Abstract
With recent advances in nanotechnology and therapeutic nucleic acids (TNAs), various nucleic acid nanoparticles (NANPs) have demonstrated great promise in diagnostics and therapeutics. However, the full realization of NANPs’ potential necessitates the development of a safe, efficient, biocompatible, stable, tissue-specific, and non-immunogenic delivery system. Exosomes, the smallest extracellular vesicles and an endogenous source of nanocarriers, offer these advantages while avoiding complications associated with manufactured agents. The lipid membranes of exosomes surround a hydrophilic core, allowing for the simultaneous incorporation of hydrophobic and hydrophilic drugs, nucleic acids, and proteins. Additional capabilities for post-isolation exosome surface modifications with imaging agents, targeting ligands, and covalent linkages also pave the way for their diverse biomedical applications. This review focuses on exosomes: their biogenesis, intracellular trafficking, transportation capacities, and applications with emphasis on the delivery of TNAs and programmable NANPs. We also highlight some of the current challenges and discuss opportunities related to the development of therapeutic exosome-based formulations and their clinical translation.
Keywords: Nucleic acid nanoparticles (NANPs), Therapeutic Nucleic Acids (TNA), nanotechnology, immunorecognition, extracellular vesicle, exosomes, drug delivery
Graphical Abstract
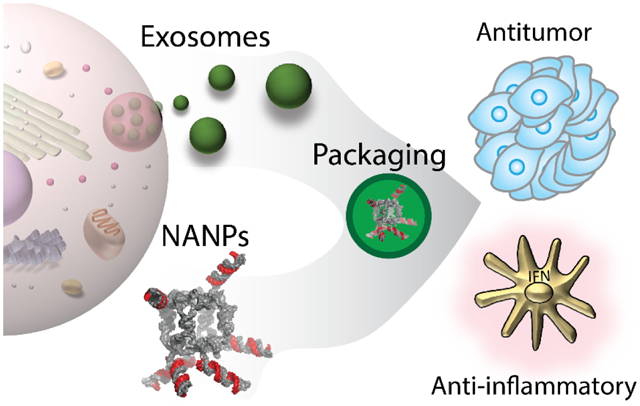
Exosome-mediated delivery of NANPs and/or other functional moieties. After isolation and purification of exosomes, cargos of interest are loaded into the exosome lumen and used for the delivery to target cells.
Nucleic acid-based therapeutics have demonstrated great potential in nanomedical applications. The 2018 release of ONPATTRO®, the first FDA (Food and Drug Administration)-approved RNAi drug1, marked a milestone and furthered the rapid development of therapeutic nucleic acids (TNAs)2, with another therapy (GIVLAARI®)3 entering the market only a year later and two more therapeutics, OXLUMO™ and LEQVIO®, just approved by the FDA and European Commission4–6. Currently, in addition to these formulations, at least 30 more different therapeutic RNAi therapies are undergoing clinical trials, and more candidates are being tested preclinically7. Other emerging classes of TNAs are exemplified by antisense oligonucleotides (KYNAMRO®) as an adjunct therapy for homozygous familial hypercholesterolemia8; mRNA vaccines against SARS-CoV-2 (BNT162b2 and mRNA-1273); aptamers (MACUGEN®) inhibiting vascular endothelial growth factor9; and a triple combination vector containing an anti-tat/rev short hairpin (sh)RNA, a nucleolar-localizing TAR decoy, and an anti-CCR5 ribozyme for autologous peripheral blood stem cell transplantation. The lattermost example (ClinicalTrials.gov study NCT01153646) has received FDA approval for treatment of AIDS and lymphoma patients in clinical trials10.
As interest in TNA development and nucleic acid technologies grows, a new class of TNAs formulated with nucleic acid nanoparticles (NANPs) becomes an expanding research focus involving more and more investigators worldwide each year11. NANPs are fully programmable nanoparticles made exclusively of nucleic acids and/or their chemical analogs that are rationally designed to self-assemble and reproducibly assume structures that can serve as nanoscaffolds capable of carrying numerous functional moieties and TNAs12–17. Functionalized NANPs offer a unique and novel platform for various applications as therapeutics, diagnostic nanodevices, and functional materials. For example, NANPs functionalized with a cocktail of RNAi inducers have been shown to simultaneously target multiple regions of the HIV-1 genome, including BS-matrix, capsid, protease, reverse transcriptase, envelope, Nef, and Rev-Tat18. Also, different techniques for the conditional intracellular activation of TNAs have been introduced to expand the therapeutic options of NANPs. One of the approaches relies on splitting the TNAs into cognate inactive pairs of RNA/DNA hybrids with their original functionalities being restored only through intracellular re-association. Using this strategy, the activation of RNA interference, NF-κB decoys, RNA transcription, immunostimulation, Förster resonance energy transfer, and RNA aptamers was demonstrated in human cells and animal models13, 15, 19–29. Another approach for the conditional intracellular activation of TNAs combines the diagnostic and therapeutic steps, as exemplified by two-stranded RNA switches. After binding to the intracellular target, the switches change their conformations, thereby releasing shRNA-like structures that act as therapies30, 31.
Besides being the carriers and actuators of TNAs, NANPs can also effectively modulate immune responses, thus providing controlled and fine-tunable immunostimulation as an additional therapeutic modality or remaining immunoquiescent to assist NANP delivery21, 24, 25, 32–45. Therefore, the relationship between NANPs’ architectural parameters and immunostimulatory properties must be clearly defined to permit their successful and safe translation into a clinical setting. Pro-inflammatory cytokines and interferons (IFNs) are key players in nucleic acid sensing by human immune cells and quantitative structure-activity relationship (QSAR) modeling can be implemented for measured physical and chemical properties of NANPs to predict their pro-inflammatory responses46. In a recent study, a set of NANPs composed of RNA and/or DNA was characterized using this process, which showed that the relative chemical stability of NANPs appeared to be the parameter which contributed the most to NANPs’ immunorecognition, followed by melting temperature, molecular weight, GC content, dissociation constant, and, finally, the size of NANPs. In addition, systematic investigations of IFNs’ induction with NANPs of various shapes, connectivity, and composition carried out in freshly collected primary human peripheral blood mononuclear cells47 demonstrated varying and specific responses from different immune cells48. It was discovered that linear NANPs elicit the lowest immune responses, while planar structures lead to higher immunostimulation and globular NANPs cause the highest immune responses. It was also shown that DNA NANPs exhibit less immune stimulation than their RNA analogs, while NANPs functionalized with multiple TNAs can be coordinated to minimize their immunorecognition through changing the extent and orientation of the functional groups. Importantly, all NANPs used without a delivery carrier appeared to be immunoquiescent and for a given delivery carrier, the dimensionality and composition of NANPs define the therapeutic efficacy, immunostimulation, and biodistribution of the formulation49. While different carriers are used to deliver NANPs, the immune cells in the peripheral blood lead to different subsequent cytokine responses50. Moreover, recent mechanistic studies revealed that chemical modifications can additionally define the mechanism and extent of the NANPs’ immune recognition51. It is now apparent that a more comprehensive understanding of how NANPs contribute to immune stimulation would allow for broader applications of this technology in various applications52. For example, while immunoquiescent NANPs can be used for TNA delivery and construction of dynamic and sensing nanodevices, the regulated activation of the immune system may become beneficial in cancer treatments. As a systemic disorder, cancers are characterized by dysfunction of various immune pathways and the cancer-immunity cycle refers to a series of events that must be initiated and expanded iteratively and efficiently in order for an anticancer immune response to effectively terminate cancer cells53. Production of type I IFNs and proinflammatory cytokines resulting from NANPs can behave as stimulatory factors that act on cancer immunity cycles and promote anticancer effects53. Immune responses elicited by NANPs may also help restore normal immune function in a host by activating antigen presenting cells and functional moieties that regulate expression of immune checkpoint proteins and homing cytokines54. This immunostimulation may become useful as a vaccine adjuvant that would aim in avoiding necrosis at the vaccine administration site, as well as an immunotherapy adjuvant that can restore and maintain immune homeostasis54. Furthermore, since NANPs’ efficacy in targeting NF-κB activity has been demonstrated55, programming NANPs with NF-κB decoys can reduce NF-κB-dependent pro-inflammatory cytokine and IFN production56.
Delivery of TNAs and therapeutic NANPs.
Nucleic acid nanomaterials and their unique and controllable properties offer a means for the specific targeting of cellular components or the scaffolded delivery of TNAs. For example, TNA-carrying RNA/DNA fibers have been shown to conditionally target the mutated BRAF gene in human melanoma cells56. High quantum yield malachite green RNA aptamers conjugated with flavin mononucleotides or theophylline have been developed as biosensors57, while aptamer-siRNA chimeras have been shown to induce apoptosis in cancer cells by targeting the eukaryotic elongation factor 2 gene58. The packaging RNA of bacteriophage phi29 DNA-packaging motors has been used to silence metallothionein-IIa and survivin in ovarian cancer59. Synthetic oligodeoxynucleotides with unmethylated CpG motifs (CpG DNA) have been shown to induce both innate immunity and specific adaptive immune responses as vaccine adjuvants60. All of these successful examples illustrate the diversity of these therapeutic platforms.
Because both TNAs and therapeutic NANPs are composed of hydrophilic and negatively charged nucleic acids, they are inefficient at crossing biological membranes. In addition, unmodified naked TNAs and NANPs are susceptible to rapid nuclease degradation in the blood. These obstacles prevent direct intracellular use of NANP-based drugs without the assistance of appropriate carriers. Over the last decade, extensive investigations of stable, efficient, and safe carriers for therapeutic NANPs have been undertaken. So far, most clinically approved carriers for therapeutics have fallen into lipid-61, 62, polymer-49, and inorganic nanoparticle-based35, 63, 64 categories. Despite their advantages, the manufactured nanomaterials often suffer from immunogenicity, cytotoxicity, rapid blood clearance, and poor biodistribution, all of which additionally hinder the clinical translation of NANPs65–71. Therefore, the development of new delivery methods aiming to overcome the aforementioned complications associated with synthetic materials remains a great challenge for nucleic acid nanotechnologists.
Extracellular vesicles.
Extracellular vesicles (EVs) are membrane-bound, cytosol-containing particles secreted into the extracellular space by almost all living cells. EVs have been found in bodily fluids including blood, urine, saliva, breast milk, cerebrospinal fluid, sputum, bile, semen, amniotic fluid, broncho-alveolar lavage fluid, and ascites72, 73. The content, size, and membrane composition of EVs depend on their cellular source and physiological conditions. At present, three subgroups of EVs have been broadly classified and generally accepted based on their sizes and biogenic pathways: apoptotic bodies, microvesicles, and exosomes. Apoptotic bodies are larger vesicles around 800–5,000 nm in diameter which cells release while undergoing programmed cell death74. Microvesicles are generally smaller (50–1,000 nm in diameter) membranous vesicles produced via plasma membrane budding and are associated with cell shedding74. The smallest EVs are exosomes, which range from 40–150 nm in diameter and undergo release during the fusion of multivesicular bodies (MVBs) with the plasma membrane. Due to their heterogeneous and dynamic nature, EV subgroup differentiation is challenging. Intercellular communication relies heavily on EVs and they contribute to numerous physiological and pathological functions. Moreover, EVs derived from cancer cells have shown to promote angiogenesis and coagulation, support tumor progression, and generate pre-metastatic niches75.
A diverse array of quantitative methods is available for vesicle characterization. Assessment of EV sizes and morphologies can be undertaken with transmission electron microscopy and cryogenic electron microscopy. Nanoparticle tracking analysis, tunable resistive pulse sensing, dynamic light scattering, and high-resolution flow cytometry not only reveal EVs’ sizes, but also provide information regarding EVs’ concentrations76. Traditional methods of EV isolation such as ultracentrifugation77, density gradient centrifugation77, size exclusion chromatography78, ultrafiltration, and gel filtration79 utilize EV size and buoyant density. A relatively new method of isolation called polymer-based precipitation utilizes the changes in solubility of exosomes using volume-excluding polymers such as polyethylene glycol (PEG) to promote aggregation80. Other novel isolation methods that have recently appeared are integrated microfluidic systems with on-chip immunoisolation81 and lipid-nanoprobe systems that enable for spontaneous labelling of EVs with rapid subsequent magnetic enrichment82.
EVs contain luminal biologically active cargo such as proteins, nucleic acids, and lipids that can be taken up by recipient cells, potentially altering their operations (Figure 1). Based on proteomic studies, EVs are highly abundant in cytoskeletal, cytosolic, heat shock, and transmembrane proteins, as well as proteins associated with intracellular trafficking83. EV protein content can be assessed by immunoblotting, immuno-gold labelling with electron microscopy, and antibody-coupled bead flow cytometry analysis83. Some transmembrane proteins found enriched in EVs are adhesion molecules such as integrins, tetraspanins (e.g., cluster of differentiation (CD) 9, CD81, CD63, CD82, CD53, CD37), intercellular adhesion molecules (ICAM, also known as CD54), globule-epidermal growth factor-factor VIII (also called lactadherin), antigen presentation proteins such as the major histocompatibility complex (MHC) class I and class II, membrane transport and fusion proteins for intracellular trafficking (such as annexins, flotillins, and Ras-associated binding), and ADP-ribosylation factor GTPases83–87. Cytosolic proteins found in the EV lumen include cytoskeletal proteins (such as actin, cofilin, moesin, and tubulin), signal transduction proteins (such as heterotrimeric G, β-catenin, and 14-3-3), enzymes (such as elongation factors, glyceraldehyde 3-phosphate dehydrogenase, peroxidases, pyruvate kinase, and enolase), chaperone proteins (such as heat shock protein (HSP)70 and HSP90), and biogenesis factors (such as programmed cell death 6-interacting protein, tumor susceptibility gene 101, syntenin, ubiquitin, clathrin, and vacuolar protein sorting 4 and 36)73, 88. Many of these proteins are also commonly considered to be specific markers for EV subgroups. CD9, CD63, CD81, Alix, Tsg101, and HSP70 are exosomal markers; integrins, flotillin-2, CD62, and CD40 are markers for microvesicles; and Annexin V and phosphatidylserine are used to identify apoptotic bodies73, 89, 90. Common EV protein signatures are critical to ensure their correct operation. The protein syntenin is a key component in MVB formation and exosome biogenesis91. The expression of glycosylphosphatidylinositol-anchored CD55 and CD59 protect EVs from complement-mediated lysis92 while surface glycosylation patterns assist EV uptake by recipient cells93. Cytokine and chemokine secretions affiliated with EVs (e.g., IL-1α94, IL-1β95, CXCL896 and CX3CL197) regulate the immune responses in target cells.
Figure 1.
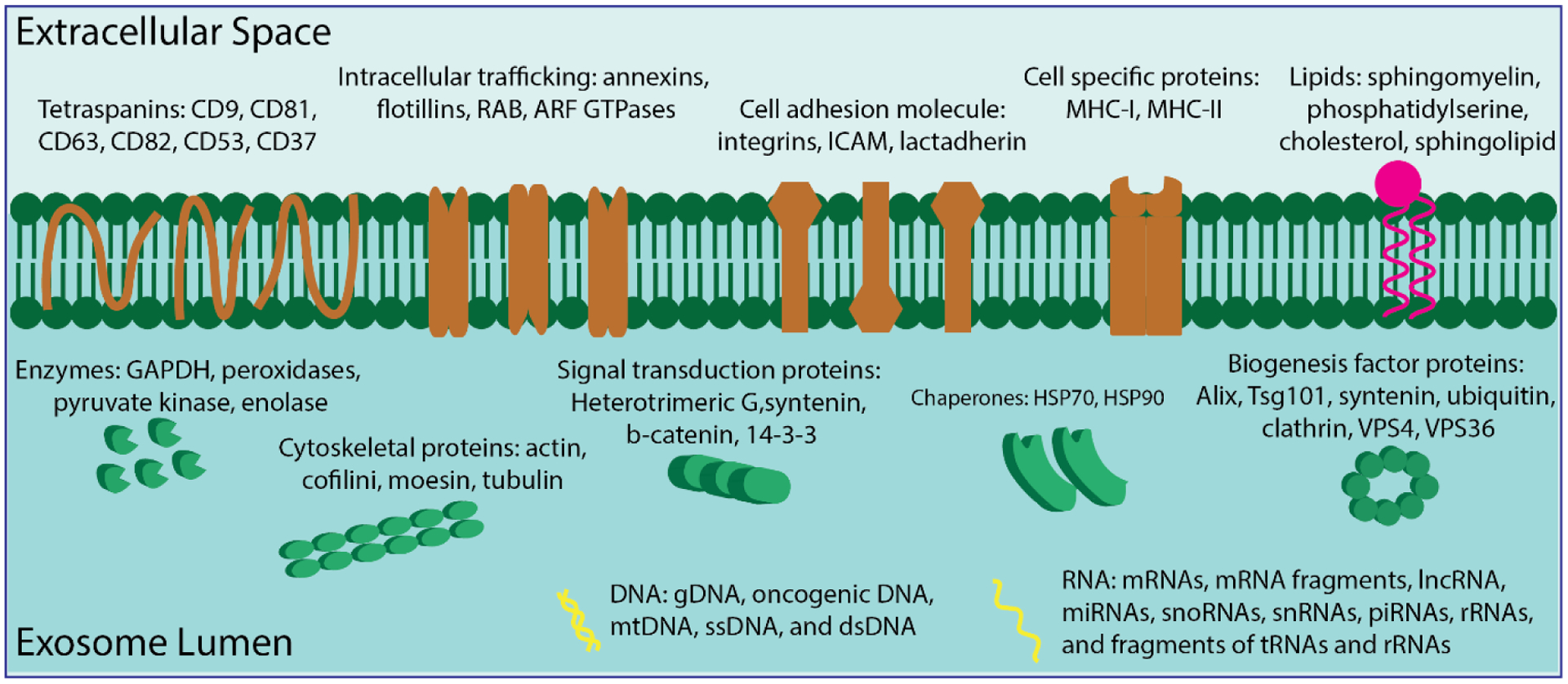
Proteins, lipids, and nucleic acids as representative biological components of exosome content.
DNA and RNA are also present in EVs. RNA pools in EVs have been identified using high-throughput RNA sequencing and verified by RT-qPCR98. These RNA populations include mRNAs, mRNA fragments, lncRNA, miRNAs, snoRNAs, snRNAs, piRNAs, rRNAs, and fragments of tRNAs and rRNAs73, 98, 99. EV RNA may regulate gene expression and protein translation in recipient cells. A 3’-untranslated region of mRNA which is rich in regulatory sequences is carried by EVs and serves as a binding site for numerous RNA-binding proteins. These “external” mRNAs may compete with the recipient cell’s mRNA for miRNA binding and specific RNA-binding proteins in the recipient cell, potentially leading to downregulation of protein production there100. Regarding the RNA sorting mechanism into EVs, growing evidence indicates that RNAs are not loaded into EVs randomly and passively. Instead, a certain population of RNAs becomes EV-enriched compared to their parental cells, suggesting that cells selectively deliver these RNAs to enhance or modify target cell functioning101. Additionally, the RNA content of EVs is regulated by cells’ physiological states. MiR-150, noted for its participation in hematopoiesis, is preferably packaged into microvesicles in lipopolysaccharide-treated human blood cells102. During immune synapse formation, miR-335 is selectively sorted into EVs derived from T lymphoblasts and unidirectionally transferred to antigen-presenting cells (APCs), resulting in downregulation of SRY-related HMG-box (SOX)-4 mRNA translation103. In contrast to RNA, little is known about EV-transported DNA’s functions, and its physiological significance in recipient cells remains under investigation83. However, genomic DNA, oncogenic DNA, mitochondrial DNA (mtDNA), ssDNA, and dsDNA have been detected in EVs104–107.
Finally, EVs possess a lipid bilayer in which the lipid distribution in the outer and inner membranes are expected to resemble the cell membrane due to similar formation mechanisms108. Compared to their cells of origin, EVs are enriched in sphingomyelin (SM), phosphatidylserine (PS), cholesterol, and sphingolipids109. However, not all EVs contain high amounts of lipids. Reticulocyte-derived EVs display no enrichment of PS or SM, whereas ceramide amounts change during reticulocyte maturation into red blood cells110. Therefore, EVs’ lipid composition is dynamic and influenced by the cell’s physiological status. Lipids perform essential functions in EVs; cholesterol appears to regulate EV trafficking by selecting membrane rafts and tetraspanin-enriched microdomains for budding111. Cholesterol, along with long saturated sphingolipid fatty acids, provides tight packaging and structural rigidity to EVs83. In cancerous conditions, SM stimulates endothelial cell migration and mediates angiogenic activity112. PS in platelet-derived EVs contributes to thrombin formation and promotes coagulation113.
Components are not consistent throughout all EVs. Instead, they are rather specific and dependent on size, the EV’s cell of origin, and purported functions114. Breast cancer cell-derived microvesicles and exosomes exhibit distinct protein profiles for extracellular matrix degradation, cancer invasion and metastasis, and cell survival115. A subpopulation of exosomes called “exomeres” lack external membranes or a spherical shape while containing different types of proteins based on proteomic analysis116. Likewise, EVs from diverse origins consist of different components of proteins, nucleic acids, and lipids that influence various physiological and pathological functions in recipient cells. Renal collecting duct cells excrete EVs that contain vasopressin-regulated water channel aquaporin-2, a protein involved in Na+ transport and control of water permeability across the nephron117. Platelet-derived EVs contain CD154 which stimulates antigen-specific IgG production to modulate inflammation and adaptive immunity at recipient cells distant from the activation site118. Saliva-derived EVs carry tissue factors that initiate thrombin formation from the zymogen prothrombin to elicit blood coagulation in target cells119. A panel of 59 well-characterized and immune-related miRNAs have been detected to be enriched in breast milk-derived EVs and are shown to assist with development of the infant immune system120. Lipidomic studies reveal a high cholesterol to phospholipid ratio in prostate gland epithelial cell-derived EVs, namely in prostasomes isolated from human semen. Fusion of the sperm plasma membrane with prostasomes contributes to sperm stability, enabling for sperm’s greater resistance to untimely acrosomal reactions121.
Compositions of specific cell type-derived EVs also change in response to fluctuations in the extracellular environment as well as different physiological conditions or differentiated cell states. For instance, EVs secreted by vascular endothelial cells under ischemia-induced hypoxia caused cytoskeletal and extracellular matrix rearrangements due to changes in EV protein and mRNA contents122. EVs secreted by cells containing the mutated Kirsten rat sarcoma viral oncogene homolog (KRAS), which occurs in 30–40% of colorectal cancer cases, dramatically affect proteomic vesicle composition, such as tumor-promoting proteins KRAS, EGFR, SRC kinase, and integrins123. Mature dendritic cell (DC)-derived EVs treated with LPS contain 50-to 100-fold more proteins, with notable enrichment of MHC class II, B7–2 (CD86), and intercellular adhesion molecule 1 (ICAM-1). Compared to EVs from immature DCs, the mature DC-derived EVs displayed greater antigen-specific T cell activation to trigger effector T-cell responses and activate naïve T cells to APCs124.
Exosome biogenesis and trafficking.
Exosomes, the smallest members of the EV family, are released by fusion of the endosome with the plasma membrane125. With exponential growth in exosome research over the past 30 years, their essential roles in healthy and pathological cells as well as their potential clinical diagnostic and therapeutic applications have been investigated. In 1987, the term “exosome” was introduced based on observations of reversed endocytotic activity in which internal cellular contents were externalized through a membrane-bound vesicle released by the same cell126. Detailed knowledge of molecular mechanisms behind the biogenesis and transport of exosomes can aid in understanding exosomal functions and further exploration of their medical utility.
Exosome biogenesis starts within the endosomal system (Figure 2A). The endocytic pathway consists of distinct membrane compartments which internalize molecules from extracellular components, recycle them to the plasma membrane, and/or sort them for degradation127. Early endosomes are the first compartments that receive molecules coming from the cell surface. They primarily function as sorting organelles; at an acidic pH, endocytosed ligands dissociate from their receptors128. Intraluminal vesicles (ILVs) are formed by the inward budding of early endosomal membranes with specifically selected molecules. During the maturation of early endosomes to late endosomes, ILVs accumulate in the lumen of transvesicular compartments, such as multivesicular bodies (MVBs), where the sorting process continues in preparation for the transcytotic pathway128. Generally, lysosomes are the last compartment of the endocytic pathway and most MVBs will fuse with lysosomes, wherein lysosomal hydrolases will degrade their contents. However, some MVBs, especially those containing high amounts of cholesterol, may fuse with the plasma membrane and the released ILVs are denoted as exosomes (Figure 2B)129. MVB populations are cell-dependent and regulated by various cellular conditions and external factors125. In addition, cells can host different subpopulations of MVBs. For example, MVBs with lysobisphosphatidic acid (LBPA) and without it coexist in human B lymphocytes, with the latter destined for degradation130.
Figure 2.
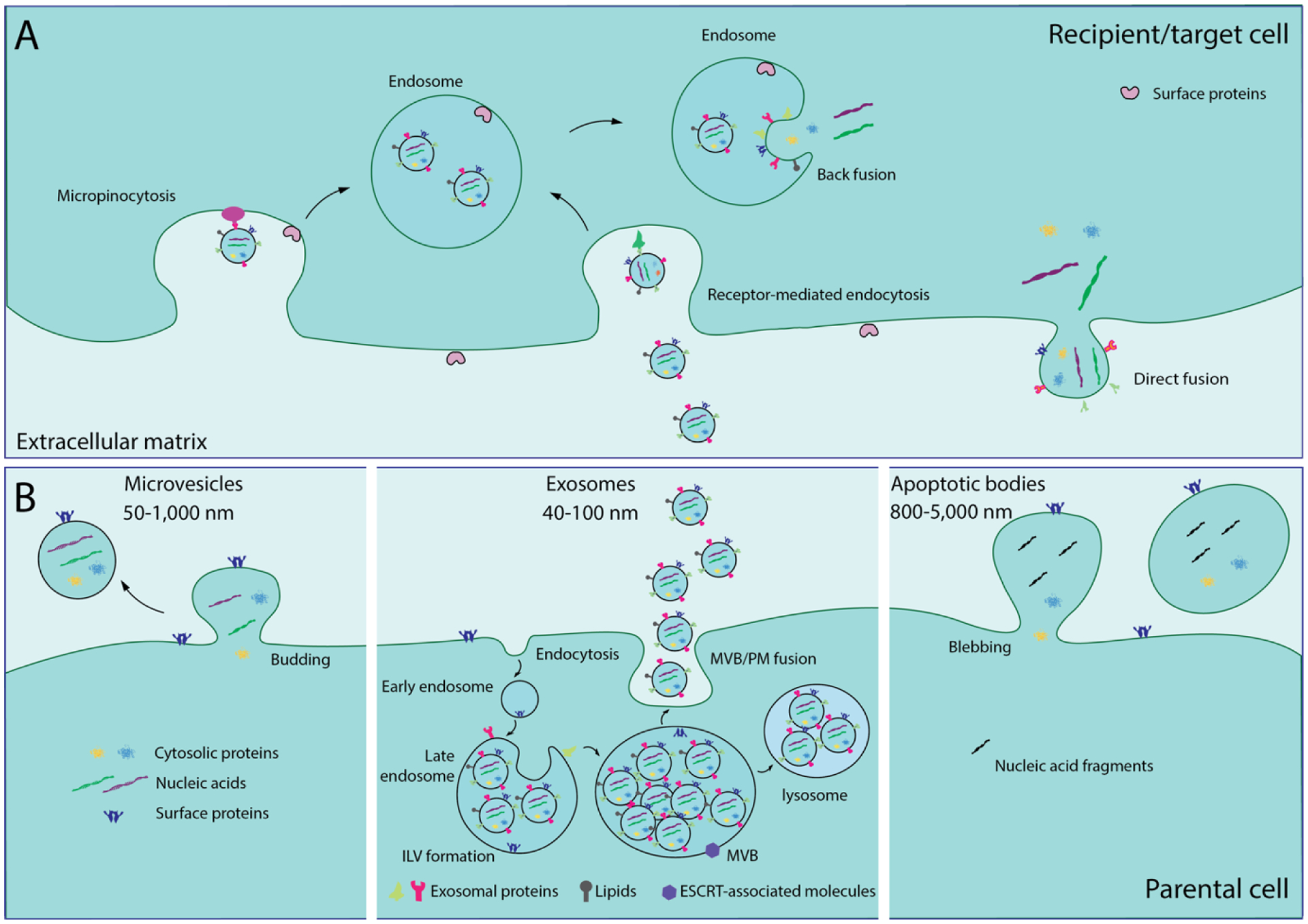
EV biogenesis and trafficking. (A) Exosomes are taken up by recipient cells via direct fusion of the exosomal membrane and the plasma membrane of the recipient cell, leading to direct release of their contents into the recipient cell’s cytoplasm. Alternatively, receptor-mediated endocytosis and micropinocytosis involve exosome uptake into endosomes, but the exosomal contents are released via back fusion of the exosomal membrane and the recipient cell’s endosomal membrane. (B) Biogenesis of microvesicles, exosomes, and apoptotic bodies. Microvesicles shed from the cell via budding of the plasma membrane. Exosome biogenesis begins with internalization of membrane proteins and lipid complexes via endocytosis and engulfment of cytosolic proteins and nucleic acids into the intraluminal vesicles (ILVs) via inward budding of the endosomal membrane. With endosome maturation, late endosomes enclose numerous ILVs to become MVBs. Some MVBs are degraded in the lysosome; exosome secretion occurs when MVB fuses with the plasma membrane. During apoptosis, cell disassembly generates apoptotic bodies, which are released via blebbing and protrusion.
The process of MVB formation involves more than 20 proteins, most of which belong to the ESCRT (endosomal sorting complex required for transport) system23. ESCRT consists of four different protein complexes—ESCRT-0, -I, -II, and -III—along with associated vacuolar protein sorting 4/suppressor of K+ transport growth defect 1 (Vps4/SKD1) and ALG-2-interacting protein X (ALIX) protein complexes131. MVB sorting involves the recognition of endocytic cargo subsets that become concentrated in endosomal membrane regions, a process which is conserved throughout eukaryotes132. The ESCRT-0 complex consists of the hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) which sorts monoubiquitinated membrane proteins into MVBs and associates with the signal transducing adaptor molecule (STAM). HRS recruits TSG101 of the ESCRT-I complex, whose roles include transferring ubiquitinated proteins between the ESCRT-0 and -II complexes and recruiting the ESCRT-III complex via ESCRT-II or ALIX. ESCRT-I and -II together induce an inward invagination from the endosomal membrane and form a neck structure of the nascent vesicle, while ESCRT-II and -III pinch off the neck and release the vesicle into the MVB lumen23. Finally, ESCRT-III associates with Vps4/SKD1 ATPase to dissociate and recycle the ESCRT machinery using energy from ATPase activity (Figure 3).
Figure 3.
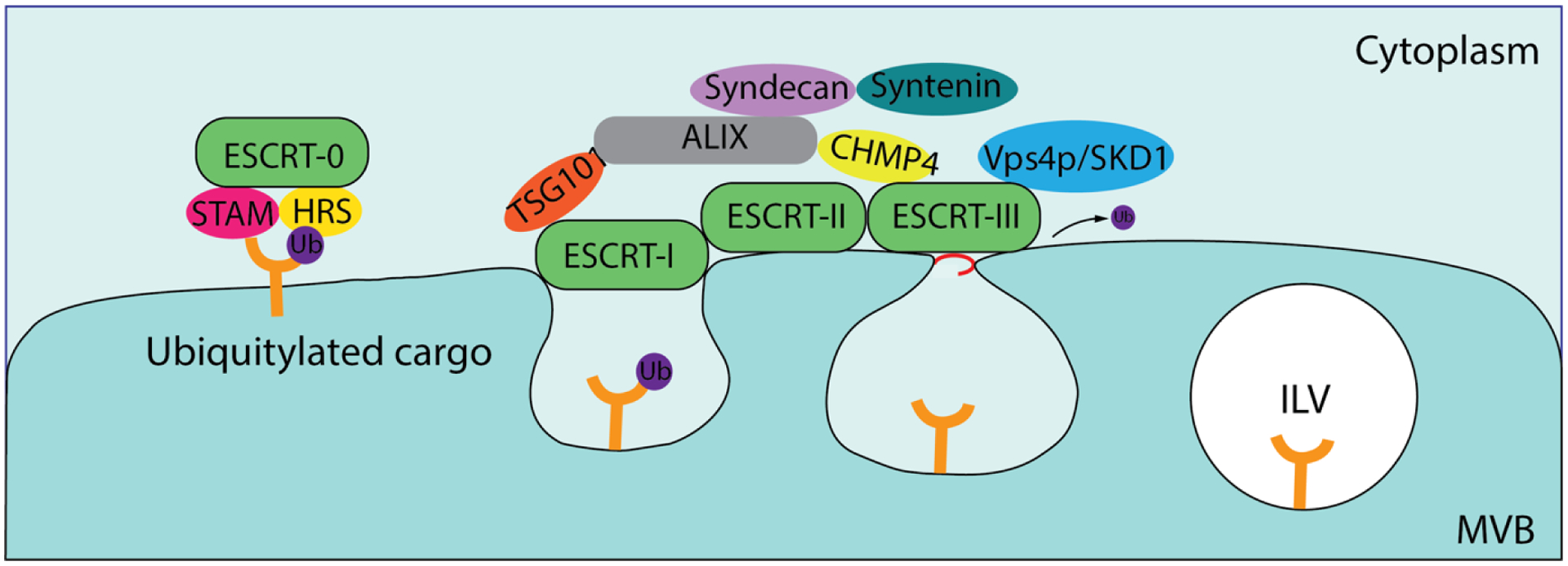
The endosomal sorting complex required for transport (ESCRT) pathway for cargo recognition and sorting. The ESCRT-0 complex recognizes and sequesters ubiquitylated cargo, whereas the ESCRT-I and -II complexes are responsible for membrane deformation which yields buds with ubiquitylated cargo. The ESCRT-III complex subsequently drives vesicle scission, resulting in ILV formation. Various ESCRT-accessory proteins participate in and assist with these processes.
In the ESCRT-dependent pathway, ubiquitin is critical for the sorting of membrane cargos into the MVB. However, MVB formation also occurs independently of ubiquitination. Heparin sulphate proteoglycans promote exosome biogenesis through syntenin, which binds syndecan with ALIX. ALIX then interacts with TSG 101 and charged MVB protein 4 (CHMP4), creating an intermediate between ESCRT-I and ESCRT-III for vesicular budding and scission processes133. Additionally, MVB formation can take place without ESCRT complexes and proteins. One alternative pathway involves the segregation of cargos associated with raft-based microdomains which possess highly enriched sphingomyelinases. Removal of the phosphocholine moiety from sphingomyelinases via hydrolysis leads to ceramide formation134. Ceramides have a cone-shaped morphology that promotes spontaneous inward curvature of the endosomal membrane and promotes domain-induced budding, facilitating exosomal lipid-sorting during exosome biogenesis135. In another case, tetraspanin-enriched microdomains serve as specialized membrane platforms for partitioning receptors and signaling proteins in the plasma membrane, aiding in the selection of receptors and intracellular components sorted into exosomes136. Many other molecules and cellular structures, including small integral membrane proteins of lysosomes and late endosomes, contribute to MVB formation using ESCRT-independent mechanisms137.
There are two outcomes of MVB formation: one is fusion with the plasma membrane and the subsequent release of internal vesicles as exosomes, while the other is lysosomal degradation. Except for the fact that high levels of cholesterol seem to promote fusion with the plasma membrane, the mechanism which controls MVBs’ route remains unclear138. The final fate of MVBs, however, is not immutable, but changes under different cellular conditions such as starvation, rapamycin treatment, ISGylation, etc139, 140. In these cases, MVBs are prone to lysosomal degradation140. Exosome release involves contributions from several Rab proteins which act as essential regulators of intracellular vesicle transport. The Rab family is composed of more than 60 GTPases that associate with membranes via geranylgeranylation to regulate cellular trafficking processes like vesicle budding, transport, and tethering, as well as mediating fusion by cycling between the active guanosine triphosphate (GTP)-bound state and the inactive guanosine diphosphate (GDP)-bound state141. Rab GTPase mechanisms are cell-specific; Rab2b, Rab4, Rab5A, Rab7, Rab9a, Rab11, Rab27a, Rab27b, and Rab35 have been implicated in various stages of exosome release among different cell types142, 143. Subcellular MVB location depends on interactions of MVBs with the microtubule cytoskeleton and actin. Cholesterol content partially controls movement of MVBs along the microtubules as well144. Once MVBs are docked to the plasma membrane, Rab and soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins participate in fusion of the MVB membrane with the plasma membrane, ultimately releasing exosomes into the extracellular space145.
As messengers of intercellular communication, exosomes secreted by their cells of origin are assumed to interact with destination cells to deliver molecular information. This exosomal intracellular trafficking and communication takes place through the coordination of several steps. First, exosome-cell surface binding is mediated by classical adhesion molecules (integrins and ICAMs) specific to cell-cell interactions. Several classical ligand/receptor pairs such as ICAM-I/lymphocyte function-associated-antigen-1 (LFA-1) are involved in exosome uptake by mature dendritic cells146, while integrin CD49d and Tspan8 support exosome binding to endothelial cells147 and T cell immunoglobulin and mucin domain-containing protein-3 (TIM-3) help mature Th1 lymphocytes capture galectin-9-bearing exosomes148. In some situations, an exosome binding to the plasma membrane of a recipient cell may be sufficient enough to initiate a signaling cascade. For example, MHC on the surface of APC-derived exosomes presents to antigen-specific T lymphocytes149. For most cases, exosomal contents must be delivered inside recipient cells. Internalization occurs via three main pathways: direct fusion150, receptor-mediated endocytosis151, and micropinocytosis152 (Figure 2A). Exosome-membrane fusion is more likely to occur at acidic endosomal sites rather than at the neutral plasma membrane144. Capabilities for endocytosis or micropinocytosis depend on recipient cells which are typically non-phagocytic. In phagocytic macrophages, neutrophils, and monocytes, exosomes are internalized via phagocytosis153.
Once internalized, exosomal contents are then released into the cytoplasm directly or via backfusion with the endosomal membrane. A subsequent effect on the recipient cell may take place154. For instance, nucleic acid-induced gene expression modification in recipient cells can be instigated by exosome delivery. Released miRNA and exogenously modified-siRNA molecules potentially inhibit mRNA translation and thus silence target genes, or released mRNA can be translated into a protein using the recipient cell’s cellular machinery. Both exosome cargo and functionality solely depend on their cell of origin. Exosome-based intercellular trafficking and communication is a dynamic system, so message modification is feasible and dependent on the physiological and pathological states of the producing cells155.
Exosomes as natural vehicles for the delivery of TNAs and NANPs.
In vivo delivery of NANPs remains a significant challenge that precludes their broader biomedical applications. Various nanocarriers have been investigated and, because most of them are chemically fabricated, the formulations suffer from issues related to immunogenicity, toxicity, rapid blood clearance, and poor biodistribution. Exosomes are natural EVs that are non-immunogenic and are not known to activate nonspecific innate immune responses such as complement system-related pseudoallergy (CARPA)156, thus offering serious advantages over many synthetic materials. A benefit of “self-generated” exosomes is the absence of any immune attack against them while they remain in circulation. As a delivery vehicle, the structure of choice should be devoid of any immunologically stimulating activity that could bring forth an inflammatory response. Exosomes’ ability to hide from the immune system comes from an “inheritance” of parental cell surface molecules which aid in their recognition as “self157.” Such surface molecules, including CD46, CD55, CD59, and CK2, effectively escape detection157. Currently, numerous cell types have been exploited as exosome factories, with some seeing more frequent use than others. Human embryonic kidney variant HEK293T cells are one of the most popular sources, and their immunostimulation is well investigated. One study using these cells measured 23 different cytokines in vivo and in detectable ranges. The results revealed no differences in cytokine production between exosomes obtained from HEK293T cells and the buffer control. Therefore, exosomes obtained from HEK293T were concluded to exhibit favorably low immune stimulation158. As drug delivery systems, exosomes loaded with therapeutic cargos must also undergo proper immunological assessment. A follow-up study using the same HEK293T cell-derived exosomes loaded with miR-199a-3p, a TNA with anti-invasion and anti-migration effects on hepatocellular carcinoma, confirmed slightly higher concentrations of several cytokines compared to free exosome levels and indicated an overall very limited immune response159.
Immature dendritic cells bear low levels of MHC-II and costimulatory molecules which reduce immune activation by transforming T cells into type 2 T helper (Th2) and regulatory T cells (Treg) or by causing T cell apoptosis, thereby promoting a tolerogenic immune response160. Exosomes derived from tumor cells are a source of tumor MHC-I molecules, tetraspanins, HSP70–80, lysosomal-associated membrane protein 1 (LAMP1), tumor rejection antigens, and various immunosuppressive molecules. These molecules can inactivate T lymphocytes or natural killer cells or promote the differentiation of regulatory T lymphocytes to suppress the immune response161, 162. Mesenchymal stem cell-derived exosomes lack MHC-I, MHC-II, and costimulatory molecules such as CD80 and CD86, rendering them less susceptible to immune rejection and more suitable for allogeneic therapeutics163.
Although toxicity is a downside of synthetic formulations, no clear evidence of exosomal cytotoxic effects exists. The spleen, as the largest lymphoid organ, plays an important role in the immune system, and it is considered a good indicator for initial immunotoxicity screening67. Data showed that splenocytes treated with HEK293T cell-derived exosomes loaded with miR-199a-3p had no effect on spleen cell composition, and neither did free exosomes158. Furthermore, no significant histopathological changes were reported in harvested spleen, heart, thymus, lung, liver, kidney, adrenal, ovary, uterus, or brain tissues ex vivo, indicating an absence of observable organ toxicity. Hematological analysis showed little effect on red blood cell, white blood cell, platelet, neutrophil, lymphocyte, and monocyte counts, or on hematocrit and hemoglobin levels between the groups treated with free and loaded exosomes and the untreated control. With a total of 14 markers tested in blood chemistry, no significant difference was observed between all exosome-treated and control groups158. However, some surface molecules present on exosomes can serve to eliminate cytotoxicity. For example, placenta-derived exosomes bear natural killer group 2 member D (NKG2D) ligands and induce the down-regulation of the NKG2D receptor on cytotoxic effector cells, leading to reduction of their cytotoxicity164. From benchtop to bedside, exosomes consistently show negligible toxicity. The first exosome Phase I clinical trial used exosomes derived from autologous monocyte derived-DC cultures pulsed with melanoma antigen 3 (MAGE 3) peptides to vaccinate stage III/IV metastatic melanoma patients. Exosomes generated with functional MHC molecules promoted T-cell immune responses including tumor rejection. No grade II toxicity was observed, and no maximal tolerated dose was achieved, demonstrating the safety of exosome administration165.
As drug carriers, exosomes, along with their behavior in vivo, must be properly understood. This includes detailed analysis of their clearance from systemic circulation. As mentioned earlier, cells of origin determine exosomes’ content, functionality, and, consequently, their biological fate. However, exogenously administered exosomes may fail to reach their targets due to very brief half-lives. Recently, a pharmacokinetic profile of intravenously injected exosomes derived from murine melanoma cells showed a circulation half-life of approximately 2 minutes with only minimal retention at 4 hours post-injection. These exosomes were rapidly cleared from circulation by macrophages in the mononuclear phagocyte system at a rate comparable to that of synthetic liposomes151. Additionally, it has been reported that exosomes derived from bone marrow-dendritic cells166, splenocytes167, and rat pancreatic adenocarcinoma168 and delivered to mice via intravenous injection ended up being engulfed by macrophages. To compensate for this problem, the exosome’s membrane may be modified using PEGylation to decrease hepatic clearance169. As naturally occurring carriers, exosomes have their own strategies to bypass the MPS. They contain transmembrane and membrane-anchored proteins that may enhance endocytosis and promote content delivery. CD47, a widely expressed integrin-associated transmembrane protein, serves as the ligand for signal regulatory protein α which produces a signal to prevent attack by macrophages. Intranasally administered monocyte- and macrophage-derived exosomes and intraperitoneally administered primary fibroblast-like mesenchymal cell-derived exosomes display CD47 on their membranes to shield them from macrophage consumption, resulting in retarded clearance170, 171.
The development of medications that act on the central nervous system to target neurodegenerative disorders and brain cancers is severely hampered by a lack of efficient drug delivery systems to carry therapeutics to the brain. It is estimated that only a small fraction (<1%) of injected antibodies enter the brain by passive diffusion, while the rest must be administered by peripheral injection or invasive intracranial procedures172. Studies have shown that exosomes derived from endothelial bEND.3 cells and dendritic cells are capable of carrying small molecule drugs and siRNAs across the blood-brain barrier173, 174. Because of this unique ability, exosomes can outcompete most current delivery systems. An understanding of in vivo biodistribution following exosome administration provides a basis for dosage prediction, route of administration, and potential off-target effects. Also, it provides indications for specific therapeutic applications to target tissues. Due to their differing cells of origin, exosomes contain specific proteins on their surfaces that mediate tissue tropism. For instance, Wnt4-associated exosomes derived from thymic epithelial cells were shown to accumulate in the thymus of mice175. Tumor-homing exosomes carrying therapeutic agents have also been employed as delivery vehicles. Hypoxic tumor-derived exosomes loaded with LYNPARZA®, a medication used for the treatment of BRCA-mutated advanced ovarian cancer in adults, displays increased apoptosis and retarded tumor growth in vivo176. Moreover, the genetic modification of exosomal surface proteins was reported to increase target-specificity, such as for rabies viral glycoprotein (RVG) for brain-targeting and human epidermal growth factor receptor 2 (HER2) or the TM domain of platelet-derived growth factor receptor (PDGFR) for cancer targeting177–179.
Among the many diverse vehicle candidates for TNA delivery, both synthetic carriers and exosomes have been studied extensively. Synthetic carriers have the advantages of high yield and straightforward, large-scale manufacturing, but their toxicity, immunogenicity, biological instability, and lack of target specificity obstruct their broader clinical applications. These impediments can be overcome by using exosomes which mediate cell-cell communication as an intrinsic function. Unlike the surface compositions of many other entities, exosomes possess well-defined proteins on their membranes that assist with target cell interactions and conceal them from the immune system. At the same time, exosomes’ inherent messenger capabilities allow them to reach their target cells and fulfill their biological fates. As with other carriers, exosome surface engineering may yield greater performance. It is hypothesized that bioengineered surface molecules like arginylglycylaspartic acid (or RGD) peptides or other targeting moieties may confer higher binding specificity and affinity when expressed on exosomal membranes as opposed to liposomes180.
Several studies have been undertaken to examine the delivery of a variety of nucleic acid-based payloads. One pioneering work found that the use of low immunostimulatory DC-derived exosomes with neuron-specific RVG peptide modifications successfully delivered exogenous siRNAs into the brains of mice181. Although the major hurdle for RNAi-based therapeutics constitutes nucleic acid delivery across the cell’s plasma membrane, exosomes derived from peripheral blood cells182, HeLa and ascites183, aortic endothelial cells184, and DCs185 have achieved success as gene delivery vectors transporting exogenous siRNAs into various target cells. The delivery of miRNAs, such as viral miRNA from Epstein-Barr virus-infected cells to uninfected ones186, let-7a miRNA to EGFR-expressing xenograft breast cancer tissue in Rag2−/− mice187, and miR-335 from T lymphocytes to APCs103, has become possible with the use of exosomes. Also, the enhanced delivery of miR-124 at an ischemic injury site using RVG fused with exosomal protein lysosome-associated membrane glycoprotein 2b was demonstarted188. Later, in another study utilizing RVG-decorated exosomes, the successful delivery of nerve growth factor (NGF) mRNA to the ischemic cortex was achieved. Subsequently, a burst in NGF production with a concomitant reduction of inflammation and improved cell survival was observed (Figure 4A)189. RNA NANPs transferred by exosomes (e.g., three-way junction (3WJ) arrowtail RNA) contain siRNA against survivin and, upon successful delivery, enhance cancer suppression without endosomal trapping (Figure 4B)190.
Figure 4.
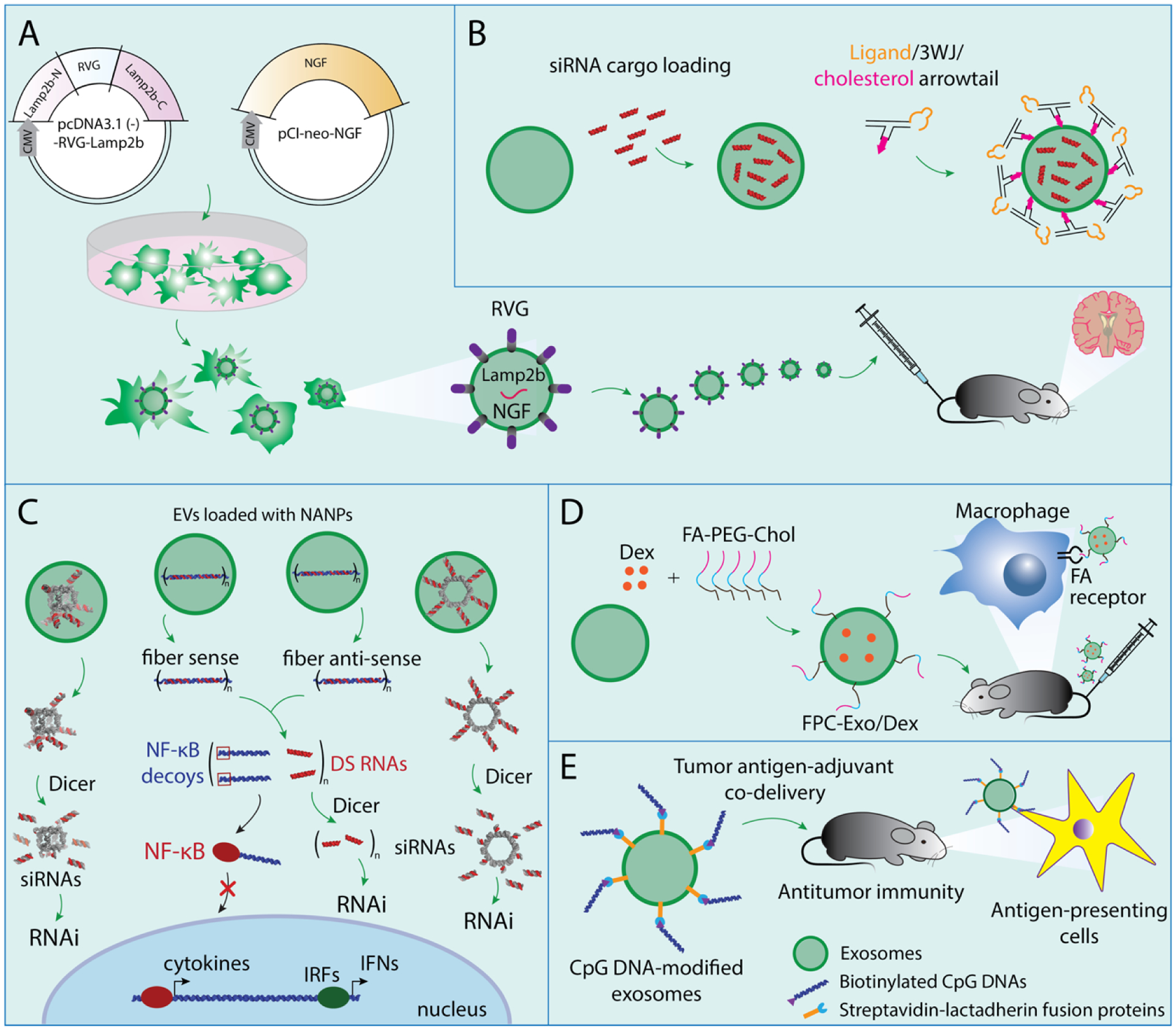
Schematic representations of some examples of exosome-mediated delivery of various cargos. (A) NGF mRNA delivered by exosomes decorated with RVG. (B) 3WJ RNA nanoparticles delivered by exosomes decorated with cholesterol and ligand. (C) RNA cube, RNA ring, fiber sense, and fiber antisense delivered by exosomes. (D) Dex delivered by exosomes decorated with Folic acid-PEG and cholesterol. (E) Biotinylated CpG DNA delivered by exosomes decorated with streptavidin-lactadherin fusion protein.
Recently, our team has successfully isolated naturally secreted exosomes further used as vehicles for the delivery of several functional NANPs to a variety of target cells. These transported NANPs exhibited different sizes, compositions, and shapes ranging from globular RNA cubes to planar RNA rings to linear RNA/DNA fibers. RNA cubes and RNA rings were designed to deliver six Dicer substrate (DS) RNAs, which upon intracellular dicing would release six siRNAs from each NANP. RNA-DNA hybrids were designed to carry several split functionalities (DS RNAs and NF-κB decoys) which would only be activated upon the intracellular reassociation of individually delivered cognate fibers. NANP-loaded exosomes were confirmed to be non-immunostimulatory and nontoxic and the released NANPs produced high silencing efficiency and immunosuppressive effects in their targets. Interestingly, the uptake efficiencies of NANP-loaded exosomes were found to be significantly higher for cancerous cells when compared to primary cells, thus potentially allowing for targeted exosome-mediated NANP delivery with reduced off-target effects in healthy cells (Figure 4C)34.
Dexamethasone sodium phosphate (Dex), one of the most frequently used glucocorticoids (GCs), plays a role in the treatment of rheumatoid arthritis. A surface-engineered exosome (modified with folic acid (FA)-PEG-cholesterol (FPC)) encapsulated Dex and its performance was compared with Dex-delivering liposomes (Lip/Dex). Exo/Dex showed better internalization and greater stability than Lip/Dex. The FPC-Exo/Dex system targeted inflamed joints, downregulated proinflammatory cytokine expression, and upregulated anti-inflammatory cytokines, leading to inhibition of macrophage-associated inflammation in collagen-induced arthritic mice (Figure 4D)191.
As an alternative to loading TNAs within the exosome interior, exosomal surface engineering can aid in the delivery of various motifs. For example, tumor cell-derived exosomes designed to express streptavidin-lactadherin on their membranes can bind to biotinylated CpG DNAs with high affinity. This exosome-based tumor antigen-adjuvant codelivery system displayed augmented antitumor effects in vivo (Figure 4E)192. Table 2 lists other cargoes delivered by exosomes.
Table 2.
Exosome-mediated delivery of various therapeutic cargos.
| Class of cargo | Cargo | Reference |
|---|---|---|
| TNAs | siRNA against RAD51 and RAD52 | Shtam et al., Commun.signal., 2013 |
| siRNA against MAPK1 | Wahlgren et al., Nucleic Acids Res., 2012 | |
| siRNA against luciferase | Banizs et al., Int J Nanomedicine, 2014 | |
| siRNA against GAPDH | Alvarez-Erviti et al., Nat. Biotec., 2011 | |
| siRNA against CD81 | Pan et al., Gut, 2012 | |
| siRNA against Huntingtin | Didiot et al., Mol Ther., 2016 | |
| miR-214 | Chen et al., Hepatology, 2013 | |
| miR-150 | Zhang et al., Mol.Cell, 2010 | |
| miR-146b | Katakowski et al., Cancer Lett., 2013 | |
| miR-143 | Kosaka et al., J.Biol. Chem., 2012 | |
| miR-133b | Xin et al., Stem Cell, 2012 | |
| miR-124 | Yang et al., Mol Ther. Nuc Acids, 2017 | |
| miR-335 | Mittelbrunn et al., Nature com., 2011 | |
| miR-138 | Li et al., Mol Ther. Nuc Acids, 2019 | |
| EBV-miR | Pegtel et al., Proc Natl Acad Sci USA, 2010 | |
| Let-7a miRNA | Ohno et al., Mol Ther., 2013 | |
| Cy5-anti-miR-9 | Munoz et al., Mol. Ther. Nuc Acids, 2013 | |
| CpG DNA | Morishita et al., Biomaterials, 2016 | |
| DNA | Lamichhane et al., Mol. Pharmaceutics, 2015 | |
| NGF mRNA | Yang et al., Mol Ther. Nuc Acids, 2020 | |
| Proteins | catalase | Haney et al., J Control Release, 2015 |
| porphyrin | Fuhrmann et al., J Control Release, 2015 | |
| Lipids | liposome | Sato et al., Sci Rep., 2016 |
| Small molecules | Curcumin | Sun et al., Mol. Ther., 2010 |
| Doxorubicin | Tian et al., Biomaterials, 2013 | |
| Paclitaxel | Agrawal et al., Nanomedicine, 2017 | |
| Paclitaxel and doxorubicin | Yang et al., Pharm Res., 2015 | |
| Olaparib | Jung et al., Biomaterials, 2018 | |
| Nanoparticles | NANPs functionalized with multiple RNAi inducers and NF-kB decoys | Nordmeier et al., Nanomedicine: Nanotechnology, Biology and Medicine, 2020 |
| superparamagnetic iron oxide nanoparticles (SPION5) | Hood et al., Analytical biochemistry, 2014 | |
| Metal-organic framework nanoparticles | Illes et al., Chem.Mater, 2017 | |
| 3WJ arrowtail RNA Nanoparticle contain siRNA against survivin | Pi et al., Nature Nanotechnology, 2018 | |
| dexamethasone sodium phosphate (Dex) nanoparticle | Yan et al., J Nanobiotechnol, 2020 | |
| Other | Adeno-associated viral vector | Maguire et al., Mol. Ther., 2012 |
| JSI-124 | Zhuang et al., Mol. Ther., 2011 | |
| CRISPR/Cas9 | Lin et al., Adv Sci., 2018 |
Current challenges in exosome research.
Exosomes’ ability to reprogram recipient cells via the delivery of functional proteins, lipids, and nucleic acids makes them viable delivery systems for therapeutic genetic materials. In addition, these structures possess an array of advantages: high safety and efficacy, bioavailability, stability, high membrane permeation capacity (including the blood-brain barrier), low toxicity, low immunostimulation, and low off-target effects. All of these properties contribute to exosomes’ outstanding potential as tools for personalized medicine. Despite their benefits, exosomes and their clinical applications still face several challenges. First, isolation and purification of exosomes suffers from underdevelopment and a lack of standardized protocols. As of right now, conventional isolation methods include ultracentrifugation, size exclusion chromatography, ultrafiltration, immunological- or magnetic-activated methods, and polymer-based precipitations (Figure 5A). Ultracentrifugation is labor-intensive, time-consuming, and its separation efficiency depends on various factors such as applied G force, rotor type, rotor k-factor, sample viscosity, etc. Due to the applied external force, morphological damage and sample aggregation often occur, resulting in low exosome recovery yield193. Size-exclusion chromatography can effectively separate albumin from purified exosomes, but this method isolates less than 5% of the entire exosome volume194. Ultrafiltration methods are rapid and achieve high purity by forcing exosomes to pass through selective pores, but the high sheer stresses may deform the exosomes’ shapes or even break them apart. Additionally, the filters may become clogged and introduce contaminating materials that interfere with downstream applications193. Immunoaffinity isolation delivers high specificity and enables for the isolation of specific exosome types. This method has the drawback of low yield, however, because it relies solely on antibodies that interact with exosomal surface proteins, and some markers may not be present or recognized on all collected exosomes116. Furthermore, the use of antibodies can be expensive193, 195. Polymer-based precipitation is easy to perform and has relatively low financial cost. Several commercial exosome isolation kits are available for carrying out exosome precipitation from various bodily fluids and culture media196. Table 3 summarizes some commercially available exosome isolation kits. Nevertheless, this technique has one significant problem: non-exosomal molecules like proteins may also precipitate. This issue, along with incomplete polymer removal, leads to low exosome isolation purity197. Both impurities and poor recovery yield hinder the production of large amounts of high-quality exosomes.
Figure 5.
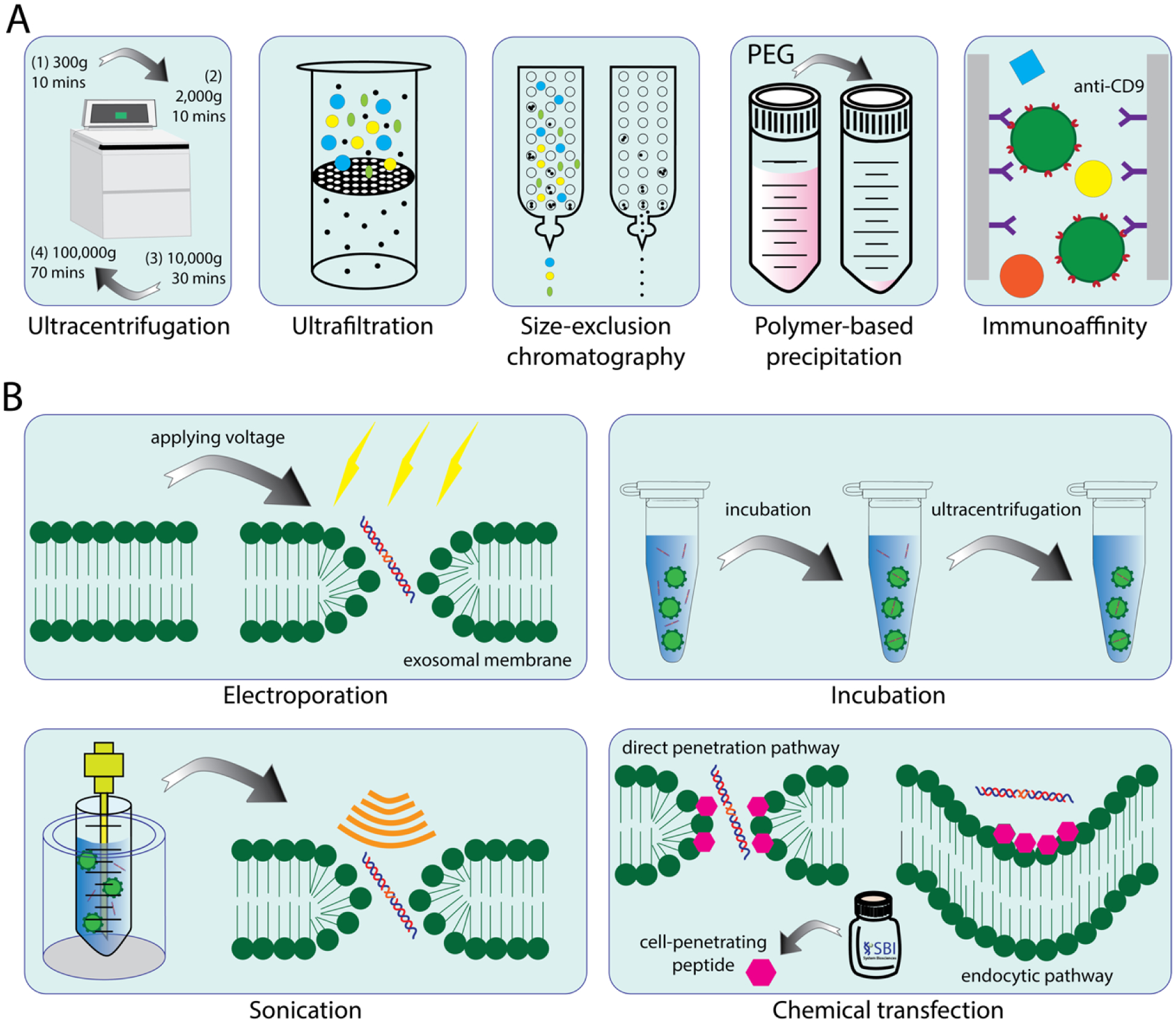
Different methods for exosome isolation and drug encapsulation. (A) Commonly used methods for exosome isolation. (B) Commonly used methods for drug loading to exosomes.
Table 3.
Commercially available exosome isolation kits.
| Product | Isolation method | Exosome source | Price estimate | Vendor | Reference |
|---|---|---|---|---|---|
| ExoQuick | Precipitation | Plasma, serum, ascites fluid, cell culture | ~$25 per reaction | System Biosciences | Nordmeier et al., Nanomedicine: Nanotechnology, Biology and Medicine, 2020 |
| Invitrogen total exosome isolation | Precipitation | Plasma, serum, cell culture, CSF, ascitic fluid, amniotic fluid, milk, saliva | ~$4 – $19 per mL of sample | Life Technologies Inc | Le Gall et al, Skeletal muscle, 2020 |
| miRCURY | Precipitation | Plasma, serum, cell culture, CSF, urine | ~$3 per mL of sample | Exiquon Inc | Mercadal et al., Int J Mol Sci, 2020 |
| ExoJuice | Precipitation | Cell culture and other media | ~$11 per reaction | ExonanoRNA | N/A |
| qEVoriginal | Size-exclusion chromatography | Plasma, cell culture, saliva | ~$21 – $50 per column | iZON Ltd | Jakubec et al., PloS one, 2020 |
| Exo-spin | Size-exclusion chromatography | Plasma, serum, cell culture, saliva and urine up to 500 ml | ~$9 – $41 per column | Cell Guidance Systems Ltd. | Kojima et al., Nat Comm, 2018 |
Heterogeneity of exosome isolation causes a polydispersity of exosome sizes and leads to challenges for exosome characterization. As mentioned earlier, exosome size ranges overlap with microvesicles, resulting in co-isolation of other EV types and problems with accurate exosome identification. The presence of exomeres, nano-sized particles with 35 nm diameters that lack external membranes and spherical shapes, also interferes with size-based exosome isolation techniques116. Exosome sizes range from 40–150 nm and display polydispersity, leading to difficulties in distinguishing exosomes from microvesicles (50–1,000 nm). Additionally, different-sized subpopulations of exosomes have been found to contain different proteins. For example, proteins involved in MVB biogenesis, membrane fusion, and vesicle budding are enriched in larger-sized exosome subpopulations, while proteins involved in RNA polymerase II complex assembly; telomere maintenance; nucleic acid, protein, and carbohydrate metabolism; and apoptosis signaling were enriched in smaller-sized groups116. Such protein content differences may lead to characterization bias of exosomal markers and proteomic analysis. Exosome shapes are also controversial―TEM morphology analyses describe cup-shaped geometries198, while SEM imaging reveals spherical shapes199.
Compared to natural cargo uptake, the therapeutic loading of exosomes with TNAs, proteins, or drugs of various sizes presents a challenge. To achieve this goal, many approaches have been considered and, as one might expect, each has its limitations. As with exosome isolation and purification, no standardized methods for drug loading exist. Some examples of techniques that encapsulate foreign materials into exosomes include incubation, sonication, electroporation, and chemical transfection (Figure 5B). Incubation simply relies on incubating foreign materials with exosomes for a certain time period200. This method is the simplest and most cost-effective, but also the most selective. Hydrophobic molecules with low to medium molecular weights such as porphyrins201 and catalase202 are more likely to be incorporated into exosomes using incubation. On the other hand, TNAs, NANPs, and other hydrophilic molecules cannot be loaded into exosomes using this strategy200.
Electroporation, a transfection method that creates temporary pores in the plasma membrane using an electrical pulse, drives charged molecules by establishing an electric potential across the plasma membrane. Electroporation voltages typically range from 150V to 700V and depend on cell type. Although the process is highly stable and efficient, high voltage pulses induce substantial lysing of the exosomes. Because of this, and the fact that membrane repair is only partially successful, a greater exosome quantity must be used in this procedure. Sonication employs ultrasonic waves to open small, transient holes in the plasma membrane, thereby increasing membrane permeability and permitting foreign material entry. Similar to electroporation, this physical transfection method inflicts damage to exosomes203. Chemical transfection represents a promising alternative method for achieving direct cargo loading. One commonly used agent, Lipofectamine, efficiently transfects nucleic acids into cell cultures. Lipofectamine contains lipid subunits that form liposomes in an aqueous environment and trap nucleic acids. The positively charged liposome surface fuses with the negatively charged plasma membrane and subsequently releases its payload. However, its toxicity limits its applications for in vivo studies204. Over the past few decades, other effective reagents have become commercially available. Exo-Fect™, for example, uses cell-penetrating peptides to transfect TNAs into exosomes. These kits are fast and easy to use, have proven high delivery efficiency, and coincide with the equipment necessary for sonication or electroporation34, 181, 205, 206. One of the potential disadvantages, however, is that the amounts of Exo-Fect™ necessary to deliver large quantities of NANPs can be prohibitively expensive.
Future directions and perspectives.
Although research has yielded significant progress over the past few decades, the mechanisms of exosomes’ biogenesis, cargo sorting, functions, and biological fates among various cell types still remain unclear. Moreover, understanding the fate of ILVs inside MVBs—whether the MVBs will be autophagically degraded in the lysosome or fuse with the cell membrane and release those ILVs as exosomes—may provide more perspectives in terms of exosome secretion. More exclusive markers which could indicate the exosome subpopulation would allow for production to be scaled up with high precision. Finally, in order to provide guidance on using exosomes as drug delivery vehicles, an advanced real-time imaging system is needed for exosome trafficking. As these limitations are overcome, a better understanding of exosome formation and functioning, along with disparities between healthy and diseased conditions, will be possible. Undoubtedly, a cost- and time-effective approach to exosome isolation, more reliable characterization, more precise targeting, and the safety of administration have to be realized before exosomes’ clinical application. Therefore, new experimental approaches and technologies must be developed to support the use of exosomes as personalized medicine. A potential workflow of such an approach is collecting and isolating exosomes from the patient at the bedside, reformulating exosomes with encapsulated therapeutic cargos, and decorating targeting moieties at the exosomal surface. These engineered multifunctional exosomes will then be challenged with the patient’s own peripheral blood mononuclear cells (PBMCs) for immunogenicity validation.Exosomes’ intrinsic low toxicity, low immunogenicity, high specificity, and high drug loading capacity represent a promising future in the field of nanomedicine. The manipulation of exosomes’ compositions, properties, secretion, biodistribution, and cell-cell interactions will effectively advance their roles in clinical applications as biomarkers and drug delivery vehicles towards the use of exosomes as potent and effective personalized medicines (Figure 6).
Figure 6.
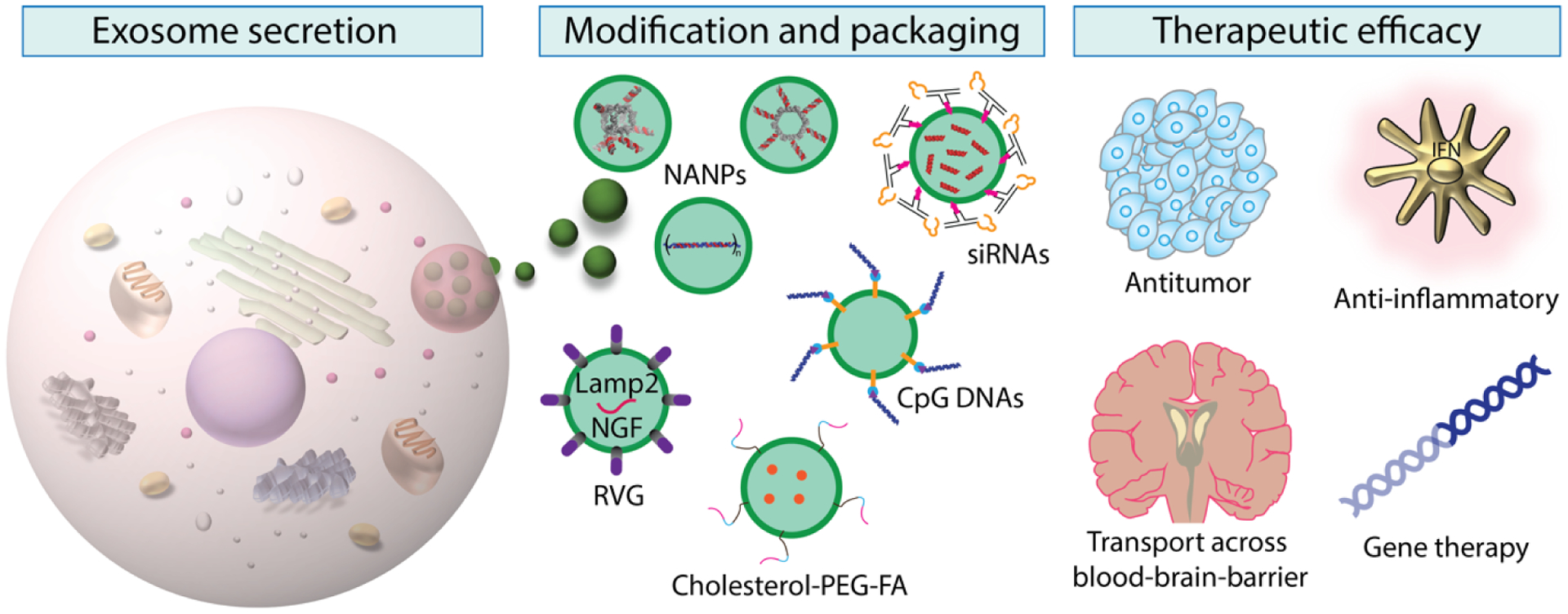
Schematic summary of the workflow of exosome-mediated delivery of different functionalized cargos. Exosomes are released by cells. After isolation and purification, exosome surfaces can be engineered for specific targeting, or modified with covalent linkages for various motifs. Therapeutic cargos are loaded into the exosome lumen or linked on the exosome surface. After uptake by the destination cells, the therapeutic motifs are able to elicit intended effects in target cells.
Table 1.
Common exosomal markers.
| Exosomal marker | Gene | Functions | Reference |
|---|---|---|---|
| Programmed cell death 6-interacting protein ALIX | PDCD6IP | vesicle trafficking | Naseri et al., Int. J. Nanomed., 2018 |
| CD9 antigen | CD9 | integral membrane protein | Naseri et al., Int. J. Nanomed., 2018 |
| Annexin V | ANX45 | phospholipid-binding | Jeppesen et al., Cell, 2019 |
| Heat shock cognate protein 71kDa | HSC70 | protein folding | Kowal et al., PNAS, 2016 |
| CD63 antigen | CD63 | endosomal cargo sorting | Hurwitz et al., Journal of Virology, 2018 |
| CD81 antigen | CD81 | cell migration | Tejera et al., Mol Biol Cell, 2013 |
| Flotillin-1 | FLOT1 | vesicle trafficking | Wang et al., Blood, 2014 |
| Heat shock protein 70kDa | HSP70 | protein folding | Wen et al., Cancer Res., 2016 |
| Tumor susceptibility gene 101 protein | TSG101 | vesicle trafficking | Goh et al., Sci. Rep., 2017 |
| Annexin A2 | ANXA2 | vesicle trafficking | Chaudhary et al., Breast Cancer Res., 2020 |
| Syntenin-1 | SDCBP | protein trafficking | Rontogianni et al., Commun Biol., 2019 |
| Peptidylprolyl isomerase A | PPIA | intracellular signaling | Gomez-Molina et al., Int J Neuropsychophar., 2019 |
| Integrin, beta-1 | ITGB1 | anchorage to ECM and cell-surface adhesion | Clayton et al., FASEB J., 2004 |
Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers R01GM120487 and R35GM139587. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Morgan Chandler (UNC Charlotte) and Gabriel Marcus for assistance with proofreading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adams D; Gonzalez-Duarte A; O’Riordan WD; Yang CC; Ueda M; Kristen AV; Tournev I; Schmidt HH; Coelho T; Berk JL; Lin KP; Vita G; Attarian S; Plante-Bordeneuve V; Mezei MM; Campistol JM; Buades J; Brannagan TH 3rd; Kim BJ; Oh J; Parman Y; Sekijima Y; Hawkins PN; Solomon SD; Polydefkis M; Dyck PJ; Gandhi PJ; Goyal S; Chen J; Strahs AL; Nochur SV; Sweetser MT; Garg PP; Vaishnaw AK; Gollob JA; Suhr OB, Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. The New England journal of medicine 2018, 379 (1), 11–21. [DOI] [PubMed] [Google Scholar]
- 2.Sridharan K; Gogtay NJ, Therapeutic nucleic acids: current clinical status. Br J Clin Pharmacol 2016, 82 (3), 659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan A; Liebow A; Yasuda M; Gan L; Racie T; Maier M; Kuchimanchi S; Foster D; Milstein S; Charisse K; Sehgal A; Manoharan M; Meyers R; Fitzgerald K; Simon A; Desnick RJ; Querbes W, Preclinical Development of a Subcutaneous ALAS1 RNAi Therapeutic for Treatment of Hepatic Porphyrias Using Circulating RNA Quantification. Mol Ther Nucleic Acids 2015, 4, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pharmaceuticals, A., Alnylam Announces U.S. Food and Drug Administration (FDA) Approval of OXLUMO™ (lumasiran), the First and Only Treatment Approved for Primary Hyperoxaluria Type 1 to Lower Urinary Oxalate Levels in Pediatric and Adult Patients.
- 5.Pharmaceuticals, A., Alnylam Receives Approval for OXLUMO™ (lumasiran) in the European Union for the Treatment of Primary Hyperoxaluria Type 1 in All Age Groups.
- 6.NOVARTIS, Novartis receives EU approval for Leqvio® (inclisiran), a first-in-class siRNA to lower cholesterol with two doses a year.
- 7.Hu B; Weng Y; Xia XH; Liang XJ; Huang Y, Clinical advances of siRNA therapeutics. The Journal of Gene Medicine 2019, 21 (7), e3097. [DOI] [PubMed] [Google Scholar]
- 8.Rader DJ; Kastelein JJ, Lomitapide and mipomersen: two first-in-class drugs for reducing low-density lipoprotein cholesterol in patients with homozygous familial hypercholesterolemia. Circulation 2014, 129 (9), 1022–1032. [DOI] [PubMed] [Google Scholar]
- 9.Shukla D; Namperumalsamy P; Goldbaum M; Cunningham ET Jr., Pegaptanib sodium for ocular vascular disease. Indian J Ophthalmol 2007, 55 (6), 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi JJ; June CH; Kohn DB, Genetic therapies against HIV. Nature biotechnology 2007, 25 (12), 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afonin KA; Dobrovolskaia MA; Church G; Bathe M, Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2020, 14 (8), 9221–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jasinski D; Haque F; Binzel DW; Guo P, Advancement of the Emerging Field of RNA Nanotechnology. ACS nano 2017, 11 (2), 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA, Multifunctional RNA Nanoparticles. Nano Letters 2014, 14 (10), 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L, Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nature Protocols 2011, 6 (12), 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A; Stepler M; Kewalramani VN; Kashlev M; Blumenthal R; Shapiro BA, Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities. Nucleic acids research 2014, 42 (3), 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler M; Panigaj M; Rolband LA; Afonin KA, Challenges to optimizing RNA nanostructures for large scale production and controlled therapeutic properties. Nanomedicine (Lond) 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panigaj M; Johnson MB; Ke W; McMillan J; Goncharova EA; Chandler M; Afonin KA, Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology. ACS Nano 2019, 13 (11), 12301–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Low JT; Knoepfel SA; Watts JM; ter Brake O; Berkhout B; Weeks KM, SHAPE-directed discovery of potent shRNA inhibitors of HIV-1. Mol Ther 2012, 20 (4), 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A; Stepler M; Kewalramani VN; Kashlev M; Blumenthal R; Shapiro BA, Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities. Nucleic Acids Res 2014, 42 (3), 2085–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN; Lu JS; Miller J; Khisamutdinov EF; Shapiro BA; Dobrovolskaia MA; Afonin KA, Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res 2017, 45 (4), 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandler M; Johnson MB; Panigaj M; Afonin KA, Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Current Opinion in Biotechnology 2020, 63, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandler M; Lyalina T; Halman J; Rackley L; Lee L; Dang D; Ke W; Sajja S; Woods S; Acharya S, Broccoli fluorets: split aptamers as a user-friendly fluorescent toolkit for dynamic RNA Nanotechnology. Molecules 2018, 23 (12), 3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandler M; Ke W; Halman JR; Panigaj M; Afonin KA, Reconfigurable nucleic acid materials for cancer therapy. In Nanooncology, Springer: 2018; pp 365–385. [Google Scholar]
- 24.Chandler M; Afonin KA, Smart-Responsive Nucleic Acid Nanoparticles (NANPs) with the Potential to Modulate Immune Behavior. Nanomaterials 2019, 9 (4), 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV, RNA–DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells. Nucleic acids research 2019, 47 (3), 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dao BN; Viard M; Martins AN; Kasprzak WK; Shapiro BA; Afonin KA, Triggering RNAi with multifunctional RNA nanoparticles and their delivery. DNA and RNA nanotechnology 2015, 1 (open-issue), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA, Activation of different split functionalities on re-association of RNA-DNA hybrids. Nature nanotechnology 2013, 8 (4), 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A, Co-transcriptional production of RNA–DNA hybrids for simultaneous release of multiple split functionalities. Nucleic acids research 2014, 42 (3), 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonin KA; Viard M; Kagiampakis I; Case CL; Dobrovolskaia MA; Hofmann J; Vrzak A; Kireeva M; Kasprzak WK; KewalRamani VN; Shapiro BA, Triggering of RNA Interference with RNA–RNA, RNA–DNA, and DNA–RNA Nanoparticles. ACS nano 2015, 9 (1), 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindewald E; Afonin KA; Viard M; Zakrevsky P; Kim T; Shapiro BA, Multistrand Structure Prediction of Nucleic Acid Assemblies and Design of RNA Switches. Nano Letters 2016, 16 (3), 1726–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zakrevsky P; Parlea L; Viard M; Bindewald E; Afonin KA; Shapiro BA, Preparation of a Conditional RNA Switch. Methods Mol Biol 2017, 1632, 303–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobrovolskaia MA; Afonin KA, Use of human peripheral blood mononuclear cells to define immunological properties of nucleic acid nanoparticles. Nature protocols 2020, 15 (11), 3678–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson MB; Halman JR; Miller DK; Cooper JS; Khisamutdinov Emil F.; Marriott I; Afonin KA, The immunorecognition, subcellular compartmentalization, and physicochemical properties of nucleic acid nanoparticles can be controlled by composition modification. Nucleic Acids Research 2020, 48 (20), 11785–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nordmeier S; Ke W; Afonin KA; Portnoy V, Exosome mediated delivery of functional nucleic acid nanoparticles (NANPs). Nanomedicine: Nanotechnology, Biology and Medicine 2020, 30, 102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juneja R; Vadarevu H; Halman J; Tarannum M; Rackley L; Dobbs J; Marquez J; Chandler M; Afonin K; Vivero-Escoto JL, Combination of Nucleic Acid and Mesoporous Silica Nanoparticles: Optimization and Therapeutic Performance In Vitro. ACS applied materials & interfaces 2020, 12 (35), 38873–38886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandler M; Panigaj M; Rolband LA; Afonin KA, Challenges to optimizing RNA nanostructures for large scale production and controlled therapeutic properties. Nanomedicine 2020, (0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson MB; Halman JR; Burmeister AR; Currin S; Khisamutdinov EF; Afonin KA; Marriott I, Retinoic acid inducible gene-I mediated detection of bacterial nucleic acids in human microglial cells. Journal of Neuroinflammation 2020, 17 (1), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halman JR; Kim K-T; Gwak S-J; Pace R; Johnson MB; Chandler MR; Rackley L; Viard M; Marriott I; Lee JS; Afonin KA, A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution. Nanomedicine: Nanotechnology, Biology and Medicine 2020, 23, 102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong E; Halman JR; Shah A; Cedrone E; Truong N; Afonin KA; Dobrovolskaia MA, Toll-like receptor-mediated recognition of nucleic acid nanoparticles (NANPs) in human primary blood cells. Molecules 2019, 24 (6), 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rackley L; Stewart JM; Salotti J; Krokhotin A; Shah A; Halman JR; Juneja R; Smollett J; Lee L; Roark K, RNA fibers as optimized nanoscaffolds for siRNA coordination and reduced immunological recognition. Advanced functional materials 2018, 28 (48), 1805959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA, Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Letters 2018, 18 (7), 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA, Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling. Small 2017, 13 (42), 1701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halman JR; Satterwhite E; Roark B; Chandler M; Viard M; Ivanina A; Bindewald E; Kasprzak WK; Panigaj M; Bui MN, Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic acids research 2017, 45 (4), 2210–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson MB; Halman JR; Burmeister AR; Currin S; Khisamutdinov EF; Afonin KA; Marriott I, Retinoic acid inducible gene-I mediated detection of bacterial nucleic acids in human microglial cells. J Neuroinflammation 2020, 17 (1), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson MB; Chandler M; Afonin KA, Nucleic acid nanoparticles (NANPs) as molecular tools to direct desirable and avoid undesirable immunological effects. Advanced Drug Delivery Reviews 2021, 173, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson MB; Halman JR; Satterwhite E; Zakharov AV; Bui MN; Benkato K; Goldsworthy V; Kim T; Hong E; Dobrovolskaia MA; Khisamutdinov EF; Marriott I; Afonin KA, Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small 2017, 13 (42). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobrovolskaia MA; Afonin KA, Use of human peripheral blood mononuclear cells to define immunological properties of nucleic acid nanoparticles. Nat Protoc 2020, 15 (11), 3678–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong E; Halman JR; Shah AB; Khisamutdinov EF; Dobrovolskaia MA; Afonin KA, Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Lett 2018, 18 (7), 4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halman JR; Kim KT; Gwak SJ; Pace R; Johnson MB; Chandler MR; Rackley L; Viard M; Marriott I; Lee JS; Afonin KA, A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (Nanps) for controlled gene silencing, immunostimulation, and biodistribution. Nanomedicine 2020, 23, 102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avila YI; Chandler M; Cedrone E; Newton HS; Richardson M; Xu J; Clogston JD; Liptrott NJ; Afonin KA; Dobrovolskaia MA, Induction of cytokines by Nucleic Acid Nanoparticles (NANPs) depends on the type of delivery carrier. Molecules 2021, 26 (3), 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson MB; Halman JR; Miller DK; Cooper JS; Khisamutdinov EF; Marriott I; Afonin KA, The immunorecognition, subcellular compartmentalization, and physicochemical properties of nucleic acid nanoparticles can be controlled by composition modification. Nucleic Acids Res 2020, 48 (20), 11785–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandler M; Johnson MB; Panigaj M; Afonin KA, Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Curr Opin Biotechnol 2020, 63, 8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen DS; Mellman I, Oncology meets immunology: the cancer-immunity cycle. Immunity 2013, 39 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- 54.Dobrovolskaia MA, Nucleic Acid Nanoparticles at a Crossroads of Vaccines and Immunotherapies. Molecules (Basel, Switzerland) 2019, 24 (24), 4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA, RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic Acids Res 2019, 47 (3), 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ke W; Hong E; Saito RF; Rangel MC; Wang J; Viard M; Richardson M; Khisamutdinov EF; Panigaj M; Dokholyan NV; Chammas R; Dobrovolskaia MA; Afonin KA, RNA–DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-κB in human cells. Nucleic Acids Research 2018, 47 (3), 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stojanovic MN; Kolpashchikov DM, Modular Aptameric Sensors. Journal of the American Chemical Society 2004, 126 (30), 9266–9270. [DOI] [PubMed] [Google Scholar]
- 58.An Aptamer–siRNA Chimera Silences the Eukaryotic Elongation Factor 2 Gene and Induces Apoptosis in Cancers Expressing αvβ3 Integrin. Nucleic Acid Ther 2013, 23 (3), 203–212. [DOI] [PubMed] [Google Scholar]
- 59.Tarapore P; Shu Y; Guo P; Ho SM, Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Mol Ther 2011, 19 (2), 386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalpke AH; Zimmermann S; Albrecht I; Heeg K, Phosphodiester CpG oligonucleotides as adjuvants: polyguanosine runs enhance cellular uptake and improve immunostimulative activity of phosphodiester CpG oligonucleotides in vitro and in vivo. Immunology 2002, 106 (1), 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Juliano RL, The delivery of therapeutic oligonucleotides. Nucleic acids research 2016, 44 (14), 6518–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim T; Viard M; Afonin KA; Gupta K; Popov M; Salotti J; Johnson PF; Linder C; Heldman E; Shapiro BA, Characterization of Cationic Bolaamphiphile Vesicles for siRNA Delivery into Tumors and Brain. Mol Ther Nucleic Acids 2020, 20, 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vivero-Escoto JL; Slowing II; Trewyn BG; Lin VSY, Mesoporous Silica Nanoparticles for Intracellular Controlled Drug Delivery. Small 2010, 6 (18), 1952–1967. [DOI] [PubMed] [Google Scholar]
- 64.Juneja R; Vaderevu H; Halman JR; Tarannum M; Rackley L; Dobbs J; Marquez J; Chandler M; Afonin KA; Vivero-Escoto JL, Combination of nucleic acid and mesoporous silica nanoparticles: optimization and therapeutic performance in vitro. ACS Appl Mater Interfaces 2020, 12 (35), 38873–38886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shah V; Taratula O; Garbuzenko OB; Patil ML; Savla R; Zhang M; Minko T, Genotoxicity of different nanocarriers: possible modifications for the delivery of nucleic acids. Curr Drug Discov Technol 2013, 10 (1), 8–15. [PMC free article] [PubMed] [Google Scholar]
- 66.Szebeni J, Complement activation-related pseudoallergy: A new class of drug-induced acute immune toxicity. Toxicology 2005, 216 (2), 106–121. [DOI] [PubMed] [Google Scholar]
- 67.Syama S; Gayathri V; Mohanan PV, Assessment of Immunotoxicity of Dextran Coated Ferrite Nanoparticles in Albino Mice. Molecular Biology International 2015, 2015, 518527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szebeni J; Moghimi SM, Liposome triggering of innate immune responses: a perspective on benefits and adverse reactions. J Liposome Res 2009, 19 (2), 85–90. [DOI] [PubMed] [Google Scholar]
- 69.Knudsen KB; Northeved H; Kumar PE; Permin A; Gjetting T; Andresen TL; Larsen S; Wegener KM; Lykkesfeldt J; Jantzen K; Loft S; Møller P; Roursgaard M, In vivo toxicity of cationic micelles and liposomes. Nanomedicine 2015, 11 (2), 467–77. [DOI] [PubMed] [Google Scholar]
- 70.Dams ET; Laverman P; Oyen WJ; Storm G; Scherphof GL; van Der Meer JW; Corstens FH; Boerman OC, Accelerated blood clearance and altered biodistribution of repeated injections of sterically stabilized liposomes. J Pharmacol Exp Ther 2000, 292 (3), 1071–9. [PubMed] [Google Scholar]
- 71.Kobayashi N; Izumi H; Morimoto Y, Review of toxicity studies of carbon nanotubes. J Occup Health 2017, 59 (5), 394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carnino JM; Lee H; Jin Y, Isolation and characterization of extracellular vesicles from Broncho-alveolar lavage fluid: a review and comparison of different methods. Respiratory Research 2019, 20 (1), 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colombo M; Raposo G; Théry C, Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014, 30, 255–89. [DOI] [PubMed] [Google Scholar]
- 74.Crescitelli R; Lässer C; Szabó TG; Kittel A; Eldh M; Dianzani I; Buzás EI; Lötvall J, Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. Journal of Extracellular Vesicles 2013, 2 (1), 20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Becker A; Thakur BK; Weiss JM; Kim HS; Peinado H; Lyden D, Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30 (6), 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konoshenko MY; Lekchnov EA; Vlassov AV; Laktionov PP, Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Research International 2018, 2018, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tauro BJ; Greening DW; Mathias RA; Ji H; Mathivanan S; Scott AM; Simpson RJ, Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56 (2), 293–304. [DOI] [PubMed] [Google Scholar]
- 78.Böing AN; Van Der Pol E; Grootemaat AE; Coumans FA; Sturk A; Nieuwland R, Single-step isolation of extracellular vesicles by size-exclusion chromatography. Journal of extracellular vesicles 2014, 3 (1), 23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor DD; Shah S, Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [DOI] [PubMed] [Google Scholar]
- 80.Andreu Z; Rivas E; Sanguino-Pascual A; Lamana A; Marazuela M; González-Alvaro I; Sánchez-Madrid F; de la Fuente H; Yáñez-Mó M, Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. Journal of extracellular vesicles 2016, 5 (1), 31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He M; Crow J; Roth M; Zeng Y; Godwin AK, Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab on a Chip 2014, 14 (19), 3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wan Y; Cheng G; Liu X; Hao S-J; Nisic M; Zhu C-D; Xia Y-Q; Li W-Q; Wang Z-G; Zhang W-L, Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nature biomedical engineering 2017, 1 (4), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yáñez-Mó M; Siljander PRM; Andreu Z; Bedina Zavec A; Borràs FE; Buzas EI; Buzas K; Casal E; Cappello F; Carvalho J; Colás E; Cordeiro-Da Silva A; Fais S; Falcon-Perez JM; Ghobrial IM; Giebel B; Gimona M; Graner M; Gursel I; Gursel M; Heegaard NHH; Hendrix A; Kierulf P; Kokubun K; Kosanovic M; Kralj-Iglic V; Krämer-Albers E-M; Laitinen S; Lässer C; Lener T; Ligeti E; Linē A; Lipps G; Llorente A; Lötvall J; Manček-Keber M; Marcilla A; Mittelbrunn M; Nazarenko I; Nolte-’T Hoen ENM; Nyman TA; O’Driscoll L; Olivan M; Oliveira C; Pállinger É; Del Portillo HA; Reventós J; Rigau M; Rohde E; Sammar M; Sánchez-Madrid F; Santarém N; Schallmoser K; Stampe Ostenfeld M; Stoorvogel W; Stukelj R; Van Der Grein SG; Helena Vasconcelos M; Wauben MHM; De Wever O, Biological properties of extracellular vesicles and their physiological functions. Journal of Extracellular Vesicles 2015, 4 (1), 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sugano G; Bernard-Pierrot I; Laé M; Battail C; Allory Y; Stransky N; Krumeich S; Lepage ML; Maille P; Donnadieu MH; Abbou CC; Benhamou S; Lebret T; Sastre-Garau X; Amigorena S; Radvanyi F; Théry C, Milk fat globule—epidermal growth factor—factor VIII (MFGE8)/lactadherin promotes bladder tumor development. Oncogene 2011, 30 (6), 642–653. [DOI] [PubMed] [Google Scholar]
- 85.Hosseinkhani B; Kuypers S; van den Akker NMS; Molin DGM; Michiels L, Extracellular Vesicles Work as a Functional Inflammatory Mediator Between Vascular Endothelial Cells and Immune Cells. Front Immunol 2018, 9, 1789–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D’Souza-Schorey C; Chavrier P, ARF proteins: roles in membrane traffic and beyond. Nature Reviews Molecular Cell Biology 2006, 7 (5), 347–358. [DOI] [PubMed] [Google Scholar]
- 87.Raposo G; Stoorvogel W, Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology 2013, 200 (4), 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao K; Bleackley M; Chisanga D; Gangoda L; Fonseka P; Liem M; Kalra H; Al Saffar H; Keerthikumar S; Ang C-S; Adda CG; Jiang L; Yap K; Poon IK; Lock P; Bulone V; Anderson M; Mathivanan S, Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun Biol 2019, 2, 305–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Borges FT; Reis LA; Schor N, Extracellular vesicles: structure, function, and potential clinical uses in renal diseases. Brazilian Journal of Medical and Biological Research 2013, 46 (10), 824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shlomovitz I; Speir M; Gerlic M, Flipping the dogma – phosphatidylserine in non-apoptotic cell death. Cell Communication and Signaling 2019, 17 (1), 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baietti MF; Zhang Z; Mortier E; Melchior A; Degeest G; Geeraerts A; Ivarsson Y; Depoortere F; Coomans C; Vermeiren E, Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nature cell biology 2012, 14 (7), 677–685. [DOI] [PubMed] [Google Scholar]
- 92.Clayton A; Harris CL; Court J; Mason MD; Morgan BP, Antigen presenting cell exosomes are protected from complement mediated lysis by expression of CD55 and CD59. European journal of immunology 2003, 33 (2), 522–531. [DOI] [PubMed] [Google Scholar]
- 93.Barrès C; Blanc L; Bette-Bobillo P; André S; Mamoun R; Gabius H-J; Vidal M, Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 2010, 115 (3), 696–705. [DOI] [PubMed] [Google Scholar]
- 94.Berda-Haddad Y; Robert S; Salers P; Zekraoui L; Farnarier C; Dinarello CA; Dignat-George F; Kaplanski G, Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin-1α. Proceedings of the National Academy of Sciences 2011, 108 (51), 20684–20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.MacKenzie A; Wilson HL; Kiss-Toth E; Dower SK; North RA; Surprenant A, Rapid secretion of interleukin-1β by microvesicle shedding. Immunity 2001, 15 (5), 825–835. [DOI] [PubMed] [Google Scholar]
- 96.Baj-Krzyworzeka M; WĘGLARCZYK K; MYTAR B; SZATANEK R; BARAN J; Zembala M, Tumour-derived microvesicles contain interleukin-8 and modulate production of chemokines by human monocytes. Anticancer research 2011, 31 (4), 1329–1335. [PubMed] [Google Scholar]
- 97.Truman LA; Ford CA; Pasikowska M; Pound JD; Wilkinson SJ; Dumitriu IE; Melville L; Melrose LA; Ogden CA; Nibbs R, CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood, The Journal of the American Society of Hematology 2008, 112 (13), 5026–5036. [DOI] [PubMed] [Google Scholar]
- 98.Kim KM; Abdelmohsen K; Mustapic M; Kapogiannis D; Gorospe M, RNA in extracellular vesicles. Wiley Interdisciplinary Reviews: RNA 2017, 8 (4), e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jenjaroenpun P; Kremenska Y; Nair VM; Kremenskoy M; Joseph B; Kurochkin IV, Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013, 1, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Batagov AO; Kurochkin IV, Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biology Direct 2013, 8 (1), 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nolte-’t Hoen ENM; Buermans HPJ; Waasdorp M; Stoorvogel W; Wauben MHM; t Hoen PAC, Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic acids research 2012, 40 (18), 9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y; Liu D; Chen X; Li J; Li L; Bian Z; Sun F; Lu J; Yin Y; Cai X; Sun Q; Wang K; Ba Y; Wang Q; Wang D; Yang J; Liu P; Xu T; Yan Q; Zhang J; Zen K; Zhang C-Y, Secreted Monocytic miR-150 Enhances Targeted Endothelial Cell Migration. Molecular Cell 2010, 39 (1), 133–144. [DOI] [PubMed] [Google Scholar]
- 103.Mittelbrunn M; Gutiérrez-Vázquez C; Villarroya-Beltri C; González S; Sánchez-Cabo F; González MÁ; Bernad A; Sánchez-Madrid F, Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature Communications 2011, 2 (1), 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Balaj L; Lessard R; Dai L; Cho Y-J; Pomeroy SL; Breakefield XO; Skog J, Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Communications 2011, 2 (1), 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thakur BK; Zhang H; Becker A; Matei I; Huang Y; Costa-Silva B; Zheng Y; Hoshino A; Brazier H; Xiang J; Williams C; Rodriguez-Barrueco R; Silva JM; Zhang W; Hearn S; Elemento O; Paknejad N; Manova-Todorova K; Welte K; Bromberg J; Peinado H; Lyden D, Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Research 2014, 24 (6), 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guescini M; Genedani S; Stocchi V; Agnati LF, Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. Journal of Neural Transmission 2010, 117 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- 107.Cai J; Han Y; Ren H; Chen C; He D; Zhou L; Eisner GM; Asico LD; Jose PA; Zeng C, Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol 2013, 5 (4), 227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Skotland T; Sagini K; Sandvig K; Llorente A, An emerging focus on lipids in extracellular vesicles. Advanced Drug Delivery Reviews 2020. [DOI] [PubMed] [Google Scholar]
- 109.Record M; Carayon K; Poirot M; Silvente-Poirot S, Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta 2014, 1841 (1), 108–20. [DOI] [PubMed] [Google Scholar]
- 110.Carayon K; Chaoui K; Ronzier E; Lazar I; Bertrand-Michel J; Roques V; Balor S; Terce F; Lopez A; Salomé L; Joly E, Proteolipidic Composition of Exosomes Changes during Reticulocyte Maturation. Journal of Biological Chemistry 2011, 286 (39), 34426–34439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Helms JB; Zurzolo C, Lipids as Targeting Signals: Lipid Rafts and Intracellular Trafficking. Traffic 2004, 5 (4), 247–254. [DOI] [PubMed] [Google Scholar]
- 112.Kim CW; Lee HM; Lee TH; Kang C; Kleinman HK; Gho YS, Extracellular Membrane Vesicles from Tumor Cells Promote Angiogenesis via Sphingomyelin. Cancer Research 2002, 62 (21), 6312–6317. [PubMed] [Google Scholar]
- 113.Tripisciano C; Weiss R; Eichhorn T; Spittler A; Heuser T; Fischer MB; Weber V, Different Potential of Extracellular Vesicles to Support Thrombin Generation: Contributions of Phosphatidylserine, Tissue Factor, and Cellular Origin. Scientific Reports 2017, 7 (1), 6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Théry C; Witwer KW; Aikawa E; Alcaraz MJ; Anderson JD; Andriantsitohaina R; Antoniou A; Arab T; Archer F; Atkin-Smith GK; Ayre DC; Bach J-M; Bachurski D; Baharvand H; Balaj L; Baldacchino S; Bauer NN; Baxter AA; Bebawy M; Beckham C; Bedina Zavec A; Benmoussa A; Berardi AC; Bergese P; Bielska E; Blenkiron C; Bobis-Wozowicz S; Boilard E; Boireau W; Bongiovanni A; Borràs FE; Bosch S; Boulanger CM; Breakefield X; Breglio AM; Brennan MÁ; Brigstock DR; Brisson A; Broekman ML; Bromberg JF; Bryl-Górecka P; Buch S; Buck AH; Burger D; Busatto S; Buschmann D; Bussolati B; Buzás EI; Byrd JB; Camussi G; Carter DR; Caruso S; Chamley LW; Chang Y-T; Chen C; Chen S; Cheng L; Chin AR; Clayton A; Clerici SP; Cocks A; Cocucci E; Coffey RJ; Cordeiro-Da-Silva A; Couch Y; Coumans FA; Coyle B; Crescitelli R; Criado MF; D’Souza-Schorey C; Das S; Datta Chaudhuri A; De Candia P; De Santana EF; De Wever O; Del Portillo HA; Demaret T; Deville S; Devitt A; Dhondt B; Di Vizio D; Dieterich LC; Dolo V; Dominguez Rubio AP; Dominici M; Dourado MR; Driedonks TA; Duarte FV; Duncan HM; Eichenberger RM; Ekström K; El Andaloussi S; Elie-Caille C; Erdbrügger U; Falcón-Pérez JM; Fatima F; Fish JE; Flores-Bellver M; Försönits A; Frelet-Barrand A; Fricke F; Fuhrmann G; Gabrielsson S; Gámez-Valero A; Gardiner C; Gärtner K; Gaudin R; Gho YS; Giebel B; Gilbert C; Gimona M; Giusti I; Goberdhan DC; Görgens A; Gorski SM; Greening DW; Gross JC; Gualerzi A; Gupta GN; Gustafson D; Handberg A; Haraszti RA; Harrison P; Hegyesi H; Hendrix A; Hill AF; Hochberg FH; Hoffmann KF; Holder B; Holthofer H; Hosseinkhani B; Hu G; Huang Y; Huber V; Hunt S; Ibrahim AG-E; Ikezu T; Inal JM; Isin M; Ivanova A; Jackson HK; Jacobsen S; Jay SM; Jayachandran M; Jenster G; Jiang L; Johnson SM; Jones JC; Jong A; Jovanovic-Talisman T; Jung S; Kalluri R; Kano S-I; Kaur S; Kawamura Y; Keller ET; Khamari D; Khomyakova E; Khvorova A; Kierulf P; Kim KP; Kislinger T; Klingeborn M; Klinke DJ; Kornek M; Kosanović MM; Kovács ÁF; Krämer-Albers E-M; Krasemann S; Krause M; Kurochkin IV; Kusuma GD; Kuypers S; Laitinen S; Langevin SM; Languino LR; Lannigan J; Lässer C; Laurent LC; Lavieu G; Lázaro-Ibáñez E; Le Lay S; Lee M-S; Lee YXF; Lemos DS; Lenassi M; Leszczynska A; Li IT; Liao K; Libregts SF; Ligeti E; Lim R; Lim SK; Linē A; Linnemannstöns K; Llorente A; Lombard CA; Lorenowicz MJ; Lörincz ÁM; Lötvall J; Lovett J; Lowry MC; Loyer X; Lu Q; Lukomska B; Lunavat TR; Maas SL; Malhi H; Marcilla A; Mariani J; Mariscal J; Martens-Uzunova ES; Martin-Jaular L; Martinez MC; Martins VR; Mathieu M; Mathivanan S; Maugeri M; McGinnis LK; McVey MJ; Meckes DG; Meehan KL; Mertens I; Minciacchi VR; Möller A; Møller Jørgensen M; Morales-Kastresana A; Morhayim J; Mullier F; Muraca M; Musante L; Mussack V; Muth DC; Myburgh KH; Najrana T; Nawaz M; Nazarenko I; Nejsum P; Neri C; Neri T; Nieuwland R; Nimrichter L; Nolan JP; Nolte-’T Hoen EN; Noren Hooten N; O’Driscoll L; O’Grady T; O’Loghlen A; Ochiya T; Olivier M; Ortiz A; Ortiz LA; Osteikoetxea X; Østergaard O; Ostrowski M; Park J; Pegtel DM; Peinado H; Perut F; Pfaffl MW; Phinney DG; Pieters BC; Pink RC; Pisetsky DS; Pogge Von Strandmann E; Polakovicova I; Poon IK; Powell BH; Prada I; Pulliam L; Quesenberry P; Radeghieri A; Raffai RL; Raimondo S; Rak J; Ramirez MI; Raposo G; Rayyan MS; Regev-Rudzki N; Ricklefs FL; Robbins PD; Roberts DD; Rodrigues SC; Rohde E; Rome S; Rouschop KM; Rughetti A; Russell AE; Saá P; Sahoo S; Salas-Huenuleo E; Sánchez C; Saugstad JA; Saul MJ; Schiffelers RM; Schneider R; Schøyen TH; Scott A; Shahaj E; Sharma S; Shatnyeva O; Shekari F; Shelke GV; Shetty AK; Shiba K; Siljander PRM; Silva AM; Skowronek A; Snyder OL; Soares RP; Sódar BW; Soekmadji C; Sotillo J; Stahl PD; Stoorvogel W; Stott SL; Strasser EF; Swift S; Tahara H; Tewari M; Timms K; Tiwari S; Tixeira R; Tkach M; Toh WS; Tomasini R; Torrecilhas AC; Tosar JP; Toxavidis V; Urbanelli L; Vader P; Van Balkom BW; Van Der Grein SG; Van Deun J; Van Herwijnen MJ; Van Keuren-Jensen K; Van Niel G; Van Royen ME; Van Wijnen AJ; Vasconcelos MH; Vechetti IJ; Veit TD; Vella LJ; Velot É; Verweij FJ; Vestad B; Viñas JL; Visnovitz T; Vukman KV; Wahlgren J; Watson DC; Wauben MH; Weaver A; Webber JP; Weber V; Wehman AM; Weiss DJ; Welsh JA; Wendt S; Wheelock AM; Wiener Z; Witte L; Wolfram J; Xagorari A; Xander P; Xu J; Yan X; Yáñez-Mó M; Yin H; Yuana Y; Zappulli V; Zarubova J; Žėkas V; Zhang J-Y; Zhao Z; Zheng L; Zheutlin AR; Zickler AM; Zimmermann P; Zivkovic AM; Zocco D; Zuba-Surma EK, Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles 2018, 7 (1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Green TM; Alpaugh ML; Barsky SH; Rappa G; Lorico A, Breast cancer-derived extracellular vesicles: characterization and contribution to the metastatic phenotype. BioMed research international 2015, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang H; Freitas D; Kim HS; Fabijanic K; Li Z; Chen H; Mark MT; Molina H; Martin AB; Bojmar L; Fang J; Rampersaud S; Hoshino A; Matei I; Kenific CM; Nakajima M; Mutvei AP; Sansone P; Buehring W; Wang H; Jimenez JP; Cohen-Gould L; Paknejad N; Brendel M; Manova-Todorova K; Magalhães A; Ferreira JA; Osório H; Silva AM; Massey A; Cubillos-Ruiz JR; Galletti G; Giannakakou P; Cuervo AM; Blenis J; Schwartz R; Brady MS; Peinado H; Bromberg J; Matsui H; Reis CA; Lyden D, Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nature Cell Biology 2018, 20 (3), 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kanno K; Sasaki S; Hirata Y; Ishikawa S.-e.; Fushimi K; Nakanishi S; Bichet DG; Marumo F, Urinary Excretion of Aquaporin-2 in Patients with Diabetes Insipidus. New England Journal of Medicine 1995, 332 (23), 1540–1545. [DOI] [PubMed] [Google Scholar]
- 118.Sprague DL; Elzey BD; Crist SA; Waldschmidt TJ; Jensen RJ; Ratliff TL, Platelet-mediated modulation of adaptive immunity: unique delivery of CD154 signal by platelet-derived membrane vesicles. Blood 2008, 111 (10), 5028–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Berckmans RJ; Sturk A; Van Tienen LM; Schaap MCL; Nieuwland R, Cell-derived vesicles exposing coagulant tissue factor in saliva. Blood 2011, 117 (11), 3172–3180. [DOI] [PubMed] [Google Scholar]
- 120.Zhou Q; Li M; Wang X; Li Q; Wang T; Zhu Q; Zhou X; Wang X; Gao X; Li X, Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci 2012, 8 (1), 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arienti G; Carlini E; Polci A; Cosmi EV; Palmerini CA, Fatty Acid Pattern of Human Prostasome Lipid. Archives of Biochemistry and Biophysics 1998, 358 (2), 391–395. [DOI] [PubMed] [Google Scholar]
- 122.de Jong OG; Verhaar MC; Chen Y; Vader P; Gremmels H; Posthuma G; Schiffelers RM; Gucek M; van Balkom BWM, Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. Journal of extracellular vesicles 2012, 1, 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Demory Beckler M; Higginbotham JN; Franklin JL; Ham A-J; Halvey PJ; Imasuen IE; Whitwell C; Li M; Liebler DC; Coffey RJ, Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics 2013, 12 (2), 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Segura E; Nicco C; Lombard BRR; VéRon P; Raposo GA; Batteux FDR; Amigorena S; ThéRy C, ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 2005, 106 (1), 216–223. [DOI] [PubMed] [Google Scholar]
- 125.Hessvik NP; Llorente A, Current knowledge on exosome biogenesis and release. Cellular and Molecular Life Sciences 2018, 75 (2), 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnstone RM; Adam M; Hammond J; Orr L; Turbide C, Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). Journal of Biological Chemistry 1987, 262 (19), 9412–9420. [PubMed] [Google Scholar]
- 127.Marsh M, Endocytosis. Oxford University Press; 2001. [Google Scholar]
- 128.Mellman I, Endocytosis and molecular sorting. Annu Rev Cell Dev Biol 1996, 12, 575–625. [DOI] [PubMed] [Google Scholar]
- 129.Möbius W; Ohno-Iwashita Y; van Donselaar EG; Oorschot VM; Shimada Y; Fujimoto T; Heijnen HF; Geuze HJ; Slot JW, Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic perfringolysin O. J Histochem Cytochem 2002, 50 (1), 43–55. [DOI] [PubMed] [Google Scholar]
- 130.Wubbolts R; Leckie RS; Veenhuizen PTM; Schwarzmann G; Mobius W; Hoernschemeyer J; Slot JW; Geuze HJ; Stoorvogel W, Proteomic and biochemical analyses of human B cell-derived exosomes - Potential implications for their function and multivesicular body formation. Journal of Biological Chemistry 2003, 278 (13), 10963–10972. [DOI] [PubMed] [Google Scholar]
- 131.Colombo M; Moita C; Van Niel G; Kowal J; Vigneron J; Benaroch P; Manel N; Moita LF; Théry C; Raposo G, Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. Journal of Cell Science 2013, 126 (24), 5553–5565. [DOI] [PubMed] [Google Scholar]
- 132.Davies BA; Lee JRE; Oestreich AJ; Katzmann DJ, Membrane protein targeting to the MVB/lysosome. Chem Rev 2009, 109 (4), 1575–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Baietti MF; Zhang Z; Mortier E; Melchior A; Degeest G; Geeraerts A; Ivarsson Y; Depoortere F; Coomans C; Vermeiren E; Zimmermann P; David G, Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol 2012, 14 (7), 677–85. [DOI] [PubMed] [Google Scholar]
- 134.Airola MV; Hannun YA, Sphingolipid Metabolism and Neutral Sphingomyelinases. In Sphingolipids: Basic Science and Drug Development, Gulbins E; Petrache I, Eds. Springer Vienna: Vienna, 2013; pp 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Castro BM; Prieto M; Silva LC, Ceramide: A simple sphingolipid with unique biophysical properties. Progress in Lipid Research 2014, 54, 53–67. [DOI] [PubMed] [Google Scholar]
- 136.Perez-Hernandez D; Gutiérrez-Vázquez C; Jorge I; López-Martín S; Ursa A; Sánchez-Madrid F; Vázquez J; Yáñez-Mó M, The Intracellular Interactome of Tetraspanin-enriched Microdomains Reveals Their Function as Sorting Machineries toward Exosomes. Journal of Biological Chemistry 2013, 288 (17), 11649–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu H; Guariglia S; Yu RY; Li W; Brancho D; Peinado H; Lyden D; Salzer J; Bennett C; Chow CW, Mutation of SIMPLE in Charcot-Marie-Tooth 1C alters production of exosomes. Mol Biol Cell 2013, 24 (11), 1619–37, s1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Villarroya-Beltri C; Baixauli F; Gutiérrez-Vázquez C; Sánchez-Madrid F; Mittelbrunn M, Sorting it out: Regulation of exosome loading. Seminars in Cancer Biology 2014, 28, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fader CM; Sánchez D; Furlán M; Colombo MI, Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 2008, 9 (2), 230–250. [DOI] [PubMed] [Google Scholar]
- 140.Villarroya-Beltri C; Baixauli F; Mittelbrunn M; Fernández-Delgado I; Torralba D; Moreno-Gonzalo O; Baldanta S; Enrich C; Guerra S; Sánchez-Madrid F, ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nature communications 2016, 7 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sönnichsen B; De Renzis S; Nielsen E; Rietdorf J; Zerial M, Distinct Membrane Domains on Endosomes in the Recycling Pathway Visualized by Multicolor Imaging of Rab4, Rab5, and Rab11. Journal of Cell Biology 2000, 149 (4), 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hsu C; Morohashi Y; Yoshimura S -i.; Manrique-Hoyos, N.; Jung, S.; Lauterbach, M. A.; Bakhti, M.; Grønborg, M.; Möbius, W.; Rhee, J.; Barr, F. A.; Simons, M., Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. Journal of Cell Biology 2010, 189 (2), 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ostrowski M; Carmo NB; Krumeich S; Fanget I; Raposo G; Savina A; Moita CF; Schauer K; Hume AN; Freitas RP; Goud B; Benaroch P; Hacohen N; Fukuda M; Desnos C; Seabra MC; Darchen F; Amigorena S; Moita LF; Thery C, Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature Cell Biology 2010, 12 (1), 19–30. [DOI] [PubMed] [Google Scholar]
- 144.Record M; Carayon K; Poirot M; Silvente-Poirot S, Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2014, 1841 (1), 108–120. [DOI] [PubMed] [Google Scholar]
- 145.Fader CM; Sánchez DG; Mestre MB; Colombo MI, TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2009, 1793 (12), 1901–1916. [DOI] [PubMed] [Google Scholar]
- 146.Segura E; Nicco C; Lombard B; Véron P; Raposo G; Batteux F; Amigorena S; Théry C, ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood 2005, 106 (1), 216–223. [DOI] [PubMed] [Google Scholar]
- 147.Nazarenko I; Rana S; Baumann A; McAlear J; Hellwig A; Trendelenburg M; Lochnit G; Preissner KT; Zöller M, Cell Surface Tetraspanin Tspan8 Contributes to Molecular Pathways of Exosome-Induced Endothelial Cell Activation. Cancer Research 2010, 70 (4), 1668–1678. [DOI] [PubMed] [Google Scholar]
- 148.Klibi J; Niki T; Riedel A; Pioche-Durieu C; Souquere S; Rubinstein E; Le Moulec S; Guigay J; Hirashima M; Guemira F; Adhikary D; Mautner J; Busson P, Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 2009, 113 (9), 1957–66. [DOI] [PubMed] [Google Scholar]
- 149.Denzer K; van Eijk M; Kleijmeer MJ; Jakobson E; de Groot C; Geuze HJ, Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. The Journal of Immunology 2000, 165 (3), 1259–1265. [DOI] [PubMed] [Google Scholar]
- 150.Parolini I; Federici C; Raggi C; Lugini L; Palleschi S; De Milito A; Coscia C; Iessi E; Logozzi M; Molinari A; Colone M; Tatti M; Sargiacomo M; Fais S, Microenvironmental pH is a key factor for exosome traffic in tumor cells. The Journal of biological chemistry 2009, 284 (49), 34211–34222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tian T; Wang Y; Wang H; Zhu Z; Xiao Z, Visualizing of the cellular uptake and intracellular trafficking of exosomes by live cell microscopy. Journal of cellular biochemistry 2010, 111 (2), 488–496. [DOI] [PubMed] [Google Scholar]
- 152.Saeedi S; Israel S; Nagy C; Turecki G, The emerging role of exosomes in mental disorders. Translational Psychiatry 2019, 9 (1), 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Feng D; Zhao WL; Ye YY; Bai XC; Liu RQ; Chang LF; Zhou Q; Sui SF, Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11 (5), 675–87. [DOI] [PubMed] [Google Scholar]
- 154.Marcus ME; Leonard JN, FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharmaceuticals (Basel) 2013, 6 (5), 659–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.De Jong OG; Van Balkom BWM; Schiffelers RM; Bouten CVC; Verhaar MC, Extracellular Vesicles: Potential Roles in Regenerative Medicine. Front Immunol 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Milosevits G; Szebeni J; Krol S, Exosomes: potential model for complement-stealth delivery systems. European Journal of Nanomedicine 2015, 7 (3), 207. [Google Scholar]
- 157.Milosevits G; Szebeni J; Krol S, Exosomes: potential model for complement-stealth delivery systems. European Journal of Nanomedicine 2015, 7 (3). [Google Scholar]
- 158.Zhu X; Badawi M; Pomeroy S; Sutaria DS; Xie Z; Baek A; Jiang J; Elgamal OA; Mo X; Perle KL; Chalmers J; Schmittgen TD; Phelps MA, Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. Journal of extracellular vesicles 2017, 6 (1), 1324730–1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu X; Badawi M; Pomeroy S; Sutaria DS; Xie Z; Baek A; Jiang J; Elgamal OA; Mo X; Perle KL; Chalmers J; Schmittgen TD; Phelps MA, Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. Journal of Extracellular Vesicles 2017, 6 (1), 1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yin W; Ouyang S; Li Y; Xiao B; Yang H, Immature dendritic cell-derived exosomes: a promise subcellular vaccine for autoimmunity. Inflammation 2013, 36 (1), 232–40. [DOI] [PubMed] [Google Scholar]
- 161.Théry C; Zitvogel L; Amigorena S, Exosomes: composition, biogenesis and function. Nature Reviews Immunology 2002, 2 (8), 569–579. [DOI] [PubMed] [Google Scholar]
- 162.Théry C, Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep 2011, 3, 15–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang B; Yeo RWY; Lai RC; Sim EWK; Chin KC; Lim SK, Mesenchymal stromal cell exosome-enhanced regulatory T-cell production through an antigen-presenting cell-mediated pathway. Cytotherapy 2018, 20 (5), 687–696. [DOI] [PubMed] [Google Scholar]
- 164.Hedlund M; Stenqvist AC; Nagaeva O; Kjellberg L; Wulff M; Baranov V; Mincheva-Nilsson L, Human placenta expresses and secretes NKG2D ligands via exosomes that down-modulate the cognate receptor expression: evidence for immunosuppressive function. J Immunol 2009, 183 (1), 340–51. [DOI] [PubMed] [Google Scholar]
- 165.Escudier B; Dorval T; Chaput N; André F; Caby M-P; Novault S; Flament C; Leboulaire C; Borg C; Amigorena S; Boccaccio C; Bonnerot C; Dhellin O; Movassagh M; Piperno S; Robert C; Serra V; Valente N; Le Pecq J-B; Spatz A; Lantz O; Tursz T; Angevin E; Zitvogel L, Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. Journal of Translational Medicine 2005, 3 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Morelli AE; Larregina AT; Shufesky WJ; Sullivan MLG; Stolz DB; Papworth GD; Zahorchak AF; Logar AJ; Wang Z; Watkins SC; Falo LD; Thomson AW, Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104 (10), 3257–3266. [DOI] [PubMed] [Google Scholar]
- 167.Saunderson SC; Dunn AC; Crocker PR; McLellan AD, CD169 mediates the capture of exosomes in spleen and lymph node. Blood 2014, 123 (2), 208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zech D; Rana S; Büchler MW; Zöller M, Tumor-exosomes and leukocyte activation: an ambivalent crosstalk. Cell Commun Signal 2012, 10 (1), 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Murphy DE; de Jong OG; Brouwer M; Wood MJ; Lavieu G; Schiffelers RM; Vader P, Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Experimental & Molecular Medicine 2019, 51 (3), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Koh E; Lee EJ; Nam GH; Hong Y; Cho E; Yang Y; Kim IS, Exosome-SIRPα, a CD47 blockade increases cancer cell phagocytosis. Biomaterials 2017, 121, 121–129. [DOI] [PubMed] [Google Scholar]
- 171.Kamerkar S; LeBleu VS; Sugimoto H; Yang S; Ruivo CF; Melo SA; Lee JJ; Kalluri R, Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546 (7659), 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Agrawal M; Ajazuddin; Tripathi DK; Saraf S; Saraf S; Antimisiaris SG; Mourtas S; Hammarlund-Udenaes M; Alexander A, Recent advancements in liposomes targeting strategies to cross blood-brain barrier (BBB) for the treatment of Alzheimer’s disease. J Control Release 2017, 260, 61–77. [DOI] [PubMed] [Google Scholar]
- 173.Alvarez-Erviti L; Seow Y; Yin H; Betts C; Lakhal S; Wood MJA, Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology 2011, 29 (4), 341–345. [DOI] [PubMed] [Google Scholar]
- 174.Yang T; Martin P; Fogarty B; Brown A; Schurman K; Phipps R; Yin VP; Lockman P; Bai S, Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharmaceutical research 2015, 32 (6), 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Banfai K; Garai K; Ernszt D; Pongracz JE; Kvell K, Transgenic Exosomes for Thymus Regeneration. Front Immunol 2019, 10, 862–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jung KO; Jo H; Yu JH; Gambhir SS; Pratx G, Development and MPI tracking of novel hypoxia-targeted theranostic exosomes. Biomaterials 2018, 177, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cui GH; Guo HD; Li H; Zhai Y; Gong ZB; Wu J; Liu JS; Dong YR; Hou SX; Liu JR, RVG-modified exosomes derived from mesenchymal stem cells rescue memory deficits by regulating inflammatory responses in a mouse model of Alzheimer’s disease. Immun Ageing 2019, 16, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Liang G; Zhu Y; Ali DJ; Tian T; Xu H; Si K; Sun B; Chen B; Xiao Z, Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology 2020, 18 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Barile L; Vassalli G, Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther 2017, 174, 63–78. [DOI] [PubMed] [Google Scholar]
- 180.Kibria G; Ramos EK; Wan Y; Gius DR; Liu H, Exosomes as a Drug Delivery System in Cancer Therapy: Potential and Challenges. Mol Pharm 2018, 15 (9), 3625–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Alvarez-Erviti L; Seow Y; Yin H; Betts C; Lakhal S; Wood MJ, Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011, 29 (4), 341–5. [DOI] [PubMed] [Google Scholar]
- 182.Wahlgren J; Karlson TDL; Brisslert M; Vaziri Sani F; Telemo E; Sunnerhagen P; Valadi H, Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Research 2012, 40 (17), e130–e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Shtam TA; Kovalev RA; Varfolomeeva E; Makarov EM; Kil YV; Filatov MV, Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Communication and Signaling 2013, 11 (1), 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Banizs AB; Huang T; Dryden K; Berr SS; Stone JR; Nakamoto RK; Shi W; He J, In vitro evaluation of endothelial exosomes as carriers for small interfering ribonucleic acid delivery. International journal of nanomedicine 2014, 9, 4223–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.El-Andaloussi S; Lee Y; Lakhal-Littleton S; Li J; Seow Y; Gardiner C; Alvarez-Erviti L; Sargent IL; Wood MJA, Exosome-mediated delivery of siRNA in vitro and in vivo. Nature Protocols 2012, 7 (12), 2112–2126. [DOI] [PubMed] [Google Scholar]
- 186.Pegtel DM; Cosmopoulos K; Thorley-Lawson DA; Van Eijndhoven MAJ; Hopmans ES; Lindenberg JL; De Gruijl TD; Wurdinger T; Middeldorp JM, Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences 2010, 107 (14), 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ohno S.-i.; Takanashi M; Sudo K; Ueda S; Ishikawa A; Matsuyama N; Fujita K; Mizutani T; Ohgi T; Ochiya T; Gotoh N; Kuroda M, Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther 2013, 21 (1), 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jialei Yang XZ, Xiangjie Chen, Lei Wang, Guodong Yang , Exosome Mediated Delivery of miR-124 Promotes Neurogenesis after Ischemia. Molecular Therapy Nucleic Acids 2017, 7, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jialei Yang SW, Lihua Hou, Shimin Yang, Guodong Yang, Yongjun Wang, Therapeutic Effects of Simultaneous Delivery of Nerve Growth Factor mRNA and Protein via Exosomes on Cerebral Ischemia. Molecular Therapy Nucleic Acids 2020, 21, 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pi F; Binzel DW; Lee TJ; Li Z; Sun M; Rychahou P; Li H; Haque F; Wang S; Croce CM; Guo B; Evers BM; Guo P, Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nature nanotechnology 2018, 13 (1), 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yan F; Zhong Z; Wang Y; Feng Y; Mei Z; Li H; Chen X; Cai L; Li C, Exosome-based biomimetic nanoparticles targeted to inflamed joints for enhanced treatment of rheumatoid arthritis. Journal of Nanobiotechnology 2020, 18 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morishita M; Takahashi Y; Matsumoto A; Nishikawa M; Takakura Y, Exosome-based tumor antigens–adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA. Biomaterials 2016, 111, 55–65. [DOI] [PubMed] [Google Scholar]
- 193.Witwer KW; Buzás EI; Bemis LT; Bora A; Lässer C; Lötvall J; Nolte-’T Hoen EN; Piper MG; Sivaraman S; Skog J; Théry C; Wauben MH; Hochberg F, Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles 2013, 2 (1), 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Baranyai T; Herczeg K; Onódi Z; Voszka I; Módos K; Marton N; Nagy G; Mäger I; Wood MJ; El Andaloussi S; Pálinkás Z; Kumar V; Nagy P; Kittel Á; Buzás EI; Ferdinandy P; Giricz Z, Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLOS ONE 2015, 10 (12), e0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Théry C; Amigorena S; Raposo G; Clayton A, Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006, Chapter 3, Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 196.Wang X, Isolation of Extracellular Vesicles from Breast Milk. Methods Mol Biol 2017, 1660, 351–353. [DOI] [PubMed] [Google Scholar]
- 197.Zarovni N; Corrado A; Guazzi P; Zocco D; Lari E; Radano G; Muhhina J; Fondelli C; Gavrilova J; Chiesi A, Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [DOI] [PubMed] [Google Scholar]
- 198.Kesimer M; Scull M; Brighton B; Demaria G; Burns K; O’Neal W; Pickles RJ; Sheehan JK, Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. The FASEB Journal 2009, 23 (6), 1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wu Y; Deng W; Klinke DJ 2nd, Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers. The Analyst 2015, 140 (19), 6631–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Das CK; Jena BC; Banerjee I; Das S; Parekh A; Bhutia SK; Mandal M, Exosome as a novel shuttle for delivery of therapeutics across biological barriers. Molecular pharmaceutics 2018, 16 (1), 24–40. [DOI] [PubMed] [Google Scholar]
- 201.Fuhrmann G; Serio A; Mazo M; Nair R; Stevens MM, Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. Journal of Controlled Release 2015, 205, 35–44. [DOI] [PubMed] [Google Scholar]
- 202.Haney MJ; Klyachko NL; Zhao Y; Gupta R; Plotnikova EG; He Z; Patel T; Piroyan A; Sokolsky M; Kabanov AV, Exosomes as drug delivery vehicles for Parkinson’s disease therapy. Journal of Controlled Release 2015, 207, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tomizawa M; Shinozaki F; Motoyoshi Y; Sugiyama T; Yamamoto S; Sueishi M, Sonoporation: Gene transfer using ultrasound. World journal of methodology 2013, 3 (4), 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Arbab AS; Yocum GT; Wilson LB; Parwana A; Jordan EK; Kalish H; Frank JA, Comparison of transfection agents in forming complexes with ferumoxides, cell labeling efficiency, and cellular viability. Molecular imaging 2004, 3 (1), 15353500200403190. [DOI] [PubMed] [Google Scholar]
- 205.Chen Z; Wang H; Xia Y; Yan F; Lu Y, Therapeutic Potential of Mesenchymal Cell-Derived miRNA-150–5p-Expressing Exosomes in Rheumatoid Arthritis Mediated by the Modulation of MMP14 and VEGF. Journal of immunology (Baltimore, Md. : 1950) 2018, 201 (8), 2472–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pegtel DM; Cosmopoulos K; Thorley-Lawson DA; van Eijndhoven MAJ; Hopmans ES; Lindenberg JL; de Gruijl TD; Würdinger T; Middeldorp JM, Functional delivery of viral miRNAs via exosomes. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (14), 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]


