Zhang et al., 2017). Interestingly, the pentameric Pox from L. delbruckii is dissolved by 6% 2-mercaptoethanol (Cornacchione & Hu, 2020). Both, S. sanguinis SpxB and S. aureus CidC contain 3 cysteine residues, while L. delbruckii Pox only has one cysteine. A structural SpxB model predicts C488 to be solvent exposed at the surface, suggesting could potentially form di-sulfide bridges to other SpxB monomers. Such an interaction would be expected to confer the complex with enhanced stability, perhaps even during SDS-PAGE (Fig. S8).
Mass spectrometry identified 5 separate 80 da mass shifts, which is consistent with phosphorylation post-translational modifications. However, we were not able to detect phosphorylation using the Phos-Tag strategy (Kinoshita et al., 2006) or via western blot analysis with a phosphorylation specific antibody (data not shown). While the phosphorylation sites were predicted by the MS/MS search program Byonic, we certainly cannot exclude the possibility that these sites are modified with alternative post-translational modifications conferring similar molecular mass shifts. Thus, the nature of the predicted post-translational modification requires more in-depth analysis in the future.
Regardless, we wanted to determine whether these residues are important for SpxB function. SpxB-M5 exhibited a slightly slower migration during SDS-PAGE. SpxB-M2 migrated identically to the wild type SpxB, suggesting that the T409D, T415D, and T508D mutations were primarily responsible for this effect. Aspartic acid residues introduce negative charges to the protein that could conceivably interfere with SDS binding and decrease SpxB mobility during SDS-PAGE.
The introduction of site-specific mutations also impacted SpxB solubility. This phenomenon was observed while comparing protein extraction results with gene expression data. Therefore, we compared wild type and mutant SpxB abundance after separating the soluble and insoluble protein fractions. Interestingly, while the total abundance of SpxB is comparable among all tested strains, the protein solubility of the mutant strains is significantly reduced. Moreover, the introduction of site-specific mutations changed several phenotypic characteristics associated with SpxB. Strains carrying the mutations exhibited significantly larger colonies compared to the wild type strain. This correlated with slightly higher optical densities observed from the mutant strains after overnight growth. We suspect this is likely a consequence of the decreased H2O2 production from the mutant strains, since H2O2 triggers some toxicity to the producer (Pericone et al., 2000). We also observed a reduction in biofilm formation for the SpxB-M2 mutant and confirmed the biofilm phenotype for a deletion of spxB in streptococci (Blanchette et al., 2016). The biofilm formation of the SpxB-M5 mutant strain was not impaired under the test conditions. This is in agreement with other phenotypes of this mutant strain resembling more the wild type. While the reason for this observation is currently not clear, overall the potential in vivo post-translational modification of SpxB might aid S. sanguinis for faster environmental adaptation under certain condition, rather than changing expression of spxB, which is a more time consuming process.
In conclusion, we have confirmed that S. sanguinis SpxB mainly localizes to the cytoplasm and we identified amino acids 212Y, 384Y, 409T, 415T and 508T as possible post-translational modification sites. These residues were shown to be important for SpxB function and stability.
Materials and Methods
Bacterial strains and culture conditions.
Streptococcus sanguinis SK36 (Xu et al., 2007) (Table 1) was routinely grown anaerobically (90% N2–5% CO2–5% H2) or aerobically (5% CO2) at 37°C in brain heart infusion broth (BHI; Difco, Sparks, MD) or on BHI agar plates. When required, 500μg ml−1 spectinomycin (Sigma-Aldrich, St. Louis, MO) or 10μg ml−1 erythromycin (Fisher Scientific, Pittsburgh, PA) was used for antibiotic selection. SK36 competence-stimulating peptide CSP (DLRGVPNPWGWIFGR) (Zhu et al., 2011) was synthesized by ELIM Biopharmaceuticals (Hayward, USA) and supplemented in cultures at a concentration of 0.3 mM for transformation reactions. For counterselection, THYE plates were supplemented with 0.4% (w/v) p-chlorophenylalanine (4-CP, Sigma).
Construction of chromosomal mutants and fusion proteins.
To construct in-frame markerless deletions, site-specific mutations and insertions, an in-frame deletion cassette (IFDC) for S. sanguinis (NCBI reference sequence NC_009009.1) was created on the basis of an IFDC for S. mutans (Xie et al., 2011a) and modified for S. sanguinis (Cheng et al., 2018). The constructs for site-specific mutations were generated via overlap extension PCR (OE-PCR). All plasmids were assembled via Gibson cloning (Gibson, 2011). pZX9 vector (Xie et al., 2013a) and pDL278 vector (Y. Y. Chen & LeBlanc, 1992) were used in this study. Detailed information can be found in the supplementary data.
Protein extraction.
For protein extraction, strains were grown in 50 ml liquid BHI medium until OD600 ~ 0.5. Cells were collected by centrifugation at 3,220 × g for 15 min at 4 °C (Eppendorf, centrifuge 5810R). Cell pellets were resuspended in 1 ml of 1 X phosphate-buffered saline (PBS) pH 7.4. 10mM phenylmethanesulfonyl fluoride (PMSF) was added to obtain the final concentration of 0.2 mM. Cells were lysed in a bead mill homogenizer (Bertin Corp, Precellys Evolution) with 0.1-mm silica beads (Thermo Fisher Scientific) at 5800 rpm for 12 cycles of 20s with 20s of break as previously described(Mu et al., 2019). Cell debris was separated by centrifugation at 16,000 × g for 15 min at 4°C (Eppendorf, centrifuge 5424R) and the supernatants were collected. Cytoplasmic and membrane fractionation were performed as previously described (Mu et al., 2019). To separate the cytoplasmic and membrane fractions, supernatants were transferred into 5 ml ultracentrifuge tubes (Beckman) and centrifuged at 105,000 × g for 1.5 h at 4 °C in an ultracentrifuge (Beckman L8–80M, 50.4Ti). After centrifugation, the supernatants were carefully removed and transferred to 1.5 ml tubes to serve as the cytoplasmic fractions. The remaining protein pellets were resuspended in an equal volume of PBS as the supernatants containing 1% Triton X-100 and then solubilized overnight at 4 °C with gentle agitation on a rotator. The next day, the solutions were centrifuged at 16,000 × g for 15 min at 4 °C before collecting the supernatants to serve as the membrane fractions. Protein concentrations were determined using the Bradford assay (Bio-Rad) according to the manufacturer’s protocol. Bovine serum albumin served as the standard.
Immunoprecipitation and mass spectrometry.
1 mg of total protein from the respective whole cell lysate was immunoprecipitated using anti-FLAGM2 Affinity Gel (Sigma) according to the manufacturer’s protocol. FLAG peptide (Sigma) with a working concentration of 100 μg ml−1 was used for the competitive elution of FLAG tagged SpxB. Eluates were loaded into polyacrylamide gels and the specific band of SpxB was cut for in-gel digestion using Trypsin Gold (Promega) and ProteaseMAX™ Surfactant (Promega) according to the manufacturer’s protocol. The tryptic peptides were extracted, dried in a vacuum dryer, resuspended in 15 μl 0.1% formic acid, and lastly analyzed by LC/MS using a Thermo Scientific Orbitrap Fusion mass spectrometer (Breitkopf & Asara, 2012).
Western immunoblotting.
Protein samples were separated by 10% SDS-PAGE gel and transferred to nitrocellulose membranes (GE Healthcare). Nitrocellulose membranes were blocked overnight at 4 °C with 5% (w/v) nonfat dry milk diluted in TBST buffer (0.1% Tween20 in Tris-buffered saline, pH 7.4). Blots were then incubated 1h with primary antibodies diluted 1:1000 in 5% milk-TBST at room temperature. Secondary antibodies were diluted 1:2000 in 5% milk-TBST and incubated with the blots for 1 h at room temperature. The blots were detected using the SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Scientific). Primary antibodies included mouse monoclonal ANTI-FLAG® M2 antibody (Sigma, F3165–1MG) while horseradish peroxidase (HRP)-conjugated secondary antibodies were goat anti-mouse IgG1 secondary antibody, HRP (Thermo Fisher Scientific, Catalog # PA1-74421).
Biofilm quantification.
Biofilm formation was determined by growing SK36 and mutants for 24 h in 96 well plates in CDM supplemented with 50mM sucrose with 5% CO2 at 37°C. Then the liquid medium was removed and the wells were gently washed with 1x PBS and stained with 0.1% safranin for 15 min. Biofilms were rinsed with 1x PBS and air-dried. The absorbance at 492 nm of safranin-stained biofilms was recorded as previously reported (Standar et al., 2010).
Fluorescent microscopy.
To determine the localization of SpxB, we generated strains containing SpxB C-terminal sfGFP-fused proteins for microscopy. Wheat Germ Agglutinin (WGA), Texas Red™-X Conjugate (Thermo Fisher Scientific) was used according to the manufacturer’s protocols and served as a counterstain for the cell wall of S. sanguinis. Planktonic exponential cells were used for cell staining and visualization. The WGA stain was added to cells suspended in Hanks’ Balanced Salt solution (HBSS) with a working concentration of 10 μg/ml and incubated for 30min at RT. Next, the cells were incubated at room temperature for 10 min, with rocking and protection from light as described previously (Cullin et al., 2017). Cells were imaged using both an Olympus IX73 microscope with a 100x oil immersion lens and Zeiss LSM 880 with Fast Airyscan (OHSU Advanced Light Microscopy Core).
Detection of gene expression with real-time RT-PCR.
Overnight cultures were diluted 1:30 in 40 ml BHI and grown until OD600 reached 0.5 or 1.0. The cells were harvested and stored at −80 °C for further processing. The procedures for RNA extraction, cDNA synthesis, and real-time RT-PCR have been described previously (Chen et al., 2012). Briefly, total RNA was extracted using Trizol (Invitrogen). 20μg RNA extract was used for DNase treatment with Turbo DNase (Ambion). The treated RNA was then purified with RNeasy kit (Qiagen) and 1 μg of total RNA was used for cDNA synthesis by using the iScript cDNA synthesis kit (Bio-Rad). The RT- PCR was then performed in CFX96 Touch Real-Time PCR Detection System (Bio-rad) using primers SK36_16s_RT_F/R and SpxB-Q-F-1/R-2.
Protein solubility detection.
Cells were grown to OD600 ~ 0.5. and were collected by centrifugation as described previously. After cell homogenization, the suspension was collected and designated as whole cell lysate (W). The whole cell lysate was further separated by centrifugation at 16,000 × g for 15 min at 4°C (Eppendorf, centrifuge 5424R). The supernatant contained the soluble proteins (S) and the pellet contained the insoluble proteins (I). The pellet was resuspended in PBS, keeping the volume the same as the supernatant. Finally, the same volume of whole cell lysate (W), soluble proteins (S) and insoluble proteins (I) were loaded on the protein gel for detection.
Detection of H2O2 Production.
Indicator plates for H2O2 production were prepared as described (Saito et al., 2007), suitable for the detection of bacterial H2O2 production via the formation of a blue precipitate (Prussian blue, ferric ferrocyanide) in the presence of H2O2. Briefly, bacteria grown in liquid BHI medium overnight were collected by centrifugation. 20ul of the bacterial suspension in PBS adjusted to OD600= 0.5 for each strain was spotted on Prussian blue agar plates. Plates were incubated aerobically in 5% CO2 at 37°C overnight.
Supplementary Material
Acknowledgements
We gratefully acknowledge the OHSU Proteomics Shared Resource for mass spectrometric support and analysis (partial support from NIH core grants P30EY010572 & P30CA069533; Orbitrap Fusion S10 OD012246). This project was supported by NIH grant DE028252 to JM, NIH grants DE028329, DE017954, DE022350 to HW and NIH grants DE029612, DE021726 and DE029492 to JK. The authors declare no conflict of interest.
Funding Information
NIH/NIDCR, Grant/Award Numbers: DE028252, DE028329, DE017954, DE022350 DE029612, DE021726, DE029492, P30EY010572, P30CA069533
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Data Availability Statement
The experimental data presented in this study are available from the corresponding author upon reasonable request.
References
- Bättig P, & Mühlemann K (2008). Influence of the spxB gene on competence in Streptococcus pneumoniae. Journal of Bacteriology, 190(4), 1184–1189. 10.1128/JB.01517-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, & Griffen AL (2002). Molecular analysis of bacterial species associated with childhood caries. Journal of Clinical Microbiology, 40(3), 1001–1009. 10.1128/JCM.40.3.1001-1009.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger AE, Clague MJ, Glass JI, & LeBlanc DJ (2004). Pyruvate oxidase is a determinant of Avery’s rough morphology. Journal of Bacteriology, 186(24), 8164–8171. 10.1128/JB.186.24.8164-8171.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M, Kil YJ, & Becker C (2012). Byonic: Advanced peptide and protein identification software. Current Protocols in Bioinformatics, CHAPTER(SUPPL.40), Unit13.20. 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette KA, Shenoy AT, Milner J, Gilley RP, McClure E, Hinojosa CA, Kumar N, Daugherty SC, Tallon LJ, Ott S, King SJ, Ferreira DM, Gordon SB, Tettelin H, & Orihuela CJ (2016). Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infection and Immunity, 84(10), 2922–2932. 10.1128/IAI.00277-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkopf SB, & Asara JM (2012). Determining in vivo phosphorylation sites using mass spectrometry. Current Protocols in Molecular Biology, 1(SUPPL.98), 1–27. 10.1002/0471142727.mb1819s98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissac T, Shenoy AT, Patterson LDA, & Orihuela CJ (2018). Cell invasion and pyruvate oxidase-derived H 2 O 2 are critical for Streptococcus pneumoniae-mediated cardiomyocyte killing. Infection and Immunity, 86(1). 10.1128/IAI.00569-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J, & Kujala U (1984). Pyruvate oxidase activity dependent on thiamine pyrophosphate, flavin adenine dinucleotide and orthophosphate in Streptococcus sanguis. FEMS Microbiology Letters, 25(1), 53–66. 10.1111/j.1574-6968.1984.tb01374.x [DOI] [Google Scholar]
- Carlsson Jan, & Edlund M-BK (1987). Pyruvate oxidase in Streptococcus sanguis under various growth conditions. Oral Microbiology and Immunology, 2(1), 10–14. 10.1111/j.1399-302X.1987.tb00263.x [DOI] [PubMed] [Google Scholar]
- Carlsson Jan, Edlund M-BK, & Lundmark SKE (1987). Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral Microbiology and Immunology, 2(1), 15–20. 10.1111/j.1399-302X.1987.tb00264.x [DOI] [PubMed] [Google Scholar]
- Caufield PW, Dasanayake AP, Li Y, Pan Y, Hsu J, & Hardin JM (2000). Natural history of Streptococcus sanguinis in the oral cavity of infants: Evidence for a discrete window of infectivity. Infection and Immunity, 68(7), 4018–4023. 10.1128/IAI.68.7.4018-4023.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, & LeBlanc DJ (1992). Genetic analysis of scrA and scrB from Streptococcus sobrinus 6715. Infection and Immunity, 60(9), 3739–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Itzek A, Malke H, Ferretti JJ, & Kreth J (2012). Dynamics of speB mRNA transcripts in streptococcus pyogenes. Journal of Bacteriology, 194(6), 1417–1426. 10.1128/JB.06612-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Redanz S, Cullin N, Zhou X, Xu X, Joshi V, Koley D, Merritt J, & Kreth J (2018). Plasticity of the pyruvate node modulates hydrogen peroxide production and acid tolerance in multiple oral streptococci. Applied and Environmental Microbiology, 84(2). 10.1128/AEM.01697-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchione LP, & Hu LT (2020). Hydrogen peroxide-producing pyruvate oxidase from Lactobacillus delbrueckii is catalytically activated by phosphotidylethanolamine. BMC Microbiology, 20(1), 128. 10.1186/s12866-020-01788-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullin N, Redanz S, Lampi KJ, Merritt J, & Kreth J (2017). Murein hydrolase LytF of Streptococcus sanguinis and the ecological consequences of competence development. Applied and Environmental Microbiology, 83(24). 10.1128/AEM.01709-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erttmann SF, & Gekara NO (2019). Hydrogen peroxide release by bacteria suppresses inflammasome-dependent innate immunity. Nature Communications, 10(1), 1–13. 10.1038/s41467-019-11169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG (2011). Enzymatic assembly of overlapping DNA fragments. In Methods in Enzymology (Vol. 498, pp. 349–361). Academic Press Inc. 10.1016/B978-0-12-385120-8.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin P, Muscariello L, Lorquet F, Stukkens A, Prozzi D, Sacco M, Kleerebezem M, & Hols P (2006). Involvement of pyruvate oxidase activity and acetate production in the survival of Lactobacillus plantarum during the stationary phase of aerobic growth. Applied and Environmental Microbiology, 72(12), 7933–7940. 10.1128/AEM.00659-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero ER, Slomka V, Bernaerts K, Boon N, Hernandez-Sanabria E, Passoni BB, Quirynen M, & Teughels W (2016). Antimicrobial effects of commensal oral species are regulated by environmental factors. Journal of Dentistry, 47, 23–33. 10.1016/j.jdent.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Ito K, & Akiyama Y (2005). CELLULAR FUNCTIONS, MECHANISM OF ACTION, AND REGULATION OF FTSH PROTEASE. Annual Review of Microbiology, 59(1), 211–231. 10.1146/annurev.micro.59.030804.121316 [DOI] [PubMed] [Google Scholar]
- Jang IA, Kim J, & Park W (2016). Endogenous hydrogen peroxide increases biofilm formation by inducing exopolysaccharide production in Acinetobacter oleivorans DR1. Scientific Reports, 6(1), 1–12. 10.1038/srep21121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, & Koike T (2006). Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Molecular and Cellular Proteomics, 5(4), 749–757. 10.1074/mcp.T500024-MCP200 [DOI] [PubMed] [Google Scholar]
- Kjos M, Aprianto R, Fernandes VE, Andrew PW, Van Strijp JAG, Nijland R, & Veening JW (2015). Bright fluorescent Streptococcus pneumoniae for live-cell imaging of host-pathogen interactions. Journal of Bacteriology, 197(5), 807–818. 10.1128/JB.02221-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Giacaman RA, Raghavan R, & Merritt J (2017). The road less traveled – defining molecular commensalism with Streptococcus sanguinis. Molecular Oral Microbiology, 32(3), 181–196. 10.1111/omi.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth Jens, Merritt J, Shi W, & Qi F (2005). Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. Journal of Bacteriology, 187(21), 7193–7203. 10.1128/JB.187.21.7193-7203.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth Jens, Vu H, Zhang Y, & Herzberg MC (2009). Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. Journal of Bacteriology, 191(20), 6281–6291. 10.1128/JB.00906-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth Jens, Zhang Y, & Herzberg MC (2008). Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. Journal of Bacteriology, 190(13), 4632–4640. 10.1128/JB.00276-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Zhai S, Xu M, Shang M, Gao Y, Liu G, Wang Q, & Zheng L (2016). SpxB-mediated H 2 O 2 induces programmed cell death in Streptococcus sanguinis. Journal of Basic Microbiology, 56(7), 741–752. 10.1002/jobm.201500617 [DOI] [PubMed] [Google Scholar]
- Lorquet F, Goffin P, Muscariello L, Baudry JB, Ladero V, Sacco M, Kleerebezem M, & Hols P (2004). Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. Journal of Bacteriology, 186(12), 3749–3759. 10.1128/JB.186.12.3749-3759.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu R, Shinde P, Zou Z, Kreth J, & Merritt J (2019). Examining the Protein Interactome and Subcellular Localization of RNase J2 Complexes in Streptococcus mutans. Frontiers in Microbiology, 10. 10.3389/fmicb.2019.02150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Weidner A, Pech A, Stubbs MT, & Tittmann K (2008). Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 105(45), 17390–17395. 10.1073/pnas.0805027105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, & Tuomanen EI (2004). Microarray analysis of pneumococcal gene expression during invasive disease. Infection and Immunity, 72(10), 5582–5596. 10.1128/IAI.72.10.5582-5596.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone CD, Overweg K, Hermans PWM, & Weiser JN (2000). Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infection and Immunity, 68(7), 3990–3997. 10.1128/IAI.68.7.3990-3997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redanz S, Cheng X, Giacaman RA, Pfeifer CS, Merritt J, & Kreth J (2018). Live and let die: Hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Molecular Oral Microbiology, 33(5), 337–352. 10.1111/omi.12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Seki M, Iida K, Nakayama H, & Yoshida S (2007). A Novel Agar Medium to Detect Hydrogen Peroxide-Producing Bacteria Based on the Prussian Blue-Forming Reaction. Microbiology and Immunology, 51(9), 889–892. 10.1111/j.1348-0421.2007.tb03971.x [DOI] [PubMed] [Google Scholar]
- Sedewitz B, Schleifer KH, & Gotz F (1984). Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum. Journal of Bacteriology, 160(1), 273–278. 10.1128/jb.160.1.273-278.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idänpään-Heikkilä I, Rosenow C, & Masure HR (1996). Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Molecular Microbiology, 19(4), 803–813. 10.1046/j.1365-2958.1996.425954.x [DOI] [PubMed] [Google Scholar]
- Standar K, Kreikemeyer B, Redanz S, Münter WL, Laue M, & Podbielski A (2010). Setup of an in vitro test system for basic studies on biofilm behavior of mixed-species cultures with dental and periodontal pathogens. PLoS ONE, 5(10). 10.1371/journal.pone.0013135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumioka R, Nakata M, Okahashi N, Li Y, Wada S, Yamaguchi M, Sumitomo T, Hayashi M, & Kawabata S (2017). Streptococcus sanguinis induces neutrophil cell death by production of hydrogen peroxide. PLoS ONE, 12(2). 10.1371/journal.pone.0172223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittmann K (2009). Reaction mechanisms of thiamin diphosphate enzymes: Redox reactions. FEBS Journal, 276(9), 2454–2468. 10.1111/j.1742-4658.2009.06966.x [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L, Lepore R, & Schwede T (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson F, Keevil CW, Wilks SA, & Chewins J (2018). Modelling vaporised hydrogen peroxide efficacy against mono-species biofilms. Scientific Reports, 8(1), 12257. 10.1038/s41598-018-30706-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Qi F, Zhang Z, & Merritt J (2011a). Cloning-independent and counterselectable markerless mutagenesis system in streptococcus mutans. Applied and Environmental Microbiology, 77(22), 8025–8033. 10.1128/AEM.06362-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Qi F, Zhang Z, & Merritt J (2011b). Cloning-independent and counterselectable markerless mutagenesis system in streptococcus mutans. Applied and Environmental Microbiology, 77(22), 8025–8033. 10.1128/AEM.06362-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Qi F, & Merritt J (2013a). Development of a tunable wide-range gene induction system useful for the study of streptococcal toxin-antitoxin systems. Applied and Environmental Microbiology, 79(20), 6375–6384. 10.1128/AEM.02320-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Qi F, & Merritt J (2013b). Development of a tunable wide-range gene induction system useful for the study of streptococcal toxin-antitoxin systems. Applied and Environmental Microbiology, 79(20), 6375–6384. 10.1128/AEM.02320-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, Manque P, Ge X, Serrano MG, Puiu D, Hendricks S, Wang Y, Chaplin MD, Akan D, Paik S, Peterson DL, Macrina FL, & Buck GA (2007). Genome of the opportunistic pathogen Streptococcus sanguinis. Journal of Bacteriology, 189(8), 3166–3175. 10.1128/JB.01808-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bayles KW, & Luca S (2017). Staphylococcus aureus CidC Is a Pyruvate:Menaquinone Oxidoreductase. 10.1021/acs.biochem.7b00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng LY, Itzek A, Chen ZY, & Kreth J (2011). Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. International Journal of Oral Science, 3(2), 82–89. 10.4248/IJOS11030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Fan J, Herzberg MC, & Kreth J (2011). Characterization of competence and biofilm development of a Streptococcus sanguinis endocarditis isolate. Molecular Oral Microbiology, 26(2), 117–126. 10.1111/j.2041-1014.2010.00602.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Lin, Xu Y, Ferretti JJ, & Kreth J (2014). Probing oral microbial functionality - Expression of spxB in plaque samples. PLoS ONE, 9(1), 1–8. 10.1371/journal.pone.0086685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The experimental data presented in this study are available from the corresponding author upon reasonable request.


